Abstract
Endozoicomonas bacteria were found highly associated with the coral Stylophora pistillata, and these bacteria are also ubiquitously associated with diverse corals worldwide. Novel Endozoicomonas-specific probes revealed that Endozoicomonas bacteria were abundant in the endodermal tissues of S. pistillata and appear to have an intimate relationship with the coral.
TEXT
Reef-building corals are “metaorganisms”; i.e., the coral animal lives in a mutualistic relationship with photosynthetic, endosymbiotic dinoflagellates of the genus Symbiodinium along with microorganisms including bacteria, archaea, fungi, and viruses. The significance of the bacterial assemblage to the coral animal is not well understood, although coral bacteria have been characterized as species specific (1) and may have roles in nitrogen fixation, carbon fixation, antibiotic production, and other features that enable their health and functioning (1–5). A substantial component of the coral bacterial community resides within the mucus layer (6, 7), and there is little understanding of which microbial partners are actually in residence within the coral tissues and potentially interacting with the coral. Here, we address this lack of knowledge by examining if and how a dominant group of bacteria frequently recovered in sequencing-based studies is located internally within the coral tissues of the Red Sea coral Stylophora pistillata. This study used Sanger and 454 pyrosequencing of the bacterial 16S rRNA gene to document the bacterial community of S. pistillata and a fluorescence in situ hybridization (FISH) approach to examine if the dominant bacteria (Endozoicomonas) associated with S. pistillata reside within the coral. Additional analyses of other Red Sea corals, as well as an in silico analysis of worldwide corals, were used to examine the prominence of Endozoicomonas in other coral species.
Five S. pistillata samples (Sp1 to Sp5) were collected in the southern Red Sea in June 2009 by scuba diving at depths between 2 and 5 m at five sites (see Fig. S1 in the supplemental material). DNA was extracted from airbrushed tissue with the PowerPlant DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA), with some modifications (8). Several primer pairs were tested prior to 454 pyrosequencing to ensure specificity to bacterial DNA (see the supplemental material), and primers 784F and 1061R (9), which include variable regions 5 and 6 of the 16S rRNA gene, were used for this analysis. Libraries were generated with the GS FLX Titanium emPCR kit (Lib-A; Roche, Branford, CT) and sequenced by Titanium FLX chemistry. Data analysis was conducted with the mothur software v.1.24.1 (10). Sequencing resulted in 287,488 reads, of which 131,421 remained (median length, 250 bp) after cleaning (Table 1). The sequences were clustered into operational taxonomic units (OTUs) at 97% similarity and classified against the 2011 version of the GreenGenes database (11) as described by Wang et al. (12), with a bootstrap cutoff of 60%. Rarefaction curves demonstrate that for samples Sp4 and Sp5, most of the diversity has been sampled but curves did not plateau for the other samples (see Fig. S2 in the supplemental material). The evenness was very low, indicating that few bacterial OTUs make up the majority of the microbiome (Table 1). In fact, two OTUs dominated the bacterial community in S. pistillata, and they were classified in the genus Endozoicomonas of the order Oceanospirillales and the genus Burkholderia of the order Burkholderiales (Fig. 1A).
Table 1.
Summary statistics for 454 sequencing of SSU rRNA genes from S. pistillataa
| Sample | No. of reads | No. of reads subsampled | No. of OTUs observed | Inverse Simpson | Shannon index | Chao1 richness | ACE richness | Simpson evenness | Sampling site |
|---|---|---|---|---|---|---|---|---|---|
| Sp1 | 18,676 | 18,676 | 418 | 2.51 | 1.88 | 845.5 | 1,250.5 | 0.006 | 5 |
| Sp2 | 29,292 | 18,676 | 465 | 3.59 | 2.02 | 895.8 | 1,361.8 | 0.0077 | 12 |
| Sp3 | 22,954 | 18,676 | 276 | 2.57 | 1.62 | 578.8 | 790.9 | 0.0093 | 14 |
| Sp4 | 35,588 | 18,676 | 139 | 1.66 | 0.80 | 387.1 | 837.6 | 0.012 | 17 |
| Sp5 | 24,911 | 18,676 | 147 | 1.54 | 0.90 | 218.9 | 205.9 | 0.0105 | 15 |
Statistics are based on 18,676 subsampled reads.
Fig 1.
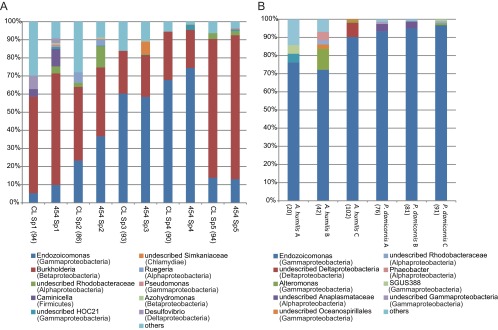
Phylogenetic distribution of coral-associated bacteria. Sequences were classified to the genus level with a minimum bootstrap value of 60%. Read counts from genera other than the 10 most common ones are summarized in the “others” category. The number of clones analyzed is shown in parentheses, and the number of reads analyzed by 454 sequencing is shown in Table 1. (A) S. pistillata (Sp1 to Sp5; n = 5)-associated bacteria detected by cloning and sequencing (CL) and 454 sequencing (454). (B) A. humilis (A to C; n = 3)- and P. damicornis (A to C; n = 3)-associated bacteria detected by cloning and sequencing.
Full-length sequences were also obtained from each sample (Sp1 to Sp5, 86 to 94 clones per sample after chimera removal; see the supplemental material) and yielded bacterial community composition results similar to those of the 454 data (Fig. 1A). Phylogenetic analyses indicated that the Endozoicomonas OTUs obtained in this study all clustered with cultivated species (Endozoicomonas numazuensis, E. montiporae, and E. elysicola), as well as other Endozoicomonas sequences from a diverse range of marine invertebrates, including many species of reef-building and gorgonian corals (see Fig. S3 in the supplemental material). The closest relative of the Burkholderia sequences was isolated from a white-rot fungus (see Fig. S4 in the supplemental material), suggesting that the Burkholderia bacteria found on S. pistillata were associated with a fungus on the coral.
In order to examine the ubiquity of Endozoicomonas bacteria in other coral species from the Red Sea, healthy specimens of Acropora humilis and Pocillopora damicornis (three of each species) were obtained from the same area as the S. pistillata samples (see Fig. S1 in the supplemental material). Bacterial small-subunit (SSU) rRNA genes were examined by cloning and sequencing (n = 20 to 102 per sample after chimera check and contaminant removal; see the supplemental material). Endozoicomonas bacteria accounted for 70 to 95% of the bacterial abundance in P. damicornis and A. humilis (Fig. 1B). Each coral species was associated with one to three Endozoicomonas OTUs (see Fig. S3 in the supplemental material).
To determine how prevalent Endozoicomonas bacteria are in other corals, we performed an in silico analysis of full- and partial-length Endozoicomonas SSU rRNA gene sequences, which revealed that these bacteria associate with 14 species of scleractinian corals (Table 2) (8, 13–19). The fact that sequences were recovered from corals over vast geographic regions suggests that Endozoicomonas bacteria probably have an important relationship with corals. Interestingly, none of the Endozoicomonas OTUs were detected in more than one species, suggesting that each coral harbors its own unique Endozoicomonas strain. Additionally, closely related OTUs may associate with the same coral species in different geographic regions. For example, Endozoicomonas sequences recovered from P. damicornis in the Red Sea (this study) formed a clade with sequences from P. damicornis from the Great Barrier Reef, Australia (13) (see Fig. S3 in the supplemental material). One of the Endozoicomonas OTUs from A. humilis (this study) also clustered with an Endozoicomonas isolate from an acroporid in the Caribbean (8) (see Fig. S3). These results suggest coevolution of Endozoicomonas bacteria and coral, an important strategy to maintain their association over time and space.
Table 2.
Summary of Endozoicomonas sequences recovered from corals
| Coral species | Location(s) | Reference(s) |
|---|---|---|
| Acropora palmata | Panama | 8 |
| Acropora hemprichii | Red Sea | 14 |
| Acropora humilis | Red Sea | This study |
| Ctenactis echinata | Red Sea | C. Roder et al.,b unpublished data |
| Ctenactis crassa | Red Sea | 16 |
| Diploria strigosa | Panama, Curacao | 8, 18 |
| Herpolithia limax | Red Sea | 16 |
| Montastrea faveolata | Panama, Puerto Rico | 8, 15 |
| Montastrea franksi | Panama | 8 |
| Montipora aequituberculata | Taiwan | 17 |
| Pocillopora damicornis | GBR,a Red Sea | 13, this study |
| Porites asteroidetes | Panama | 8 |
| Porites compressa | Hawaii | 19 |
| Stylophora pistillata | Red Sea | This study |
GBR, Great Barrier Reef.
C. Roder, T. Bayer, M. Aranda, M. Kruse, C. Walcher, and C. R. Voolstra.
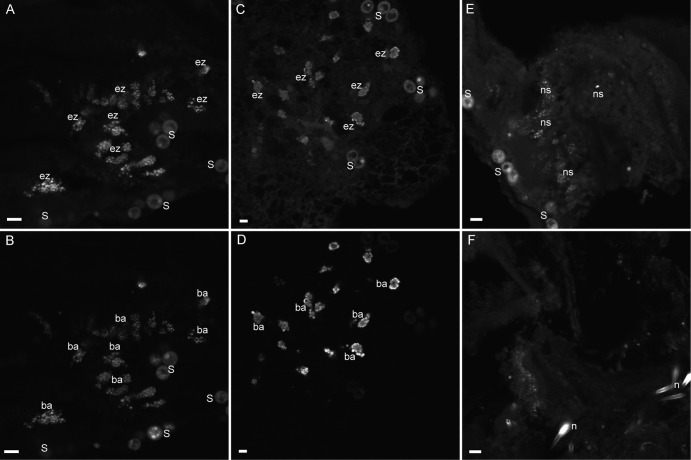
To further investigate the relationship of Endozoicomonas bacteria and S. pistillata, FISH experiments were conducted with novel Endozoicomonas-specific oligonucleotide probes (see the supplemental material). Five or six specimens from colonies Sp1 to Sp5 were examined by imaging in three or four areas of each specimen and compared with control specimens (no probe, NON338 nonsense probe) of each respective colony. In all of the colonies examined, the Endozoicomonas probes were found to hybridize to cells located in close proximity to Symbiodinium cells (Fig. 2A and C) and within the autofluorescent coral tissues, suggesting that they reside within the coral endoderm. Endozoicomonas cells were arranged in multiple dense aggregates generally containing 10 to 50 cells and represented the majority of the cells that hybridized to the general bacterial probe (Fig. 2B and D). These are the first observations of Endozoicomonas in association with corals, but the aggregations do resemble the “ovoid bacterial clusters” that have been previously identified with corals (20–22). Hybridization of the S. pistillata samples with the NON338 probe resulted in the probe hybridizing to aggregates similar to the Endozoicomonas probed cells (Fig. 2E), but the intensity and abundance of the apparent nonspecific probe binding were much lower than those of the specific probes (Fig. 2A to D). The nonspecific binding to portions of the aggregates, as well as to coral nematocysts (Fig. 2F), suggests that an adhesive-type substance may surround the aggregates. Aggregates were also dimly visible in the no-probe controls at the same intensity as the autofluorescent coral tissues (not shown), and this suggests that the cells are embedded within the host endoderm.
Fig 2.
FISH analysis. The mixed Endozoicomonas probes (Cy3, selected aggregates designated “ez”) within the tissues of S. pistillata (A, C) display a hybridization pattern similar to that of the general EUB338 bacterial probe (Cy5, selected aggregates designated “ba”) (B, D). Some cells in aggregates from similar specimens were also illuminated by the nonspecific probe NONEUB (Cy3, selected nonspecificity labeled “ns”) (E), and nematocyst cells (labeled “n”) were also illuminated by the NONEUB probe (F). Symbiodinium cells (S) are autofluorescent at the imaging wavelengths used (except in panel D, where cells faded under imaging). Each image is a compilation of two to four optical slices. The scale bars are approximately 8 μm.
Endozoicomonas bacteria appear to have an intimate and established relationship with many Red Sea corals and other corals worldwide. One feature of Endozoicomonas bacteria associated with S. pistillata is that they may produce quorum-sensing molecules (reviewed in reference 23). The dense cell aggregations found suggest that reaching a critical mass may provide an advantage to the cells. Despite our limited knowledge of Endozoicomonas bacteria, they appear to be an important group of bacteria that require further investigation of their potential role in the functional system of the coral holobiont, as well as their interactions with other invertebrate associates.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the NCBI Sequence Read Archive under accession number PRJNA189184 and in the GenBank database under accession numbers KC668414 to KC669277.
Supplementary Material
ACKNOWLEDGMENTS
We thank Whitney Bernstein, Kate Furby, Jessie Kneeland, and Justin Ossolinski for sample collection; Karie Holterman for technical support; and Aubrie O'Rourke for help in primer testing, cloning, and sequencing.
M.N. is supported by a KAUST-WHOI Special Academic Partnership Fellows award. Support for A.A. was provided by WHOI's Coastal Research Fund in Support of Scientific Staff, WHOI's Penzance Endowed Fund in Support of Assistant Scientists, and a grant from the National Science Foundation (OCE-1233612).
Footnotes
Published ahead of print 24 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00695-13.
REFERENCES
- 1. Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1–10 [Google Scholar]
- 2. Williams WM, Viner AB, Broughton WJ. 1987. Nitrogen fixation (acetylene reduction) associated with the living coral Acropora variabilis. Mar. Biol. 94:531–535 [Google Scholar]
- 3. Shashar N, Cohen Y, Loya Y, Sar N. 1994. Nitrogen fixation (acetylene reduction) in stony corals: evidence for coral-bacteria interactions. Mar. Ecol. Prog. Ser. 111:259–264 [Google Scholar]
- 4. Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. 2004. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305:997–1000 [DOI] [PubMed] [Google Scholar]
- 5. Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. 2007. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9:2707–2719 [DOI] [PubMed] [Google Scholar]
- 6. Daniels CA, Zeifman A, Heym K, Ritchie KB, Watson CA, Berzins I, Breitbart M. 2011. Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar. Ecol. Prog. Ser. 426:29–40 [Google Scholar]
- 7. Kooperman N, Ben-Dov E, Kramarsky-Winter E, Barak Z, Kushmaro A. 2007. Coral mucus-associated bacterial communities from natural and aquarium environments. FEMS Microbiol. Lett. 276:106–113 [DOI] [PubMed] [Google Scholar]
- 8. Sunagawa S, Woodley CM, Medina M. 2010. Threatened corals provide underexplored microbial habitats. PLoS One 5:e9554. 10.1371/journal.pone.0009554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. 10.1371/journal.pone.0002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bourne DG, Munn CB. 2005. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 7:1162–1174 [DOI] [PubMed] [Google Scholar]
- 14. Jessen C, Villa Lizcano JF, Bayer T, Roder C, Aranda M, Wild C, Voolstra CR. 2013. In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the Red Sea coral Acropora hemprichii. PLoS One 8:e62091. 10.1371/journal.pone.0062091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sunagawa S, DeSantis TZ, Piceno YM, Brodie EL, DeSalvo MK, Voolstra CR, Weil E, Andersen GL, Medina M. 2009. Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3:512–521 [DOI] [PubMed] [Google Scholar]
- 16. Apprill A, Hughen K, Mincer T. 21 March 2013. Major similarities in the bacterial communities associated with lesioned and healthy Fungiidae corals. Environ. Microbiol. [Epub ahead of print.]. 10.1111/1462-2920.12107 [DOI] [PubMed] [Google Scholar]
- 17. Yang C-S, Chen M-H, Arun AB, Chen CA, Wang J-T, Chen W-M. 2010. Endozoicomonas montiporae sp. nov., isolated from the encrusting pore coral Montipora aequituberculata. Int. J. Syst. Evol. Microbiol. 60:1158–1162 [DOI] [PubMed] [Google Scholar]
- 18. Klaus JS, Janse I, Fouke BW. 2011. Coral black band disease microbial communities and genotypic variability of the dominant cyanobacteria (CD1C11). Bull. Mar. Sci. 87:795–821 [Google Scholar]
- 19. Speck MD, Donachie SP. 2012. Widespread Oceanospirillaceae bacteria in Porites spp. J. Mar. Biol. 2012:1–7 [Google Scholar]
- 20. Peters EC. 1984. A survey of cellular reactions to environmental stress and disease in Caribbean scleractinian corals. Helgoland Mar. Res. 37:113–137 [Google Scholar]
- 21. Santavy D, Peters E. 1997. Microbial pests: coral disease research in the western Atlantic, p 607–612 In Proc. 8th Int. Coral Reef Symp. The WorldFish Center ReefBase Project, Penang, Malaysia [Google Scholar]
- 22. Ainsworth TD, Hoegh-Guldberg O. 2009. Bacterial communities closely associated with coral tissues vary under experimental and natural reef conditions and thermal stress. Aquat. Biol. 4:289–296 [Google Scholar]
- 23. Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319–346 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.