Abstract
Lymph nodes (mandibular, mesenteric, mediastinal, and subiliac; n = 68) and fecal (n = 68) and hide (n = 35) samples were collected from beef carcasses harvested in an abattoir in Mexico. Samples were analyzed for Salmonella, and presumptive colonies were subjected to latex agglutination. Of the isolates recovered, a subset of 91 was characterized by serotyping, pulsed-field gel electrophoresis (PFGE), and antimicrobial susceptibility phenotyping. Salmonella was isolated from 100% (hide), 94.1% (feces), 91.2% (mesenteric), 76.5% (subiliac), 55.9% (mandibular), and 7.4% (mediastinal) of samples. From the 87 typeable isolates, eight Salmonella enterica serotypes, including Kentucky (32.2%), Anatum (29.9%), Reading (17.2%), Meleagridis (12.6%), Cerro (4.6%), Muenster (1.1%), Give (1.1%), and Mbandaka (1.1%), were identified. S. Meleagridis was more likely (P = 0.03) to be recovered from lymph nodes than from feces or hides, whereas S. Kentucky was more likely (P = 0.02) to be recovered from feces and hides than from lymph nodes. The majority (59.3%) of the Salmonella isolates were pansusceptible; however, multidrug resistance was observed in 13.2% of isolates. Typing by PFGE revealed that Salmonella strains generally clustered by serotype, but some serotypes (Anatum, Kentucky, Meleagridis, and Reading) were comprised of multiple PFGE subtypes. Indistinguishable PFGE subtypes and, therefore, serotypes were isolated from multiple sample types, and multiple PFGE subtypes were commonly observed within an animal. Given the overrepresentation of some serotypes within lymph nodes, we hypothesize that certain Salmonella strains may be better at entering the bovine host than other Salmonella strains or that some may be better adapted for survival within lymph nodes. Our data provide insight into the ecology of Salmonella within cohorts of cattle and offer direction for intervention opportunities.
INTRODUCTION
The food-borne pathogen Salmonella causes substantial morbidity and mortality within the United States and elsewhere. Moreover, the public health burden imposed by Salmonella, measured by the number of illnesses attributed to Salmonella, actually increased between 2006 and 2008 (1) despite substantial efforts to control this food-borne pathogen. Of the pathogens included in FoodNet surveillance and detectable with laboratory procedures, Salmonella was the most common laboratory-confirmed infectious agent and accounted for 43.3% of infections, 53.9% of hospitalizations, and 42.6% of deaths caused by an identified pathogen (1). Salmonella infections are also an economic burden, often resulting in substantial medical costs and loss of productivity (2). Because 95% of salmonellosis cases are attributed to a food-borne route of infection (3–5), improved efforts to control this pathogen in the food supply are warranted. However, control of Salmonella in food is challenging given that it is ubiquitous in the environment (6) and associated with diverse food types (1). Furthermore, numerous serotypes have been observed across a variety of animal reservoirs (1), and many are commonly shed in the feces of seemingly healthy animals (6).
Cattle are an important reservoir for Salmonella (6–11), and consumption of ground beef has been linked to outbreaks of salmonellosis (7, 12–14). Moreover, Salmonella can be recovered somewhat routinely from preevisceration beef carcasses in the United States, although the reported recovery rate varied widely from 3.0% to 50.2% (15–17). Interventions used in abattoirs are highly efficacious at removing Salmonella from the carcass surface in that it is very uncommon to recover Salmonella from the surface of beef carcasses at the end of the slaughter process (15–18). Yet despite effective control of Salmonella surface contamination on beef carcasses, it is not altogether uncommon to isolate this pathogen from ground beef. Salmonella was recovered from 4.2% of 4,136 ground beef samples from commercial retailers across the United States (7), and in 2011, the USDA Food Safety Inspection Service recovered Salmonella from 2.4% of 13,517 25-gram ground beef samples. From samples collected in Mexico, Salmonella was detected in 8.0% of 262 ground beef samples purchased from butcher shops, street vendors, city markets, and supermarkets in Mexico City, Monterrey, and Guadalajara (19).
A recent but growing body of evidence demonstrates that cattle lymph nodes commonly harbor Salmonella (20–23) and that, when incorporated into ground beef, lymph nodes may contribute to the observed discrepancy between effective control on carcass surfaces and subsequent isolation from ground beef. A more complete understanding of the ecology of cattle and Salmonella is needed. In particular, important data gaps include an understanding of the diversity of Salmonella within and among lymph nodes of cattle, the routes by which Salmonella infects lymph nodes, and whether certain Salmonella serotypes are overrepresented within lymph nodes in comparison to other sites. Closing these knowledge gaps will aid development and application of targeted interventions to reduce the incidence at which Salmonella enters lymph nodes and/or the duration of survival within lymph nodes. The objective of this study was, therefore, to evaluate the diversity of Salmonella isolates recovered from lymph nodes, feces, and hide swabs collected from healthy cattle at harvest in a commercial abattoir.
MATERIALS AND METHODS
Sample collection.
Samples were acquired from 68 beef cattle presented for harvest at an abattoir in Mexico. From each beef carcass, lymph nodes (mandibular, mesenteric, mediastinal, and subiliac) and feces were collected. From a subset of these carcasses (n = 35), hide samples were also collected. Feces were sampled from the rectocolon portion of the intestinal tract of each beef carcass. An approximately 100-cm2 area of hide over the foreshank was swabbed using a sterile sponge prehydrated with 10 ml of buffered peptone water (World Bio Products, Mundelein, IL). All samples were stored on ice and transported to Texas Tech University for processing. United States Department of Agriculture APHIS permits (permit number 114031) were used to import the samples into the United States.
Lymph node analysis.
All lymph nodes were processed and analyzed for the presence of Salmonella as previously described (21). Briefly, fat and fascia were trimmed and lymph nodes were surface sterilized in boiling water. A modified incubation in tryptic soy broth (TSB; EMD, Darmstadt, Germany) at 42°C for 6 h was employed, and samples were subjected to immunomagnetic separation (IMS). Recovered IMS beads were enriched in Rappaport-Vasiliadis (RV; Remel, St. Louis, MO) broth and then streaked onto xylose lysine desoxycholate (XLD; Remel, St. Louis, MO) and brilliant green sulfa (BGS) agars. Salmonella latex agglutination kits (Remel, Lenexa, KS) were used on presumptively positive colonies (colonies with black centers, XLD; pink colonies, BGS). Colonies were transferred to 9 ml of TSB or brain heart infusion (BHI; BD Difco, Sparks, MD) containing 10% glycerol (EMD, Darmstadt, Germany) prior to incubation at 37°C for 18 to 24 h. From each isolate tube, 1-ml aliquots were frozen in duplicate at −80°C.
Fecal and hide analysis.
Fecal samples were enriched for Salmonella by inoculating 1.0 g ± 0.1 g into 9 ml each of tetrathionate (TT) broth (Difco, Sparks, MD) and RV broth (EMD, Darmstadt, Germany). Hide samples were homogenized (Seward model 400; Bohemia, NY) for 30 s, and 1 ml was transferred to 9 ml each of TT and RV. All fecal and hide TT and RV tubes were vortexed and then incubated at 42°C for 24 h. Following incubation, TT and RV broths were streaked for isolation onto xylose lysine tergitol-4 (XLT4; BD Difco, Sparks, MD) agar plates and incubated at 37°C for 24 h. Salmonella latex agglutination kits were used on presumptively positive colonies. Colonies were transferred to 9 ml of TSB or BHI containing 10% glycerol prior to incubation at 37°C for 18 to 24 h. From each isolate tube, 1-ml aliquots were frozen in duplicate at −80°C.
PFGE.
Carcasses (n = 18) harboring Salmonella within multiple (3 or 4 per carcass) lymph nodes, in feces, and/or on the hide were selected for pulsed-field gel electrophoresis (PFGE) typing. A total of 91 isolates were subjected to a PFGE protocol for Salmonella (24) using XbaI (Roche Applied Science, Indianapolis, IN) enzyme for restriction and electrophoresis (CHEF Mapper XA Chiller System; Bio-Rad Laboratories, Hercules, CA). To optimize pattern images and improve band intensity, cell lysis of agarose plugs was performed for up to 18 h and plug slices were allowed to restrict for a maximum of 7 h. Banding patterns were inspected by visual confirmation and then further analyzed and compared (BioNumerics 6.6; Applied Maths, Austin, TX) to identify genetic similarities among sample types both within an animal and among animals. The Dice similarity coefficient (band based), with 2% band tolerance and relaxed doublet matching options, was used to determine genetic similarity. Cluster analyses were calculated, and dendrograms were produced based upon pairwise similarities by means of the unweighted-pair group method using average linkages (UPGMA).
Salmonella serotyping.
Isolates analyzed by PFGE were subjected to molecular serotyping methods (25). Resulting genotypes were confirmed by traditional slide agglutination (O typing) and tube agglutination (flagellar H typing) methods using commercial antisera (Difco, BD Diagnostic Systems, Sparks, MD) by following the manufacturer's guidelines.
Antimicrobial susceptibility.
Isolates were streaked onto tryptic soy agar (TSA; EMD, Darmstadt, Germany) containing 5% defibrinated sheep blood (bioMérieux, Inc., Durham, NC) and incubated at 37°C for 18 to 24 h. Broth microdilution (Sensititre CMV2AGNF test plates; TREK Diagnostic Systems, Inc., Cleveland, OH) was used according to the manufacturer's guidelines. Salmonella isolates were classified as susceptible, intermediate, or resistant to each antimicrobial agent based upon breakpoints established by the Clinical and Laboratory Standards Institute (CLSI) or the National Antimicrobial Resistance Monitoring System (NARMS) (26, 27). Isolates exhibiting resistance to three or more classes of antimicrobials were classified as multidrug resistant (MDR).
Statistical analysis.
Data were analyzed as a binomial response distribution to estimate Salmonella prevalence and 95% confidence limits (CL) using commercially available software (SAS version 9.3; The SAS Institute, Cary, NC).
RESULTS
Salmonella was recovered from every sample type evaluated. Among lymph nodes, prevalence was 55.9% (95% CL, 43.7 to 67.4%), 91.2% (81.6 to 96.0%), 7.4% (3.1 to 16.2%), and 76.5% (64.8 to 85.2%) for mandibular, mesenteric, mediastinal, and subiliac nodes, respectively. Salmonella was recovered from 94.1% (95% CL, 85.3 to 97.8%) and 100.0% (90.1 to 100%) of fecal and hide samples, respectively.
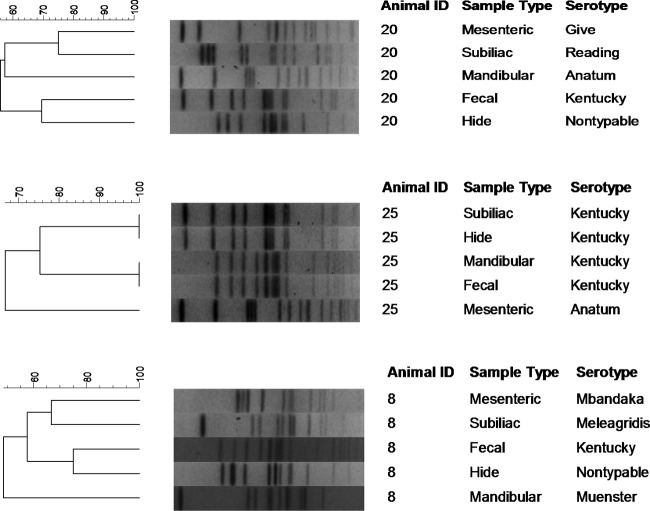
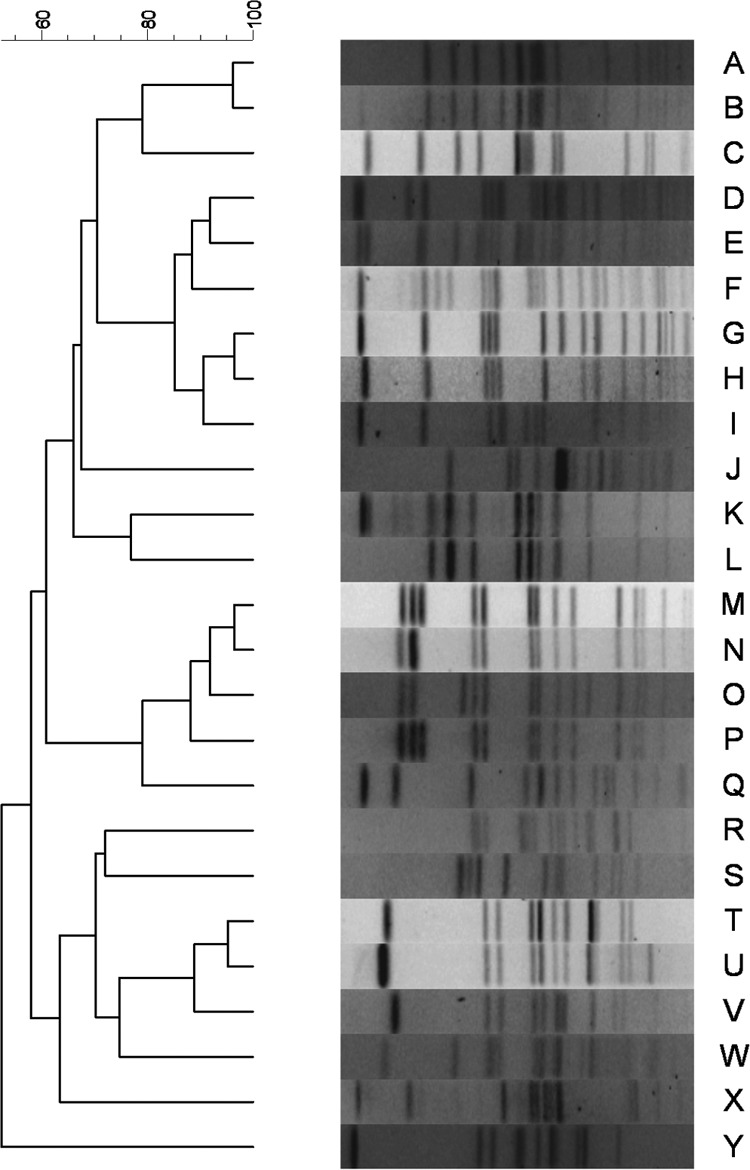
All beef carcasses (n = 68) harbored Salmonella within at least one sample type collected. Eighteen carcasses harbored Salmonella in five different sample types, and these isolates were selected for further characterization by PFGE subtyping, serotyping, and antimicrobial susceptibility testing (one isolate per sample type where available, for a total of 90 isolates). One hide isolate was found to be a mixed culture consisting of two different serotypes, and each serotype was treated as a separate isolate, for a total of 17 hide isolates and an overall total of 91 isolates. Of the 91 isolates subjected to serotyping, 87 isolates were typeable and 8 serotypes were identified (Table 1), including Salmonella enterica serovar Kentucky (32.2%), S. enterica serovar Anatum (29.9%), S. enterica serovar Reading (17.2%), S. enterica serovar Meleagridis (12.6%), S. enterica serovar Cerro (4.6%), S. enterica serovar Muenster (1.1%), S. enterica serovar Give (1.1%), and S. enterica serovar Mbandaka (1.1%). Four isolates (4.4%) were nontypeable given the methods employed. S. Reading was the only serotype isolated from all sample types. Salmonella Kentucky and S. Anatum were isolated from all sample types with the exception of the mediastinal lymph nodes. Salmonella Meleagridis was not recovered from mediastinal lymph nodes or hides; it was, however, recovered from feces and mandibular, mesenteric, and subiliac lymph nodes. Salmonella Give and S. Mbandaka were isolated from one mesenteric lymph node each, while S. Muenster was recovered from one mandibular lymph node. Salmonella Cerro was not recovered from hides or within the mesenteric and subiliac lymph nodes. Nontypeable isolates were recovered from feces, hides, and one mesenteric lymph node. In general, PFGE subtypes clustered by Salmonella serotype (Fig. 1 and Table 2). Salmonella Cerro was the only serotype with indistinguishable banding patterns among all isolates analyzed (n = 4). Salmonella Give, S. Mbandaka, and S. Muenster were each comprised of a single isolate that produced a banding pattern unique from all others. The remaining serotypes (S. Anatum, S. Kentucky, S. Meleagridis, and nontypeable) were represented by both indistinguishable and dissimilar (difference of 1 or more bands) subtypes (Fig. 1 and Table 2).
Table 1.
Salmonella enterica subspecies enterica serotypes isolated from lymph nodes, feces, and hides of beef cattle presented for harvest at an abattoir in Mexico
| Serotype | No. of each sample type with each serotype |
Total no. (n = 91a,b) | Overall % | |||||
|---|---|---|---|---|---|---|---|---|
| Mandibular (n = 18) | Mesenteric (n = 18) | Mediastinal (n = 2) | Subiliac (n = 18) | Feces (n = 18) | Hide (n = 17a) | |||
| Kentucky | 2 | 5 | 0 | 5 | 12 | 4 | 28 | 30.8 |
| Anatum | 9 | 2 | 0 | 5 | 3 | 7 | 26 | 28.6 |
| Reading | 5 | 2 | 2 | 3 | 1 | 2 | 15 | 16.5 |
| Meleagridis | 1 | 5 | 0 | 4 | 1 | 0 | 11 | 12.1 |
| Cerro | 0 | 1 | 0 | 1 | 0 | 2 | 4 | 4.4 |
| Muenster | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1.1 |
| Give | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1.1 |
| Mbandaka | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1.1 |
| Nontypeable | 0 | 1 | 0 | 0 | 1 | 2 | 4 | 4.4 |
One hide isolate was identified as a mixed culture of two serotypes, thereby increasing the number of hide isolates to 17 and the total isolate number to 91.
Only those isolates analyzed by PFGE were serotyped.
Fig 1.
Partial dendrogram representing the distribution of Salmonella enterica subspecies enterica pulsed-field gel electrophoresis subtypes among the lymph nodes, feces, and hides of cattle presented for harvest at a Mexico slaughter facility.
Table 2.
Salmonella isolate characterization by PFGE subtype, serotype, and antimicrobial susceptibility phenotype
| PFGE subtype | No. of isolates | No. of animals | Serotype | Antimicrobial susceptibilitya | No. of isolates of each sample type with each subtype |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mandibular | Mesenteric | Mediastinal | Subiliac | Fecal | Hide | |||||
| A | 11 | 8 | Kentucky | Pansusceptible | 1 | 2 | 0 | 1 | 5 | 2 |
| B | 1 | 1 | Kentucky | Pansusceptible | 0 | 0 | 0 | 1 | 0 | 0 |
| C | 14 | 9 | Kentucky | Pansusceptible | 1 | 1 | 0 | 3 | 7 | 2 |
| D | 1 | 1 | Anatum | (F), (C), Su | 0 | 0 | 0 | 0 | 0 | 1 |
| E | 1 | 1 | Kentucky | Pansusceptible | 0 | 0 | 0 | 0 | 0 | 1 |
| F | 1 | 1 | Anatum | Am, Ap, F, Su, Te, TS | 1 | 0 | 0 | 0 | 0 | 0 |
| G | 21 | 13 | Anatum | Pansusceptible; Te | 8 | 1 | 0 | 4 | 3 | 5 |
| H | 2 | 2 | Anatum | Pansusceptible; S, Su, Te | 0 | 1 | 0 | 0 | 0 | 1 |
| I | 1 | 1 | Anatum | Am, NA, Su, Te, TS | 0 | 0 | 0 | 1 | 0 | 0 |
| J | 1 | 1 | Kentucky | Te | 0 | 1 | 0 | 0 | 0 | 0 |
| K | 2 | 1 | Nontypeable | Pansusceptible; Ap, Su, Te, TS | 0 | 1 | 0 | 0 | 1 | 0 |
| L | 2 | 2 | Nontypeable | Pansusceptible | 0 | 0 | 0 | 0 | 0 | 2 |
| M | 10 | 8 | Reading | Te | 5 | 1 | 0 | 2 | 1 | 1 |
| N | 2 | 2 | Reading | Te | 0 | 0 | 2 | 0 | 0 | 0 |
| O | 1 | 1 | Reading | Te | 0 | 1 | 0 | 0 | 0 | 0 |
| P | 1 | 1 | Reading | Te | 0 | 0 | 0 | 1 | 0 | 0 |
| Q | 1 | 1 | Give | S, Te | 0 | 1 | 0 | 0 | 0 | 0 |
| R | 1 | 1 | Kentucky | Pansusceptible | 0 | 1 | 0 | 0 | 0 | 0 |
| S | 1 | 1 | Mbandaka | Pansusceptible | 0 | 1 | 0 | 0 | 0 | 0 |
| T | 6 | 5 | Meleagridis | S, Su, Te; Su, Te | 0 | 4 | 0 | 2 | 0 | 0 |
| U | 2 | 2 | Meleagridis | S, Su, Te | 0 | 0 | 0 | 1 | 1 | 0 |
| V | 2 | 2 | Meleagridis | Pansusceptible | 1 | 0 | 0 | 1 | 0 | 0 |
| W | 1 | 1 | Meleagridis | Am, Ap, F, S, Su, Te | 0 | 1 | 0 | 0 | 0 | 0 |
| X | 4 | 3 | Cerro | Pansusceptible; (F), (Te) | 0 | 1 | 0 | 1 | 0 | 2 |
| Y | 1 | 1 | Muenster | NA | 1 | 0 | 0 | 0 | 0 | 0 |
Am, amoxicillin-clavulanic acid; Ap, ampicillin; F, cefoxitin; C, chloramphenicol; NA, nalidixic acid; S, streptomycin; Su, sulfisoxazole; Te, tetracycline; TS, trimethoprim-sulfamethoxazole. Parentheses indicate intermediate resistance.
Dendrograms were constructed to examine the similarity of Salmonella strains isolated from samples collected from the same animal. Within a single beef carcass, indistinguishable subtypes were often found in two or more sample types throughout the carcass (Fig. 1 and 2 and Table 2). Three beef carcasses harbored a unique PFGE subtype within each sample type that was analyzed (Fig. 1 and 2 and Table 2).
Fig 2.
Animal-level distribution of Salmonella enterica subspecies enterica serotypes and pulsed-field gel electrophoresis subtypes among the lymph nodes, feces, and hides of 3 cattle presented for harvest at a Mexico slaughter facility.
Serotype-level comparisons suggest the possibility that certain serotypes may be better adapted for particular sample types. S. Meleagridis was significantly more likely to be recovered from lymph nodes than from feces or hides (P = 0.03). Conversely, S. Kentucky was more likely to be recovered from feces and hides than from lymph nodes (P = 0.02). While S. Reading appeared to be more commonly isolated from lymph nodes than from feces or hides, this relationship was not statistically significant (P = 0.20).
The majority (n = 54; 59.3%) of isolates were susceptible to all antibiotics (Table 3). Tetracycline resistance was observed in 22.0% (n = 20) of Salmonella strains. Multidrug resistance was observed for 13.2% (n = 12) of isolates. Of the isolates recovered from lymph nodes (n = 56), feces (n = 18), and hide samples (n = 17), 30 (53.6%), 3 (16.7%), and 4 (23.5%) exhibited resistance to one or more drugs, respectively. Antimicrobial susceptibility phenotypes generally clustered together with indistinguishable or similar PFGE subtypes and, therefore, serotypes (Fig. 1 and Table 2). Salmonella Reading, S. Meleagridis, and S. Anatum were the most commonly resistant serotypes.
Table 3.
Salmonella serotypes and observed antimicrobial resistance phenotypes recovered from lymph nodes (mandibular, mesenteric, mediastinal, and subiliac), feces, and hides of beef cattle at harvest in a Mexico slaughter facility
| Sample type (no. of isolates) | Serotype | Antimicrobial resistance phenotypea | No. of samples |
|---|---|---|---|
| Mandibular (18) | Reading | Te | 5 |
| Anatum | Te | 1 | |
| Muenster | NA | 1 | |
| Anatum | Am, Ap, F, Su, Te, TS | 1 | |
| Mediastinal (2) | Reading | Te | 2 |
| Mesenteric (18) | Reading | Te | 2 |
| Kentucky | Te | 1 | |
| Anatum | Te | 1 | |
| Meleagridis | Su, Te | 1 | |
| Give | S, Te | 1 | |
| Meleagridis | S, Su, Te | 3 | |
| Meleagridis | Am, Ap, F, S, Su, Te | 1 | |
| Subiliac (18) | Anatum | Te | 2 |
| Reading | Te | 3 | |
| Cerro | (F), (Te) | 1 | |
| Meleagridis | S, Su, Te | 3 | |
| Anatum | Am, NA, Su, Te, TS | 1 | |
| Fecal (18) | Reading | Te | 1 |
| Meleagridis | S, Su, Te | 1 | |
| Nontypeable | Ap, Su, Te, TS | 1 | |
| Hide (17) | Reading | Te | 1 |
| Anatum | Te | 1 | |
| Anatum | S, Su, Te | 1 | |
| Anatum | (F), (C), Su | 1 |
Am, amoxicillin-clavulanic acid; Ap, ampicillin; F, cefoxitin; C, chloramphenicol; NA, nalidixic acid; S, streptomycin; Su, sulfisoxazole; Te, tetracycline; TS, trimethoprim-sulfamethoxazole. Parentheses indicate intermediate resistance.
DISCUSSION
The data reported herein demonstrate that Salmonella can be recovered from various lymph nodes that are disseminated widely throughout the animal. Because lymph nodes are commonly included in beef trimmings intended for further manufacturing, such as grinding, lymph nodes may be an important source of Salmonella in ground beef. Koohmaraie et al. (28) concluded that cattle lymph nodes and hides are likely the sources of ground beef contamination upon comparison of Salmonella PFGE subtypes obtained from hides, prescapular lymph nodes, trim, and ground beef of 100 dairy cattle at slaughter. Similar to prescapular lymph nodes, the subiliac is positioned in fat trim such that it is commonly included in ground beef. Because Salmonella isolates within lymph nodes are protected from interventions employed by commercial abattoirs, current approaches to Salmonella control within abattoirs may need further exploration and development. One approach may be removal of certain lymph nodes during carcass disassembly. However, given the extensive number of lymph nodes throughout the body, complete removal of these tissues prior to ground beef production is not a feasible solution. Guo and colleagues (29) adopted the Hald Salmonella Bayesian model to evaluate food source attribution of human salmonellosis in the United States and estimated that ground beef contributes to 28% of infections. Recognizing that lymph nodes are likely a contributing factor to ground beef contamination, identification of effective interventions is necessary to mitigate the potential risk to public health.
In the data reported herein, we recovered Salmonella from 76.5% of the subiliac lymph nodes. Haneklaus and colleagues (23) reported similar rates of recovery in that Salmonella was detected in 88.2% (n = 85) of the subiliac and prescapular lymph nodes collected from cattle from one feedlot. In contrast, Arthur et al. (20) recovered Salmonella from 1.6% of subiliac and prescapular lymph nodes (n = 1,140) collected from cull and feedlot cattle in the United States. However, Salmonella prevalence appears to be seasonal among cattle, with reduced prevalence throughout colder months; thus, sample season may have contributed to this low prevalence. More-recent findings suggest a distinct regional and seasonal effect on Salmonella prevalence in bovine peripheral lymph nodes, with the highest prevalence occurring among feedlot cattle sampled throughout the summer and fall months in the southern United States (21). The present study supports these findings with an elevated prevalence of 76.5% for subiliac lymph nodes. Given the early fall sampling period and southern proximity of Mexico to the United States, our data provide additional evidence of the seasonal and regional trends previously described for Salmonella contamination in bovine lymph nodes.
Salmonella was isolated from the mandibular lymph node of over half (55.9%) of the beef carcasses sampled, a finding which is not necessarily surprising given that the mouth in particular, or head area in general, is a point of entry for Salmonella. It seems plausible, although it is not evaluated here, that other lymph nodes of the head and neck occasionally harbor Salmonella. If so, incision of these nodes at slaughter, during postslaughter government inspection, for example, may be a source of cross-contamination of Salmonella, both within a carcass and between carcasses, to other tissues, such as the Masseter muscle, which also is frequently incised during inspection. This route of cross-contamination might be an additional avenue by which Salmonella of lymph node origin may enter the food supply.
All serotypes recovered from lymph nodes in this study, with the possible exception of S. Give, have been previously isolated from ground beef (30). Additionally, given that each serotype identified in this study has been linked to laboratory-confirmed human illnesses (though none ranked among the top 20 serotypes associated with human illness in 2009 [31]), the potential impact of Salmonella carriage in lymph nodes on human health must be considered. The serotypes noted for causing the majority of laboratory-confirmed Salmonella outbreaks are commonly associated with chicken, eggs, and pork and include S. enterica serovar Enteritidis, S. enterica serovar Typhimurium, and S. enterica serovar Heidelberg (32); however, S. Typhimurium (33, 34) and S. Enteritidis (35) have also been implicated in ground beef outbreaks.
In general, PFGE subtypes clustered based upon serotype, with multiple PFGE patterns observed for S. Anatum, S. Kentucky, S. Meleagridis, and S. Reading. While serotypes were recovered across multiple sample types, we found evidence of sample type dependency among S. Meleagridis and S. Kentucky, a finding which may suggest a genetic adaptation associated with preferential colonization or improved survivability in the lymph node, fecal, or hide environments. Conversely, serotype Anatum was common in this study but lacked sample type dependency (P = 0.91). It is worth highlighting that different enrichment and isolation techniques were employed for fecal and hide samples than for lymph nodes, a result which may have contributed to the observed variability in serotypes and PFGE subtypes among the sample types. For example, if the IMS beads differ in terms of their affinity to various serotypes, it may explain the observed disparity in some serotypes between matrices because IMS beads were used in isolation of Salmonella from lymph nodes but not from feces or hides. In other words, the different culture methods may favor recovery of certain serotypes over others. Further, it is possible that the duration of infection varies between different sites (i.e., lymph nodes versus within the gastrointestinal tract [GIT]). If so, then the serotype recovered may be more a reflection of this duration of infection than a consequence of tissue tropism. Additionally, only one isolate per sample was characterized, and if mixed cultures were present, the probability of selection is related to the serotype abundance within the site. If, for example, S. Meleagridis were present in feces but at lower concentrations than S. Kentucky, it would be less likely to be selected. Thus, this finding of differences among some serotypes recovered from various matrices should be interpreted with caution. We believe, however, that this observation warrants further investigation to explore potential genetic adaptations that might contribute to tissue tropism and, therefore, the site of colonization.
Given the ubiquity of Salmonella in feedlot environments, isolation of diverse serotypes from fecal and hide samples is not surprising (36). It is generally believed that Salmonella enters the body from the gastrointestinal tract (GIT) and may disseminate systemically via the cardiovascular system (i.e., subsequent to bacteremia or septicemia). While this is distinctly possible, our data warrant exploration of an alternative hypothesis for other, non-GIT routes of entry into the animal. If, for instance, Salmonella disseminates systemically via the vasculature, then it would be expected that Salmonella would be recovered from multiple nodes. In other words, statistical dependency would be observed for the likelihood of recovering Salmonella among lymph nodes within an animal. This was, however, not observed in the data described herein. Of the 68 animals from which lymph node and fecal samples were collected, Salmonella was recovered from all four nodes as well as feces from 2.9% (n = 2) of animals. The joint probability that all of these samples would be positive is 2.7% (i.e., almost identical to that observed). Saliently, joint probability assumes statistical independence in terms of the outcome (Salmonella recovery) among the various samples within a carcass. These data, therefore, suggest that Salmonella entered the lymph nodes (within an animal) following different and independent events. Furthermore, different serotypes and PFGE subtypes were routinely isolated within a single carcass. If so, these different and independent events may include multiple translocations from the GIT or translocations across various points of the integument and capture by regionally draining lymph nodes. Given these observations, we hypothesize that Salmonella recovered from peripheral lymph nodes—at least the subiliac lymph node—may have entered the body transdermally and then drained to the regional peripheral lymph node. For example, Salmonella may be introduced transdermally from the hide on the ventral abdomen by biting flies, which can harbor Salmonella (37), or skin abrasions and then enter the lymphatic vessels from the interstitial spaces and ultimately end up in the subiliac lymph node. If so, the ecology of Salmonella within cattle populations may be far more complex than that implied by a simple fecal-oral route of transmission and may require a different approach for the identification and evaluation of control strategies. Further research is warranted to evaluate this alternative route of transmission hypothesis.
Our data illustrate the frequent isolation of Salmonella from lymph nodes destined for ground beef production. Antimicrobial resistance phenotypes were observed among 40.7% (n = 37) of the 91 Salmonella isolates characterized, 13.2% of which exhibited MDR phenotypes. These data are not overly dissimilar from data reported elsewhere in that other investigators reported that 14% (n = 266) of feedlot and cull cattle subiliac lymph nodes harbored antimicrobial-resistant Salmonella and 8.3% harbored MDR Salmonella (21). Arthur and colleagues (20) isolated Salmonella (n = 18) from subiliac and superficial cervical lymph nodes and reported resistance among 22.2% (n = 4) of isolates, with 16.7% (n = 3) demonstrating MDR phenotypes. Salmonella antimicrobial resistance poses a challenge to human medicine, as ceftriaxone, an extended-spectrum cephalosporin, is a common choice for treating salmonellosis in children (38). Adult salmonellosis is often treated with nalidixic acid and ciprofloxacin, a quinolone and fluoroquinolone, respectively (38). In our study, one subiliac lymph node exhibited resistance to nalidixic acid while resistance to ceftriaxone or ciprofloxacin was not observed. Thus, based upon these data, we conclude that the consequence to public health is likely low at present time.
Conclusions.
Salmonella isolates were frequently recovered from a variety of lymph nodes collected from animals presented for harvest in a Mexico abattoir. Salmonella strains harbored within peripheral lymph nodes are frequently included in beef trimmings that are processed into ground beef. We hypothesize that Salmonella may enter the body transdermally through skin abrasions or biting insects and then drain to the regional lymph node. If so, this route of entry provides insight into control of Salmonella within preharvest environments.
ACKNOWLEDGMENTS
This work was supported by the International Center for Food Industry Excellence at Texas Tech University, The Beef Checkoff, and USDA National Integrated Food Safety Initiative contract number 2011-51110-31081.
We thank Kim Kucera, Alexandra Calle, Diana Ayala, and Ashley Hartzog-Hawkins for their excellent technical support.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
Footnotes
Published ahead of print 21 June 2013
REFERENCES
- 1. CDC 2011. Vital signs: incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 1996–2010. MMWR Morb. Mortal. Wkly. Rep. 60:749–755 [PubMed] [Google Scholar]
- 2. Adhikari B, Angulo F, Meltzer M. 2004 Economic burden of Salmonella infections in the United States, abstr 20050. Abstr. 2004 Annu. Meet. Am. Agric. Econ. Assoc., 1 to 4 August 2004 American Agricultural Economics Association, Denver, CO [Google Scholar]
- 3. Bacon RT, Sofos JN, Belk KE, Hyatt DR, Smith GC. 2002. Prevalence and antibiotic susceptibility of Salmonella isolated from beef animal hides and carcasses. J. Food Prot. 65:284–290 [DOI] [PubMed] [Google Scholar]
- 4. Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tauxe RV. 1991. Salmonella: a postmodern pathogen. J. Food Prot. 54:563–568 [DOI] [PubMed] [Google Scholar]
- 6. Fedorka-Cray PJ, Dargatz DA, Thomas LA, Gray JT. 1998. Survey of Salmonella serotypes in feedlot cattle. J. Food Prot. 61:525–530 [DOI] [PubMed] [Google Scholar]
- 7. Bosilevac JM, Guerini MN, Kalchayanand N, Koohmaraie M. 2009. Prevalence and characterization of Salmonella in commercial ground beef in the United States. Appl. Environ. Microbiol. 75:1892–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bean NH, Griffin PM. 1990. Foodborne disease outbreaks in the United States, 1973–1987: pathogens, vehicles, and trends. J. Food Prot. 53:804–817 [DOI] [PubMed] [Google Scholar]
- 9. Lammerding AM, Garcia MM, Mann ED, Robinson Y, Dorward WJ, Truscott RB, Tittiger F. 1988. Prevalence of Salmonella and thermophilic Campylobacter in fresh pork, beef, veal and poultry in Canada. J. Food Prot. 51:47–52 [DOI] [PubMed] [Google Scholar]
- 10. Fegan N, Vanderlinde P, Higgs G, Desmarchelier P. 2005. A study of the prevalence and enumeration of Salmonella enterica in cattle and on carcasses during processing. J. Food Prot. 68:1147–1153 [DOI] [PubMed] [Google Scholar]
- 11. Loneragan GH, Thomson DU, McCarthy RM, Webb HE, Daniels AE, Edrington TS, Nisbet DJ, Trojan SJ, Rankin SC, Brashears MM. 2012. Salmonella diversity and burden in cows on and culled from dairy farms in the Texas high plains. Foodborne Pathog. Dis. 9:549–555 [DOI] [PubMed] [Google Scholar]
- 12. CDC 2002. Outbreak of multidrug-resistant Salmonella Newport—United States, January–April 2002. MMWR Morb. Mortal. Wkly. Rep. 51:545–548 [PubMed] [Google Scholar]
- 13. CDC 2006. Multistate outbreak of Salmonella Typhimurium infections associated with eating ground beef—United States, 2004. MMWR Morb. Mortal. Wkly. Rep. 55:180–182 [PubMed] [Google Scholar]
- 14. McLaughlin JB, Castrodale LJ, Gardner MJ, Ahmed R, Gessner BD. 2006. Outbreak of multidrug-resistant Salmonella Typhimurium associated with ground beef served at a school potluck. J. Food Prot. 69:666–670 [DOI] [PubMed] [Google Scholar]
- 15. Barkocy-Gallagher GA, Arthur TM, Rivera-Betancourt M, Nou X, Shackelford SD, Wheeler TL, Koohmaraie M. 2003. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. J. Food Prot. 66:1978–1986 [DOI] [PubMed] [Google Scholar]
- 16. Brichta-Harhay DM, Guerini MN, Arthur TM, Bosilevac JM, Kalchayanand N, Shackelford SD, Wheeler TL, Koohmaraie M. 2008. Salmonella and Escherichia coli O157:H7 contamination on hides and carcasses of cull cattle presented for slaughter in the United States: an evaluation of prevalence and bacterial loads by immunomagnetic separation and direct plating methods. Appl. Environ. Microbiol. 74:6289–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rivera-Betancourt M, Shackelford SD, Arthur TM, Westmoreland KE, Bellinger G, Rossman M, Reagan JO, Koohmaraie M. 2004. Prevalence of Escherichia coli O157:H7, Listeria monocytogenes, and Salmonella in two geographically distant commercial beef processing plants in the United States. J. Food Prot. 67:295–302 [DOI] [PubMed] [Google Scholar]
- 18. Rose BE, Hill WE, Umholtz R, Ransom GM, James WO. 2002. Testing for Salmonella in raw meat and poultry products collected at federally inspected establishments in the United States, 1998 through 2000. J. Food Prot. 65:937–947 [DOI] [PubMed] [Google Scholar]
- 19. Pond AR, Miller MF, Echeverry A, Loneragan GH, Rubio M, Chavez A, Huerta N, Rupnow J, Brashears MT, Brashears MM. 2010. Salmonella and pathogenic E. coli prevalence and generic E. coli quantitative baselines in raw pork and beef at retail outlets in Mexico. J. Food Prot. 73(Suppl A):175 [Google Scholar]
- 20. Arthur TM, Brichta-Harhay DM, Bosilevac JM, Guerini MN, Kalchayanand N, Wells JE, Shackelford SD, Wheeler TL, Koohmaraie M. 2008. Prevalence and characterization of Salmonella in bovine lymph nodes potentially destined for use in ground beef. J. Food Prot. 71:1685–1688 [DOI] [PubMed] [Google Scholar]
- 21. Gragg SE, Loneragan GH, Brashears MM, Arthur TM, Bosilevac JM, Kalchayanand N, Wang R, Schmidt JW, Brooks JC, Shackelford SD, Wheeler TL, Brown TR, Edrington TS, Brichta-Harhay DM. 2013. Salmonella enterica carriage in subiliac lymph nodes of cull and feedlot cattle at harvest. Foodborne Pathog. Dis. 10:368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Samuel JL, O'Boyle D, Mathers WJ, Frost AJ. 1980. Distribution of Salmonella in the carcases of normal cattle at slaughter. Res. Vet. Sci. 28:368–372 [PubMed] [Google Scholar]
- 23. Haneklaus AN, Harris KB, Griffin DB, Edrington TS, Lucia LM, Savell JW. 2012. Salmonella prevalence in bovine lymph nodes differs among feedyards. J. Food Prot. 75:1131–1133 [DOI] [PubMed] [Google Scholar]
- 24. CDC 2009. One-day (24–28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, Salmonella serotypes, Shigella sonnei, and Shigella flexneri by pulsed field gel electrophoresis (PFGE). PulseNet International, Atlanta, GA [Google Scholar]
- 25. Herrera-León S, McQuiston JR, Usera MA, Fields PI, Garaizar J, Echeita MA. 2004. Multiplex PCR for distinguishing the most common phase-1 flagellar antigens of Salmonella spp. J. Clin. Microbiol. 42:2581–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. CDC 2012. National Antimicrobial Monitoring System for enteric bacteria (NARMS): human isolates final report, 2010. US Department of Health and Human Services, Atlanta, GA [Google Scholar]
- 27. CDC 2012. National Antimicrobial Resistance Monitoring System for enteric bacteria (NARMS): manual of laboratory methods. US Department of Health and Human Services, Atlanta, Georgia [Google Scholar]
- 28. Koohmaraie M, Scanga JA, De La Zerda MJ, Koohmaraie B, Tapay L, Beskhlebnaya V, Mai T, Greeson K, Samadpour M. 2012. Tracking the sources of Salmonella in ground beef produced from nonfed cattle. J. Food Prot. 75:1464–1468 [DOI] [PubMed] [Google Scholar]
- 29. Guo C, Hoekstra RM, Schroeder CM, Pires SM, Ong KL, Hartnett E, Naugle A, Harman J, Bennett P, Cieslak P, Scallan E, Rose B, Holt KG, Kissler B, Mbandi E, Roodsari R, Angulo FJ, Cole D. 2011. Application of Bayesian techniques to model the burden of human salmonellosis attributable to U.S. food commodities at the point of processing: adaptation of a Danish model. Foodborne Pathog. Dis. 8:509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. FSIS 2011. Serotypes profile of Salmonella isolates from meat and poultry products: January 1998 through December 2010. Food Safety and Inspection Service, US Department of Agriculture, Washington, DC: http://www.fsis.usda.gov/PDF/Serotypes_Profile_Salmonella_2010.pdf [Google Scholar]
- 31. CDC 2011. National Salmonella surveillance annual data summary, 2009. US Department of Health and Human Services, Atlanta, GA [Google Scholar]
- 32. CDC 2012. Foodborne Outbreak Online Database (FOOD). US Department of Health and Human Services, Atlanta, GA: http://wwwn.cdc.gov/foodborneoutbreaks/Default.aspx [Google Scholar]
- 33. CDC 15 March 2013, posting date Multistate outbreak of Salmonella Typhimurium infections linked to ground beef (final update). Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/salmonella/typhimurium-01-13/index.html [Google Scholar]
- 34. CDC 1 February 2012, posting date Investigation update: multistate outbreak of human Salmonella Typhimurium infections linked to ground beef. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/salmonella/typhimurium-groundbeef/020112/index.html [Google Scholar]
- 35. CDC 13 September 2012, posting date Multistate outbreak of Salmonella Enteritidis infections linked to ground beef (final update). Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/salmonella/enteritidis-07-12/index.html [Google Scholar]
- 36. Kunze DJ, Loneragan GH, Platt TM, Miller MF, Besser TE, Koohmaraie M, Stephens T, Brashears MM. 2008. Salmonella enterica burden in harvest-ready cattle populations from the southern high plains of the United States. Appl. Environ. Microbiol. 74:345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mian LS, Maag H, Tacal JV. 2002. Isolation of Salmonella from muscoid flies at commercial animal establishments in San Bernardino County, California. J. Vector Ecol. 27:82–85 [PubMed] [Google Scholar]
- 38. Brichta-Harhay DM, Arthur TM, Bosilevac JM, Kalchayanand N, Shackelford SD, Wheeler TL, Koohmaraie M. 2011. Diversity of multidrug-resistant Salmonella enterica strains associated with cattle at harvest in the United States. Appl. Environ. Microbiol. 77:1783–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]