Abstract
Central core disease is a myopathy often arising from mutations in the type 1 ryanodine receptor (RYR1) gene, encoding the sarcoplasmic reticulum calcium release channel RyR1. No treatment is currently available for this disease. We studied the pathological situation of a severely affected child with two recessive mutations, which resulted in a massive reduction in the amount of RyR1. The paternal mutation induced the inclusion of a new in-frame pseudo-exon in RyR1 mRNA that resulted in the insertion of additional amino acids leading to the instability of the protein. We hypothesized that skipping this additional exon would be sufficient to restore RyR1 expression and to normalize calcium releases. We therefore developed U7-AON lentiviral vectors to force exon skipping on affected primary muscle cells. The efficiency of the exon skipping was evaluated at the mRNA level, at the protein level, and at the functional level using calcium imaging. In these affected cells, we observed a decreased inclusion of the pseudo-exon, an increased RyR1 protein expression, and a restoration of calcium releases of normal amplitude either upon direct RyR1 stimulation or in response to membrane depolarization. This study is the first demonstration of the potential of exon-skipping strategy for the therapy of central core disease, from the molecular to the functional level.
Rendu and colleagues identify a patient with a recessive central core myopathy in which one of the disease-causing mutations results in pseudo-exon inclusion in the type 1 ryanodine receptor calcium channel mRNA. They demonstrate that a lentivirus vector expressing a U7 snRNA-linked antisense sequence can induce skipping of the pseudo-exon in primary patient myocytes, resulting in rescue of ryanodine receptor expression and correction of abnormal calcium release.
Introduction
Type 1 ryanodine receptor (RyR1), encoded by the RYR1 gene (MIM#180901), is the main sarcoplasmic reticulum calcium channel in skeletal muscles. RyR1 and its major partner in the calcium release complex, the voltage gated calcium channel—dihydropyridine receptor (DHPR)—are responsible for the excitation–contraction coupling process (Lanner et al., 2010).
Mutations in the RYR1 gene have been associated with congenital myopathies: central core disease (CCD; MIM#117000) (Lynch et al., 1999; Monnier et al., 2001; Davis et al., 2003) and multiminicore disease (MIM#602771) (Feirrero et al., 2002; Jungbluth et al., 2002; Monnier et al., 2003). CCD is characterized by the presence of cores in type 1 muscle fibers. Those are large areas of abnormal myofibrillar architecture characterized by sarcomeric disorganization and absence of mitochondria (Monnier et al., 2001; De Cauwer et al., 2002). CCD presents either an autosomal dominant or a recessive transmission pattern and is clinically heterogeneous. Clinical presentation ranges from mild phenotype, with moderate hypotonia during early childhood, delayed motor abilities, and slowly progressive proximal muscle weakness, to severe phenotype, including fetal akinesia, respiratory insufficiency at birth, and generalized muscle weakness (Romero et al., 2003; Jungbluth, 2007). No treatment is currently available for this disease.
We have previously described a patient affected by a recessive form of the disease with a very severe phenotype (Monnier et al., 2008). He presented at birth with a major respiratory insufficiency associated with hypotonia. At six, he showed a generalized muscle weakness and significant scoliosis. He was unable to walk and was under respiratory assistance. A muscle biopsy, performed when he was 1 year old, showed the presence of well-delimited large cores in type 1 fibers. He was diagnosed as being affected with CCD. Genetic analysis showed that he was compound heterozygous for the RYR1 gene, with one mutation on the maternal allele of incomplete penetrance leading to a STOP codon, and a deep intronic mutation on the paternal allele leading to the introduction of an additional exon, called pseudo-exon 101bis. The consequence of both mutations was a massive loss in the amount of the RyR1 protein, although RyR1 transcript levels were unaffected (Monnier et al., 2008).
The exon-skipping therapeutic strategy has been successfully applied to different genetic diseases (Wein et al., 2010; Hammond and Wood, 2011; Taniguchi-Ikeda et al., 2011; Spitali and Aartsma-Rus, 2012). The principle of exon skipping is to block some specific splice sites during mRNA maturation with antisense oligonucleotides (AONs) in order to restore the reading frame at the transcript level. This approach has been used in mouse and dog animal models of Duchenne muscular dystrophy (Goyenvalle et al., 2004; Vulin et al., 2012), demonstrating its ability to restore an internally truncated but functional protein, and is now used in clinical trials (Cirak et al., 2011). Initially focused on Duchenne muscular dystrophy, exon skipping has been further applied to other muscle diseases, for example, the Fukuyama muscular dystrophy and dysferlinopathy (Wein et al., 2010; Taniguchi-Ikeda et al., 2011). In the present study, we have developed an exon-skipping strategy to force the skipping of the additional 101bis pseudo-exon in RyR1 pre-mRNA and thus restore the expression of a normal protein. This strategy, applied to human primary muscle cultures having the mutation, led to an increase in the amount of the RyR1 protein. In addition, the restored protein was functional, as observed by normalization of the amplitude of calcium releases in treated cells. This study is the first demonstration of a possible treatment for an RyR1-related myopathy.
Materials and Methods
Ethics statement
Investigations on patient material were performed after signature of an informed consent by the patients according to French regulation and have received approval from the local ethics committee (Comité de Protection des Personnes Sud-Est).
RYR1 reference sequence
The RYR1 sequence used in this work is accessible as NCI RefSeq NM_000540.2.
Quantitative Western blot analysis
The amount of RyR1 present in the muscle sample was determined by quantitative Western blot analysis using antibodies directed against RyR1 (Marty et al., 1994) and normalized to the amount of γ-sarcoglycan (antibody from Novo Castra) as described previously (Monnier et al., 2008). Signals were detected using a chemiluminescent HRP substrate and quantified using a ChemiDoc XRS apparatus (Biorad) and the Quantity One software (Biorad).
Cell culture, cell sorting, and differentiation
HEK293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen).
Fetal muscle biopsies were obtained from 14-week-old fetuses after medical abortion in the case of the mutant fetal (MF) cells and legal in-request abortion in the case of the control fetal (CF) cells. Primary cultures obtained from these biopsies were further enriched in myoblasts using a MACS sorting kit (CD56 microbeads; Miltenyi Biotec SAS), according to manufacturer's protocol. The amount of myoblasts in each culture was evaluated by desmin immunolabeling and was estimated to 83% for MF cells and 46% for CF cells.
MF and CF myoblasts were grown in proliferation medium (HamF10 supplemented with 20% fetal bovine serum, 2% Ultroser [Pall Biosepra], and 1% penicillin/streptomycin). Differentiation was induced by shifting cells to differentiation medium (DMEM 1% glucose supplemented with 2% horse serum), during 4 days for reverse transcriptase–polymerase chain reaction (RT-PCR) experiments, or 8–10 days for RyR1 protein detection and calcium imaging experiments.
Reverse transcriptase–polymerase chain reaction
Cells were lyzed in TRIzol reagent (Invitrogen, Life Technologies SAS) and total RNA was extracted with the Ambion PureLink kit (Life Technologies SAS) according to the manufacturer's instructions. Complementary DNA was synthesized from 500 to 750 ng of total RNA using 0.5 μl of Transcriptor (Roche) and 0.3 μg of oligo-dT as described (Monnier et al., 2001). PCR with 25 amplification cycles were then performed with the following primers: pCIneoF and T3R for the minigene splicing assay; CCL28/2F and CCL28/2R for amplification of the fragment of RYR1 comprising exons 101 and 102. Primers (see sequences in Table 1) were produced and purified by Eurogentec. After PCR, the amplified products were separated by electrophoresis on agarose gels and quantified with a Chemidoc XRS apparatus and Quantity One software (Bio-Rad).
Table 1.
Sequences of Primers and Antisense Oligonucleotides Used in This Study
| Primer | Sequence | Use |
|---|---|---|
| pCIneoF | 5′ gttcaattacagctcttaag 3′ | RT-PCR |
| T3R | 5′ aattaaccctcactaaaggg 3′ | RT-PCR |
| CCL28/2F | 5′ tctggaagttcggggtcatctt 3′ | RT-PCR |
| CCL28/2R | 5′ gccatgcggtgtcgtatcaa 3′ | RT-PCR |
| MluI-101bis-F | 5′ gggacgcgtagagacacaggccacaatcc 3′ | Minigene |
| SalI-101bis-R | 5′ ggggtcgacgagtgcaatggcgtgatct 3′ | Minigene |
| U7-D+E antisense | 5′ ccacagcgcccggcauacacuuguucuuc 3′ | Lentivirus |
| U7-CTRL antisense | 5′ gguguauugcaugauaugu 3′ | Lentivirus |
| AON-A | 5′ ucauccuagaguaaaaacua 3′ | Exon skipping |
| AON-B | 5′ gaacacagucauccaagagu 3′ | Exon skipping |
| AON-C | 5′ augugagagaacacagucau 3′ | Exon skipping |
| AON-D | 5′ guucuuuccguguccccagg 3′ | Exon skipping |
| AON-E | 5′ ccacagcgcccggcauacac 3′ | Exon skipping |
| AON-F | 5′ ggauuaaaagcgugagccac 3′ | Exon skipping |
| AON-G | 5′ augauccaccuaccuuggcc 3′ | Exon skipping |
RT-PCR, reverse transcriptase–polymerase chain reaction.
Minigene splicing assay
A PCR was performed on the genomic DNA of individual III.1 with MluI-101bis-F and SalI-101bis-R primers (Table 1) to amplify a region of 496 bp encompassing the c.14646+2563C>T mutation, and the amplified fragments from both alleles were inserted in a pCIneo plasmid modified as described previously (Roux-Buisson et al., 2011). Two minigene plasmids were cloned and sequenced, one with the wild-type sequence, called C construct, and one with the c.14646+2563C>T mutation, called M construct. Transfections were performed on 3.105 HEK293 cells using ExGen500 (Euromedex) with a DNA:ExGen ratio of 3:10, according to manufacturer's instructions. Forty hours after transfection, cells were collected by trypsination and processed for RT-PCR, using primers pCIneoF and T3R.
AON design and transfection
AONs were chosen according to several criteria: type and accessibility of the target and stability of the AON-pre mRNA binding energy (Aartsma-Rus et al., 2009). The 101bis pseudo-exon and its intronic junctions in the RYR1 sequence were analyzed by the Human splicing Finder software (Desmet et al., 2009), in order to identify the splice sites and to find potential exonic splicing enhancers (ESE). The accessibility was determined by analysis of the pre-mRNA with Mfold software (Zuker, 2003): open structures in the pre-mRNA strand were considered to have a better accessibility than apparied ones. Within these regions defined as interesting targets (splice sites or ESE in the pseudo-exon), seven AONs were designed according to their binding energy using the “Oligowalk” option in the RNA structure 4.5 software (Reuter and Mathews, 2010). The sequences of the AONs are indicated in Table 1. The seven 2′O-methyl AONs with a phosphorothioate backbone were produced and purified by Alpha DNA.
MF or CF myoblasts (105 cells) were seeded in 35 mm dishes and were transfected with 8.5 μl of Exgen500 and 500 nM AONs (Aartsma-Rus et al., 2003). After 2 days in the proliferation medium, the cells were switched to a differentiation medium. After 24 hr of differentiation, cells underwent a second transfection following the same protocol and were collected 3 days after for RT-PCR analyses (4 days post-differentiation).
U7 small nuclear RNA lentiviral vectors and transduction
The U7 DNA fragments were engineered from the previously described U7SmOPT (Gorman et al., 1998; Wein et al., 2010), with antisense sequence targeting the histone pre-mRNA replaced by the specific antisense sequences (Table 1) targeting the human pre-mRNA of RYR1. The U7-Ctrl and U7-D+E fragments were subcloned into a plasmid pRRL–cPPT–mcs–WPRE, derived from the pRRL–cPPT–hPGK–eGFP–WPRE construct (Naldini, 1998), and sequences of constructs were checked by sequencing. Lentiviral vectors were produced at Genethon as previously described by transient quadri-transfection into HEK293T cells. Viral titers (infectious particles) were determined by transduction of HCT116 cells and assayed by quantitative real-time PCR on genomic DNA (Charrier et al., 2005). For myoblast transduction, 3×105 MF or CF cells were seeded in a 500 μl drop of a proliferating medium in 60 mm culture dishes. Lentiviral particles (MOI 300) were added in the drop, and after 8 hr of incubation at 37°C, 2 ml of proliferating medium was added and cells were allowed to grow for 48 hr. After amplification to 107 cells, cells were trypsinized and split into several dishes to perform RT-PCR, immunofluorescence and calcium imaging assays in parallel.
Immunofluorescence microscopy
Myotubes (8–10 days of differentiation) grown on plastic Ibidi dishes (Biovalley) coated with laminin were washed with PBS and fixed 6 min in methanol at −20°C. Myotubes were then processed for immunolabeling with a monoclonal antibody against myosin heavy chain (MyHC), as a marker of differentiation (MF20; Developmental Studies Hybridoma Bank, NICHD, University of Iowa) and a polyclonal antibody against RyR1 (Marty et al., 1994). After a 15 min permeabilization with PBS-0.1% Tween 20 (PBS-T), cells were incubated for 1 hr 30 min with primary antibodies in PBS-T at room temperature. After three washes with PBS-T, cells were incubated for 30 min at room temperature with fluorescent secondary antibodies, and washed again before mounting in Fluorsave (Merck Chemicals). The secondary antibodies were either coupled to Alexa-488 (Molecular Probes, Life Technologies SAS) or Cy3 (Jackson ImmunoResearch Europe). The samples were analyzed using a Leica SPE confocal laser scanning microscope (Leica Microsystemes SAS).
RyR1 quantification and myotubes width measurements by image analysis
For RyR1 quantification in myotubes, images of five focal planes separated by 1.5 μm were acquired with identical confocal settings between the different images and the different experimental conditions (transduced and nontransduced myotubes). Settings were adjusted in order to avoid any pixel saturation, both for RyR1 and for MyHC staining. Z-projections of the five planes were performed and the Sum images were used for quantification, as described below. Relative expression levels of the RyR1 protein in myotubes were quantified using ImageJ software (version 1.44i, NIH) and a home-made macro. The macro performed different operations: myotubes were first delineated in regions of interest (ROIs) using the MyHC image; the mean pixel intensities of the MyHC staining were measured inside ROIs and MyHC background was measured outside the ROIs; the same ROIs were reported on the RyR1 staining image, and the RyR1 signal mean intensity and the RyR1 background were measured. The ratio of RyR1 to MyHC intensities was calculated as follows: (RyR1 mean pixel intensity − RyR1 global background)/(MyHC mean pixel intensity − MyHC global background). For myotube width measurements, the maximal width per myotube was measured on MyHC z-projections images, using ImageJ software.
Calcium imaging assays
Changes in intracellular calcium were measured using the calcium-dependent fluorescent dye Fluo4-AM (Molecular Probes, Life Technologies SAS), as described before (Rezgui et al., 2005). Myotubes were incubated for 1 hr at room temperature with Fluo-4-AM 10 μM, in Krebs buffer (136 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES pH 7.4). Uptake of the dye was facilitated by the addition of 0.02% pluronic F-127 acid (Sigma). After loading, myotubes were washed for 1 hr at 37°C to allow de-esterification of the dye. Calcium imaging was performed in Krebs buffer. 4-Chloro-m-Cresol (CmC) was prepared as stock solution in Krebs buffer and used at final concentrations of 500 μM. In the depolarization solution, NaCl was replaced by KCl (140 mM final concentration). Fluorescence was measured by confocal laser scanning microscopy, using a Leica TCS-SPE operating system, in the xyt mode. The fluorescence images were collected every 0.9 sec, during 2–4 min, and then analyzed frame by frame. Fluorescence curves are expressed as a function of time, as ΔF/F0, where F0 represents the baseline fluorescence immediately before depolarization and ΔF represents the fluorescence changes from baseline. Data are given as mean±SEM, and n represents the number of myotubes in each condition.
Statistics
Statistics were performed using Student's t-test or analysis of variance followed by Bonferroni t-test for multiple comparisons, on Prism 6.0 software (GraphPad), with n=number of myotubes in at least three different experiments, and after checking the normal distribution of the samples.
Results
Case description
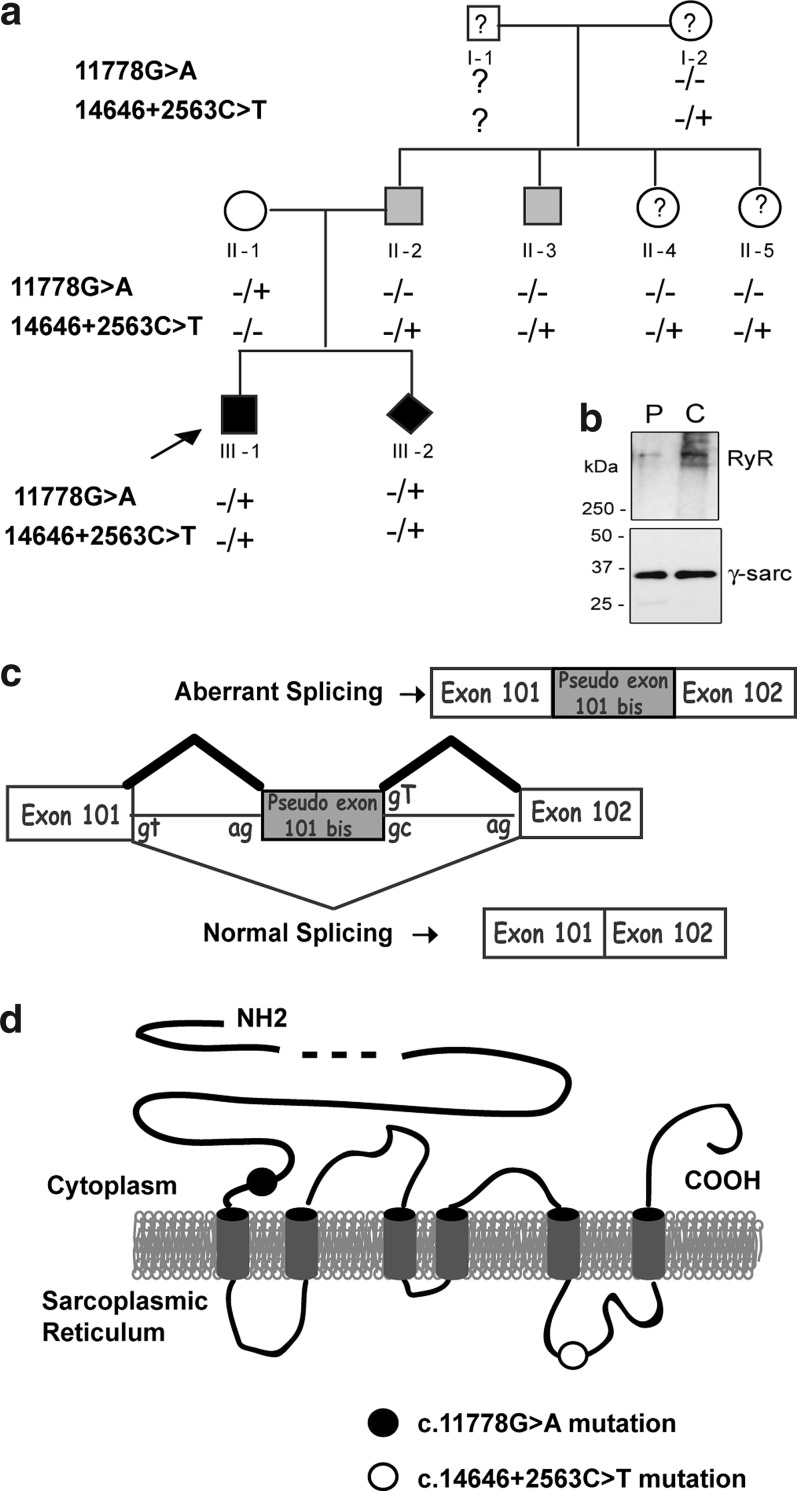
The proband (individual III:1; Fig. 1a), first described in Monnier et al. (2008), harbored two RYR1 mutations, one inherited from his father (c.14646+2563C>T), and one from his mother (c.11778G>A). The c.1464+2563C>T change in intron 101 of the paternal allele created a donor splice site and activated an upstream acceptor site responsible for the insertion of a 99 bp pseudo-exon, called 101bis, at the junction of exons 101 and 102 (Fig. 1c). Because the 101bis pseudo-exon was in frame, this led to the insertion of 33 amino acids within the last luminal loop of RyR1 (Fig. 1d) that is involved in the interaction between RyR1 and its protein partner triadin (Lee et al., 2004). The mutation was found in the proband's father (individual II:2) and paternal uncle (individual II:3), both mildly affected. On the maternal allele, a silent c.11778G>A change of the last nucleotide of exon 85 (p.Gln3926Gln) unveiled a cryptic donor site downstream from the regular donor site. As a consequence of an incomplete penetrance, the maternal mutation generated two transcripts. In the first transcript, the splicing was unaffected and the presence of a CAA instead of a CAG codon did not modify the glutamine at position 3926. In the second transcript, the use of the cryptic donor site resulted in a premature STOP codon at position p.3926+4 (Monnier et al., 2008). The mother (individual II:1) was considered healthy.
FIG. 1.
Case description. (a) Pedigree of the family. Black symbols denote the symptomatic individuals, gray symbols correspond to pauci-symptomatic individuals, open symbols indicate asymptomatic individuals, and question marks refer to individuals with an unknown clinical status. The arrow indicates the proband. Diamond symbol represents the fetus. Signs+and − refer to the presence and absence of a mutation in the RYR1 gene, respectively. (b) Western blot analysis of the amount of RyR1 in a muscle biopsy from the patient (P, ind III.1) compared with an age-matched control (C). Amount of RyR1 was normalized to the amount of γ-sarcoglycan. (c) Representation of the consequences of the c.14646+2563C>T paternal intronic mutation on the splicing of the RYR1 pre-mRNA. The additional pseudo-exon inserted in the mRNA after aberrant splicing is in gray. (d) Schematic representation of the RyR1 protein with the approximate localizations of the maternal c.11778G>A and paternal c.14646+2563C>T mutations. The six transmembrane domains of RyR1 are represented in gray, in the sarcoplasmic reticulum membrane. RyR1, type 1 ryanodine receptor.
A protein analysis performed on a muscle biopsy from the proband's uncle (individual II:3, who had only the c.14646+2563C>T paternal mutation at heterozygous state) showed a residual amount of RyR1 of 57%±3% of a control, without alteration of the amount of the RyR1 transcript (Monnier et al., 2008). As this mutation induced the addition of 33 amino acids in the channel part of RyR1, it can be assumed that this most probably led to an abnormal folding of the protein and to its degradation. Because the maternal mutation induced the presence of a premature STOP codon, the amount of the RyR1 protein in muscles of the proband (III:1) with the two mutations was expected be much lower than that of his uncle (II:3). This very low amount of protein could explain the severe phenotype of the proband (Monnier et al., 2008). The amount of the RyR1 protein in a biopsy of the proband III:1 was analyzed in the present study (Fig 1b). Although evaluation of the RyR1 amount by Western blot analysis could be performed only twice because of the limited biopsy available, it confirmed a major reduction in the amount of RyR1 for the proband (35% of protein remaining compared with an age-matched control biopsy) (Fig. 1b).
Mutation analysis
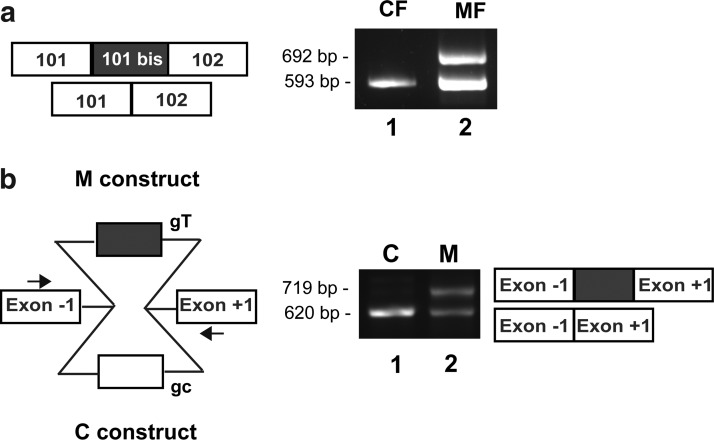
A prenatal diagnosis was subsequently performed in this family and led to a medical abortion because the fetus (ind. III:2; Fig. 1a) was bearing the two very same mutations identified in the proband. Affected fetal primary muscle cells, called MF cells, were produced from a muscle biopsy of the fetus. CF primary muscle cells were produced from an age-matched normal fetus. The effect of the c.14646+2563C>T intronic mutation on RYR1 expression was studied by RT-PCR using MF and CF cell cultures. An amplification of RyR1 mRNA was performed on the region spanning nucleotides 14438–15030 that contained exons 101 and 102. Two amplicons were obtained in MF cells (Fig. 2a, lane 2): one of normal 593 bp size, as in control CF cells (lane 1), and one of 692 bp size, which corresponded to the insertion of the 99 bp of exon 101bis. This result on MF cells confirmed the insertion of the pseudo-exon that was described for patient III:1 (Monnier et al., 2008). However, it was not possible to know if the residual normal-sized 593 bp fragment was because of incomplete penetrance of maternal or paternal mutation. The penetrance of the paternal mutation was analyzed using a minigene assay. The DNA sequence encoding the pseudo-exon and its flanking 5′ and 3′ regions was inserted into a pCIneo plasmid (Fig. 2b). Two constructs were developed, one with the normal sequence as a control (C construct) and one with the c.14646+2563C>T mutation (M construct). Each minigene was transfected in HEK293 cells, which do not express any endogenous RyR1. Two days after transfection, an RT-PCR analysis of minigene splicing was performed (Fig. 2b). Only one fragment was amplified from cells transfected with the control C construct (Fig.2b, lane 1), while two fragments were amplified from cells transfected with the mutant M construct (Fig. 2b, lane 2). This confirmed that the paternal intronic c.14646+2563C>T mutation by itself was sufficient to induce the insertion of a 99 bp sequence and that the mutation was of incomplete penetrance, at least in HEK293 cells.
FIG. 2.
Characterization of the paternal c.14646+2563C>T mutation. (a) RT-PCR performed on CF and MF cells differentiated during 4 days. RT-PCR amplification was performed on the region c.14438–15030 of RYR1 including exons 101 and 102. Size analysis of the amplified fragments showed the presence of a normal-sized fragment of 593 bp and of an abnormal 692 bp fragment in MF cells (lane 2), while only the normal-sized fragment is present in CF cells (lane 1). (b) Scheme of the minigene constructs used for the analysis of the c.14646+2563C>T mutation. RYR1 intronic regions containing sequence of the pseudo-exon, with (gray) or without (white) the mutation, were inserted into a pCIneo vector between exons −1 and +1. An RT-PCR amplification using a forward primer in exon −1 and a reverse primer in exon +1 (arrows) was performed on HEK 293 cells transfected with each minigene construct. Size analysis of the amplified fragments showed the presence of a unique fragment of 620 bp in HEK293 cells transfected with the C construct (lane 1), corresponding to the normal splicing, and of two fragments of 620 and 719 bp in HEK293 cells transfected with M construct (lane 2), corresponding to the inclusion of the pseudo-exon. The identity of these fragments was confirmed by sequencing. CF, control fetal; MF, mutant fetal; RT-PCR, reverse transcriptase–polymerase chain reaction.
Exon-skipping strategy for removal of the 101bis pseudo-exon
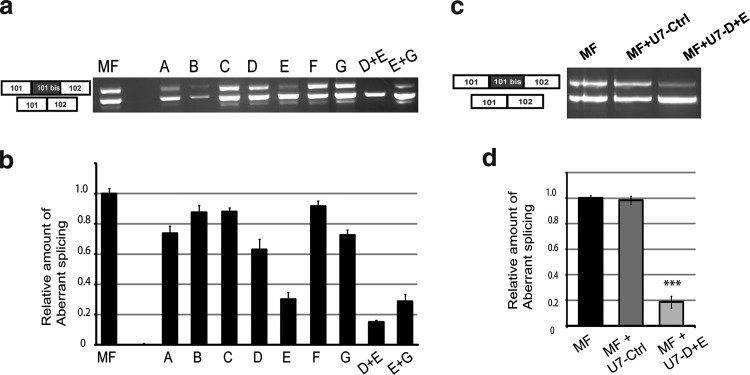
We have previously shown that the insertion of the 99 bp pseudo-exon in RyR1 mRNA considerably lowered the amount of the RyR1 protein in skeletal muscles (ind II:3) without affecting the total amount of RyR1 transcripts (Monnier et al., 2008). In the present study, we tested whether a removal of the pseudo-exon by exon skipping would restore the normal transcript expression, increase the RyR1 protein expression and restore Ca2+ releases. We designed seven 2′O-methyl AONs (A to G; Table 1) with a phosphorothioate backbone. The ability of these AONs to force the skipping of the pseudo-exon was evaluated by transfection of MF cells with each AON followed by their differentiation into myotubes. Total RNA was extracted from transfected myotubes and RT-PCR was performed on RyR1 mRNA to amplify the region including exons 101 and 102 (Fig. 3a). The efficiency of each AON, evaluated by quantification of the relative amount of abnormal splicing compared with the total amount of PCR products (normal transcript plus abnormal transcript), is presented on Figure 3b. Results showed that AON-D and AON-E, targeting putative ESE of pseudo-exon 101bis, were the most efficient in reducing the relative amount of abnormal splicing (respectively, 37% and 70% of aberrant transcript reduction) (Fig. 3b). These AONs were tested pair-wised by simultaneous transfection, and the D+E pair induced the most effective exon skipping (Fig. 3b, 82% reduction of aberrant transcript).
FIG. 3.
Test of AONs and lentiviral vectors to skip the 101bis pseudo-exon. (a) Seven AONs (A to G) were tested on MF cells for their capacity to skip the pseudo-exon. MF cells were either not transfected (lane MF) or transfected with each of the AONs (A to G) or with pairs of AONs (D+E or E+G). Cells were then induced to differentiate for 4 days and RT-PCR was performed using same primers as in Fig. 2a. (b) Ratios of the amount of pseudo-exon-containing fragment versus the total amount of amplified fragments (normal+pseudo-exon containing fragments) were calculated for each condition and normalized to 1 for nontransfected MF cells. Results are given as mean±SEM of three experiments. (c) Lentiviral U7-snRNA vectors containing antisense sequences corresponding to a nonrelevant control AON (U7-Ctrl) or to AONs D+E (U7-D+E) were tested for their efficacy to skip the pseudo-exon. MF cells were either not transduced (lane MF), or transduced with U7-Ctrl or U7-D+E vectors. Cells were then induced to differentiate for 4 days and RT-PCR was performed as in (a). (d) Quantifications were performed as in (b), and the mean±SEM of four experiments is plotted. ***p<0.001, Student's t-test comparison of MF and MF U7-D+E cells. AONs, antisense oligonucleotides; snRNA, small nuclear RNA.
Lentiviral-induced skipping of pseudo-exon 101bis
Cell transfection with the different AONs allowed us to select the most efficient ones. However, transfection induced a large mortality of the cells incompatible with their subsequent characterization by calcium imaging. We therefore decided to use lentiviral vectors to insert the AONs into the cells. Recent advances have been made in antisense approaches using small nuclear RNAs (snRNAs) and U7 snRNP has been used for splicing modulation as it offers many advantages: stability of the antisense sequence, specific subcellular co-localization with target pre-mRNAs, and the possibility of a long-term correction when introduced with viral vectors (for review, cf. Benchaouir and Goyenvalle, 2012). U7-snRNAs with antisense sequences corresponding to AONs D+E (called U7-D+E), and with a fluorescein-isothiocyanate-labeled scramble AON (U7-Ctrl; Table 1) were developed and inserted in lentiviral vectors. MF cells were transduced with these vectors, differentiated, and analyzed for RyR1 transcripts as previously described. As expected, RT-PCR analysis showed that U7-D+E was very efficient in skipping the 101bis pseudo-exon at the mRNA level (reduction of 80% of the relative amount of aberrant transcript), while the U7-Ctrl had no effect (Fig. 3c and d). To assess that no modification in the total amount of RyR1 transcripts was observed between the three conditions (MF, MF+U7-CTRL, and MF+U7-D+E), a PCR amplification of exons 103–105 of RyR1 mRNA was performed and normalized to the reference gene human acidic phosphoprotein—PO (data not shown).
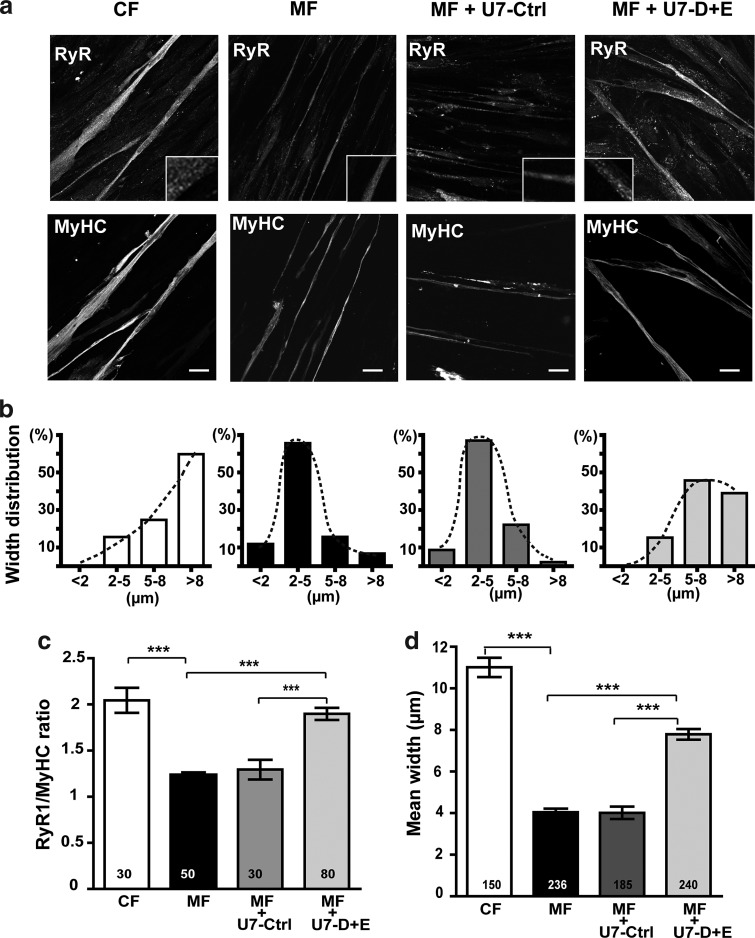
Skipping of the 101bis pseudo-exon in MF myotubes increases amount of the RyR1 protein and has a trophic effect
Quantification of the RyR1 protein in differentiated myotubes could not be assessed using Western blot because RyR1 levels in whole cell homogenates were below the detection limits of the technique. Therefore, we used image analysis after immunofluorescent staining of the myotubes to evaluate the relative amount of RyR1 compared with MyHC. Similar image analysis methods have been used to quantify dystrophin in dystrophinopathy muscle specimens (Taylor et al., 2012). MyHC is a marker of differentiation whose expression increases during differentiation as observed for RyR1. Hence, the quantification of RyR1 compared with MyHC allowed to get rid of a possible altered differentiation related to RyR1 reduction (Porter et al., 2002). The relative amounts of RyR1 were evaluated in differentiated control (CF) or mutant (MF) cells with or without transduction with U7-Ctrl or with U7-D+E (Fig. 4a and c). RyR1 presented the typical dotted pattern found in immunfluorescent staining of muscle cells (Fig 4a insets). The relative amounts of RyR1 in MF myotubes were about twofold lower than those in control CF myotubes (Fig. 4c), in good correlation with the results obtained in Western blot on muscle biopsy from ind III:1 (Fig 1b). Treatment of MF cells with U7-Ctrl did not change the level of RyR1 in myotubes. In contrast, treatment of MF cells with U7-D+E raised the relative amounts of RyR1 by approximately 1.5-fold (Fig. 4c).
FIG. 4.
Lentiviral vector treatments restore amounts of the RyR1 protein and affect the size of differentiated MF cells. (a) Control CF cells, untreated MF cells, or MF cells treated with U7-Ctrl vector or with U7-D+E vector were induced to differentiate for 8 days, and immunolabeled with antibodies against RyR1 and myosin heavy chain (MyHC). A magnification of RyR1 labeling is presented in the insets. Scale bars=20 μm. (b) Width distribution of myotubes. Each myotube was classified in one of the four categories according to its largest measured width (<2 μm, 2–5 μm, 5–8 μm, and >8 μm). The distribution of the myotube population in the four categories is represented for each condition (number of myotubes indicated in (d). (c) Quantification by image analysis of the mean fluorescence intensity of RyR1 staining in myotubes relative to the mean fluorescence intensity of MyHC staining, presented as mean±SEM. The mean fluorescence intensity of MyHC staining is not statistically different between the different conditions. The number of myotubes used for quantification is indicated in each bar of the plot. (d) Mean width of myotubes was calculated and results given in μm as mean±SEM of the number of myotubes indicated in each bar. ***p<0.001, ANOVA analysis followed by Bonferroni multiple comparison test. ANOVA, analysis of variance.
In addition, an effect on the morphology of myotubes concomitant to the increase in the relative amount of RyR1 was observed (Fig. 4a, b, and d). CF myotubes were significantly larger than MF myotubes (mean width 11±0.4 μm for CF myotubes and 4±0.3 μm for MF myotubes, p<0.001; Fig.4d). Treatment of MF cells with U7-D+E induced a significant increase in the width of myotubes compared with untreated MF myotubes (mean width after treatment 7.8±0.3 μm, p<0.001; Fig. 4d), with a redistribution of widths toward those of normal CF myotubes (Fig. 4b). Treatment of MF cells with U7-Ctrl had no effect on myotubes' width. Altogether, the lentiviral vector U7-D+E increased RyR1 protein levels in MF cells, and favored a restoration of the myotubes' width.
Skipping of the 101bis pseudo-exon restores calcium release capacities in affected cells
Functionality of the RyR1 protein was further tested by calcium imaging. RyR1 is specifically expressed in differentiated muscle cells, where it is the main intracellular calcium channel involved in excitation–contraction coupling (Brillantes et al., 1994; Kyselovic et al., 1994; Tarroni et al., 1997). Depolarization of the muscle plasma membrane activates the DHPR, which induces the opening of RyR1 and the release of calcium from the sarcoplasmic reticulum. Therefore, activity of RyR1 calcium channel was tested using calcium imaging on cultured myotubes, by direct RyR1 stimulation using its agonist CmC to assay the channel function of the restored protein, and by plasma membrane depolarization to assay its functional coupling to DHPR.
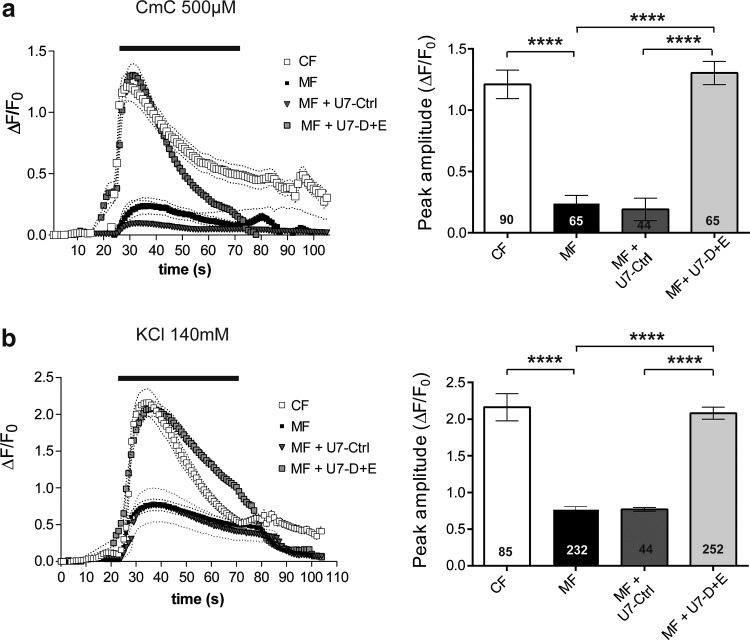
Calcium imaging experiments were performed on myotubes produced from control CF cells, untreated MF cells, or MF cells transduced with U7-Ctrl or U7-D+E (Fig. 5). Direct RyR1 stimulation was first assayed by 4-Chloro-m-Cresol (Fig. 5a), at a concentration resulting in maximal calcium release (EC50≈100 μM; Hermann-Frank et al., 1996). Addition of 500 μM CmC to MF myotubes induced a calcium release of reduced amplitude compared with CF myotubes (ΔF/F0=0.24±0.06, n=65 and ΔF/F0=1.21±0.11, n=90, for MF and CF myotubes, respectively, p<0.0001), the peak amplitude in MF myotubes being 20% of control CF myotubes (Fig. 5a). This reduced calcium release was not significantly modified by transduction by U7-Ctrl, and fully restored by transduction with U7-D+E (p<0.0001). Function of the whole calcium release complex, and functional coupling between DHPR and RyR1 were further tested by membrane depolarization. Addition of 140 mM KCl in the presence of extracellular calcium induces a membrane depolarization close to 0 mV, and leads to an almost maximal stimulation of DHPR (Rezgui et al., 2005). It resulted in a calcium release of much lower amplitude in MF cells (ΔF/F0=0.77±0.03, n=232) than in control CF cells (ΔF/F0=2.18±0.18, n=85, p<0.0001), the amplitude of the peak in MF myotubes corresponding to 35% of the amplitude observed in CF myotubes (Fig. 5b). Transduction of MF cells with U7-Ctrl had no effect, the amplitude of depolarization-induced calcium release being identical to MF cells. On the opposite, transduction with U7-D+E almost fully restored normal depolarization, the amplitude of the calcium release being not statistically different from control cells (ΔF/F0=2.08±0.08, n=252 for MF cells transduced with U7-D+E, p=0.64 between CF and MF U7-D+E, and p<0.0001 between MF and MF U7-D+E). Using both stimulations (CmC and KCl) the levels of calcium release in MF myotubes treated with U7-D+E were therefore comparable to those in control CF myotubes. These results demonstrated that the RyR1 calcium channel restored by exon skipping using the lentiviral vector U7-D+E was fully functional, correctly coupled to DHPR, and able to sustain calcium release from the sarcoplasmic reticulum to the cytoplasm.
FIG. 5.
Lentiviral vector treatments restore the RyR1 calcium release activity in differentiated MF cells. Calcium imaging performed on control CF cells (empty square), untreated MF cells (black square), and MF cells treated with U7-Crtl vector (black triangle) or with U7-D+E vector (gray square), and differentiated for 8–10 days before calcium imaging. (a) Fluorescence variation curves induced by application during 50 sec (black bar) of CmC (500 μM) in the presence of 2 mM external calcium, presented as mean (symbols)±SEM (dotted lines). The maximal amplitude of the peak for each curve is presented in the bar plots on the right, with the number of myotubes analyzed in each bar. (b) Fluorescence variation curves induced by membrane depolarization (KCl 140 mM) applied during 50 sec (black bar) in the presence of 2 mM external calcium, presented as mean (symbols)±SEM (dotted lines). ****p<0.0001, ANOVA analysis followed by Bonferroni multiple comparison test. CmC, 4-Chloro-m-Cresol.
Discussion
This study is the first example of an exon-skipping strategy applied to a congenital core myopathy—in this case, a severe form of CCD. The disease was caused by compound heterozygous mutations on the RYR1 gene, inducing an important drop in the amount of the RyR1 protein. The intronic paternal mutation resulted in a reduction in the amount of protein to 57% of a control muscle, the affected heterozygous individuals being pauci symptomatic (Monnier et al., 2008). In the severely affected child, the maternal mutation induced a further reduction in RyR1 to 35% of the amount observed in a control. We applied an exon-skipping strategy to skip the paternal pseudo-exon in order to restore an asymptomatic phenotype as observed for the mother. The goal of this study was thus to increase the amount of the RyR1 protein in affected muscle cells, and to test the functionality of the restored protein.
Exon skipping was performed on fetal muscle MF cells with the two mutations, using U7-AON vectorized in lentiviral vectors. The efficiency of exon skipping, shown by RT-PCR at the mRNA level, was further evaluated at the protein level by immunofluorescent staining and image analysis. The relative amount of the RyR1 protein in MF myotubes was reduced compared with CF myotubes, and significantly restored after treatment of MF cells by U7-D+E lentiviral vector. The precise evaluation of the amount of RyR1 in treated MF cells was not possible, but theoretically could not reach the amount in control cells because of the conjugated effect of the uncorrected maternal mutation and the incomplete skipping of pseudo-exon 101bis. Nevertheless, we showed that the abnormal RyR1 transcript was reduced five times by exon skipping, which suggested that the amount of the RyR1 protein in corrected cells was probably almost similar to the amount in the asymptomatic mother.
Functional studies using calcium imaging on MF myotubes allowed us to evaluate whether the RyR1 protein recovered by exon skipping was functional. Amplitude of calcium releases in untreated MF cells was about 20–35% of that in control cells, in good correlation with the massive reduction in the RyR1 amount observed in the proband III.1 (remaining RyR1 quantity evaluated to 35%). Treatment of MF myotubes with the U7-D+E vector resulted in calcium releases of amplitude equivalent to that observed in control myotubes. This increase in calcium releases of amplitude showed that restored RyR1 was functional. Moreover, it suggested that the restored level of RyR was sufficient to trigger calcium release of normal amplitude, raising the question of the quantity of RyR1 necessary for a correct muscle function. Interestingly, individual II-3, with an RyR1 amount of 57% of a control, showed only mild signs of muscle weakness. Patients with null mutations, that is, resulting in the absence of expression of one allele of RYR1 and leading to a 50% reduction in the amount of RyR1 at a protein level, have been shown to be asymptomatic or to present with mild phenotype (Zhou et al., 2007; Monnier et al., 2008). In the same way, and although no muscle biopsy was available to precisely quantify RyR1, the presence of a heterozygous splice mutation in the asymptomatic mother (ind II-1) was likely to result in a reduced expression of RyR1. On the basis of this observation, it can thus be hypothesized that reaching an RyR1 expression of 100% is not necessary to sustain a normal muscle function. Such an observation has already been done when exon-skipping strategy was applied to the chloride channel in myotonic dystrophy (Wheeler et al., 2007).
Our data demonstrate that exon skipping could be a relevant therapeutic strategy for an RYR1 splicing mutation resulting in the inclusion of a pseudo-exon and involved in CCD, because (i) the restored protein is a completely normal one, in contrast with the truncated dystrophin or the mini-dysferlin re-expressed after exon skipping in Duchenne muscular dystrophy (DMD) or dysferlinopathy (Goyenvalle et al., 2004; Wein et al., 2010), and (ii) it is not necessary to reach a full correction of the abnormal transcript and to restore 100% of the normal protein to reach normal levels of calcium release. Splice mutations account for 10–15% of RYR1 mutations involved in CCD, and pseudo-exon creating mutations constitute about 1–2% of RyR1 mutations identified up to now. This proportion is probably underestimated because these mutations are difficult to identify and need the analysis of RyR1 mRNA from a muscle biopsy. Exon-skipping strategy could most probably not be applied to any RYR1 mutation, and it would not be possible to suppress a whole exon affected by a point mutation, as it has been done for dystrophin or dysferlin. Indeed, a number of the central exons of dystrophin have been shown to be skippable, with no or only mild effect. This is not the case for RyR1, in which no exon has been identified as needless up to now.
An unexpected observation was the effect of exon skipping on myotube morphology: MF myotubes were very thin compared with control myotubes, and this morphologic feature was reversed by transduction with U7-D+E lentivirus. This effect on myotube morphology was specific of the vector inducing a skipping of the aberrant RyR1 pseudo-exon. An effect of the aberrant transcript per se cannot be ruled out (either a toxic effect on myotube differentiation or an induction of a degradation pathway), but it was most probably the increase in the normal RyR1 transcript/protein that was the cause of this trophic effect. The molecular mechanisms underlying the link between restoration of the RyR1 normal mRNA/protein and the trophic effect on myotubes are not yet understood. They could be related to an altered differentiation in the absence of RyR1. Even if expressed mainly in differentiated muscle cells with a major function in the calcium releases for contraction, other functions could be associated to this calcium channel in early maturation steps. The RyR1 protein has already been described as important for normal myofiber morphogenesis and muscle development in vivo. Indeed, the RyR1-null mice exhibited muscular degeneration associated with excitation–contraction uncoupling (Takeshima et al., 1994; Barone et al., 1998). Similar muscle degeneration was observed in the mdg mouse line, devoid of the alpha-1 subunit of DHPR (Banker, 1977; Chaudhari, 1992). Therefore, deletion of RyR1 or alpha-1 could alter the first steps of muscle formation. We have also recently observed that in vivo downregulation of the alpha-1 subunit of skeletal muscle DHPR induced massive muscle atrophy (Pietri-Rouxel et al., 2010). The situation is less clear for in vitro studies using cultured cells. Primary cultures produced from atrophic RyR1-null muscles or alpha-1-null muscles are, nevertheless, still able to form myotubes in vitro. In contrast, perturbation of intracellular calcium levels by several drugs targeting either RyR1 or its partner the DHPR inhibits myoblast differentiation (Seigneurin-Venin et al., 1996; Porter et al., 2002). Therefore, it is tempting to speculate that the presence of RyR1 and DHPR, or the precise control of intracellular calcium level, is important not only for muscle contraction during the excitation–contraction coupling, but also for myotube formation and/or for maintenance of muscle mass. In this case, the trophic effect of U7 exon-skipping vectors on MF cells reflects the restoration of a functional RyR1 protein in an indirect manner.
Overall, the present study describes a successful exon-skipping strategy for the correction of the genetic cause of a core myopathy in affected human cells. In the case studied here, the severe form of CCD was caused by a massive drop of RyR1 calcium channel expression, and the exon-skipping strategy was efficient both to increase the amount and to restore the activity of RyR1. This is the first time that such an efficient functional restoration is observed after exon skipping. In DMD, the ongoing clinical trials demonstrate an increase in dystrophin expression up to 18% of normal level after exon skipping, but this level seems too low to restore a correct muscle function. It has been shown very recently that molecules able to block RyR1 (dantrolene, ryanodine, S107) improve the efficiency of exon skipping in DMD patients cells (Kendall et al., 2012). The underlying mechanisms are still unknown, but it can be hypothesized that the improved exon-skipping efficiency in presence of these chemicals is related to their ability to reduce the amount of calcium released in the cytoplasm, which could affect the splicing through the calcium regulation of spliceosomal components (Krebs, 2009). In the case presented here, the cytosolic calcium reduction is a direct consequence of the mutation, and therefore could also be responsible for the high efficiency of the exon skipping. Therefore, this model study paves the way for therapies for core myopathies, even beyond similar RYR1 splicing alterations. Indeed, other therapies targeting mRNA such as trans-splicing or exon exchange are extremely promising, but as they have a much lower efficiency (Lorain et al., 2010) their use in future therapies is related to improvement of splicing. If improved splicing in skeletal muscle is correlated to reduced function of RyR1, then therapies based on exon exchange could be assayed for patients with any RYR1 mutation leading to a reduced RyR1 function, for whom it would be possible to successfully exchange the mutant exon(s) for normal one(s). All the tools developed in this work (calcium imaging, myotube morphology, etc.) will help for evaluation of any gene therapy aimed at the restoration of functional RyR1.
Acknowledgments
We thank all family members for their contribution to this study. We thank the Myocastor study group for fruitful discussions and the Genethon (Evry, France) for lentiviral production. This work was supported by grants from the “Institut National de la Santé et de la Recherche Médicale” (INSERM), the “Association Française contre les Myopathies” (AFM), the “Fondation Daniel Ducoin,” the “Direction de la Recherche Clinique du CHU Grenoble,” and the “Vivier de la Recherche de la Faculté de Médecine de Grenoble.”
Author Disclosure Statement
No competing financial interests exist for this study.
References
- Aartsma-Rus A. Janson A.A. Kaman W.E., et al. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum. Mol. Genet. 2003;12:907–914. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A. van Vliet L. Hirschi M., et al. Guidelines for antisense oligonucleotide design and insight into splice-modulating mechanisms. Mol. Ther. 2009;17:548–553. doi: 10.1038/mt.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker B.Q. Muscular dysgenesis in the mouse (mdg/mdg). I. Ultrastructural study of skeletal and cardiac muscle. J. Neuropathol. Exp. Neurol. 1977;36:100–127. doi: 10.1097/00005072-197701000-00010. [DOI] [PubMed] [Google Scholar]
- Barone V. Bertocchini F. Bottinelli R., et al. Contractile impairment and structural alterations of skeletal muscles from knockout mice lacking type 1 and type 3 ryanodine receptors. FEBS Lett. 1998;422:160–164. doi: 10.1016/s0014-5793(98)00003-9. [DOI] [PubMed] [Google Scholar]
- Benchaouir R. Goyenvalle A. Splicing modulation mediated by small nuclear RNAs as therapeutic approaches for muscular dystrophies. Curr. Gene Ther. 2012;12:179–191. doi: 10.2174/156652312800840586. [DOI] [PubMed] [Google Scholar]
- Brillantes A.M. Bezprozvannaya S. Marks A.R. Developmental and tissue-specific regulation of rabbit skeletal and cardiac muscle calcium channels involved in excitation-contraction coupling. Circ. Res. 1994;75:503–510. doi: 10.1161/01.res.75.3.503. [DOI] [PubMed] [Google Scholar]
- Charrier S. Stockholm D. Seye K., et al. A lentiviral vector encoding the human Wiskott-Aldrich syndrome protein corrects immune and cytoskeletal defects in WASP knockout mice. Gene Ther. 2005;12:597–606. doi: 10.1038/sj.gt.3302440. [DOI] [PubMed] [Google Scholar]
- Chaudhari N. A single nucleotide deletion in the skeletal muscle-specific calcium channel transcript of muscular dysgenesis (mdg) mice. J. Biol. Chem. 1992;267:25636–25639. [PubMed] [Google Scholar]
- Cirak S. Arechavala-Gomeza V. Guglieri M., et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.R. Haan E. Jungbluth H., et al. Principal mutation hotspot for central core disease and related myopathies in the C-terminal transmembrane region of the RYR1 gene. Neuromuscul. Disord. 2003;13:151–157. doi: 10.1016/s0960-8966(02)00218-3. [DOI] [PubMed] [Google Scholar]
- De Cauwer H. Heytens L. Martin J.J. Workshop report of the 89th ENMC International Workshop: Central Core Disease, 19–20 January 2001, Hilversum, The Netherlands. Neuromuscul. Disord. 2002;12:588–595. doi: 10.1016/s0960-8966(02)00002-0. [DOI] [PubMed] [Google Scholar]
- Desmet F.O. Hamroun D. Lalande M., et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro A. Monnier N. Romero N.B., et al. A recessive form of central core disease, transiently presenting as multi-minicore disease, is associated with a homozygous mutation in the ryanodine receptor type 1 gene. Ann. Neurol. 2002;51:750–759. doi: 10.1002/ana.10231. [DOI] [PubMed] [Google Scholar]
- Gorman L. Suter D. Emerick V., et al. Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc. Natl. Acad. Sci. USA. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyenvalle A. Vulin A. Fougerousse F., et al. Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306:1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- Hammond S.M. Wood M.J. Genetic therapies for RNA mis-splicing diseases. Trends Genet. 2011;27:196–205. doi: 10.1016/j.tig.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Herrmann-Frank A. Richter M. Sarközi S., et al. 4-Chloro-m-cresol, a potent and specific activator of the skeletal muscle ryanodine receptor. Biochim. Biophys. Acta. 1996;1289:31–40. doi: 10.1016/0304-4165(95)00131-x. [DOI] [PubMed] [Google Scholar]
- Jungbluth H. Central core disease. Orphanet J. Rare Dis. 2007;2:25. doi: 10.1186/1750-1172-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungbluth H. Muller C.R. Halliger-Keller B., et al. Autosomal recessive inheritance of RYR1 mutations in a congenital myopathy with cores. Neurology. 2002;59:284–287. doi: 10.1212/wnl.59.2.284. [DOI] [PubMed] [Google Scholar]
- Kendall G.C. Mokhonova E.I. Moran M., et al. Dantrolene enhances antisense-mediated exon skipping in human and mouse models of Duchenne muscular dystrophy. Sci. Transl. Med. 2012;4:164ra160. doi: 10.1126/scitranslmed.3005054. [DOI] [PubMed] [Google Scholar]
- Kyselovic J. Leddy J.J. Ray A., et al. Temporal differences in the induction of dihydropyridine receptor subunits and ryanodine receptors during skeletal muscle development. J. Biol. Chem. 1994;269:21770–21777. [PubMed] [Google Scholar]
- Krebs J. The influence of calcium signaling on the regulation of alternative splicing. Biochim. Biophys. Acta. 2009;1793:979–984. doi: 10.1016/j.bbamcr.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Lanner J.T. Georgiou D.K. Joshi A.D. Hamilton S.L. Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harb. Perspect. Biol. 2010;2:a003996. doi: 10.1101/cshperspect.a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M. Rho S.H. Shin D.W., et al. Negatively charged amino acids within the intraluminal loop of ryanodine receptor are involved in the interaction with triadin. J. Biol. Chem. 2004;279:6994–7000. doi: 10.1074/jbc.M312446200. [DOI] [PubMed] [Google Scholar]
- Lorain S. Peccate C. Le Hir M. Garcia L. Exon exchange approach to repair Duchenne dystrophin transcripts. PLoS One. 2010;5:e10894. doi: 10.1371/journal.pone.0010894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch P.J. Tong J. Lehane M., et al. A mutation in the transmembrane/luminal domain of the ryanodine receptor is associated with abnormal Ca2+ release channel function and severe central core disease. Proc. Natl. Acad. Sci. USA. 1999;96:4164–4169. doi: 10.1073/pnas.96.7.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty I. Robert M. Villaz M., et al. Biochemical evidence for a complex involving dihydropyridine receptor and ryanodine receptor in triad junctions of skeletal muscle. Proc. Natl. Acad. Sci. USA. 1994;91:2270–2274. doi: 10.1073/pnas.91.6.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier N. Romero N.B. Lerale J., et al. Familial and sporadic forms of central core disease are associated with mutations in the C-terminal domain of the skeletal muscle ryanodine receptor. Hum. Mol. Genet. 2001;10:2581–2592. doi: 10.1093/hmg/10.22.2581. [DOI] [PubMed] [Google Scholar]
- Monnier N. Ferreiro A. Marty I., et al. A homozygous splicing mutation causing a depletion of skeletal muscle RYR1 is associated with multi-minicore disease congenital myopathy with ophthalmoplegia. Hum. Mol. Genet. 2003;12:1171–1178. doi: 10.1093/hmg/ddg121. [DOI] [PubMed] [Google Scholar]
- Monnier N. Marty I. Faure J., et al. Null mutations causing depletion of the type 1 ryanodine receptor (RYR1) are commonly associated with recessive structural congenital myopathies with cores. Hum. Mutat. 2008;29:670–678. doi: 10.1002/humu.20696. [DOI] [PubMed] [Google Scholar]
- Naldini L. Lentiviruses as gene transfer agents for delivery to non-dividing cells. Curr. Opin. Biotechnol. 1998;9:457–463. doi: 10.1016/s0958-1669(98)80029-3. [DOI] [PubMed] [Google Scholar]
- Pietri-Rouxel F. Gentil C. Vassilopoulos S., et al. DHPR alpha1S subunit controls skeletal muscle mass and morphogenesis. EMBO J. 2010;29:643–654. doi: 10.1038/emboj.2009.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter G.A., Jr. Makuck R.F. Rivkees S.A. Reduction in intracellular calcium levels inhibits myoblast differentiation. J. Biol. Chem. 2002;277:28942–28947. doi: 10.1074/jbc.M203961200. [DOI] [PubMed] [Google Scholar]
- Reuter J.S. Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezgui S.S. Vassilopoulos S. Brocard J., et al. Triadin (Trisk 95) overexpression blocks excitation-contraction coupling in rat skeletal myotubes. J. Biol. Chem. 2005;280:39302–39308. doi: 10.1074/jbc.M506566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero N.B. Monnier N. Viollet L., et al. Dominant and recessive central core disease associated with RYR1 mutations and fetal akinesia. Brain. 2003;126:2341–2349. doi: 10.1093/brain/awg244. [DOI] [PubMed] [Google Scholar]
- Roux-Buisson N. Rendu J. Denjoy I., et al. Functional analysis reveals splicing mutations of the CASQ2 gene in patients with CPVT: implication for genetic counselling and clinical management. Hum. Mutat. 2011;32:995–999. doi: 10.1002/humu.21537. [DOI] [PubMed] [Google Scholar]
- Seigneurin-Venin S. Parrish E. Marty I., et al. Involvement of the dihydropyridine receptor and internal Ca2+ stores in myoblast fusion. Exp. Cell Res. 1996;223:301–307. doi: 10.1006/excr.1996.0085. [DOI] [PubMed] [Google Scholar]
- Spitali P. Aartsma-Rus A. Splice modulating therapies for human disease. Cell. 2012;148:1085–1088. doi: 10.1016/j.cell.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Takeshima H. Iino M. Takekura H., et al. Excitation-contraction uncoupling and muscular degeneration in mice lacking functional skeletal muscle ryanodine-receptor gene. Nature. 1994;369:556–559. doi: 10.1038/369556a0. [DOI] [PubMed] [Google Scholar]
- Taniguchi-Ikeda M. Kobayashi K. Kanagawa M., et al. Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature. 2011;478:127–131. doi: 10.1038/nature10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarroni P. Rossi D. Conti A. Sorrentino V. Expression of the ryanodine receptor type 3 calcium release channel during development and differentiation of mammalian skeletal muscle cells. J. Biol. Chem. 1997;272:19808–19813. doi: 10.1074/jbc.272.32.19808. [DOI] [PubMed] [Google Scholar]
- Taylor L.E. Kaminoh Y.J. Rodesch C.K. Flanigan K.M. Quantification of dystrophin immunofluorescence in dystrophinopathy muscle specimens. Neuropathol. Appl. Neurobiol. 2012;38:591–601. doi: 10.1111/j.1365-2990.2012.01250.x. [DOI] [PubMed] [Google Scholar]
- Vulin A. Barthelemy I. Goyenvalle A., et al. Muscle function recovery in golden retriever muscular dystrophy after AAV1-U7 exon skipping. Mol. Ther. 2012;20:2120–2133. doi: 10.1038/mt.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein N. Avril A. Bartoli M., et al. Efficient bypass of mutations in dysferlin deficient patient cells by antisense-induced exon skipping. Hum. Mutat. 2010;31:136–142. doi: 10.1002/humu.21160. [DOI] [PubMed] [Google Scholar]
- Wheeler T.M. Lueck J.D. Swanson M.S., et al. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. Jungbluth H. Sewry C.A., et al. Molecular mechanisms and phenotypic variation in RYR1-related congenital myopathies. Brain. 2007;130:2024–2036. doi: 10.1093/brain/awm096. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]