Abstract
A novel arsenate-reducing bacterium, designated strain PSR-1, was isolated from arsenic-contaminated soil. Strain PSR-1 was phylogenetically closely related to Anaeromyxobacter dehalogenans 2CP-1T with 16S rRNA gene similarity of 99.7% and coupled the oxidation of acetate with the reduction of arsenate. Arsenate reduction was inhibited almost completely by respiratory inhibitors such as dicumarol and 2-heptyl-4-hydroxyquinoline N-oxide. Strain PSR-1 also utilized soluble Fe(III), ferrihydrite, nitrate, oxygen, and fumarate as electron acceptors. Strain PSR-1 catalyzed the release of arsenic from arsenate-adsorbed ferrihydrite. In addition, inoculation of washed cells of strain PSR-1 into sterilized soil successfully reproduced arsenic release. Arsenic K-edge X-ray absorption near-edge structure (XANES) analysis revealed that the proportion of arsenite in the soil solid phase actually increased from 20% to 50% during incubation with washed cells of strain PSR-1. These results suggest that strain PSR-1 is capable of reducing not only dissolved arsenate but also arsenate adsorbed on the soil mineral phase. Arsenate reduction by strain PSR-1 expands the metabolic versatility of Anaeromyxobacter dehalogenans. Considering its distribution throughout diverse soils and anoxic sediments, Anaeromyxobacter dehalogenans may play a role in arsenic release from these environments.
INTRODUCTION
The predominant chemical forms of arsenic in the environment are inorganic arsenate [As(V)] and arsenite [As(III)]. Arsenate is thermodynamically stable under oxic conditions, while arsenite is stable under reducing conditions and is much more toxic than arsenate (1, 2). Elevated levels of arsenic in groundwater threaten the health of people worldwide. Bangladesh and West Bengal have the most serious groundwater arsenic problem, and approximately 60 to 100 million people are exposed to more than 10 μg liter−1 of arsenic in drinking water (1). Arsenic sorption on metal oxide minerals, especially on iron (hydr)oxides, is an important process controlling the dissolved concentration of arsenic in various environments. Generally, arsenate is strongly associated with soil minerals, including iron, aluminum, and manganese (hydr)oxides, whereas arsenite predominantly adsorbs to iron (hydr)oxides and is more mobile than arsenate (3–5).
Microorganisms can reduce arsenate either by a detoxification pathway or by an energy-conserving respiratory pathway. The former is catalyzed by the arsenic-resistant system, in which dissolved arsenate is taken up into cytosol by a phosphate transporter (such as Pit in Escherichia coli), reduced to arsenite by a soluble arsenate reductase, ArsC, and extruded outside the cell by an efflux pump ArsB. However, the arsenic-resistant system may play a relatively minor role in the release of arsenate adsorbed on soil minerals, since it can reduce dissolved arsenate only in the liquid phase (6). Thus, the latter microorganisms, i.e., dissimilatory arsenate-reducing bacteria, are considered to be much more important in the release of arsenic in flooded soils and anoxic sediments (7, 8). Dissimilatory arsenate-reducing bacteria are capable of utilizing arsenate as a terminal electron acceptor for respiration and are phylogenetically diverse, including members of Firmicutes, Gamma-, Delta-, and Epsilonproteobacteria (9, 10). In these bacteria, arsenate reduction is considered to be catalyzed by the dissimilatory arsenate reductase complex, which consists of a large molybdenum-containing subunit (ArrA) and a small iron-sulfur-containing subunit (ArrB) (11, 12).
Anaeromyxobacter dehalogenans was first isolated by Cole et al. (13) and was later described by Sanford et al. (14) as a chlororespiring facultative anaerobic myxobacterium within the Deltaproteobacteria. It can utilize a wide variety of electron acceptors for growth, including ortho-substituted halophenols (such as 2-chlorophenol), nitrate, oxygen, fumarate, and Fe(III) (14, 15). Cell suspensions of A. dehalogenans were also found to reduce U(VI) and Se(IV) (16, 17). The metabolic versatility of A. dehalogenans makes it a promising candidate for bioremediation, such as the in situ biostimulation of U(VI) immobilization (18). In 16S rRNA gene-based community analysis of an in situ bioremediation experiment, A. dehalogenans-related microorganisms have been found to be important metal-reducing bacteria together with Geobacter-related microorganisms (19, 20).
In this study, we isolated strain PSR-1, a novel dissimilatory arsenate-reducing bacterium, from arsenic-contaminated soil and found that it is phylogenetically closely related to A. dehalogenans, with 16S rRNA gene sequence similarity of 99.7%. To our knowledge, arsenate reduction by Anaeromyxobacter sp. has not been reported so far. For detailed understanding of the metabolic versatility of A. dehalogenans, physiological characterization of strain PSR-1 was performed with special attention to its arsenic release from the soil mineral phase.
MATERIALS AND METHODS
Enrichment and isolation.
A strict anaerobic technique (21) was used in the preparation of the minimal medium and manipulation of the enrichments. Arsenic-contaminated soil was collected from a site formerly used as a chemical plant in Japan. It contained approximately 13,000 μmol kg−1 of arsenic. The enrichment culture was prepared by inoculating 1 ml soil slurry (a mixture of 20 g wet weight of the soil and 60 ml distilled water) into 19 ml minimal medium. The medium contained the following (per liter): NH4Cl (0.535 g), KH2PO4 (0.136 g), MgCl2·6H2O (0.204 g), CaCl2·2H2O (0.147 g), trace mineral element solution (1 ml), vitamin solution (1 ml), 1 g liter−1 resazurin solution (1 ml), and NaHCO3 (2.52 g). The medium was dispensed into 60-ml serum bottles under an N2:CO2 (80:20) atmosphere and autoclaved. Acetate (final concentration of 2 mM), arsenate (5 mM), and cysteine-HCl (1 mM) were added separately from sterile anaerobic stock solutions. We added 1 mM cysteine to the medium as a reducing agent, but this concentration was not sufficient for the formation of a highly reducing condition, which was judged from the color of resazurin in the medium. Addition of more than 1 mM cysteine was inappropriate, since abiotic reduction of arsenate occurred significantly. The enrichment was incubated at 30°C in the dark without shaking. The concentration of arsenate and arsenite in the culture supernatant was determined by high-performance liquid chromatography (HPLC) (L-7000 system; Hitachi, Tokyo, Japan) with an Aminex HPX-87H ion exclusion column (Bio-Rad Laboratories, Hercules, CA). After complete reduction of arsenate was confirmed, 1 ml of the enrichment was transferred to 19 ml of fresh liquid medium.
After several rounds of subculturing, the enrichment was serially diluted and inoculated into anaerobic shake tubes prepared with the minimal medium and 2% Bacto agar (Difco, Sparks, MD). After incubation at 30°C, a single colony was picked and inoculated into new shake tubes, and the resulting single colony was picked again to ensure purity. The pure culture was then grown anaerobically on acetate and arsenate in liquid medium.
Growth experiments.
When potential electron donors were tested, 5 mM arsenate was used as the sole electron acceptor, and the electron donors being tested were added to the minimal medium at 5 mM. The strain was considered positive for electron donor utilization if more than 3 mM arsenate consumption as well as arsenite production was confirmed by HPLC.
Fe(III) chelated with nitrilotriacetic acid [Fe(III)-NTA], ferrihydrite, nitrate, nitrite, oxygen, manganese oxide (MnO2), selenate, fumarate, malate, sulfate, thiosulfate, sulfite, and elemental sulfur were tested as potential electron acceptors. Acetate (5 mM) was used as the electron donor and carbon source, and the electron acceptors being tested were added to the medium at 5 mM, except for ferrihydrite (2 mmol liter−1) and oxygen (0.6% to 21%). The strain was considered positive for electron acceptor utilization if more than 1 mM acetate consumption was confirmed by HPLC. In cases of mineral reduction, utilization was assessed by observation of color changes from black to white (MnO2), from clear to red [Se(VI)], and from dark brown to black [Fe(III)-NTA]. Reduction of ferrihydrite was confirmed by colorimetric detection of dissolved Fe(II) by the ferrozine method (22). Two-line ferrihydrite was synthesized using the method of Schwertmann and Cornell (23). When growth on oxygen was tested, cysteine was omitted from the minimal medium.
Sequencing and phylogenetic analysis of 16S rRNA gene.
Genomic DNA of strain PSR-1 was isolated as described previously (24). The 16S rRNA gene was amplified by PCR using the bacterial consensus primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′, Escherichia coli positions 8 to 27) and 1491R (5′-GGTTACCTTGTTACGACTT-3′, Escherichia coli positions 1509 to 1491). PCR products were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced using a BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI Prism 3100 Genetic Analyzer (Applied Biosystems) and appropriate sequencing primers (25). The obtained 16S rRNA gene sequences were subjected to a BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/) to determine sequence similarity. The retrieved sequences were aligned using the ClustalX program, version 2.0. The phylogenetic tree was constructed using the neighbor-joining method (26). Bootstrap values were obtained for 1,000 replicates to estimate the confidence level of the tree topologies.
PCR-DGGE.
DNA was extracted from the enrichment using a FastDNA Spin kit (MP Biomedicals, Irvine, CA) according to the manufacturer's instructions. PCR-denaturing gradient gel electrophoresis (PCR-DGGE) was performed according to the method reported by Muyzer et al. using the primers 341fGC and 534r (27). The PCR protocol used was as follows: (i) initial denaturation at 94°C for 10 min; (ii) 35 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 45 s, and extension at 72°C for 30 s; and (iii) final extension at 72°C for 30 min. DGGE was performed using a DCode universal mutation detection system (Bio-Rad Laboratories) as described previously (28). The major bands were excised and used for reamplification with the primers 341f and 534r; the products obtained were sequenced as described previously (28).
Arsenic release from arsenate-adsorbed ferrihydrite.
Arsenate-adsorbed ferrihydrite was prepared by adding 3 g of ferrihydrite to 50 ml of KH2AsO4 solution (500 mM) and stirring for 24 h. After centrifugation, the precipitate was washed twice with distilled water and freeze-dried. The atomic ratio of As to Fe in arsenate-adsorbed ferrihydrite was 0.4. If it dissolved completely in the minimal medium, As and Fe concentrations in the liquid phase theoretically equaled 7.2 and 17.8 mM, respectively.
Strain PSR-1 was pregrown with either arsenate or Fe(III)-NTA as the electron acceptor. After the culture (1 ml) was centrifuged (15,000 × g, 10 min), the cells were washed with sterile 0.8% NaCl and resuspended in 1 ml of 0.8% NaCl. The cell suspension (1 ml) was inoculated into the minimal medium (19 ml) containing 50 mg of arsenate-adsorbed ferrihydrite as the sole electron acceptor. Acetate (2 mM) was also included in the minimal medium as the electron donor. Shewanella oneidensis MR-1 (ATCC 700550) was pregrown in the minimal medium supplemented with 0.1 g liter−1 Casamino Acids, with 10 mM lactate and 5 mM Fe(III)-NTA as the electron donor and acceptor, respectively. Washed cells of MR-1 were prepared and inoculated in the same manner as strain PSR-1, except that 2 mM lactate was used as the electron donor.
Arsenic release from sterile soil inoculated with washed cells of strain PSR-1.
To reproduce arsenic release from soil, washed cells of strain PSR-1 were inoculated into sterile soil slurry. Soil collected from a fallow paddy field containing approximately 527 μmol kg−1 of total arsenic (29) was used in this experiment. Soil slurry (a mixture of 20 g wet weight of the soil and 40 ml of distilled water) was dispensed into 60-ml serum bottles, and the bottles were flushed with a N2 gas stream and sealed with butyl rubber stoppers and aluminum caps. The bottles were then sterilized with gamma ray irradiation at 50 kGy. After irradiation, 3 mM acetate was added to the slurry as the electron donor, and it was flushed with H2 gas aseptically to maintain an Eh value of less than −200 mV. Strain PSR-1 was pregrown either on arsenate or on Fe(III)-NTA as the electron acceptor. After the culture (13.3 ml) was centrifuged (10,000 × g, 20 min), the cells were washed 3 times with sterile 0.8% NaCl and resuspended in 1 ml of 0.8% NaCl. The cell suspension (1 ml) was inoculated into the sterile soil slurry. A background (no cells inoculated) control was also prepared. The slurries were incubated for 1 week at 30°C in the dark without shaking. After the incubation, arsenate and arsenite in the solution phase were analyzed with an HPLC system (PU 712i; GL Science, Tokyo, Japan) connected with a quadrupole inductively coupled plasma mass spectrometer (ICP-MS) (Elan DRC-e; PerkinElmer, Waltham, MA) as described previously (29).
For analysis of arsenic speciation in the soil solid phase, arsenic K-edge (11,867-eV) X-ray absorption near-edge structure (XANES) spectra were acquired at BL12C at the Photon Factory (High-Energy Accelerator Research Organization, KEK, Tsukuba, Japan) as described previously (29, 30). Briefly, the XANES spectra of soil samples were collected in the fluorescence detection mode using a 19-element Ge semiconductor detector, whereas those of the reference materials (Na2AsO3 and NaHAsO4) were collected in the transmission mode. The soil samples were analyzed as a wet paste at an ambient temperature. Compositions of arsenic species in the soil solid phases were evaluated by linear combination fitting (LCF) of XANES spectra with reference compounds. The REX 2000 ver. 2.5 program packages (Rigaku, Tokyo, Japan) were used to subtract the pre-edge background and normalize the spectra and LCFs.
Amplification of putative arrA.
Attempts to amplify a putative arrA gene were done using conventional or nested PCR according to the PCR protocols described by Song et al. (31). The amplified products (approximately 630 to 640 bp) were confirmed on 2% agarose gels by electrophoresis. Other PCR primers, including ArrAfwd and ArrArev (32), HAArrA-D1F and HAArrA-G2R (33), and ArrAUF1 and ArrAUR3 (34), were also tested. We also designed a new degenerate PCR primer based on the codon usage by A. dehalogenans 2CP-C. The designed primers, ArrPSRfwd (5′-AGTTCGTSCCSATCWSSTGGGAC-3′) and ArrPSRrev (5′-ACTCSGGSGTSYKGTCCTTSAG-3′), target two conserved regions of ArrA [KFVPISWD and LKD(K/R)TPEW], whose sequences have been frequently used for design of degenerate PCR primers (31–33).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences identified in this study have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers AB795400 (strain PSR-1) and AB821349 through AB821352 (DGGE bands).
RESULTS
Isolation of arsenate-reducing bacterium strain PSR-1.
An arsenate-reducing enrichment was prepared by inoculating arsenic-contaminated soil containing approximately 13,000 μmol kg−1 of arsenic into the minimal medium. After complete reduction of arsenate and concomitant production of arsenite were observed, the enrichment (1 ml) was transferred to the fresh medium. After 3 rounds of subculturing, the microbial community in the enrichment was analyzed by PCR-DGGE targeting the 16S rRNA gene. As shown in Fig. 1 and Table 1, bacteria closely related to Geobacter sp., Sedimentibacter spp., and Anaeromyxobacter dehalogenans were predominant in the enrichment.
Fig 1.
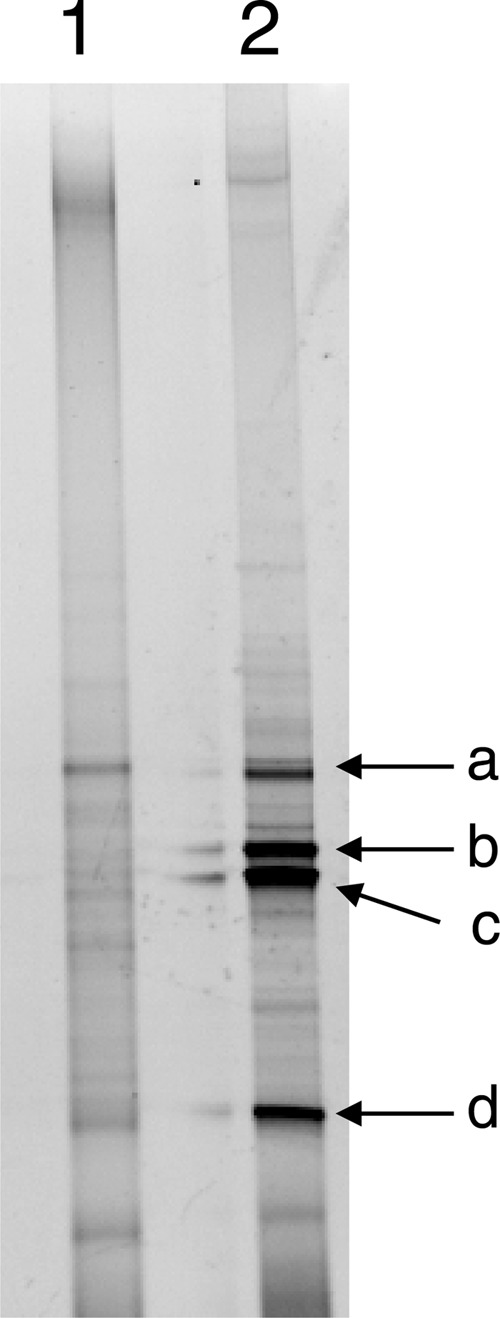
PCR-DGGE analysis of the arsenate-reducing enrichment prepared from arsenic-contaminated soil. The DGGE profiles of the original soil (lane 1) and the enrichment (lane 2) are shown. Arrows indicate bands recovered for DNA sequencing (see Table 1).
Table 1.
Sequence analysis of 16S rRNA genes recovered from DGGE bands
| Band | Length (no. of bases) | Most closely related organism in GenBank database | Phylum | Accession no. | % similarity |
|---|---|---|---|---|---|
| a | 198 | Geobacter sp. EB1 | Deltaproteobacteria | JX287365 | 94 |
| b | 174 | Sedimentibacter sp. JN18_V27_I | Firmicutes | EF059533 | 96 |
| c | 139 | Sedimentibacter sp. JLN1 | Firmicutes | JQ918080 | 96 |
| d | 198 | Anaeromyxobacter dehalogenans 2CP-1 | Deltaproteobacteria | CP001359 | 98 |
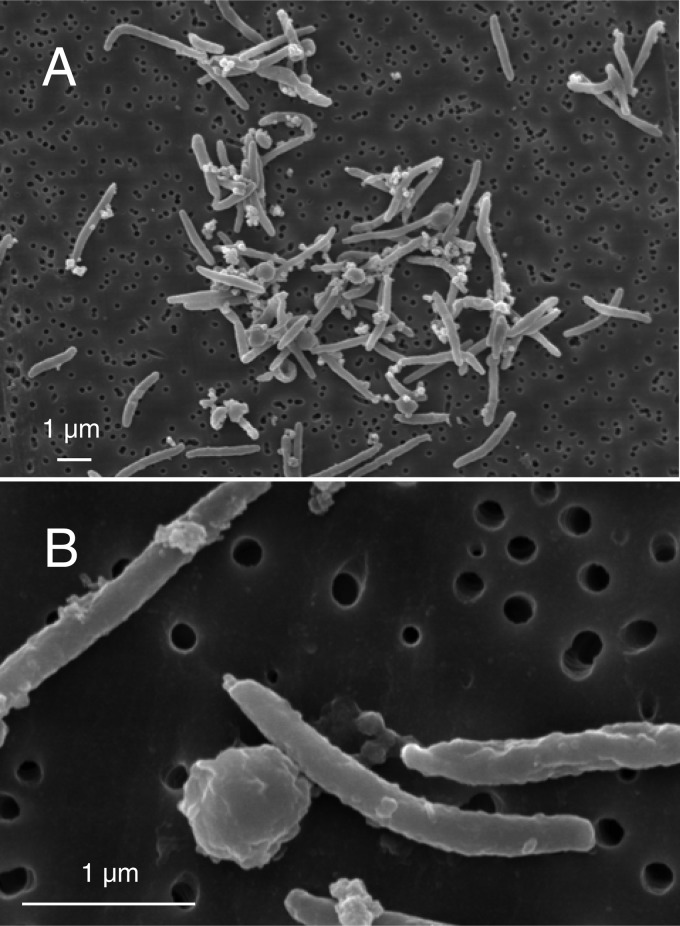
The arsenate-reducing enrichment culture was serially diluted and inoculated into anaerobic shake tubes solidified with Bacto agar. After incubation for 3 days, white bacterial colonies appeared, and the colonies turned red after 7 days. The red colony was then purified by repeated inoculation and picking the single colony from the shake tube at least 5 times. Finally, the resulting colony was inoculated into anaerobic liquid medium containing acetate and arsenate. After complete reduction of arsenate was confirmed, this anaerobic microorganism was designated strain PSR-1. Culture purity was determined by PCR-DGGE (data not shown) and microscopy. Cells of strain PSR-1 were straight or curved rods 2 to 4 μm long and 0.25 μm in diameter (Fig. 2). The Gram-stain reaction result was negative. Spores or cyst formation was observed. A bleb-like structure was also found in the terminal ends of the cells.
Fig 2.
(A) Scanning electron micrograph of strain PSR-1. (B) Cells and spore- or cyst-like structure. A bleb-like structure is also found in the terminal ends of the cells. In both cases, the strain was grown using acetate as the electron donor and arsenate as the electron acceptor.
Phylogenetic analysis of the 16S rRNA gene.
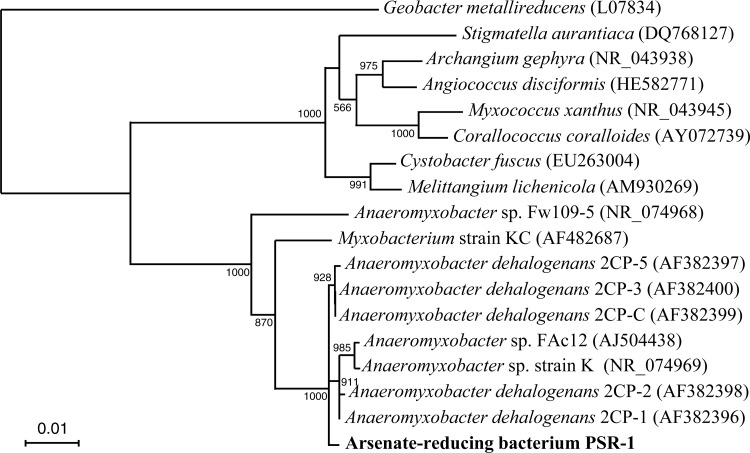
BLAST and similarity analyses of the 16S rRNA gene sequence showed that strain PSR-1 was most similar to known strains of A. dehalogenans, such as strains 2CP-1T, 2CP-2, 2CP-3, 2CP-5, and 2CP-C, with sequence similarities of 99.6% (strain 2CP-2) to 99.7% (strains 2CP-1, 2CP-3, 2CP-5, and 2CP-C). Phylogenetic comparison of the 16S rRNA gene sequence of strain PSR-1 with those of selected representatives was performed using approximately 1,500 bases (Fig. 3). The result indicated that strain PSR-1 is most closely related to A. dehalogenans within the Deltaproteobacteria.
Fig 3.
Phylogenetic tree showing the relationship between strain PSR-1 and related Myxococcales species within the Deltaproteobacteria on the basis of 16S rRNA gene sequences. The tree was constructed using the neighbor-joining method. Numbers at nodes show bootstrap values obtained from 1,000 resamplings, but bootstrap values below 500 were omitted. The GenBank accession number for each reference strain is shown in parentheses. The scale bar indicates 1% estimated sequence divergence.
Growth of strain PSR-1 on arsenate and various electron acceptors.
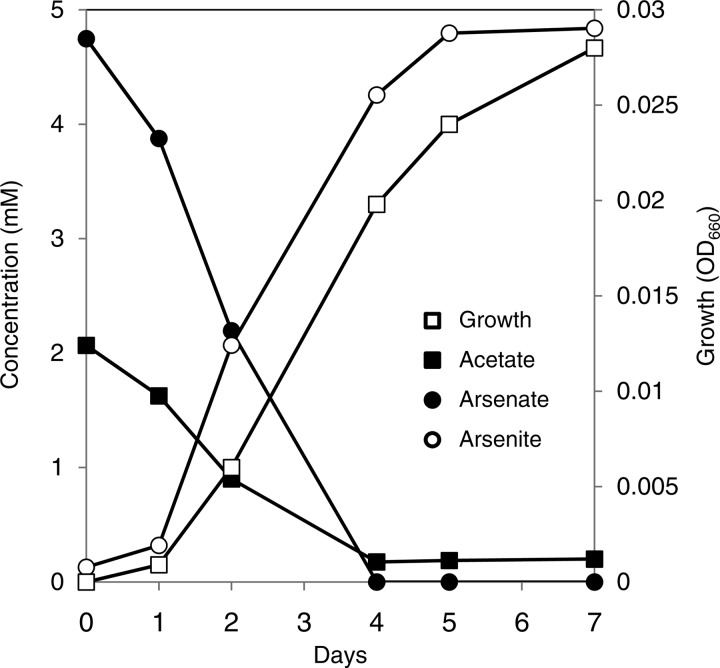
Strain PSR-1 grew by the oxidation of acetate coupled to the reduction of arsenate (Fig. 4). In the presence of acetate, the growth of strain PSR-1 coincided with the reduction and production of arsenate and arsenite, respectively. In the absence of arsenate or acetate, no significant growth was observed (OD660 <0.005). The ratios of arsenate consumed and arsenite produced to acetate consumed were 2.51 and 2.49, respectively. The reduction of arsenate by strain PSR-1 was almost completely inhibited by respiratory inhibitors such as dicumarol (100 μM) and 2-heptyl-4-hydroxyquinoline N-oxide (HQNO; 10 μM) (see Fig. S1 in the supplemental material). These results suggest that arsenate reduction by strain PSR-1 is a respiratory process.
Fig 4.
Growth of strain PSR-1 on arsenate as the sole electron acceptor. The strain was grown on 2 mM acetate as the electron donor and 5 mM arsenate as the electron acceptor. A representative result from 3 independent experiments is shown.
With arsenate (5 mM) as the electron acceptor, strain PSR-1 was capable of oxidizing acetate, propionate, pyruvate, succinate, and malate. Lactate, formate, butyrate, glucose, fructose, glycerol, and yeast extract did not serve as electron donors. With acetate (5 mM) as the electron donor, the strain was capable of reducing the following electron acceptors: 5 mM arsenate, 5 mM Fe(III)-NTA, 2 mmol liter−1 ferryhydrite, 5 mM nitrate, 5 mM fumarate, 5 mmol liter−1 manganese oxide (MnO2), and 5 mM selenate. The strain did not utilize 5 mM nitrite but did utilize it at 1 mM. Malate, sulfate, thiosulfate, sulfite, and elemental sulfur did not serve as electron acceptors.
Oxygen consumption by strain PSR-1 was determined according to the method of Sanford et al. (14) by injecting 3% (vol/vol) air (0.6% oxygen) into the headspace of the anaerobic liquid medium. Acetate was used as the electron donor, and cysteine in the medium was omitted. Although the resazurin included in the medium was pink at the start of the experiment, it turned clear after cultivation for 2 to 3 days due to oxygen consumption by strain PSR-1. The injection of 3% air was then repeated several times. After oxygen was consumed 3 times, visible turbidity was confirmed. When air was injected to give oxygen concentrations in the headspace of 2%, 10%, and 21%, the growth (i.e., optical density [OD660]) of strain PSR-1 after 2 weeks of cultivation was 0.068 ± 0.0021, 0.12 ± 0.0032, and 0.20 ± 0.0038, respectively; however, the strain did not show significant growth under aerobic conditions (on minimum medium solidified with agar). These results suggest that strain PSR-1 was a microaerophilic bacterium, as has been proposed for A. dehalogenans (14, 35).
Release of arsenic adsorbed on ferrihydrite.
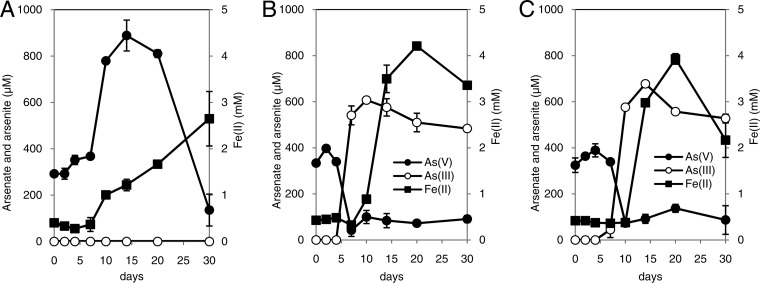
Bacteria were cultured with arsenate-adsorbed ferrihydrite as the sole electron acceptor. As was observed in a recent study (36), approximately 300 to 350 μM arsenate was detected in the liquid phase at time zero, probably because partial desorption of arsenate had occurred (7, 8, 37). In the culture inoculated with S. oneidensis MR-1, a representative dissimilatory iron-reducing bacterium without the capacity for arsenate reduction, soluble Fe(II) increased to 2.5 mM during 30 days of incubation (Fig. 5A). Concomitant with Fe(II) production, arsenate was released into the liquid phase, which showed the highest value of 890 μM at 14 days. In the last half of the incubation period, however, the arsenate level gradually decreased. No arsenite was detected throughout the duration of the experiment. In the cultures inoculated with strain PSR-1, however, arsenate levels decreased to <100 μM within 7 to 10 days, and 610 to 680 μM arsenite was simultaneously produced (Fig. 5B and C). Soluble Fe(II) production followed arsenite production, and the maximum value was approximately 4 mM. The maximum levels of arsenite in the liquid phase were significantly higher than the arsenate levels at time zero, indicating that strain PSR-1 is capable of reducing and releasing arsenic adsorbed on ferrihydrite. Cells pregrown on arsenate showed relatively much faster release of arsenic than those pregrown on Fe(III).
Fig 5.
Bacterial dissolution of arsenic from arsenate-adsorbed ferrihydrite. The minimal medium containing arsenate-adsorbed ferrihydrite as the sole electron acceptor was prepared and was inoculated with washed cells of Shewanella oneidensis MR-1 (A) and with washed cells of strain PSR-1 pregrown on arsenate (B) or on Fe(III)-NTA (C). Symbols represent the mean values obtained for triplicate determinations, and bars indicate standard deviations.
Arsenic release from sterile soil inoculated with cells of strain PSR-1.
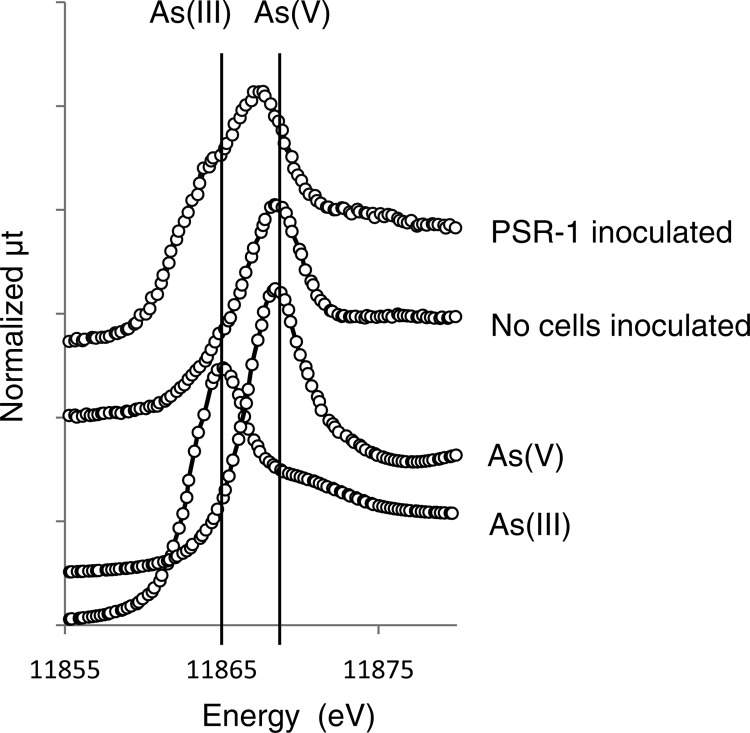
The gamma ray-irradiated sterile soil slurry was incubated for 1 week with acetate and washed cells of strain PSR-1. As shown in Table 2, 1,500 nM and 500 μM arsenite and Fe, respectively, were detected in the supernatant of the slurry incubated with washed cells pregrown on arsenate. When the cells pregrown on Fe(III)-NTA were inoculated, similar levels of arsenite and Fe (1,800 nM arsenite and 560 μM Fe) were observed. In the absence of the inoculated cells, little arsenite and Fe were observed (Table 2). Arsenic K-edge XANES analysis revealed that the proportion of arsenite in the soil solid phase after incubation was approximately 50%, while that in the absence of cells was only 20% (Table 2 and Fig. 6). The latter value was the same as that of the original soil before incubation (29).
Table 2.
Arsenic release from sterile paddy soil slurry inoculated with cells of strain PSR-1a
| Strain | Mean (± SD) level of: |
% As(III) in the soil solid phaseb | ||
|---|---|---|---|---|
| As(III) (nM) | As(V) (nM) | Fe (μM) | ||
| No cells inoculated | 77.7 ± 5.62 | 11.2 ± 0.334 | 29.1 ± 4.34 | 20 |
| PSR-1 (As)c | 1,526 ± 29.4 | 57.6 ± 13.8 | 500 ± 19.7 | 49 |
| PSR-1 (Fe)d | 1,798 ± 115 | 57.6 ± 7.17 | 560 ± 33.5 | 52 |
All values except for those of XANES analysis are expressed as mean values ± SD of triplicate determinations.
Arsenic speciation in the soil solid phase was determined by XANES analysis.
Cells grown on arsenate as the electron acceptor were inoculated.
Cells grown on Fe(III)-NTA as the electron acceptor were inoculated.
Fig 6.
Arsenic K-edge XANES spectra of reference materials and sterile paddy soil incubated with washed cells of strain PSR-1. The result in which cells grown on Fe(III)-NTA were inoculated is shown. A similar result was obtained when cells grown on arsenate were inoculated. Dotted lines represent the linear combination of XANES spectra from reference compounds to reproduce the experimental spectra.
Amplification of putative arrA gene.
We attempted to amplify the putative arrA gene from genomic DNA of strain PSR-1 using degenerate primer pairs AS1F-AS1R and AS2F-AS1R according to the PCR protocols described by Song et al. (31). Although a single band of expected size (630 bp) was observed in a positive control (genomic DNA of Geobacter sp. OR-1), strain PSR-1 did not show any band or showed only nonspecific bands (data not shown). We then tried to amplify the arrA gene by the nested PCR technique, in which the first PCR was performed with primers AS1F and AS1R and the second PCR was performed with primers AS2F and AS1R; however, no amplification product was obtained from strain PSR-1. Amplification with other PCR primers, ArrAfwd and ArrArev (32), HAArrA-D1F and HAArrA-G2R (33), ArrAUF1 and ArrAUR3 (34), and ArrPSRfwd and ArrPSRrev (newly designed in this study), was also unsuccessful (data not shown).
DISCUSSION
A. dehalogenans is a member of the order Myxococcales within the Deltaproteobacteria and is able to utilize a wide range of electron acceptors, including halogenated phenols, soluble and insoluble Fe(III), nitrate, oxygen, fumarate, U(VI), and Se(IV) (14–17); however, there have been no reports on arsenate reduction by this microorganism. In this study, we isolated a novel dissimilatory arsenate-reducing bacterium strain, PSR-1, and found that it is closely related to A. dehalogenans, with 16S rRNA gene sequence similarity of 99.7%. Furthermore, strain PSR-1 was capable of releasing arsenic from arsenate-adsorbed ferrihydrite or from sterile soil. Since arsenic in flooded soils and anoxic sediments is commonly associated with the mineral phase such as ferrihydrite, the ability of A. dehalogenans-related bacterium to release arsenic could have significant environmental impact.
It is apparent that arsenate reduction by strain PSR-1 is a respiratory process. The strain coupled the oxidation of acetate with the reduction of arsenate, and respiratory inhibitors such as dicumarol (an inhibitor of menaquinone electron transport) and HQNO (an inhibitor of NADH dehydrogenase complex) strongly inhibited its arsenate reduction. In addition, strain PSR-1 did not grow in the absence of arsenate or acetate. Oxidation of acetate coupled to arsenate reduction can be expressed in the following equation: CH3COO− + 2HAsO42− + 2H2AsO4− + 5H+ → 4H3AsO3 + 2HCO3−.
Thus, theoretical consideration predicts a 1:4 ratio of acetate consumed to arsenate reduced; however, as shown in Fig. 4, the molar ratio of arsenate reduced to acetate consumed by strain PSR-1 was 2.51. When we plotted acetate consumed versus arsenate consumed during the growth of strain PSR-1, the slope of the regression line was 2.41 with an R2 value of 0.990. Thus, the lower molar ratio can be explained by the incorporation of some carbon into the cell biomass. Similar results were reported in other A. dehalogenans strains (2CP-C and 2CP-1) grown under dechlorination conditions, in which 34% of the electrons from acetate incorporated into biomass (14, 15).
Phylogenetic analysis based on the 16S rRNA gene revealed that strain PSR-1 was closely related to known strains of A. dehalogenans, such as strain 2CP-1, with high sequence similarity (more than 99.6%). Strain PSR-1 also showed high sequence similarity with A. dehalogenans-related microorganisms such as strain FAc12 (99.2% for an approximately 1,000-bp comparison) (38) and strain KC (97.7% for an approximately 1,500-bp comparison) (39). The former is a dissimilatory iron-reducing bacterium, and the latter is an anaerobic bacterium capable of utilizing humic substances as the electron donor for respiration. Strain PSR-1 also showed morphological and phenotypic characteristics that are similar to those observed in A. dehalogenans, i.e., long rod-shaped cells, formation of red pigmentation, formation of spores or cysts, preferred utilization of acetate over lactate, susceptibility to nitrite, microaerophilic growth, and capacity for soluble- and insoluble-Fe(III) reduction (14, 15). These results strongly suggest that strain PSR-1 is the first A. dehalogenans-related bacterium with the capacity for dissimilatory arsenate reduction. It is still unclear if strain PSR-1 possesses other important features of A. dehalogenans, gliding motility and chlororespiring ability (14). In addition, it is of great importance to determine if known strains of A. dehalogenans are capable of dissimilatory arsenate reduction. To date, complete genome sequences of at least 4 strains of Anaeromyxobacter spp. (strains 2CP-1, 2CP-C, K, and Fw109-5) have been released. Although these strains code a large number of molybdopterin oxidoreductases in their genomes, no proteins show high sequence similarity with known ArrA. We purchased A. dehalogenans 2CP-1 (ATCC BAA-258) and attempted to grow it with arsenate as the sole electron acceptor; however, strain 2CP-1 did not grow on arsenate, while it showed good growth on fumarate (data not shown). Our results suggest that not all A. dehalogenans strains have the capacity for dissimilatory arsenate reduction.
Strain PSR-1 was capable of reducing and releasing arsenic adsorbed on ferrihydrite (Fig. 5B and C). In addition, solid-state speciation by XANES analysis revealed that the proportion of arsenite in the soil solid phase actually increased from 20% to 50% after inoculation with washed cells of PSR-1 (Table 2 and Fig. 6). There are two possible explanations for these observations. One is that reductive dissolution of iron stimulated arsenate release from the solid phase and that arsenate was reduced to arsenite in the liquid phase. Finally, arsenite was readsorbed on the soil solid phase. A second possibility is that arsenate was reduced directly on the soil solid phase and that part of it was released into the liquid phase. In this case, iron reduction may also stimulate arsenic release, as observed in the culture of S. oneidensis MR-1 (Fig. 5A). Previously, we anaerobically incubated soil slurry that had been prepared from same soil used in this study for 60 days (29). Significant release of arsenite from the slurry occurred, and the proportion of arsenite in the solid phase was 80% at 60 days. Nevertheless, only 3.6% of the total arsenic in soil was released into the liquid phase, while most arsenite was still retained in the soil solid phase (29). Weber et al. (40) also reported a similar observation that less than 3.9% of arsenite was released into the liquid phase after flooding of soil for 52 days. Therefore, we consider that the first possibility is unlikely because such a large proportion (50% to 80%) of arsenic should not be released into the liquid phase. In addition, previous studies suggested that arsenate reduction in the liquid phase does not play a role in arsenic release from the solid phase (6) and that direct reduction on soil solid phase is a significant process (8). Therefore, it is considered that strain PSR-1 is able to reduce arsenate directly in the soil solid phase.
The representative iron-reducing bacterium S. oneidensis MR-1 released arsenate adsorbed on ferrihydrite, although it could not reduce arsenate to arsenite (Fig. 5A). Our result is consistent with that observed in a previous study by Cummings et al. (37) in which an iron-reducing bacterium, Shewanella alga BrY, did not reduce but released arsenic (as arsenate) from crystalline ferric arsenate. The concentration of dissolved arsenate, however, decreased again in the late incubation period of S. oneidensis MR-1. This was probably because the dissolved arsenate was reassociated with Fe(II)-bearing secondary minerals that had been formed as a result of iron reduction by S. oneidensis MR-1. Islam et al. (41) reported that such minerals, including siderite (FeCO3), vivianite (iron phosphate), and magnetite, were all able to adsorb arsenate efficiently and removed arsenate from a culture of G. sulfurreducens. Since our growth medium was buffered with bicarbonate, siderite formation could have been very favorable. Recently, we performed a similar experiment using another representative iron-reducing bacterium, Geobacter metallireducens GS-15; however, this bacterium released neither arsenate nor arsenite in the liquid phase (36). The reason why Shewanella spp. can efficiently release arsenate adsorbed on ferrihydrite is still unclear.
Multiple attempts to amplify the putative arrA gene from genomic DNA of strain PSR-1 were unsuccessful. Most of the degenerate primers used in this study were designed by comparing conserved regions in the arrA genes from Firmicutes, Gamma- and Epsilonproteobacteria, and Chrysiogenetes, although deltaproteobacterial arrA genes were not involved. However, the fact that a putative arrA of Geobacter sp. OR-1, which is also one of the Deltaproteobacteria (36), could be amplified by primer pair AS1F and AS1R or AS2F and AS1R suggests that some of these primers sufficiently amplify phylogenetically diverse arrA genes. It is possible that strain PSR-1 harbors an atypical arrA gene that cannot be amplified by previously designed degenerate primers, as has been observed recently in nosZ genes of several A. dehalogenans strains (42). In order to identify the putative arrA gene, genome sequencing of strain PSR-1 is under way in our laboratory.
In conclusion, arsenate reduction by strain PSR-1 expands the metabolic versatility of Anaeromyxobacter dehalogenans. Considering its distribution throughout diverse soils and anoxic sediments, Anaeromyxobacter dehalogenans may play a role in arsenic release from these environments.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Y. Takahashi (Hiroshima University) for XANES measurements.
This work was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Research Project for Ensuring Food Safety from Farm to Table AC-1122) and from JSPS KAKENHI (grant number 23580103).
The XANES measurements were performed under approval of the High-Energy Accelerator Research Organization, KEK (proposal no. 2011G016).
Footnotes
Published ahead of print 24 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00693-13.
REFERENCES
- 1. Ng JC, Wang J, Shraim A. 2003. A global health problem caused by arsenic from natural sources. Chemosphere 52:1353–1359 [DOI] [PubMed] [Google Scholar]
- 2. Smedley PL, Kinniburgh DG. 2002. A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 17:517–568 [Google Scholar]
- 3. Dixit S, Hering JG. 2003. Comparison of arsenic(V) and arsenic(III) sorption onto iron oxide minerals: implications for arsenic mobility. Environ. Sci. Technol. 37:4182–4189 [DOI] [PubMed] [Google Scholar]
- 4. Goldberg S. 2002. Competitive adsorption of arsenate and arsenite on oxides and clay minerals. Soil Sci. Soc. Am. J. 66:413–421 [Google Scholar]
- 5. Manning BA, Fendorf SE, Bostick B, Suarez DL. 2002. Arsenic(III) oxidation and arsenic(V) adsorption reactions on synthetic birnessite. Environ. Sci. Technol. 36:976–981 [DOI] [PubMed] [Google Scholar]
- 6. Langner HW, Inskeep WP. 2000. Microbial reduction of arsenate in the presence of ferrihydrite. Environ. Sci. Technol. 34:3131–3136 [Google Scholar]
- 7. Ahmann D, Krumholz LR, Hemond HF, Lovley DR, Morel FMM. 1997. Microbial mobilization of arsenic from sediments of the Aberjona Watershed. Environ. Sci. Technol. 31:2923–2930 [Google Scholar]
- 8. Zobrist J, Dowdle PR, Davis JA, Oremland RS. 2000. Mobilization of arsenite by dissimilatory reduction of adsorbed arsenate. Environ. Sci. Technol. 34:4747–4753 [Google Scholar]
- 9. Oremland RS, Stolz JF. 2005. Arsenic, microbes and contaminated aquifers. TRENDS Microbiol. 13:45–49 [DOI] [PubMed] [Google Scholar]
- 10. Stolz JF, Basu P, Santini JM, Oremland RS. 2006. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60:107–130 [DOI] [PubMed] [Google Scholar]
- 11. Krafft T, Macy JM. 1998. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur. J. Biochem. 255:647–653 [DOI] [PubMed] [Google Scholar]
- 12. Saltikov CW, Newman DK. 2003. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U. S. A. 100:10983–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cole JR, Cascarelli AL, Mohn WW, Tiedje JM. 1994. Isolation and characterization of a novel bacterium growing via reductive dehalogenation of 2-chlorophenol. Appl. Environ. Microbiol. 60:3536–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sanford RA, Cole JR, Tiedje JM. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 68:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He Q, Sanford RA. 2003. Characterization of Fe(III) reduction by chlororespiring Anaeromyxobacter dehalogenans. Appl. Environ. Microbiol. 69:2712–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He Q, Yao K. 2010. Microbial reduction of selenium oxyanions by Anaeromyxobacter dehalogenans. Bioresour. Technol. 101:3760–3764 [DOI] [PubMed] [Google Scholar]
- 17. Wu Q, Sanford RA, Löffler FE. 2006. Uranium(VI) reduction by Anaeromyxobacter dehalogenans strain 2CP-C. Appl. Environ. Microbiol. 72:3608–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomas SH, Padilla-Crespo E, Jardine PM, Sanford RA, Löffler FE. 2009. Diversity and distribution of Anaeromyxobacter strains in a uranium-contaminated subsurface environment with a nonuniform groundwater flow. Appl. Environ. Microbiol. 75:3679–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. North NN, Dollhopf SL, Petrie L, Istok JD, Balkwill DL, Kostka JE. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrie L, North NN, Dollhopf SL, Balkwill DL, Kostka JE. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sedoments contaminated with uranium(VI). Appl. Environ. Microbiol. 69:7467–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller TL, Wolin MJ. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27:985–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lovley DR, Phillips EJ. 1986. Availability of ferric iron for microbial reduction in bottom sediments of the freshwater tidal Potomac River. Appl. Environ. Microbiol. 52:751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwertmann U, Cornell RM. 2000. Ferrihydrite, p 103–112 In Iron oxides in the laboratory: preparation and characterization. Willey-VCH, Weinheim, Germany [Google Scholar]
- 24. Hiraishi A. 1992. Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett. Appl. Microbiol. 15:210–213 [DOI] [PubMed] [Google Scholar]
- 25. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 27. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suda W, Oto M, Amachi S, Shinoyama H, Shishido M. 2008. A direct method to isolate DNA from phyllosphere microbial communities without disrupting leaf tissues. Microbes Environ. 23:248–252 [DOI] [PubMed] [Google Scholar]
- 29. Yamaguchi N, Nakamura T, Dong D, Takahashi Y, Amachi S, Makino T. 2011. Arsenic release from flooded paddy soils is influenced by speciation, Eh, pH, and iron dissolution. Chemosphere 83:925–932 [DOI] [PubMed] [Google Scholar]
- 30. Takahashi Y, Minamikawa R, Hattori KH, Kurishima K, Kihou N, Yuita K. 2004. Arsenic behavior in paddy fields during the cycle of flooded and non-flooded periods. Environ. Sci. Technol. 38:1038–1044 [DOI] [PubMed] [Google Scholar]
- 31. Song B, Chyun E, Jaffé PR, Ward BB. 2009. Molecular methods to detect and monitor dissimilatory arsenate-respiring bacteria (DARB) in sediments. FEMS Microbiol. Ecol. 68:108–117 [DOI] [PubMed] [Google Scholar]
- 32. Malasarn D, Saltikov CW, Campbell KM, Santini JM, Hering JG, Newman DK. 2004. arrA is a reliable marker for As(V) respiration. Science 306:455. [DOI] [PubMed] [Google Scholar]
- 33. Kulp TR, Hoeft SE, Miller LG, Saltikov C, Murphy JN, Han S, Lanoil B, Oremland RS. 2006. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl. Environ. Microbiol. 72:6514–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fisher E, Dawson AM, Polshyna G, Lisak J, Crable B, Perera E, Ranganathan M, Thangavelu M, Basu P, Stolz JF. 2008. Transformation of inorganic and organic arsenic by Alkaliphilus oremlandii sp. nov. strain OhILAs. Ann. N. Y. Acad. Sci. 1125:230–241 [DOI] [PubMed] [Google Scholar]
- 35. Thomas SH, Sanford RA, Amos BK, Leigh MB, Cardenas E, Löffler FE. 2010. Unique ecophysiology among U(VI)-reducing bacteria as revealed by evaluation of oxygen metabolism in Anaeromyxobacter dehalogenans strain 2CP-C. Appl. Environ. Microbiol. 76:176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohtsuka T, Yamaguchi N, Makino T, Sakurai K, Kimura K, Kudo K, Homma E, Dong DT, Amachi S. 13 May 2013. Arsenic dissolution from Japanese paddy soil by a dissimilatory arsenate-reducing bacterium Geobacter sp. OR-1. Environ. Sci. Technol. [Epub ahead of print.] 10.1021/es400231x [DOI] [PubMed] [Google Scholar]
- 37. Cummings DE, Caccavo F, Jr, Fendorf S, Rosenzweig RF. 1999. Arsenic mobilization by the dissimilatory Fe(III)-reducing bacterium Shewanella alga BrY. Environ. Sci. Technol. 33:723–729 [Google Scholar]
- 38. Treude N, Rosencrantz D, Liesack W, Schnell S. 2003. Strain FAc12, a dissimilatory iron-reducing member of the Anaeromyxobacter subgroup of Myxococcales. FEMS Microbiol. Ecol. 44:261–269 [DOI] [PubMed] [Google Scholar]
- 39. Coates JD, Cole KA, Chakraborty R, O'Connor SM, Achenbach LA. 2002. Diversity and ubiquity of bacteria capable of utilizing humic substrates as electron donors for anaerobic respiration. Appl. Environ. Microbiol. 68:2445–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weber F-A, Hofacker AF, Voegelin A, Kretzschmar R. 2010. Temperature dependence and coupling of iron and arsenic reduction and release during flooding of a contaminated soil. Environ. Sci. Technol. 44:116–122 [DOI] [PubMed] [Google Scholar]
- 41. Islam FS, Pederick RL, Gault AG, Adams LK, Polya DA, Charnock JM, Lloyd JR. 2005. Interactions between the Fe(III)-reducing bacterium Geobacter sulfurreducens and arsenate, and capture of the metalloid by biogenic Fe(II). Appl. Environ. Microbiol. 71:8642–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-García C, Rodríguez G, Massol-Deyá A, Krishnani KK, Ritalahti KM, Nissen S, Konstantinidis T, Löffler FE. 2012. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl. Acad. Sci. U. S. A. 109:19709–19714 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.