Abstract
SUMMARY
Within the past decade, clinical microbiology laboratories experienced revolutionary changes in the way in which microorganisms are identified, moving away from slow, traditional microbial identification algorithms toward rapid molecular methods and mass spectrometry (MS). Historically, MS was clinically utilized as a high-complexity method adapted for protein-centered analysis of samples in chemistry and hematology laboratories. Today, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS is adapted for use in microbiology laboratories, where it serves as a paradigm-shifting, rapid, and robust method for accurate microbial identification. Multiple instrument platforms, marketed by well-established manufacturers, are beginning to displace automated phenotypic identification instruments and in some cases genetic sequence-based identification practices. This review summarizes the current position of MALDI-TOF MS in clinical research and in diagnostic clinical microbiology laboratories and serves as a primer to examine the “nuts and bolts” of MALDI-TOF MS, highlighting research associated with sample preparation, spectral analysis, and accuracy. Currently available MALDI-TOF MS hardware and software platforms that support the use of MALDI-TOF with direct and precultured specimens and integration of the technology into the laboratory workflow are also discussed. Finally, this review closes with a prospective view of the future of MALDI-TOF MS in the clinical microbiology laboratory to accelerate diagnosis and microbial identification to improve patient care.
INTRODUCTION
Timely and accurate identification of microorganisms is the underlying function of any clinical microbiology laboratory and is accomplished through a consistently evolving repertoire of laboratory techniques. Historically, confirmation of microbial identification was dependent upon a hierarchy of assays separated into stages: (i) stain-based methodologies for classification of microscopic morphology to support early diagnostic and therapeutic decisions; (ii) microbial culture for propagation of the offending organism on agar or in liquid medium; (iii) biochemical or antigenic techniques for the subsequent metabolic and phenotypic analysis of the microorganism, ultimately leading to microbe identification; and (iv) antimicrobial susceptibility testing to confirm therapeutic choices or tailor therapy (1). While historical sentiment among both microbiologists and clinicians accepted these established protocols as reference standards (in terms of accuracy, speed, and costs), microbiologists, clinicians, and patients were at the mercy of the microorganism's growth rate. Robust growth and active microbial biochemistry were usually required for most determinative phenotypic assays, thus extending the time to result by days and, in some cases, weeks.
As new technology emerged, prevailing expectations defined standards for more rapid, accurate, and sensitive methods aimed at optimizing patient care and therapy. For example, genetic sequencing uncovered numerous errors inherent to phenotypic identification and became part of the new standard for microbial identification (2, 3). Real-time PCR (4, 5) and fluorescence in situ hybridization (FISH) (6–8) methods established new norms for speed and sensitivity.
While new technology is often necessary for optimal patient care and therapy, reagent and quality control costs often exceed those of historical methods, thus placing an additional burden on laboratories to define and monitor quantitative measures of cost benefit for patients and in some cases the entire health care system. In addition, since the technological complexity and laboratory space design requirements can hinder test performance in resource-poor settings, laboratory leadership must be mindful not to create an imbalance in the standards for laboratory practice with societal implications.
Among the last pieces of clinical data to be reported, antibiotic susceptibility testing further extends the time necessary for a final determinative report for therapeutic purposes. Often, an additional 24 h is necessary for susceptibility testing to be completed by using Kirby-Bauer disc diffusion testing, broth microdilution methods, automated susceptibility testing (AST), and Etests. While methods for rapid susceptibility testing, such as those reported for the Vitek-2 (bioMérieux) instrument, are directly from positive blood culture broths, these adapted methods are not always accurate enough to be reported without additional confirmatory analysis, making them useful in some cases but not applicable to all clinical situations (9).
The integration of molecular testing methodologies into determinative microbial identification algorithms supported critical advances in analytical sensitivity, allowing microbiologists to explore options other than routine culture and to begin testing patient specimens directly for the presence or absence of particular organisms. Additionally, the ability to rapidly and uniformly test both direct patient specimens and cultured organisms in near real time by molecular methods transformed the microbiology laboratory (1). Nucleic acid-based methods such as FISH and PCR-based strategies drastically decreased the time to result and provided significant improvements to both laboratory workflow and patient prognoses (6, 121). However, a significant limitation of these molecular methods is that a majority of these assays require advance knowledge of the characteristics of the microorganism(s), or a likelihood of that particular organism being present, in order to select the correct assay to fit the testing application. Moreover, in the case of polymicrobial infections, multiple molecular assays, preliminary culture and separation, or additional downstream testing is sometimes required for full characterization of the clinical specimen, adding to the result turnaround time and the overall financial cost.
Gene sequencing provided an attractive option for universal identification of fungi and bacteria. Since its implementation, it is considered among the most definitive of all molecular microbiological analyses. Although 16S rRNA and 18S rRNA gene sequencing (for bacterial and fungal identifications, respectively) are powerful diagnostic tools with high discriminatory power for species- and strain-level determinations (10), these methods are employed primarily by large high-complexity clinical and reference laboratories for reflex and confirmatory testing. As with several other molecular methodologies, rRNA gene sequencing often requires specialized instrumentation and dedicated laboratory space and staff. These constraints often render rRNA gene sequencing impractical for most laboratories; therefore, the use of automated instruments for the phenotypic analysis of bacterial isolates still predominates as the basis for routine microbial identification, despite imperfections in accuracy, robustness, and time to identification. In spite of nearly 20 years of clinical evidence depicting molecular methods to be significantly faster and often more accurate with respect to diagnoses, many laboratories have not yet adopted them as part of their routine practice. Clearly, the development and validation of alternative rapid and universal identification methods are warranted; MALDI-TOF MS methods may fill some of these critical gaps.
One significant challenge faced by clinical microbiologists and the diagnostic industry is the sheer breadth of testing associated with the discipline. The diversity of etiological agents of disease encountered in the microbiology section of the clinical laboratory is staggering. Many pathogenic agents require dedicated diagnostic testing platforms for accurate diagnosis of infection (i.e., specialized culture, molecular methods for noncultivatable or difficult-to-recover organisms, and microscopic inspection and special staining for the characterization of some organisms, including parasites). Add to this the above-mentioned need for rapid and accurate results, and laboratories are left to sort through a complex interplay between traditional and molecular methods to achieve robust, rapid, and accurate identifications for the wide range of organisms potentially encountered. In response, scientists searched for a method which would prove to be standardized and nearly universal in scope and which could identify pathogens and commensals alike, using streamlined workflow with minimal costs and expertise—that method could well be mass spectrometry (MS).
While automated phenotypic and molecular methods received heavy use in clinical laboratories throughout the previous decade, MS-based methods quietly began to develop. Originally confined to basic research laboratories, MS methods were used consistently to address questions that were applicable to the clinical laboratory, including microbiological identification, taxonomy, and bacterial cell composition. Due to its high resolving power and analytical sensitivity, MS is mechanistically well suited to serve as a basis for microbial identification in the clinical laboratory. Similar to high-complexity molecular methods, the technology may at first be relegated to large reference laboratories due to high instrument costs; however, it is likely that MS technology could be utilized on a routine basis, even in small laboratories if instrument costs decrease. If modified to allow for the analysis of a larger variety of microbes and molecules, standardized to allow ease of use for a highly varied workforce, and integrated into laboratory information systems, MS methods are destined to become an integral tool for most diagnostic microbiology laboratories.
Across the globe, the trend of the use of diagnostic MS methods is apparent (11, 12), and while laboratory scientists await FDA approval of the technology in the United States, some are self-verifying the use of the technology. Sample preparation is both simple and reproducible. Most medical laboratory scientists can easily perform analysis of raw MS data and determine microbial identifications with the aid of associated software. Finally, MS technology can interface directly with the laboratory information system (LIS) and reflex to other diagnostic testing. Thus, as MS continues to be implemented into modern clinical microbiology laboratories, it is important that laboratorians and clinicians alike become familiar with this paradigm-shifting technology. In short, MS technology is rapid, robust, customizable pursuant to the needs of the laboratory, more cost-effective than current phenotypic testing methods despite the initial cost of the instrument, and, perhaps most importantly, easy to use. In this review, the mechanics and processes underlying MS for microbial identification will be described and demystified to make the technology more familiar and understandable (see Table 1 for a list of definitions).
Table 1.
Common terms used in mass spectrometry
| Term | Definition |
|---|---|
| Adduct | Ion formed by the interaction of an ion with one or more atoms or molecules to form an ion containing all the constituent atoms of the precursor ion as well as the additional atoms from the associated atoms or molecules |
| Analyte | Biomolecule or sample that is being analyzed |
| Chromophore | Functional group in a molecule that is known to absorb light; this is necessary for the MALDI matrix in order to absorb the energy of the laser beam |
| Desorption | The opposite of absorption; here a substance is released from or through the surface rather than going into it |
| Detector | The ions generated in a mass spectrometer after traveling through the flight tube ultimately hit the analyzer, where they are detected and converted into a digital output signal |
| Mass analyzer | Chamber having an electrostatic field; its purpose is to separate the ions coming from the source depending on their mass-to-charge ratio so that they can be detected by the detector |
| Matrix | Compound that is mixed with the sample that is being analyzed; the matrix protects the sample molecules from being destroyed by direct focus of the laser beams and facilitates the sample's vaporization and ionization |
| Sublimation | Passing from solid to gas without going through a liquid phase |
| TOF | Time taken by the ions to travel through the flight tube when an electrostatic potential is applied at its ends |
MECHANICS OF MS FOR IDENTIFICATION OF MICROBES
Mass spectrometry was historically utilized as an analytical tool of the clinical chemist, making use of its high levels of sensitivity and specificity in routine processes, in the diagnosis of some cancers (13), inherited disorders (14), and novel biomarkers for disease diagnostics (15). The earliest attempts at the use of mass spectrometry for the identification of bacteria predate the first description of matrix-assisted laser desorption ionization (MALDI) mass spectrometry (16). The ability to analyze large biomolecules was first realized close to 3 decades ago through the application of so-called “soft ionization” techniques that gently ionize target molecules in the sample, called analytes, to generate a spectrum of components. The MALDI method was first introduced in 1987 (17) and subsequently reported in similar experiments in 1988 (18) and was honored with a shared Nobel Prize in 2002. Since then, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) evolved into a rapid and highly reliable analytical tool for the characterization of a diverse collection of microbes encountered in the clinical laboratory (19, 20). Currently, a variety of analysis methods and MS instruments are available, and while not all of them are currently used in commercial MALDI-TOF MS diagnostic applications, it is useful to understand the diversity and modern iterations of MALDI-TOF MS technology.
Mechanisms and Components Leading to Sample Ionization in MALDI-TOF MS
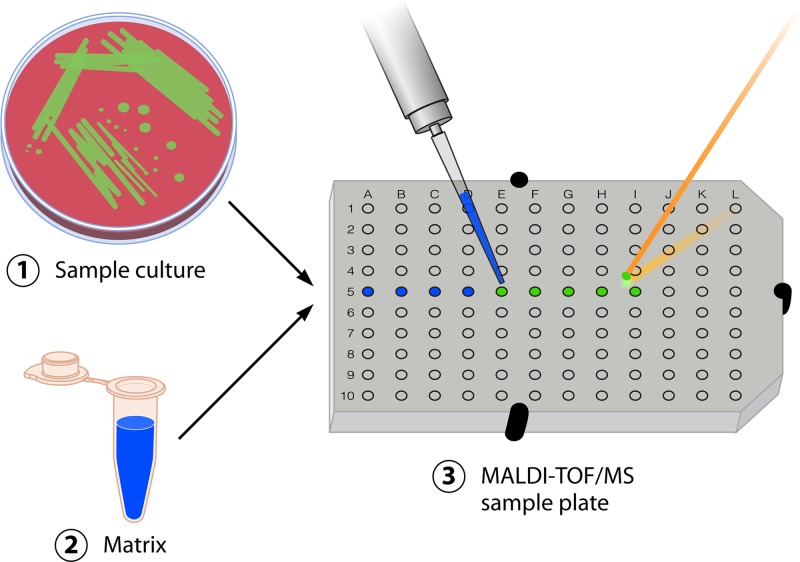
In an MS analysis utilizing MALDI as a soft ionization mechanism, a saturated solution of a low-mass organic compound, called a matrix, is added to the sample, and the mixture is then spotted onto a metal target plate for analysis (Fig. 1). In the case of bacterial or fungal identification, a microbial colony is analyzed, or in some cases, direct blood culture material, urine, cerebrospinal fluid (CSF), or protein extract is used. Upon drying, the clinical material and the matrix cocrystallize and form a solid deposit of sample embedded into the matrix. The matrix is essential for the successful ionization of the clinical sample, as it acts both as a scaffold by which ionization can occur and as a supplier of protons for the ionization of the clinical material. This sample-matrix crystal, now present on the surface of the metal plate, is irradiated by using a UV laser beam (usually, an N2 laser beam with a wavelength of 337 nm is utilized in commercial instruments). Irradiation occurs for a short time to avoid damage or degradation of the sample embedded in the matrix, which could be caused by excess heating.
Fig 1.
General schematic for the identification of bacteria and yeast by MALDI-TOF MS using the intact-cell method. Bacterial or fungal growth is isolated from plated culture media (or can be concentrated from broth culture by centrifugation in specific cases) and applied directly onto the MALDI test plate. Samples are then overlaid with matrix and dried. The plate is subsequently loaded into the MALDI-TOF MS instrument and analyzed by software associated with the respective system, allowing rapid identification of the organism.
The laser beam is focused on a small spot on the matrix-clinical sample crystalline surface (typically 0.05 to 0.2 mm in diameter), and a beam attenuator is employed in the laser optics to adjust the irradiance (defined as the intensity per unit of surface). This laser attenuation can be individually adjusted for each measurement, depending upon the sample type, but is usually standardized by the manufacturer for routine applications. The interaction among the photons from the laser and matrix molecules caused by uptake of energy from the beam triggers a sublimation of the matrix into a gas phase, forming a plume, which is directly followed by the ionization of the clinical sample (Fig. 2). Other wavelengths of the laser ranging from UV to infrared are also used in MALDI experiments. UV lasers are most commonly used and include those from most nitrogen lasers (337 nm), followed by excimer lasers, neodymium-doped yttrium aluminum garnet (Nd:YAG) lasers (355 nm), and, more recently, infrared lasers such as erbium-doped yttrium aluminum garnet (Er:YAG) lasers (2.94 μm) and transversely excited atmospheric (TEA-CO2) lasers (10.6 μm).
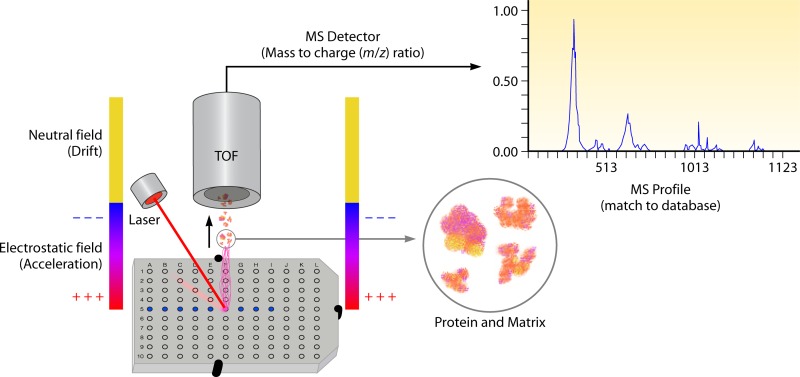
Fig 2.
General schematic for MS analysis of ionized microbiological isolates and clinical material. Once appropriately processed samples are added to the MALDI plate, overlaid with matrix, and dried, the sample is bombarded by the laser. This bombardment results in the sublimation and ionization of both the sample and matrix. These generated ions are separated based on their mass-to-charge ratio via a TOF tube, and a spectral representation of these ions is generated and analyzed by the MS software, generating an MS profile. This profile is subsequently compared to a database of reference MS spectra and matched to either identical or the most related spectra contained in the database, generating an identification for bacteria or yeast contained within the sample.
Ion Formation within the Crystalline Deposit on the MALDI Plate
Intense focusing of the laser beam on the sample material, mixed with the crystalline matrix, causes irradiation by the laser and rapid heating of matrix crystals and the dried clinical specimen due to absorption of a large amount of energy from the beam. Pulsation by the laser provokes both the matrix and clinical sample to rapidly sublimate from the solid phase into the gas phase (without passing through a liquid phase), forming a plume containing ions from both the matrix and the clinical sample. Although the exact mechanism of ionization is not well understood, it can be best explained by a simplified two-step mechanism consisting of primary and secondary ionization events.
Soft ionization of proteins is critical for bacterial identification methods, as it allows the analysis of large biomolecules, including ribosomal proteins, with sizes measured up to 100 kDa (17, 18, 21, 22). Common soft ionization techniques include both electrospray ionization (ESI) and MALDI, both of which are techniques currently used for the identification of bacteria and fungi based on either proteomic fingerprints or amplification of genetic material (11, 23).
Once ionized, proteins within the clinical specimen are analyzed by a component of the mass spectrometer called the mass analyzer to reveal characteristic information about the composition of the sample in the context of a spectrum of mass-to-charge (m/z) ratios (Fig. 2). The m/z ratios are electrodynamic measurements of how quickly charged ions from the clinical sample material move through the time of flight (TOF) tube and reach a detector. Once spectra are generated, comparison to a database of defined reference spectra leads to microbial identification. As the protein compositions differ between bacterial species (and even between bacterial strains and subspecies), different spectra will be generated, allowing for discrimination between closely related organisms. In general, the m/z ratios that are generated and considered when formulating a microbial identification are ribosomal proteins that are unique to their respective bacterial groups or species (17, 18, 21, 22).
The first step of the hypothesized ionization mechanism involves multiphoton ionization of the matrix molecule to produce a radical cation. In short, photons from the laser bombard the clinical sample-matrix mixture and remove an electron from a molecule of matrix material, generating a radical cation of matrix (M+/+·). The process is illustrated by the following chemical formula:
Two photons are required for this reaction because the irradiances of the laser are too low to allow for additional absorption in this time scale (24).
Within this hypothesized mechanism, a caveat exists. The ionization potentials (IPs) for the matrix are too high for two-photon absorption (9 to 10 eV IP, 7.36 eV photon energy for a N2 laser, and 6.98 eV for an Nd:YAG laser); therefore, an alternate two-step theory for the generation of the matrix radical, named the energy pooling theory, is proposed. In the energy pooling model, two or more excited-state matrix molecules produce one matrix radical cation. This reaction is possible as the matrix molecules are densely packed in close proximity when dried on the MALDI plate in the solid phase (Fig. 1). This is exemplified by the presence of matrix clusters or aggregates in the gas phase (25). The energy pooling theory results in the following mechanism, whereby the absorbed photon from the excited-state matrix molecule (M*) is transferred to the second excited matrix molecule, resulting in the formation of a cationic matrix radical (M+·), a nonradical matrix molecule (M), and a free electron (e−):
The second step of this two-part reaction involves a proton transfer event from the excited matrix molecule to the clinical sample (A), resulting in ionization of a molecule of the clinical sample:
Additional ions of the clinical specimen are formed by secondary ion-molecule reactions between matrix-matrix and matrix-specimen interactions. These reactions are thermodynamically favorable because the proton affinity of MALDI matrices is typically lower than that of peptides and proteins to be analyzed in clinical material. This is modeled by the following equations:
and
Types of Matrices Used in MALDI-TOF MS Experiments
Matrices used in MALDI-TOF MS experiments are generally crystalline solids with low vapor pressure that can easily become volatilized to form ions in a vacuum (as in the context of MALDI-TOF MS). The chemical matrix is mixed in excess with a clinical sample and allows for production of intact, gas-phase ions from large, nonvolatile, and thermally labile compounds such as proteins. The matrix plays a key role by absorbing the laser light energy and causing a small part of the target substrate to vaporize. Matrices should possess certain characteristics, such as having a strong absorbance at laser wavelengths used to facilitate ionization, stability in a vacuum to force an interaction with the coionized clinical specimen, an ability to ionize the clinical specimen, solubility in solvents that are compatible with the clinical specimen in order to create an effective matrix-specimen mixture, and a complete lack of any chemical reactivity with the clinical specimen, to avoid unwanted alterations or damage to peptides contained within the sample. In the case of MALDI-TOF MS, which uses a UV laser, the matrix molecule must also have a strong chromophore as part of its composition to help absorb energy, thus preserving the protein fragmentation. Chromophores are selected based on their ability to absorb specific laser wavelengths, resulting in electronic excitation of the matrix. A list of matrices commonly used for analyzing biomolecules by MALDI-TOF MS is provided in Table 2.
Table 2.
List of common matrices used for UV-MALDI methods
| Chromophore matrix(es)a | Sample type(s) analyzed |
|---|---|
| PA, HPA, 3-aminopicolinic acid | Oligonucleotides, DNA, and biopolymers |
| DHB | Oligosaccharides |
| CCA | Peptides and triacylglycerol |
| SA | Proteins |
| HABA | Peptides, proteins, glycoproteins |
| MBT | Peptides, proteins, synthetic polymers |
| DHAP | Glycopeptides, phosphopeptides |
| THAP | Oligonucleotides |
PA, picolinic acid; HPA, 3-hydroxypicolinic acid; SA, 3,5-dimethoxy-4-hydroxycinnamic acid; HABA, 2-(−4-hydroxyphenylazo)benzoic acid; MBT, 2-mercaptobenzothiazole; DHAP, 2,6-dihyroxyacetophenone; THAP, 2,4,6-trihydroxyacetophenone.
Laboratories involved in basic life science research will often vary the matrix that is utilized in order to more completely and accurately analyze a subset of molecules from biological specimens. Commonly used for analyzing proteins and triacylglycerols, α-cyano-4-hydroxycinnamic acid (CCA) and its derivate 4-chloro-α-cyanocinnamic acid (Cl-CCA) have been shown to be more efficient in proteomic analysis than other matrices (26). Sinapinic acid is also popular due to its ability to reduce photochemically generated adducts, greatly improving the mass resolution for proteins (27). Finally, 2,5-dihydroxybenzoic acid (DHB) is another commonly used matrix for general analysis of protein digests, carbohydrates, oligosaccharides, glycopeptides, and both proteins and peptides below 10 kDa. This matrix is also well suited for the negative ion MALDI-TOF MS glycolipids. With regard to clinical identification of infectious microorganisms, a number of matrices were investigated, with various levels of success, and are reported elsewhere (21).
With respect to analyte ionization, MALDI has proven to be a useful tool in molecular analysis of large compounds. Sample preparation is simple, and it shows more tolerance to salts and detergents than other mechanisms of soft ionization such as ESI, aspects that are of consequence to the clinical microbiologist, as microbial growth medium is often rich in salts, and detergents are sometimes formed during bacterial growth. Additionally, MALDI is often found to be more sensitive than other ionization techniques, as the laser beam is focused on a small portion of the matrix, allowing efficient energy transfer and preventing destruction of the clinical sample. Moreover, the analyte molecules are widely separated within the matrix mixture, preventing the clustering of molecular ions that can hamper analysis.
Mass Analyzers Used for Characterization of Ionized Clinical Specimens
Following laser bombardment, ions generated from both the matrix and clinical material must be analyzed to determine their respective masses and identities. The mass analyzer is the component of a mass spectrometer that functions to determine these representative masses, aiding in the identification of the proteins being analyzed. A variety of mass analyzers exist for measuring ionized proteins from biological samples. In theory, no single analyzer is ideal for all applications, and instruments must be selected on the basis of experimental necessities. In the case of microbial diagnostics, commercial systems have been developed for MALDI-TOF MS-based identification, but it is still necessary to perform instrument calibration and quality control. MALDI utilizes a pulsed ionization source, where a pulse of ions from the clinical specimen is produced by an instantaneous exposure to the laser beam. The pulsed nature of the MALDI process pairs naturally with the TOF mass analyzer, which requires that all ions enter the flight tube simultaneously (28). Additionally, the TOF mass analyzer is ideal for MALDI, due to its virtually unlimited mass range, which is advantageous because MALDI typically produces singly charged molecular ions that can have a high mass-to-charge (m/z) ratio. The implementation of mass spectrometric techniques into the clinical laboratory is highly dependent upon method standardization and reproducibility; therefore, mass analyzers are often preselected, optimized, and marketed as part of an instrument package dedicated to microbial identification. Common mass analyzers used in MS analysis are listed in Table 3.
Table 3.
Common mass analyzers and their properties
| Mass analyzer | Separation property | Resolutionb | Mass accuracy (Da) | m/z range |
|---|---|---|---|---|
| Quadrupole | Ion trajectory stability | 1,000–2,000 | 0.1 | 200–4,000 Da |
| Time of flighta | Drift velocity | 2,000–100,000 | 0.001 | Up to 10 MDa |
| Quadrupole ion trap | Ion trajectory stability | 1,000–2,000 | 0.1 | 200–4,000 Da |
| Ion cyclotron resonance | Orbital frequency | 5,000–5,000,000 | 0.0001 | 200–20,000 Da |
Commonly used in clinical microbiology.
A unitless measure used to describe resolution of peptides or proteins.
The Time of Flight Analyzer
The time of flight (TOF) analyzer is dependent upon the principle that applying an electrostatic field (eV) to the ionized clinical material causes a generated ion with a charge (z) to accelerate, imparting to it some amount of kinetic energy (KE). The ions then move into a field-free drift region, where the only force affecting ionic movement is the kinetic energy from the acceleration step. The velocity (v) of the ionized molecule from a clinical specimen can therefore be calculated by using the following equation, where KE is kinetic energy, m is mass, v is velocity, z is the charge of the ion (+1 for MALDI), eV is the voltage applied, D is the distance to the detector, and t is time:
In this context, D and eV are constant and t is measured, allowing the m/z ratio to be determined. A simple mathematical rearrangement results in the following equation (29, 30):
This equation demonstrates that drift time is directly proportional to the m/z ratio. Larger ions will have a longer drift time and smaller molecules will have a shorter drift time, demonstrating separation of molecules based on mass (31). This allows for separation of ions originating from clinical material based on the m/z ratio.
Linear Time of Flight Mass Spectrometry
In linear TOF, the method most commonly used for the MS analysis of microbial specimens, ions generated from the source are accelerated into the flight tube and enter a field-free region where they are separated according to their velocities (and subsequently size, as discussed above), before hitting the detector located at the other end of the tube. The linear TOF method has high sensitivity and high efficiency, with the ability to analyze molecules in femtomolar (10−15 mol/liter) and attomolar (10−18 mol/liter) concentrations (32). However, a limitation of the linear TOF method is that it provides a poor resolution due to the peak broadening that can occur due to the spatial distribution of analyte molecules on the surface and the unequal distribution of energies from the laser pulse. This results in ions with the same m/z having different kinetic energies.
Pulsed-ion extraction (PIE) was designed to resolve limitations associated with peak broadening. In PIE, there is a delay in the application of the acceleration voltage following ionization. Ions that gain more kinetic energy in the ionization process will drift away from the target plate. When the acceleration voltage is applied, there is a gradient between the target plate and ground, and the ions farther from the target will experience lower voltage and will therefore have a lower deposited kinetic energy. This phenomenon results in averaging with ions that received less kinetic energy in the ionization process, thus normalizing the kinetic energy of ions having the same m/z ratio.
Reflectron
A reflectron is a focusing element at the end of the TOF instrument that changes the direction of ion travel. A voltage is applied to these lenses and causes a change in the trajectory of that ion. Ions with a higher kinetic energy will penetrate the reflectron deeper than those with a lower kinetic energy, such that the flight path is elongated, allowing for averaging of flight times and decreasing peak broadening. Although a reflectron is effective at reducing peak broadening, it essentially doubles the ion path; therefore, when sensitivity is an issue, it is necessary to use linear TOF due to the potential for ion scattering. For this reason, when analyzing high-mass ions with MALDI-TOF MS, as in the case of clinical material utilizing commercial MALDI-TOF MS platforms, linear TOF is most commonly used.
ISSUES AND IMPLICATIONS FOR USE IN CLINICAL MICROBIOLOGY LABORATORIES
Standardization
Although innovative and greatly informative, many initial studies using MALDI-TOF MS were limited in scope and lacked databases, standardized reagents, and protocols for the analysis of intact bacterial cells. Many early investigations found that the spectra generated from microorganisms exhibited a high degree of variation under different culture conditions and among studies performed in different laboratories (33). Early databases utilized for microbial identification and characterization by MALDI-TOF MS were often developed “in-house” to fit the needs of the laboratory responsible for their design (34) and thus contained a high percentage of organisms from strain collections of individual investigators, making comparisons between the results from different laboratories difficult. While in-house databases are still constructed and provide valuable information toward more discriminatory analysis (i.e., serotype, subspecies, and epidemiological analyses), routine analysis is generally performed by using proprietary databases marketed with commercial MALDI-TOF MS systems.
Following the publication of conflicting results in some early investigations, the issue of standardization was preliminarily addressed with regard to bacterial culture conditions (35), MS conditions (36, 37), and preanalytical processing (34, 38). Additionally, a shift in analytical focus away from bacterial surface components (39), which can vary in levels of expression under different culture conditions, toward the analysis of ribosomal proteins that are ubiquitously expressed throughout all phases of growth added to the stabilization and robustness of the generated spectra and supported enhanced analytical capabilities. The adaptation of these defined methods sharply reduced the variation among spectral profiles of isolates being analyzed, vastly improving the accuracy and reliability of MALDI-TOF MS for bacterial identification in inter- and intralaboratory evaluations (12).
Evolution of Intact-Cell MALDI-TOF MS
Early studies evaluating the use of MALDI-TOF MS for microbial identification focused on the ability of the technology to accurately determine the identity of whole microorganisms isolated from agar-based culture. MALDI-TOF MS provided the capability to eliminate protein extraction methods prior to analysis, allowing intact microorganisms to be simply spotted onto a solid plate and mixed or overlaid with a matrix compound and cocrystallized, which facilitates the dissociation and ionization of bacterial proteins (40, 41). The intact-cell (IC) method, as it is sometimes called, provided a new and simple mechanism for rapid analysis of bacterial components based on the generation of specific spectral fingerprints that facilitated accurate microbial identification and characterization (19, 41, 42).
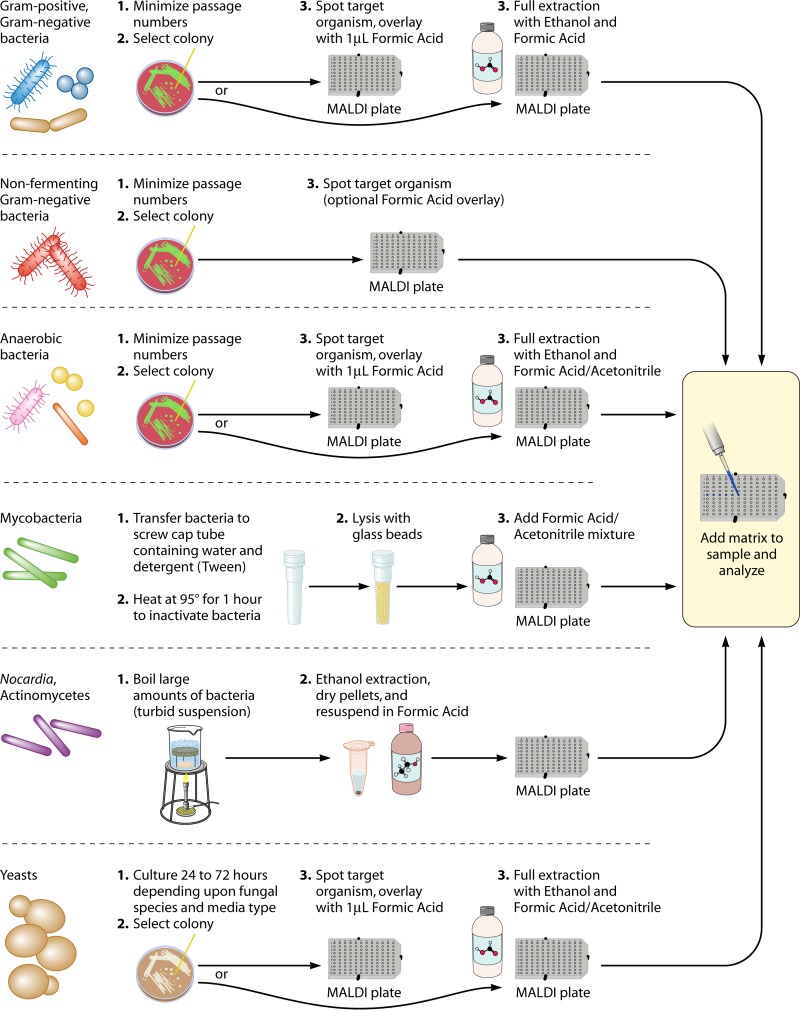
Due to the simple mechanism of sample preparation, IC MS became an attractive alternative to phenotypic and genetic methods of microorganism identification. Several preliminary studies supported the observation that IC MALDI-TOF MS was indeed sensitive enough to differentiate closely related organisms (43) and perhaps even discriminate between different strains of the same or phenotypically similar organisms (44–46), providing new avenues for genus-, species-, and strain-level identifications. However, as analysis of microorganisms by MALDI-TOF MS became more commonplace, it became apparent that the IC method was not always appropriate for all specimen types in spite of its relative simplicity; problems with spectral generation from some microbes (47, 48) and biosafety issues (49, 50) arose. In an effort to improve spectral generation and be compliant with biosafety regulations, modified versions of sample preparation methods have been reported for different groups of microorganisms and range from on-plate inactivation using formic acid (FA) and matrix to full-scale protein extraction using ethanol-based methods (Fig. 3).
Fig 3.
Additional suggestions for MALDI-TOF MS sample preparations for use with different classes of microbes. Different groups of microorganisms vary fundamentally in their cellular composition and architecture. These differences have been demonstrated to affect the quality of spectra generated during MS experiments and, thus, the accuracy of MALDI-TOF MS-derived identifications. As such, investigators from a number of independent studies have evaluated different methods for sample preparation of different groups of microorganisms, ranging directly from intact-cell to full-protein-extraction-based methodologies. Results from these studies are summarized here. Proper biological safety precautions should be followed with respect to dangerous members of these groups of organisms.
CURRENTLY AVAILABLE COMMERCIAL PLATFORMS FOR MALDI-TOF MS IDENTIFICATION OF MICROBES
Multiple platforms from a number of well-established commercial manufacturers are available for MALDI-TOF MS identification of bacteria and yeast. Spectral databases are often marketed as part of a proprietary system, as opposed to a publicly accessible open platform, and are constructed and maintained by their representative manufacturers. A majority of these databases can be expanded to accommodate spectral entries that are not included in marketed versions. The ability to add spectra and construct custom databases is important for further discriminatory analysis using MALDI-TOF MS, including strain typing and epidemiological investigations. As each proprietary system uses its own algorithms, databases, software, and interpretive criteria for microbial identification, numerical data (i.e., spectral scores) from different commercial systems are not directly comparable (51). Comparative analysis between MS systems is therefore usually performed by using final identifications in the context of each system's interpretive algorithms. In this section, MS platforms from different manufacturers are reviewed, and data from investigations examining their ability to identify microbes in clinical settings are summarized.
Andromas
The Andromas system is a database manufactured and maintained by Andromas SAS (Paris, France). Utilized predominantly for clinical diagnostics in Europe, the database is available in North America for research use only. The Andromas system uses multiple species-specific spectral profiles for each organism in the database to increase the robustness of identification. These spectra are derived from either members of the same strain of bacterium with divergent spectral profiles or the same bacterium cultured on different types of growth media. The software separates entries into three separate databases dedicated to the identification of bacteria, mycobacteria, yeasts, and Aspergillus spp. When constructed, this database was populated with spectra generated from direct-colony ionization only, with protein extraction not being performed (52). Sample results are reported as a percentage of similarity between the spectra generated by the microorganism in question and the reference spectra that are in the database. Identifications made by Andromas are then grouped into one of three categories, “good identification,” “identification to be confirmed,” and “no identification,” based upon statistical cutoff values determined by the manufacturer and by the operator.
The current and earlier iterations of Andromas were utilized in a number of studies for the identification of both bacteria and yeast from culture media (53–55) as well as directly from patient specimens (53, 56). Limitations of the Andromas database have been highlighted by Bille et al. Discrimination between species that are closely related is problematic, a shortcoming experienced by a number of MS and non-MS methods alike. Of particular interest was the noted inability of the database to differentiate Streptococcus pneumoniae from Streptococcus mitis, Escherichia coli from Shigella, and Listeria monocytogenes from Listeria innocua and Listeria ivanovii. In these cases, the authors used additional downstream testing, in many cases utilizing serum agglutination assays, to identify the bacteria of interest (53). Farfour et al. additionally evaluated a set of 659 Gram-positive rods by using Andromas and compared these identifications to identifications derived by reference methods. The database performed well but still exhibited problems in providing species-level identifications for members of Listeria spp. (52) despite the reported ability of other commercial databases to discriminate between Listeria spp. by incorporating a protein extraction method (57).
SARAMIS Evolves to Vitek-MS
The SARAMIS database was previously created and maintained by AnagnosTec GmbH prior to being purchased by bioMérieux for incorporation into the Vitek-MS platform. Prior to its integration into the Vitek-MS platform, the database was marketed and sold by Shimadzu along with Axima mass spectrometers as the Axima-iD Plus system, which featured linear/reflectron combinations and high-energy collision-induced dissociation modes, allowing for greater resolution and greater mass accuracy than simple linear ion mode analyzers The database uses SuperSpectra, which contained a conglomerate of biomarkers from at least 15 individual isolates, indicative of specific genera, species, and strains from a variety of geographical locations. These spectra were obtained following variations in growth conditions and growth media, generating a robust diagnostic for microbial identification. Spectra that are generated are consolidated into a list of peaks and intensities, which is then queried against the database to identify potential matches with archived SuperSpectra. If no statistically satisfactory match can be obtained, an expanded database containing a broader collection of spectral information is then queried in order to generate identification. The software is also able to perform hierarchical analysis of the spectral data to examine changes in a population of similar microorganisms, determine relatedness of different isolates, and generate additional SuperSpectra for in-house database expansion (Axima-iD Plus brochure [http://www.ssi.shimadzu.com/products/literature/biotech/mo347_v1.pdf {accessed 19 April 2013}]). Following acquisition of SARAMIS by bioMérieux, the manufacturers developed their own algorithms for MS-based microbial identification and have begun marketing the database and associated software and hardware under the name Vitek-MS (51).
BioTyper
The BioTyper system was conceived and marketed exclusively by Bruker Daltonics and is capable of analyzing specimens including bacterial, mycobacterial, and fungal samples in addition to samples recovered directly from positive blood culture bottles. Of all the mass spectral analysis software programs, the BioTyper platform is perhaps the most heavily utilized software package in the United States. The software is marketed as a versatile tool for the clinical microbiologist, including both options for batched specimens and the ability to interrupt routine runs for the analysis of specimens whose results are urgently needed as well as automatic calibration and integration into existing laboratory information systems. The BioTyper software package is currently sold along with the Flex line of benchtop MALDI mass spectrometers.
Like other software systems discussed above, the BioTyper software is an open platform allowing the user to save runs to expand the database of stored spectra by utilizing tools included in the software. Mass spectra are generated, and data are analyzed with regard to spectrum peak frequency, position, and intensity. These spectra are then compared against a library of main spectra encoded in the BioTyper database. These main spectra are derived again from replicative measurements of the type strain with the goal of generating representative spectra of the organism across a range of biological variables. The user also has the ability to create main spectra with the assistance of the software and to populate the database with entries derived from microorganisms isolated in-house.
Two distinct criteria are used to analyze the results of the spectral database search: a score value and a consistency category. Log score values range from 0.000 to 3.000 and are correlated with an explanation of genus and species consistency within the database. A score ranging from 2.3 to 3.000 is interpreted by the software as a highly probable species-level identification. Log scores of between 2.00 and 2.299 represent secure genus identification and probable species-level identification. In both cases, the results can usually be released as a positive identification pursuant to the testing algorithms implemented in the laboratory performing the testing. Log scores ranging from 1.70 to 1.999 represent a probable genus identification, with additional testing being required for a positive reportable identification. Log scores ranging from 1.699 to 0 are not considered to be a reliable identification, and further sample processing, analysis, and testing are warranted (MALDI BioTyper brochure [http://maldibiotyper.com/literature.html {accessed 10 June 2013}]).
PERFORMANCE AND COMPARISON OF COMMERCIAL METHODS IN ROUTINE CLINICAL MICROBIOLOGY
MALDI-TOF MS is an accurate method for routine bacterial identification, even with changing conditions such as culture medium or pH, and interlaboratory comparison is usually good, provided that minimal common reagents are used (35, 58–60). Most errors in published reports are attributed to an incomplete population of databases associated with the instruments, clerical error in database assembly or during data acquisition, or an inability of the MS spectra to differentiate similar species.
Seng et al. were the first to report the feasibility of MALDI-TOF MS as the first-line system for routine bacterial identification in clinical microbiology laboratories using bacterial colonies growing on agar plates (61). A total of 1,660 bacterial isolates were identified, and discrepancies between the MALDI-TOF MS results were verified via gene sequencing. At that time, the authors correctly identified only 84.1% of isolates to the species level by a direct analysis of bacteria without additional protein extraction. Stenotrophomonas maltophilia and Shigella sonnei were frequently misidentified (7/10 [70%] 5/5 [100%] isolates, respectively). In the case of S. maltophilia, false identification resulted because the references of Pseudomonas hibiscicola and Pseudomonas beteli entered into the BioTyper database are actually S. maltophilia. Most of the other unidentified species were absent from the database at the time of study.
In the first comparison of two commercially available systems for clinical laboratory use, Cherkaoui et al. compared the Bruker BioTyper and bioMérieux systems with their respective databases, BioTyper and an early version of SARAMIS (62). In this study, 16S rRNA gene sequencing was used as the gold standard for comparison. Using these systems, 720 clinical isolates were identified to the species level. Of these, 99.1% were identified with the Bruker MALDI-TOF MS spectrometer and 88.8% were identified with the Shimadzu MALDI-TOF MS spectrometer (of note, this database was comprehensively revised since that report). Not surprisingly, anaerobes were among the species most frequently not identified, probably due to the lack of reference spectra in the databases. As in nearly all other studies, poor identification of streptococci was observed, with an identification rate of 41% with both systems. Mellmann et al. identified 1,116 clinical isolates by MALDI-TOF MS using the same database and using manual and automated phenotypic methods as the reference standard with 16S rRNA gene sequencing for discrepant testing; for Enterobacteriaceae, nonfermenting Gram-negative rods, staphylococci, enterococci, and streptococci, they achieved correct identifications to the species level for 95.5, 79.7, 99.5, 100, and 93.7% of isolates, respectively (12). Shigella spp. and Streptococcus mitis/Streptococcus oralis were misidentified by MALDI-TOF MS, with 0/7 and 0/6 correct identifications, respectively. In contrast, correct identification was obtained for staphylococci, enterococci, and Enterobacteriaceae; correct identifications to the species level for 100, 95.7, and 83.2% of isolates, respectively, were reported (63).
van Veen et al. reported similar results for 980 clinical microbial isolates, including 61 yeast isolates; the overall identification rate at the species level was 92%. After identification was verified by using 16S rRNA gene sequencing in cases of discrepancies between MALDI-TOF MS-based identification and biochemical identification, correct identifications were obtained at the species level for Enterobacteriaceae, nonfermenting Gram-negative rods, staphylococci, streptococci, and yeasts for 97.7, 92, 94.3, 84.8, and 85.2% of isolates, respectively (64). Misidentifications were associated with a lack of spectra for some rare species and problems in identification of viridans group streptococci (VGS) and S. pneumoniae; 12/21 (57.1%) isolates of viridans group streptococci were falsely identified as S. pneumoniae.
In another study, Prod'hom et al. reported correct identification of 1,278/1,371 clinical isolates (93.2%) to the species level (65). The 56 discordant results were analyzed, and most errors were due to false identification of Enterobacter cloacae as Enterobacter hormaechei and S. maltophilia as Pseudomonas hibiscicola and Pseudomonas beteli. Problems with identification of Shigella spp. and Propionibacterium acnes were also observed. In another study, the same limitations of the database were also observed for the identification of some species, including anaerobic bacteria; the authors proposed that an extraction step may be necessary to improve identification of some species (66). The identification level improved from 82.6 to 97.3% when an extraction step was added.
Finally, Martiny et al. compared the three commercial databases, including the first report of the database planned for in vitro diagnostics (IVD) use supported by Vitek-MS (bioMérieux). In this study, 1,129 isolates were examined, including 73 anaerobes. The Bruker LT BioTyper and the Vitek-MS databases performed equally well, with correct identification of 93% of routine isolates (67).
COMPARISONS BY MICROBE CATEGORY
With the introduction of any new technology for clinical diagnostics, comprehensive reviews are undertaken to evaluate the method against methods currently used as reference standards to determine reproducibility, accuracy, and robustness. MALDI-TOF MS is no exception, as a myriad of studies have appeared in the literature in the past 5 years examining the diagnostic accuracy of the method against phenotypic and molecular methods such as sequencing-based approaches, Vitek 2 and API-based methodologies, and serological approaches. The focus of this section is to highlight and summarize key findings from this large repertoire of studies in the hopes of providing a comprehensive examination of the dynamic clinical utility of MALDI-TOF MS for the identification of specific groups of microbes as well as to explore the ability of the technology and its uses for specialized testing pertaining to specific microbial genera. Published reports of MALDI-TOF accuracy for a wide variety of microbes are listed in Table 4.
Table 4.
Genus-specific studies utilizing MALDI-TOF MS for bacterial identification and characterization
| Genus | Species or group evaluated | Reference(s) |
|---|---|---|
| Gram-positive organisms | ||
| Staphylococcus | Coagulase-negative staphylococci | 75, 79, 80 |
| S. aureus | 44, 81–86, 88, 97 | |
| Coagulase positive, non-S. aureus | 89 | |
| Mixed staphylococcal species | 55, 90, 91 | |
| Micrococcus | Micrococcus spp. | 94 |
| Streptococcus | Beta-hemolytic species | 45, 99 |
| Group A streptococci | 101 | |
| Group B streptococci | 103, 104 | |
| Streptococcus pneumoniae | 105 | |
| Viridans group streptococci | 112, 113 | |
| Nutritionally variant streptococci | 70, 118 | |
| Enterococcus | Enterococcus spp. | 122, 123 |
| Lactococcus | Lactococcus spp. | 128 |
| Bacillus | Bacillus spp. | 129–131 |
| Listeria | Listeria spp. | 57 |
| Corynebacterium | Corynebacterium spp. | 133–135 |
| Arcanobacterium/Trueperella | Trueperella spp./A. haemolyticum | 136, 137 |
| Nocardia/mycobacteria | ||
| Nocardia | Nocardia spp. | 48 |
| Mycobacterium | Mycobacterium spp. | 46, 152–156 |
| Gram-negative bacteria | ||
| Enterobacteriaceae | ||
| Salmonella | Salmonella spp. | 161–165 |
| Escherichia/Shigella | E. coli/Shigella spp. | 167 |
| Cronobacter | Cronobacter spp. | 173–176 |
| Enterobacter | Enterobacter cloacae complex | 177 |
| Pantoea | Pantoea spp. | 178 |
| Plesiomonas | P. shigelloides | 179 |
| Klebsiella/Raoultella | K. oxytoca/Raoultella spp. | 180 |
| Yersinia | Yersinia spp. | 181–183 |
| Y. enterocolitica | 184 | |
| Y. pestis/Y. pseudotuberculosis | 185 | |
| Nonfermenting rods | ||
| Acinetobacter | Acinetobacter | 190–194 |
| Burkholderia | B. cepacia complex | 187, 198, 199 |
| B. mallei/B. pseudomallei | 204, 205, 207 | |
| Pseudomonas | Pseudomonas spp. | 208 |
| Stenotrophomonas | Stenotrophomonas maltophilia | 209 |
| Fastidious organisms | ||
| Brucella | Brucella spp. | 211, 213 |
| Bartonella | Bartonella spp. | 214 |
| Francisella | Francisella spp. | 215, 216 |
| Haemophilus | Haemophilus spp. | 217, 220 |
| Vibrio | Vibrio spp. | 221–223 |
| Aeromonas | Aeromonas spp. | 224–227 |
| Campylobacter | Campylobacter spp. | 228–233 |
| Helicobacter | Helicobacter spp. | 228, 231, 234, 235 |
| Neisseria | Neisseria gonorrhoeae/N. meningitidis | 234, 236, 237 |
| Moraxella | Moraxella catarrhalis | 238 |
| Legionella | Legionella spp. | 239–242 |
| Anaerobic bacteria | ||
| Propionibacterium | P. acnes | 256 |
| Bacteroides | Bacteroides spp. | 244, 257 |
| Clostridium | Clostridium spp. | 258 |
| C. difficile | 261 | |
| Fungi | ||
| Yeasts | ||
| Candida | Candida spp. | 264, 210, 266–269 |
| Cryptococcus | Cryptococcus spp. | 271–273 |
| Filamentous fungi/molds | ||
| Aspergillus | Aspergillus spp. | 277–280 |
| Fusarium | Fusarium spp. | 281–283 |
| Dermatophytes | 286–289 | |
| Pseudallescheria-Scedosporium | Pseudallescheria-Scedosporium | 291 |
| Penicillium | Penicillium spp. | 292 |
| Lichtheimia | Lichtheimia spp. | 295 |
GRAM-POSITIVE BACTERIA
Gram-positive bacterial species include a large number of pathogenic bacteria both frequently and infrequently encountered in the clinical laboratory. In general, these organisms possess large quantities of peptidoglycan at their cell wall, which is used for the display of a number of surface proteins involved in adhesion to and interaction with host tissues. This thick layer of peptidoglycan can sometimes render these bacteria more resistant to lysis than their Gram-negative counterparts, and a number of pretreatment or enhancement strategies were devised to counteract this, including the addition of lysostaphin, lysozyme, mutanolysin, and proteinase K to bacterial suspensions.
In a thorough and well-designed study, Alatoom et al. described the comparison of the application of whole cells (direct colony) to protein extraction for the identification of Gram-positive cocci using the Bruker BioTyper software. A significant improvement in the number of isolates able to be identified to both the genus and species levels was seen when protein extraction with FA was performed prior to analysis (68).
The Andromas MALDI-TOF MS system was evaluated for the routine identification of Gram-positive bacilli. In a comprehensive analysis of 659 isolates, Farfour et al. reported that 594 (98.5%) of these isolates could be identified to the species level, with most members of the genus Listeria unidentifiable beyond the species level due to spectral peak similarity. MALDI-TOF MS was reported to perform as well as routine identification mechanisms supplemented with extended diagnostic techniques that would have been needed to definitively identify some bacterial isolates within the collection (52), thus demonstrating savings of both time and resources.
Sample Processing for Identification of Gram-Positive Bacteria by MALDI-TOF MS
The processing of Gram-positive isolates for routine identification by MALDI-TOF MS was recently evaluated. Sample processing for Gram-positive organisms included the application of both whole cells as well as a protein extraction step prior to analysis. In place of a time-consuming full protein extraction, TeKippe et al. investigated a method using a 1-μl fluoroacetic acid (FA) overlay. Four different smearing methods were compared with respect to plate processing: heavy and light smears of bacterial culture with and without an overlay of FA added to that respective smear. Using the Bruker BioTyper 3.0 software, 239 aerobic Gram-positive clinical isolates were analyzed, and results were compared to previously obtained phenotypic identifications; discrepant results were resolved by 16S rRNA gene sequencing. Results of this study conclusively determined that the modified FA addition significantly enhanced identification compared to the use of whole cells. Other important conclusions included the finding that frequent passaging of organisms led to an increase in the number of unidentified isolates but that incubation temperature and medium type (blood, chocolate, etc.) did not influence MALDI-TOF MS identification (69).
In conclusion, while more labor-intensive, protein extraction prior to MALDI-TOF MS analysis consistently increases the ability of the technology to identify Gram-positive species. The modified FA extraction procedure described by TeKippe et al. represents a viable alternative more suited to routine clinical workflow, but full protein extraction may still need to be utilized in cases where the microbe is particularly difficult to process (69). Additionally, while commercial software databases were demonstrated to be suitable for routine clinical use, the results of Christensen et al. highlight the importance of a continuously evolving collection of reference spectra in addition to members of the clinical laboratory becoming familiar with methods of in-house generation of spectral collections for the identification of uncommon species not currently contained within commercial databases (70).
Staphylococci
The staphylococci represent a genus of bacteria commonly encountered in the clinical microbiology laboratory in a wide variety of specimen types. Like other bacterial genera, certain species of staphylococci are more commonly associated with disease than others, so accurate identification of these microorganisms to the species level often aids in making a distinction as to whether they are clinically relevant, normal flora, or culture contaminants. Compared to other bacterial genera, staphylococcal taxonomy remains generally straightforward due to the clonal nature of the organism. For most specimen sources, determination of major distinctions between species can be accomplished in an acceptable time frame by using traditional biochemical methods; however, for body fluids, blood cultures, and tissues, additional testing is sometimes necessary. Staphylococci cultivated from these normally sterile body sites or from an immunocompromised host are increasingly known to cause infections (71–74); therefore, they may require highly accurate species-level identification to rule out contamination by skin flora. Some less frequently encountered staphylococcal species can bear phenotypic similarities to more commonly encountered species (i.e., coagulase activity) or share significant genetic similarity, which can result in misidentification by phenotypic and molecular methods, respectively.
Coagulase-negative staphylococci.
Coagulase-negative staphylococci (CoNS) are among the most frequently isolated bacteria in clinical microbiology laboratories (75) and are important opportunistic and device-related pathogens. The organisms are well equipped for this niche, by virtue of their capacity to produce strong biofilms, allowing them to persist in various environments and develop increased resistance to antibiotics (76). In addition, the group maintains a large number of molecules that function to provide protection from the defenses of the host immune system (77). The term “coagulase-negative staphylococci” is somewhat a collective grouping commonly reserved for nonhemolytic staphylococcal species which are not Staphylococcus aureus; however, this group also includes a small number of other coagulase-positive, non-S. aureus staphylococci. As such, the group is populated with a large number of species, many of which share significant genetic and phenotypic homology, rendering definitive identification challenging. The correct species determination of CoNS can be particularly difficult due to a high degree of genetic similarity between species, and phenotypic tests for identification do not always provide reliable results (78).
Currently, a number of methods are used to identify coagulase-negative staphylococci, including both phenotypic (RapID 32) and molecular (tufB and sodA sequencing) assays. Molecular methods were utilized for the characterization of CoNS, with identifications being made on the basis of housekeeping gene sequencing or 16S rRNA sequencing. Automated bacterial identification systems have also been used to identify CoNS, but molecular methods are often preferred due to higher levels of discriminatory accuracy for determinative identification to the species level.
MALDI-TOF MS has been compared to automated phenotypic bacterial identification systems for the identification of CoNS. Dupont et al. compared MALDI-TOF MS linked to a constructed database to the Phoenix (Becton, Dickinson) and Vitek-2 (bioMérieux) systems for the identification of 234 CoNS (20 species total) isolates from clinical laboratories, using the sequence of the superoxide dismutase gene sodA as a reference. In all, MALDI-TOF MS identified 93.2% of isolates correctly, with only 75.6% and 75.2% of isolates being correctly identified with the Phoenix and Vitek-2 systems, respectively (75). A second study, this time using the BioTyper 2.0 software in place of an in-house database, analyzed MALDI-TOF MS compared to tuf gene sequencing for the identification of 62 CoNS reference isolates to the species level. All isolates were identified to the species level, demonstrating 100% concordance with tuf gene sequencing for CoNS identification (79).
In perhaps one of the most conclusive comparisons of phenotypic, molecular, and proteomic methodologies for the identification of coagulase-negative staphylococci to date, Loonen et al. compared the Vitek-2 and RapID 32 phenotypic methods for CoNS to two different sequencing methods (tuf and 16S rRNA gene sequencing) and MALDI-TOF MS using the Bruker BioTyper database. The results of this study determined that the MALDI-TOF MS platform had a 99.3% correct identification rate when a collection of 142 strains consisting of both clinical and reference isolates was analyzed. The Vitek-2 system used in combination with tuf gene sequencing was suggested by those authors to be a suitable alternative for laboratories without access to MALDI-TOF MS, an approach which would result in additional time to definitive diagnosis and higher cost to the patient (80). In summary, MALDI-TOF MS demonstrates high diagnostic accuracy for CoNS and allows for a simple, rapid, and cost-effective mechanism for identification of CoNS.
Staphylococcus aureus.
In contrast to CoNS, Staphylococcus aureus, the predominant pathogenic species from the genus Staphylococcus, is routinely identified with high accuracy in laboratories using long-established and well-standardized phenotypic protocols; however, misidentifications can occur, typically confused with coagulate-positive non-S. aureus species. Perhaps the more pressing clinical challenge posed by S. aureus isolates is not their identification per se but the determination of antimicrobial resistance (i.e., methicillin-resistant S. aureus [MRSA] versus methicillin-sensitive S. aureus [MSSA]) and the identification of certain clonal lineages in outbreak situations.
Automated methods are available to determine antimicrobial susceptibility patterns for S. aureus but often require additional time for the cultivation of the organism in the presence of the antibiotic. Rapid protocols for the determination of methicillin resistance were developed for S. aureus and are usually based upon the detection of a variety of targets, including the mecA gene product, the penicillin binding protein PPB2a, via slide agglutination (Oxoid PBP2′ latex agglutination test; Thermo Fisher), detection of S. aureus-specific bacteriophage via the KeyPath MRSA/MSSA Blood Culture Test-BT (Microphage, Longmont, CA), or detection of the staphylococcal cassette chromosome mec (SCCmec) genetic element insertion site via real-time PCR (Cepheid, Sunnyvale, CA, and Becton, Dickinson, Franklin Lakes, NJ). Methods for the genetic typing of S. aureus isolates rely on molecular approaches that are costly, labor-intensive, highly complex, and variable and, as such, are not performed in all laboratories.
While accurate genus and species identification of S. aureus can be accomplished by MALDI-TOF MS systems, other aspects of S. aureus characterization by MALDI-TOF MS are being investigated in research settings. In 2000, MALDI-TOF MS spectral peaks specific for different species of staphylococci and methicillin-resistant isolates were identified (81). By 2002, three independent studies utilizing MALDI-TOF MS for the analysis of S. aureus were published (82–84).
Using two type strains of S. aureus, one MRSA and one MSSA, and a number of clinical isolates of S. aureus, one study analyzed the use of MALDI-TOF MS for identifying and discriminating between different S. aureus strains. It was concluded that stable, strain-specific spectra could be derived from the two type strains, which could be used for both identification and clonal analysis of the clinical isolates, but a uniform profile could not be elucidated for S. aureus (82).
Another study, utilizing 76 organisms identified as S. aureus by the Vitek system (bioMérieux, Durham, NC) and nuc gene PCR, reported that only 74% of these organisms could be identified by MALDI-TOF MS and the MicrobeLynx software package as S. aureus. The determination of methicillin resistance was also evaluated by MALDI-TOF MS, with varied results. Potential explanations for the low accuracy of the MALDI-TOF MS identification were listed as the incompleteness of the database and variation in bacterial culture conditions used during the course of the study (83).
Once MALDI-TOF MS was determined to be a viable technology for the characterization and analysis of S. aureus, studies examining the reproducibility and standardization of testing methodologies were undertaken. An inter- and intralaboratory reproducibility study of MRSA strains determined that variation of culture media used for propagation of S. aureus generated different spectral profiles but found that the variation among mass spectrometers from different manufacturers was negligible, and it was concluded that MALDI-TOF MS provided a rapid method for the identification of MRSA (84). Three years later, the same group, working with scientists from the mass spectrometer manufacturer Shimadzu, reported a detailed optimized protocol for MALDI-TOF MS fingerprinting of MRSA, including incubation periods, bacterial passage analysis, preparation times, and mass spectrometer settings, among other variables (44).
As MALDI-TOF MS accuracy and sensitivity improved, additional studies investigating its use for the characterization of S. aureus were reported, with more complex aims outside the realm of bacterial identification. One study focused on antibiotic resistance and strain heterogeneity and demonstrated that MALDI-TOF MS was suitable to characterize a small set of isogenic isolates of MRSA and spontaneously arising MSSA isolates with different susceptibility results for the glycopeptide antibiotic teichoplanin. Interestingly, these isolates were considered identical by pulsed-field gel electrophoresis (PFGE) analysis, highlighting the discriminatory power of MALDI-TOF MS (85). A high-throughput study using 134 clinical isolates of S. aureus confirmed by 16S rRNA gene sequencing and an expanded database (MicrobeLynx) of spectral profiles also demonstrated the high accuracy of MALDI-TOF MS for the identification of S. aureus but advocated for the further standardization of culture and diagnostic parameters. These authors were unable to identify unique spectral markers for MRSA, in contrast to previous reports (86).
Finally, a large sample set of 602 molecularly defined S. aureus and 412 CoNS isolates was used to assess the accuracy of BioTyper 2.0 to identify these isolates to the species level. MALDI-TOF MS performed well for all isolates tested, with 100% concordance with sodA or tufB sequencing for the identification of CoNS and with a mean time to result of 22 min (87). The ability of MALDI-TOF MS to identify MRSA lineages from ionized samples has also been investigated, demonstrating the ability of mass spectrometry to organize isolates into groups with high concordance compared to the clonal complex (CC) classifications routinely used for MRSA characterization (88). These studies demonstrate the utility of MALDI-TOF MS and its ability to not only accurately and rapidly identify S. aureus isolates but also potentially classify and provide valuable epidemiological data for MRSA isolates.
Non-S. aureus, coagulase-positive staphylococci.
Although the presence of coagulase is conventionally believed to be a hallmark of S. aureus, other staphylococcal species do indeed produce this enzyme and can exhibit positive results for both slide and tube coagulase as well as protein A and staphylococcal latex. Staphylococci belonging to the Staphylococcus intermedius group (S. intermedius, S. pseudintermedius, and S. delphini) share many biochemical similarities with S. aureus, including coagulase production, which can hamper identification when encountered in the clinical laboratory. Molecular methods are the most reliable mechanism for identification of these staphylococci, but not all clinical laboratories have on-site access to sequencing instruments.
An important analysis of staphylococci belonging to the S. intermedius group was recently reported (89). Using the Shimadzu instrument and the SAMARIS database, reference spectra (SuperSpectra) were created based on unique peaks present for each species. Sixty-nine strains were analyzed, and reference identification was performed by using the hsp60 gene sequence. Using the constructed database, MALDI-TOF MS demonstrated 95% sensitivity and 100% specificity for S. intermedius identification (95% confidence interval [CI], 0.68 to 0.99), 78% sensitivity and 97% specificity for S. pseudintermedius (95% CI, 0.60 to 0.90), and 64% sensitivity and 100% specificity for S. delphini (95% CI, 0.41 to 0.83), demonstrating a relatively reliable method for identification of S. intermedius group staphylococci, with some improvement being warranted for S. pseudintermedius and S. delphini. In this case, MALDI-TOF MS may be utilized as a confirmatory mechanism to differentiate different coagulase-positive staphylococci when S. aureus is not suspected.
Testing of clinical samples for mixed staphylococcal species.
A number of studies have examined the use of MALDI-TOF MS for the identification of multiple species of staphylococci simultaneously in clinical samples. Multiple clinical laboratory investigations comparing MALDI-TOF MS identifications to various molecular and phenotypic methods for the identification of both coagulase-positive staphylococci and CoNS were reported. By identifying unique peaks in the spectra of reference strains belonging to the family Micrococcaceae, a database was constructed and queried to identify a set of 196 staphylococcal clinical isolates that had previously been identified to the species level by both phenotypic (coagulase and agglutination, etc.) and molecular (sodA sequencing) testing. In all cases, the generated MALDI-TOF MS spectra best matched the spectra of the reference organism of the same species (55). In another study of 450 staphylococcal isolates from blood cultures, representing 18 species, MALDI-TOF MS and the BioTyper 2.0 database were compared to rpoB sequencing for the identification of staphylococci to the species level. MALDI-TOF MS identified 99.3% (447/450) of isolates correctly to the species level and correctly identified all subspecies included in the study. Also, using the BioTyper 2.0 database, a third study of 152 staphylococcal isolates consisting of 22 species reported 99.3% agreement (151/152) with the StaphArray microarray-based staphylococcal identification system and identified clonal lineages among environmental and clinical isolates (90).
Conversely, using an early version of the SAMARIS database, a collection of 186 strains consisting of 35 species and subspecies of staphylococci of human and animal origins was compared to sequencing of the tuf and gap genes for identification. In this study, 81.5% of identifications made by the MALDI-TOF MS were correct, compared to 98.9% of isolates identified correctly by tuf sequencing (91). Importantly, a database of 47 type strains was used to analyze sequence data, exceeding the number of entries present in the SAMARIS database at that time. Those authors rightfully noted that of the 45 known staphylococcal species and 21 subspecies, only half were cultured from human specimens. Those authors further elaborate, citing that although the SAMARIS database contains 38 species and subspecies, only 15 have SuperSpectra, which appears to be a requirement for reliable identification of staphylococci. This example demonstrates that completeness of the database is paramount to the ability of MALDI-TOF MS to correctly identify culture isolates; therefore, expansion of current commercially available databases is warranted to provide broader coverage of the Staphylococcus genus, including opportunistic veterinary pathogens (91).
Micrococci
Similar to CoNS, the identification of micrococci also presents a situation where an important determination must be made to categorize the isolate as either clinically relevant or a contaminant from the skin microflora. In a majority of cases, micrococci are considered to be culture contaminants that are not clinically significant; however, reports of micrococci causing serious, life-threatening infections, particularly among patients with intravascular devices (92, 93), are present in the literature. Despite the high frequency at which these organisms are encountered in the microbiology laboratory, identification to the genus level is generally sufficient for micrococci, and species-level identification is rarely performed. Micrococci are often reliably differentiated from staphylococci by virtue of the performance of a modified oxidase (microdase) test. With respect to MALDI-TOF MS, Micrococcus spp. are able to be reliably identified by using the BioTyper software. In a study of environmental micrococci, isolates were identified correctly to the genus level, but no species-level identification could be reached due to limited entries in the database (94). It is likely that with the appropriate database, species-level discriminations could be made when necessary.
Staphylococci and MALDI-TOF MS: Future Uses and Implications
The implementation of MALDI-TOF MS systems in microbiology laboratories will change the way in which staphylococci are identified when isolated in routine culture and will provide physicians and researchers with more information about CoNS and their roles in human infections. One ongoing problem encountered by clinical laboratories is the identification of CoNS to the species level. As reported in the above-mentioned studies (75, 79), MALDI-TOF MS provides an excellent tool for the accurate and rapid identification of CoNS species and can potentially supplant labor-intensive and high-complexity molecular testing, thus increasing in-house capabilities and allowing information to be provided to physicians in a more timely manner. As more laboratories opt for identification of CoNS to the species level, clinical researchers can better define the impact of these important opportunistic pathogens. While other mass spectrometry-based systems can provide epidemiological data to group MRSA isolates into clonal complexes (88, 95), further investigations are warranted to determine if MALDI-TOF can be used to provide simultaneous identification and epidemiological data.
Although a powerful tool for bacterial analysis, the use of MALDI-TOF MS generates some controversy regarding the characterization of S. aureus susceptibility to antibiotics. There are conflicting reports describing the ability of MALDI-TOF MS to discriminate MRSA from MSSA (44, 84, 86), making strain characterization an important area of interest for clinical microbiologists, physicians, and pharmacists. By providing a mechanism to determine methicillin resistance in tandem with bacterial identification, patient isolation and infection control measures could be implemented faster, thus reducing time to diagnosis and allowing the timely administration of more appropriate antibiotic therapy. An additional debate surrounds the capacity of MALDI-TOF MS to identify Panton-Valentine leukocidin (PVL) from staphylococci. Conflicting publications argue both for (96) and against (97) the use of MALDI-TOF MS to identify PVL based on the presence of unique peaks in the mass spectra.
Streptococci
The streptococci are some of the most dynamic and medically relevant human bacterial pathogens identified in the clinical laboratory. These facultative Gram-positive bacteria are capable of causing a variety of diseases ranging from uncomplicated pharyngitis to multiple manifestations of invasive disease, including meningitis, sepsis, and necrotizing fasciitis. The genomes of streptococcal species encode a number of virulence determinants that mediate adhesion to and invasion of both localized and systemic tissues as well as a diverse repertoire of exotoxins and superantigens, depending upon the species in question. The genus Streptococcus is separated into many species but has undergone considerable taxonomic changes in the past decades with the availability of more sensitive testing methodologies. Phenotypically, the streptococci are often classified into larger groups based on their hemolytic activity on blood-containing media and antigenic differences in their associated surface components. This practice has resulted in streptococci often being referred to as either alpha-hemolytic (viridans group streptococci/S. pneumoniae), betahemolytic, or nonhemolytic when describing their appearance and being able to be grouped into different serogroups via typing of Lancefield antigens. Species identification of cultured streptococci is traditionally performed either by biochemical methods, including bile solubility and antibiotic resistance patterns (optochin/bacitracin sensitivity), or by agglutination testing (98). While streptococcal taxonomy has been in a state of flux over the previous decade, the accurate identification of these organisms in infectious processes is essential in most cases. Here we review the ability of MALDI-TOF MS to discriminate between different streptococcal species and the potential pitfalls or deficiencies that should be addressed in the near future. MALDI-TOF MS identification of Abiotrophia, a member of the so-called “nutritionally variant streptococci,” will also be briefly considered.
Beta-hemolytic streptococci.
Beta-hemolytic streptococci (BHS) are a group of species comprised of different serogroups. These organisms are important human pathogens, and their timely and accurate identification is paramount in the context of infection and is crucial to clinical diagnosis. As such, a number of phenotypic and molecular methods were developed for their identification (98). BHS include Lancefield antigenic group A (S. pyogenes), antigenic group B (S. agalactiae), antigenic group C (S. dysgalactiae and others), and group G (S. canis and others). The timely identification of these organisms, particularly those of groups A and B, is highly important to patient care. Left untreated for prolonged periods of time, group A streptococcal infections can manifest as serious autoimmune sequelae, including glomerulonephritis or rheumatic fever.
The rapid identification of group B streptococci is an important component of prenatal care in antepartum units due to the ability of S. agalactiae to cause serious infections in newborn infants. Therefore, the ability to quickly identify these organisms when encountered in clinical specimens directly influences patient care.
The ability of MALDI-TOF MS to discriminate between bacterial species that appear phenotypically identical, such as some Streptococcus spp., demonstrates the power of this tool. Studies of beta-hemolytic streptococci depict recent improvements to MALDI-TOF MS identification. A preliminary investigation from 2004 utilized MALDI-TOF MS to characterize the spectra generated from ionization of intact cells from three of the four major groups of beta-hemolytic streptococci isolated from hospital patients and community members. Although the study was performed before automated databases of bacterial spectra were widely available, the investigators were able to identify differences in the spectra of group A, C, and D streptococci with good reproducibility as well as roughly begin to examine spectral differences between strains of the same group (45).
More recently, Cherkaoui et al. described clinical microbiology on the cusp of monumental change in the way in which clinical laboratories analyze and routinely identify bacteria. Using the Bruker BioTyper 2.0 database (library v. 3.1.1.0), results from a broad sample set were compared to traditional phenotypic analysis by the Vitek-2 system coupled with a latex agglutination test (Bio-Rad) for the identification of 386 beta-hemolytic streptococcal isolates from clinical specimens (99). The MALDI-TOF MS system proved to be superior to traditional phenotypic methods for identifying beta-hemolytic streptococcal isolates. Fifty-two isolates of group A streptococci, 306 isolates of S. agalactiae, 10 isolates of S. dysgalactiae (Lancefield group C), and 18 isolates of S. dysgalactiae (Lancefield group G) that originated from a variety of anatomical sites were analyzed in the clinical laboratory. MALDI-TOF MS identification gave high-confidence identification to the species level for all organisms tested, whereas the phenotypic methodology used (Vitek-2) identified only 85% of the isolates with high confidence. Discrepant analysis was performed by using 16S rRNA gene sequencing; all discrepant cases resulted in confirmation of the identification determined by MALDI-TOF MS and the Bruker BioTyper database (99). This study demonstrated not only the superior discriminatory power of MALDI-TOF MS for beta-hemolytic streptococcal species compared to traditional phenotypic identification methods but also the adaptability of the instrumentation to the workflow of the clinical microbiology laboratory.
(i) Group A streptococci.
The group A streptococcus (GAS) S. pyogenes is capable of causing a variety of pathologies; the two most severe include streptococcal toxic shock syndrome and necrotizing fasciitis (100). GAS is identified by the intense zone of beta-hemolysis surrounding catalase-negative colonies grown on media containing blood and by agglutination with antisera against the Lancefield A antigen. Although rapid antigen detection tests and molecular methods are available for throat swabs to diagnose streptococcal pharyngitis, many of these direct and rapid assays are not designed or approved for use with other clinical specimens such as body fluids, tissue samples, blood, and wound specimens, and therefore, bacterial culture is the current method of streptococcal identification from these sources.
In a small preliminary study, Moura et al. successfully demonstrated the potential discriminatory power of MALDI-TOF MS for identifying and characterizing GAS isolates (101). Eight clinical isolates and one GAS strain were analyzed. Strains included in the study were phenotypically identified as GAS and typed by both PCR and sequencing of the emm genetic locus. Whole bacterial cells were utilized for MALDI-TOF MS, and the spectra were compared. Those authors demonstrated that gamma irradiation of the cells prior to MS analysis did not interfere with the generated spectra and could be potentially introduced into the MS sample preparation methodology as a safety control step for virulent isolates. Using a combination of MS and statistical analysis, GAS generated unique spectra that enabled them to be unequivocally identified compared to the spectra generated by the panel of other control organisms in the study. Additionally, GAS isolates from invasive necrotizing fasciitis cases were grouped together in a unique clade away from other clinical isolates based on their spectral profiles and irrespective of their emm type (101).
(ii) Group B streptococci.
Group B streptococci (GBS), comprised of S. agalactiae, are a leading cause of infectious morbidity and mortality among neonates in the United States. Maternal colonization with GBS is the primary risk factor for transmission to newborns, either in utero or during delivery; therefore, screening mechanisms exist to identify at-risk patients and provide them with prophylactic options. Rapid identification mechanisms for GBS include agglutination assays, chromogenic media, and molecular testing including PCR with direct specimens. Nevertheless, culture and subsequent identification from enrichment broth remain the reference standard for the recovery and identification of GBS for prenatal patients (102). In the clinical laboratory, GBS is identified by the CAMP test (so named for scientists Christie, Atkins, and Munch-Petersen), hippurate hydrolysis assays, PYR, or molecular methods.
Two studies from Lartigue et al. focused exclusively on GBS identification by MALDI-TOF MS analysis (103, 104). The first study investigated the ability of MALDI-TOF MS to identify 110 isolates of molecularly and serologically characterized GBS using cellular extracts. This study also included an important phylogenetic component, determining that variations among proteins expressed between different serogroups of GBS influenced spectra generated by MALDI-TOF MS but not significantly enough to alter organism identification. All 110 GBS isolates were identified to the species level with good confidence by using the Bruker BioTyper database v.1.1, irrespective of their associated serogroup (103). The second study investigated the ability of MALDI-TOF MS to identify GBS and differentiate “highly virulent” lineages of GBS. An expanded sample set of 197 GBS isolates from pregnant females, infections of nonpregnant adults, and neonatal meningitis patients was selected for analysis. Cell extracts were obtained and analyzed by using the Bruker BioTyper v2.0 database. Consistent with previous results, MALDI-TOF MS was able to identify each of the isolates with high confidence. Moreover, the technique was also able to discriminate highly virulent isolates of the ST-17 and ST-1 serotypes from other isolates with strong predictive values, indicating the potential of this method for future use with respect to strain identification and infection management in clinical practice (104).
Streptococcus pneumoniae.
S. pneumoniae is an alpha-hemolytic streptococcus that is distinct from the viridans group streptococci despite a close genetic relationship. In contrast to GAS and GBS, there are fewer investigations to identify and characterize isolates of S. pneumoniae. One study examined the ability of the technique to differentiate conjunctivitis outbreak isolates of S. pneumoniae based on proteomic analysis (105). The publication demonstrates the ability of MALDI-TOF MS to identify S. pneumoniae isolates as well as biomarkers useful in strain typing that could potentially offer valuable information for epidemiological studies. In contrast, other studies have reported difficulties in identification of S. pneumoniae, sometimes misidentified as S. mitis by MALDI-TOF MS (23). This misidentification is a serious limitation given the importance of rapid and accurate identification of S. pneumoniae. Werno et al. reported improvements to the database that would more reliably distinguish between these two species (106), and Neville et al. showed similar corrections (107).
Viridans group streptococci.
Accurate identification of viridans group streptococci (VGS) to the species level poses a considerable challenge for many clinical laboratories. This heterogeneous group of commensal organisms colonizes the gastrointestinal, respiratory, and genitourinary tracts as well as the oral mucosa and can cause localized infections. Conversely, these organisms are known to cause systemic infections, leading to sepsis, meningitis, and endocarditis, among others, at a variety of anatomical sites. These bacteria were quite accurately referred to as a “grab bag” of leftover organisms once the beta-hemolytic streptococci, pneumococci, and enterococci were excluded (108). VGS can be separated into five major classifications, the mutans, salivarius, anginosus, mitis, and bovis groups (109). A heavy reliance on a combination of molecular methods and phenotypic methods is often required for complete identification to the species level, as many of these organisms are genetically similar (110). Further confounding the situation is a confusing nomenclature and continually changing taxonomy within the VGS. While a thorough analysis of the VGS is outside the scope of this review, the reader is referred to two excellent reviews on VGS characterization, taxonomy, and identification (108, 111). Although confounded by taxonomical and methodological challenges, the need for appropriate identification of VGS to the species level remains, particularly to help identify strains from blood culture bottles, where species identification may help sort out differences between skin contaminants and pathogens (110).
MALDI-TOF MS was utilized for the accurate identification of VGS to the species level (43, 112). In a large study of mutans group streptococci, MALDI-TOF MS was demonstrated to be capable of species-level differentiation. The discriminatory power of MALDI-TOF MS was further demonstrated by the ability of the technique to correctly identify reference isolates that had been misidentified previously (43). Similar results were also found in another study based on VGS, highlighting the application of MALDI-TOF MS analysis as a necessary quality control measure for existing culture collections (113).
Friedrichs et al. used protein extracts from reference strains and clinical isolates to assemble a database that was used to compare the spectra generated from protein extracts from 99 consecutive VGS clinical isolates isolated from a variety of anatomical sites. Importantly, it was demonstrated that members of the VGS generated unique spectra when analyzed by MALDI-TOF MS and that spectra from members of the same group of VGS were more similar than those from other groups. All strains used in the study were identified in parallel by the phenotypic RapID 32 Strep system (bioMérieux) and by molecular methods such as 16S rRNA analysis or by species-specific PCR. Twenty-three strains, identified as either S. oralis or S. mitis by molecular methods, were analyzed by MALDI-TOF MS, and all but two were unequivocally identified to the species level. The remaining strains were analyzed with a different set of statistical parameters and were subsequently identified by MALDI-TOF MS. The authors reported that MALDI-TOF MS demonstrated 100% consistency with the results obtained by phenotypic and molecular testing and concluded that MALDI-TOF MS was a rapid and accurate strategy for VGS identification (112).
Nonenterococcal group D streptococci are important members of the viridans group streptococci capable of causing a variety of infections in humans, resulting in systemic bacteremia and endocarditis. One member of the group D streptococci, S. bovis, has been associated with colorectal cancer and reviewed by others (114). S. bovis is included in a large complex of group D streptococci (named the S. bovis-S. equinus complex), which has undergone recent taxonomic revision (115). Identification of members of the S. bovis-S. equinis complex is possible through the use of traditional biochemical analysis, but resolution to the appropriate species and subspecies is not always possible. Molecular testing is also challenging due to the high level of 16S rRNA sequence identity among members of this group (113). Currently, the most reliable method for species identification of the S. bovis-S. equinus complex is sequence analysis of the sodA gene (113, 116). Using both sodA sequencing and MALDI-TOF MS analysis of whole bacterial cells, Hinse and colleagues described a high level of concordance between the two methods and reliable identification to the species level of S. bovis-S. equinus complex organisms by MALDI-TOF MS. Importantly, the investigators utilized the commercially available SARAMIS database software for spectral analysis in this study (113). Other groups have reported difficulty discriminating between members of this complex of organisms using the BioTyper version 3.0 software, with identifications being further hampered by recent taxonomic revisions that have occurred within the complex (117).
Nutritionally variant streptococci and related genera.
Nutritionally variant streptococci (NVS) represent a challenging group of organisms to identify due to their fastidious nature and divergent biochemical profiles. Once classified as members of the genus Streptococcus, these organisms were reclassified into different genera based on their genetic divergence from the genus Streptococcus. Abiotrophia represents one genus within this classification that was identified as the causative infectious agent in a number of pathological processes, including infective endocarditis, sepsis, and implant-related infections. Recently, MALDI-TOF MS was utilized in parallel with traditional biochemical methods for the rapid identification of Abiotrophia defectiva from a patient with infective endocarditis. MALDI-TOF MS correctly identified the isolated bacteria as A. defectiva, with the result confirmed by 16S rRNA gene PCR (118).
In an analysis of catalase-negative Gram-positive cocci not belonging to either Streptococcus or Enterococcus spp., Christensen et al. reported the application of MALDI-TOF MS-based identification to a collection of 51 isolates representing 16 genera, all of unique species, by using the Bruker BioTyper 2.0 software. Samples were processed by using 70% formic acid for protein extraction. Although low scores were obtained initially due to the lack of the presence of these specific genera in the database, the creation of a dedicated spectral database allowed for robust results, with no misidentifications occurring at the genus level (70).
Enterococcus spp.
The enterococci are important human pathogens capable of causing serious infections and are often resistant to numerous antibiotics. There is evidence to suggest that enterococci possess specific traits which allow them to persist in the nosocomial environment, colonize patients, and contribute to serious infections, including bacteremia, peritonitis, endocarditis, wound infections, and infections of the urinary tract and medical devices (119). Additionally, the ability of enterococci to obtain mobile genetic elements encoding antibiotic resistance genes has contributed to their emergence as important pathogens in the hospital environment (120). The identification of Enterococcus spp. in the clinical laboratory relies heavily on phenotypic analysis; however, rapid identifications based on molecular techniques have also been developed (121).
MALDI-TOF MS has been used for the characterization of enterococcal isolates from both human and environmental sources (122, 123). Böhme et al. documented the accuracy of the Bruker BioTyper system for identification of 30 Enterococcus spp. isolated from blood cultures (124). In a study of E. faecalis dental isolates from European countries, MALDI-TOF MS was able to be used to perform cluster analysis by using MicrobeLynx software (Waters). Based on the spectral signatures of their surface components, a collection of 58 isolates was grouped by geographical location. Evidence of quinupristin-dalfopristin (Synercid) susceptibility was also provided (122). A second study utilized MALDI-TOF MS as a bacterial source-tracking tool to characterize environmental isolates of Enterococcus spp. Animal and human enterococcal isolates were collected and analyzed to determine if MS spectra could be used to group isolates according to their respective sources (123). MALDI-TOF MS showed promise as a source-tracking tool for environmental studies, but the technique required further refinement, particularly in the sample-processing phase of experimentation.
Fang et al. recently completed a large comparative analysis of molecular, phenotypic, and MS methods for the species-level identification of 132 clinical Enterococcus isolates. Importantly, two MS platforms (Bruker BioTyper version 3.0 and Vitek-MS) were compared. Both MS systems provided identical results for all isolates tested, and both performed equally as well as multiplex PCR for enterococcal identification (125). In summary, these studies demonstrate the ability of MALDI-TOF MS to generally characterize enterococci from clinical and nonclinical sources and provide information regarding clonal relationships and potential distribution mechanisms.
Lactococcus spp.
Lactococci are important organisms for the maintenance of the gastrointestinal tracts of humans and ruminant animals and for the industrial production of fermented dairy products but are rarely identified in clinical infections. Although perhaps less clinically significant than other Gram-positive species reviewed in this section, the lactococci are capable of causing human infections (126, 127). In a study examining the identification and subspecies determination of 30 strains of lactococci, Tanigawa et al. demonstrated that analysis by MALDI-TOF MS was suitable for the identification of members of the genus Lactococcus to the species level and was the most effective method for the discrimination of subspecies compared to molecular methods such as housekeeping gene sequencing by amplified fragment length polymorphism (AFLP) analysis (128).
Bacillus spp.
Members of the genus Bacillus are Gram-positive, spore-forming, rod-shaped bacteria that are normal inhabitants of soil and other environmental niches. The two major pathogenic species are B. cereus and B. anthracis, both of which are genetically related to each other. B. cereus is responsible for a self-limiting food-borne illness that is toxin mediated and may cause line-related bacteremia in neutropenic patients. In contrast, B. anthracis is a category A bioterrorism agent that requires rapid identification and reporting to federal authorities upon isolation. For identification processes, strict adherence to the Laboratory Response Network (LRN) classification of one's laboratory is required, following rule-out and referral steps designated in the particular laboratory's classification for B. anthracis and other select agents. B. anthracis and B. cereus are highly related to each other in terms of genetic content, sharing approximately 90% of their core genomes. In the laboratory, the two species are differentiated by the production of capsule by B. anthracis on blood-containing media as well as variant biochemical profiles. As the identification of B. anthracis in patient samples has serious implications for both the patient and the general public, rapid and accurate identification of this organism is essential.
Early mass spectrometry-based studies of Bacillus spp. were centered on the identification of organisms by proteomic characterization of bacterial spores. Studies examining the ability of MALDI-TOF MS to differentiate vegetative cells of B. anthracis from B. cereus and other members of the genus have recently been undertaken. Early investigations analyzed both vegetative cells and spores by MS to identify unique biomarkers that could potentially be used to detect their presence. As spores are the metabolically dormant form of the organism, the detection of this form of the Bacillus species life cycle would have important health implications, as the spores are also infectious. With respect to spore proteins, a number of unique biomarkers were discovered in spore samples, which were demonstrated to aid in identification of different species of Bacillus.
In contrast, the clinical microbiology laboratory more commonly deals with the presence of vegetative cells in patient specimens and traditionally uses these cells as the basis for bacterial identification. A 1996 study examining extracted proteins from four different species of Bacillus identified genus-, species-, and strain-specific biomarkers, highlighting the potential of MALDI-TOF MS for use in the identification of Bacillus spp. (129). In a study examining 374 strains of Bacillus spp., a combination of MALDI-TOF MS and artificial neural networks for spectral analysis was used to identify unique biomarkers for both species, and sensitivity and specificity of 100% were achieved for differentiating B. anthracis and B. cereus. Similar to analyses with other species, the authors noted the importance of a standardization of culture conditions to generate reproducible spectra for analysis (130). Recently, an expanded MALDI-TOF MS-based analysis of the genus Bacillus was undertaken by using a database constructed based on ribosomal protein markers (131). B. cereus group organisms were differentiated easily with this database, whereas 16S rRNA gene sequence analysis proved to be more difficult. In all, MALDI-TOF MS appears to provide a rapid and reliable method for the identification of B. anthracis, B. cereus, and other bacilli to the species level.
Listeria spp.
Listeriae are a group Gram-positive, rod-shaped bacteria with low G+C content that are divided into six species, only two of which are pathogenic (132). Readily found in the environment, the predominant pathogenic species of the genus, L. monocytogenes, is responsible for contamination and food-borne illness that can manifest as serious, life-threatening infections, including meningitis, sepsis, and encephalitis. Immunocompromised patients or patients of advanced age have increased susceptibility to infection (74). L. monocytogenes poses a risk to pregnant patients, as the bacteria have the ability to cross the placental barrier and infect the fetus in utero, resulting in serious complications including neonatal meningitis, premature labor, and stillbirth. Thus, rapid identification of Listeria spp. is necessary for successful patient outcomes.
Listeria spp. are readily identified by a number of biochemical methods. Identification of these organisms to the species level relies on detection of growth at reduced temperatures, serodiagnosis, hemolytic activity, and carbohydrate utilization patterns. Testing procedures often require additional incubations and additional subculture, prolonging the time to definitive diagnosis. Additionally, subtyping of different Listeria species isolates requires specialized testing, including serological, phage-dependent, or molecular analysis. Accurate identification to the species level and subtyping of the organism are important, as the identification of L. monocytogenes is a reportable agent to public health authorities for food-borne outbreaks.
MALDI-TOF MS has been demonstrated to be a rapid alternative to current phenotypic and molecular methods for species identification and subtyping of Listeria spp. In a sentinel study by Barbuddhe and colleagues, 146 strains representing each of the six species of Listeria were analyzed by MALDI-TOF MS and included reference isolates in addition to isolates from outbreaks and clinical isolates (57). Species-level analysis was performed, and a Listeria-specific reference library of MS spectra was constructed by using the Bruker BioTyper software (v.1.1). In all but 10 discrepant cases, the species-level identification generated by MALDI-TOF MS matched the previously determined identification. Upon analysis of discrepant identifications by 16S rRNA gene sequencing, it was determined that the identification provided by the MALDI-TOF MS system was correct and that the original species designation of the isolate was in error. Importantly, MALDI-TOF MS was also able to resolve clonal lineages, all of which were in agreement with the reference method, PFGE, demonstrating the ability of MALDI-TOF MS to be able to simultaneously provide rapid and accurate species-level identification and genetic subtyping of Listeria species isolates (57).
Corynebacterium spp.
Corynebacteria represent a diverse group of pleomorphic bacteria that in some cases are normal inhabitants of the skin microflora and in other cases have the potential to cause either opportunistic or severe disease (74). These organisms are Gram-positive, non-spore-forming rods that are often eliminated as causative agents of infection upon routine microbiological examination due to their innocuous presence on the skin. In some cases, they are demonstrated to be clinically relevant. Corynebacterium diphtheriae is perhaps the most medically significant member of the genus and the causative agent of diphtheria. Other toxigenic corynebacterial species, including C. ulcerans and C. pseudotuberculosis, are also capable of causing human infections. Although the prevalence of infection with C. diphtheriae is low due to current vaccination practices, rapid identification and reporting of the organism are important due to public health concerns. Currently, a number of identifications systems are available for the identification of corynebacteria, including that API RapID Coryne system (bioMérieux) and the BBL Crystal system (Becton Dickinson), among others. These systems require additional incubation time in order to develop the biochemical reactions needed for proper species-level identification of corynebacteria. Additional testing of Corynebacterium species isolates is sometimes performed to determine toxin production, including Elek immunodiffusion testing and PCR-based methods.
An analysis of 116 isolates of Corynebacterium spp. of clinical and veterinary origins by MALDI-TOF MS was undertaken to investigate the discriminatory power of the technology with respect to this genus (133). Isolates were collected over a period of 13 years. The reference database utilized for the study contained 138 reference spectra from 71 different Corynebacterium isolates in addition to reference spectra for the toxigenic species derived from type strains. Compared to rpoB sequencing, 115/116 (99.1%) isolates were correctly identified to the species level by MALDI-TOF MS, with only 1 isolate restricted to genus-level identification. The authors included a key point in their discussion, describing the current inability of MALDI-TOF MS to differentiate between different biovars of C. diphtheriae, but suggested that this type of testing may be possible upon the generation of an appropriate database (133).
Moving theory into practice, a second study utilized MALDI-TOF MS to analyze outbreak strains of C. pseudodiphtheriticum in France among cystic fibrosis (CF) patients (134). Eighteen isolates from pediatric patients were collected over a period of almost 3 years. MALDI-TOF MS identifications of these isolates by using the BioTyper database (Bruker) matched identifications made by rpoB sequencing with 100% concordance and generated similar phylogenetic relationships among corynebacterial isolates (134).
MALDI-TOF MS was also utilized as the primary mechanism for definitive species-level identification of uncommonly encountered corynebacterial isolates (135). As databases become better populated and more refined, studies such as these will play an important role in both pathogen detection and the identification of novel emerging pathogens, particularly for groups of often overlooked organisms, such as the corynebacteria.
Arcanobacterium and Trueperella spp.
The genera Arcanobacterium and Trueperella contain Gram-positive, non-spore-forming, facultative anaerobic rods. Trueperella was, until 2011, classified within the genus Arcanobacterium but has since been reclassified into its own genus. These two genera contain clinically important human and veterinary pathogens alike. A. haemolyticum is a significant cause of pharyngitis and is often overlooked within the clinical laboratory due to its slow-growing and fastidious nature as well as its delayed hemolytic phenotype. Trueperella (Arcanobacterium) pyogenes is an important veterinary pathogen and is a rare cause of infection in humans.
Using the BioTyper software, Hijazin et al. analyzed the ability of MALDI-TOF MS to identify members of both Arcanobacterium and Trueperella. All 98 isolates that were analyzed were correctly identified to the species level with scores of >2.0, indicating strong matches to the database (136). The method was also utilized to identify a novel member of the genus Trueperella, T. abortisuis, in a publication by the same research group (137).
Nocardia and Mycobacteria
Mycobacteria, Nocardia, and aerobic actinomycetes can be significant diagnostic challenges to the clinical laboratory. Moreover, due to the complex nature of their representative cell walls, bacteria from this group of organisms may require specialized processing procedures prior to their analysis by MALDI-TOF MS in order to obtain the most accurate results.
Sample preparation methods for MALDI-TOF MS identification of mycobacteria and Nocardia spp.
Verroken et al. described a modified extraction method for attaining material for robust MALDI-TOF MS material from Nocardia isolates (48). Briefly, 10 colonies of biomass were resuspended in water and boiled to promote cellular lysis. This lysate was then centrifuged to remove cellular debris, followed by the addition of ethanol to precipitate proteins contained within the supernatant. Precipitated proteins were then centrifuged and dried, resuspended in 70% FA and acetonitrile, centrifuged a final time, and then analyzed by MALDI-TOF MS. This method provided material that could be accurately used to analyze Nocardia isolates. Authors not utilizing this method have reported problems analyzing actinomycetes such as Streptomyces spp. (138).
As of now, no consensus mechanism exists for the processing of mycobacteria for MALDI-TOF MS. Previously, direct whole cells and FA-treated cells were utilized with success by a number of groups, with safety being a substantial concern during routine analysis. El Khéchine et al. rightfully described an investigation of different methods for the inactivation and processing of mycobacterial samples for MALDI-TOF MS. Solubilization of bacterial aggregates, which can impair MALDI-TOF MS analysis, was critical in their investigation, as was the avoidance excessive centrifugation to minimize the potential of aerosolized exposure to laboratory personnel. Through rigorous testing, the authors arrived at a final procedure representing a synthesis of inactivation and processing methods. Colonies were collected in screw-cap tubes containing water and 0.5% Tween 20 and inactivated by heating at 95°C for 1 h. Inactivated samples were centrifuged, washed twice with water, and then vortexed with glass beads to facilitate complete cellular disruption. Following centrifugation, the pellet was resuspended in FA-acetonitrile and centrifuged again. Finally, the supernatant was deposited onto the MALDI test plate and overlaid with matrix (50).
Nocardia spp.
Nocardia spp. are ubiquitous bacteria from numerous environmental sources and are not considered normal flora when isolated from patient samples. Some species are capable of infecting humans, with a majority of infections reported in immunocompromised patients including those with HIV/AIDS and transplant patients (74). Diagnosis of nocardiosis with patient specimens often requires isolation of the organism from patient specimens, which can require extended incubation times, up to 2 weeks in some cases, for cultivation alone. Differentiation from other filamentous bacteria is often achieved by differential staining including a modified acid-fast stain. Identification of Nocardia organisms to the species level relies on a combination of biochemical evaluation and molecular testing, with 16S rRNA sequencing typically used as a determinative method for final identification to the species level (139, 140). Agreement between molecular methods and biochemical analysis is between 70 and 90%, indicating the need for more accurate and reliable testing methodologies for the identification of Nocardia spp. (48, 140).
A single study evaluated the use of MALDI-TOF MS for the species-level identification of Nocardia spp. (48). A panel of 153 clinical isolates was analyzed by MALDI-TOF MS and the BioTyper database (v.2.0), utilizing an expanded database generated by the investigators. This panel consisted of both clinical isolates and reference strains of the most frequently isolated species. One hundred ten isolates were used to generate a Nocardia-specific database, while the remaining 43 isolates were blinded and used as a challenge set. During database construction, distinct divisions between different species of Nocardia could be visualized with the MALDI-TOF MS software. The unmodified BioTyper database was able to correctly identify 19 isolates (44%) to the genus level, of which 10 (23%) were correctly identified to the species level. The addition of the Nocardia-specific database significantly improved identification scores such that 38/43 (88%) isolates were identified to the genus level and 34/43 (79%) were identified correctly to the species level (48). This study provided additional evidence that through the use of expanded databases populated with entries of various clinical significances, the sensitivity of MALDI-TOF MS can be dramatically improved. It may be important in the future to include both organisms of environmental origin as well as those of clinical origin to be able to accurately identify organisms responsible for opportunistic infections and other emerging pathogens.
Mycobacteria.
Identification of Mycobacterium tuberculosis and other mycobacteria from clinical specimens often requires a number of diverse techniques for observation, recovery, growth, and characterization and space specifically engineered for biosafety. As such, not all clinical laboratories are equipped for the culturing of mycobacterial samples; therefore, the referral of specimens to other laboratories for testing is a common practice that can result in delays in testing, reporting, and treatment initiation among patients (141). The average turnaround time for specimens submitted for mycobacterial testing is often reported to be days to weeks (142–144).
Historically, bacterial culture remains the most sensitive method for the detection of M. tuberculosis in clinical specimens and serves as the starting point for downstream applications, including the identification of mycobacteria to the species level and the determination of antibiotic resistance (142). The identification of mycobacteria to the species level cannot be performed with traditional automated identification systems and instead requires more complex and labor-intensive molecular assays, including nucleic acid amplification strategies, molecular probes, high-performance liquid chromatography (HPLC) analysis of mycolic acids from the bacterial cell wall, or DNA sequencing methods (142, 145–148). Moreover, the general biology of the organism slows the time to diagnosis due to its requirements for specialized growth media, fastidious nature, and exceedingly slow generation time. Only recently have real-time PCR methods emerged as routine, practical methods for detection of M. tuberculosis directly from clinical specimens (149), but rapid detection of other species from colonies is common, though not all inclusive, via GenProbe methods (150, 151). The use of MALDI-TOF MS for the identification of mycobacteria thus offers an attractive option for laboratories both to expand their testing menus and to perform their own testing in-house in place of sending out specimens for mycobacterial identification.
Early investigations into the use of MALDI-TOF MS for the identification of mycobacteria proved to be successful before the generation of standardized spectral databases (46, 152, 153). A preliminary investigation utilizing MALDI-TOF MS for the identification of six species of Mycobacterium demonstrated that each species could be unequivocally identified based on their unique mass-to-charge (m/z) ratios (152). Additionally, biomarkers thought to be unique to the genus Mycobacterium could also be identified among tested species. This study also demonstrated that both whole cells and protein extracts could be used to generate comparable spectra, thus decreasing the biosafety risks of working with whole cells. A follow-up to this investigation successfully demonstrated the potential for the use of MALDI-TOF MS for the classification of mycobacteria (n = 16) at the strain level (46). A larger investigation followed, this time using intact cells for analysis, and demonstrated that species-specific spectra could be generated for all but 1 of the 37 isolates tested (153). In each of the above-mentioned studies, multiple replicates were utilized to populate a database for the identification of different species of Mycobacterium at the species and strain levels.
Database refinement is key for identification of Mycobacterium spp. In a recent study by Lotz et al., the investigators extended a database population technique to construct a database for analysis of mycobacteria from commercially available type strains (154). Importantly, the study also analyzed the ability of the database to identify mycobacterial strains cultured both on Löwenstein-Jensen (LJ) plates and in mycobacterium growth indicator tube (MGIT) medium. The database was then challenged with 311 isolates from 31 distinct species of Mycobacterium from LJ plates and demonstrated 97% concordance with genus and species identifications and no discordant identifications. Three percent of the tested strains from LJ plates could not be identified due to insufficient spectral information. Eighty-two strains were grown in MGIT broth and analyzed by MALDI-TOF MS. In contrast to LJ plates, only 77% of samples could be identified by MALDI-TOF MS using MGIT samples due to insufficient spectra, potentially due to additives in the MGIT medium. Despite fewer successful identifications, no discordant results were found by using MALDI-TOF MS and MGIT medium. Although only one type of liquid medium was tested, it is important to consider the reduced capabilities of MALDI-TOF MS using broth cultures, as liquid medium is preferable to solid medium when isolating mycobacteria from direct specimens (155). Lotz et al. reviewed the capabilities of MALDI-TOF MS compared to molecular methods of mycobacterial identification. The authors noted that MALDI-TOF MS can distinguish between members of various mycobacterial complexes and noted superiority of the mass spectrometry-based technique compared to nucleic acid probe and DNA strip methodologies (154).
As commercial databases become available for the identification of microorganisms by MALDI-TOF MS, it will become important to thoroughly evaluate their utility in the clinical setting. It will also be important to be able to modify or add entries to the database as required with regard to geographical variation among strains. A recent study by investigators at the National Institutes of Health examined the performance of MALDI-TOF MS for identification of Mycobacterium spp. from protein extracts using an expanded database of 42 type and reference strains representing 37 species. The Bruker database (v. 3.0.2.0) contained 50 strains representing only 18 species (156). The constructed database was then challenged with 104 clinical isolates representing 17 species. All M. tuberculosis complex (MTC) isolates obtained strong MS scores and were easily differentiated from nontuberculous mycobacteria, but the organisms M. tuberculosis and M. bovis, comprising the MTC, could not be identified to the species level. An earlier study reported that these organisms could be differentiated at the species level (152) but utilized a different database containing fewer strains and a different exaction methodology to make these species determinations. Other difficulties in species identification were reported for the identification of genetically similar mycobacteria, as these strains often require sequencing of single or multiple targets to be differentiated. These closely related strains aside, the rest of the isolates could be easily and accurately identified to the species level, demonstrating the utility of MALDI-TOF MS for the identification of mycobacteria in the routine clinical laboratory (156).
The implementation of MALDI-TOF MS in the routine clinical laboratory will provide a powerful and accurate tool to quickly identify mycobacteria from culture. This implementation will also change testing algorithms for mycobacteria and provide a mechanism for enhanced surveillance and epidemiological data worldwide. The implementation of MALDI-TOF MS technology will reduce testing costs for mycobacterial identification, with consumable costs per specimen estimated to be less than $1 per isolate (156). In sum, these studies demonstrate the feasibility and robust accuracy associated with MALDI-TOF MS for mycobacterial identification.
GRAM-NEGATIVE BACTERIA
Gram-negative bacteria are encountered in the clinical laboratory in all sample types analyzed and are ubiquitous members of the normal human flora. Saffert et al. analyzed the ability of the Bruker BioTyper version 2.0 MALDI-TOF MS software to identify Gram-negative bacilli compared to identifications obtained by using the BD Phoenix system, using their collection of 440 common and infrequently encountered bacterial species. There was no significant difference between the two systems for commonly encountered species of Gram-negative bacilli; however, the BioTyper system was better than the Phoenix system for the identification of infrequently isolated Gram-negative bacilli (157).
Sample Preparation for Gram-Negative Bacteria
With respect to sample processing for accurate identification and the analysis of variables potentially influencing MALDI-TOF MS identification, Ford and Burnham recently reviewed methods for optimizing routine Gram-negative identification using the BioTyper system. Using a collection of 208 enteric Gram-negative organisms and 252 nonfermenting Gram-negative organisms, the organisms were spotted onto a MALDI plate using either a light or heavy smear (correlating to the amount of inoculum) and either overlaid with 1 μl of 100% formic acid or not overlaid, as reported previously for the processing of Gram-positive organisms (69). Samples were finally mixed with matrix solution and analyzed by using the BioTyper 3.0 software (Bruker). These identifications were compared to those derived phenotypically and to those derived by utilizing a full ethanol-based protein extraction similar to those necessary for analysis of mycobacteria and Nocardia, again with 16S rRNA gene sequencing being used as a reference standard to resolve discrepant results. For enteric organisms, a heavy smear with an FA overlay provided preferential results compared to a light smear of FA or to specimens without FA treatment. In contrast, heavy smears provided better identifications, but the addition of FA did not significantly influence MALDI-TOF MS identification of nonfermenting organisms, allowing the authors to conclude that in the case of nonfermenting bacilli, FA treatment was not necessary for optimal identification by MALDI-TOF MS (158).
The Enterobacteriaceae
The Enterobacteriaceae represent a dynamic group of organisms encountered in the clinical laboratory that are responsible for a wide range of pathologies. This broad group of organisms is often characterized by the ability of the organisms to ferment lactose, as determined via biochemical testing. MALDI-TOF MS was utilized by investigators to identify members of this large group of bacteria and classify them as such while the technique was still in its infancy. Prior to the advent of comprehensive databases that can be easily queried with acquired spectra, early work using MALDI-TOF MS for the identification of the Enterobacteriaceae was focused on the identification of suitable biomarkers for group-level and genus-level identification. One early study from 1999 using intact cells lysed by one cycle of freeze-thaw from storage at −20°C examined the use of MALDI-TOF MS to distinguish between members of this group of organisms: E. coli O157:H7, Klebsiella pneumoniae, Salmonella enterica subsp. enterica serovar Typhimurium, Salmonella enterica subsp. enterica serovar Dublin, and Providencia rettgeri. For these organisms, the authors reported MS spectral peaks representing family-specific biomarkers for the Enterobacteriaceae, genus- and species-specific biomarkers for the two serovars of Salmonella analyzed, and strain-specific biomarkers for the two strains of E. coli (159).
In 2005, Pribil and Fenselau used cultures of E. coli, Enterobacter cloacae, Erwinia herbicola, and Salmonella Typhimurium from the ATCC and analyzed them by using trypsin-digested whole cells. Spectra were searched against the NCBInr database using a taxonomy restricted to eubacteria. The returned peptide hits consisted of a number of cell surface-associated outer membrane proteins (OMPs), leading the authors to suggest that a comparison of OMPs from the members of the Enterobacteriaceae might lead to a mechanism for species-specific identification of members of this group of bacteria (160). Although these first studies were limited to a small sample size and had restricted diversity with respect to the genera analyzed, these were important steps toward demonstrating the utility of MALDI-TOF MS for the identification of members of the Enterobacteriaceae.
In a study examining the ability of MALDI-TOF MS to differentiate Gram-negative species involved in seafood spoilage, Böhme et al. reported the creation of a library of mass spectra associated with the main pathogenic species of Gram-negative bacteria associated with food spoilage. In this study, a collection of 29 isolates spanning 15 genera, including members of the Enterobacteriaceae, were selected for analysis by MALDI-TOF MS. Bacterial colonies were harvested and mixed with trifluoroacetic acid (TFA) and acetonitrile and analyzed by MALDI-TOF MS. The resultant spectra were added to a library that contained peak data for peptides in the 2,000- to 10,000-Da range. The spectral peaks generated could be easily grouped, with members of the Enterobacteriaceae generating spectra that could easily be concluded to be similar, yet differences could be identified, allowing genus- and species-specific identifications. Of note, spectra derived from members of the genus Serratia were different from those of other members of the Enterobacteriaceae. The authors also noted important similarities in spectra between organisms that are genetically closely related as well as organisms found in similar niches (i.e., marine environments) (124).
Salmonella spp.
Salmonella spp. are important organisms often associated with food-borne disease and gastrointestinal pathology. Detection of Salmonella spp. in the clinical laboratory is a multistep process often involving multiple selective medium types, subculture, and serology before a final identification can be reached. The identification of Salmonella spp. from stool cultures can take upwards of 2 days when utilizing traditional biochemical methods. While molecular methods are available, serological protocols predominate as the method of choice for many laboratories for serovar determination, further adding to the time to definitive diagnosis. MALDI-TOF MS has since been utilized to aid in both the detection and species-level identification of Salmonella.
MALDI-TOF MS was identified early on as an attractive option for the species and subspecies typing of Salmonella. Before the widespread use of MALDI-TOF MS for the identification of clinical bacterial isolates or the establishment of comprehensive databases, Lynn et al. identified genus- and species-specific biomarkers dedicated specifically to the identification of Enterobacteriaceae for MALDI-TOF MS identification of Salmonella, but their study lacked an appropriate system for complex profile analysis (159). They defined consensus peaks specific to six Salmonella serovars but found discrepancies with other studies. Many of these consensus peaks identified by Lynn et al. were unable to be confirmed by a second group that performed a similar analysis using members of the genus Salmonella, but key differences between the studies (including differences in the type of mass spectrometer and laser source utilized) were partially attributed to the inability of the same consensus spectral peaks to be identified (161). Expanding on the need for a more defined set of biomarkers for the identification of the salmonellae, Dieckmann et al. examined variations in housekeeping gene levels among a large collection of Salmonella isolates in order to generate a more comprehensive phylogenetic classification mechanism using whole-cell MALDI-TOF MS (162). Although these preliminary studies utilized nonstandardized methods for the analysis and generation of their respective mass spectra, they represent some of the first important steps toward a uniform ability to differentiate members of the genus Salmonella on species- and serovar-specific levels.
One of the greatest challenges associated with Salmonella identification is perhaps not the identification of the organism as a member of the genus but the further identification to the subspecies level and taxonomy associated with definitive identification. In a second publication, Dieckmann and Malorny made significant contributions toward the identification of serovar-specific biomarkers by MALDI-TOF MS utilizing an exhaustive collection of 913 Salmonella enterica subsp. enterica strains comprising 89 unique serovars and the SAMARIS (release 3.4) database. While some serovar-specific spectral peaks could be identified, the authors concluded that MALDI-TOF MS would be suitable only for the rapid screening of isolates, with subsequent serovar identification being reliant on traditional serotyping methods for a majority of strains (163).
While Salmonella enterica serovar Typhi has been essentially eradicated in the United States, it remains a significant health concern in many African countries and the developing world. Returning to the issue of Salmonella serovar determination using MALDI-TOF MS, Kuhns et al. recently evaluated the ability of the technology to discriminate S. enterica serovar Typhi from other Salmonella serovars. By using clinical blood culture isolates collected from epidemiological studies and reference strains representing 160 S. enterica subsp. enterica isolates and 12 serovars, intact-cell MALDI-TOF MS using the Bruker BioTyper 3.0 database was evaluated and compared to conventional identification methods. In all cases, Salmonella spp. were readily distinguished from other members of the Enterobacteriaceae. The authors also reported their ability to identify biomarkers specific to S. enterica serovar Typhi compared to spectra generated for other Salmonella isolates included in their study and showed that the spectra generated by S. enterica serovar Typhi isolates in this particular collection were significantly different from those of the other serovars present. Furthermore, the S. enterica serovar Typhi isolates could be identified based on their respective mass spectral profiles (164). Well-planned, large-scale studies similar to those performed by Kuhns et al. and Dieckmann et al. will further enhance the ability of MALDI-TOF MS to accurately determine subspecies- and serovar-level identifications for members of the genus Salmonella.
Sparbier et al. recently examined the ability of MALDI-TOF MS to identify S. enterica serovar Typhimurium from spiked stool specimens obtained from healthy volunteers and from hospital patients. Samples were treated with formic acid-acetonitrile and analyzed by using the BioTyper 2.0 software database. Not surprisingly, MALDI-TOF MS was demonstrated to be able to definitively identify Salmonella spp. from spiked samples versus other bacteria from unspiked controls enriched in selenite enrichment broth. Importantly, the authors also performed a measurement of the sensitivity of MALDI-TOF MS for the identification of Salmonella by spiking serially diluted samples of S. enterica serovar Typhimurium into stool and enriching the samples for bacterial growth. Spiking of samples with 800 CFU or greater followed by enrichment led to a clear identification of Salmonella spp. by the software, whereas inoculation of the enrichment broth with >80 CFU led to ambiguous MALDI-TOF MS identifications, likely due to overgrowth by fecal flora. An examination of this enrichment method followed by MALDI-TOF MS on 4,847 routine clinical specimens found that of the 108 Salmonella-positive specimens identified by traditional biochemical analysis, MALDI-TOF MS identified 100 of them correctly 24 h earlier. The 8 specimens missed by MALDI-TOF MS could be identified as Salmonella positive only after plating of the enrichment medium, indicating that the bacterial concentration was likely too low to be detected by MALDI-TOF MS (165).
In sum, the current utility of MALDI-TOF MS for the clinical diagnosis of Salmonella infections appears to be best exemplified by the ability of the technology to rapidly identify members of the genus, in some cases up to 24 h sooner. MALDI-TOF MS fits easily into the routine workflow associated with Salmonella identification but as of now still requires enrichment and supplemental culture for the most accurate diagnostic results with respect to specimens containing a low bacterial inoculum. As a tool for subspecies and serovar typing, MALDI-TOF MS shows significant promise but will require additional studies and modifications to existing protocols before the method can be used as a stand-alone mechanism.
Escherichia coli and Shigella spp.
The identification of pathogenic Escherichia coli and Shigella species is critical and often challenging for the clinical laboratory because the organisms are closely related on the genetic level. Growth on sorbitol-containing MacConkey medium is sometimes used as a preliminary mechanism to differentiate pathogenic species of E. coli from nonpathogenic species, requiring the use of specialized media and additional culture time. This method is not definitive for all Shiga-toxin-producing strains, and therefore, additional reflex testing with serological and molecular methods is often required.
As early as 2001, the use of MALDI-TOF MS was evaluated for the identification of pathogenic E. coli and Shigella species. Conway et al. reported good identification of E. coli in a preliminary study in which spectra from 25 clinical isolates were compared to reference spectra from an in-house database developed for other members of the Enterobacteriaceae, including Salmonella and Shigella. The authors also undertook exhaustive studies examining the effects of culture conditions, medium selection, and biomarker identification in one of the earliest studies examining the feasibility of the technology for the identification of this group of organisms. Finally, the authors reported that cluster analysis of the spectra generated from these isolates by MALDI-TOF MS allowed for the construction of a phylogenetic dendrogram, which was comparable to that generated by molecular methods. At the time, the authors conceded that the technology was not suitable for strain-specific identification (166).
A more recent study from the realm of basic proteomic research sought to determine specific biomarkers for E. coli O157:H7 by using a combination of MALDI-TOF/TOF (tandem time of flight) and a top-down proteomics approach (167). By using this approach, six protein biomarkers were able to be identified, and the associated genes encoding these biomarkers in strains of different lipopolysaccharide (LPS) and flagellar antigenic types were sequenced. A majority of these biomarkers were determined to be proteins involved in stress responses, with four of the six proteins containing a signal sequence indicating their function at the membrane. While these biomarkers were unable to distinguish between an O157:H7 strain and an O55:H7 strain, they were suitable for discrimination between an O157:H7 strain and a nonpathogenic E. coli strain by virtue of a single-amino-acid change. Subsequently, a second biomarker was also identified as the HdeB stress chaperone-like protein, which was present in the mass spectra of the non-O157:H7 isolate but was absent in the spectra of O157:H7 strains identified (167).
Similarly, Karger et al. sought to use an intact-cell strategy for the discrimination of Shiga toxin-producing E. coli strains representing selected serotypes, with mixed results (168). These works demonstrate the significant sensitivity of mass spectrometry and its use as a research tool for biomarker discovery; however, in a situation similar to that for Salmonella isolates, additional research is warranted to adapt MALDI-TOF/TOF technology to be suitable for strain-specific identifications of E. coli isolates.
In addition to the current inability of MALDI-TOF MS to reliably distinguish pathogenic from nonpathogenic E. coli isolates, numerous reports describe the difficulty encountered when trying to discriminate E. coli from Shigella spp. Differentiation of pathogenic E. coli strains from Shigella spp. is challenging because of the close genetic relatedness of the organisms. In some instances, even molecular analysis, such as 16S rRNA gene sequencing, is unable to distinguish the organisms. In 2010, He et al. reported that the BioTyper 2.0 software misidentified 39 Shigella isolates and 3 enterohemorrhagic E. coli (EHEC) isolates as E. coli, with the authors rightfully noting the absence of Shigella sonnei from the database. Based upon the spectra generated, the authors concluded that redundant ribosomal proteins could not be used to accurately distinguish Shigella and EHEC from nonpathogenic E. coli (169). Some investigators concluded that MALDI-TOF MS, at its current point in development, is inappropriate for the identification of Shigella species (170) or have implemented supplementary testing for definitive determinations (171). Additional in-depth analysis of the spectral output of pathogenic E. coli and Shigella species isolates will obviously be required for enhanced discrimination between these organisms.
Proteus spp.
In a study of Proteus mirabilis isolates totally or intermediately resistant to amoxicillin-clavulanic acid, cefoxitin, cefotaxime, or ceftazidime, MALDI-TOF MS was utilized in comparison to repetitive element sequence-based PCR (rep-PCR) (Diversilab; bioMérieux) for the typing and classification of these organisms. Colonies were cultured on blood agar and overlaid by using DHB as a matrix material for MALDI-TOF analysis. The SARAMIS database was utilized for identification and characterization of the P. mirabilis isolates. Among these strains, high levels of genomic variability were detected by both rep-PCR and MALDI-TOF MS. Common clustering of strains could be achieved with rep-PCR and MALDI-TOF MS with some overlap; however, the authors concluded that a determination as to which technique was more suited to this type of analysis could not be elucidated due to the small sample size analyzed by these techniques (172).
Cronobacter spp.
Bacteria within the genus Cronobacter are closely related to members of the genus Enterobacter and are the causative agents of opportunistic food-borne outbreaks. Using the SARAMIS database software, Stephan et al. generated a comprehensive library of spectra generated by using 54 Cronobacter strains spanning the six species included in the Cronobacter genus. Importantly, a number of non-Cronobacter strains were also included in the construction of this library, particularly those belonging to the closely related genus Enterobacter. The authors used these 54 strains to determine SuperSpectra for defining this genus. During library construction, biomarker masses were determined for protein targets that were Enterobacteriaceae specific, Cronobacter specific, and Cronobacter species specific. Whole cells were utilized without previous protein extractions and analyzed by MALDI-TOF MS in this study. The generated library was subsequently challenged with 36 additional Cronobacter isolates gathered from a field study whose identities were previously confirmed by PCR and 8 non-Cronobacter isolates. MALDI-TOF MS was able to derive genus- and species-level identifications for these isolates or identify them as non-Cronobacter isolates in the case of nontarget strains. Cluster analysis was performed by using whole spectra derived from these Cronobacter isolates, with the resultant dendrogram strongly resembling dendrograms derived by using molecular techniques (16S rRNA sequencing, fluorescent amplified fragment length polymorphism analysis, and ribotyping) (173).
Two studies recently examined the use of MALDI-TOF MS for the identification of Cronobacter compared to other diagnostic procedures. Zhu et al. analyzed the ability of MALDI-TOF MS to identify Cronobacter spp. and closely related members of the Enterobacteriaceae compared to molecular (16S rRNA sequencing) and phenotypic (API 32E; bioMérieux) methods of definitive identification. By using phenotypic approaches, only 22% of isolates could be identified to the species level, 66% of isolates could be identified on a genus-specific level, and 14% could not be identified. 16S rRNA gene sequencing and MALDI-TOF MS were able to distinguish all isolates on the species level, with the mass spectrometry-derived identifications being more discriminating (174). As sequencing methodologies would likely require additional time due to sequence generation and analysis time along with time needed for the extraction and processing of DNA, MALDI-TOF MS represents a logical choice for the analysis of these types of isolates due to its strong discriminatory power and rapid analysis time.
Cetinkaya et al. described different findings when analyzing Cronobacter species isolates by MALDI-TOF MS. Using a very small sample set of 6 isolates, MALDI-TOF MS was performed and analyzed by using an in-house database. All strains were identified as C. sakazakii by MALDI-TOF analysis and as Enterobacter sakazakii by phenotypic approaches (API 20E and API ID32E), as Cronobacter sp. is not included in their databases. 16S rRNA and fusA gene sequencing revealed that three distinct species were present in this small sample set. The authors concluded that molecular techniques such as 16S rRNA and fusA gene sequencing and multilocus sequence typing (MLST) are more reliable mechanisms of Cronobacter identification (175); however, the use of an unverified in-house database often raises questions related to the robustness and accuracy of the results obtained by their respective MALDI-TOF analyses (173). Biogrouping analysis among the same species was possible by using a database created in-house (176). As comprehensive molecular analysis such as gene sequencing is not available to a large number of clinical laboratories, nor is it appropriate for use in situations where rapid identifications are necessary, a more complete analysis of MALDI-TOF MS for the identification of species within this sample set of organisms is warranted prior to drawing conclusions regarding the clinical utility of this technique.
Further demonstrating the discriminatory power of MALDI-TOF MS for the characterization of Cronobacter, Karamonová et al. investigated the use of the technique to identify different biogroups (biovars) within the C. sakazakii species. A collection of 29 C. sakazakii isolates with biovars previously determined and 5 Enterobacter isolates of various species were processed by using a whole-cell technique and analyzed by MALDI-TOF MS using the Bruker BioTyper 2.0 system to verify the genus Cronobacter, and a specific database was then created in order to analyze members of each biovar. The database was then challenged with 10 C. sakazakii isolates of undetermined biovars, which were able to be grouped into biovars by MALDI-TOF MS. The groupings were confirmed by biochemical testing, and the authors concluded that MALDI-TOF MS was a rapid and acceptable method for the discrimination of biogroups of bacteria within the same species (176).
Enterobacter cloacae complex.
The Enterobacter cloacae complex of organisms is composed of six species of Enterobacter and is responsible for significant numbers of nosocomial infections. A number of biochemical and molecular methods were utilized in an attempt to separate members of this complex of organisms into single species, but identification to the species level remains difficult even when using some of the most discriminatory molecular methods. Pavlovic et al. sought to evaluate the ability of MALDI-TOF MS to generate species-level identifications from separate members of this complex of bacteria. The authors utilized MALDI-TOF MS in conjunction with an in-house-developed multiplex PCR to determine species-level identifications. MALDI-TOF MS was found not to be able to identify 11 of 56 isolates determined to be members of the Enterobacter cloacae complex by biochemical methods, highlighting a shortcoming of the technology. However, as MALDI-TOF MS was able to determine species-level identifications for some members of the complex or characterize the isolates as either Enterobacter cloacae or a member of the Enterobacter cloacae complex, use of dnaJ duplex real-time PCR in combination with MALDI-TOF MS was suggested for cases where a definitive species-level identification is necessary (177). It remains to be determined if creation of a MALDI-TOF MS database compiled with spectra derived exclusively from members of the Enterobacter cloacae complex of organisms would allow for the identification of species-specific biomarkers to enhance the resolving power of MALDI-TOF MS toward rapid species-specific identification of members of the Enterobacter cloacae complex without the need for additional confirmatory molecular verification.
Pantoea spp.
Members of the genus Pantoea are infrequently encountered in clinical settings but have been reported as etiological agents in sporadic cases. By far the most encountered clinical species, Pantoea (Enterobacter) agglomerans is often associated with wound infections, polymicrobial infections, or infections of immunocompromised patients. A number of commercially available identification systems have difficulty reaching a determinative identification of Pantoea spp. The overlapping taxonomy of the Pantoea genus, which is comprised of former members of the Erwinia and Enterobacter genera, adds further complication.
Rezzonico et al. sought to utilize MALDI-TOF MS to characterize a collection of isolates previously identified as Pantoea spp. The authors described a current classification scheme in need of serious overhaul, noting significant inaccuracies with respect to 16S rRNA gene sequence data and the use of outdated classification methodologies in the identification of Pantoea spp. The authors used a combination of biochemical (BD Phoenix), molecular (gyrB and 16S rRNA gene sequencing), and MALDI-TOF MS approaches to characterize a collection of 73 isolates comprised of Pantoea spp. and other closely related isolates from the family Enterobacteriaceae. Whole cells harvested from LB plates were overlaid with matrix, and the generated spectra were imported into the SAMARIS database. While gyrB sequencing provided a more robust discriminatory mechanism than phenotypic methods, MALDI-TOF MS provided results almost equivalent to those of gyrB sequencing and was also able to sort strains within the genus into separate species and clades. The authors continued to refine their search parameters and concluded that MALDI-TOF MS was both accurate and suitable for the identification of Pantoea spp. but noted that the database would benefit from additional entries being added to further populate it with entries of both environmental and clinical interest (178).
Plesiomonas shigelloides.
Plesiomonas shigelloides is an uncommonly encountered member of the Enterobacteriaceae implicated in cases of sporadic and epidemic travelers' diarrhea. The organism is often isolated from water, soil, and other environmental sources and represents a challenge to clinical microbiologists due to familiarity of the organism. The organism is usually identified by its biochemical profile, with serological and molecular methods being available as extended options for definitive identification. MALDI-TOF MS was evaluated for the identification of Plesiomonas spp. from clinical specimens by using whole cells. Seventy-four isolates were identified as Plesiomonas shigelloides by biochemical methods and serologically typed prior to analysis by MALDI-TOF MS, with Aeromonas and Shigella isolates being used as outgroups for comparison. A database was constructed de novo, and the authors determined that MALDI-TOF MS was suitable for the identification of Plesiomonas shigelloides, but there was no correlation between the spectral profiles generated and serogrouping results (179).
Klebsiella/Raoultella spp.
Members of the genus Klebsiella are encapsulated Gram-negative organisms commonly encountered in the clinical laboratory. K. oxytoca is an important nosocomical pathogen that is closely related to K. pneumoniae and is phenotypically distinguished by a positive indole reaction. Members of the genus Raoultella are closely related to Klebsiella but are infrequently isolated from clinical specimens. de Jong et al. evaluated the use of MALDI-TOF MS to verify isolates identified as K. oxytoca by the BD Phoenix system (phenotypic identification) compared to 16S rRNA gene sequencing. Ninety-nine presumptive K. oxytoca clinical isolates were typed by MALDI-TOF MS using the Bruker BioTyper 3.0 database. Eight identifications were discordant, with these isolates being identified as members of the genus Raoultella by MALDI-TOF MS. Indeed, 16S rRNA gene sequencing identified five of these questionable isolates as Raoultella spp., while the additional three were identified as K. oxytoca. Due to the high level of spectral similarity between many Gram-negative species, a 10% difference in the MALDI-TOF score between the best and second-best results is necessary to accurately determine species-level identifications. The authors concluded that applying this rule to MALDI-TOF MS analysis increases the accuracy of the technique for the genus-level discrimination of Raoultella from Klebsiella oxytoca (180).
Yersinia spp.
The yersiniae represent a clinically important group of organisms infrequently encountered in the diagnostic workup of clinical specimens. The determination of pathogenic species within this genus is important during routine clinical diagnostics, as the identification of Y. pestis is reportable. The use of MALDI-TOF MS for the broad species-level identification of members of the yersiniae was investigated by two independent research groups. In 2010, Lasch et al. developed a spectral database consisting of Enterobacteriaceae, focusing on members of the genus Yersinia. As many members of the genus included in the study represent virulent pathogens, inactivation of the organisms by the addition of trifluoroacetic acid (TFA) was rightfully performed prior to matrix overlay and MALDI-TOF MS analysis as well as liquid chromatography-MALDI tandem mass spectrometry. Importantly, and similar to other studies, the authors described the presence of Enterobacteriaceae-specific biomarker peaks, which can be used to determine, in a general sense, members of this family of bacteria. Moreover, Yersinia-specific genus- and species-level peaks were also described. The authors discuss the identification of candidate biomarker peaks to discriminate between Y. pestis and Y. pseudotuberculosis, two clinically relevant and highly genetically similar organisms with identical 16S rRNA gene sequences (181).
Ayyadurai et al. also investigated the use of MALDI-TOF MS for rapid identification and typing of environmental and clinical Yersinia isolates using a database constructed from spectra of 39 Yersinia strains representing 12 species and 3 biotypes of Y. pestis. MALDI-TOF MS and the Bruker BioTyper 2.0 software were able to identify Y. pestis and Y. enterocolitica isolates using the constructed database, in addition to being able to discriminate correctly between Y. pestis biotypes (182). Thus, MALDI-TOF MS represents a robust and accurate method for the identification and characterization of pathogenic and nonpathogenic yersiniae in addition to providing epidemiological information regarding Y. pestis biotypes.
One important aspect of analysis that is sometimes overlooked is the protocols by which pathogenic organisms are inactivated. In the case of a biosafety level 3 (BSL-3) organism such as Yersinia, it is important to choose a mechanism of inactivation that will have a minimal influence on the MALDI-TOF MS spectra generated in order to garner optimal results. Couderc et al. compared ethanol and TFA inactivation protocols in parallel with MALDI-TOF MS to examine which inactivation methodology allowed the most robust spectra to be obtained for identification. In their study, ethanol inactivation yielded spectra of higher quality than spectra obtained by using TFA extraction. Despite the fact that ethanol inactivation of the organisms took substantially longer than TFA inactivation, the authors concluded that it was still compatible for routine use in the clinical laboratory for accurate identification of Yersinia spp. (183).
Two Yersinia species, Y. pestis and Y. enterocolitica, have had additional studies focused on the ability of MALDI-TOF MS to identify and characterize these organisms. The following sections will focus on these particular studies, the contribution of MALDI-TOF MS to the identification and subtyping of Y. enterocolitica isolates, and the ability of MALDI-TOF MS to discriminate between the highly related species Y. pestis and Y. pseudotuberculosis.
(i) Yersinia enterocolitica.
Y. enterocolitica is an important etiological agent of food-borne infections, with disease being associated with specific biotypes of the organism. Serological and biochemical analysis can be performed to differentiate different biotypes within the species, but this testing requires additional time and cost to the patient, in addition to testing reagents which might not be compatible with determinative algorithms of nonspecialized laboratories. Stephan et al. used MALDI-TOF MS in combination with SARAMIS SuperSpectra analysis to identify and subtype Y. enterocolitica isolates. A collection of yersiniae including 19 Y. enterocolitica isolates and representative members of other species were used to define the SuperSpectra for the different species of Yersinia as well as the biotypes associated with Y. enterocolitica isolates. The authors noted that different biotypes of Y. enterocolitica displayed high levels of spectral similarity, but key differences in the mass patterns among strains of different biotypes were observed. In all, 15 genus-identifying, 25 species-identifying, and 48 biotype-identifying biomarkers were elucidated by using this analysis. This collection of SuperSpectra was then rigorously challenged by using 117 additional Y. enterocolitica clinical isolates of previously defined biotypes using an intact-cell method. Cells were overlaid with sinapic acid-acetonitrile and TFA, air dried, and analyzed by MALDI-TOF MS. All 117 strains were correctly identified to the species level and were assigned biotypes with 100% correlation to assignments made by traditional biotyping methods (184).
(ii) Yersinia pestis and Yersinia pseudotuberculosis.
The species Y. pestis and Y. pseudotuberculosis are highly related and have recently been proposed to represent two lineages of a single species rather than two different species. The ability of MALDI-TOF MS to discriminate between these species was investigated previously (181), with the study concluding that the technology was able to accurately identify members of each genus. Wittwer et al. conducted similar experiments using a collection of 61 well-characterized Yersinia species isolates to generate a collection of spectra using the SARAMIS software package for analysis. Importantly, the authors reported that by using an unsupervised clustering approach, MALDI-TOF MS was able to identify all isolates to the genus level, but 7 of the 11 Y. pestis isolates were identified as Y. pseudotuberculosis at the species level. Once a supervised classification mechanism was utilized by using algorithms derived by the authors, MALDI-TOF MS was able to accurately distinguish Y. pseudotuberculosis from Y. pestis isolates (185).
Nonfermenting Gram-Negative Bacteria
A number of studies have focused on the identification of nonfermenting Gram-negative bacilli using MALDI-TOF MS methods, many of which examined strains isolated from cystic fibrosis (CF) patients. In this section, we review the identification of this group of bacteria by MALDI-TOF MS, with emphasis on both group-wide and genus-specific studies (Table 4).
Teramoto et al. reported that MALDI-TOF MS identification was accurate for Pseudomonas putida isolates, allowing identification to the strain level with a cluster analysis and showing a phylogeny comparable to that of the DNA gyrase subunit B gene sequences (186). Mellmann et al. created a database containing the spectra of 248 strains of 37 genera of human-pathogenic nonfermenting bacteria and used 16S rRNA gene sequencing as a reference standard for identification. In this study, MALDI-TOF MS identified 82.5% of 80 clinical isolates to the species level by using the BioTyper software (58). After improvements, they compared the MALDI results to results obtained by eight international clinical laboratories and reported accurate identification to the species level for 98.75% of their isolates (12).
In a comparable study, MADLI-TOF methods accurately identified 512 clinical isolates and 47 reference strains of nonfermenting Gram-negative bacilli (54). In this work, P. aeruginosa, S. maltophilia, and Alcaligenes (now Achromobacter) xylosoxidans were correctly identified to the species level, but the Burkholderia cepacia complex (BCC) isolates were poorly identified. Enrichment of the spectrum database allowed 98% correct identification of all strains tested at the species level. Discrepancies in these results were clarified by analysis performed on B. cepacia at approximately the same time using two data analysis algorithms: SARAMIS (Shimadzu and AnagnosTec) and BioNumerics (Applied Maths) (187). The authors accurately identified 65 and 69 out of 75 isolates of Burkholderia spp. to the species level with SARAMIS and BioNumerics, respectively. In studies not specifically dedicated to P. aeruginosa and other nonfermenting Gram-negative species, identification to the species level was 100% for P. aeruginosa and ranged from 56.6 to 100% for other species.
More recently, Fernández-Olmos et al. further investigated the use of MALDI-TOF MS for the identification of nonfermenting Gram-negative organisms from cystic fibrosis patients. In their study, the authors utilized a collection of 182 isolates collected from CF patients and stored over a 15-year period, representing a wide range of diverse genera, as identified by phenotypic methods. MALDI-TOF analysis was performed directly on colonies by using the Bruker BioTyper 2.0 database, with discordant identifications or nonidentifications being resolved by 16S rRNA gene sequencing. MALDI-TOF MS was determined to be significantly better for the identification of this collection of isolates. MALDI-TOF MS proved to be more discriminatory for members of the genus Achromobacter and Pandoraea, in addition to better identifying specific members of the Burkholderia cepacia complex of organisms. Organisms not identified by MALDI-TOF analysis (defined as a “no identification” result) included members of the genus Ralstonia, Bordetella petrii, Chryseobacterium, and Sphingobacterium spiritivorum, which were determined to be due to the absence of suitable reference spectra within the database (188).
Finally, a comparative analysis of two different MS platforms and databases was recently performed to evaluate their respective abilities to identify nonfermenting bacilli from CF patients. Two hundred isolates were tested by using both the Bruker BioTyper 3.0 software and bioMérieux Vitek-MS (formally SARAMIS database 3.62), using single-spot analysis for the Bruker analysis and double-spot analysis for the Vitek-MS study. Identifications were compared to biochemical- and molecular-derived identifications made previously. 16S rRNA gene sequencing was utilized to resolve discordant results. In all, the Bruker system identification was concordant with reference testing for 72.5% of isolates tested to the species level of identification, with 3% of isolates being unable to be identified, but required ethanol and FA extraction steps more frequently than the Vitek-MS system. The Vitek-MS system identified 80% of isolates to the species level, with 7% of isolates not being able to be identified. Both systems were determined to be better than conventional phenotypic methods for the identification of organisms within this set of isolates (189).
In the following sections, we review studies dedicated to the identification of individual species within specific genera contained with the nonfermenting Gram-negative bacilli.
Acinetobacter spp.
Prior to the advent of specialized microbial databases for the identification and analysis of different microbial genera and species, an analysis of environmental bacteria by Ruelle and colleagues demonstrated that MALDI-TOF MS could be utilized to identify the genus Acinetobacter versus E. coli and Salmonella (190). Since that time, the standardization of culture and MS techniques facilitated significant progress in the characterization of Acinetobacter species isolates with MALDI-TOF MS. Three main types of studies have been undertaken. Studies examining the antibiotic resistance profiles of these isolates are reviewed elsewhere in this article, whereas here we examine works directed toward the species-level identification of members of the genus in addition to MALDI-TOF MS-based typing methods to analyze Acinetobacter baumannii.
Two studies examining the ability of MALDI-TOF MS to discriminate between different species within the genus Acinetobacter were published. The first, by Šedo et al., sought to determine a method of sample processing which would enhance MS-mediated analysis of Acinetobacter spp. (191). The second study, by Álvarez-Buylla et al., examined the use of the Bruker BioTyper as an alternative to molecular methods for the species-specific identification of Acinetobacter spp. One hundred nine isolates were routinely identified as Acinetobacter spp. by phenotypic methods (Vitek-2; bioMérieux). MALDI-TOF MS analysis was performed by using the Bruker BioTyper 2.0 software, and identifications were compared to rpoB sequencing used in conjunction with PCR to detect the presence or absence of the blaOXA-51-like gene. Importantly, the authors concluded that MALDI-TOF MS was able to accurately distinguish A. baumannii from other members of Acinetobacter spp.; however, using this software version, the technique was not able to discriminate well among non-A. baumannii isolates compared to rpoB sequencing (192) without elaborating on the spectra currently available in that version of the BioTyper software.
The Acinetobacter baumannii group of organisms is a collection of three closely related species, A. baumannii, A. pittii, and A. nosocomialis, which are difficult to resolve by using phenotypic methods. Espinal et al. investigated the ability of MALDI-TOF MS to discriminate between these species using the BioTyper 2.0 software and an extended database constructed in-house. Strong discrimination between these highly related species was observed, making MALDI-TOF MS a rapid and attractive alternative to costly molecular analyses needed for species-level determination among these isolates (193). Finally, Mencacci et al. recognized the potential of MALDI-TOF MS for real-time detection of pathogens and chose to analyze the technology for the detection of nosocomial Acinetobacter baumannii outbreaks compared with rep-PCR (Diversilab). The authors concluded that the technology could be applicable to real-time evaluation of Acinetobacter baumannii outbreaks, providing results well before established molecular methods (194).
Burkholderia cepacia complex.
The Burkholderia cepacia complex (BCC) represents a group of closely related, nonfermenting, Gram-negative rods that are important opportunistic pathogens, particularly in patients with cystic fibrosis (CF). Members of this group of bacteria can be isolated from both the environment and infected patients and can be readily transmitted between CF patients, resulting in outbreaks (195). Differentiation of the species contained within the complex is challenging by biochemical analysis alone, with phylogenetic techniques such as recA 16S rRNA gene sequencing and MLST predominating for species-specific analysis of members of the complex (196). Additionally, the large genomes associated with members of the complex can hinder both molecular- and phenotypic-based identification (197), and in some cases, multiple assays are necessary to discern specific species. Thus, the implementation of proteome-based identification is an attractive option for determinative analysis of members of this complex of bacteria.
A number of recent studies have examined the ability of MALDI-TOF MS to fill the role of a rapid and automated system for the species-specific discrimination of members of the B. cepacia complex of organisms (187, 198, 199). In addition to studies evaluating the technology for the routine identification of bacteria from CF patients compared to phenotypic methods (200), one of the first investigations of the technology included an analysis of 75 isolates of BCC or BCC-like organisms and used MALDI-TOF MS to generate MS spectra analyzed by SARAMIS and BioNumerics (version 4.5) software. MALDI-TOF MS with cluster analysis provided good discrimination between members of the BCC and outliers, in addition to phylogenetic analysis of species within the complex (187).
A second study evaluated two different intact-cell MS methods for the analysis of spectra for the discrimination of BCC species, including members of taxon K, representing a relatively new and loosely important classification of organisms within the BCC demonstrated to consist of novel species (201). Analyzing 26 reference and 146 clinical isolates from sputum samples of CF patients, Minan et al. reported the identification of specific biomarkers for the discrimination of members within the BCC from other nonfermenting Gram-negative rods. Species-level discrimination using biomarkers visualized by using the in-gel view of the software was also described. Clustering analysis also allowed the discrimination of closely related species within the BCC (199).
The most recent study involving the application of MALDI-TOF MS for the discrimination of members of the BCC from CF patients was reported by Lambiase et al., who compared MS data to identifications made by PCR-restriction length polymorphism (RFLP) analysis of the recA gene. Each of 57 isolates was identified by phenotypic methods (BD Phoenix with API 20 NE used for confirmation) and MALDI-TOF MS using the BioTyper version 1.0 software. DNA was also prepared from each sample, and the recA gene was PCR amplified and subjected to RFLP. Both methods performed amicably for the identification of isolates to the species level, with RFLP providing slightly more discriminatory results in its ability to discriminate between two different lineages of B. cenocepacia but conversely being more expensive than MALDI-TOF MS (198).
Burkholderia mallei and Burkholderia pseudomallei.
Both Burkholderia mallei and Burkholderia pseudomallei are closely related Gram-negative organisms with high pathogenic potential in animals and humans. B. pseudomallei is the etiological agent of melioidosis, a condition acquired through the inhalation, ingestion, or invasion of the organism, with disease manifesting as abscess formation in internal organs and occasionally as septic shock. Conversely, B. mallei is the causative agent of equine glanders and infrequently infects humans in areas of endemicity (202). The organism cannot persist in the environment outside its host, with both species being facultative intracellular bacteria that are capable of replication inside epithelial and phagocytic cells (203). When isolated, both species are considered reportable and represent virulent and dangerous biosafety level 3 (BSL-3) organisms.
Identification of B. pseudomallei can be difficult in the clinical laboratory, as both molecular and phenotypic assays may not be discriminatory enough to differentiate it from closely related species, including B. thailandensis (204). Additionally, full analysis and definitive identification of Burkholderia isolates can take up to 1 week (205).
Lau et al. evaluated the use of MALDI-TOF MS for the identification of B. pseudomallei using the BioTyper version 3.0 software. As reported in this study and elsewhere, the routine database does not contain reference spectra for select agents, and identification would normally require the use of an extended commercial database available from Bruker (206, 207). In this instance, the authors supplemented the version 3.0 database with spectra from B. pseudomallei and the closely related organism B. thailandensis. MALDI-TOF MS performed well in the accurate identification of B. pseudomallei, with the authors noting that the inclusion of additional validated spectra would enhance the accuracy of identification of these and other related species (204).
Two additional studies have examined the identification of both B. mallei and B. pseudomallei by MALDI-TOF MS, one of which examined the ability of MALDI-TOF MS to identify these species both directly from positive blood culture bottles as well as from plated media by using an in-house library generated from collected spectra of Burkholderia strains (207). The second study sought to further evaluate the technology for the identification of both species, where MALDI-TOF MS was able to accurately discriminate between members of both species after careful selection of reference spectra. Additionally, the authors reported that spectra generated from B. mallei exhibited higher levels of homogeneity than did those from B. pseudomallei, with the type strain of B. pseudomallei being separated from more recent isolates of B. pseudomallei. This may be due to genetic modification of the type strain due to significant passage or inappropriate medium combinations (205). This finding represents a potential challenge with regard to standardization of specimen age and levels of manipulation of microbes submitted for MALDI-TOF MS identification.
Pseudomonas spp.
The genus Pseudomonas is perhaps one of the most complex genera, representing an ever-growing number of species. Recently, a comparison of MALDI-TOF MS and multilocus sequence typing (MLST) was performed to evaluate the ability of MS to accurately identify species of Pseudomonas. A total of 141 Pseudomonas strains (133 of which were type strains, further illustrating the diversity of bacteria included in this genus) were processed for MALDI-TOF MS and MLST. MS spectra were obtained and compiled by using SARAMIS software, and a phylogram was created to examine the relatedness of the strains included in the study. Compared to each other, both MLST and MALDI-TOF MS allowed for resolution of the included Pseudomonas species to the group or subgroup level. The authors concluded that while MLST should be used for more in-depth phylogenetic studies, MALDI-TOF MS represents a rapid and viable option for species-level differentiation and identification in clinical and environmental microbiology settings (208).
Stenotrophomonas maltophilia.
The most clinically relevant species of the genus Stenotrophomonas is S. maltophilia. This organism is responsible for an increasing number of infections resulting in diverse clinical presentations such as sepsis, urinary tract infections (UTIs), and endocarditis. Vasileuskaya-Schulz et al. recently evaluated the use of the MALDI-TOF MS BioTyper system in concert with multilocus sequence analysis for the analysis of 21 strains belonging to different species within the genus. Good conformity was observed between the two techniques, with intra- and interstrain differences being observed with MALDI-TOF MS spectra generated from these isolates (209). Additional analysis of members of this group of nonfermenting bacteria will be important to further examine the discriminatory ability of MALDI-TOF MS for Stenotrophomonas spp.
Fastidious Gram-Negative Bacteria
Fastidious Gram-negative bacteria are a group of microorganisms that require additional nutritional requirements for their cultivation. Many of these bacteria are substantial pathogens and significant concerns for public health. As such, both the MALDI-TOF MS spectral libraries used to analyze these pathogens and sample preparation mechanisms associated with these organisms need to be standardized. As mentioned on numerous occasions throughout this review and others, the construction of refined spectral databases greatly enhances the capacity of MALDI-TOF MS to provide rapid and accurate species-level identifications for a wide variety of bacterial pathogens. These spectral databases are usually constructed from in-house collections of isolates from clinical material or from established collections which have had significant analysis previously performed on them to serve as references in comparative studies.
Sample preparation methods for MALDI-TOF MS analysis of fastidious Gram-negative rods, including dangerous bacteria.
Cunningham and Patel raise an excellent point in their recent publication examining the use of MALDI-TOF MS when analyzing select agents. They point out that while database enhancement is demonstrated to improve MS performance, access to select agents is quite limited, and as such, construction of specialized databases for species such as Francisella tularensis, Burkholderia pseudomallei, and Brucella spp. would be difficult for most laboratories (206). The BioTyper reference library does not contain spectra for the identification of select agents; such spectra are contained in the security-relevant library, which can be queried in tandem with the reference library, as previously reported. The authors evaluated the MALDI-TOF MS system for the identification of 20 isolates representing select agents. Spectra were evaluated by the BioTyper software (version 3.0) with and without the use of the extended security-relevant library (version 1.0) (206).
Using the BioTyper software alone, 18 isolates returned spectra with scores of >1.7 but were identified as having “no reliable identification” due to a lack of references in the database, and 2 Burkholderia pseudomallei isolates returned identifications of “Burkholderia thailandensis” with scores that would be acceptable for genus-level identification, along with a comment mentioning that B. thailandensis is related to B. pseudomallei. The addition of the security-relevant database resulted in species-level identification of five F. tularensis isolates, with two additional F. tularensis isolates being identified to the genus level. The extended database was also used to identify Brucella and Burkholderia to the genus level, with species-level identifications being achieved in some cases. Importantly, the authors warn that a result of “no reliable identification” could potentially lead to additional testing of these problem isolates, increasing the potential for exposure to laboratory personnel. Finally, the authors rightfully conclude that for optimal patient care and laboratory safety, spectra for members of these genera should be included in the standard BioTyper database, and the direct-colony approach for analysis of Burkholderia, Francisella, and Brucella species isolates should not be performed (206).
Drevinek et al. recently evaluated sample preparation methods for highly dangerous bacteria in an attempt to propose a processing scheme to streamline and ensure safe manipulation of these organisms for MALDI-TOF MS (195). While direct analysis of intact cells was proposed for a number of organisms as the optimal approach for analysis by MALDI-TOF MS, work with BSL-3 organisms often dictates their inactivation prior to manipulation in the laboratory to minimize exposure risks. Four different specimen preparation methods were analyzed for BSL-3 agents, including Bacillus anthracis, Clostridium botulinum, Brucella melitensis, Burkholderia mallei, Francisella tularensis, Shigella dysenteriae, Vibrio cholerae, Yersinia pestis, and Legionella pneumophila. Analysis to determine the most appropriate method focused on different aspects of sample preparation and MALDI-TOF MS analysis, including inactivation of the organism, concentration of protein obtained from the specimen, and quality and stability of the MS spectra obtained. Importantly, the authors found that ethanol pretreatment and the use of formic acid with acetonitrile for protein extraction, as described by Marklein et al. (210), could be used as a rapid and universal method for the MALDI-TOF MS processing of these sample types. Additionally, inactivated bacterial lysates in 300 μl water and 900 μl ethanol were recommended for interlaboratory evaluations, although the quality and robustness of spectra obtained from these samples decrease, making strain-level identifications more difficult over time (49).
Brucella spp.
Ferreira et al. used a collection of 131 Brucella human isolates (Brucella abortus, Brucella melitensis, and Brucella suis) and obtained 100% identification to the genus level (211). The identification to the species level was not reliable. A spectrum bank was created from 12 Brucella strains from six species by using the MALDI BioTyper 2.0 system. Species identification may not represent a serious limitation for the technology, since some taxonomists propose the grouping of classical Brucella species into one species, as biovars of Brucella melitensis. Others argue that despite their high level of DNA identity, Brucella species can be distinguished by distinct biochemical and fatty acid characters as well as by a marked host range (e.g., B. suis for swine, B. melitensis for sheep and goats, and B. abortus for cattle) (212). It is reasonable that identification as Brucella spp. would be useful for clinical laboratories who will be alerted to handling of the select agent with proper biosafety precautions according to LRN guidelines.
Since the previous publication, Lista et al. reported the construction of a spectral database to be used for the species-level identification of Brucella isolates using strains that demonstrate genetic variation at the species and biovar levels by multilocus variable-number tandem-repeat analysis (MLVA). This database was then used to analyze 152 Brucella isolates also preliminarily characterized by MLVA using the BioTyper 2.0 software. Using the data analysis parameters reported in their article, the authors concluded that MALDI-TOF MS could indeed discriminate between and accurately identify different species and biovars of Brucella (213).
Bartonella spp.
Members of the genus Bartonella are zoonotic bacteria that are becoming increasingly recognized as etiological agents of human disease. The phenotypic analysis of these organisms is difficult and not well defined due to poor biochemical reactivity associated with the genus. MALDI-TOF MS has recently been proposed as a mechanism to determine species-level identifications for members of this genus. Using type strains representing 17 species of Bartonella, Fournier et al. utilized the BioTyper software (which did not contain spectra generated from Bartonella) for spectral analysis. The generated spectra were then used to identify an additional 39 isolates. MALDI-TOF MS was able to correctly provide species-level identifications for all strains tested, with most generating scores of ≥2.0. Moreover, subspecies-level identifications of B. vinsonii isolates could be determined, in addition to genotype discrimination between the two genotypes associated with B. henselae (214).
Francisella spp.
For identification of Francisella spp. and Francisella tularensis, the causative agent of tularemia, the phenotypic discrimination of closely related, but differently virulent, Francisella tularensis subspecies is difficult and often produces ambiguous results. Seibold et al. reported the testing of reference spectra from five representative strains of various Francisella spp. and subspecies, including F. tularensis, establishing spectral references and evaluating the capability of MALDI-TOF MS to correctly identify 45 blind-coded Francisella strains against a database (215). Identity was confirmed by 23S rRNA sequencing. In this study, all strains were correctly identified, with both methods showing perfect agreement at the species level as well as at the subspecies level. The identification of Francisella strains was reproducible using replicate testing examining different culture media, cultivation times, serial passages of the same strain, preparation protocols, and mass spectrometers. In an additional study, Muller et al. used MALDI-TOF MS to identify F. tularensis isolates from a field study involving European brown hares. The authors reported that isolates generated strong spectra and that MALDI-TOF MS was able to rapidly identify isolates to the subspecies level (216).
Haemophilus spp.
Members of the genus Haemophilus are nonmotile bacilli that are identified based on their nutritional requirements for X and V factors found in blood. Recovery and identification from culture can be challenging, as most species are not readily isolated from culture on blood-containing media, often requiring chocolate agar for their isolation. There are currently 10 recognized species of Haemophilus, some of which can be further classified into distinct serotypes that can provide valuable epidemiological data in outbreak situations by either serological or molecular methods. Members of the species H. influenzae can be further separated into biotypes that were historically associated with disease presentations, antibiotic resistance patterns, and sources of infection.
MALDI-TOF MS was utilized in the identification and characterization of different members of the genus Haemophilus. Haemophilus species, with an emphasis on Haemophilus ducreyi, were identified by MALDI-TOF MS (217). Strains of Aggregatibacter (formerly Actinobacillus) actinomycetemcomitans were also identified (217). Additional studies demonstrated good genus-level identification of Haemophilus, Aggregatibacter, Cardiobacterium, Eikenella, and Kingella (so-called HACEK species) by MALDI-TOF MS. Couturier et al. tested 103 species of HACEK group microbes and achieved 93% genus-level identification but only 66% species-level identification with the Bruker database (218). During a study focusing on the differentiation between H. parahaemolyticus and H. paraphrohaemolyticus, a distinct taxon of uncharacterized Haemophilus spp. was described and validated by 16S rRNA gene sequencing as unique. This unique species, now named Haemophilus sputorum, was additionally confirmed by MALDI-TOF MS using the BioTyper version 2.0 software due to the generation of a spectrum unique from those of other bacterial species analyzed (219).
Nontypeable H. influenzae causes a number of serious etiologies, including pneumonia, meningitis, and sinus and ear infections. Microbiological identification of this particular serotype of H. influenzae is difficult, as it is phenotypically similar to H. haemolyticus, a species that is not usually associated with serious infections. Several molecular techniques, such as gene-specific PCR and sequence analysis of outer membrane protein genes, have proven to be unsuitable for definitive discrimination between these two species, with multilocus sequence typing (MLST) providing good discrimination. Zhu et al. recently utilized MALDI-TOF MS using the BioTyper version 2.0 software to discriminate between these species. Rightfully noting that the commercial database contained only 10 H. influenzae reference spectra, the authors constructed an in-house version with additional spectra derived from 10 nontypeable H. influenzae and H. haemolyticus isolates and then evaluated the database against 42 nontypeable H. influenzae and 10 H. haemolyticus isolates. The authors concluded MALDI-TOF MS, using the extended database constructed in-house, was an excellent method for differentiating these two closely related organisms (220).
Vibrio spp.
The use of MALDI-TOF MS for the characterization of Vibrio species was investigated. MALDI-TOF MS also accurately identified 67 isolates of Vibrio spp. from 16 different species to the species level and discriminated between closely related species, such as Aeromonas spp., Photobacterium damselae, and Grimontia hollisae (221). In another study, 20 strains of Vibrio parahaemolyticus were successfully identified to the species level from nine different species of Vibrio (222). In a survey of 30 environmental isolates from wastewater sources, MALDI-TOF MS was successfully used to identify closely related Vibrio species using the BioTyper software (223).
Aeromonas spp.
In two independent studies, 32 isolates of Aeromonas from 17 different species and 34 out of 52 environmental isolates of Aeromonas spp. were correctly identified (224, 225). In a more recent examination of 171 Aeromonas isolates representing type strains and clinical and environmental isolates, the Bruker BioTyper version 2.0 software was evaluated for genus- and species-level identifications. MALDI-TOF MS was able to determine genus-level identification with 100% accuracy and species-level identifications with 90% (clinical isolates), 93.9% (environmental isolates), and 90.6% (type strains) accuracies (226).
Benagli et al. described difficulties in determining the taxonomic standing of some isolates of Aeromonas spp. using phenotypic methods and described the development of a database for the rapid characterization of such isolates. Despite complex and evolving taxonomy within the genus, gene sequencing and other time-consuming and labor-intensive molecular methods have predominated when attempting specific identification of Aeromonas isolates. Using SARAMIS, the authors assembled a database containing information from 94 Aeromonas isolates and generated SuperSpectra to provide identifications for 11 different species, and these identifications were compared to results from gyrB sequencing and generated useful data regarding Aeromonas taxonomy. The representative database was then used to characterize 741 strains of Aeromonas, with 93% of these isolates being identified successfully and with unidentified isolates not having adequate spectra represented in the database (227).
Campylobacter spp.
Members of the Campylobacter genus have also been studied by MALDI-TOF MS and represent one of the earliest genera to be evaluated for species-level identification by the technology. In 1999, Winkler et al. published findings of a study of the direct analysis of Campylobacter and Helicobacter using methanol-inactivated isolates. The study determined that MALDI-TOF MS could be used to differentiate between the two genera as well as related members of different species through the identification of specific spectral peaks corresponding to biomarkers (228). Subsequent studies (229, 230) further examined the ability of MALDI-TOF MS to provide rapid and accurate species-level identifications for members of the genus, with encouraging results.
More recent attempts to identify and characterize Campylobacter isolates have also been successful. After constructing a spectral database using the BioTyper 1.1 software package, Alispahic et al. reported 100% identification to the species level for 144 clinical isolates of Campylobacter spp. and the related genera Arcobacter and Helicobacter. Additionally, it was noticed that mass spectral fingerprints obtained from thermophilic Campylobacter species (C. jejuni, C. coli, and C. lari) were distinct from nonthermophilic species (C. fetus and C. hyointestinalis) (231), potentially providing preliminary targets for biomarker analysis for more discriminatory typing methods using MALDI-TOF MS. Interestingly, the choice of medium used for cultivation of Campylobacter (in addition to Helicobacter and Arcobacter) in this study proved to have bearing on MS spectral integrity, as bacteria grown on modified charcoal-cefoperazone-deoxycholate agar generated poor spectral output. As this agar is routinely used for the identification of Campylobacter spp., additional culturing on supplemental agar may be necessary prior to definitive identification by MALDI-TOF.
In a rigorous study by Bessède et al., 1,003 successive Campylobacter-like strains from the French National Reference Center for Campylobacter and Helicobacter were analyzed by MALDI-TOF MS, with these identifications being compared to those made by using phenotypic and molecular methods, including real-time PCR. The Bruker BioTyper 2.0 software package was utilized to analyze the spectra collected from these isolates. Compared with molecular methods, MALDI-TOF MS provided similar results, with the exception of a notable discrepancy in the misidentification of four isolates of C. jejuni as either C. coli or C. fetus with identification scores of >2. Four of the cultures analyzed were determined to contain a mixture of organisms, for which MALDI-TOF MS was not able to identify each of the organisms present (232).
Finally, a comparison between routine clinical diagnostic methods relying on biochemical profiling/molecular typing and MALDI-TOF MS was undertaken by Martiny et al. This comparative study examined the ability of the API Campy (bioMérieux) biochemical-based assay, the Vitek-2 system using the Neisseria-Haemophilus card, and MALDI-TOF MS using the Bruker BioTyper database to identify 224 clinical and 10 reference isolates representing Campylobacter, Helicobacter, and Arcobacter. For C. coli and C. jejuni isolates, MALDI-TOF MS correctly identified 100% of these strains, with an overall sensitivity of 98.3% for all isolates tested. In addition to significantly outperforming the other methods with respect to identification of isolates at the species level, MALDI-TOF MS did not require any additional testing for the identification of these isolates, in contrast to the phenotypic identification systems. The authors concluded that MALDI-TOF MS should be the method of choice for the routine identification of these organisms (233).
Helicobacter spp.
In comparison with Campylobacter, few studies have specifically evaluated MALDI-TOF MS for the characterization of Helicobacter species. Winkler et al. previously reported that members of the genus Helicobacter could be differentiated from Campylobacter by MALDI-TOF MS and that species within the genus could be differentiated from each other (228). In a study involving Neisseria gonorrhoeae (described below) and H. pylori, it was reported that due to high intraspecies diversity accumulated among a collection of 2 reference and 22 clinical strains of H. pylori, the use of MALDI-TOF MS for the identification of H. pylori appeared difficult (234). However, Alispahic et al. later confirmed the use of MALDI-TOF MS for the identification of Helicobacter pullorum and H. pametensis using the BioTyper software (231), indicating that perhaps species-specific discrimination between members of this genus was possible.
Preliminary cluster analysis was able to determine some level of relatedness between these 24 isolates, indicating the potential for strain-specific analysis of H. pylori isolates (234). These studies were expanded in a 2010 publication examining the application of MALDI-TOF MS for the species-specific identification of H. pylori (235). Using a collection of 2 reference strains and 17 clinical isolates, MALDI-TOF MS analysis was performed with the Bruker BioTyper 2.0 software. Despite significant differences observed in MALDI spectra analyzed both in this study and previously, the BioTyper database identified all isolates tested as H. pylori. Thus, MALDI-TOF MS was deemed suitable for the identification of H. pylori, even in the case of an organism which exhibits high levels of genetic plasticity (234, 235).
Neisseria spp.
Commercial databases for the identification of Neisseria species isolates have also been undertaken. By using BioTyper software, members of Neisseria spp. were accurately identified, with 100% correct identification of 57 strains of Neisseria spp., including 29 Neisseria meningitidis and 13 Neisseria gonorrhoeae strains. No differences in the spectra were observed between different serogroups, leaving no evidence to support the use of MADLI-TOF MS for epidemiological serotyping purposes (236). Strains of N. gonorrhoeae were successfully identified by MALDI-TOF MS by a comparison of selected peaks in the obtained spectra without using any software for comparison (217).
With respect to intraspecies analysis, the variable combination of the mass-to-charge (m/z) ratios of three ribosomal proteins was utilized to group strains into one of four groups in an analysis of 278 N. gonorrhoeae isolates (234). Additionally, Lowe et al. reported that the detection of a single-nucleotide polymorphism within the fumC gene could be utilized to differentiate a hypervirulent ET-15 lineage of N. meningitidis from other isolates by using MALDI-TOF MS (237). Thus, while few studies were conducted to assess the ability of MALDI-TOF MS to definitively characterize strains of Neisseria, MALDI-TOF MS has exhibited promise toward the intraspecies analysis of this group of diverse bacteria.
Moraxella catarrhalis.
Currently, no definitive studies examining the typing of M. catarrhalis isolates have been undertaken using commercially available MS databases. Schaller et al. reported the description of two distinct 16S rRNA gene types of M. catarrhalis and information regarding the profiling of OMPs by MALDI-TOF MS. In their analysis, a collection of 18 characterized M. catarrhalis isolates was utilized for analysis by an intact-cell approach. Results were compared to 16S rRNA typing methods, which classified the isolates into either RNA type 1, 2, or 3, which have strong associations with specific OMPs expressed by different M. catarrhalis strains. Strains belonging to rRNA type 2 or 3 exhibit a decreased or lack of expression of certain OMPs, which represent potential vaccine candidates. Thus, M. catarrhalis isolates can be broadly classified into two distinct groups. The authors found agreement between results of MALDI-TOF MS and 16S rRNA typing methods in 15 of the 18 cases, with the 3 discrepant isolates being misclassified within 16S rRNA groups 2 and 3. Additionally, it was determined that each 16S rRNA group differentially expressed additional OMPs (238). Although preliminary, this study demonstrated the usefulness of MALDI-TOF MS for typing of M. catarrhalis isolates, which could provide useful information for future epidemiological studies and in outbreak situations.
Legionella spp.
Moliner et al. were one of the first groups to evaluate MALDI-TOF MS for the identification of Legionella spp. Noting that the early version of the Bruker database used in this study contained only a few entries for Legionella spp. (one spectrum per species), the authors analyzed representative strains of all Legionella species recognized at the time, including members of the same species but different serogroups, and constructed their own database in order to more robustly analyze members of the genus. Spectra were obtained for all strains tested, and no differences were observed between members of the same species belonging to different serogroups. The assembled database was then evaluated by using 237 additional isolates. MALDI-TOF MS was found to be a good method for discrimination at the strain level, particularly for L. pneumophila isolates, but as observed during the construction of the database, serogroup-specific identification was not possible (239).
In a subsequent study, the ability of MALDI-TOF MS to identify and group Legionella isolates was compared to that of mip (macrophage infectivity poteniator) gene phylogenetic analysis utilizing SARAMIS software. Using a collection of 453 clinical and environmental samples representing 38 species of Legionella, Gaia et al. used 216 of these isolates to construct a Legionella-specific database of SuperSpectra and the remaining 237 isolates to evaluate that database. MALDI-TOF MS identifications were compared to identifications derived by mip sequence analysis and agglutination assays for Legionella species identification. Assembled dendrograms revealed good correlation between mip analysis and MALDI-TOF MS to determine relatedness between members of the genus. All but two isolates could be identified to the species level by MALDI-TOF MS. The authors concluded that by using their constructed database, spectral differences between members of the same genus could be identified; however, identification and classification of strains into different serogroups were still not possible (240).
Fujinami et al. addressed the question of further examining intraspecies differences in MALDI-TOF MS spectra between different L. pneumophila strains. Using 23 reference L. pneumophila strains as references, mass spectra were generated and analyzed by using the Bruker BioTyper version 1.1 software and compared to results from PFGE analysis for phylogenetic classification of isolates. PFGE analysis revealed a strong association between the geographical location of strain acquisition and organism relatedness, a finding also observed in the MALDI-TOF MS clustering of organisms (241).
Svarrer and Uldum evaluated the ability of MALDI-TOF MS to identify species other than L. pneumophila from samples of both clinical and environmental origins. In their study, 33 isolates of nonpneumophila Legionella species were obtained from respiratory patients, and 42 isolates were obtained from environmental origins. All isolates were identified to the species level by mip sequencing and subsequently processed for MALDI-TOF MS analysis with the Bruker BioTyper 2.0 software. Isolates yielding discrepant identifications were analyzed by 16 rRNA gene sequencing. Here the authors reported that 3 of the 33 clinical isolates had scores of <2.0, yielding an overall sensitivity of 90.6%; however, these isolates were identified correctly to the species level compared with mip sequencing. Thirty-four of the 42 environmental isolates were correctly identified by MALDI-TOF MS to the species level, and 8 isolates yielded identification scores of <2.0, with these isolates being either not represented or represented by only a single spectrum in the database (242).
Thus, MALDI-TOF MS represents an accurate method for the species-level identification of Legionella species, with the level of accuracy being strongly dependent upon the number of species-specific spectra populating the database. Many of the authors of the above-mentioned reviews call for databases with expanded numbers of Legionella-specific entries to more accurately discriminate between different species within the genus. However, serogroup-level determinations are still not possible using this technology.
ANAEROBIC BACTERIA
The MALDI-TOF method has special importance in routine identification of pathogens that require long incubation times for isolation and are biochemically inactive, such as anaerobic bacteria. MALDI-TOF MS has also been utilized to evaluate subsets of anaerobic microorganisms isolated from defined biological niches. The applicability of this method for routine identification of important human-pathogenic anaerobic bacteria was evaluated by Shah et al., who showed that through the use of intact-cell MALDI-TOF MS, Porphyromonas strains could be differentiated (243). Nagy et al. reported 97% accuracy at the species level for 277 isolates and 9 species of Bacteroides (244). Stîngu et al. analyzed 84 strains of oral anaerobic bacteria in a study designed to streamline the identification of microbes from patients with periodontal disease. A database was created, and analysis of results revealed identifications consistent with phenotypic and 16S rRNA gene sequencing data. The technology was able to differentiate between two species of Prevotella, P. intermedia and P. nigrescens, a task which would usually be relegated to costly and time-consuming 16S rRNA gene analysis (245).
Nagy et al. tested 283 clinically relevant anaerobic isolates and compared the results derived from the MALDI BioTyper 3.0 software to conventional identification. In this study, 218 (77%) of 283 isolates were identified to the species level when a log score of >2.0 was used as a cutoff, 31 isolates (10.95%) were identified to the genus level (log score of 1.7 to 2.0), and 34 (12%) produced a nonreliable identification (log score of <1.7). Of 31 isolates with a log score of 1.7 to 2.0, a total of 24 isolates were noted to have the same species name as that determined by classical identification. For the 44 discordant results, 16S rRNA gene sequencing confirmed the MALDI-TOF MS identification in 41 cases, leaving 3 isolates (0.7%) that were misidentified by MALDI-TOF MS (246).
In a 2011 article, La Scola et al. surveyed the ability of MALDI-TOF MS to identify anaerobic microorganisms isolated from a large clinical laboratory (247). Culture isolates from positive blood culture bottles and other clinical specimens were subcultured and grown in an anaerobic chamber. In contrast to other studies, MALDI-TOF MS was utilized for the routine identification of anaerobic isolates, with colonies not able to be identified by MS methods being identified by 16S rRNA gene sequencing. Of the 554 isolates tested, MALDI-TOF MS was able to identify 61% of them, with the authors highlighting genus- and species-specific trends likely due to a combination of the quality and number of spectra available in the MALDI-TOF MS database. Of the 212 remaining isolates, 95% were able to be identified to the species level by 16S rRNA gene sequencing. The authors concluded that 16S rRNA gene sequencing represents a strong technique to complement MALDI-TOF MS for the identifications of anaerobic bacteria, with MS technologies likely to be implemented as the primary determinative method in the future as the technology continues to develop (247). It is also important to emphasize that the ability of MALDI-TOF MS to identify anaerobic species currently is not as robust as it is for the routine species-level identification of other groups of bacteria; therefore, the use of additional confirmatory testing will likely be necessary for some time to come.
In a series of evaluations of MALDI-TOF MS technology from the same group, Veloo et al. examined 79 clinical isolates representing 19 anaerobic genera by comparing the Bruker and Shimadzu systems with 16S rRNA sequencing, achieving correct identifications to the genus level for 61% and 71% of isolates and to the species level for 51% and 61% of isolates, respectively. When species not present in the database were excluded, correct identification was achieved for 75% of isolates with the Bruker system, versus 76.7% for the Shimadzu system (248), leading the authors to conclude that database optimization was necessary. In a subsequent evaluation of the technology for the identification of Gram-positive anaerobic cocci, the authors used a specialized database constructed in-house and compared these identifications to those of 16S rRNA gene sequencing. MALDI-TOF MS performed well, identifying 96 of 107 clinical isolates (249). Finally, in an excellent review from the same year, the authors again highlight the importance of expanded databases for the identification of anaerobic bacteria (250).
More recently, the use of MALDI-TOF MS for the routine identification of anaerobic bacteria from clinical specimens was evaluated and compared to 16S rRNA gene sequencing by using the Bruker BioTyper v2.0 software. Following the accurate identification of reference strains, 152 clinical isolates were analyzed. MALDI-TOF MS correctly identified 130 (86%) isolates when the threshold for confident identification was lowered from >2.0 to >1.8 and when an expanded database and protein extracts rather than whole cells were utilized (251). This report demonstrated higher levels of correct identifications than those reported in previous studies due to the fact that the authors rigorously optimized both their analysis and identification protocols. Thus, sample preparation and depth of the database to be utilized are important considerations when utilizing MALDI-TOF MS for the identification of anaerobic organisms.
Comparisons between different MALDI-TOF MS systems for the identification of anaerobic microbes have also been undertaken. Justesen et al. used both the Bruker BioTyper version 3.1 and SARAMIS software to examine 290 clinically relevant anaerobic isolates compared to 16S rRNA gene sequencing and reported that the Bruker system generated more correct identifications at the species level but also more incorrect identifications overall (252). This study also highlights important questions regarding database content and expansion and rightfully proposed that cutoff values associated with a confident identification may need to be adjusted on a genus- and species-specific basis. Of the isolates that could not be reliably identified, a large proportion of them belonged to the group of metronidazole-resistant Gram-positive rods, despite spectra being available for members of these groups within the system database. The authors concluded that this either could represent an issue with species diversity, which could be solved with database optimization, or could be due to a biological issue with these species, which may result in the need for an optimized extraction protocol for MALDI-TOF MS analysis of these organisms (252).
MALDI-TOF MS Sample Preparation for Identification of Anaerobic Bacteria
Specific protocols for sample preparation for the analysis of anaerobic bacteria have also been investigated. Fournier et al. compared three preanalytical processing methods for MALDI-TOF MS analysis of anaerobic bacteria: direct smear of intact cells and chemical extraction (253). Two hundred thirty-eight consecutive clinical isolates previously identified by conventional phenotypic methods were used, with both chemical extraction and direct-colony analysis being performed following 48 h of growth by MS and the BioTyper version 3.0 software. The authors concluded that there was no significant difference in the numbers of species-level identifications derived between direct-smear and chemical extraction after adjusting the acceptable cutoff score to 1.7, as others have suggested for the identification of anaerobic bacteria. Moreover, some isolates which were not able to be identified following chemical extraction were able to be identified by using direct-smear methods, arguing that in some cases, protein extraction may be detrimental to optimal spectral generation (253).
Similar to published work examining protein extraction for yeast and corynebacteria from the same group (254), Schmitt et al. continued their evaluation of an on-plate method of FA extraction for MALDI-TOF MS identification of anaerobic bacteria. Bacterial isolates were smeared directly onto the MALDI plate, with 1 μl of 70% FA being directly deposited onto the smear. The samples were subsequently overlaid with matrix and analyzed by MALDI-TOF MS. Samples were analyzed with the Bruker BioTyper version 3.0 software, with manufacturer-recommended cutoff log score values being used for genus- and species-level determinations. Identifications determined by MALDI-TOF MS were compared to identifications derived from phenotypic methods, with discrepant samples being resolved by 16S rRNA gene sequencing. Of the 253 isolates included in the study, the BioTyper correctly identified 232 (91.7%) to the genus level and 179 (70.8%) to the species level, with expansion of the database being suggested as a mechanism to improve species-level identifications (255).
Propionibacterium spp.
Propionibacteria represent a significant diagnostic challenge for the clinical microbiology laboratory due to their slow-growing nature and reduced biochemical activity. Despite past challenges with routine identification, the use of MALDI-TOF MS was investigated for the phylogenetic analysis of isolates of Propionibacterium acnes (256). P. acnes classification is based upon the presence of different phylotypes present within the genus, as identified by differences in cell wall sugars, carbohydrate fermentation tests, molecular analysis, and serological methods. Some of these genetic groups were clearly more associated with disease than others. The authors used MALDI-TOF MS to evaluate the ability of MS-based methods to rapidly classify P. acnes isolates into distinct phylotypes compared to MLST. In total, 12 reference and 49 clinical strains were analyzed. Of note, samples processed for MALDI-TOF MS were processed by using ethanol precipitation and formic acid prior to MALDI-TOF MS and analysis by the Bruker BioTyper 3.0 software and database. The MS platform was able to accurately discriminate P. acnes from other Propionibacterium spp. tested. In an evaluation of culture conditions, the authors found that P. acnes was able to be accurately identified to the species level following 24 h of incubation when only relatively little biomass (pinpoint colonies) was available. MALDI-TOF MS was able to group the collection of isolates tested into distinct phylotypes, with more discriminatory MLST methods grouping the isolates into 28 different sequence types. However, considering advantages in time to result and relative assay costs, MALDI-TOF MS represents an attractive mechanism for discrimination between the main phylotypes of P. acnes (256).
Bacteroides spp.
The species-level identification of members of the Bacteroides genus is important, as the species of the isolate encountered can have a substantial bearing on the antibiotic resistance profile of that particular isolate. As such, a number of species-specific investigations using MALDI-TOF MS on members the genus Bacteroides have been undertaken. Nagy et al. utilized a collection of 277 clinical isolates characterized by phenotypic methods including RapID 32A and API20 ANA (bioMérieux) (244). Proteins were additionally extracted and analyzed by MALDI-TOF MS using the BioTyper 2.0 software package. Discrepant identifications between the MS and phenotypic methods were resolved by 16S rRNA gene sequence analysis. All but 7 isolates generated scores of ≥2.0. Of the 270 isolates identified, 23 were discrepant with biochemical testing, and 11 of these isolates were analyzed by 16S rRNA gene sequencing. The MALDI-TOF MS identification matched the 16S rRNA sequencing identification in 10 out of the 11 cases, with the 11th isolate being identified as B. vulgatus by MALDI-TOF MS, but a reliable species identification could not be determined by 16S rRNA gene sequencing. Additionally, biomarker peaks for rapid and accurate discrimination between subtypes of B. fragilis could be identified and were reproducible (244).
A subsequent study of 193 B. fragilis group isolates previously identified by RapID 32A (bioMérieux) was undertaken by Culebras et al. with the goal to further evaluate MALDI-TOF MS with the Bruker BioTyper 2.0 software for the routine identification of Bacteroides spp. (257). When 16S rRNA gene sequencing was used as a reference, MALDI-TOF MS correctly identified 168 (87%) isolates to the species level, whereas the phenotypic method identified 101 (52%). Misidentifications or no identifications by MALDI-TOF MS were consistent with an absence of spectra from the database or with new or closely related species (257). Thus, as with other groups of bacteria analyzed, database expansion could aid in the species-level identification of Bacteroides spp. and perhaps enhance MALDI-TOF MS performance such that more discriminatory types of analysis could be performed, such as grouping of subspecies typing and antibiotic resistance determination among clinical isolates.
Clostridium spp.
Members of the genus Clostridium are very diverse and medically important. In a preliminary analysis, Grosse-Herrenthey et al. evaluated the use of MALDI-TOF MS for the identification of members of the genus Clostridium (258). Sixty-four strains representing 31 distinct species were analyzed with the Bruker BioTyper version 1.1 software. Cells were harvested from plated medium, resuspended in 80% TFA for 5 min to inactivate spores, and then processed for MALDI-TOF MS analysis. A spectral database was compiled and used to analyze a set of clinical isolates from the genus. Media used for bacterial cultivation had minimal bearing on the spectra produced in this study, whereas the spores' presence had more of an impact, leading the investigators to use younger cultures in their analysis to minimize spectral interference. The robustness and accuracy of the spectra generated allowed phylogenetic analysis of the genus, in addition to enhancing the ability of MALDI-TOF MS to provide species-level identifications for a collection of clostridial clinical isolates. In this early study, MALDI-TOF MS provided a rapid and efficient method for determining species-level identifications of members of the genus Clostridium using a dedicated database (258).
Clostridium difficile is a worldwide problem, often causing large nosocomial outbreaks of antibiotic-associated diarrhea in health care settings (259), with disease being mediated by the use of several important virulence determinants (260). A number of typing methodologies were used to examine genetic lineages of C. difficile associated with outbreaks, including ribotyping, PFGE, MLST, and restriction enzyme analysis (REA). Ribotyping represents an analysis of the sizes of fragments generated by PCR, constituting intragenic spacer regions between conserved genes. These PCR fragments are visualized via gel or capillary electrophoresis to generate a pattern that can then be compared to patterns from other strains, resulting in a classification scheme.
Recently, the SARAMIS MALDI-TOF MS system was evaluated as a tool to discern different ribotypes among isolates of C. difficile (261). Direct cells from 24-h cultures were deposited onto the MALDI-TOF MS plate, and matrix solution was applied. A recognized collection of isolates (ECDC-Brazier) encompassing 25 different ribotypes of C. difficile was used to generate a dedicated database using derived SuperSpectra. By using this method, four specific ribotypes were identified, including ribotype 027 (NAP1), which is associated with epidemic outbreaks of C. difficile. A collection of 355 PCR-ribotyped clinical isolates was tested against the database. Using parameters determined in-house for database matching, all ribotype 001 isolates (n = 248), all ribotype 027 isolates (n = 7), and 6 of 7 ribotype 126/078 isolates could not be further separated due to high levels of similarity. Importantly, the authors concluded that this method may represent a mechanism for the rapid identification and characterization of C. difficile isolates in the future, but more complete databases encompassing a wider variety of ribotypes, including isolates from diverse geographical locations, will be needed to assess MALDI-TOF MS capabilities of discriminating between closely related strains (261).
FUNGI: YEASTS
The rapid analysis of fungi by MALDI-TOF MS has the potential to revolutionize medical mycology. Identification of fungi in the clinical laboratory is most traditionally associated with the use of selective or differential media in addition to both manual and automated biochemical identification systems. By implementing a rapid system which functions on a universal platform for the definitive identification of fungal specimens, significant reductions in time to diagnosis and laboratory costs could be realized, in addition to benefits with respect to patient care.
An appropriate drug regimen with antifungal treatment is a critical component to successfully cure serious fungal infections, but targeted therapy is difficult. It requires several days for growth and identification by conventional biochemical and morphological approaches, and the results may sometimes be inconclusive. Species-specific susceptibility patterns can help clinicians make therapeutic decisions, but determination of antifungal susceptibility can take weeks in some cases. Protein content and expression levels of fungal isolates may be affected by growth states and mycological life cycles; therefore, standardization of medium and growth phase will be important when using MS technology for evaluation of therapeutics. Still, MALDI-TOF MS holds the promise of significantly accelerating these processes, substantially improving fungal diagnostics and patient treatment.
For 138 common and 103 archived strains of yeast, MALDI-TOF MS was compared to phenotypic testing for yeast identification, with 96.3% and 84.5% accurate species-level identifications (spectral scores, ≥1.8) for common and archived strains, respectively. The authors additionally performed a cost analysis to determine if MALDI-TOF MS was financially competitive with other rapid identification systems for fungal identification, determining that operating costs were lower than those of most conventional and molecular testing systems in terms of reagent costs and hands-on time required for sample processing (262).
Putignani et al. analyzed spectra by the Bruker BioTyper software from 303 clinical isolates using standard pattern matching. Identifications were compared to identifications by a reference biochemical-based system (Vitek-2), and when results were discordant, BioTyper identifications were verified with genotyping identifications obtained by sequencing of the 25S-28S rRNA hypervariable D2 region. Of the 26 discordant results, only 5 appeared to be real once further determinative testing was performed. The BioTyper showed high analytical performance and was able to discriminate patterns for strain typing of some species (263).
The Bruker BioTyper has the most publications describing yeast identification by MALDI-TOF MS. In perhaps the largest analysis published to date, a diverse collection of 1,192 yeast and yeast-like isolates was tested by Bader et al., who compared BioTyper and SARAMIS databases to a classical differentiation scheme based on microscopic and biochemical characteristics. For 95.1% of the isolates, all three methods produced the correct species identification, with improved identification noted for both MALDI-TOF MS systems. Closely related species, such as Candida orthopsilosis, C. metapsilosis, and C. parapsilosis or Candida glabrata and C. bracarensis, could be resolved by both MALDI-TOF MS systems but not by the biochemical methods (264).
Yeast Sample Preparation for MALDI-TOF MS
In a manner similar to bacterial studies, the preanalytical steps associated with processing of fungi were previously investigated. Theel et al. evaluated an FA-based on-plate method for the MALDI-TOF MS-based identification of fungi from the clinical laboratory. In total, 90 clinical yeast isolates were analyzed by directly smearing a small amount of biomass onto a MALDI plate and overlaying the smear with 1 μl 70% FA, followed by drying and addition of matrix. MALDI-TOF MS analysis was performed with the Bruker BioTyper 3.0 database, and a log score of 1.7 or higher was accepted for a species-level identification. MS identifications were compared to those derived by standard phenotypic methods, with discrepant results being resolved by 28S rRNA gene sequencing. Eighty-six of 90 (95.6%) isolates were identified to the genus level, and 73/90 (81.1%) were identified to the species level, with a single misidentification being reported. Finally, in comparison to a commonly used ethanol-based extraction method described as being more time-consuming and requiring additional consumables, the direct on-plate method performed better for species-level identification of clinical yeast isolates (254). A second comparison of this modified method determined that the modified method was suitable for rapid identification of yeast isolates; however, the log score values for fungal identification needed to be lowered to accommodate the lower spectral scores generated by using this method (265).
The age and amount of fungal culture, a second aspect of preanalytical processing of yeast for MALDI-TOF MS analysis, have also been investigated by Goyer et al. The authors determined that only a single colony of yeast isolated from CHROMAgar needed be isolated to provide accurate identification of yeast isolates, instead of five colonies, which had been reported elsewhere (210). Forty-eight-hour and 72-h cultures were also compared to determine if culture age had a bearing on identification. No significant difference was found when using 48- or 72-h cultures. Good identification was also achieved by using 24-h cultures, but this method was deemed not suitable for routine practice, as 48 h is necessary for full color development with chromogenic medium in order to accurately detect fungal mixtures (38).
In the following sections, we review the literature pertaining to the identification of specific groups of fungal pathogens by MALDI-TOF MS.
Candida spp.
Candida species infections are a major health problem worldwide. The epidemiology of candidemia has substantially changed over the last decades with the emergence of individual species formerly classified into the nonalbicans Candida species group, known for variability in susceptibility to antifungals and typically isolated from those most unlikely to battle the infection, the immunocompromised and other compromised subpopulations. This inherent variability highlights the need for the proper identification of Candida spp. to enhance regional choices for prophylaxis and empirical treatment and to further characterize the epidemiology of infections. For example, recent studies of Candida species outbreaks showed an increased incidence of bloodstream infections in neonatal intensive care units (NICUs) caused by C. parapsilosis. Species-specific differentiation of two closely related yeasts, Candida albicans and C. dubliniensis, is important to better understand the epidemiology and virulence of C. dubliniensis.
MALDI-TOF MS shows its potential for the rapid identification of C. albicans and related species. In a study by Pinto et al., MALDI-TOF MS was performed on a 264-strain library composed of clinical and reference strains. Discordant and unreliable identifications were resolved by sequencing of the internal transcribed spacer (ITS) region of the rRNA gene cluster. In this analysis, 20 (67%; 16 species) and 24 (80%) of 30 reference strains were identified to the species (spectral score, ≥2.0) and genus (score, ≥1.70) levels, respectively. Of clinical isolates, 140/167 (84%) strains were correctly identified with scores of ≥2.0, and 160/167 (96%) strains were correctly identified with scores of ≥1.70; among Candida spp. (n = 148), correct species assignment with scores of ≥2.0, and ≥1.70 were obtained for 86% and 96% of isolates, respectively (versus 76.4% correct assignments by biochemical methods). MALDI-TOF MS identified uncommon Candida spp., differentiated C. parapsilosis from C. orthopsilosis and C. metapsilosis, and distinguished between C. glabrata, C. nivariensis, and C. bracarensis. Yeasts with scores of <1.70 included 4/12 Cryptococcus neoformans isolates. When protein extraction was used, there were no misidentifications in the data set (266).
A total of 18 type collection strains and 267 recent clinical isolates (Candida [n = 250], Cryptococcus, Saccharomyces, Trichosporon, Geotrichum, Pichia, and Blastoschizomyces spp.) were identified by BioTyper analysis. The results were compared with those obtained by conventional phenotyping and biochemical tests including the API ID 32C system (bioMérieux) and other biochemical tests. After complementation of the database, with species identification from 26S rRNA gene sequencing, accurate species identification by MALDI-TOF MS was achieved for all isolates. In contrast, the API ID 32C biochemical diagnostic system identified 244 isolates (210).
Discrimination between C. albicans and C. dubliniensis is possible by using MS methods, as demonstrated by analysis of reference strains from type culture collections and other well-characterized isolates. The spectra of C. albicans and C. dubliniensis easily differentiated species, and further study revealed that each species consists of several clades, which could be distinguished by MALDI-TOF MS (267).
Dermatological yeast isolates were tested by Seyfarth et al. using MALDI-TOF MS, the API ID 32C system, and sequencing of the ITS regions of ribosomal DNA. The accuracy of MALDI-TOF MS compared to the results derived from ITS sequence analysis was 94%, whereas API ID 32C was accurate for only 84.3% of the isolates. Species tested included C. albicans (41.9%), C. parapsilosis (20.3%), C. glabrata (10.8%), and C. krusei (6 isolates) (8.1%). Rarely isolated yeasts, including C. colliculosa, C. famata, C. guilliermondii, C. lusitaniae, and C. tropicalis, as well as Geotrichum candidum, Rhodotorula mucilaginosa, and Trichosporon mucoides were also correctly identified. For the API ID system, C. krusei was incorrectly identified as C. inconspicua/C. norvegensis, Candida tropicalis, or Geotrichum capitatum. In contrast, all C. krusei strains were correctly identified with discriminatory power comparatively similar to that of ITS sequence analysis (268).
Similar to the BioTyper, accurate results were observed by using the Confidence Axima system (Shimadzu) with Shimadzu Launchpad software and the SARAMIS database (AnagnosTec GmbH). Nonalbicans Candida spp. (n = 73) isolated from noninvasive samples were tested by using the Vitek-2 systems YST and API CAUX, identifying 67 yeast isolates to the species level and 6 to the genus level. Discrepancies were resolved by SeptiFast LightCycler multiplex PCR, C. glabrata-specific PCR, and enzymatic digestion (269).
For the Andromas system, the accuracy for identification of Candida spp. is also quite high. Using the Andromas software and MALDI-TOF MS, Bille et al. analyzed 162 yeast isolates and found 96.3% accuracy for the first acquisition of spectra and 98.8% accuracy after a second acquisition (53).
After constructing an in-house database (270), Marklein et al. used the BioTyper system to identify 267 clinical isolates and 18 collection strains of yeast and yeast-like fungi (210). Candida spp. were correctly identified for 240/250 (96%) clinical isolates on the first attempt, with no false-positive results. Investigation of the discrepancies between biochemical MALDI-TOF MS identifications by 26S rRNA gene sequencing resulted in 100% identification to the species level. The same procedure used for identification of the 17 clinical isolates of yeast-like fungi identified 100% of isolates to the species level.
Stevenson et al. reported accurate identification of 194 clinical isolates after implementation of a database containing 109 type and reference strains of yeasts from 44 different species. Correct identification to the species level with a score of ≥1.8 was obtained for 192 (99%) isolates (271). Finally, in the largest set of yeasts ever tested for analysis of MALDI-TOF MS performance for identification, Bader et al. tested two systems, BioTyper and SARAMIS (264). Twenty-one species, representing 1,148 isolates of yeast, were tested, with large sample sizes for C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis and fewer samples for other Candida and non-Candida species. The results obtained with these two systems were comparable, with identification rates at the species level of ≥99% for isolates that were represented in the respective databases. Both methods each misidentified two single isolates absent from the respective databases as a wrong species, while the biochemical approach (ID 32C; bioMérieux) misidentified 30 isolates as a wrong species, instead of reporting them as “unknown.” In contrast, the success rate for classical identification techniques was 96.7%. Closely related species (e.g., Candida orthopsilosis, C. metapsilosis, and C. parapsilosis or Candida glabrata and C. bracarensis) were resolved by both MALDI-TOF MS systems but not by the biochemical approach.
Cryptococcus spp.
The Cryptococcus neoformans-C. gattii species complex comprises two sibling species that are divided into eight major molecular types that differ in host range, epidemiology, virulence, antifungal susceptibility, and geographic distribution. Protein extracts obtained from 164 C. neoformans-C. gattii isolates by the formic acid extraction method were tested, including four interspecies hybrids. The mass spectra correctly identified 100% of isolates, grouped each isolate according to the currently recognized species C. neoformans and C. gattii, and detected potential hybrids. In addition, all isolates were clearly separated according to their major molecular type (272). In another study, Stevenson et al. used Bruker Daltonics MALDI BioTyper software and created a spectral database library for 109 type and reference strains of yeast (44 species in 8 genera). The database was 99.0% accurate for 194 clinical isolates (23 species in 6 genera) (271).
Compared to molecular-based methods, MALDI-TOF MS fares very well. Kaleta et al. evaluated identification of yeast to the species level with both the Ibis T5000 PCR-ESI-MS and the Bruker MALDI BioTyper 2.0 platforms, versus Vitek-2 analysis as the reference standard identification. PCR-ESI-MS and MALDI-TOF MS were equivalent in their abilities to characterize yeast with respect to the reference standard (23). Compared to DNA sequence analysis, MALDI-TOF MS correctly identified 100% of Cryptococcus species, distinguishing the notable pathogens C. neoformans and C. gattii. Identification was greatly enhanced by supplementing a commercial spectral library with additional entries to account for subspecies variability (273).
FILAMENTOUS FUNGI AND MOLDS
Delayed and incorrect diagnoses are potential risk factors leading to high mortality rates due to invasive aspergillosis and other systemic fungal infections. Because of variability of growth patterns and difficulty obtaining a standardized inoculum, mold identification proves to be more difficult than identification of bacteria and yeast. Phenotypic identification of molds requires experienced and skilled mycologists, who are not available to all laboratories, while molecular methods experience limitations with respect to difficulties associated with lysis and PCR inhibition of mold specimens (274). Moreover, phenotypic identification is slow and labor-intensive, with many species being phenotypically similar but genotypically distinct or different in their propensity to cause disease (275). Therefore, a rapid mechanism requiring minimal sample preparation and analysis by the technician for the identification of these organisms is appealing. MALDI-TOF MS has been evaluated to potentially fill this niche (reviewed here and in reference 276). Initial yet promising reports are available; however, more optimization will need to occur before the breadth of clinical mold infection diagnoses can be accomplished by using this technology.
A number of studies have examined the ability of MALDI-TOF MS to identify molds, and here we highlight those which have focused on mold identification in a general sense. Cassagne et al. examined a number of sample preparation conditions (discussed below) prior to their construction of a dedicated spectral library for the identification of clinical mold isolates using 143 reference spectra. Clinical testing of 177 sequential isolates using the constructed database followed, with MALDI-TOF MS-derived identifications being compared to phenotypic identifications. 28S rRNA gene sequencing was used to resolve discrepant samples. MALDI-TOF MS correctly identified 87% (154/177) of isolates, with a 12% (21/177) failure rate for species not included in the reference library, and two misidentifications (274).
In an evaluation of the ability of the technology to identify molds, Lau et al. also constructed a MALDI-TOF MS database (named the NIH Mold Database) to facilitate the identification of clinical isolates. This database was comprised of spectra from 249 reference isolates and members of the collection of the ATCC. When challenged with spectra from a collection of 421 clinical isolates, the NIH Mold Database provided acceptable species-level identification for 88.9% (370/421) of isolates tested. Importantly, when the isolates were tested by using the Bruker BioTyper library (version 3.1) alone, only 3 (0.7%) isolates were identified, highlighting the need for database expansion for the identification of clinical mold isolates. No isolates were misidentified by MALDI-TOF MS. Implementation of the database has improved laboratory efficiency and turnaround time in the experience of the authors, and the database could be made available to other laboratories for subsequent evaluations (275).
Mold Sample Preparation for MALDI-TOF MS
Sample preparation methods for MALDI-TOF MS analysis of fungal hyphae and spores have been investigated. Cassagne et al. evaluated five methods for the preparation of samples from this material, using various combinations of culture conditions (agar versus broth), ethanol and no ethanol treatments, heat treatments, and mechanical lysis procedures to determine the method which yielded the best results. Following these procedures, extraction with FA was performed, and the sample was analyzed by MALDI-TOF MS. Following rigorous statistical analysis of spectra and evaluation of the amount of labor necessary to complete the procedure, a protocol was elucidated, which was easily performed and provided robust results. Fungi were cultivated on a Sabouraud gentamicin-chloramphenicol agar plate for 72 h at 27°C and then extracted with FA following incubation in ethanol. Acetonitrile was added, and the mixture was centrifuged, with the resulting supernatant being analyzed by MALDI-TOF MS (274).
Lau et al. reported a mechanical lysis method for the preparation of samples from mold specimens for MALDI-TOF MS analysis. In their study, a small piece of mold isolate was resuspended in ethanol and zirconia-silica beads, emulsified thoroughly, and vortexed. Following centrifugation, the supernatant was removed, and the remaining pellet was resuspended in 70% FA, vortexed a second time, and centrifuged. The resulting supernatant was then either analyzed immediately or stored for up to 1 week at −20°C for subsequent analysis by MALDI-TOF MS (275). Both methods worked well for the investigators in their respective studies, and both protocols will add value toward the future standardization of sample preparation methods for mold specimens. Irrespective of which method(s) is chosen in the future, the use of a biosafety cabinet during sample processing and the minimization of aerosols will be important considerations during protocol selection to ensure the safety of laboratory personnel working in close proximity.
Aspergillus spp.
MALDI-TOF MS-based identification of Aspergillus has been undertaken in the context of both clinical evaluations of the technology in a routine setting as well as focused studies evaluating the technology for species- and strain-level identifications. In a clinical evaluation of the Vitek-MS system, Iriart et al. compared the performance of the Vitek-MS system to those of both routine laboratory techniques and Vitek-2. The Vitek-MS system performed well in the identification of yeasts and a limited number of Aspergillus spp. (93.2% correct identifications) (277). Bille et al. reported a similar overall identification rate for Aspergillus spp. of 98.4% (63/64) in their study using the Andromas strategy (53).
In an analysis of Aspergillus, Fusarium, and Mucorales, De Carolis reported the construction of a specific database dedicated to the identification of members of these fungal classifications by using a collection of 55 reference strains and the BioTyper 2.0 software. The database was challenged with 103 blind-coded isolates to determine discriminatory ability. Excluding isolates that were not contained in the database, MALDI-TOF MS successfully identified 96.8% (91/94) of isolates to the species level (278). When examining invasive Aspergillus isolates, Pan et al. reported that more of the differential spectral peaks were present at the second stage of sporulation, which contains differentiated structures, than at the first stage, which is comprised primarily of vegetative hyphae (279). Finally, Hettick et al. used MALDI-TOF MS to derive spectra for 12 species of Aspergillus and five different strains of A. flavus. Classification of each species and strain of Aspergillus tested was accomplished with 100% accuracy in their analysis (280).
Fusarium spp.
Fusarium spp. are significant pathogens, causing infection primarily in immunocompromised patients. Identification of these species is important for patient management, but conventional identification strategies can be difficult due to phenotypic polymorphism. Aspects of MALDI-TOF MS analysis have been investigated for Fusarium, including preanalytical sample preparation (281, 282). Marinach-Patrice et al. studied 62 Fusarium strains representing nine species. Molecular identification using tef1 gene sequencing was the reference standard compared to MALDI-TOF MS. The most frequently isolated species, including F. solani, F. oxysporum, F. verticillioides, F. proliferatum, and F. dimerum, were tested, with MALDI-TOF MS correctly identifying 57/61 strains. Four species not represented in the MS database were not identified. MALDI-TOF MS yielded results within 1 h, demonstrating its utility for identification of Fusarium spp. (283).
Dermatophytes
Dermatophytes are keratinophilic fungi that cause superficial infections in humans by infecting nails, hair, and the stratum corneum, the outermost layer of the epidermis (284). Dermatophyte taxonomy is complex and has undergone recent revision with the advent of more sophisticated molecular assays for identification as well as genomic characterization. Molecular analysis is the gold standard for identification of these fungi at the species level (285). Phenotypic identifications are based upon growth on selective media, colony morphology, and microscopic observation of conidia (286). A number of studies have been dedicated to the identification and characterization of these fungi by MALDI-TOF MS. Erhard et al. were the first to evaluate MALDI-TOF MS specifically with the aim of identifying species causing onychomycosis and tinea pedis with an early version of the SARAMIS database. Although the sample size was small, distinct differences in generated spectra could be identified, and results of MALDI-TOF MS identification were consistent with the reference standard (sequencing) for identification (287).
Theel et al. reported the use of the Bruker BioTyper version 3.0 for identification of dermatophyte species compared to 28S rRNA gene sequencing. Performance of the BioTyper database was compared using an unmodified version and a version supplemented with in-house-generated dermatophyte spectra. For optimal performance of MALDI-TOF MS for dermatophyte identification in this study, supplementation of the commercial library was necessary, as was reducing the log score cutoff values for genus-level (from ≥1.7 to ≥1.5) and species-level (from ≥2 to ≥1.7) identifications, as has been reported for other fungi and the identification of bacteria directly from patient specimens (286).
Work from Alshawa and colleagues described the generation of their own spectral database for dermatophyte identification, constructed from 50 reference strains comprising 12 dermatophyte species and 2 species of Neoscytalidium, and its subsequent evaluation using a collection of 381 clinical isolates. MALDI-TOF MS was performed, and spectral evaluation was undertaken by using the Andromas system. Samples were harvested and resuspended in FA prior to analysis, reflecting a relatively simple method compared to those reported by other studies highlighted in this section. Compared to conventional methods, MALDI-TOF MS identified 331 of 360 (91.9%) dermatophyte isolates and 18 of 21 (85.7%) Neoscytalidium isolates tested. The authors concluded that MALDI-TOF MS could replace conventional methods of dermatophyte identification in the near future in both routine and specialized diagnostic laboratories (288).
Nenoff et al. reported a recent substantial study in which the authors analyzed 285 isolates comprising 21 different species from the genera Trichophyton, Microsporum, Epidermophyton, and Arthroderma using MALDI-TOF MS and the SARAMIS system as well as gene sequencing following the construction of an in-house database. Identifications derived by using conventional algorithms matched MS-derived identifications in 78.2% of cases. Importantly, MALDI-TOF MS matched identifications derived by gene sequencing in 99.3% (283/285) of cases, demonstrating the high level of accuracy the MS system using the in-house database (289) as well as the need for a rapid and accurate mechanism of dermatophyte identification to enhance conventional mechanisms.
Pseudallescheria-Scedosporium Complex
Members of the Pseudallescheria-Scedosporium complex (PSC) are commonly associated with opportunistic mold infections in both immunocompetent and immunocompromised patients. These molds present a significant diagnostic challenge, as they cannot be distinguished from other filamentous fungi by histological examination alone, and rapid identification is necessary due to their poor response to certain antifungal agents (290). Twenty-two PSC reference strains and three clinical isolates were tested by using MALDI-TOF MS and the BioTyper software (Bruker). MS identification was stable after the fungi were subcultured over a 1-month period. While neither culture medium (Sabouraud versus malt extract) nor protein extraction methods (FA versus TFA) significantly influenced the quality of the MS identifications, identification was considerably enhanced between days 3 and 6 of incubation. Further analysis is necessary to evaluate the ability of MALDI-TOF MS to discriminate between recently identified species within the Pseudallescheria boydii group of organisms (291).
Penicillium spp.
Hettick et al. used MALDI-TOF MS to identify 12 species within the genus Penicillium. Analysis of undisrupted cells generated consistently poorer spectra, so a disruption protocol was utilized. Conidia and spores were collected from cultures, resuspended in TFA-acetonitrile with glass beads, vortexed, and centrifuged, with the resulting supernatant being used for MALDI-TOF MS analysis. Although a small collection of isolates was utilized and spectral analysis was done in-house without the aid of standardized software, Penicillium species within this collection were able to be discriminated to the species level with 100% accuracy (292).
Lichtheimia spp.
The genus Lichtheimia is separated into five species, with only three (L. corymbifera, L. ramosa, and L. ornata) being associated with clinical disease (293). These organisms are saprophytic fungi that inhabit soil and decaying plant matter, which can cause relatively rare yet dramatic infections requiring medical intervention in immunocompromised patients (294). Schrödl et al. recently utilized MALDI-TOF MS to identify members of the genus Lichtheimia. Samples were prepared by using a combination of ethanol inactivation, freeze-thawing, and FA extraction prior to matrix overlay. MALDI-TOF MS identifications were compared to those derived by rRNA gene sequencing. A spectral database was constructed by using 12 strains of Lichtheimia and seven related genera, which was subsequently challenged with 34 additional clinical and environmental isolates. MALDI-TOF MS was able to identify the Lichtheimia genus in 100% of cases and was also able to discriminate specific species within the genus for 32 out of 34 isolates (295).
Extended Testing of Fungal Isolates by MALDI-TOF MS: Antifungals and Epidemiology
Recently, MALDI-TOF MS profiling of fungal isolates has been taken in exciting new directions beyond standard microbial identification. Although in its infancy, antifungal testing using MALDI-TOF MS is a possibility. MALDI-TOF MS was evaluated for testing susceptibility to caspofungin of wild-type and fks mutant isolates of Candida spp. Caspofungin functions by inhibition of the synthesis of cell wall components of medically important fungi, including Candida spp. and Aspergillus spp. (296), with mutations in fks genes conferring reduced susceptibility to echinocandin antifungals, including caspofungin (297). Complete essential agreement was observed with the CLSI reference method, with categorical agreement for 94.1% of the Candida spp. Thus, MALDI-TOF MS is a reliable and accurate method to detect fungal isolates with reduced caspofungin susceptibility (278).
Epidemiological testing of fungal isolates has also been investigated using MALDI-TOF MS. Strain typing of yeast was attempted when 19 strains of C. parapsilosis isolated from blood cultures of neonates were genotyped by the amplification of eight simple sequence repeat (SSR) markers and by MALDI-TOF MS. Both methods were rapid and effective in highlighting identical strains and studying microevolutionary changes in the yeast population (298). Further investigations are warranted prior to using MALDI-TOF MS for outbreak investigations or rapid antifungal susceptibility testing of yeast isolates; however, these preliminary investigations provide promising results and highlight the diverse utility of the technology.
USE OF MALDI-TOF MS IN EPIDEMIOLOGY
Although rapid and accurate microbial identification by MALDI-TOF MS will be what primarily attracts laboratories to mass spectrometry-based analysis methods, the potential for MALDI-TOF MS to provide critical epidemiological data cannot be understated. One of the great challenges faced by clinical laboratory scientists (CLSs), infection prevention practitioners, clinicians, and public health laboratories is determining strain-specific data for representative taxonomy of clinical isolates in outbreak situations. In some specific cases (i.e., Salmonella spp. and Streptococcus spp.), microbial typing is particularly challenging and often requires additional time-consuming second-stage testing in order to correctly determine serotype, subspecies, or other taxonomic classifications. This challenge can be further confounded when the specific implications of a serotype or subspecies in the context of an infection is ambiguous.
Due to their complexity, many strain typing methods require a specialized laboratory test method and additional instrumental or material demands. Therefore, these analyses are not routinely performed in every laboratory, resulting in an increased amount of “send-out” testing and delays in investigation of hospital-associated infections and outbreaks. This can result in delayed time to detection, loss of important epidemiological data, increased testing costs, and potentially inaccurate or inconsistent results. These minute but important details may be performed in real time on clinical specimens by the CLS in the laboratory of origin. Having this information readily available will assist physicians, nurses, pharmacists, infection control personnel, and local and state authorities in treating and tracking infectious microbes and their associated epidemiology.
Much of the strain typing research has been completed in applied research settings outside the clinical laboratory, using specialized databases constructed through the analysis of expanded sample collections of isolates consisting primarily of a specific group of organisms, genus, or species. The ability of MALDI-TOF MS to discriminate between these highly related organisms, as demonstrated in these studies, has profound implications for the clinical microbiology laboratory. As databases used for the species- and strain-specific identification of organisms mature, a second tier of epidemiological capabilities may emerge, using the same or similar MALDI-TOF MS instrumentation as that used for microbial identification. The implementation of an instrument into the clinical laboratory workflow that can not only identify microorganisms but also provide rapid and accurate epidemiological data about identified clinical isolates has the potential to expand the role of the microbiology laboratory beyond microbial diagnostics and further refine critical roles in hospital infection control, national surveillance of microbial outbreaks, and biodefense.
While the use of MALDI-TOF MS for strain typing is still in its infancy, the advantages that this type of data could provide to both hospital and public health officials are profound. As microbes continue to change at the genetic level and evolve new mechanisms for infection and resistance to antibiotics, the microbiology laboratory must too evolve, implementing new mechanisms for increased accuracy, speed, and sensitivity for the identification and characterization of pathogenic microbes.
MS IDENTIFICATION OF BACTERIA DIRECTLY FROM PATIENT SPECIMENS
While MALDI-TOF MS has been extensively evaluated as a universal platform for the proteomic analysis and identification of bacteria and yeasts from culture media, the technology is also being exploited to analyze patient specimens directly, completely bypassing the need for culture by detecting the presence or absence of pathogens in the clinical specimen proper. This type of direct analysis proved impossible before the advent of molecular analysis (Fig. 4).
Fig 4.
Current position of MALDI-TOF MS in the workflow of the clinical microbiology laboratory, including the current options for analysis of bacteria directly from patient specimens. The MALDI-TOF MS instrument fits easily into the clinical microbiology workflow, occupying the position once held by instruments for automated phenotypic-based identifications (blue arrows). Evaluated mechanisms for the processing of samples directly from patient specimens are included (hatched red arrows), as are options for the use of traditional (green arrows) and MALDI-TOF MS (hatched green arrows) mechanisms. Finally, results are imported into the laboratory information system from the MADLI-TOF MS instrument or other instruments and reported to physicians and pharmacists as indicated. BAL, bronchoalveolar lavage.
Due to its exquisite sensitivity, MALDI-TOF MS provides an attractive mechanism to be used either in place of or in concert with PCR-based strategies for direct detection of pathogens from clinical material, as no amplification of the target material is required. The ability of both molecular and proteomic approaches to identify targets in these types of samples can also be enhanced by preliminary processing of these samples, removing some of the elements (proteins, nucleic acids, and cellular debris, etc.) that can inhibit analysis. In this section, we review the current knowledge regarding the testing of patient specimens directly in conjunction with MALDI-TOF MS.
Urine
Proteomic profiling of human urine has been utilized for quite some time to identify disease-specific biomarkers, and previous analysis has demonstrated that a number of factors, including storage time, pH, and the number of freeze-thaw cycles, influenced analysis. Prior to the use of MALDI-TOF MS for the diagnosis of urinary tract infections (UTIs), the presence of blood and bacteria in the urine was known to interfere with urinary proteomic analysis, specifically altering key peptide mass signals in the sample (299). Additional studies similarly suggested that bacterial overgrowth of the urine could hamper proteomic analysis and recommended that samples be centrifuged and stored immediately at 4°C and have boric acid or NaN3 added to prevent bacteria from overgrowing (300). It remains to be seen which specimen handling conditions are necessary for optimal identification of bacteria in urinary samples, as higher bacterial burdens within the specimen could potentially simplify the detection of the bacteria in urine samples at the expense of the rest of the urinary proteome.
One of the first studies to directly compare MALDI-TOF MS to conventional methods of bacterial identification in urine specimens concluded that MALDI-TOF MS was a promising method for this type of analysis following differential centrifugation. Low-speed centrifugation was utilized to remove leukocytes, followed by high-speed centrifugation to collect bacteria in the sample, and these intact cells were analyzed by MALDI-TOF MS. In an analysis of 269 samples detected as positive by urine particle analysis (urine microscopy), 20 were positive in the screening device and negative by both culture and MALDI-TOF MS analysis using the Bruker BioTyper 2.0 database. Two hundred twenty of these samples showed high levels of bacterial growth (>105 CFU/ml), and identifications for this group were consistent at the genus level in 204 (92.7%) cases and consistent at the species level in 202 (91.8%) cases compared to identifications made by standard methods (Wider MIC/ID [Microscan] and Vitek-2 [bioMérieux]). The authors concluded that MALDI-TOF MS provided good identification from urine, particularly in cases where Gram-negative bacteria were present at high levels (301). A second study by the same group further addressed the issue of sample preparation with respect to direct urine samples prior to MALDI-TOF MS analysis. Ferreira et al. reported that following differential centrifugation to remove cell debris and leukocytes, ethanol precipitation of proteins followed by formic acid and acetonitrile resuspension of proteins significantly improved the performance of MALDI-TOF MS for the identification of urinary pathogens compared to the intact-cell method, as previously reported, leading to the reporting of analytical results within minutes (138, 301).
An additional study by Kohling et al. investigated the ability of MALDI-TOF MS to identify bacteria directly from urine specimens and compared the accuracy of this method to identifications derived by either phenotypic (Vitek-2, API, and Microscan Walk Away) or molecular (long-chain fatty acid pattern analysis by gas chromatography) methods. Although a smaller sample set than those used in studies discussed above in this section (n = 107), the authors similarly determined that MALDI-TOF MS was a reliable methodology for detection of bacteria directly from urinary specimens. Additionally, the authors concluded that this reliability was applicable to samples with bacterial densities as low as 103 CFU/ml (302).
The most recent studies to date regarding the direct analysis of urinary specimens were aimed at incorporating MALDI-TOF MS into the laboratory workflow in conjunction with the urinalysis section of the clinical laboratory. In a comprehensive study by Wang et al., urine flow cytometry was utilized as a prescreening method to eliminate negative samples. Samples determined to be positive for the presence of bacteria (>105 CFU/ml) by flow cytometry were processed for MALDI-TOF MS for bacterial identification. Samples were differentially centrifuged to remove leukocytes from the suspension, followed by high-speed centrifugation to pellet bacteria. The bacterial pellet isolated from the aqueous urine was subsequently treated with formic acid and acetonitrile, and extracted proteins were analyzed by MALDI-TOF MS (BioTyper database) for bacterial identification. These identifications were compared to identifications derived from cultured bacteria by using phenotypic methods (Vitek-2), with discrepant identifications being resolved by 16S rRNA sequencing. Of the 1,456 samples from patients with UTI symptoms included in the study, 932 (64%) were determined to be negative for the presence of bacteria. The correct result (no bacteria present or correct bacterial identification) was obtained for 1,381 of the 1,456 cases (94.8%). Among the 430 positive samples, 8 were found to be discrepant between the MALDI-TOF MS and the Vitek-2 results, and all 8 identifications by MALDI-TOF MS were confirmed by 16S rRNA gene sequencing (303).
It is clear that MALDI-TOF MS has the potential to be used with great success for the direct identification of bacteria in urine samples, potentially negating the need for urine culture in complicated cases. However, while tremendous work has been dedicated to the direct analysis of urine specimens by MALDI-TOF MS, there are number of questions remaining to be examined. Both Ferreria et al. and Wang et al. reported that MALDI-TOF MS could not accurately identify mixed bacteria present in urinary specimens (138, 303). It remains to be determined if improvements to the respective databases will allow the accurate identification of mixed bacteria in urinary specimens. Additionally, no standardized methodology is currently available regarding the processing of specimens prior to analysis by MALDI-TOF MS. Simple protein extraction (i.e., formic acid or ethanol-acetonitrile) is demonstrated to be cost-effective and fast and significantly enhances the ability of MALDI-TOF MS to correctly identify bacteria from urine specimens, which would be a reasonable step to include in a standardization of processing methods prior to MALDI-TOF MS analysis. Moreover, many clinical laboratories are already performing urine flow cytometry, with urine cultures often being submitted for reflex testing following a positive microscopic observation. There is a possibility that MALDI-TOF MS could be added to the testing menu and performed following a positive urine microscopic test, as described previously (303). Irrespective of future standardization in the specimen-processing workflow, MALDI-TOF MS currently represents a robust and accurate technology for the identification and characterization of single bacterial species present in direct urine specimens.
Cerebrospinal Fluid
Similar to urine, cerebrospinal fluid (CSF) has been used for proteomic profiling for the diagnosis of disease. The presence or absence of specific proteins in patient CSF was used as a biomarker for a number of neurological disorders. Bacterial meningitis represents one of the most serious and clinically significant manifestations of bacterial infection and represents a situation where fast and accurate detection of the offending bacterial agent is paramount. Currently, the detection of bacterial pathogens responsible for meningitis is accomplished via Gram staining of the CSF and looking for the presence of bacteria. Samples generating negative smears are cultured to rule out the presence of circulating bacteria, while patients with positive cultures are treated immediately with broad-spectrum antibiotics based upon the Gram stain result until a definitive identification can be reached and targeted antimicrobial therapy can be administered.
While CSF can be analyzed by MALDI-TOF MS, there are very few reports in the literature describing the use of MALDI-TOF MS for the direct identification of bacteria from CSF. One such report describes the diagnosis of pneumococcal meningitis in a 46-year-old man (304). Following sample acquisition, the CSF was processed in a manner similar to that discussed above for urine samples: low-speed centrifugation to remove leukocytes followed by high-speed centrifugation to pellet bacteria. The pelleted debris was then solubilized in formic acid-acetonitrile as described above, centrifuged, and analyzed by MALDI-TOF MS using the BioTyper 2.0 database. The identification generated by MALDI-TOF MS analysis was interpreted as being valid species-level identification following manual manipulation of the data, although the automatic analysis would have allowed for only a statistically confident genus-level identification. Irrespective of the genus or species level of identification, the use of this technique in combination with Gram stain and traditional bacterial culture could represent an important turning point in the diagnosis of bacterial meningitis, increasing sensitivity and decreasing time to diagnosis and allowing for targeted and aggressive antibiotic therapy for a patient population that is critically ill. This is an application of MALDI-TOF MS that warrants significant further investigation.
Identification of Bacteria Directly from Blood Cultures
Direct identification of bacteria and yeast from blood culture bottle broth is a promising option for MADLI-TOF methods with the potential to speed the identification process (305, 306). After preprocessing of the blood culture broth to limit interference from blood cells and hemoglobin and to concentrate the microbes present, the procedure is similar to that used for testing of bacterial colonies.
Identification by MALDI-TOF MS depends on an adequate concentration of the inoculum (56). Experiments using S. aureus and Escherichia coli spiked into blood culture broth indicate that bacteria can be successfully identified from 107 CFU/ml of organisms when the median CFU in a positive blood culture broth is 108 CFU/ml (307). The problem with inoculum size is worsened for bacteria with poor spectral quality, such as streptococci (112). There is concern that mixed infections would be impossible to identify; therefore, Gram staining would still be required to mitigate that risk. Contrary to most protocols that try to identify infection in blood culture broth as soon as growth is detected by the automated system (65, 306), some authors have proposed to maintain positive vials for 3 to 10 h at room temperature to allow for storage and transport if required (308).
A variety of different protocols have been reported to accurately identify the microorganisms present in positive blood culture broth; however, a lack of standardized protocols and the use of different software for mass analysis and different blood culture bottles make it difficult to compare the performances of the different methods. One study reported that protocols using extraction are more effective than the intact-cell method (138). Bact/Alert bottles without charcoal were tested, and the authors reported accurate identification with a quick preparation procedure (56); however, preliminary tests performed on Bact/Alert vials with charcoal (309) produced poorer results than those obtained by using Bactec vials, probably due to the presence of charcoal.
In the first large published study of blood culture broth testing, 584 positive blood cultures were tested, and 562 contained a unique bacterial species (306). Two extraction protocols were used, and good results were reported for Gram-negative bacteria at the species level. In the same study, Gram-positive bacteria were poorly identified, with only 37 to 67% being identified to the species level. To accommodate errors in MALDI-TOF identification when mixed cultures were observed, Gram staining was recommended to optimize detection of mixed species.
In a study by Prod'hom et al., identification to the species level was obtained for only 79% of 122 positive blood cultures, and identification problems were observed for streptococci and staphylococci. S. epidermidis was identified only 26% of the time (65). Some microorganisms, such as Klebsiella pneumoniae and Haemophilus influenzae, were not accurately identified, suggesting a specific problem for testing of encapsulated microorganisms.
Stevenson et al. used the BioTyper software to test 212 positive cultures, and correct identification was obtained at the species level with scores of ≥1.9 for 138 (65%) isolates and at the genus level with scores of ≥1.7 for 162 (76%) isolates (308). The eight isolates of S. mitis led to the inaccurate results, being identified as S. pneumoniae. Among 373 monomicrobial-positive blood culture bottles, correct identification was achieved for 98% of them at the species level; however, the authors included an additional test for S. pneumoniae identification to differentiate it from S. mitis. In addition, they reported 100% identification of the 11 Candida albicans samples.
Christner et al. reported accurate identification to the species level (95% of 277 samples). Of 15 nonidentified isolates, 3 were bacteria for which spectra were not present in the database (307). In a recent study, the authors confirmed that MALDI-TOF MS accurately identified, in most cases, one of the species present in a polymicrobial vial and achieved excellent identification at the species level (90% of 497 monomicrobial samples) (307). Ferroni et al. described the only study reporting good results for blood culture bottles with polymicrobial infections; however, this study uniquely used an Andromas-specific database (56). Problems with identification of S. mitis and with polymicrobial vials were still described with this database.
Vlek et al. reported that the implementation of MALDI-TOF MS in the laboratory has resulted in significant improvements to patient care when used for the analysis of positive blood cultures. In their trial, MALDI-TOF MS with Bruker BioTyper version 2.0 software was performed on blood culture broths. This reduced the time to result by 28.8 h and increased the proportion of patients receiving targeted antimicrobials within 24 h of the receipt of the sample by 11.3% (310).
Recently, results of the first study to compare two different MS platforms for the identification of microorganisms directly from positive blood cultures were published. Chen et al. compared the Vitek-MS (bioMérieux) system to the Bruker BioTyper version 3.0 for the identification of microorganisms from 202 positive Bactec bottles. Sample processing was performed with the Bruker SepsiTyper kit according to the manufacturer's instructions. Identifications by the MS system were compared to identifications derived from 16S rRNA gene sequencing and phenotypic (Vitek-2) methods. The BioTyper system was able to make a higher number of accurate identifications to the species level than the Vitek-MS system and demonstrated better performance than Vitek-MS with regard to Gram-positive bacteria at the genus and species levels. Both systems performed poorly when analyzing polymicrobial specimens (311).
Identification of Yeast Directly from Blood Culture Broth
The identification of yeast isolates directly from positive blood culture broth has also been evaluated by a number of groups. Two studies with a limited number of isolates, 20 and 18, respectively, demonstrated that the identification of yeasts is possible when using sample aliquots removed directly from blood culture bottles (56, 138); however, accuracy varied widely between these two studies, with correct identification levels of 5 and 100%, respectively. It is likely that the different protocols, software, and databases used to perform data analysis explain this discrepancy (23). In addition, Kaleta et al. described accurate characterization of yeast colonies isolated from blood cultures using the Bruker MALDI BioTyper 2.0 (MALDI-TOF MS) compared to direct identification with the Ibis T5000 PCR-ESI-MS system (23).
In 2010, an evaluation of species-level identification of Candida isolates from positive blood culture broth was reported. Marinach-Patrice et al. spiked blood culture bottles with different Candida species. The authors noted that a direct identification from the positive bottle would allow bypass of subculture and subsequent identification steps, allowing results to be obtained up to 3 days sooner. Extraction protocols were optimized by using SDS and ethanol, and MS spectra were analyzed for each of the different species of Candida utilized in that study, C. glabrata, C. krusei, C. lusitaniae, C. parapsilosis, and C. tropicalis, with verifiable differences between the generated spectra observed. The method was tested on one routine positive blood culture from a patient, with correct identification being provided (312), allowing the authors to conclude that the method for the direct identification of Candida species from blood culture bottles using MALDI-TOF MS was a rapid and accurate mechanism that could lower costs and hasten appropriate antifungal therapy.
Finally, Spanu et al. evaluated the Bruker BioTyper version 2.0 software for rapid identification of Candida species causing bloodstream infections in a large routine setting. Similar to previous reports, preliminary testing using ATCC isolates yielded 100% accuracy when determining species-level identifications. Polyfungal isolates that were analyzed were unable to be reliably identified, a drawback that many users noted with polybacterial infections as well. Although identification of isolates directly from blood culture bottles generated log score values that were consistently lower than those for identification of isolates from plated media, the software performed well compared to traditional culture-based identifications (187/195 [95.9% concordant for C. albicans] and 128/148 [86.5% concordant for nonalbicans Candida isolates]). Perhaps most importantly, 80% of the positive blood cultures included in the study were reported to be positive ≤24 h after sample entry, allowing in many cases species-level identifications to be reported to physicians within 24 h after blood draw (313).
Extraction Methods for Identification of Microbes Directly from Blood Culture Bottles
Similar to bacterial identifications, the standardization of methods for the extraction of proteins from blood culture bottles will need refinement prior to the widespread utilization of MALDI-TOF MS for direct blood culture analysis. A number of reports have identified that preanalytical steps can greatly influence the quality of spectra derived from sample processing; however, in this case, in addition to the type of organism(s) present, the sample itself can pose difficult challenges when trying to identify the bacteria or yeast contained within. Components from human blood can cause interference or generation of noisy spectra by MALDI-TOF MS when analyzing blood culture specimens directly (271). Prior to the availability of a commercial kit for the processing of specimens directly, the technology was evaluated by using a number of in-house methods (65, 138, 306, 314).
Bruker has made significant steps in standardizing the method of sample extraction with the introduction of the SepsiTyper kit for direct analysis of bacteria from positive blood culture broth samples. In the first analysis of the SepsiTyper kit, an analysis of 507 mono- and polymicrobial blood cultures was undertaken to investigate the utility of the kit prior to MALDI-TOF MS. More Gram-negative organisms were accurately identified than Gram-positive organisms, with significant difficulties being reported for the identification of anaerobic bacteria, alpha-hemolytic streptococci, and polymicrobial mixtures and with ongoing technical development aiming toward addressing these issues (314). A number of comparative evaluations of in-house methods and the SepsiTyper kit soon followed (315–319).
Following the initial description of the SepsiTyper kit, Buchan et al. sought to evaluate the performance of the kit in combination with MALDI-TOF MS for the identification of blood culture isolates using standard MS parameters. One hundred sixty-four isolates composed of Gram-positive, Gram-negative, and fungal hematopathogens were analyzed. The MALDI-TOF MS system with preanalytical preparation by the SepsiTyper system was able to identify 85.5% of isolates directly from blood culture. Gram-negative bacteria still provided better log scores than Gram-positive bacteria, but genus and species concordance was comparable with the reference methods (Vitek-2 and Phoenix) for both groups. Fourteen polymicrobial blood cultures were also evaluated, with the MALDI-TOF MS system identifying at least one organism with high confidence in nine blood cultures. However, no blood cultures containing yeast generated acceptable scores by using standard MS parameters (320).
In a smaller study, Lagacé-Wiens et al. evaluated the SepsiTyper system in tandem with the BioTyper for the identification of bacteria from blood culture bottles and also included a thorough analysis of turnaround time and laboratory costs associated with the testing. Limitations similar to those previously documented by other groups were also reported here with respect to identification of Gram-positive organisms (alpha-hemolytic streptococci) and polymicrobial cultures. A mean reduction in turnaround time of 34.3 h was reported compared to conventional analysis where no additional microbial testing was necessary, and an estimated 26.5-h reduction was reported where some additional characterization of the organism was necessary prior to final identification. With the cost of the SepsiTyper kit being found to be comparable to that of commercial panels, laboratories using primarily commercial panels were determined to see a marginal change in operating costs, whereas laboratories using in-house panels were likely to see an increase in costs per sample with MALDI-TOF MS. These costs might be offset by the use of MALDI-TOF MS in other areas of the microbiology laboratory aside from blood culture analysis (321).
Nonnemann et al. also evaluated the SepsiTyper for routine use in the diagnostic laboratory but included 19 bottles that were spiked with fungi in their study. Decreasing the log score cutoff to 1.5 increased the number of species-level identifications for both Gram-positive and Gram-negative blood cultures; however, the number of fungal cultures identified remained the same at both cutoff values. Seventy-seven percent of fungal isolates were identified to the species level by MALDI-TOF MS using the SepsiTyper kit for preanalytical processing (322). Adjustment of the log score cutoff value to 1.5 improves identification of bacteria directly from blood culture, as reported elsewhere (318).
Significantly less information is available to describe standardization of preparatory methods for pathogenic yeast isolates recovered from blood culture. In an analysis of randomly selected clinical samples, Yan et al. evaluated the SepsiTyper sample-processing system in conjunction with the BioTyper version 2.0 software for the analysis of yeast species directly from blood specimens. The SepsiTyper kit was adapted for use with yeast specimens, and MALDI-TOF MS-derived identifications were compared to phenotypic identifications (germ tube, urease, and API 20C AUX), with 23S rRNA gene sequencing being used to resolve discrepant identifications. In all, 42 cultures were analyzed, which included C. albicans, C. parapsilosis, C. tropicalis, and one isolate of Cryptococcus neoformans, with all samples yielding correct species-level identifications. The MALDI-TOF MS identifications derived directly from blood samples did not change when plated cultures from the same blood culture bottle were analyzed by MALDI-TOF MS. Moreover, the entire procedure (including time for specimen extraction) was reportedly completed in an hour (323).
The analysis of bacteria and fungi directly from blood specimens shows promise for drastically improving the time to result and allowing faster implementation of targeted therapy for patients with sepsis. These technologies will continue to develop to allow greater sensitivity and discrimination among bacteria, particular Gram-positive species, and yeast. Continued evaluation of MALDI-TOF MS by clinicians and laboratory personnel will aid in the development and adaptation of the technology.
MS IDENTIFICATION OF ANTIMICROBIAL RESISTANCE
In addition to the accurate identification of bacteria in clinical material, another critical responsibility of the clinical microbiology laboratory is to determine antibiotic susceptibilities of organisms and to report these findings to both physicians and pharmacists for the implementation of targeted antimicrobial therapy. Mechanisms used for the determination of antimicrobial susceptibilities can vary depending upon the type of organism in question but can include culture-based methods such as Kirby-Bauer disc diffusion testing, Etests, selective growth media, and broth microdilution analysis. Additionally, many automated technologies used for the phenotypic identification of bacteria, including the Vitek-2 (bioMérieux) and the Phoenix (Becton, Dickinson) systems, also function to determine antimicrobial susceptibilities. The drawback of these technologies is the increased time to result associated with them. Rapid testing mechanisms for commonly encountered resistant organisms (i.e., MRSA) were developed as well, including agglutination and enzyme immunoassay (EIA) testing, but are limited to a small number of bacterial species. What is not currently available is a universal platform for the rapid determination of antimicrobial resistance covering an extended spectrum of bacterial genera that can be implemented into the workflow of the clinical laboratory.
Detection of Resistance to Beta-Lactam Antibiotics in Enteric and Nonfermenting Gram-Negative Rods
As MALDI-TOF MS gains acceptance as an improvement over most phenotypic methods utilized for bacterial identification, its exquisite sensitivity with regard to the type of molecules that can be detected during the analysis of bacterial samples is evolving. It is no wonder that a number of investigators have sought to adapt the technology for the identification of bacterial proteins associated with antimicrobial resistance: laboratorians are anxious to have antimicrobial assays run in parallel with bacterial identification. The speed of MALDI-TOF MS identification will no doubt be helpful for treatment of some microbes that have clearly delineated antibiograms; however, for many microbes, no clearly defined antimicrobial changes will be possible without rapid and parallel detection of genetic or phenotypic recognition of antimicrobial resistance. Although in its early stages, several publications are summarized and describe the successful use of MALDI-TOF MS for discriminating antibiotic-resistant bacterial strains. While a comprehensive description of the use of MALDI-TOF MS for the identification of microbes in the clinical laboratory is expertly reviewed elsewhere (324) and is outside the scope of this review, we will instead highlight some of the uses of MALDI-TOF MS for the detection of different groups of clinically important antibiotic-resistant bacteria.
Due to the heavy use of cephalosporin antibiotics worldwide, the spread of Enterobacteriaceae capable of producing extended-spectrum beta-lactamases (ESBLs) is a cause for concern. With the exception of carbapenems, ESBL-producing organisms are resistant to cephalosporins, and as such, strains capable of producing carbapenemase enzymes are closely monitored (325). As MALDI-TOF MS is demonstrated to be a sensitive method for the analysis of cellular products from bacteria, it makes sense that it could potentially be used to characterize products of antibiotic resistance mechanisms. This may be accomplished by observations based simply on the change of molecular mass of antibiotic compounds due to their alteration as a mechanism of resistance (i.e., beta-lactam antibiotics and beta-lactamase production).
In 2007, Camara and Hays used MALDI-TOF MS to discriminate between ampicillin-resistant and -sensitive strains of E. coli by MALDI-TOF MS. While largely a control study of MS sensitivity, their investigation demonstrated that the technology could identify the presence of the beta-lactamase enzyme in spectra generated from protein extracts obtained from broth-grown cultures (326). Since then, a number of additional studies were undertaken to investigate beta-lactam resistance among bacteria, albeit focusing more on the fate of the antibiotic (hydrolysis and degradation, etc.) than on the physical presence of the resistance determinant.
Sparbier et al. detected hydrolysis of the beta-lactam ring after a few hours of incubation by detecting a mass shift of +18 Da. Different enterobacteria were screened for resistance to ampicillin, piperacillin, cefotaxime, ceftazidime, ertapenem, imipenem, and meropenem (327). Similar studies have also been performed to examine carbapenemase activity in enteric organisms and members of the genus Pseudomonas, with good results and rapid turnaround times reported to be as quick as 1 to 4 h following the initiation of testing (328, 329).
Carbapenem-Resistant Acinetobacter baumannii
Carbapenem-resistant Acinetobacter baumannii is a concern for clinicians, as it drastically limits options for antibiotic intervention (330). Kempf et al. analyzed 63 well-characterized carbapenem-resistant A. baumannii strains for carbapenemase production using a 4-h preincubation with imipenem, centrifugation, and MALDI-TOF assessment of supernatant for imipenem metabolite-specific peaks. In addition, 43 non-carbapenemase-producing strains and 43 control strains (7 carbapenem-resistant and 36 carbapenem-sensitive strains) were studied. For this strain set of clinical isolates, sensitivity and specificity were 100.0% (331). Another method used meropenem and this approach was successfully validated on 108 carbapenemase-producing members of the Enterobacteriaceae, 2 NDM-1-producing A. baumannii isolates, and 35 carbapenem-resistant enterobacteria producing no carbapenemase (332).
Results of rapid MALDI-TOF MS analysis for the detection of carbapenemase activity are promising, and investigators have begun to establish standardized procedures for the identification of antibiotic resistance among isolates in a similar manner to what has been done for routine bacterial identification. Recently, isolates of A. baumannii identified by rpoB sequencing were included in a study to standardize a sample-processing workflow for the determination of carbapenem resistance by MALDI-TOF MS. MICs of imipenem and meropenem were determined, and MALDI-TOF MS was performed by using samples treated with four different concentrations of imipenem diluted in three different testing solutions with and without the addition of SDS. Comparison of the peaks generated by MALDI-TOF MS to the spectrum of imipenem allowed for the selection of peaks that could be related to antibiotic hydrolysis. A standard protocol was derived, 1 mg/ml imipenem with 2.5 × 1010 CFU/ml inoculum incubated at 35°C for 1 h with shaking (500 rpm) and then validated against A. baumannii isolates that produced different types of carbapenemases (333).
Carbapenem-Resistant Klebsiella spp.
The spread of carbapenem-resistant Klebsiella spp. is of significant concern to health care practitioners, as K. pneumoniae is known to be a significant cause of nosocomial infections and a reservoir for the accumulation and dissemination of antibiotic resistance determinants among endogenous bacteria (334). Recently, MALDI-TOF MS was evaluated for the detection of carbapenem resistance among strains of Klebsiella spp.
Porins serve as channels at the Gram-negative outer membrane that allow the diffusion of molecules (including beta-lactam antibiotics) into the cell. The loss or reduced expression of specific outer membrane proteins (OMPs) is implicated as a mechanism for carbapenem resistance. One of these OMPs, OmpK36, has been identified as a major membrane porin of K. pneumoniae (335). Porin analysis of bacterial isolates is often relegated to specialized laboratories, with analysis being labor-intensive and requiring techniques not found in most routine clinical microbiology laboratories (typically SDS-PAGE). In a study of eight carbapenem-resistant K. pneumoniae isolates and one K. oxytoca isolate, loss of OmpK36 was investigated by using MALDI-TOF MS. Strains previously determined to be deficient in OmpK36 were identified and compared to strains that were known to express the protein, although more robust analysis will be necessary to validate this method (336).
Carbapenem-Resistant Bacteroides fragilis
Resistance to carbapenem antibiotics in B. fragilis strains is mediated through the carriage and expression of the cfiA gene, which produces an imipenem-hydrolyzing metallo-β-lactamase (337). Using a number of molecular techniques, including MLST and ribotyping, B. fragilis strains characterized as cfiA positive or negative were found to belong to two genotypically distinct groups (338). Due to the ability of MALDI-TOF MS to be able to discriminate between strains of bacteria that are closely related, a number of studies have examined the ability of the technology to differentiate strains that are cfiA positive from those that are cfiA negative. A study by Wybo et al. determined that MALDI-TOF MS provided a suitable mechanism to distinguish between cfiA-positive and -negative B. fragilis isolates. Cluster analysis of B. fragilis clinical isolates clearly delineated these two groups, and this delineation was found not to be dependent upon the presence or absence of a single spectral peak (338). Nagy et al. reported similar results with a set of 40 clinical isolates. Using a subset of 12 of these isolates with known cfiA status, the authors compared these results to spectra derived from 28 B. fragilis isolates of unknown cfiA status. The two groups of B. fragilis were again clearly separated by MALDI-TOF MS spectral analysis (339), suggesting that MALDI-TOF MS may be a useful tool for the rapid identification of these resistant bacteria.
MRSA and Vancomycin-Intermediate Staphylococcus aureus
Some investigations into the ability of MALDI-TOF MS to rapidly identify methicillin-resistant isolates of S. aureus within in the context of bacterial identification and lineage determination have been described elsewhere in this review (83, 85). Here we describe additional studies examining the characterization of resistant isolates, including vancomycin intermediately resistant S. aureus (VISA) strains.
An examination of the use of MALDI-TOF MS for the identification of biomarkers that could be used to rapidly identify hospital-acquired MRSA (HA-MRSA), community-acquired MRSA (CA-MRSA), heterogeneous VISA (hVISA), and VISA was performed with a collection of reference and clinical isolates by using ethanol-extracted proteins for analysis. Following optimization of bacterial culture and MS procedures, spectra were analyzed for the identification of specific biomarkers that could be used to discriminate between HA-MRSA and CA-MRSA isolates. Relationships between the generated spectra and SCCmec types of the strains could be discerned, with the peptide profiles from SCCmec types I to III differing from the profiles from SCCmec types IV and V. Two distinct spectral peaks belonging to members of the phenol-soluble modulin family of proteins were reported, which could potentially be used to differentiate between HA-MRSA and CA-MRSA strains following bacterial identification. Additional peaks were also identified in hVISA and VISA strains belonging to processed forms of acyl carrier protein, which could potentially be useful for determination of vancomycin resistance in S. aureus isolates by MALDI-TOF MS (340).
Wolters et al. established a method of typing MRSA isolates using MALDI-TOF MS analysis for the most abundant HA-MRSA strains. A total of 85 MRSA strains belonging to the five major HA-MRSA clonal complexes (CCs) were analyzed in the course of the study, with 25 being utilized for preliminary spectral analysis and the remaining 60 being used for downstream evaluation of the derived method. Unique spectral peaks obtained from the 25 preliminary isolates were identified and used to classify the remaining 60 isolates. Fifteen different groups were identified by MALDI-TOF analysis and had good concordance with CC groupings, as determined by spa typing. The authors used FA extraction of proteins in order to streamline their method into most laboratory MS workflows, and results could be provided for up to 19 isolates in 2.5 h (88).
In a similar investigation regarding the use of MALDI-TOF MS for characterizing specific lineages of S. aureus, Josten et al. chose to examine peptides identified in the MS spectra of S. aureus and MRSA isolates that exhibited mass shifts in order to evaluate the technology for lineage-specific identification. MRSA isolates could be differentiated into specific CCs, as could MSSA isolates. The method was then tested by using 33 isolates from an outbreak. MALDI-TOF MS could identify the CC associated with a majority of isolates responsible for the outbreak and delineate between different outliers within this collection that were not associated with the outbreak of MSSA and borderline oxacillin-resistant S. aureus (BORSA) (341).
Each of the above-mentioned studies demonstrates the strong discriminatory power that MALDI-TOF/MS exhibits for identification and characterization of drug-resistant S. aureus. Because of these findings, further research should be aimed at enhancing these studies with more streamlined methods and evaluating determined typing schemes with large collections of geographically diverse isolates from HA-MRSA and CA-MRSA lineages with the ultimate goal of realizing a real-time typing method using MALDI-TOF MS instrumentation for use in the clinical laboratory.
Vancomycin-Resistant Enterococcus
Glycopeptide antibiotics are used as a cornerstone for the treatment of severe or resistant Gram-positive infections. Vancomycin-resistant enterococci pose a significant burden to health care systems worldwide. The acquisition of resistance genes and the protein components that they encode was proposed to be a potential mechanism whereby vancomycin-resistant enterococci could be differentiated from vancomycin-sensitive isolates by MALDI-TOF MS. In fact, MALDI-TOF MS was demonstrated to be able to rapidly and accurately discriminate vanB-positive Enterococcus faecium isolates from susceptible strains, in addition to examining relatedness between strains during an outbreak situation (342). Extension of such rigorous analysis could be beneficial, especially when considering organisms that rely on similar mechanisms for glycopeptide resistance.
FUTURE VIEW AND IMPLICATIONS
Although some of the methods described here are currently still restricted to clinical research laboratories and large reference laboratories, the potential of these testing algorithms to move into routine clinical microbiology and public health laboratories seems imminent. The implementation of MALDI-TOF MS in the routine clinical laboratory will provide a powerful and accurate tool to quickly identify bacteria, mycobacteria, fungi, and Nocardia from culture. Further improvements in specimen processing of blood culture broth and urine will be required prior to implementation in clinical laboratories that will be faced with the challenge of selecting between MALDI-TOF methods and emerging molecular methods to identify bacteria from broth or directly from specimens. Improvements to spectral databases and analysis software should optimize the use of MALDI-TOF methods and should reduce the turnaround time for identification of nearly all microbes. In the future, mass spectrometers will be linked with automated antimicrobial susceptibility systems, thus allowing partial or complete automation of routine microbial characterization. The first generation of this automation, the BD Kiestra system, is available from Becton Dickinson. Integration of MALDI-TOF analysis should greatly impact the current processes used in clinical microbiology laboratories. Additionally, research dedicated to adapting MALDI-TOF MS technology to the direct analysis of patient specimens shows promise and may eliminate the need for culture in some cases, thus streamlining diagnostic analysis, which could result in shorter hospital stays, improved prognoses, and decreased financial burden to both the hospital and patient. Rapid and accurate identification of microorganisms directly from clinical specimens is becoming an expectation for clinical laboratories, essential for optimal diagnosis and treatment of patients with infections, and MALDI-TOF technology may support these efforts in the future, but direct-from-specimen testing methods are not currently available.
As the number of studies examining the accuracy and applicability of MALDI-TOF MS in routine clinical situations continues to grow, so too will publications examining the financial and clinical benefits of implementing this rapid technology. An exemplary study by Tan et al. prospectively examined the implementation of MALDI-TOF MS in terms of time to identification in a specimen-based, bench-to-bench approach. This facilitated the real-time evaluation of the technology within the clinical workflow and allowed direct evaluation of instrument performance compared with traditional methodologies. Reduced laboratory costs and time to results were reported when using MALDI-TOF MS for routine testing. This study should serve as a model for future investigations regarding the implementation of MALDI-TOF MS in different laboratories and evaluations of associated benefits and drawbacks (171).
Newer determinative technologies are constantly evolving for clinical microbiology laboratories, aiding the evolution of the microbiology laboratory from one of the slowest of all laboratory services to one that is a plastic and dynamic entity able to directly influence patient outcomes with speed and robust accuracy. As we usher in an era of improved response and higher expectation of accuracy, clinical laboratories are consistently striving to extend the capabilities of these new methods, often in partnership with developmental scientists, resulting in novel technologies, such as MALDI-TOF MS, which should shape and define the diagnostic landscape for years.
ACKNOWLEDGMENTS
We thank Kevin A. Lewis for assistance with generation and rendering of figures and Linda Hilbert, Nina Nguyen, and Natalie Whitfield for critical reading of the manuscript.
Biographies

Andrew E. Clark, M.S., is a Ph.D. student in the Department of Veterinary Science and Microbiology at the University of Arizona, Tucson, AZ. He is a microbiologist with interests in applied clinical, diagnostic, and molecular microbiology. He has worked extensively in both clinical and basic research laboratories, with special interest in the development and evaluation of molecular techniques for the diagnosis and characterization of bacterial infections. His current research is directed toward understanding the molecular events leading to Clostridium difficile colonization and the interplay between pathogen and subsequent host immune responses.

Erin J. Kaleta, Ph.D., received her bachelor's degree in biochemistry from Cedar Crest College and doctorate in chemistry from the University of Arizona. She is currently a clinical chemistry fellow in the Department of Laboratory Medicine and Pathology at the Mayo Clinic. She has been involved in a diverse range of mass spectrometry research for about 10 years, having worked in clinical microbiology as well as analytical chemistry, clinical chemistry, and proteomics.

Amit Arora, M.D., is currently pursuing his master's of science (M.S.) in Epidemiology at the Mel and Enid Zukerman College of Public Health at the University of Arizona (UA). He completed his medical education in India, where he also served as the lead facilitator and analyst for multiple phase I and II clinical trials for various respiratory diseases. As an M.S. candidate, Dr. Arora is currently collaborating with researchers at the UA BIO5 Institute on genomic analysis of quantitative traits among type II diabetic patients. His further research interests lie in analytical methods in bioinformatics and high-throughput genomic and quantitative proteomics.

Donna M. Wolk, M.H.A., Ph.D., D(ABMM), is an Associate Professor at the Arizona Health Science Center and a Clinical Research Scholar at the University of Arizona's BIO5 Institute, which houses her clinical and translational research efforts in the Infectious Disease Research Core Laboratory. She currently serves as the System Director of Clinical Microbiology at Geisinger Health System in Danville, PA, and as a principal investigator at the Weis Center for Research. Her translational research efforts focus on evaluation of new technology for infectious disease diagnostics, thus her interest in mass spectrometry as a tool in clinical microbiology. She received her Ph.D. in Pathobiology at the University of Arizona and rejoined the faculty in 2001, after completing a postdoctoral Medical and Public Health Microbiology Laboratory fellowship at Mayo Clinic in Rochester, MN. She is a Diplomate of the American Board of Medical Microbiology (ABMM) and earned a master's degree in Health Administration.
REFERENCES
- 1. van Belkum A, Durand G, Peyret M, Chatellier S, Zambardi G, Schrenzel J, Shortridge D, Engelhardt A, Dunne WM., Jr 2013. Rapid clinical bacteriology and its future impact. Ann. Lab. Med. 33:14–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clarridge JE. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Petti CA, Polage CR, Schreckenberger P. 2005. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J. Clin. Microbiol. 43:6123–6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wassenberg MWM, Kluytmans JAJW, Box ATA, Bosboom RW, Buiting AGM, Van Elzakker EPM, Melchers WJG, Van Rijen MML, Thijsen SFT, Troelstra A, Vandenbroucke-Grauls CMJE, Visser CE, Voss A, Wolffs PFG, Wulf MWH, Van Zwet AA, De Wit GA, Bonten MJM. 2010. Rapid screening of methicillin-resistant Staphylococcus aureus using PCR and chromogenic agar: a prospective study to evaluate costs and effects. Clin. Microbiol. Infect. 16:1754–1761 [DOI] [PubMed] [Google Scholar]
- 5. Gaydos CA, Van Der Pol B, Jett-Goheen M, Barnes M, Quinn N, Clark C, Daniel GE, Dixon PB, Hook EW, III, CT/NG Study Group 2013. Performance of the Cepheid CT/NG Xpert Rapid PCR Test for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 51:1666–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forrest GN. 2007. PNA FISH: present and future impact on patient management. Expert Rev. Mol. Diagn. 7:231–236 [DOI] [PubMed] [Google Scholar]
- 7. Stone NRH, Gorton RL, Barker K, Ramnarain P, Kibbler CC. 2013. Evaluation of PNA-FISH Yeast Traffic Light for rapid identification of yeast directly from positive blood cultures and assessment of clinical impact. J. Clin. Microbiol. 51:1301–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Almeida C, Azevedo NF, Bento JC, Cerca N, Ramos H, Vieira MJ, Keevil CW. 2013. Rapid detection of urinary tract infections caused by Proteus spp. using PNA-FISH. Eur. J. Clin. Microbiol. Infect. Dis. 32:781–786 [DOI] [PubMed] [Google Scholar]
- 9. Gherardi G, Angeletti S, Panitti M, Pompilio A, Di Bonaventura G, Crea F, Avola A, Fico L, Palazzo C, Sapia GF, Visaggio D, Dicuonzo G. 2012. Comparative evaluation of the Vitek-2 Compact and Phoenix systems for rapid identification and antibiotic susceptibility testing directly from blood cultures of Gram-negative and Gram-positive isolates. Diagn. Microbiol. Infect. Dis. 72:20–31 [DOI] [PubMed] [Google Scholar]
- 10. Woo PCY, Lau SKP, Teng JLL, Tse H, Yuen KY. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908–934 [DOI] [PubMed] [Google Scholar]
- 11. Emonet S, Shah HN, Cherkaoui A, Schrenzel J. 2010. Application and use of various mass spectrometry methods in clinical microbiology. Clin. Microbiol. Infect. 16:1604–1613 [DOI] [PubMed] [Google Scholar]
- 12. Mellmann A, Bimet F, Bizet C, Borovskaya AD, Drake RR, Eigner U, Fahr AM, He Y, Ilina EN, Kostrzewa M, Maier T, Mancinelli L, Moussaoui W, Prevost G, Putignani L, Seachord CL, Tang YW, Harmsen D. 2009. High interlaboratory reproducibility of matrix-assisted laser desorption ionization–time of flight mass spectrometry-based species identification of nonfermenting bacteria. J. Clin. Microbiol. 47:3732–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diamandis EP. 2004. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool. Mol. Cell. Proteomics 3:367–378 [DOI] [PubMed] [Google Scholar]
- 14. Chace DH, Millington DS, Terada N, Kahler SG, Roe CR, Hofman LF. 1993. Rapid diagnosis of phenylketonuria by quantitative analysis for phenylalanine and tyrosine in neonatal blood spots by tandem mass spectrometry. Clin. Chem. 39:66–71 [PubMed] [Google Scholar]
- 15. Ackermann BL, Hale JE, Duffin KL. 2006. The role of mass spectrometry in biomarker discovery and measurement. Curr. Drug Metab. 7:525–539 [DOI] [PubMed] [Google Scholar]
- 16. Anhalt JP, Fenselau C. 1975. Identification of bacteria using mass spectrometry. Anal. Chem. 47:219–225 [Google Scholar]
- 17. Karas M, Bachmann D, Bahr U, Hillenkamp F. 1987. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int. J. Mass Spectrom. Ion Process. 78:53–68 [Google Scholar]
- 18. Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, Matsuo T. 1988. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2:151–153 [Google Scholar]
- 19. Fenselau C, Demirev PA. 2001. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20:157–171 [DOI] [PubMed] [Google Scholar]
- 20. Lay JO. 2001. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20:172–194 [DOI] [PubMed] [Google Scholar]
- 21. Albrethsen J. 2007. Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin. Chem. 53:852–858 [DOI] [PubMed] [Google Scholar]
- 22. Hillenkamp F, Karas M. 1990. Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Methods Enzymol. 193:280–295 [DOI] [PubMed] [Google Scholar]
- 23. Kaleta EJ, Clark AE, Cherkaoui A, Wysocki VH, Ingram EL, Schrenzel J, Wolk DM. 2011. Comparative analysis of PCR-electrospray ionization/mass spectrometry (MS) and MALDI-TOF/MS for the identification of bacteria and yeast from positive blood culture bottles. Clin. Chem. 57:1057–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knochenmuss R, Dubois F, Dale MJ, Zenobi R. 1996. The matrix suppression effect and ionization mechanisms in matrix-assisted laser desorption/ionization. Rapid Commun. Mass Spectrom. 10:871–877 [Google Scholar]
- 25. Karbach V, Knochenmuss R. 1998. Do single matrix molecules generate primary ions in ultraviolet matrix-assisted laser desorption/ionization. Rapid Commun. Mass Spectrom. 12:968–974 [Google Scholar]
- 26. Jaskolla TW, Lehmann W-D, Karas M. 2008. 4-Chloro-alpha-cyanocinnamic acid is an advanced, rationally designed MALDI matrix. Proc. Natl. Acad. Sci. U. S. A. 105:12200–12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beavis RC, Chait BT, Fales HM. 1989. Cinnamic acid derivatives as matrices for ultraviolet laser desorption mass spectrometry of proteins. Rapid Commun. Mass Spectrom. 3:432–435 [DOI] [PubMed] [Google Scholar]
- 28. Cotter RJ. 1987. Laser mass spectrometry: an overview of techniques, instruments and applications. Anal. Chim. Acta 195:45–59 [Google Scholar]
- 29. Vékey K. 1996. Internal energy effects in mass spectrometry. J. Mass Spectrom. 31:445–463 [Google Scholar]
- 30. Drahos L, Vékey K. 2001. MassKinetics: a theoretical model of mass spectra incorporating physical processes, reaction kinetics and mathematical descriptions. J. Mass Spectrom. 36:237–263 [DOI] [PubMed] [Google Scholar]
- 31. Cotter RJ. 1997. Time-of-flight mass spectrometry: instrumentation and applications in biological research. American Chemical Society, Washington, DC [Google Scholar]
- 32. Önnerfjord P, Nilsson J, Wallman L, Laurell T, Marko-Varga G. 1998. Picoliter sample preparation in MALDI-TOF MS using a micromachined silicon flow-through dispenser. Anal. Chem. 70:4755–4760 [DOI] [PubMed] [Google Scholar]
- 33. Wunschel SC, Jarman KH, Petersen CE, Valentine NB, Wahl KL, Schauki D, Jackman J, Nelson CP, White E. 2005. Bacterial analysis by MALDI-TOF mass spectrometry: an inter-laboratory comparison. J. Am. Soc. Mass Spectrom. 16:456–462 [DOI] [PubMed] [Google Scholar]
- 34. Keys CJ, Dare DJ, Sutton H, Wells G, Lunt M, McKenna T, McDowall M, Shah HN. 2004. Compilation of a MALDI-TOF mass spectral database for the rapid screening and characterisation of bacteria implicated in human infectious diseases. Infect. Genet. Evol. 4:221–242 [DOI] [PubMed] [Google Scholar]
- 35. Valentine N, Wunschel S, Wunschel D, Petersen C, Wahl K. 2005. Effect of culture conditions on microorganism identification by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 71:58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williams TL, Andrzejewski D, Lay JO, Jr, Musser SM. 2003. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass Spectrom. 14:342–351 [DOI] [PubMed] [Google Scholar]
- 37. Evason DJ, Claydon MA, Gordon DB. 2000. Effects of ion mode and matrix additives in the identification of bacteria by intact cell mass spectrometry. Rapid Commun. Mass Spectrom. 14:669–672 [DOI] [PubMed] [Google Scholar]
- 38. Goyer M, Lucchi G, Ducoroy P, Vagner O, Bonnin A, Dalle F. 2012. Optimization of the preanalytical steps of matrix-assisted laser desorption ionization–time of flight mass spectrometry identification provides a flexible and efficient tool for identification of clinical yeast isolates in medical laboratories. J. Clin. Microbiol. 50:3066–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evason DJ, Claydon MA, Gordon DB. 2001. Exploring the limits of bacterial identification by intact cell-mass spectrometry. J. Am. Soc. Mass Spectrom. 12:49–54 [DOI] [PubMed] [Google Scholar]
- 40. Holland RD, Wilkes JG, Rafii F, Sutherland JB, Persons CC, Voorhees KJ, Lay JO., Jr 1996. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:1227–1232 [DOI] [PubMed] [Google Scholar]
- 41. Claydon MA, Davey SN, Edwards-Jones V, Gordon DB. 1996. The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol. 14:1584–1586 [DOI] [PubMed] [Google Scholar]
- 42. Ryzhov V, Fenselau C. 2001. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 73:746–750 [DOI] [PubMed] [Google Scholar]
- 43. Rupf S, Breitung K, Schellenberger W, Merte K, Kneist S, Eschrich K. 2005. Differentiation of mutans streptococci by intact cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Oral Microbiol. Immunol. 20:267–273 [DOI] [PubMed] [Google Scholar]
- 44. Jackson KA, Edwards-Jones V, Sutton CW, Fox AJ. 2005. Optimisation of intact cell MALDI method for fingerprinting of methicillin-resistant Staphylococcus aureus. J. Microbiol. Methods 62:273–284 [DOI] [PubMed] [Google Scholar]
- 45. Kumar MP, Vairamani M, Raju RP, Lobo C, Anbumani N, Kumar CP, Menon T, Shanmugasundaram S. 2004. Rapid discrimination between strains of beta haemolytic streptococci by intact cell mass spectrometry. Indian J. Med. Res. 119:283–288 [PubMed] [Google Scholar]
- 46. Hettick JM, Kashon ML, Slaven JE, Ma Y, Simpson JP, Siegel PD, Mazurek GN, Weissman DN. 2006. Discrimination of intact mycobacteria at the strain level: a combined MALDI-TOF MS and biostatistical analysis. Proteomics 6:6416–6425 [DOI] [PubMed] [Google Scholar]
- 47. Bizzini A, Jaton K, Romo D, Bille J, Prod'hom G, Greub G. 2011. Matrix-assisted laser desorption ionization–time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol. 49:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Verroken A, Janssens M, Berhin C, Bogaerts P, Huang TD, Wauters G, Glupczynski Y. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of Nocardia species. J. Clin. Microbiol. 48:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drevinek M, Dresler J, Klimentova J, Pisa L, Hubalek M. 2012. Evaluation of sample preparation methods for MALDI-TOF MS identification of highly dangerous bacteria. Lett. Appl. Microbiol. 55:40–46 [DOI] [PubMed] [Google Scholar]
- 50. El Khéchine A, Couderc C, Flaudrops C, Raoult D, Drancourt M. 2011. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS One 6:e24720. 10.1371/journal.pone.0024720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Patel R. 16 May 2013. Matrix assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin. Infect. Dis. [Epub ahead of print.] 10.1093/cid/cit247 [DOI] [PubMed] [Google Scholar]
- 52. Farfour E, Leto J, Barritault M, Barberis C, Meyer J, Dauphin B, Le Guern A-S, Leflèche A, Badell E, Guiso N, Leclercq A, Le Monnier A, Lecuit M, Rodriguez-Nava V, Bergeron E, Raymond J, Vimont S, Bille E, Carbonnelle E, Guet-Revillet H, Lécuyer H, Beretti J-L, Vay C, Berche P, Ferroni A, Nassif X, Join-Lambert O. 2012. Evaluation of the Andromas matrix-assisted laser desorption ionization–time of flight mass spectrometry system for identification of aerobically growing gram-positive bacilli. J. Clin. Microbiol. 50:2702–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bille E, Dauphin B, Leto J, Bougnoux ME, Beretti JL, Lotz A, Suarez S, Meyer J, Join-Lambert O, Descamps P, Grall N, Mory F, Dubreuil L, Berche P, Nassif X, Ferroni A. 2012. MALDI-TOF MS Andromas strategy for the routine identification of bacteria, mycobacteria, yeasts, Aspergillus spp. and positive blood cultures. Clin. Microbiol. Infect. 18:1117–1125 [DOI] [PubMed] [Google Scholar]
- 54. Degand N, Carbonnelle E, Dauphin B, Beretti JL, Le Bourgeois M, Sermet-Gaudelus I, Segonds C, Berche P, Nassif X, Ferroni A. 2008. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carbonnelle E, Beretti JL, Cottyn S, Quesne G, Berche P, Nassif X, Ferroni A. 2007. Rapid identification of Staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 45:2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ferroni A, Suarez S, Beretti JL, Dauphin B, Bille E, Meyer J, Bougnoux ME, Alanio A, Berche P, Nassif X. 2010. Real-time identification of bacteria and Candida species in positive blood culture broths by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barbuddhe SB, Maier T, Schwarz G, Kostrzewa M, Hof H, Domann E, Chakraborty T, Hain T. 2008. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5402–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mellmann A, Cloud J, Maier T, Keckevoet U, Ramminger I, Iwen P, Dunn J, Hall G, Wilson D, Lasala P, Kostrzewa M, Harmsen D. 2008. Evaluation of matrix-assisted laser desorption ionization–time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wunschel DS, Hill EA, McLean JS, Jarman K, Gorby YA, Valentine N, Wahl K. 2005. Effects of varied pH, growth rate and temperature using controlled fermentation and batch culture on matrix assisted laser desorption/ionization whole cell protein fingerprints #93. J. Microbiol. Methods 62:259–271 [DOI] [PubMed] [Google Scholar]
- 60. Liu H, Du Z, Wang J, Yang R. 2007. Universal sample preparation method for characterization of bacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 73:1899–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 62. Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, Schrenzel J. 2010. Comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Blondiaux N, Gaillot O, Courcol RJ. 2010. MALDI-TOF mass spectrometry to identify clinical bacterial isolates: evaluation in a teaching hospital in Lille. Pathol. Biol. (Paris) 58:55–57 (In French.) [DOI] [PubMed] [Google Scholar]
- 64. van Veen SQ, Claas EC, Kuijper EJ. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization–time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prod'hom G, Bizzini A, Durussel C, Bille J, Greub G. 2010. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J. Clin. Microbiol. 48:1481–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bessède E, Angla-Gre M, Delagarde Y, Sep HS, Menard A, Megraud F. 2011. Matrix-assisted laser-desorption/ionization biotyper: experience in the routine of a university hospital. Clin. Microbiol. Infect. 17:533–538 [DOI] [PubMed] [Google Scholar]
- 67. Martiny D, Busson L, Wybo I, El Haj RA, Dediste A, Vandenberg O. 2012. Comparison of the Microflex LT and Vitek MS systems for the routine identification of bacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:1313–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of Gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. TeKippe ME, Shuey S, Winkler DW, Butler MA, Burnham C-AD. 2013. Optimizing identification of clinically relevant Gram-positive organisms by use of the Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry system. J. Clin. Microbiol. 51:1421–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Christensen JJ, Dargis R, Hammer M, Justesen US, Nielsen XC, Kemp M, Danish MALDI-TOF MS Study Group 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis of Gram-positive, catalase-negative cocci not belonging to the Streptococcus or Enterococcus genus and benefits of database extension. J. Clin. Microbiol. 50:1787–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. von Eiff C, Proctor RA, Peters G. 2001. Coagulase-negative staphylococci. Pathogens have major role in nosocomial infections. Postgrad. Med. 110:63–64, 69-70, 73-76 [PubMed] [Google Scholar]
- 72. Kosmidis CI, Chandrasekar PH. 2012. Management of gram-positive bacterial infections in patients with cancer. Leuk. Lymphoma 53:8–18 [DOI] [PubMed] [Google Scholar]
- 73. Tokars JI, Richards C, Andrus M, Klevens M, Curtis A, Horan T, Jernigan J, Cardo D. 2004. The changing face of surveillance for health care-associated infections. Clin. Infect. Dis. 39:1347–1352 [DOI] [PubMed] [Google Scholar]
- 74. Hayden RT, Carroll KC, Tang Y-W, Wolk DM. (ed). 2009. Diagnostic microbiology of the immunocompromised host. ASM Press, Washington, DC [Google Scholar]
- 75. Dupont C, Sivadon-Tardy V, Bille E, Dauphin B, Beretti JL, Alvarez AS, Degand N, Ferroni A, Rottman M, Herrmann JL, Nassif X, Ronco E, Carbonnelle E. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin. Microbiol. Infect. 16:998–1004 [DOI] [PubMed] [Google Scholar]
- 76. Kiedrowski MR, Horswill AR. 2011. New approaches for treating staphylococcal biofilm infections. Ann. N. Y. Acad. Sci. 1241:104–121 [DOI] [PubMed] [Google Scholar]
- 77. Otto M. 2012. Molecular basis of Staphylococcus epidermidis infections. Semin. Immunopathol. 34:201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kim M, Heo SR, Choi SH, Kwon H, Park JS, Seong MW, Lee DH, Park KU, Song J, Kim EC. 2008. Comparison of the MicroScan, VITEK 2, and Crystal GP with 16S rRNA sequencing and MicroSeq 500 v2.0 analysis for coagulase-negative staphylococci. BMC Microbiol. 8:233. 10.1186/1471-2180-8-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Carpaij N, Willems RJ, Bonten MJ, Fluit AC. 2011. Comparison of the identification of coagulase-negative staphylococci by matrix-assisted laser desorption ionization time-of-flight mass spectrometry and tuf sequencing. Eur. J. Clin. Microbiol. Infect. Dis. 30:1169–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Loonen AJM, Jansz AR, Bergland JNB, Valkenburg M, Wolffs PFG, van den Brule AJC. 2012. Comparative study using phenotypic, genotypic, and proteomics methods for identification of coagulase-negative staphylococci. J. Clin. Microbiol. 50:1437–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Edwards-Jones V, Claydon MA, Evason DJ, Walker J, Fox AJ, Gordon DB. 2000. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 49:295–300 [DOI] [PubMed] [Google Scholar]
- 82. Bernardo K, Pakulat N, Macht M, Krut O, Seifert H, Fleer S, Hunger F, Kronke M. 2002. Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2:747–753 [DOI] [PubMed] [Google Scholar]
- 83. Du Z, Yang R, Guo Z, Song Y, Wang J. 2002. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 74:5487–5491 [DOI] [PubMed] [Google Scholar]
- 84. Walker J, Fox AJ, Edwards-Jones V, Gordon DB. 2002. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J. Microbiol. Methods 48:117–126 [DOI] [PubMed] [Google Scholar]
- 85. Majcherczyk PA, McKenna T, Moreillon P, Vaudaux P. 2006. The discriminatory power of MALDI-TOF mass spectrometry to differentiate between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 255:233–239 [DOI] [PubMed] [Google Scholar]
- 86. Rajakaruna L, Hallas G, Molenaar L, Dare D, Sutton H, Encheva V, Culak R, Innes I, Ball G, Sefton AM, Eydmann M, Kearns AM, Shah HN. 2009. High throughput identification of clinical isolates of Staphylococcus aureus using MALDI-TOF-MS of intact cells. Infect. Genet. Evol. 9:507–513 [DOI] [PubMed] [Google Scholar]
- 87. Szabados F, Woloszyn J, Richter C, Kaase M, Gatermann S. 2010. Identification of molecularly defined Staphylococcus aureus strains using matrix-assisted laser desorption/ionization time of flight mass spectrometry and the Biotyper 2.0 database. J. Med. Microbiol. 59:787–790 [DOI] [PubMed] [Google Scholar]
- 88. Wolters M, Rohde H, Maier T, Belmar-Campos C, Franke G, Scherpe S, Aepfelbacher M, Christner M. 2011. MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant Staphylococcus aureus lineages. Int. J. Med. Microbiol. 301:64–68 [DOI] [PubMed] [Google Scholar]
- 89. Decristophoris P, Fasola A, Benagli C, Tonolla M, Petrini O. 2011. Identification of Staphylococcus intermedius group by MALDI-TOF MS. Syst. Appl. Microbiol. 34:45–51 [DOI] [PubMed] [Google Scholar]
- 90. Dubois D, Leyssene D, Chacornac JP, Kostrzewa M, Schmit PO, Talon R, Bonnet R, Delmas J. 2010. Identification of a variety of Staphylococcus species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bergeron M, Dauwalder O, Gouy M, Freydiere AM, Bes M, Meugnier H, Benito Y, Etienne J, Lina G, Vandenesch F, Boisset S. 2011. Species identification of staphylococci by amplification and sequencing of the tuf gene compared to the gap gene and by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 30:343–354 [DOI] [PubMed] [Google Scholar]
- 92. Ramos ER, Hachem R, Youssef S, Fang X, Jiang Y, Raad I. 2009. The crucial role of catheters in micrococcal bloodstream infections in cancer patients. Infect. Control Hosp. Epidemiol. 30:83–85 [DOI] [PubMed] [Google Scholar]
- 93. Oudiz RJ, Widlitz A, Beckmann XJ, Camanga D, Alfie J, Brundage BH, Barst RJ. 2004. Micrococcus-associated central venous catheter infection in patients with pulmonary arterial hypertension. Chest 126:90–94 [DOI] [PubMed] [Google Scholar]
- 94. Fox K, Fox A, Elssner T, Feigley C, Salzberg D. 2010. MALDI-TOF mass spectrometry speciation of staphylococci and their discrimination from micrococci isolated from indoor air of schoolrooms. J. Environ. Monit. 12:917–923 [DOI] [PubMed] [Google Scholar]
- 95. Wolk DM, Blyn LB, Hall TA, Sampath R, Ranken R, Ivy C, Melton R, Matthews H, White N, Li F, Harpin V, Ecker DJ, Limbago B, McDougal LK, Wysocki VH, Cai M, Carroll KC. 2009. Pathogen profiling: rapid molecular characterization of Staphylococcus aureus by PCR/electrospray ionization-mass spectrometry and correlation with phenotype. J. Clin. Microbiol. 47:3129–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bittar F, Ouchenane Z, Smati F, Raoult D, Rolain JM. 2009. MALDI-TOF-MS for rapid detection of staphylococcal Panton-Valentine leukocidin. Int. J. Antimicrob. Agents 34:467–470 [DOI] [PubMed] [Google Scholar]
- 97. Szabados F, Becker K, von Eiff C, Kaase M, Gatermann S. 2011. The matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS)-based protein peaks of 4448 and 5302 Da are not associated with the presence of Panton-Valentine leukocidin. Int. J. Med. Microbiol. 301:58–63 [DOI] [PubMed] [Google Scholar]
- 98. Poppert S, Nickel D, Berger A, Yildiz T, Kaestner N, Mauerer S, Spellerberg B. 2009. Rapid identification of beta-hemolytic streptococci by fluorescence in situ hybridization (FISH). Int. J. Med. Microbiol. 299:421–426 [DOI] [PubMed] [Google Scholar]
- 99. Cherkaoui A, Emonet S, Fernandez J, Schorderet D, Schrenzel J. 2011. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid identification of beta-hemolytic streptococci. J. Clin. Microbiol. 49:3004–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Johansson L, Thulin P, Low DE, Norrby-Teglund A. 2010. Getting under the skin: the immunopathogenesis of Streptococcus pyogenes deep tissue infections. Clin. Infect. Dis. 51:58–65 [DOI] [PubMed] [Google Scholar]
- 101. Moura H, Woolfitt AR, Carvalho MG, Pavlopoulos A, Teixeira LM, Satten GA, Barr JR. 2008. MALDI-TOF mass spectrometry as a tool for differentiation of invasive and noninvasive Streptococcus pyogenes isolates. FEMS Immunol. Med. Microbiol. 53:333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Verani JR, McGee L, Schrag SJ. 2010. Prevention of perinatal group B streptococcal disease—revised guidelines from CDC, 2010. MMWR Recommend. Rep. 59(RR10):1–36 [PubMed] [Google Scholar]
- 103. Lartigue MF, Hery-Arnaud G, Haguenoer E, Domelier AS, Schmit PO, van der Mee-Marquet N, Lanotte P, Mereghetti L, Kostrzewa M, Quentin R. 2009. Identification of Streptococcus agalactiae isolates from various phylogenetic lineages by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 47:2284–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lartigue MF, Kostrzewa M, Salloum M, Haguenoer E, Hery-Arnaud G, Domelier AS, Stumpf S, Quentin R. 2011. Rapid detection of “highly virulent” group B Streptococcus ST-17 and emerging ST-1 clones by MALDI-TOF mass spectrometry. J. Microbiol. Methods 86:262–265 [DOI] [PubMed] [Google Scholar]
- 105. Williamson YM, Moura H, Woolfitt AR, Pirkle JL, Barr JR, Carvalho MDG, Ades EP, Carlone GM, Sampson JS. 2008. Differentiation of Streptococcus pneumoniae conjunctivitis outbreak isolates by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5891–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Werno AM, Christner M, Anderson TP, Murdoch DR. 2012. Differentiation of Streptococcus pneumoniae from nonpneumococcal streptococci of the Streptococcus mitis group by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:2863–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Neville SA, Lecordier A, Ziochos H, Chater MJ, Gosbell IB, Maley MW, van Hal SJ. 2011. Utility of matrix-assisted laser desorption ionization–time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J. Clin. Microbiol. 49:2980–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Doern CD, Burnham CA. 2010. It's not easy being green: the viridans group streptococci, with a focus on pediatric clinical manifestations. J. Clin. Microbiol. 48:3829–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406–408 [DOI] [PubMed] [Google Scholar]
- 110. Ikryannikova LN, Lapin KN, Malakhova MV, Filimonova AV, Ilina EN, Dubovickaya VA, Sidorenko SV, Govorun VM. 2011. Misidentification of alpha-hemolytic streptococci by routine tests in clinical practice. Infect. Genet. Evol. 11:1709–1715 [DOI] [PubMed] [Google Scholar]
- 111. Facklam R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Friedrichs C, Rodloff AC, Chhatwal GS, Schellenberger W, Eschrich K. 2007. Rapid identification of viridans streptococci by mass spectrometric discrimination. J. Clin. Microbiol. 45:2392–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hinse D, Vollmer T, Erhard M, Welker M, Moore ER, Kleesiek K, Dreier J. 2011. Differentiation of species of the Streptococcus bovis/equinus-complex by MALDI-TOF mass spectrometry in comparison to sodA sequence analyses. Syst. Appl. Microbiol. 34:52–57 [DOI] [PubMed] [Google Scholar]
- 114. Herrera P, Kwon YM, Ricke SC. 2009. Ecology and pathogenicity of gastrointestinal Streptococcus bovis. Anaerobe 15:44–54 [DOI] [PubMed] [Google Scholar]
- 115. Schlegel L, Grimont F, Ageron E, Grimont PAD, Bouvet A. 2003. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int. J. Syst. Evol. Microbiol. 53:631–645 [DOI] [PubMed] [Google Scholar]
- 116. Poyart C, Quesne G, Trieu-Cuot P. 2002. Taxonomic dissection of the Streptococcus bovis group by analysis of manganese-dependent superoxide dismutase gene (sodA) sequences: reclassification of ‘Streptococcus infantarius subsp. coli’ as Streptococcus lutetiensis sp. nov. and of Streptococcus bovis biotype 11.2 as Streptococcus pasteurianus sp. nov. Int. J. Syst. Evol. Microbiol. 52:1247–1255 [DOI] [PubMed] [Google Scholar]
- 117. Schulthess B, Brodner K, Bloemberg GV, Zbinden R, Böttger EC, Hombach M. 2013. Identification of Gram-positive cocci by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry: comparison of different preparation methods and implementation of a practical algorithm for routine diagnostics. J. Clin. Microbiol. 51:1834–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Holler JG, Pedersen LK, Calum H, Nielsen JB, Tvede M, Schonning K, Knudsen JD. 2011. Using MALDI-TOF mass spectrometry as a rapid and accurate diagnostic tool in infective endocarditis: a case report of a patient with mitral valve infective endocarditis caused by Abiotrophia defectiva. Scand. J. Infect. Dis. 43:234–237 [DOI] [PubMed] [Google Scholar]
- 119. Sava IG, Heikens E, Huebner J. 2010. Pathogenesis and immunity in enterococcal infections. Clin. Microbiol. Infect. 16:533–540 [DOI] [PubMed] [Google Scholar]
- 120. Palmer KL, Kos VN, Gilmore MS. 2010. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 13:632–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Forrest GN, Roghmann MC, Toombs LS, Johnson JK, Weekes E, Lincalis DP, Venezia RA. 2008. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob. Agents Chemother. 52:3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Reynaud af Geijersstam A, Culak R, Molenaar L, Chattaway M, Roslie E, Peciuliene V, Haapasalo M, Shah HN. 2007. Comparative analysis of virulence determinants and mass spectral profiles of Finnish and Lithuanian endodontic Enterococcus faecalis isolates. Oral Microbiol. Immunol. 22:87–94 [DOI] [PubMed] [Google Scholar]
- 123. Giebel RA, Fredenberg W, Sandrin TR. 2008. Characterization of environmental isolates of Enterococcus spp. by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Water Res. 42:931–940 [DOI] [PubMed] [Google Scholar]
- 124. Böhme K, Fernández-No IC, Barros-Velázquez J, Gallardo JM, Calo-Mata P, Cañas B. 2010. Species differentiation of seafood spoilage and pathogenic Gram-negative bacteria by MALDI-TOF mass fingerprinting. J. Proteome Res. 9:3169–3183 [DOI] [PubMed] [Google Scholar]
- 125. Fang H, Ohlsson AK, Ullberg M, Ozenci V. 2012. Evaluation of species-specific PCR, Bruker MS, VITEK MS and the VITEK 2 system for the identification of clinical Enterococcus isolates. Eur. J. Clin. Microbiol. Infect. Dis. 31:3073–3077 [DOI] [PubMed] [Google Scholar]
- 126. Glikman D, Sprecher H, Chernokozinsky A, Weintraub Z. 2010. Lactococcus lactis catheter-related bacteremia in an infant. Infection 38:145–146 [DOI] [PubMed] [Google Scholar]
- 127. Chan JF, Woo PC, Teng JL, Lau SK, Leung SS, Tam FC, Yuen KY. 2011. Primary infective spondylodiscitis caused by Lactococcus garvieae and a review of human L. garvieae infections. Infection 39:259–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Tanigawa K, Kawabata H, Watanabe K. 2010. Identification and typing of Lactococcus lactis by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 76:4055–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Krishnamurthy T, Ross PL, Rajamani U. 1996. Detection of pathogenic and non-pathogenic bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:883–888 [DOI] [PubMed] [Google Scholar]
- 130. Lasch P, Beyer W, Nattermann H, Stammler M, Siegbrecht E, Grunow R, Naumann D. 2009. Identification of Bacillus anthracis by using matrix-assisted laser desorption ionization–time of flight mass spectrometry and artificial neural networks. Appl. Environ. Microbiol. 75:7229–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hotta Y, Sato J, Sato H, Hosoda A, Tamura H. 2011. Classification of the genus Bacillus based on MALDI-TOF MS analysis of ribosomal proteins coded in S10 and spc operons. J. Agric. Food Chem. 59:5222–5230 [DOI] [PubMed] [Google Scholar]
- 132. Liu D. 2006. Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J. Med. Microbiol. 55:645–659 [DOI] [PubMed] [Google Scholar]
- 133. Konrad R, Berger A, Huber I, Boschert V, Hormansdorfer S, Busch U, Hogardt M, Schubert S, Sing A. 2010. Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry as a tool for rapid diagnosis of potentially toxigenic Corynebacterium species in the laboratory management of diphtheria-associated bacteria. Euro Surveill. 15(43):pii=19699 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19699 [DOI] [PubMed] [Google Scholar]
- 134. Bittar F, Cassagne C, Bosdure E, Stremler N, Dubus JC, Sarles J, Reynaud-Gaubert M, Raoult D, Rolain JM. 2010. Outbreak of Corynebacterium pseudodiphtheriticum infection in cystic fibrosis patients, France. Emerg. Infect. Dis. 16:1231–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Díez-Aguilar M, Ruiz-Garbajosa P, Fernández-Olmos A, Guisado P, Campo R, Quereda C, Cantón R, Meseguer MA. 2013. Non-diphtheriae Corynebacterium species: an emerging respiratory pathogen. Eur. J. Clin. Microbiol. Infect. Dis. 32:769–772 [DOI] [PubMed] [Google Scholar]
- 136. Hijazin M, Hassan AA, Alber J, Lämmler C, Timke M, Kostrzewa M, Prenger-Berninghoff E, Zschöck M. 2012. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for species identification of bacteria of genera Arcanobacterium and Trueperella. Vet. Microbiol. 157:243–245 [DOI] [PubMed] [Google Scholar]
- 137. Hijazin M, Ulbegi-Mohyla H, Alber J, Lammler C, Hassan AA, Timke M, Kostrzewa M, Prenger-Berninghoff E, Weiss R, Zschock M. 2012. Identification of Arcanobacterium (Trueperella) abortisuis, a novel species of veterinary importance, by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Berl. Munch. Tierarztl. Wochenschr. 125:32–37 [PubMed] [Google Scholar]
- 138. Ferreira L, Sanchez-Juanes F, Munoz-Bellido JL, Gonzalez-Buitrago JM. 2011. Rapid method for direct identification of bacteria in urine and blood culture samples by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: intact cell vs. extraction method. Clin. Microbiol. Infect. 17:1007–1012 [DOI] [PubMed] [Google Scholar]
- 139. Ambrosioni J, Lew D, Garbino J. 2010. Nocardiosis: updated clinical review and experience at a tertiary center. Infection 38:89–97 [DOI] [PubMed] [Google Scholar]
- 140. Cloud JL, Conville PS, Croft A, Harmsen D, Witebsky FG, Carroll KC. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 42:578–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Shinnick TM, Iademarco MF, Ridderhof JC. 2005. National plan for reliable tuberculosis laboratory services using a systems approach. Recommendations from CDC and the Association of Public Health Laboratories Task Force on Tuberculosis Laboratory Services. MMWR Recommend. Rep. 54(RR6):1–12 [PubMed] [Google Scholar]
- 142. Parrish NM, Carroll KC. 2011. Role of the clinical mycobacteriology laboratory in diagnosis and management of tuberculosis in low-prevalence settings. J. Clin. Microbiol. 49:772–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. McNerney R, Maeurer M, Abubakar I, Marais B, McHugh TD, Ford N, Weyer K, Lawn S, Grobusch MP, Memish Z, Squire SB, Pantaleo G, Chakaya J, Casenghi M, Migliori GB, Mwaba P, Zijenah L, Hoelscher M, Cox H, Swaminathan S, Kim PS, Schito M, Harari A, Bates M, Schwank S, O'Grady J, Pletschette M, Ditui L, Atun R, Zumla A. 2012. Tuberculosis diagnostics and biomarkers: needs, challenges, recent advances, and opportunities. J. Infect. Dis. 205(Suppl 2):S147–S158. 10.1093/infdis/jir860 [DOI] [PubMed] [Google Scholar]
- 144. Shingadia D. 2012. The diagnosis of tuberculosis. Pediatr. Infect. Dis. J. 31:302–305 [DOI] [PubMed] [Google Scholar]
- 145. Garcia-Sierra N, Lacoma A, Prat C, Haba L, Maldonado J, Ruiz-Manzano J, Gavin P, Samper S, Ausina V, Dominguez J. 2011. Pyrosequencing for rapid molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 49:3683–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bao JR, Master RN, Schwab DA, Clark RB. 2010. Identification of acid-fast bacilli using pyrosequencing analysis. Diagn. Microbiol. Infect. Dis. 67:234–238 [DOI] [PubMed] [Google Scholar]
- 147. Bravo LT, Tuohy MJ, Ang C, Destura RV, Mendoza M, Procop GW, Gordon SM, Hall GS, Shrestha NK. 2009. Pyrosequencing for rapid detection of Mycobacterium tuberculosis resistance to rifampin, isoniazid, and fluoroquinolones. J. Clin. Microbiol. 47:3985–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Heller LC, Jones M, Widen RH. 2008. Comparison of DNA pyrosequencing with alternative methods for identification of mycobacteria. J. Clin. Microbiol. 46:2092–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Pauls RJ, Turenne CY, Wolfe JN, Kabani A. 2003. A high proportion of novel mycobacteria species identified by 16S rDNA analysis among slowly growing AccuProbe-negative strains in a clinical setting. Am. J. Clin. Pathol. 120:560–566 [DOI] [PubMed] [Google Scholar]
- 151. Evans KD, Nakasone AS, Sutherland PA, de la Maza LM, Peterson EM. 1992. Identification of Mycobacterium tuberculosis and Mycobacterium avium-M. intracellulare directly from primary BACTEC cultures by using acridinium-ester-labeled DNA probes. J. Clin. Microbiol. 30:2427–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Hettick JM, Kashon ML, Simpson JP, Siegel PD, Mazurek GH, Weissman DN. 2004. Proteomic profiling of intact mycobacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 76:5769–5776 [DOI] [PubMed] [Google Scholar]
- 153. Pignone M, Greth KM, Cooper J, Emerson D, Tang J. 2006. Identification of mycobacteria by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry. J. Clin. Microbiol. 44:1963–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lotz A, Ferroni A, Beretti JL, Dauphin B, Carbonnelle E, Guet-Revillet H, Veziris N, Heym B, Jarlier V, Gaillard JL, Pierre-Audigier C, Frapy E, Berche P, Nassif X, Bille E. 2010. Rapid identification of mycobacterial whole cells in solid and liquid culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:4481–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Dinnes J, Deeks J, Kunst H, Gibson A, Cummins E, Waugh N, Drobniewski F, Lalvani A. 2007. A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol. Assess. 11:1–196 [DOI] [PubMed] [Google Scholar]
- 156. Saleeb PG, Drake SK, Murray PR, Zelazny AM. 2011. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:1790–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Saffert RT, Cunningham SA, Ihde SM, Jobe KE, Mandrekar J, Patel R. 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J. Clin. Microbiol. 49:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Ford BA, Burnham C-AD. 2013. Optimization of routine identification of clinically relevant Gram-negative bacteria by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry and the Bruker Biotyper. J. Clin. Microbiol. 51:1412–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Lynn EC, Chung MC, Tsai WC, Han CC. 1999. Identification of Enterobacteriaceae bacteria by direct matrix-assisted laser desorptiom/ionization mass spectrometric analysis of whole cells. Rapid Commun. Mass Spectrom. 13:2022–2027 [DOI] [PubMed] [Google Scholar]
- 160. Pribil P, Fenselau C. 2005. Characterization of enterobacteria using MALDI-TOF mass spectrometry. Anal. Chem. 77:6092–6095 [DOI] [PubMed] [Google Scholar]
- 161. Leuschner RGK, Beresford-Jones N, Robinson C. 2004. Difference and consensus of whole cell Salmonella enterica subsp. enterica serovars matrix-assisted laser desorption/ionization time-of-flight mass spectrometry spectra. Lett. Appl. Microbiol. 38:24–31 [DOI] [PubMed] [Google Scholar]
- 162. Dieckmann R, Helmuth R, Erhard M, Malorny B. 2008. Rapid classification and identification of salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 74:7767–7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Dieckmann R, Malorny B. 2011. Rapid screening of epidemiologically important Salmonella enterica subsp. enterica serovars by whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 77:4136–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kuhns M, Zautner AE, Rabsch W, Zimmermann O, Weig M, Bader O, Groß U. 2012. Rapid discrimination of Salmonella enterica serovar Typhi from other serovars by MALDI-TOF mass spectrometry. PLoS One 7:e40004. 10.1371/journal.pone.0040004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Sparbier K, Weller U, Boogen C, Kostrzewa M. 2012. Rapid detection of Salmonella sp. by means of a combination of selective enrichment broth and MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis. 31:767–773 [DOI] [PubMed] [Google Scholar]
- 166. Conway GC, Smole SC, Sarracino DA, Arbeit RD, Leopold PE. 2001. Phyloproteomics: species identification of Enterobacteriaceae using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mol. Microbiol. Biotechnol. 3:103–112 [PubMed] [Google Scholar]
- 167. Fagerquist CK, Garbus BR, Miller WG, Williams KE, Yee E, Bates AH, Boyle S, Harden LA, Cooley MB, Mandrell RE. 2010. Rapid identification of protein biomarkers of Escherichia coli O157:H7 by matrix-assisted laser desorption ionization-time-of-flight-time-of-flight mass spectrometry and top-down proteomics. Anal. Chem. 82:2717–2725 [DOI] [PubMed] [Google Scholar]
- 168. Karger A, Ziller M, Bettin B, Mintel B, Schares S, Geue L. 2011. Determination of serotypes of Shiga toxin-producing Escherichia coli isolates by intact cell matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 77:896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. He Y, Li H, Lu X, Stratton CW, Tang Y-W. 2010. Mass spectrometry biotyper system identifies enteric bacterial pathogens directly from colonies grown on selective stool culture media. J. Clin. Microbiol. 48:3888–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Steensels D, Verhaegen J, Lagrou K. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of bacteria and yeasts in a clinical microbiological laboratory: a review. Acta Clin. Belg. 66:267–273 [DOI] [PubMed] [Google Scholar]
- 171. Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. 2012. Prospective evaluation of a matrix-assisted laser desorption ionization–time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J. Clin. Microbiol. 50:3301–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Trevino M, Navarro D, Barbeito G, Areses P, Garcia-Riestra C, Regueiro BJ. 2012. Plasmid-mediated AMPc producing Proteus mirabilis in the health care area of Santiago de Compostela: molecular and epidemiological analysis by rep-PCR and MALDI-TOF. Rev. Esp. Quimioter. 25:122–128 (In Spanish.) [PubMed] [Google Scholar]
- 173. Stephan R, Ziegler D, Pflüger V, Vogel G, Lehner A. 2010. Rapid genus- and species-specific identification of Cronobacter spp. by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zhu S, Ratering S, Schnell S, Wacker R. 2011. Matrix-assisted laser desorption and ionizationtime-of-flight mass spectrometry, 16S rRNA gene sequencing, and API 32E for identification of Cronobacter spp.: a comparative study. J. Food Prot. 74:2182–2187 [DOI] [PubMed] [Google Scholar]
- 175. Cetinkaya E, Joseph S, Ayhan K, Forsythe SJ. 2013. Comparison of methods for the microbiological identification and profiling of Cronobacter species from ingredients used in the preparation of infant formula. Mol. Cell. Probes 27:60–64 [DOI] [PubMed] [Google Scholar]
- 176. Karamonová L, Junková P, Mihalová D, Javůrková B, Fukal L, Rauch P, Blažková M. 2013. The potential of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the identification of biogroups of Cronobacter sakazakii. Rapid Commun. Mass Spectrom. 27:409–418 [DOI] [PubMed] [Google Scholar]
- 177. Pavlovic M, Konrad R, Iwobi AN, Sing A, Busch U, Huber I. 2012. A dual approach employing MALDI-TOF MS and real-time PCR for fast species identification within the Enterobacter cloacae complex. FEMS Microbiol. Lett. 328:46–53 [DOI] [PubMed] [Google Scholar]
- 178. Rezzonico F, Vogel G, Duffy B, Tonolla M. 2010. Application of whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid identification and clustering analysis of Pantoea species. Appl. Environ. Microbiol. 76:4497–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Kolínská R, Dřevínek M, Aldová E, Žemličková H. 2010. Identification of Plesiomonas spp.: serological and MALDI-TOF MS methods. Folia Microbiol. 55:669–672 [DOI] [PubMed] [Google Scholar]
- 180. de Jong E, de Jong AS, Smidts-van den Berg N, Rentenaar RJ. 2013. Differentiation of Raoultella ornithinolytica/planticola and Klebsiella oxytoca clinical isolates by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Diagn. Microbiol. Infect. Dis. 75:431–433 [DOI] [PubMed] [Google Scholar]
- 181. Lasch P, Drevinek M, Nattermann H, Grunow R, Stämmler, Dieckmann MR, Schwecke T, Naumann D. 2010. Characterization of Yersinia using MALDI-TOF mass spectrometry and chemometrics. Anal. Chem. 82:8464–8475 [DOI] [PubMed] [Google Scholar]
- 182. Ayyadurai S, Flaudrops C, Raoult D, Drancourt M. 2010. Rapid identification and typing of Yersinia pestis and other Yersinia species by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. BMC Microbiol. 10:285. 10.1186/1471-2180-10-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Couderc C, Nappez C, Drancourt M. 2012. Comparing inactivation protocols of Yersinia organisms for identification with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 26:710–714 [DOI] [PubMed] [Google Scholar]
- 184. Stephan R, Cernela N, Ziegler D, Pflüger V, Tonolla M, Ravasi D, Fredriksson-Ahomaa M, Hächler H. 2011. Rapid species specific identification and subtyping of Yersinia enterocolitica by MALDI-TOF mass spectrometry. J. Microbiol. Methods 87:150–153 [DOI] [PubMed] [Google Scholar]
- 185. Wittwer M, Heim J, Schär M, Dewarrat G, Schürch N. 2011. Tapping the potential of intact cell mass spectrometry with a combined data analytical approach applied to Yersinia spp.: detection, differentiation and identification of Y. pestis. Syst. Appl. Microbiol. 34:12–19 [DOI] [PubMed] [Google Scholar]
- 186. Teramoto K, Sato H, Sun L, Torimura M, Tao H, Yoshikawa H, Hotta Y, Hosoda A, Tamura H. 2007. Phylogenetic classification of Pseudomonas putida strains by MALDI-MS using ribosomal subunit proteins as biomarkers. Anal. Chem. 79:8712–8719 [DOI] [PubMed] [Google Scholar]
- 187. Vanlaere E, Sergeant K, Dawyndt P, Kallow W, Erhard M, Sutton H, Dare D, Devreese B, Samyn B, Vandamme P. 2008. Matrix-assisted laser desorption ionisation-time-of of-flight mass spectrometry of intact cells allows rapid identification of Burkholderia cepacia complex. J. Microbiol. Methods 75:279–286 [DOI] [PubMed] [Google Scholar]
- 188. Fernández-Olmos A, García-Castillo M, Morosini M-I, Lamas A, Máiz L, Cantón R. 2012. MALDI-TOF MS improves routine identification of non-fermenting Gram negative isolates from cystic fibrosis patients. J. Cyst. Fibros. 11:59–62 [DOI] [PubMed] [Google Scholar]
- 189. Marko DC, Saffert RT, Cunningham SA, Hyman J, Walsh J, Arbefeville S, Howard W, Pruessner J, Safwat N, Cockerill FR, Bossler AD, Patel R, Richter SS. 2012. Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry systems for identification of nonfermenting Gram-negative bacilli isolated from cultures from cystic fibrosis patients. J. Clin. Microbiol. 50:2034–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Ruelle V, Moualij BE, Zorzi W, Ledent P, Pauw ED. 2004. Rapid identification of environmental bacterial strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 18:2013–2019 [DOI] [PubMed] [Google Scholar]
- 191. Šedo O, Voráč A, Zdráhal Z. 2011. Optimization of mass spectral features in MALDI-TOF MS profiling of Acinetobacter species. Syst. Appl. Microbiol. 34:30–34 [DOI] [PubMed] [Google Scholar]
- 192. Álvarez-Buylla A, Culebras E, Picazo JJ. 2012. Identification of Acinetobacter species: is Bruker biotyper MALDI-TOF mass spectrometry a good alternative to molecular techniques? Infect. Genet. Evol. 12:345–349 [DOI] [PubMed] [Google Scholar]
- 193. Espinal P, Seifert H, Dijkshoorn L, Vila J, Roca I. 2012. Rapid and accurate identification of genomic species from the Acinetobacter baumannii (Ab) group by MALDI-TOF MS. Clin. Microbiol. Infect. 18:1097–1103 [DOI] [PubMed] [Google Scholar]
- 194. Mencacci A, Monari C, Leli C, Merlini L, De Carolis E, Vella A, Cacioni M, Buzi S, Nardelli E, Bistoni F, Sanguinetti M, Vecchiarelli A. 2013. Typing of nosocomial outbreaks of Acinetobacter baumannii by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:603–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 16:821–830 [DOI] [PubMed] [Google Scholar]
- 196. Mahenthiralingam E, Baldwin A, Dowson CG. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104:1539–1551 [DOI] [PubMed] [Google Scholar]
- 197. Vandamme P, Dawyndt P. 2011. Classification and identification of the Burkholderia cepacia complex: past, present and future. Syst. Appl. Microbiol. 34:87–95 [DOI] [PubMed] [Google Scholar]
- 198. Lambiase A, Del Pezzo M, Cerbone D, Raia V, Rossano F, Catania MR. 2013. Rapid identification of Burkholderia cepacia complex species recovered from cystic fibrosis patients using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Microbiol. Methods 92:145–149 [DOI] [PubMed] [Google Scholar]
- 199. Minan A, Bosch A, Lasch P, Stammler M, Serra DO, Degrossi J, Gatti B, Vay C, D'Aquino M, Yantorno O, Naumann D. 2009. Rapid identification of Burkholderia cepacia complex species including strains of the novel taxon K, recovered from cystic fibrosis patients by intact cell MALDI-ToF mass spectrometry. Analyst 134:1138–1148 [DOI] [PubMed] [Google Scholar]
- 200. Desai AP, Stanley T, Atuan M, McKey J, Lipuma JJ, Rogers B, Jerris R. 2012. Use of matrix assisted laser desorption ionisation–time of flight mass spectrometry in a paediatric clinical laboratory for identification of bacteria commonly isolated from cystic fibrosis patients. J. Clin. Pathol. 65:835–838 [DOI] [PubMed] [Google Scholar]
- 201. Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102–111 [DOI] [PubMed] [Google Scholar]
- 202. Majerczyk C, Kinman L, Han T, Bunt R, Greenberg EP. 2013. Virulence of Burkholderia mallei quorum-sensing mutants. Infect. Immun. 81:1471–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Galyov EE, Brett PJ, DeShazer D. 2010. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu. Rev. Microbiol. 64:495–517 [DOI] [PubMed] [Google Scholar]
- 204. Lau SKP, Tang BSF, Curreem SOT, Chan T-M, Martelli P, Tse CWS, Wu AKL, Yuen K-Y, Woo PCY. 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid identification of Burkholderia pseudomallei: importance of expanding databases with pathogens endemic to different localities. J. Clin. Microbiol. 50:3142–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Karger A, Stock R, Ziller M, Elschner M, Bettin B, Melzer F, Maier T, Kostrzewa M, Scholz H, Neubauer H, Tomaso H. 2012. Rapid identification of Burkholderia mallei and Burkholderia pseudomallei by intact cell matrix-assisted laser desorption/ionisation mass spectrometric typing. BMC Microbiol. 12:229. 10.1186/1471-2180-12-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Cunningham SA, Patel R. 2013. Importance of using Bruker's Security-Relevant Library for Biotyper identification of Burkholderia pseudomallei, Brucella species, and Francisella tularensis. J. Clin. Microbiol. 51:1639–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Inglis TJJ, Healy PE, Fremlin LJ, Golledge CL. 2012. Use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis for rapid confirmation of Burkholderia pseudomallei in septicemic melioidosis. Am. J. Trop. Med. Hyg. 86:1039–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Mulet M, Gomila M, Scotta C, Sánchez D, Lalucat J, García-Valdés E. 2012. Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analysis approaches in species discrimination within the genus Pseudomonas. Syst. Appl. Microbiol. 35:455–464 [DOI] [PubMed] [Google Scholar]
- 209. Vasileuskaya-Schulz Z, Kaiser S, Maier T, Kostrzewa M, Jonas D. 2011. Delineation of Stenotrophomonas spp. by multi-locus sequence analysis and MALDI-TOF mass spectrometry. Syst. Appl. Microbiol. 34:35–39 [DOI] [PubMed] [Google Scholar]
- 210. Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG. 2009. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Ferreira L, Vega Castano S, Sanchez-Juanes F, Gonzalez-Cabrero S, Menegotto F, Orduna-Domingo A, Gonzalez-Buitrago JM, Munoz-Bellido JL. 2010. Identification of Brucella by MALDI-TOF mass spectrometry. Fast and reliable identification from agar plates and blood cultures. PLoS One 5:e14235. 10.1371/journal.pone.0014235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Chain PS, Comerci DJ, Tolmasky ME, Larimer FW, Malfatti SA, Vergez LM, Aguero F, Land ML, Ugalde RA, Garcia E. 2005. Whole-genome analyses of speciation events in pathogenic brucellae. Infect. Immun. 73:8353–8361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Lista F, Reubsaet F, De Santis R, Parchen R, de Jong A, Kieboom J, van der Laaken A, Voskamp-Visser I, Fillo S, Jansen H-J, Van der Plas J, Paauw A. 2011. Reliable identification at the species level of Brucella isolates with MALDI-TOF-MS. BMC Microbiol. 11:267. 10.1186/1471-2180-11-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Fournier P-E, Couderc C, Buffet S, Flaudrops C, Raoult D. 2009. Rapid and cost-effective identification of Bartonella species using mass spectrometry. J. Med. Microbiol. 58:1154–1159 [DOI] [PubMed] [Google Scholar]
- 215. Seibold E, Maier T, Kostrzewa M, Zeman E, Splettstoesser W. 2010. Identification of Francisella tularensis by whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry: fast, reliable, robust, and cost-effective differentiation on species and subspecies levels. J. Clin. Microbiol. 48:1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Muller W, Hotzel H, Otto P, Karger A, Bettin B, Bocklisch H, Braune S, Eskens U, Hormansdorfer S, Konrad R, Nesseler A, Peters M, Runge M, Schmoock G, Schwarz B-A, Sting R, Myrtennas K, Karlsson E, Forsman M, Tomaso H. 2013. German Francisella tularensis isolates from European brown hares (Lepus europaeus) reveal genetic and phenotypic diversity. BMC Microbiol. 13:61. 10.1186/1471-2180-13-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Haag AM, Taylor SN, Johnston KH, Cole RB. 1998. Rapid identification and speciation of Haemophilus bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 33:750–756 [DOI] [PubMed] [Google Scholar]
- 218. Couturier MR, Mehinovic E, Croft AC, Fisher MA. 2011. Identification of HACEK clinical isolates by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:1104–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Nørskov-Lauritsen N, Bruun B, Andersen C, Kilian M. 2012. Identification of haemolytic Haemophilus species isolated from human clinical specimens and description of Haemophilus sputorum sp. nov. Int. J. Med. Microbiol. 302:78–83 [DOI] [PubMed] [Google Scholar]
- 220. Zhu B, Xiao D, Zhang H, Zhang Y, Gao Y, Xu L, Lv J, Wang Y, Zhang J, Shao Z. 2013. MALDI-TOF MS distinctly differentiates nontypable Haemophilus influenzae from Haemophilus haemolyticus. PLoS One 8:e56139. 10.1371/journal.pone.0056139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Dieckmann R, Strauch E, Alter T. 2010. Rapid identification and characterization of Vibrio species using whole-cell MALDI-TOF mass spectrometry. J. Appl. Microbiol. 109:199–211 [DOI] [PubMed] [Google Scholar]
- 222. Hazen TH, Martinez RJ, Chen Y, Lafon PC, Garrett NM, Parsons MB, Bopp CA, Sullards MC, Sobecky PA. 2009. Rapid identification of Vibrio parahaemolyticus by whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 75:6745–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Eddabra R, Prévost G, Scheftel J-M. 2012. Rapid discrimination of environmental Vibrio by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Microbiol. Res. 167:226–230 [DOI] [PubMed] [Google Scholar]
- 224. Donohue MJ, Smallwood AW, Pfaller S, Rodgers M, Shoemaker JA. 2006. The development of a matrix-assisted laser desorption/ionization mass spectrometry-based method for the protein fingerprinting and identification of Aeromonas species using whole cells. J. Microbiol. Methods 65:380–389 [DOI] [PubMed] [Google Scholar]
- 225. Donohue MJ, Best JM, Smallwood AW, Kostich M, Rodgers M, Shoemaker JA. 2007. Differentiation of Aeromonas isolated from drinking water distribution systems using matrix-assisted laser desorption/ionization-mass spectrometry. Anal. Chem. 79:1939–1946 [DOI] [PubMed] [Google Scholar]
- 226. Lamy B, Kodjo A, Laurent F. 2011. Identification of Aeromonas isolates by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Diagn. Microbiol. Infect. Dis. 71:1–5 [DOI] [PubMed] [Google Scholar]
- 227. Benagli C, Demarta A, Caminada A, Ziegler D, Petrini O, Tonolla M. 2012. A rapid MALDI-TOF MS identification database at genospecies level for clinical and environmental Aeromonas strains. PLoS One 7:e48441. 10.1371/journal.pone.0048441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Winkler MA, Uher J, Cepa S. 1999. Direct analysis and identification of Helicobacter and Campylobacter species by MALDI-TOF mass spectrometry. Anal. Chem. 71:3416–3419 [DOI] [PubMed] [Google Scholar]
- 229. Mandrell RE, Harden LA, Bates A, Miller WG, Haddon WF, Fagerquist CK. 2005. Speciation of Campylobacter coli, C. jejuni, C. helveticus, C. lari, C. sputorum, and C. upsaliensis by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 71:6292–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Kolínská R, Dřevínek M, Jakubů V, Žemličková H. 2008. Species identification of Campylobacter jejuni ssp. jejuni and C. coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and PCR. Folia Microbiol. 53:403–409 [DOI] [PubMed] [Google Scholar]
- 231. Alispahic M, Hummel K, Jandreski-Cvetkovic D, Nobauer K, Razzazi-Fazeli E, Hess M, Hess C. 2010. Species-specific identification and differentiation of Arcobacter, Helicobacter and Campylobacter by full-spectral matrix-associated laser desorption/ionization time of flight mass spectrometry analysis. J. Med. Microbiol. 59:295–301 [DOI] [PubMed] [Google Scholar]
- 232. Bessède E, Solecki O, Sifré E, Labadi L, Mégraud F. 2011. Identification of Campylobacter species and related organisms by matrix assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry. Clin. Microbiol. Infect. 17:1735–1739 [DOI] [PubMed] [Google Scholar]
- 233. Martiny D, Dediste A, Debruyne L, Vlaes L, Haddou NB, Vandamme P, Vandenberg O. 2011. Accuracy of the API Campy system, the Vitek 2 Neisseria-Haemophilus card and matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the identification of Campylobacter and related organisms. Clin. Microbiol. Infect. 17:1001–1006 [DOI] [PubMed] [Google Scholar]
- 234. Ilina EN. 2009. Direct matrix-assisted laser desorption-ionisation (MALDI) mass-spectrometry bacteria profiling for identifying and characterizing pathogens. Acta Naturae 1:115–120 [PMC free article] [PubMed] [Google Scholar]
- 235. Ilina EN, Borovskaya AD, Serebryakova MV, Chelysheva VV, Momynaliev KT, Maier T, Kostrzewa M, Govorun VM. 2010. Application of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the study of Helicobacter pylori. Rapid Commun. Mass Spectrom. 24:328–334 [DOI] [PubMed] [Google Scholar]
- 236. Ilina EN, Borovskaya AD, Malakhova MM, Vereshchagin VA, Kubanova AA, Kruglov AN, Svistunova TS, Gazarian AO, Maier T, Kostrzewa M, Govorun VM. 2009. Direct bacterial profiling by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for identification of pathogenic Neisseria. J. Mol. Diagn. 11:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Lowe CA, Diggle MA, Clarke SC. 2004. A single nucleotide polymorphism identification assay for the genotypic characterisation of Neisseria meningitidis using MALDI-TOF mass spectrometry. Br. J. Biomed. Sci. 61:8–10 [DOI] [PubMed] [Google Scholar]
- 238. Schaller A, Troller R, Molina D, Gallati S, Aebi C, Stutzmann Meier P. 2006. Rapid typing of Moraxella catarrhalis subpopulations based on outer membrane proteins using mass spectrometry. Proteomics 6:172–180 [DOI] [PubMed] [Google Scholar]
- 239. Moliner C, Ginevra C, Jarraud S, Flaudrops C, Bedotto M, Couderc C, Etienne J, Fournier P-E. 2010. Rapid identification of Legionella species by mass spectrometry. J. Med. Microbiol. 59:273–284 [DOI] [PubMed] [Google Scholar]
- 240. Gaia V, Casati S, Tonolla M. 2011. Rapid identification of Legionella spp. by MALDI-TOF MS based protein mass fingerprinting. Syst. Appl. Microbiol. 34:40–44 [DOI] [PubMed] [Google Scholar]
- 241. Fujinami Y, Kikkawa HS, Kurosaki Y, Sakurada K, Yoshino M, Yasuda J. 2011. Rapid discrimination of Legionella by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Microbiol. Res. 166:77–86 [DOI] [PubMed] [Google Scholar]
- 242. Svarrer CW, Uldum SA. 2012. The occurrence of Legionella species other than Legionella pneumophila in clinical and environmental samples in Denmark identified by mip gene sequencing and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 18:1004–1009 [DOI] [PubMed] [Google Scholar]
- 243. Shah HN, Keys CJ, Schmid O, Gharbia SE. 2002. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and proteomics: a new era in anaerobic microbiology. Clin. Infect. Dis. 35:S58–S64. 10.1086/341922 [DOI] [PubMed] [Google Scholar]
- 244. Nagy E, Maier T, Urban E, Terhes G, Kostrzewa M. 2009. Species identification of clinical isolates of Bacteroides by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 15:796–802 [DOI] [PubMed] [Google Scholar]
- 245. Stîngu CS, Rodloff AC, Jentsch H, Schaumann R, Eschrich K. 2008. Rapid identification of oral anaerobic bacteria cultivated from subgingival biofilm by MALDI-TOF-MS. Oral Microbiol. Immunol. 23:372–376 [DOI] [PubMed] [Google Scholar]
- 246. Nagy E, Becker S, Kostrzewa M, Barta N, Urban E. 2012. The value of MALDI-TOF MS for the identification of clinically relevant anaerobic bacteria in routine laboratories. J. Med. Microbiol. 61:1393–1400 [DOI] [PubMed] [Google Scholar]
- 247. La Scola B, Fournier P-E, Raoult D. 2011. Burden of emerging anaerobes in the MALDI-TOF and 16S rRNA gene sequencing era. Anaerobe 17:106–112 [DOI] [PubMed] [Google Scholar]
- 248. Veloo AC, Knoester M, Degener JE, Kuijper EJ. 2011. Comparison of two matrix-assisted laser desorption ionisation-time of flight mass spectrometry methods for the identification of clinically relevant anaerobic bacteria. Clin. Microbiol. Infect. 17:1501–1506 [DOI] [PubMed] [Google Scholar]
- 249. Veloo ACM, Erhard M, Welker M, Welling GW, Degener JE. 2011. Identification of Gram-positive anaerobic cocci by MALDI-TOF mass spectrometry. Syst. Appl. Microbiol. 34:58–62 [DOI] [PubMed] [Google Scholar]
- 250. Veloo ACM, Welling GW, Degener JE. 2011. The identification of anaerobic bacteria using MALDI-TOF MS. Anaerobe 17:211–212 [DOI] [PubMed] [Google Scholar]
- 251. Fedorko DP, Drake SK, Stock F, Murray PR. 2012. Identification of clinical isolates of anaerobic bacteria using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 31:2257–2262 [DOI] [PubMed] [Google Scholar]
- 252. Justesen US, Holm A, Knudsen E, Andersen LB, Jensen TG, Kemp M, Skov MN, Gahrn-Hansen B, Møller JK. 2011. Species identification of clinical isolates of anaerobic bacteria: a comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry systems. J. Clin. Microbiol. 49:4314–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Fournier R, Wallet F, Grandbastien B, Dubreuil L, Courcol R, Neut C, Dessein R. 2012. Chemical extraction versus direct smear for MALDI-TOF mass spectrometry identification of anaerobic bacteria. Anaerobe 18:294–297 [DOI] [PubMed] [Google Scholar]
- 254. Theel ES, Schmitt BH, Hall L, Cunningham SA, Walchak RC, Patel R, Wengenack NL. 2012. Formic acid-based direct, on-plate testing of yeast and Corynebacterium species by Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:3093–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Schmitt BH, Cunningham SA, Dailey AL, Gustafson DR, Patel R. 2013. Identification of anaerobic bacteria by Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry with on-plate formic acid preparation. J. Clin. Microbiol. 51:782–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Nagy E, Urbán E, Becker S, Kostrzewa M, Vörös A, Hunyadkürti J, Nagy I. 2013. MALDI-TOF MS fingerprinting facilitates rapid discrimination of phylotypes I, II and III of Propionibacterium acnes. Anaerobe 20:20–26 [DOI] [PubMed] [Google Scholar]
- 257. Culebras E, Rodríguez-Avial I, Betriu C, Gómez M, Picazo JJ. 2012. Rapid identification of clinical isolates of Bacteroides species by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. Anaerobe 18:163–165 [DOI] [PubMed] [Google Scholar]
- 258. Grosse-Herrenthey A, Maier T, Gessler F, Schaumann R, Böhnel H, Kostrzewa M, Krüger M. 2008. Challenging the problem of clostridial identification with matrix-assisted laser desorption and ionization–time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe 14:242–249 [DOI] [PubMed] [Google Scholar]
- 259. Viswanathan VK, Mallozzi M, Vedantam G. 2010. Clostridium difficile infection: an overview of the disease and its pathogenesis, epidemiology and interventions. Gut Microbes 1:234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK. 2012. Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes 3:121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Reil M, Erhard M, Kuijper EJ, Kist M, Zaiss H, Witte W, Gruber H, Borgmann S. 2011. Recognition of Clostridium difficile PCR-ribotypes 001, 027 and 126/078 using an extended MALDI-TOF MS system. Eur. J. Clin. Microbiol. Infect. Dis. 30:1431–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization–time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Putignani L, Del Chierico F, Onori M, Mancinelli L, Argentieri M, Bernaschi P, Coltella L, Lucignano B, Pansani L, Ranno S, Russo C, Urbani A, Federici G, Menichella D. 2011. MALDI-TOF mass spectrometry proteomic phenotyping of clinically relevant fungi. Mol. Biosyst. 7:620–629 [DOI] [PubMed] [Google Scholar]
- 264. Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. 2011. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17:1359–1365 [DOI] [PubMed] [Google Scholar]
- 265. Van Herendael BH, Bruynseels P, Bensaid M, Boekhout T, De Baere T, Surmont I, Mertens AH. 2012. Validation of a modified algorithm for the identification of yeast isolates using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS). Eur. J. Clin. Microbiol. Infect. Dis. 31:841–848 [DOI] [PubMed] [Google Scholar]
- 266. Pinto A, Halliday C, Zahra M, van Hal S, Olma T, Maszewska K, Iredell JR, Meyer W, Chen SC. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One 6:e25712. 10.1371/journal.pone.0025712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Hof H, Eigner U, Maier T, Staib P. 2012. Differentiation of Candida dubliniensis from Candida albicans by means of MALDI-TOF mass spectrometry. Clin. Lab. 58:927–931 [PubMed] [Google Scholar]
- 268. Seyfarth F, Wiegand C, Erhard M, Graser Y, Elsner P, Hipler UC. 2012. Identification of yeast isolated from dermatological patients by MALDI-TOF mass spectrometry. Mycoses 55:276–280 [DOI] [PubMed] [Google Scholar]
- 269. Martínez-Lamas L, Pérez del Molino ML, Pardo F, Varela E, Regueiro BJ. 2011. Espectrometría de masas matrix-assisted laser desorption ionization time-of-flight vs. metodología convencional en la identificación de Candida no-albicans. Enferm. Infecc. Microbiol. Clin. 29:568–572 [DOI] [PubMed] [Google Scholar]
- 270. Qian J, Cutler JE, Cole RB, Cai Y. 2008. MALDI-TOF mass signatures for differentiation of yeast species, strain grouping and monitoring of morphogenesis markers. Anal. Bioanal. Chem. 392:439–449 [DOI] [PubMed] [Google Scholar]
- 271. Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of clinically important yeast species. J. Clin.Microbiol. 48:3482–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Firacative C, Trilles L, Meyer W. 2012. MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS One 7:e37566. 10.1371/journal.pone.0037566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. McTaggart LR, Lei E, Richardson SE, Hoang L, Fothergill A, Zhang SX. 2011. Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:3050–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Cassagne C, Ranque S, Normand A-C, Fourquet P, Thiebault S, Planard C, Hendrickx M, Piarroux R. 2011. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One 6:e28425. 10.1371/journal.pone.0028425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Lau AF, Drake SK, Calhoun LB, Henderson CM, Zelazny AM. 2013. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:828–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Santos C, Paterson RRM, Venâncio A, Lima N. 2010. Filamentous fungal characterizations by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Appl. Microbiol. 108:375–385 [DOI] [PubMed] [Google Scholar]
- 277. Iriart X, Lavergne RA, Fillaux J, Valentin A, Magnaval JF, Berry A, Cassaing S. 2012. Routine identification of medical fungi by the new Vitek MS matrix-assisted laser desorption ionization–time of flight system with a new time-effective strategy. J. Clin. Microbiol. 50:2107–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. De Carolis E, Vella A, Florio AR, Posteraro P, Perlin DS, Sanguinetti M, Posteraro B. 2012. Use of matrix-assisted laser desorption ionization–time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J. Clin. Microbiol. 50:2479–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Pan Y-L, Chow N-H, Chang TC, Chang H-C. 2011. Identification of lethal Aspergillus at early growth stages based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Diagn. Microbiol. Infect. Dis. 70:344–354 [DOI] [PubMed] [Google Scholar]
- 280. Hettick JM, Green BJ, Buskirk AD, Kashon ML, Slaven JE, Janotka E, Blachere FM, Schmechel D, Beezhold DH. 2008. Discrimination of Aspergillus isolates at the species and strain level by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting. Anal. Biochem. 380:276–281 [DOI] [PubMed] [Google Scholar]
- 281. Kemptner J, Marchetti-Deschmann M, Mach R, Druzhinina IS, Kubicek CP, Allmaier G. 2009. Evaluation of matrix-assisted laser desorption/ionization (MALDI) preparation techniques for surface characterization of intact Fusarium spores by MALDI linear time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 23:877–884 [DOI] [PubMed] [Google Scholar]
- 282. Dong H, Kemptner J, Marchetti-Deschmann M, Kubicek C, Allmaier G. 2009. Development of a MALDI two-layer volume sample preparation technique for analysis of colored conidia spores of Fusarium by MALDI linear TOF mass spectrometry. Anal. Bioanal. Chem. 395:1373–1383 [DOI] [PubMed] [Google Scholar]
- 283. Marinach-Patrice C, Lethuillier A, Marly A, Brossas JY, Gene J, Symoens F, Datry A, Guarro J, Mazier D, Hennequin C. 2009. Use of mass spectrometry to identify clinical Fusarium isolates. Clin. Microbiol. Infect. 15:634–642 [DOI] [PubMed] [Google Scholar]
- 284. Baldo A, Monod M, Mathy A, Cambier L, Bagut ET, Defaweux V, Symoens F, Antoine N, Mignon B. 2012. Mechanisms of skin adherence and invasion by dermatophytes. Mycoses 55:218–223 [DOI] [PubMed] [Google Scholar]
- 285. Jensen RH, Arendrup MC. 2012. Molecular diagnosis of dermatophyte infections. Curr. Opin. Infect. Dis. 25:126–134 [DOI] [PubMed] [Google Scholar]
- 286. Theel ES, Hall L, Mandrekar J, Wengenack NL. 2011. Dermatophyte identification using matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:4067–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Erhard M, Hipler U-C, Burmester A, Brakhage AA, Wöstemeyer J. 2008. Identification of dermatophyte species causing onychomycosis and tinea pedis by MALDI-TOF mass spectrometry. Exp. Dermatol. 17:356–361 [DOI] [PubMed] [Google Scholar]
- 288. Alshawa K, Beretti J-L, Lacroix C, Feuilhade M, Dauphin B, Quesne G, Hassouni N, Nassif X, Bougnoux M-E. 2012. Successful identification of clinical dermatophyte and Neoscytalidium species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:2277–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Nenoff P, Erhard M, Simon JC, Muylowa GK, Herrmann J, Rataj W, Graser Y. 2013. MALDI-TOF mass spectrometry—a rapid method for the identification of dermatophyte species. Med. Mycol. 51:17–24 [DOI] [PubMed] [Google Scholar]
- 290. Tammer I, Tintelnot K, Braun-Dullaeus RC, Mawrin C, Scherlach C, Schlüter D, König W. 2011. Infections due to Pseudallescheria/Scedosporium species in patients with advanced HIV disease—a diagnostic and therapeutic challenge. Int. J. Infect. Dis. 15:e422–e429. 10.1016/j.ijid.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 291. Coulibaly O, Marinach-Patrice C, Cassagne C, Piarroux R, Mazier D, Ranque S. 2011. Pseudallescheria/Scedosporium complex species identification by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Med. Mycol. 49:621–626 [DOI] [PubMed] [Google Scholar]
- 292. Hettick JM, Green BJ, Buskirk AD, Kashon ML, Slaven JE, Janotka E, Blachere FM, Schmechel D, Beezhold DH. 2008. Discrimination of Penicillium isolates by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting. Rapid Commun. Mass Spectrom. 22:2555–2560 [DOI] [PubMed] [Google Scholar]
- 293. Schwartze VU, Hoffmann K, Nyilasi I, Papp T, Vágvölgyi C, de Hoog S, Voigt K, Jacobsen ID. 2012. Lichtheimia species exhibit differences in virulence potential. PLoS One 7:e40908. 10.1371/journal.pone.0040908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Alastruey-Izquierdo A, Hoffmann K, de Hoog GS, Rodriguez-Tudela JL, Voigt K, Bibashi E, Walther G. 2010. Species recognition and clinical relevance of the zygomycetous genus Lichtheimia (syn. Absidia pro parte, Mycocladus). J. Clin. Microbiol. 48:2154–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Schrödl W, Heydel T, Schwartze VU, Hoffmann K, Große-Herrenthey A, Walther G, Alastruey-Izquierdo A, Rodriguez-Tudela JL, Olias P, Jacobsen ID, de Hoog GS, Voigt K. 2012. Direct analysis and identification of pathogenic Lichtheimia species by matrix-assisted laser desorption ionization–time of flight analyzer-mediated mass spectrometry. J. Clin. Microbiol. 50:419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Ngai AL, Bourque MR, Lupinacci RJ, Strohmaier KM, Kartsonis NA. 2011. Overview of safety experience with caspofungin in clinical trials conducted over the first 15 years: a brief report. Int. J. Antimicrob. Agents 38:540–544 [DOI] [PubMed] [Google Scholar]
- 297. Shields RK, Nguyen MH, Press EG, Kwa AL, Cheng S, Du C, Clancy CJ. 2012. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob. Agents Chemother. 56:4862–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Pulcrano G, Roscetto E, Iula VD, Panellis D, Rossano F, Catania MR. 2012. MALDI-TOF mass spectrometry and microsatellite markers to evaluate Candida parapsilosis transmission in neonatal intensive care units. Eur. J. Clin. Microbiol. Infect. Dis. 31:2919–2928 [DOI] [PubMed] [Google Scholar]
- 299. Fiedler GM, Baumann S, Leichtle A, Oltmann A, Kase J, Thiery J, Ceglarek U. 2007. Standardized peptidome profiling of human urine by magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin. Chem. 53:421–428 [DOI] [PubMed] [Google Scholar]
- 300. Thongboonkerd V, Saetun P. 2007. Bacterial overgrowth affects urinary proteome analysis: recommendation for centrifugation, temperature, duration, and the use of preservatives during sample collection. J. Proteome Res. 6:4173–4181 [DOI] [PubMed] [Google Scholar]
- 301. Ferreira L, Sánchez-Juanes F, González-Ávila M, Cembrero-Fuciños D, Herrero-Hernández A, González-Buitrago JM, Muñoz-Bellido JL. 2010. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:2110–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Kohling HL, Bittner A, Muller KD, Buer J, Becker M, Rubben H, Rettenmeier AW, Mosel F. 2012. Direct identification of bacteria in urine samples by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and relevance of defensins as interfering factors. J. Med. Microbiol. 61:339–344 [DOI] [PubMed] [Google Scholar]
- 303. Wang XH, Zhang G, Fan YY, Yang X, Sui WJ, Lu XX. 2013. Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry. J. Microbiol. Methods 92:231–235 [DOI] [PubMed] [Google Scholar]
- 304. Nyvang Hartmeyer G, Kvistholm Jensen A, Böcher S, Damkjaer Bartels M, Pedersen M, Engell Clausen M, Abdul-Redha R, Dargis R, Schouenborg P, Højlyng N, Kemp M, Christensen JJE. 2010. Mass spectrometry: pneumococcal meningitis verified and Brucella species identified in less than half an hour. Scand. J. Infect. Dis. 42:716–718 [DOI] [PubMed] [Google Scholar]
- 305. La Scola B. 2011. Intact cell MALDI-TOF mass spectrometry-based approaches for the diagnosis of bloodstream infections. Expert Rev. Mol. Diagn. 11:287–298 [DOI] [PubMed] [Google Scholar]
- 306. La Scola B, Raoult D. 2009. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One 4:e8041. 10.1371/journal.pone.0008041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Christner M, Rohde H, Wolters M, Sobottka I, Wegscheider K, Aepfelbacher M. 2010. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J. Clin. Microbiol. 48:1584–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Stevenson LG, Drake SK, Murray PR. 2010. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:444–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Szabados F, Michels M, Kaase M, Gatermann S. 2011. The sensitivity of direct identification from positive BacT/ALERT (bioMerieux) blood culture bottles by matrix-assisted laser desorption ionization time-of-flight mass spectrometry is low. Clin. Microbiol. Infect. 17:192–195 [DOI] [PubMed] [Google Scholar]
- 310. Vlek ALM, Bonten MJM, Boel CHE. 2012. Direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry improves appropriateness of antibiotic treatment of bacteremia. PLoS One 7:e32589. 10.1371/journal.pone.0032589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Chen JHK, Ho P-L, Kwan GSW, She KKK, Siu GKH, Cheng VCC, Yuen K-Y, Yam W-C. 2013. Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization–time of flight mass spectrometry systems. J. Clin. Microbiol. 51:1733–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Marinach-Patrice C, Fekkar A, Atanasova R, Gomes J, Djamdjian L, Brossas J-Y, Meyer I, Buffet P, Snounou G, Datry A, Hennequin C, Golmard J-L, Mazier D. 2010. Rapid species diagnosis for invasive candidiasis using mass spectrometry. PLoS One 5:e8862. 10.1371/journal.pone.0008862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Spanu T, Posteraro B, Fiori B, D'Inzeo T, Campoli S, Ruggeri A, Tumbarello M, Canu G, Trecarichi EM, Parisi G, Tronci M, Sanguinetti M, Fadda G. 2012. Direct MALDI-TOF mass spectrometry assay of blood culture broths for rapid identification of Candida species causing bloodstream infections: an observational study in two large microbiology laboratories. J. Clin. Microbiol. 50:176–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Kok J, Thomas LC, Olma T, Chen SCA, Iredell JR. 2011. Identification of bacteria in blood culture broths using matrix-assisted laser desorption-ionization Sepsityper and time of flight mass spectrometry. PLoS One 6:e23285. 10.1371/journal.pone.0023285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Juiz PM, Almela M, Melción C, Campo I, Esteban C, Pitart C, Marco F, Vila J. 2012. A comparative study of two different methods of sample preparation for positive blood cultures for the rapid identification of bacteria using MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis. 31:1353–1358 [DOI] [PubMed] [Google Scholar]
- 316. Loonen AJM, Jansz AR, Stalpers J, Wolffs PFG, Brule AJC. 2012. An evaluation of three processing methods and the effect of reduced culture times for faster direct identification of pathogens from BacT/ALERT blood cultures by MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis. 31:1575–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Martiny D, Dediste A, Vandenberg O. 2012. Comparison of an in-house method and the commercial Sepsityper kit for bacterial identification directly from positive blood culture broths by matrix-assisted laser desorption-ionisation time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 31:2269–2281 [DOI] [PubMed] [Google Scholar]
- 318. Saffert RT, Cunningham SA, Mandrekar J, Patel R. 2012. Comparison of three preparatory methods for detection of bacteremia by MALDI-TOF mass spectrometry. Diagn. Microbiol. Infect. Dis. 73:21–26 [DOI] [PubMed] [Google Scholar]
- 319. Meex C, Neuville F, Descy J, Huynen P, Hayette M-P, De Mol P, Melin P. 2012. Direct identification of bacteria from BacT/ALERT anaerobic positive blood cultures by MALDI-TOF MS: MALDI Sepsityper kit versus an in-house saponin method for bacterial extraction. J. Med. Microbiol. 61:1511–1516 [DOI] [PubMed] [Google Scholar]
- 320. Buchan BW, Riebe KM, Ledeboer NA. 2012. Comparison of the MALDI Biotyper system using sepsityper specimen processing to routine microbiological methods for identification of bacteria from positive blood culture bottles. J. Clin. Microbiol. 50:346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Lagacé-Wiens PRS, Adam HJ, Karlowsky JA, Nichol KA, Pang PF, Guenther J, Webb AA, Miller C, Alfa MJ. 2012. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J. Clin. Microbiol. 50:3324–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Nonnemann B, Tvede M, Bjarnsholt T. 18 January 2013. Identification of pathogenic microorganisms directly from positive blood vials by matrix-assisted laser desorption/ionization time of flight mass spectrometry. APMIS [Epub ahead of print.] 10.1111/apm.12050 [DOI] [PubMed] [Google Scholar]
- 323. Yan Y, He Y, Maier T, Quinn C, Shi G, Li H, Stratton CW, Kostrzewa M, Tang Y-W. 2011. Improved identification of yeast species directly from positive blood culture media by combining Sepsityper specimen processing and Microflex analysis with the matrix-assisted laser desorption ionization biotyper system. J. Clin. Microbiol. 49:2528–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Hrabák J, Chudáčková E, Walková R. 2013. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin. Microbiol. Rev. 26:103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Camara J, Hays F. 2007. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 389:1633–1638 [DOI] [PubMed] [Google Scholar]
- 327. Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. J. Clin. Microbiol. 50:927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 49:3321–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Hrabák J, Walková R, Študentová V, Chudáčková E, Bergerová T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836 [DOI] [PubMed] [Google Scholar]
- 331. Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel J-M, Drissi M, Mesli E, Touati A, Rolain J-M. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676. 10.1371/journal.pone.0031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Hrabák J, Študentová V, Walková R, Žemličková H, Jakubů V, Chudáčková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerová T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:2441–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Álvarez-Buylla A, Picazo JJ, Culebras E. 2013. Optimized method for Acinetobacter species carbapenemase detection and identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:1589–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 335. Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55:1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Cai JC, Hu YY, Zhang R, Zhou HW, Chen G-X. 2012. Detection of OmpK36 porin loss in Klebsiella spp. by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 50:2179–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Yamazoe K, Kato N, Kato H, Tanaka K, Katagiri Y, Watanabe K. 1999. Distribution of the cfiA gene among Bacteroides fragilis strains in Japan and relatedness of cfiA to imipenem resistance. Antimicrob. Agents Chemother. 43:2808–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Wybo I, De Bel A, Soetens O, Echahidi F, Vandoorslaer K, Van Cauwenbergh M, Piérard D. 2011. Differentiation of cfiA-negative and cfiA-positive Bacteroides fragilis isolates by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:1961–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Nagy E, Becker S, Soki J, Urban E, Kostrzewa M. 2011. Differentiation of division I (cfiA-negative) and division II (cfiA-positive) Bacteroides fragilis strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Med. Microbiol. 60:1584–1590 [DOI] [PubMed] [Google Scholar]
- 340. Lu J-J, Tsai F-J, Ho C-M, Liu Y-C, Chen C-J. 2012. Peptide biomarker discovery for identification of methicillin-resistant and vancomycin-intermediate Staphylococcus aureus strains by MALDI-TOF. Anal. Chem. 84:5685–5692 [DOI] [PubMed] [Google Scholar]
- 341. Josten M, Reif M, Szekat C, Al-Sabti N, Roemer T, Sparbier K, Kostrzewa M, Rohde H, Sahl H-G, Bierbaum G. 2013. Analysis of the matrix-assisted laser desorption ionization–time of flight mass spectrum of Staphylococcus aureus identifies mutations that allow differentiation of the main clonal lineages. J. Clin. Microbiol. 51:1809–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Griffin PM, Price GR, Schooneveldt JM, Schlebusch S, Tilse MH, Urbanski T, Hamilton B, Venter D. 2012. Use of matrix-assisted laser desorption ionization–time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J. Clin. Microbiol. 50:2918–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]