Abstract
SUMMARY
The AIDS pandemic that started in the early 1980s is due to human immunodeficiency virus type 1 (HIV-1) group M (HIV-M), but apart from this major group, many divergent variants have been described (HIV-1 groups N, O, and P and HIV-2). The four HIV-1 groups arose from independent cross-species transmission of the simian immunodeficiency viruses (SIVs) SIVcpz, infecting chimpanzees, and SIVgor, infecting gorillas. This, together with human adaptation, accounts for their genomic, phylogenetic, and virological specificities. Nevertheless, the natural course of non-M HIV infection seems similar to that of HIV-M. The virological monitoring of infected patients is now possible with commercial kits, but their therapeutic management remains complex. All non-M variants were principally described for patients linked to Cameroon, where HIV-O accounts for 1% of all HIV infections; only 15 cases of HIV-N infection and 2 HIV-P infections have been reported. Despite improvements in our knowledge, many fascinating questions remain concerning the origin, genetic evolution, and slow spread of these variants. Other variants may already exist or may arise in the future, calling for close surveillance. This review provides a comprehensive, up-to-date summary of the current knowledge on these pathogens, including the historical background of their discovery; the latest advances in the comprehension of their origin and spread; and clinical, therapeutic, and laboratory aspects that may be useful for the management and the treatment of patients infected with these divergent viruses.
INTRODUCTION
The first human immunodeficiency virus (HIV) to be isolated, in 1983, was the prototype of what was later designated HIV type 1 (HIV-1) group M (HIV-M) and is the virus responsible for the current pandemic (1). The existence and circulation of other major HIV variants were first suspected in 1985, based on atypical biological profiles of infection among prostitutes in Dakar, Senegal (2). This led to the characterization of a new variant in 1986 (3), designated HIV-2, as it showed marked genetic differences from HIV-M, including over 50% sequence divergence in the genes encoding the envelope proteins.
Other variants exhibiting less marked genetic divergence from the HIV-M prototype were subsequently identified and are currently divided into three groups based on sequence similarities. Each group arose from independent transmissions of great ape viruses to humans. The first of these variant groups to be identified was HIV-1 group O (HIV-O) in 1990, followed by HIV-1 group N (HIV-N) in 1998, both in patients of Cameroonian origin. In 2009, a new variant was isolated in France, also from a Cameroonian woman, and represented the prototype of a new group, HIV-P.
Although these variants all cause a similar disease in humans, they have specific phylogenetic, virological, and epidemiological characteristics.
DISCOVERY
HIV-O
The prototype strain of HIV-O, ANT70, was isolated in 1990 at the Institute of Tropical Medicine in Antwerp, Belgium, from a Cameroonian couple living in Belgium who presented with generalized lymphadenopathies (4). This virus had particular antigenic and genetic characteristics but was more closely related to HIV-1 than to HIV-2. Subsequent serological studies demonstrated its presence in Cameroon and Gabon (5). In 1994, a new divergent strain (MVP5180), similar to strain ANT70, was isolated in Germany by Gurtler et al. from a Cameroonian man with AIDS (6). In the same year, another variant, VAU, was identified in a French patient with AIDS. The sequence of its env gene was similar to those of ANT70 and MVP5180 (7), but phylogenetic analyses showed that these three viruses were as different from one another as the different HIV-M subtypes (7). Nucleotide sequencing showed that the pol gene of strains ANT70 and MVP5180 shared 73% homology with HIV-1 variants of European and African origins, whereas the env gene shared only 50% homology (8). The overall difference between the genomes was less than 50%, excluding the creation of a new HIV type but requiring HIV-1 to be split into two groups: group M (major) and group O (outlier).
HIV-N
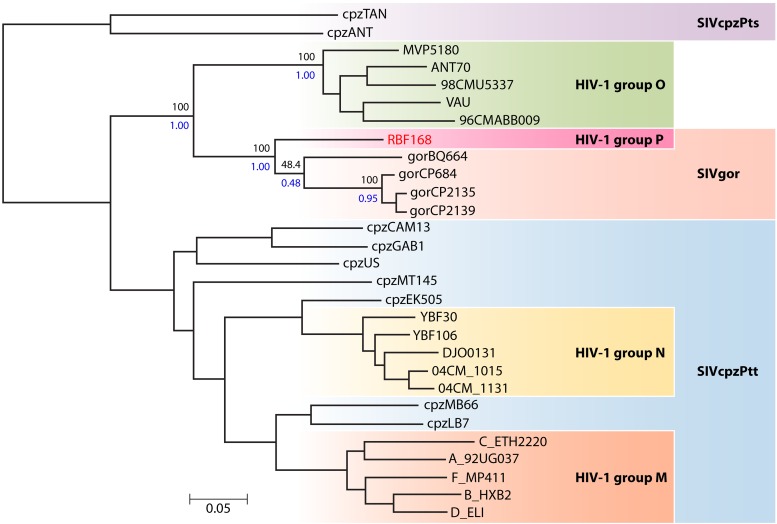
In 1998, a Franco-Cameroonian team identified a new HIV-1 variant strain (YBF30) isolated from a Cameroonian woman who had died of AIDS in 1995 (9), leading to the definition of a new branch in the HIV-1 lineage (Fig. 1). This patient's serum reacted with an envelope antigen from a simian immunodeficiency virus (SIV) isolated from a chimpanzee (SIVcpz), rather than with representative group M and O antigens. Sequence analysis of this strain showed that the phylogenetic position depended on the gene: YBF30 clustered with SIVcpz variants in env and between SIVcpz and HIV-M in gag and pol. A contemporary seroepidemiological study of 700 HIV-positive sera identified 3 that reacted with an antigen representative of the YBF30 prototype strain. Molecular characterization also showed a phylogenetic link between these samples and YBF30, confirming the circulation of these variants in Cameroon. This led to the creation of a new HIV-1 group, group N (HIV-N), for “non-M, non-O.”
Fig 1.
Phylogenetic relationships between the HIV-1, SIVcpz, and SIVgor lineages. (Reprinted from reference 10 with permission of the publisher.)
HIV-P
An atypical variant (RBF168) was first identified in 2009 in a 62-year-old Cameroonian woman who was confirmed to be HIV-1 seropositive in 2004, soon after arriving in France (10). HIV-O infection was initially suspected on the basis of negative specific tests for HIV-M along with the patient's origin and the results of a serotyping assay. A high level of viral replication, detected with a technique specific for HIV-O and a group-nonspecific technique, reinforced these suspicions. However, PCR tests specific for group O were negative. Whole-genome sequencing revealed that this virus was clearly distinct from HIV-M, -N, and -O but very similar to a simian immunodeficiency virus (SIVgor) identified in a gorilla population in Cameroon (11) (Fig. 1). This strain was therefore considered the first representative of a new HIV-1 branch, designated group “P” according to the nomenclature. Subsequently, an American team confirmed the presence of this variant in Cameroon after examining a sample taken in 2006 from a 54-year-old Cameroonian man (12).
ORIGIN, ADAPTATION, AND DIVERSIFICATION
Simian Origin
After the discovery in 1989 of an SIV linked to HIV-1, the chimpanzee species Pan troglodytes troglodytes (SIVcpzPtt) was suspected of being the original source of HIV-1 (13, 14). The structure of a phylogenetic tree including sequences of SIVcpzPtt and HIV-1 suggested that groups M, N, and O arose from three distinct cross-species transmission events (15, 16). Studies in Cameroon have since demonstrated the strong endemicity and diversity of SIVcpz in wild chimpanzees as well as differences in the geographic distribution of chimpanzees infected by the SIV variants that gave rise to HIV-M and HIV-N (17, 18).
In 2006, the Western lowland gorilla of Cameroon was also shown to carry an SIV (11). The phylogenetic position of SIVgor in the SIVcpzPtt lineage implies the existence of one or several transmission events between chimpanzees and gorillas, without clearly identifying the mode of transmission (19). A phylogenetic link was also established between SIVgor and HIV-O (Fig. 1), but the genetic distance was too large to conclude that HIV-O originated directly from SIVgor. The simian reservoir of HIV-O therefore remains to be identified.
However, the close phylogenetic relationship between HIV-P (represented by the RBF168 prototype strain) and SIVgor makes gorillas the source of this new HIV-1 group (Fig. 2) (10) and suggests that they may be an intermediate host between chimpanzees and humans. This discovery reopens the debate on the origin, evolution, and complex interrelationships between SIV and HIV. A large number of SIV variants, representing potential sources of new HIV variants and also potentially responsible for past transmission events that remain to be determined, probably remain to be identified in African nonhuman primates.
Fig 2.
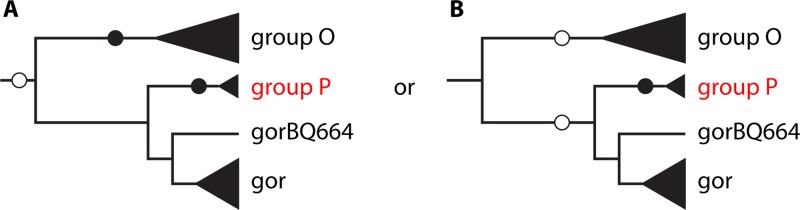
Scenarios for transmission of SIV from chimpanzees and gorillas to humans, explaining the emergence of HIV-O and HIV-P. Empty circles, SIV transmission events from chimpanzees; solid circles, SIV transmission events from gorillas. (A) SIVcpz transmitted to gorillas on one occasion, followed by transmission of SIVgor to humans on two occasions (groups O and P). (B) SIVcpz transmitted to humans on one occasion (group O) and to gorillas on one occasion and then transmission of SIVgor to humans on one occasion (group P). (Modified from reference 10 with permission of the publisher.)
It is difficult to date the initial interspecies transmission event, but molecular-based calculations suggest that the most recent common ancestors (MRCAs) responsible for the current epidemics arose at the beginning of the 20th century (1884 to 1924) for HIV-M (20), in the 1920s (1890 to 1940) for HIV-O (21), and in 1963 (1948 to 1977) for HIV-N (22). Too few HIV-P and SIVgor sequences are available for precise dating of the HIV-P MRCAs, but a recent work estimated the group P MRCA to have arisen in the 1980s and the HIV-P/SIVgor MRCA to have arisen in the second half of the 19th century (23).
The evolutionary time scale of HIV-O thus appears to be similar to that of HIV-M but with slower growth of the viral population during the 20th century. This would partly explain the lesser dissemination of HIV-O strains (21). The age of the HIV-O epidemic has been confirmed by a study of necropsy specimens from a Norwegian family who died of AIDS in 1976, with up to 10 years of clinical manifestations of HIV infection (24). HIV-O DNA was detected in the father (a sailor who had visited several African countries during the 1960s) and his daughter, while anti-HIV-O antibodies were detected in both the father and the mother. The father represents the oldest documented case of AIDS in Europe. The VAU strain was isolated from a French woman who had never traveled to Africa but who died of AIDS in 1992 (7). This patient was most likely infected before 1980 (her child born in 1980 died in 1981 with clinical features of AIDS). These data suggest that HIV-O was already present in Europe over 40 years ago and that its low prevalence is therefore not due to recent emergence.
Adaptation to Humans
One plausible mode of simian-to-human transmission is human contact with monkey blood during activities with a risk of skin trauma, such as hunting and preparation of bushmeat, or through bites of captive or domesticated monkeys. Once the interspecies barrier was crossed, various biological factors (cellular restriction factors and replicative capacity in the new host) and also environmental, sociological, demographic, behavioral, and medical factors (injections and transfusions, etc.) would have facilitated epidemic spread in human populations. The slow spread of non-M variants could thus be due not only to epidemiological factors but also to poorer human adaptation of these viruses.
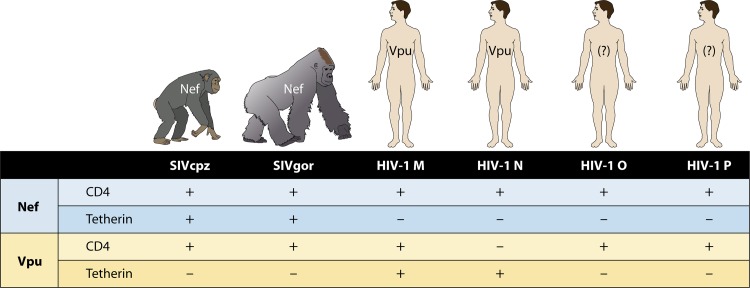
Wain et al., upon comparing the entire genomic sequences of HIV-1 and SIVcpz, identified residue 30R (Arg, basic) in the matrix protein p17Gag as an amino acid signature shared by HIV groups M, N, and O (25), whereas all SIVcpzPtt and SIVgor isolates harbor 30M (Met, hydrophobic) (19). The M30R mutation would thus be specific to human adaptation. The HIV-P prototype strain described in France by Plantier et al. carries 30M and is thus more closely related to SIVcpz/SIVgor than to other human viruses (10). This lack of the Gag-30R residue has not prevented human HIV-P transmission but may result in poor adaptation. Paradoxically, the second, recently identified group P strain carries a different adaptive change (Gag-30K) (12).
Among the cellular restriction factors with antiviral activity, tetherin seems to have played a major role in human adaptation of SIV. Tetherin inhibits the release of enveloped viruses by “attaching” them to the cell membrane. In chimpanzees and gorillas, the SIVcpz/SIVgor protein Nef counteracts the action of tetherin (Fig. 3). In humans, tetherin is resistant to the action of the HIV-M Nef protein, the role of which is replaced by the viral protein Vpu. Furthermore, Vpu degrades CD4 receptors at the surface of infected cells, thus avoiding CD4 interference during virion release. Interestingly, distinct studies recently demonstrated that the HIV-O and HIV-P Vpu proteins do not antagonize tetherin but degrade CD4 receptors (23, 26, 27). HIV-N Vpu proteins gained some modest antitetherin activity (26), except for that of the unique strain recently described outside Cameroon (28) that fully counteracted this antiviral factor as efficiently as the Vpu proteins of pandemic HIV-M strains (29), but none of them can degrade CD4 receptors, thus leading to reinternalization of excreted viruses (26) (Fig. 3). Thus, only HIV-M appears to have evolved a Vpu protein allowing efficient infection of human cells. This may partly explain the success of HIV-M.
Fig 3.
Adaptive evolution of the Nef and Vpu viral proteins in primate lentiviruses. The degradation of CD4 receptors by the Nef and Vpu viral proteins and their antagonistic activity against the restriction factor tetherin are represented by +, and the absence of these properties or very low activity is represented by −. The Nef proteins of SIVcpz and SIVgor degrade CD4 receptors and antagonize the factor tetherin in their respective hosts, whereas those of HIV-1 do not have any activity against human tetherin. Only the Vpu protein from HIV-1 group M, through adaptive evolution, possesses antitetherin properties and degrades CD4 receptors. (Reprinted from reference 26 with permission from Elsevier.)
Despite these possible adaptive limitations, non-M variants are pathogenic for humans and produce high plasma viral loads, contrary to HIV-2 (derived from another SIV variant [SIVsmm] infecting the sooty mangabey [Cercocebus atys]). The genetic proximity between chimpanzees, gorillas, and humans could explain why HIV-1 maintains a high replicative capacity (fitness) in humans. However, Arien et al. reported results of competitive in vitro experiments suggesting that the replicative fitness of HIV-O could be inferior to that of HIV-2, which itself had lower replicative fitness than HIV-M (30). Although this “fitness pyramid” matches current differences in the prevalences of these variants, it does not correspond to differences between HIV-2 and HIV-M or HIV-O in terms of viral load, pathogenicity, or transmissibility.
Genetic Diversification in Humans
Following simian-to-human transmission events, HIV evolution has been influenced by biological selective pressure and epidemiological factors.
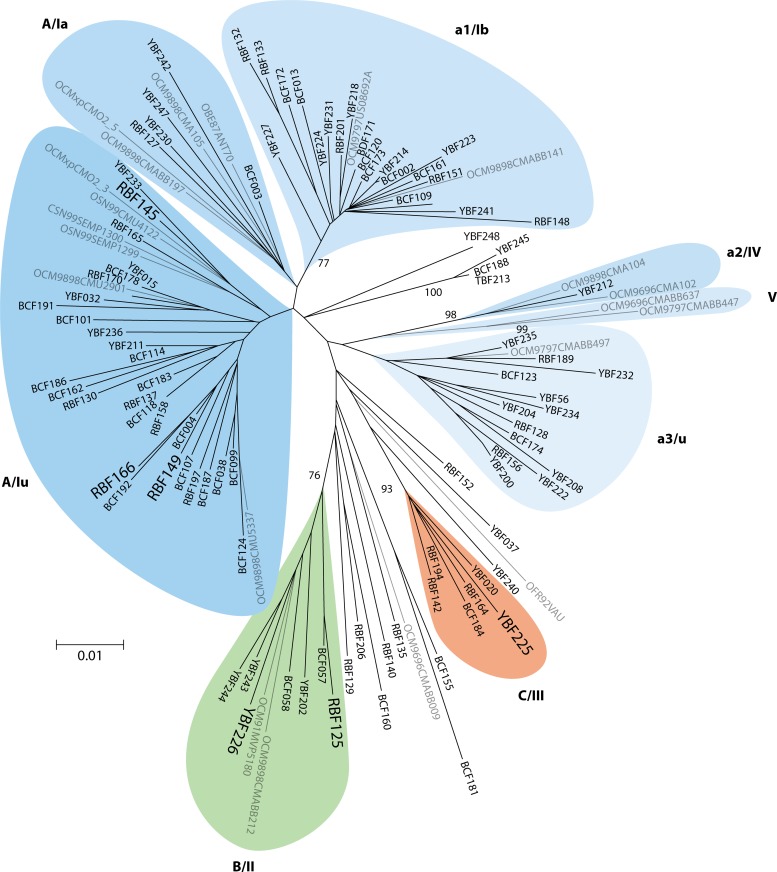
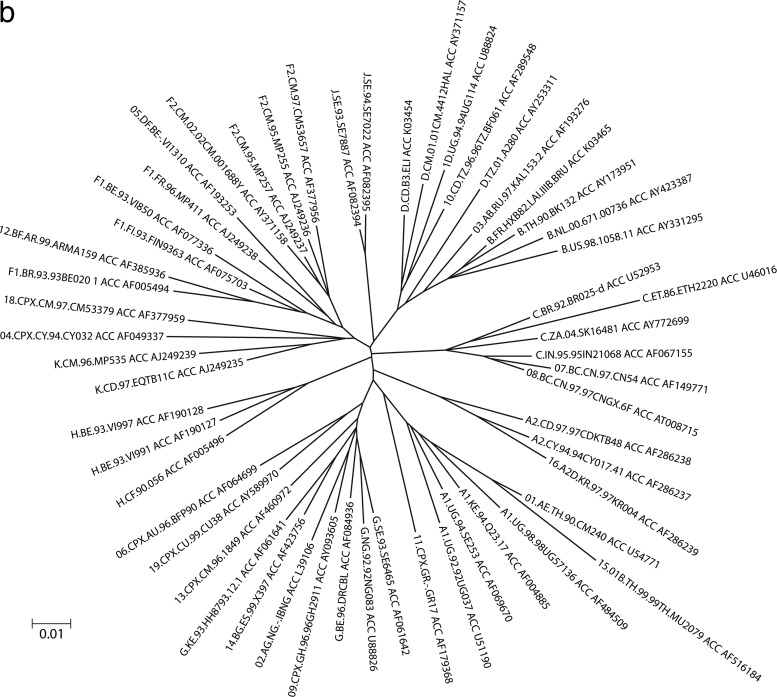
HIV-O strains show substantial intragroup genetic diversity. Phylogenetic trees constructed from group O genomic sequences show a comet-like symmetry (Fig. 4a), intermediate between the double-star symmetry of HIV-M pandemic strains (Fig. 4b) and the topology of HIV-M sequences from the Democratic Republic of Congo, considered to be among the oldest HIV variants (31). This intragroup diversity has led to HIV-O being divided into clades or clusters that are less closely related to one another than are HIV-M subtypes. Three clades (clades A, B, and C) (32) and five clusters (clusters I to V) (33) have been described, along with numerous divergent unclassified strains (Fig. 4a). These clades and clusters overlap, and a consensus classification is still pending. Among 202 viruses characterized in France and Cameroon, 79% belong to clade A and 4% to 5% belong to clades B and C, while 12% belong to none of these three clades (our unpublished data). This distribution is presumably due to human-to-human transmission in restricted geographic areas, with founder effects of microepidemics represented today by clades A, B, and C, or to a different diversification and evolutionary history of group O relative to those of group M (32).
Fig 4.
Comparison of phylogenetic trees constructed from 117 HIV-O sequences (a) and 55 HIV-M sequences (b) in the integrase region (603 bp). ACC, GenBank accession number. (Modified from reference 104 with permission of the publisher.)
Data on the genetic diversity of HIV-N are more limited. To date, nine near-complete genomic sequences and five partial sequences have been determined in 14 cases of HIV-N infection confirmed by molecular biology (28, 34). These genomes all have the same profile, including an env gene derived from an SIVcpz variant and gag and pol genes closely related to those of HIV-M. This points to previous dual infection by two SIVcpz variants, probably in a chimpanzee, followed by their recombination (17). Sequence analysis shows a high level of HIV-N intragroup homogeneity (34, 35), which, together with the low prevalence of this group, confirms a relatively recent introduction and very slow spread in the human population.
The two HIV-P strains characterized to date are phylogenetically closely related, yet there is no evidence of any link between the two patients (12). Comparison with SIVs from chimpanzees and gorillas shows no evidence of recombination between these variants and SIV (10). Pending the discovery and characterization of other strains, it is impossible to speculate on the evolution of HIV-P.
NATURAL HISTORY OF INFECTION
The natural history of primate lentivirus infections depends on the virus and its host. Many studies in captive monkeys have indicated that SIV infections were generally considered nonpathogenic in their natural hosts (36). This was particularly demonstrated for sooty mangabey infection by SIVsmm (the original source of HIV-2), characterized by active viral replication but no disease progression (36). However, recently, progression to AIDS has been described in nonhuman primates infected over long periods and having lived long enough, beyond the average life span, to make visible the disease. In particular, infection in chimpanzees by their own SIV (SIVcpzPtt and SIVcpzPts, for the Pan troglodytes troglodytes and Pan troglodytes schweinfurthii species, respectively) was thought to be harmless until studies demonstrated that SIV infection of P. troglodytes schweinfurthii was associated with a 10- to 16-fold increase in age-corrected risk of death (37) and that observations of a naturally SIV-infected P. troglodytes troglodytes chimpanzee suggested clinical progression to an AIDS-like disease (38).
In humans, pathogenicity and natural history are largely known for HIV-M pandemic infection, but very few data are available for infections with HIV-1 non-M variants.
HIV-O
Nkengasong et al. described a 10-year follow-up study of a couple infected by strain ANT70 (39). Clinical progression without antiretroviral (ARV) therapy resembled that of HIV-M infection. The woman was still asymptomatic 8 years after seroconversion, and no switch from a non-syncytium-inducing (NSI) to a syncytium-inducing (SI) phenotype had occurred, although her CD4 cell count fell markedly, as in her partner, whose virus switched to an SI phenotype. More recently, serial blood samples collected from an ARV-naive patient during a 7-year period, from diagnosis to symptom onset, were studied retrospectively (40). Clinical progression occurred in the absence of an SI phenotype switch, with viruses using only the CCR5 receptor. A recent ex vivo study examined the replicative capacity and cytopathogenicity of eight primary HIV-O isolates and seven HIV-M isolates in models based on peripheral blood mononuclear cells (PBMCs) and excised tonsils. The replicative “fitness” of the HIV-O isolates was five to eight times lower than that of the HIV-M isolates. However, the HIV-O strains showed variable cytopathogenicity in terms of CD4 cell depletion. Two isolates with R5 tropism caused massive depletion, comparable to that caused by X4 isolates and superior to that caused by group M R5 isolates (41).
HIV-O infection thus appears to have a natural history similar to that of HIV-M infection, in keeping with the limited data on in vivo replication (42, 43). In the absence of a specific cohort, most data were obtained from evaluations of assay techniques or from follow-up studies of limited numbers of patients and indicated a level of replication similar to that of HIV-M. This was confirmed by a recent study of 18 ARV-naive Cameroonian patients showing a median plasma viral load of 4.1 log copies/ml (42).
Vertical (mother-to-child) transmission of HIV-O has rarely been reported (44, 45), but this may simply be due to the low prevalence of these variants and their difficult detection.
HIV-N
Despite the small number of cases described to date, HIV-N appears to be a pathogenic virus with in vitro and in vivo replication similar to that of HIV-M (9, 35). Patients develop clinical manifestations similar to those seen in HIV-M infection, even during the acute phase of the primary infection, and progress to AIDS (9, 28, 35, 46, 47). The identification of HIV-N in a 7-year-old orphan suggests that mother-to-child transmission can occur (35, 46), and phylogenetic studies have shown the capacity for human-to-human HIV-N transmission (47, 48).
HIV-P
The HIV-P prototype strain RBF168 was easily isolated from the patient's plasma and cells. Immunological follow-up of this patient, who was ARV naive for 5 years, revealed high plasma viral loads (between 4.5 and 5 log copies/ml) and immunosuppression (mean CD4 cell count, 326 cells/mm3; nadir, 260 cells/mm3) characteristic of HIV infection (10). However, without knowledge of the date of infection, it is impossible to estimate the kinetics of disease progression. No data on the second case are available (12).
EPIDEMIOLOGY
Situation in Western Central Africa
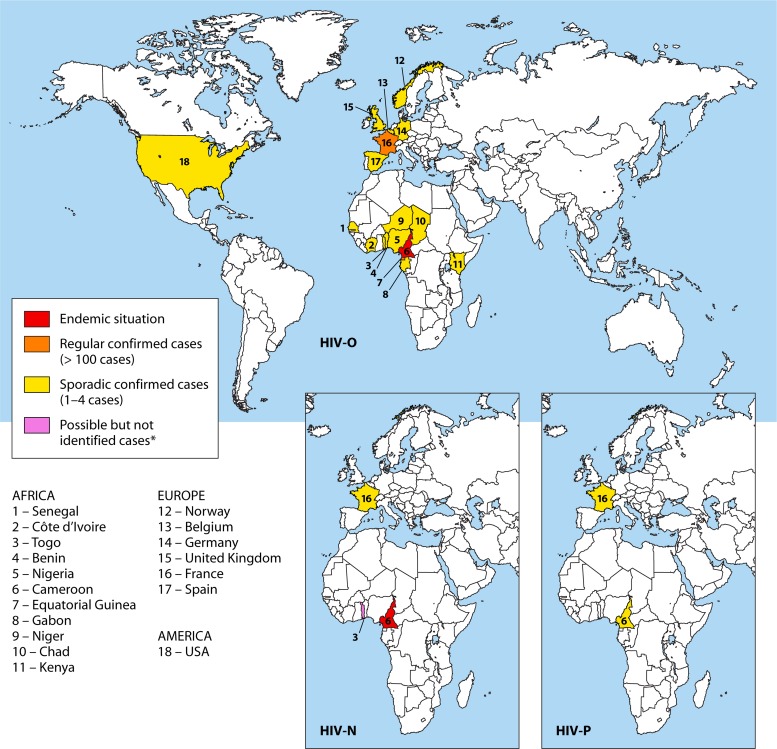
All group M subtypes and most circulating recombinant forms (CRFs) are present in Central Africa, reflecting the antiquity of the epidemic in this region. However, HIV-1 non-M variants are mainly endemic to Cameroon (and, possibly, bordering regions), suggesting that this region is the origin of all existing HIV-1 groups (Fig. 5). The existence of SIV reservoirs in close proximity to a large human population has undoubtedly contributed to the diversity of the epidemic, through repeated interspecies transmission events.
Fig 5.
Molecular epidemiology of HIV-1 non-M variants. Shown is a representation of the worldwide distribution of HIV-1 non-M variants according to the number of reported cases published. Three maps are presented for the HIV-O, HIV-N, and HIV-P variants. Only Europe and Africa are represented for the distribution of the variants N and P, as no infection was reported outside these regions. ∗, Togo is the most likely place of contamination of the HIV-N case reported by Delaugerre et al. (28), but no case was reported from this country.
Sporadic cases of HIV-O were reported in different countries of western Central Africa, such as Chad, Equatorial Guinea, and Gabon (49–52). However, to date, HIV-O is most prevalent in Cameroon, accounting for about 1% of all HIV infections in Cameroon (42). The prevalence has been stable over the last 10 years, while the prevalence of HIV-M has progressed exponentially (53, 54). Cocirculation of groups M and O in Cameroon has resulted in cases of dual M and O (M-O) infections, associated or not with recombinant forms (54–57). The prevalence of dual M-O infection and M/O recombinants is difficult to assess. A seromolecular survey conducted at the Centre Pasteur du Cameroun between 2006 and 2009 suggested that the prevalence of dual infection was about 10% among patients with HIV-O infection (58). This study also identified five unique M/O recombinants, two in patients with no evidence of dual infection, suggesting direct transmission. This confirmed the circulation of M/O recombinants in Cameroon (55–57), despite a low prevalence of M-O dual infection. The complex epidemiological situation in Central Africa, from where M/O CRFs may emerge in the future, must be taken into account when designing therapeutic strategies and future vaccination programs.
To date, only 14 cases of HIV-N infection have been reported in this region, 13 of which have been confirmed by molecular biology, most of them in Cameroon (9, 34, 35, 46–48). Analyses of samples collected between 1997 and 1999 (46) and between 2002 and 2006 (34) in Cameroon showed a very low prevalence of HIV-N infection of 0.1% among all HIV-positive samples.
With only two documented cases of HIV-P infection, both in patients of Cameroonian origin, the prevalence is impossible to estimate. However, a retrospective analysis of 1,736 HIV-positive sera by Vallari et al. suggested a prevalence of 0.06% in Cameroon (12). Regular seroprevalence studies are needed to document the spread of this strain.
Situation outside Western Central Africa
Sporadic cases of HIV-O infection have been described in other regions of West and East Africa, including Senegal, Niger, Benin, Togo, Côte d'Ivoire, Nigeria, and Kenya, (59–64), and in Europe and the United States (4, 65–69) but always in patients or partners of patients originating from or linked to western Central Africa (Fig. 5). In France, little epidemiological information is available, apart from recent data from mandatory notifications, which indicate a prevalence of 0.1% among new diagnoses, representing 35 cases identified since 2003 (according to updated HIV National Reference Center data) (70). This is why a network (RES-O) has been set up by the National HIV Reference Center to monitor these variants. One hundred thirty-six HIV-O-infected patients have been identified since the first case was described in 1992 (43, 71). This is the largest series studied outside Cameroon, including two M-O dual infections (72), one HIV-M superinfection in a patient already infected by HIV-O (73), and one unique M/O recombinant (74).
Regarding HIV-N, it was thought until recently that these variants circulated only in Cameroon. However, in 2011, a primary infection was diagnosed in a Togolese patient living in France after returning from Togo, where he probably became infected. This report indicates that HIV-N is circulating outside Cameroon (28).
All these data, and the discovery of the HIV-P prototype strain in a Cameroonian woman living in France, illustrate the spread and diversity of the HIV pandemic. Regular surveillance of the diversity of strains circulating in Central Africa and also in countries linked to this region is therefore crucial.
LABORATORY TOOLS
Serological Diagnosis
HIV-O.
The marked genetic differences between HIV-O and HIV-M are reflected at the antigen level. The poor sensitivity of commercial serological tests for HIV-O variants was first noted in 1994 (75, 76), as all these tests are based on group M antigens. The addition of group O-specific antigens and development of fourth-generation enzyme-linked immunosorbent assay (ELISA) detection tests (combining the detection of p24 antigens and antibodies) have substantially improved the situation (77), but there is still a risk of false-negative HIV-O results (78–84), because test performance is evaluated by using restricted panels that do not cover the broad antigenic diversity of group O and because specific antigens are lacking in some kits.
Western blot profiles of HIV-O infection are variable, ranging from complete profiles to absent or weak reactivity with envelope glycoproteins and stronger reactivity against Gag and Pol proteins (Fig. 6). Serological confirmation of HIV-O infection requires an ELISA with group-specific peptides derived from the immunodominant region of the transmembrane glycoprotein (Gp41) and the V3 region of the surface glycoprotein (Gp120) (85, 86). No such commercial assays are available, and testing is currently performed only in specialized laboratories and reference centers.
Fig 6.
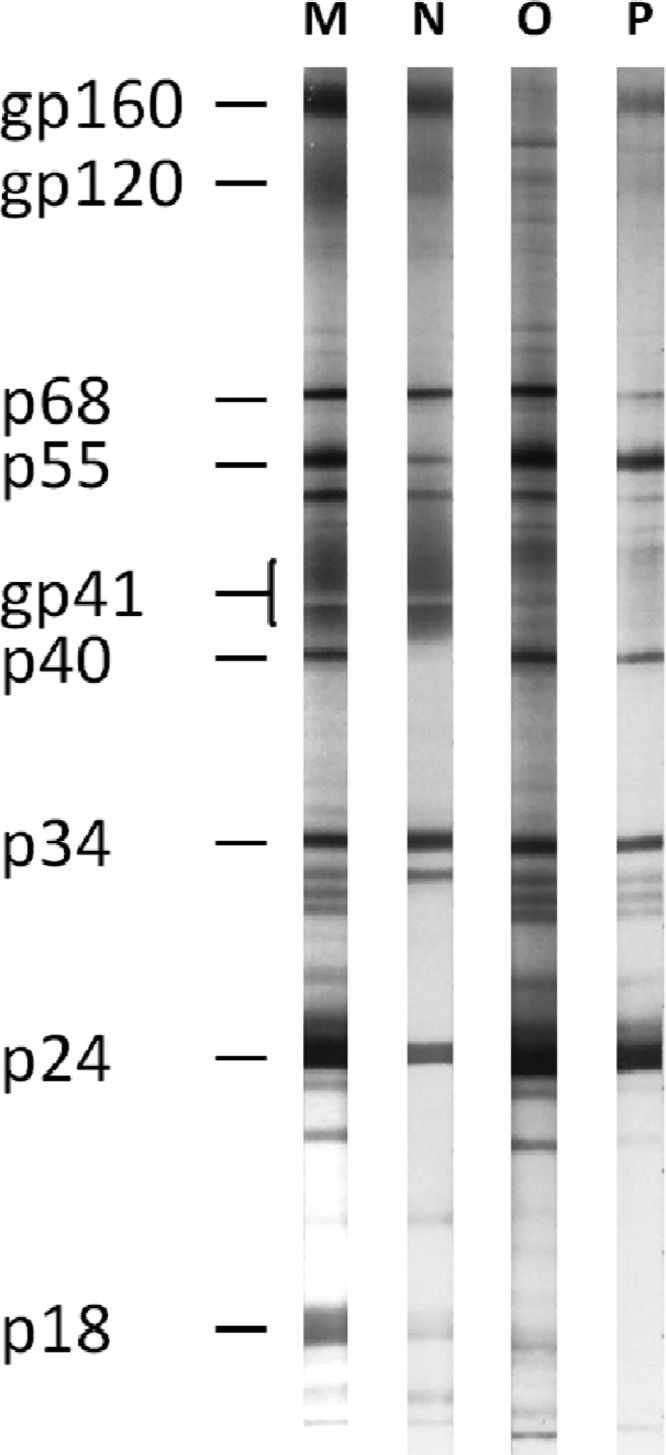
Western blot profiles of HIV-1 groups M, N, O, and P. The figure shows examples of Western blot reactivities with sera positive for the four groups. The group P-positive sample comes from the patient infected by the prototype strain RBF168. Of note, the group N-positive serum does not present a complete profile, due to advanced-stage disease (see reference 35 for complete profiles). The molecular masses of HIV-1 glycoproteins (gp) and proteins (p) are indicated in kDa.
HIV-N.
Although derived from SIVcpz, the HIV-N envelope elicits antibodies detectable by commercial tests and gives complete profiles on HIV-1 Western blots (35). The gp41/IDR and gp120/V3 epitopes are well conserved among HIV-N strains (34), theoretically allowing the detection of all such infections. Vallari et al. reported that none of eight HIV-N plasma samples tested with five commercial tests were negative (34). Of note, the primary infection identified in France was well detected by the tests used for routine screening (28).
HIV-P.
Although highly divergent from other HIV-1 strains, the two HIV-P strains identified so far raised no problems for serological detection (10, 12). Antibodies directed against the HIV-P prototype strain RBF168 were detected by all six kits tested and gave ratios or intensities (for rapid tests) similar to those of HIV-M strains (10). The Western blot profile (Fig. 6) does not point to a particular variant, while serotyping suggests HIV-O infection. Molecular analyses are thus needed to confirm HIV-P infection.
In conclusion, current screening tests for HIV infection appear to cover non-M variants (except for possible false-negative results with group O), because of the cross-reactivity of group M antigens or the inclusion of group O antigens, but test performance must be continuously monitored. The failure of screening tests to discriminate among HIV groups may partly explain why the number of non-M cases reported in regions where the disease is endemic is so small and calls for molecular investigations in these countries.
Measurement of Plasma Viral Load
Commercial viral load assays were initially not suitable for quantifying HIV-O variants, until the development by Abbott Molecular (Des Plaines, IL) of assays that can quantify group M and O variants albeit nonspecifically (87–89). Thus, alternative techniques were developed to allow specific molecular diagnosis and quantification of these variants (45, 90, 91). These specific assays are particularly adapted for the management of patients treated with ARV therapies, for the prevention of mother-to-child transmission, and for the proper quantification of HIV-O in patients with dual HIV-M–HIV-O infections or where both HIV-1 groups cocirculate. Comparison of the Abbott RealTime HIV-1 assay with a recent version of an alternative test showed a good correlation, although neither method is perfect (91). The marked genetic diversity of HIV-O means that these methods cannot reliably quantify some strains. Significantly lower values have been observed with the Abbott technique than with the alternative test, due to marked underquantifications for some patients. Particular care is necessary concerning the detection limits of the techniques, with some samples being undetectable with one technique and giving more than 500 copies/ml with another method. This means that residual replication can be overlooked, with a risk of resistance onset.
More recently, Roche Molecular Diagnostics (Rotkreuz, Switzerland) developed a new version of its real-time RT-PCR Cobas TaqMan HIV-1 assay. This v2.0 assay was improved (92), allowing nonspecific quantification of group M and O variants. Initial evaluation with a limited number of samples gave quantitative results for HIV-O equivalent to those of the Abbott assay (93), but these preliminary results need to be confirmed.
Proviral DNA screening for early diagnosis of mother-to-child transmission, especially during ARV therapy of newborns, is possible only with the alternative assays (45, 91).
Few studies are available for groups N and P. The new version of the Roche Cobas v2.0 kit and the Abbott assay can be used to quantify variants belonging to both groups (10, 93, 94), although the results for HIV-N may be underestimated with the latter (28, 94).
Paradoxically, these group-nonspecific assays can be used for virologic follow-up of patients with non-M infection but can no longer be used to detect these viruses. This can lead to delays in appropriate treatment and to a failure to identify dual infection or the presence of recombinant viruses, which cocirculate in regions where the disease is endemic or in countries linked to these regions. Indeed, these viruses were previously detected on the basis of discrepancies between different techniques or between immunologic and virologic results. Now, only genome amplification failure during tests for resistance mutations can suggest the presence of a variant, as current methods are not suited to these strains.
ANTIRETROVIRAL AGENTS
Natural Resistance Based on the Agence Nationale de Recherches sur Le SIDA et Les Hépatites Virales (ANRS) Algorithm
There are no specific recommendations on the therapeutic management of patients infected by non-M variants. However, as the clinical course appears to be similar to that of HIV-M infection, the same criteria for starting treatment are currently used. There is no HIV-O-specific algorithm for the interpretation of genotypic resistance, and results obtained by using algorithms designed for HIV-M are therefore only indicative and must be interpreted with care (95).
Data on HIV-O resistance are far from exhaustive (43, 95). One particularity of most HIV-O strains is their natural resistance to first-generation nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) (i.e., nevirapine and efavirenz), due to the natural presence of the Y181C mutation (clade dependent) in the RT region (96, 97). Although 40% of strains possess “wild-type” residue Y181Y, the variable phenotypic sensitivity of these strains to NNRTIs excludes the use of these drugs in all HIV-O-infected patients (98, 99). Etravirine, a second-generation NNRTI, seems to be effective because of its distinct genetic barrier, but the natural presence of the mutations A98G, V106I, E138A, and Y181C described for HIV-M could lead to a lesser sensitivity of a minority of strains (43, 95), although this needs to be confirmed in vitro. Concerning nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs), there are few polymorphic positions, except for the systematic presence of 210Y, but its influence on drug resistance in HIV-M is not known (43, 95).
The most important polymorphism is found in the protease region. HIV-O does not naturally possess major protease inhibitor (PI) resistance mutations, but the marked polymorphism of these strains is associated with a large number of minor mutations (at least 10) described for group M (42, 43, 95, 100). These mutations lead to possible genotypic resistance to saquinavir and resistance to tipranavir. The phenotypic consequences and impact on the rate of acquisition of PI resistance are not clearly established, owing to the lack of large clinical cohort studies, but these mutations could play a role in the onset of resistance, as suggested for HIV-2 (101–103).
The integrase active site seems to be well conserved; however, marked polymorphism has been noted at known resistance positions in vitro (104, 105), but their impact remains to be determined in vivo. Recent data on the use of raltegravir combined with other drugs suggest that this class is effective (95, 105–107).
In the Gp41 region, the presence of the N42D mutation in nearly all HIV-O strains, conferring genotypic resistance to enfuvirtide in HIV-M (depending on the algorithms used), seems to rule out the use of this drug (108). However, phenotypic studies and in vivo findings in treated patients show that HIV-O strains can be sensitive (95, 108–110).
These limited data therefore suggest that natural polymorphisms in HIV-O have different impacts depending on the target region, with probably little or no consequences for NRTI and integrase strand transfer inhibitor (INSTI) efficacy, uncertainty concerning PIs, and a genotype-phenotype discrepancy for enfuvirtide.
Finally, no resistance data on CCR5 antagonists have yet been published. A recent report on seven HIV-O primary isolates suggests that genotypic tools for determining tropism are poorly predictive (111), but this needs to be confirmed on a larger number of strains.
None of the HIV-N isolates characterized so far has harbored natural polymorphisms leading to resistance to NRTIs, NNRTIs, and PIs (34, 47).
Response to Antiretroviral Therapy and Virologic Failure
Data on the treatment response in vivo and on the selection of mutations upon virologic failure are extremely sparse. Table 1 summarizes the current clinical experience with treatment of these non-M HIV-1 infections.
Table 1.
Current clinical experience with treatment of non-M infectionsa
| Virus group | No. of treated patients | Country(ies) of monitoring | Treatment line(s) usedb | Reference |
|---|---|---|---|---|
| HIV-O | 22 | France | 3 NRTI (n = 2) | 43 |
| 2 NRTI + PI (n = 18) | ||||
| 2 NRTI + PI + RAL (n = 1) | ||||
| 2 NRTI + RAL (n = 1) | ||||
| 12 | France | Multiple lines including NRTI ± NNRTI ± PI ± T20 + RALc | 106 | |
| 9 | France | Multiple lines including NRTI ± NNRTI ± PI ± RAL + T20c | 108 | |
| 6 | Spain and England | Multiple lines including 2 NRTI + PIc | 113 | |
| 2 | Belgium and The Netherlands | Multiple lines including 2 NRTI + PI (d4T + 3TC + IDVc) | 112 | |
| 1 | Cameroon | Multiple lines including 2 NRTI + PI + RAL (TDF + 3TC + DRV-r + RALc) | 107 | |
| 1 | France | 2 NRTI + PI (AZT + 3TC + LPV-rc) | 73 | |
| 1 | Spain | Multiple lines including 2 NRTI + PI + RAL (ddI + FTC + DRV-r + RALc) | 105 | |
| 1 | Spain | Multiple lines including NRTI + PI + T20 (d4T + 3TC + TDF + TPV-r + T20c) | 110 | |
| HIV-N | 1 | Cameroon | 2 NRTI + NNRTI (3TC + d4T + NVPc) | 47 |
| 1 | France | 2 NRTI + PI + RAL + MVC (TDF + FTC + DRV-r + RAL + MVCc) | 28 | |
| HIV-P | 1 | France | 2 NRTI + PI (ABC + 3TC + LPV-rc) | Our unpublished data |
As the clinical course of non-M infections appears to be similar to that of HIV-M infection, the same criteria for starting treatment are currently used. Data on the treatment response in vivo are extremely sparse. Presented is a summary of current clinical experiences with treatment of infections with HIV-O, -N, and -P.
The drugs are as follows: 3TC, lamivudine; ABC, abacavir; AZT, zidovudine; d4T, stavudine; ddI, didanosine; DRV, darunavir; FTC, emtricitabine; IDV, indinavir; LPV, lopinavir; MVC, maraviroc; NVP, nevirapine; r, ritonavir; RAL, raltegravir; T20, enfuvirtide; TDF, tenofovir; TPV, tipranavir. The drug classes are as follows: NRTIs, nucleoside/nucleotide reverse transcriptase inhibitors; NNRTIs, nonnucleoside reverse transcriptase inhibitors; PIs, protease inhibitors.
Late-known treatment at the time of publication.
For HIV-O, the data are anecdotal or relatively old, and no cohort follow-up studies are available. In France, limited follow-up of 22 patients with documented treatment (all lines) showed therapeutic success (undetectable viral load) in 13 cases (59%) (43). The paucity of data and the lack of long-term follow-up studies rule out firm conclusions on the efficacy of the different lines of treatment. The same applies to data on virologic failure, which has been linked to mutations described for HIV-M (especially for NRTIs) and also to more specific mutations of HIV-O (112, 113). The impact of the marked protease gene polymorphism on the virologic response has not yet been determined, nor has the predictive value of resistance interpretation algorithms designed for HIV-M been determined, except in a recent study that reported imperfect adaptation (95). However, recent findings showed that even if the management of virological failure in HIV-O-infected patients is difficult, a successful response could be achieved, even for antiretroviral-experienced patients, with combinations using more recent molecules (95, 106).
For HIV-N, data for only two patients treated with ART have been reported. One study reported data for a patient treated with a three-drug regimen (stavudine, lamivudine, and nevirapine) for 18 months, which suggested efficacy of the treatment based on low viral loads and the absence of emergence of resistance mutations in target regions (47). The other study corresponds to the primary infection detected in France, for which the patient was successfully treated by antiretroviral combination therapy with tenofovir, emtricitabine, darunavir-ritonavir, raltegravir, and maraviroc (28).
For HIV-P, only one (RBF168) of the two patients was treated, with a regimen containing abacavir, lamivudine, and lopinavir-ritonavir for 42 months, and this patient presents undetectable viral loads (our unpublished data).
CONCLUSION
The discovery of these HIV-1 non-M variants, as well as HIV-2, and our knowledge of SIV reservoirs show that the HIV pandemic results from multiple transmission events, themselves subject to dynamic processes, including interspecies passages and more or less successful spread in humans. Their emergence is therefore multifactorial, being dependent on specific virological properties of each variant as well as host-related phenomena and factors (biological, historical, epidemiological, and social).
Studies of the molecular mechanisms responsible for the epidemiological differences between these variants and the pandemic group M are needed to better understand the characteristics of replication and transmission of these simian retroviruses to humans. Prospective cohort studies are also necessary to establish the pathophysiology of these infections.
While the emergence of these variants resulted from interspecies transmission in Africa, their circulation has no boundaries, as reflected by the recent detection of HIV-1 group N associated with the initial identification of HIV-1 groups O and P in patients living in Europe. Although current commercial tests cover most of this genetic diversity, vigilance must be maintained, especially in cases of immunologic-virologic discrepancies or conflicting results of serological diagnosis and virologic follow-up (undetectable viral load or failure to amplify antiretroviral drug target regions). This must be completed by effective global surveillance of genetic diversity, as is currently performed in the United Kingdom, France, and, recently, the United States (114), to ensure the performances of laboratory tests, to dispense the adapted treatment regimens, and to survey the extent of HIV diversity to support the development of effective vaccines.
Despite improvements in our knowledge of these rare variants, many fascinating questions remain regarding their origin and genetic evolution and the reasons for their relatively inefficient spread in the human population.
ACKNOWLEDGMENTS
We thank the entire team of the Virology Laboratory of Rouen University Hospital for their involvement in studies on HIV-O, and we also thank all the biologists and physicians participating in the French RES-O network. We thank Fabienne De Oliveira, Marie Gueudin, Véronique Lemée, and Marie Leoz of the Rouen laboratory for their active and valuable participations in HIV non-M studies. We also thank Elodie Alessandri-Gradt for useful comments and critical reading of the manuscript.
We thank the Institut de Veille Sanitaire (InVS), the Agence Nationale de Recherches sur le SIDA et les hépatites virales (ANRS), and Rouen University Hospital for financial support. We have no conflicts of interest.
T.M. and J.-C.P. wrote the different manuscripts. T.M., F.S., and J.-C.P. revised the final version.
Biographies

Thomas Mourez (Pharm.D., Ph.D.) studied pharmacy at the University of Normandy and Paris-Descartes and biology at the University of Paris-Diderot and Paris-Sud. He performed his Ph.D. research at the Pasteur Institute in Paris and specialized in the development of artificial chimeras between SIV and measles virus and their potential use as HIV vaccines. As an associate professor at the University of Normandy, his research interest is now directed at the study of intra- and intergroup recombinant HIVs and their potential as emerging viruses.

Francois Simon (M.D., Ph.D.) is Professor at the Faculty of Medicine of Paris-Diderot and Head of the Microbiology Department at Saint-Louis Hospital in Paris, France. He specializes in virology, in the field of molecular epidemiology, with special focus on identification and characterization of HIV variant strains (HIV-2 and HIV-1 groups N, O, and P). Professor Simon is former head of the Retrovirology Department at CIRMF in Gabon and former CEO of the Pasteur Institute in Dakar, Senegal. His current work focuses on the consequences of HIV diversity in the diagnosis and management of patients. Professor Simon has actively participated in numerous collaborative studies and training/education programs in Africa. This has provided him with a unique understanding of the specific needs of such countries in fighting the HIV epidemic. Professor Simon has 188 scientific publications in peer-reviewed journals and has spoken at numerous international scientific conferences.

Jean-Christophe Plantier (Pharm.D., Ph.D.) is Professor of Virology at the Faculty of Medicine of Rouen and Head of the Microbiology Department at Rouen University Hospital, France. He is head of the HIV quantification group at ANRS and head of the laboratory affiliated with the French National Reference Center for HIV, which is in charge of the identification, characterization, and monitoring of HIV variant strains. Professor Plantier is involved in the molecular survey of HIV-1 genetic diversity circulating in France. His research team focuses on the virological, phenotypic, diagnostic, and therapeutic consequences of HIV-1 genetic diversity and its phylogenetic and phylodynamic evolution. He is currently working on the genetic evolution of non-M variants; the impact of their divergence on diagnosis, susceptibility, and virological response to antiretroviral treatment; as well as genetic evolution by recombination between strains of group M and between group M and highly divergent group O strains.
REFERENCES
- 1. Barré-Sinoussi F, Chermann JC, Rey P, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Brun-Vezinet F, Rouzioux C, Rozenbaum W, Montagnier L. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–871 [DOI] [PubMed] [Google Scholar]
- 2. Barin F, M'Boup S, Denis F, Kanki P, Allan JS, Lee TH, Essex M. 1985. Serological evidence for virus related to simian T-lymphotropic retrovirus III in residents of West Africa. Lancet ii:1387–1389 [DOI] [PubMed] [Google Scholar]
- 3. Clavel F, Guétard D, Brun-Vézinet F, Chamaret S, Rey MA, Santos-Ferreira MO, Laurent AG, Dauguet C, Katlama C, Rouzioux C, Klatzmann D, Champalimaud JL, Montagnier L. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233:343–346 [DOI] [PubMed] [Google Scholar]
- 4. De Leys R, Vanderborght B, Vanden Haesevelde M, Heyndrickx L, van Geel A, Wauters C, Bernaerts R, Saman E, Nijs P, Willems B, Taelman H, van der Groen G, Piot P, Tersmette T, Huisman JG, van Heuverswyn H. 1990. Isolation and partial characterization of an unusual human immunodeficiency retrovirus from two persons of West-Central African origin. J. Virol. 64:1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nkengasong JN, Peeters M, vanden Haesevelde M, Musi SS, Willems B, Ndumbe PM, Delaporte E, Perret JL, Piot P, van den Groen G. 1993. Antigenic evidence of the presence of the aberrant HIV-1ant70 virus in Cameroon and Gabon. AIDS 7:1536–1538 [PubMed] [Google Scholar]
- 6. Gurtler LG, Hauser PH, Eberle J, von Brunn A, Knapp S, Zekeng L, Tsague JM, Kaptue L. 1994. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J. Virol. 68:1581–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charneau P, Borman AM, Quillent C, Guetard D, Chamaret S, Cohen J, Remy G, Montagnier L, Clavel F. 1994. Isolation and envelope sequence of a highly divergent HIV-1 isolate: definition of a new HIV-1 group. Virology 205:247–253 [DOI] [PubMed] [Google Scholar]
- 8. Vanden Haesevelde M, Decourt JL, De Leys RJ, Vanderborght B, van der Groen G, van Heuverswijn H, Saman E. 1994. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J. Virol. 68:1586–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin MC, Saragosti S, Georges-Courbot MC, Barre-Sinoussi F, Brun-Vezinet F. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032–1037 [DOI] [PubMed] [Google Scholar]
- 10. Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V, Damond F, Robertson DL, Simon F. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871–872 [DOI] [PubMed] [Google Scholar]
- 11. Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, Ngolle EM, Sharp PM, Shaw GM, Delaporte E, Hahn BH, Peeters M. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. 10.1038/444164a [DOI] [PubMed] [Google Scholar]
- 12. Vallari A, Holzmayer V, Harris B, Yamaguchi J, Ngansop C, Makamche F, Mbanya D, Kaptue L, Ndembi N, Gurtler L, Devare S, Brennan CA. 2011. Confirmation of putative HIV-1 group P in Cameroon. J. Virol. 85:1403–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peeters M, Honore C, Huet T, Bedjabaga L, Ossari S, Bussi P, Cooper RW, Delaporte E. 1989. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS 3:625–630 [DOI] [PubMed] [Google Scholar]
- 14. Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356–359 [DOI] [PubMed] [Google Scholar]
- 15. Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 [DOI] [PubMed] [Google Scholar]
- 16. Hahn B, Shaw GM, De Cock K, Sharp PM. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614 [DOI] [PubMed] [Google Scholar]
- 17. Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Heuverswyn F, Li Y, Bailes E, Neel C, Lafay B, Keele BF, Shaw KS, Takehisa J, Kraus MH, Loul S, Butel C, Liegeois F, Yangda B, Sharp PM, Mpoudi-Ngole E, Delaporte E, Hahn BH, Peeters M. 2007. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368:155–171 [DOI] [PubMed] [Google Scholar]
- 19. Takehisa J, Kraus MH, Ayouba A, Bailes E, Van Heuverswyn F, Decker JM, Li Y, Rudicell RS, Learn GH, Neel C, Ngole EM, Shaw GM, Peeters M, Sharp PM, Hahn BH. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JM, Kalengayi RM, Van Marck E, Gilbert MT, Wolinsky SM. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455:661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lemey P, Pybus OG, Rambaut A, Drummond AJ, Robertson DL, Roques P, Worobey M, Vandamme A-M. 2004. The molecular population genetics of HIV-1 group O. Genetics 167:1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wertheim JO, Worobey M. 2009. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput. Biol. 5:e1000377. 10.1371/journal.pcbi.1000377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sauter D, Hue S, Petit SJ, Plantier JC, Towers GJ, Kirchhoff F, Gupta RK. 2011. HIV-1 group P is unable to antagonize human tetherin by Vpu, Env or Nef. Retrovirology 8:103. 10.1186/1742-4690-8-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jonassen TO, Stene-Johansen K, Berg ES, Hungnes O, Lindboe CF, Froland SS, Grinde B. 1997. Sequence analysis of HIV-1 group O from Norwegian patients infected in the 1960s. Virology 231:43–47 [DOI] [PubMed] [Google Scholar]
- 25. Wain LV, Bailes E, Bibollet-Ruche F, Decker JM, Keele BF, Van Heuverswyn F, Li Y, Takehisa J, Ngole EM, Shaw GM, Peeters M, Hahn BH, Sharp PM. 2007. Adaptation of HIV-1 to its human host. Mol. Biol. Evol. 24:1853–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang SJ, Lopez LA, Exline CM, Haworth KG, Cannon PM. 2011. Lack of adaptation to human tetherin in HIV-1 group O and P. Retrovirology 8:78. 10.1186/1742-4690-8-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Delaugerre C, De Oliveira F, Lascoux-Combe C, Plantier JC, Simon F. 2011. HIV-1 group N: travelling beyond Cameroon. Lancet 378:1894. 10.1016/S0140-6736(11)61457-8 [DOI] [PubMed] [Google Scholar]
- 29. Sauter D, Unterweger D, Vogl M, Usmani SM, Heigele A, Kluge SF, Hermkes E, Moll M, Barker E, Peeters M, Learn GH, Bibollet-Ruche F, Fritz JV, Fackler OT, Hahn BH, Kirchhoff F. 2012. Human tetherin exerts strong selection pressure on the HIV-1 group N Vpu protein. PLoS Pathog. 8:e1003093. 10.1371/journal.ppat.1003093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arien KK, Abraha A, Quinones-Mateu ME, Kestens L, Vanham G, Arts EJ. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979–8990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Archer J, Robertson DL. 2007. Understanding the diversification of HIV-1 groups M and O. AIDS 21:1693–1700 [DOI] [PubMed] [Google Scholar]
- 32. Roques P, Robertson DL, Souquiere S, Damond F, Ayouba A, Farfara I, Depienne C, Nerrienet E, Dormont D, Brun-Vezinet F, Simon F, Mauclere P. 2002. Phylogenetic analysis of 49 newly derived HIV-1 group O strains: high viral diversity but no group M-like subtype structure. Virology 302:259–273 [DOI] [PubMed] [Google Scholar]
- 33. Yamaguchi J, Vallari AS, Swanson P, Bodelle P, Kaptue L, Ngansop C, Zekeng L, Gurtler LG, Devare SG, Brennan CA. 2002. Evaluation of HIV type 1 group O isolates: identification of five phylogenetic clusters. AIDS Res. Hum. Retroviruses 18:269–282 [DOI] [PubMed] [Google Scholar]
- 34. Vallari A, Bodelle P, Ngansop C, Makamche F, Ndembi N, Mbanya D, Kaptue L, Gurtler LG, McArthur CP, Devare SG, Brennan CA. 2010. Four new HIV-1 group N isolates from Cameroon: prevalence continues to be low. AIDS Res. Hum. Retroviruses 26:109–115 [DOI] [PubMed] [Google Scholar]
- 35. Roques P, Robertson DL, Souquiere S, Apetrei C, Nerrienet E, Barre-Sinoussi F, Muller-Trutwin M, Simon F. 2004. Phylogenetic characteristics of three new HIV-1 N strains and implications for the origin of group N. AIDS 18:1371–1381 [DOI] [PubMed] [Google Scholar]
- 36. Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. 2012. Natural SIV hosts: showing AIDS the door. Science 335:1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, Li Y, Learn GH, Beasley TM, Schumacher-Stankey J, Wroblewski E, Mosser A, Raphael J, Kamenya S, Lonsdorf EV, Travis DA, Mlengeya T, Kinsel MJ, Else JG, Silvestri G, Goodall J, Sharp PM, Shaw GM, Pusey AE, Hahn BH. 2009. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460:515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Etienne L, Nerrienet E, LeBreton M, Bibila GT, Foupouapouognigni Y, Rousset D, Nana A, Djoko CF, Tamoufe U, Aghokeng AF, Mpoudi-Ngole E, Delaporte E, Peeters M, Wolfe ND, Ayouba A. 2011. Characterization of a new simian immunodeficiency virus strain in a naturally infected Pan troglodytes troglodytes chimpanzee with AIDS related symptoms. Retrovirology 8:4. 10.1186/1742-4690-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nkengasong JN, Fransen K, Willems B, Karita E, Vingerhoets J, Kestens L, Colebunders R, Piot P, van der Groen G. 1997. Virologic, immunologic, and clinical follow-up of a couple infected by the human immunodeficiency virus type one, group O. J. Med. Virol. 51:202–209 [PubMed] [Google Scholar]
- 40. Andreoletti L, Reveil B, Moret H, Brodard V, Philbert F, Tabary T, Cohen JH. 2007. Significant genetic and antigenic variability within the env gene of systemic human immunodeficiency virus type 1 group O populations during the natural course of a heterosexual infection: a pilot study. J. Clin. Microbiol. 45:1319–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Geuenich S, Kaderali L, Allespach I, Sertel S, Keppler OT. 2009. Biological signature characteristics of primary isolates from human immunodeficiency virus type 1 group O in ex vivo human tonsil histocultures. J. Virol. 83:10494–10503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vessiere A, Rousset D, Kfutwah A, Leoz M, Depatureaux A, Simon F, Plantier JC. 2010. Diagnosis and monitoring of HIV-1 group O-infected patients in Cameroun. J. Acquir. Immune Defic. Syndr. 53:107–110 [DOI] [PubMed] [Google Scholar]
- 43. Depatureaux A, Leoz M, De Oliveira F, Gueudin M, Damond F, Descamps D, Brun-Vezinet F, Lemee V, Simon F, Barin F, Plantier JC. 19 June 2010. Specific diagnosis and follow-up of HIV-1 group O infection: RES-O data. Med. Mal. Infect. (In French.) 10.1016/j.medmal.2010.1004.1011 [DOI] [PubMed] [Google Scholar]
- 44. Chaix-Baudier ML, Chappey C, Burgard M, Letourneur F, Igual J, Saragosti S, Rouzioux C. 1998. First case of mother-to-infant HIV type 1 group O transmission and evolution of C2V3 sequences in the infected child. French HIV Pediatric Cohort Study Group. AIDS Res. Hum. Retroviruses 14:15–23 [DOI] [PubMed] [Google Scholar]
- 45. Gueudin M, Lemee V, Ferre V, Beby-Defaux A, Pathe JP, Guist'hau O, Braun J, Simon F, Plantier JC. 2004. Virologic diagnosis and follow-up of children born to mothers infected by HIV-1 group O. J. Acquir. Immune Defic. Syndr. 36:639–641 [DOI] [PubMed] [Google Scholar]
- 46. Ayouba A, Souquieres S, Njinku B, Martin PM, Muller-Trutwin MC, Roques P, Barre-Sinoussi F, Mauclere P, Simon F, Nerrienet E. 2000. HIV-1 group N among HIV-1-seropositive individuals in Cameroon. AIDS 14:2623–2625 [DOI] [PubMed] [Google Scholar]
- 47. Yamaguchi J, Coffey R, Vallari A, Ngansop C, Mbanya D, Ndembi N, Kaptue L, Gurtler LG, Bodelle P, Schochetman G, Devare SG, Brennan CA. 2006. Identification of HIV type 1 group N infections in a husband and wife in Cameroon: viral genome sequences provide evidence for horizontal transmission. AIDS Res. Hum. Retroviruses 22:83–92 [DOI] [PubMed] [Google Scholar]
- 48. Yamaguchi J, McArthur CP, Vallari A, Coffey R, Bodelle P, Beyeme M, Schochetman G, Devare SG, Brennan CA. 2006. HIV-1 group N: evidence of ongoing transmission in Cameroon. AIDS Res. Hum. Retroviruses 22:453–457 [DOI] [PubMed] [Google Scholar]
- 49. Delaporte E, Janssens W, Peeters M, Buve A, Dibanga G, Perret JL, Ditsambou V, Mba JR, Courbot MC, Georges A, Bourgeois A, Samb B, Henzel D, Heyndrickx L, Fransen K, van der Groen G, Larouze B. 1996. Epidemiological and molecular characteristics of HIV infection in Gabon, 1986-1994. AIDS 10:903–910 [DOI] [PubMed] [Google Scholar]
- 50. Zekeng L, Obiang Sima J, Hampl H, Ndemesogo JM, Ntutumu J, Sima V, Devare S, Kaptue L, Gurtler L. 1997. Update on HIV-1 group O infection in Equatorial Guinea, Central Africa. AIDS 11:1410–1412 [PubMed] [Google Scholar]
- 51. Hunt JC, Brennan CA, Golden AM, Yamaguchi J, Lund JK, Vallari AS, Hickman RK, Zekeng L, Gurtler LG, Hampl H, Kaptue L, Devare SG. 1997. Molecular analyses of HIV-1 group O and HIV-2 variants from Africa. Leukemia 11(Suppl 3):138–141 [PubMed] [Google Scholar]
- 52. Bibollet-Ruche F, Peeters M, Mboup S, Ekaza E, Gandji R, Torimiro J, Mpoudi EN, Amblard J, Dibanga G, Saidou M, Esu-Williams E, Vanden Haesevelde M, Saman E, Delaporte E. 1998. Molecular characterization of the envelope transmembrane glycoprotein of 13 new human immunodeficiency virus type 1 group O strains from six different African countries. AIDS Res. Hum. Retroviruses 14:1281–1285 [DOI] [PubMed] [Google Scholar]
- 53. Ayouba A, Mauclere P, Martin PM, Cunin P, Mfoupouendoun J, Njinku B, Souquieres S, Simon F. 2001. HIV-1 group O infection in Cameroon, 1986 to 1998. Emerg. Infect. Dis. 7:466–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vergne L, Bourgeois A, Mpoudi-Ngole E, Mougnutou R, Mbuagbaw J, Liegeois F, Laurent C, Butel C, Zekeng L, Delaporte E, Peeters M. 2003. Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology 310:254–266 [DOI] [PubMed] [Google Scholar]
- 55. Takehisa J, Zekeng L, Ido E, Yamaguchi-Kabata Y, Mboudjeka I, Harada Y, Miura T, Kaptue L, Hayami M. 1999. Human immunodeficiency virus type 1 intergroup (M/O) recombination in Cameroon. J. Virol. 73:6810–6820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Peeters M, Liegeois F, Torimiro N, Bourgeois A, Mpoudi E, Vergne L, Saman E, Delaporte E, Saragosti S. 1999. Characterization of a highly replicative intergroup M/O human immunodeficiency virus type 1 recombinant isolated from a Cameroonian patient. J. Virol. 73:7368–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yamaguchi BP, Vallari AS, Coffey R, McArthur CP, Schochetman G, Devare SG, Brennan CA. 2004. HIV infections in northwestern Cameroon: identification of HIV type 1 group O and dual HIV type 1 group M and group O infections. AIDS Res. Hum. Retroviruses 20:944–955 [DOI] [PubMed] [Google Scholar]
- 58. Vessiere A, De Oliveira F, Leoz M, Rousset D, Kfutwah A, Simon F, Plantier JC. 2011. HIV-1 M/O dual infections and recombinant forms circulating in Cameroon and France, abstr 461. Abstr. 18th Conf. Retroviruses Opportun. Infect., Boston, MA [Google Scholar]
- 59. Kabeya CM, Esu-Williams E, Eni E, Peeters M, Saman E, Delaporte E. 1995. Evidence for HIV-1 group O infection in Nigeria. Lancet 346:308. [DOI] [PubMed] [Google Scholar]
- 60. Songok EM, Libondo DK, Rotich MC, Oogo SA, Tukei PM. 1996. Surveillance for HIV-1 subtypes O and M in Kenya. Lancet 347:1700. [DOI] [PubMed] [Google Scholar]
- 61. Heyndrickx L, Alary M, Janssens W, Davo N, van der Groen G. 1996. HIV-1 group O and group M dual infection in Benin. Lancet 347:902–903 [DOI] [PubMed] [Google Scholar]
- 62. Peeters M, Gaye A, Mboup S, Badombena W, Bassabi K, Prince-David M, Develoux M, Liegeois F, van der Groen G, Saman E, Delaporte E. 1996. Presence of HIV-1 group O infection in West Africa. AIDS 10:343–344 [DOI] [PubMed] [Google Scholar]
- 63. Peeters M, Gueye A, Mboup S, Bibollet-Ruche F, Ekaza E, Mulanga C, Ouedrago R, Gandji R, Mpele P, Dibanga G, Koumare B, Saidou M, Esu-Williams E, Lombart JP, Badombena W, Luo N, Vanden Haesevelde M, Delaporte E. 1997. Geographical distribution of HIV-1 group O viruses in Africa. AIDS 11:493–498 [DOI] [PubMed] [Google Scholar]
- 64. Nkengasong J, Sylla-Koko F, Peeters M, Ellenberger D, Sassan-Morokro M, Ekpini RA, Msellati P, Greenberg AE, Combe P, Rayfield M. 1998. HIV-1 group O virus infection in Abidjan, Cote d'Ivoire. AIDS 12:1565–1566 [DOI] [PubMed] [Google Scholar]
- 65. Hampl H, Sawitzky D, Stoffler-Meilicke M, Groh A, Schmitt M, Eberle J, Gurtler L. 1995. First case of HIV-1 subtype 0 infection in Germany. Infection 23:369–370 [DOI] [PubMed] [Google Scholar]
- 66. Rayfield MA, Sullivan P, Bandea CI, Britvan L, Otten RA, Pau CP, Pieniazek D, Subbarao S, Simon P, Schable CA, Wright AC, Ward J, Schochetman G. 1996. HIV-1 group O virus identified for the first time in the United States. Emerg. Infect. Dis. 2:209–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Soriano V, Gutierrez M, Garcia-Lerma G, Aguilera O, Mas A, Bravo R, Perez-Labad ML, Baquero M, Gonzalez-Lahoz J. 1996. First case of HIV-1 group O infection in Spain. Vox Sang. 71:66. [DOI] [PubMed] [Google Scholar]
- 68. Gould K. 1997. Infection with HIV-1 group O. AIDS Patient Care STDs 11:399–405 [DOI] [PubMed] [Google Scholar]
- 69. Sullivan PS, Do AN, Ellenberger D, Pau CP, Paul S, Robbins K, Kalish M, Storck C, Schable CA, Wise H, Tetteh C, Jones JL, McFarland J, Yang C, Lal RB, Ward JW. 2000. Human immunodeficiency virus (HIV) subtype surveillance of African-born persons at risk for group O and group N HIV infections in the United States. J. Infect. Dis. 181:463–469 [DOI] [PubMed] [Google Scholar]
- 70. Barin F, Cazein F, Lot F, Pillonel J, Brunet S, Thierry D, Damond F, Brun-Vezinet F, Desenclos JC, Semaille C. 2007. Prevalence of HIV-2 and HIV-1 group O infections among new HIV diagnoses in France: 2003-2006. AIDS 21:2351–2353 [DOI] [PubMed] [Google Scholar]
- 71. Agut H, Rabanel B, Candotti D, Huraux JM, Remy G, Tabary T, Ingrand D, Chippaux C, Chamaret S, Dauguet C, Guetard D, Montagnier L. 1992. Isolation of atypical HIV-1-related retrovirus from AIDS patient. Lancet 340:681–682 [DOI] [PubMed] [Google Scholar]
- 72. Brand D, Beby-Defaux A, Mace M, Brunet S, Moreau A, Godet C, Jais X, Cazein F, Semaille C, Barin F. 2004. First identification of HIV-1 groups M and O dual infections in Europe. AIDS 18:2425–2428 [PubMed] [Google Scholar]
- 73. Plantier JC, Lemee V, Dorval I, Gueudin M, Braun J, Hutin P, Ruffault A, Simon F. 2004. HIV-1 group M superinfection in an HIV-1 group O-infected patient. AIDS 18:2444–2446 [PubMed] [Google Scholar]
- 74. Vessiere A, Leoz M, Brodard V, Strady C, Lemee V, Depatureaux A, Simon F, Plantier JC. 2010. First evidence of a HIV-1 M/O recombinant form circulating outside Cameroon. AIDS 24:1079–1082 [DOI] [PubMed] [Google Scholar]
- 75. Loussert-Ajaka I, Ly TD, Chaix ML, Ingrand D, Saragosti S, Courouce AM, Brun-Vezinet F, Simon F. 1994. HIV-1/HIV-2 seronegativity in HIV-1 subtype O infected patients. Lancet 343:1393–1394 [DOI] [PubMed] [Google Scholar]
- 76. Simon F, Ly TD, Baillou-Beaufils A, Fauveau V, De Saint-Martin J, Loussert-Ajaka I, Chaix ML, Saragosti S, Courouce AM, Ingrand D, Janot C, Brun-Vézinet F. 1994. Sensitivity of screening kits for anti-HIV-1 subtype O antibodies. AIDS 8:1628–1629 [DOI] [PubMed] [Google Scholar]
- 77. Ly TD, Laperche S, Brennan C, Vallari A, Ebel A, Hunt J, Martin L, Daghfal D, Schochetman G, Devare S. 2004. Evaluation of the sensitivity and specificity of six HIV combined p24 antigen and antibody assays. J. Virol. Methods 122:185–194 [DOI] [PubMed] [Google Scholar]
- 78. Zouhair S, Roussin-Bretagne S, Moreau A, Brunet S, Laperche S, Maniez M, Barin F, Harzic M. 2006. Group O human immunodeficiency virus type 1 infection that escaped detection in two immmunoassays. J. Clin. Microbiol. 44:662–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Henquell C, Jacomet C, Antoniotti O, Chaib A, Regagnon C, Brunet S, Peigue-Lafeuille H, Barin F. 2008. Difficulties in diagnosing group O human immunodeficiency virus type 1 acute primary infection. J. Clin. Microbiol. 46:2453–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gautheret-Dejean A, Mesmin-Poho S, Birguel J, Lemee V, Huraux JM, Plantier JC. 2008. Unequal detection of HIV type 1 group O infection by simple rapid tests. Clin. Infect. Dis. 46:1936–1937 [DOI] [PubMed] [Google Scholar]
- 81. Plantier JC, Djemai M, Lemee V, Reggiani A, Leoz M, Burc L, Vessiere A, Rousset D, Poveda JD, Henquell C, Gautheret-Dejean A, Barin F. 2009. Census and analysis of persistent false-negative results in serological diagnosis of human immunodeficiency virus type 1 group O infections. J. Clin. Microbiol. 47:2906–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Aghokeng AF, Mpoudi-Ngole E, Dimodi H, Atem-Tambe A, Tongo M, Butel C, Delaporte E, Peeters M. 2009. Inaccurate diagnosis of HIV-1 group M and O is a key challenge for ongoing universal access to antiretroviral treatment and HIV prevention in Cameroon. PLoS One 4:e7702. 10.1371/journal.pone.0007702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Laperche S, Leballais L, Ly TD, Plantier JC. 2012. Failures in the detection of HIV p24 antigen with the Determine HIV-1/2 Ag/Ab Combo rapid test. J. Infect. Dis. 206:1946–1947 [DOI] [PubMed] [Google Scholar]
- 84. Ly TD, Plantier JC, Leballais L, Gonzalo S, Lemee V, Laperche S. 2012. The variable sensitivity of HIV Ag/Ab combination assays in the detection of p24Ag according to genotype could compromise the diagnosis of early HIV infection. J. Clin. Virol. 55:121–127 [DOI] [PubMed] [Google Scholar]
- 85. Simon F, Souquiere S, Damond F, Kfutwah A, Makuwa M, Leroy E, Rouquet P, Berthier JL, Rigoulet J, Lecu A, Telfer PT, Pandrea I, Plantier JC, Barre-Sinoussi F, Roques P, Muller-Trutwin MC, Apetrei C. 2001. Synthetic peptide strategy for the detection of and discrimination among highly divergent primate lentiviruses. AIDS Res. Hum. Retroviruses 17:937–952 [DOI] [PubMed] [Google Scholar]
- 86. Barin F, Plantier JC, Brand D, Brunet S, Moreau A, Liandier B, Thierry D, Cazein F, Lot F, Semaille C, Desenclos JC. 2006. Human immunodeficiency virus serotyping on dried serum spots as a screening tool for the surveillance of the AIDS epidemic. J. Med. Virol. 78(Suppl 1):S13–S18. 10.1002/jmv.20600 [DOI] [PubMed] [Google Scholar]
- 87. Plantier JC, Gueudin M, Damond F, Braun J, Mauclere P, Simon F. 2003. Plasma RNA quantification and HIV-1 divergent strains. J. Acquir. Immune Defic. Syndr. 33:1–7 [DOI] [PubMed] [Google Scholar]
- 88. Swanson P, de Mendoza C, Joshi Y, Golden A, Hodinka RL, Soriano V, Devare SG, Hackett J., Jr 2005. Impact of human immunodeficiency virus type 1 (HIV-1) genetic diversity on performance of four commercial viral load assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v1.5, VERSANT HIV-1 RNA 3.0, and NucliSens HIV-1 QT. J. Clin. Microbiol. 43:3860–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gueudin M, Plantier JC, Lemee V, Schmitt MP, Chartier L, Bourlet T, Ruffault A, Damond F, Vray M, Simon F. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44:500–505 [DOI] [PubMed] [Google Scholar]
- 90. Gueudin M, Plantier JC, Damond F, Roques P, Mauclere P, Simon F. 2003. Plasma viral RNA assay in HIV-1 group O infection by real-time PCR. J. Virol. Methods 113:43–49 [DOI] [PubMed] [Google Scholar]
- 91. Gueudin M, Leoz M, Lemee V, De Oliveira F, Vessiere A, Kfutwah A, Plantier JC. 2012. A new real-time quantitative PCR for diagnosis and monitoring of HIV-1 group O infection. J. Clin. Microbiol. 50:831–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Damond F, Avettand-Fenoel V, Collin G, Roquebert B, Plantier JC, Ganon A, Sizmann D, Babiel R, Glaubitz J, Chaix ML, Brun-Vezinet F, Descamps D, Rouzioux C. 2010. Evaluation of an upgraded version of the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test for HIV-1 load quantification. J. Clin. Microbiol. 48:1413–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sire JM, Vray M, Merzouk M, Plantier JC, Pavie J, Maylin S, Timsit J, Lascoux-Combe C, Molina JM, Simon F, Delaugerre C. 2011. Comparative RNA quantification of HIV-1 group M and non-M with the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 v2.0 and Abbott real-time HIV-1 PCR assays. J. Acquir. Immune Defic. Syndr. 56:239–243 [DOI] [PubMed] [Google Scholar]
- 94. Tang N, Huang S, Salituro J, Mak WB, Cloherty G, Johanson J, Li YH, Schneider G, Robinson J, Hackett J, Jr, Swanson P, Abravaya K. 2007. A RealTime HIV-1 viral load assay for automated quantitation of HIV-1 RNA in genetically diverse group M subtypes A-H, group O and group N samples. J. Virol. Methods 146:236–245 [DOI] [PubMed] [Google Scholar]
- 95. Depatureaux A, Charpentier C, Leoz M, Unal G, Damond F, Kfutwah A, Vessiere A, Simon F, Plantier JC. 2011. Impact of HIV-1 group O genetic diversity on genotypic resistance interpretation by algorithms designed for HIV-1 group M. J. Acquir. Immune Defic. Syndr. 56:139–145 [DOI] [PubMed] [Google Scholar]
- 96. Depatureaux A, Leoz M, Vessière A, Rousset D, Damond F, Simon F, Plantier JC. 2009. Natural presence of resistance mutation Y181C in HIV-1 group O variants is clade-dependent, p A93 In XVIII International HIV Drug Resistance Workshop: basic principles & clinical implications, vol 14, suppl 1 Antiviral Therapy, Fort Myers, FL [Google Scholar]
- 97. Tebit DM, Lobritz M, Lalonde M, Immonen T, Singh K, Sarafianos S, Herchenroder O, Krausslich HG, Arts EJ. 2010. Divergent evolution in reverse transcriptase (RT) of HIV-1 group O and M lineages: impact on structure, fitness, and sensitivity to nonnucleoside RT inhibitors. J. Virol. 84:9817–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Descamps D, Collin G, Letourneur F, Apetrei C, Damond F, Loussert-Ajaka I, Simon F, Saragosti S, Brun-Vezinet F. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J. Virol. 71:8893–8898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tuaillon E, Gueudin M, Lemee V, Gueit I, Roques P, Corrigan GE, Plantier JC, Simon F, Braun J. 2004. Phenotypic susceptibility to nonnucleoside inhibitors of virion-associated reverse transcriptase from different HIV types and groups. J. Acquir. Immune Defic. Syndr. 37:1543–1549 [DOI] [PubMed] [Google Scholar]
- 100. Luk KC, Kaptue L, Zekeng L, Soriano V, Gurtler L, Devare SG, Schochetman G, Hackett J., Jr 2001. Naturally occurring sequence polymorphisms within HIV type 1 group O protease. AIDS Res. Hum. Retroviruses 17:1555–1561 [DOI] [PubMed] [Google Scholar]
- 101. Witvrouw M, Pannecouque C, Switzer WM, Folks TM, De Clercq E, Heneine W. 2004. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir. Ther. 9:57–65 [PubMed] [Google Scholar]
- 102. Parkin NT, Schapiro JM. 2004. Antiretroviral drug resistance in non-subtype B HIV-1, HIV-2 and SIV. Antivir. Ther. 9:3–12 [PubMed] [Google Scholar]
- 103. Desbois D, Roquebert B, Peytavin G, Damond F, Collin G, Benard A, Campa P, Matheron S, Chene G, Brun-Vezinet F, Descamps D. 2008. In vitro phenotypic susceptibility of human immunodeficiency virus type 2 clinical isolates to protease inhibitors. Antimicrob. Agents Chemother. 52:1545–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Leoz M, Depatureaux A, Vessiere A, Roquebert B, Damond F, Rousset D, Roques P, Simon F, Plantier JC. 2008. Integrase polymorphism and HIV-1 group O diversity. AIDS 22(10):1239–1243 [DOI] [PubMed] [Google Scholar]
- 105. Briz V, Garrido C, Poveda E, Morello J, Barreiro P, de Mendoza C, Soriano V. 2009. Raltegravir and etravirine are active against HIV type 1 group O. AIDS Res. Hum. Retroviruses 25:225–227 [DOI] [PubMed] [Google Scholar]
- 106. Depatureaux A, Leoz M, Le Moal G, Pathe JP, Pavie J, Batisse D, Daneluzzi V, Genet P, Gerard L, Lascaux-Cametz AS, Lambolez T, Chennebault JM, Plantier JC. 2012. Raltegravir-based regimens are effective in HIV-1 group O-infected patients. J. Acquir. Immune Defic. Syndr. 61:e1–e3. 10.1097/QAI.0b013e31826327c4 [DOI] [PubMed] [Google Scholar]
- 107. Aghokeng AF, Kouanfack C, Peeters M, Mpoudi-Ngole E, Delaporte E. 2013. Successful integrase inhibitor-based highly active antiretroviral therapy for a multidrug-class-resistant HIV type 1 group O-infected patient in Cameroon. AIDS Res. Hum. Retroviruses 29:1–3 [DOI] [PubMed] [Google Scholar]
- 108. Depatureaux A, Charpentier C, Collin G, Leoz M, Descamps D, Vessiere A, Damond F, Rousset D, Brun-Vezinet F, Plantier JC. 2010. Baseline genotypic and phenotypic susceptibilities of HIV-1 group O to enfuvirtide. Antimicrob. Agents Chemother. 54:4016–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chinnadurai R, Munch J, Dittmar MT, Kirchhoff F. 2005. Inhibition of HIV-1 group M and O isolates by fusion inhibitors. AIDS 19:1919–1922 [DOI] [PubMed] [Google Scholar]
- 110. Poveda E, Barreiro P, Rodes B, Soriano V. 2005. Enfuvirtide is active against HIV type 1 group O. AIDS Res. Hum. Retroviruses 21:583–585 [DOI] [PubMed] [Google Scholar]
- 111. Rupp D, Geuenich S, Keppler OT. 2010. Poor performance of bioinformatics programs for genotypic prediction of coreceptor usage of HIV-1 group O isolates. J. Acquir. Immune Defic. Syndr. 53:412–413 [DOI] [PubMed] [Google Scholar]
- 112. de Baar MP, Janssens W, de Ronde A, Fransen K, Colebunders R, Kestens L, van der Groen G, Goudsmit J. 2000. Natural residues versus antiretroviral drug-selected mutations in HIV type 1 group O reverse transcriptase and protease related to virological drug failure in vivo. AIDS Res. Hum. Retroviruses 16:1385–1394 [DOI] [PubMed] [Google Scholar]
- 113. Rodes B, de Mendoza C, Rodgers M, Newell A, Jimenez V, Lopez-Brugada RM, Soriano V. 2005. Treatment response and drug resistance in patients infected with HIV type 1 group O viruses. AIDS Res. Hum. Retroviruses 21:602–607 [DOI] [PubMed] [Google Scholar]
- 114. Pieniazek D, Ziebell R, Ocfemia CB, Saduvala N, Kline R, Prejean J, Kim D, Hall I. 2012. Phylogenetic surveillance of HIV-1 non-B subtypes among US-born and foreign-born individuals living in the US revealed similar viral genetic diversity but different race and male transmission patterns, abstr 515. Abstr. 19th Conf. Retroviruses Opportun. Infect., Seattle, WA [Google Scholar]