Abstract
Propionic acid (PA) is an important chemical building block and is widely applied for organic synthesis, food, feedstuff, and pharmaceuticals. To date, the strains that can efficiently produce PA have included Propionibacterium thoenii, P. freudenreichii, and P. acidipropionici. In this report, we show that P. jensenii ATCC 4868 is also able to produce PA in much higher yields than the previously reported strains. To further improve the production capacity, a P. jensenii-Escherichia coli shuttle vector was developed for the metabolic engineering of P. jensenii. Specifically, a 6.9-kb endogenous plasmid, pZGX01, was isolated from P. acidipropionici ATCC 4875 and sequenced. Since the sequencing analysis indicated that pZGX01 could encode 11 proteins, the transcriptional levels of the corresponding genes were also investigated. Then, a P. jensenii-Escherichia coli shuttle vector was constructed using the pZGX01 plasmid, the E. coli pUC18 plasmid, and a chloramphenicol resistance gene. Interestingly, not only could the developed shuttle vector be transformed into P. jensenii ATCC 4868 and 4870, but it also could be transformed into freudenreichii ATCC 6207 subspecies of P. freudenreichii. Finally, the glycerol dehydrogenase gene (gldA) from Klebsiella pneumoniae was expressed in P. jensenii ATCC 4868 with the constructed shuttle vector. In a 3-liter batch culture, the PA production by the engineered P. jensenii ATCC 4868 strain reached 28.23 ± 1.0 g/liter, which was 26.07% higher than that produced by the wild-type strain (22.06 ± 1.2 g/liter). This result indicated that the constructed vector can be used a useful tool for metabolic engineering of P. jensenii.
INTRODUCTION
Propionic acid (PA) is a valuable C3 platform chemical (three-carbon compound that can be used to synthesize a series of products with high added value) that is widely used as a preservative in animal feed and human foods. It is also an important chemical intermediate for the synthesis of cellulose fibers, herbicides, perfumes, and pharmaceuticals (1, 2). According to the U.S. Department of Energy, PA is among the top 30 candidate platform chemicals used for a variety of applications (3). Industrially, PA is produced from petrochemical raw materials (e.g., ethylene and carbon monoxide) via oxo-synthesis (4). Due to exhaustion of petroleum resources and the serious environmental pollution caused by the utilization of fossil resources, the production of PA by microbial fermentation from renewable resources has attracted increasing attention (5). To date, the species used for PA production have included Propionibacterium freudenreichii (6), P. acidipropionici (5, 7–14), and P. thoenii (15). Almost all the reports on this topic mainly focused on the optimization of the process (6–8, 11, 14, 16) and helped to significantly improve the PA production levels. However, the biotechnological production of PA cannot be achieved at industrial scale due to the weak economic competiveness compared with the conventional chemical synthesis route. Therefore, it seems that there is not much room for further enhancement of PA production by process optimization, and thus, it is necessary to develop metabolic engineering strategies aiming at further enhancing PA production.
Several vectors have been isolated from dairy propionibacteria such as P. freudenreichii and P. acidipropionici (17–20). The reported Propionibacterium plasmids include pLME108 from P. freudenreichii DF2, pLME106 from P. jensenii DF1 (21), pRGO1 from P. acidipropionici E214 (19), and p545 from P. freudenreichii LMG 16545 (18). To date, four Propionibacterium-Escherichia coli shuttle vectors have been developed by using these plasmids (18, 19, 21, 22). Kiatpapan et al. (19) developed the Propionibacterium-E. coli shuttle vector pPK705 by using plasmids pRG01 and pUC18 and the hygromycin B resistance gene. Jore et al. (18) established a reproducible transformation approach for P. freudenreichii with plasmid p545 from P. freudenreichii itself and tested the erythromycin resistance gene and the chloramphenicol resistance gene as the selection marker. The successful transformation of P. freudenreichii was also achieved with two different vectors based on the plasmids pLME106 and pLME108 (21). Brede et al. (22) constructed the Propionibacterium-E. coli shuttle vector pSL104 by using plasmid pLME108 and a propionicin F bacteriocin immunity gene. Unfortunately, these reported vectors can only be transformed into P. freudenreichii (17–19, 21) and P. acnes (23), and thus, the genetic manipulation has been limited to these two organisms. For example, the hemA, hemB, and choA genes were overexpressed for the production of vitamin B12 in P. freudenreichii (24, 25). The only report about the metabolic engineering of Propionibacterium for the enhancement of PA production focused on the deletion of the byproduct acetic acid-encoding gene in P. acidipropionici (12). The slow development of genetic engineering of Propionibacterium is mainly due to the lack of detailed genome information and transformable plasmids (26), although several species, including P. acidipropionici, P. propionicum, and P. freudenreinchii, have already been completely sequenced.
Although P. jensenii belongs to the dairy propionibacteria, most of the studies reporting on this organism were focused on the production of bacteriocin, antimicrobial peptides, and the polyene pigment granadaene (27–30). In this work, we reported that P. jensenii ATCC 4868 is capable of efficiently producing PA in large amounts. Currently, there is no transformable plasmid for P. jensenii to be used for strengthening the PA synthesis capacity of P. jensenii. Therefore, we constructed a P. jensenii-Escherichia coli shuttle vector as an engineering tool for gene expression in P. jensenii. Moreover, the glycerol dehydrogenase-encoding gene (gldA) from Klebsiella pneumoniae was expressed in P. jensenii ATCC 4868, resulting in PA production that was noticeably higher than that of the wild-type strain. This work may contribute to the development of genetic manipulation of Propionibacterium strains for highly efficient production of PA.
MATERIALS AND METHODS
Bacterial strains, vectors, and media.
Propionibacterium strains were purchased from American Type Culture Collection (ATCC). E. coli JM109 and pUC18 plasmid were purchased from TaKaRa (Dalian, China). E. coli JM110 was purchased from Stratagene (La Jolla, CA). pXZ10145, a plasmid containing a chloramphenicol resistance gene (cml/cmx), was provided by Z. X. Zheng (Fudan University, Shanghai, China). The bacterial strains and vectors used in this work are shown in Table 1. E. coli was cultured at 37°C in Luria-Bertani (LB) medium containing 1% (wt/vol) NaCl, 1% (wt/vol) peptone, and 0.5% (wt/vol) yeast extract supplemented with 100 μg/ml ampicillin. Propionibacterium was grown anaerobically at 30°C in sodium lactate broth (SLB) medium containing 1% (wt/vol) sodium lactate, 1% (wt/vol) yeast extract, and 1% (wt/vol) Trypticase soy broth supplemented with 10 μg/ml chloramphenicol.
Table 1.
Plasmids and strains used in this studya
| Plasmid or strain | Relevant characteristic(s) | Source |
|---|---|---|
| Plasmids | ||
| pUC18 | E. coli cloning vector; Apr | TaKaRa |
| pZGX01 | Wild plasmid in P. acidipropionici ATCC 4875 | This study |
| pZGX04 | E. coli-PAB shuttle vector Apr in E. coli and Cmr in PAB; derived from pUC18, pZGX02, and cml/cmx PCR product | This study |
| pZGX04-gldA | gldA expression vector | This study |
| Strains | ||
| E. coli JM109 | Clone host | TaKaRa |
| E. coli JM110 | dam and dcm mutant conferring no methylation of DNA | Stratagene |
| K. pneumonia subsp. pneumoniae ATCC 12657 | Carries gldA | ATCC |
| P. acidipropionici ATCC 4875 | Carries plasmid pZGX01 | ATCC |
| P. acidipropionici ATCC 4965 | No native plasmid; electrotransformation | ATCC |
| P. acidipropionici ATCC 25562 | No native plasmid; electrotransformation | ATCC |
| P. jensenii ATCC 4868 | No native plasmid; electrotransformation; transformable by shuttle vector pZGX04 | ATCC |
| P. jensenii ATCC 4868 (pZGX04-gldA) | P. jensenii ATCC 4868 carries pZGX04-gldA | This study |
| P. jensenii ATCC 4870 | No native plasmid; electrotransformation; transformable by shuttle vector pZGX04 | ATCC |
| P. freudenreichii subsp. freudenreichii ATCC 6207 | No native plasmid; electrotransformation; transformable by shuttle vector pZGX04 | ATCC |
| P. freudenreichii subsp. shermanii ATCC 9614 | No native plasmid; electrotransformation | ATCC |
| P. thoenii ATCC 4872 | No native plasmid; electrotransformation | ATCC |
| P. thoenii ATCC 4874 | No native plasmid; electrotransformation | ATCC |
Apr, ampicillin selection; Cmr, chloramphenicol selection; cml/cmx, complete cml(A) and cmx(A) genes; ATCC, American Type Culture Collection, Manassas, VA.
DNA isolation and manipulation.
The plasmids from Propionibacterium and E. coli were extracted using a MiniBEST plasmid purification kit (TaKaRa, Dalian, China) or a Sangong Biotech large-scale plasmid purification kit (Sangong, Shanghai, China). The genomic DNA of Propionibacterium strains was prepared using a UNlQ-10 column bacterial genomic DNA isolation kit (Sangong, Shanghai, China). The Propionibacterium cells were treated with 10 mg/ml lysozyme and 100 U/ml mutanolysin for 30 min before plasmid extraction. Restriction enzymes and T4 DNA ligase (TaKaRa, Dalian, China) were used according to the manufacturer's instructions.
Reverse transcription-quantitative PCR (RT-qPCR).
To conduct transcriptional analysis of the isolated plasmids, total RNA was extracted from the cells when their optical density at 600 nm (OD600) reached 0.7 in SLB medium. The RNA was isolated using an RNAisoPlus kit (TaKaRa, Dalian, China), and the cells were suspended in a lysozyme solution (40 mg/ml) for pretreatment. The total RNA was treated with a PrimeScript II first-strand cDNA synthesis kit (TaKaRa, Dalian, China). The obtained cDNA was subjected to quantitative PCR (qPCR) using the LightCycler 2.0 system (Roche, Basel, Switzerland) and SYBR Premix Ex Taq (TaKaRa, Dalian, China). Primers were designed by using Primer Premier software (version 5.00; PREMIER Biosoft International, Palo Alto, CA) and are listed in Table S1 in the supplemental material.
Transformation of Propionibacterium strains.
The transformation optimization was conducted based on several previous reports (17, 19, 21). Propionibacterium strains were cultured up to the stationary-growth phase, and then the cultures were diluted 50 times with SLB medium. After incubation for 20 h, the cells in the exponential-growth phase (OD600 = 0.7) were kept on ice for 30 min. After centrifugation at 5,000 × g for 4 min, the cells were washed twice with ice-cold sucrose (0.5 M). Finally, the cells were resuspended in a 0.01 volume of ice-cold electroporation buffer (0.5 M sucrose, 1 mM potassium acetate, pH 5.5). The transformation was conducted by electroporation using a Gene Pulser apparatus (Bio-Rad, Hercules, CA). Eighty microliters of the cell suspension was mixed with 1.5 μg of plasmid in an ice-cold electroporation cuvette and kept on ice for 30 min. Then, an electric pulse was delivered at a field strength of 12.5 kV/cm with a resistance of 200 Ω. After the pulse, 900 μl of ice-cold SLB medium containing 0.5 M sucrose was added. After 3 h of incubation at 30°C, the cells were spread on SLB agar plates containing 10 μg/ml chloramphenicol. The transformants could be observed after incubation at 30°C for 7 to 10 days under anaerobic conditions.
DNA sequencing.
To determine the complete nucleotide sequence of pZGX01, the plasmid was linearized by BamHI and subcloned in pUC18 for sequencing (fold coverage, 30×), which was performed by TaKaRa (Dalian, China). The sequencing data were assembled and analyzed by using Vector NTI (version 11; Invitrogen, New York, NY), GLIMMER (version 3.2; Center for Bioinformatics and Computational Biology, University of Maryland [http://www.cbcb.umd.edu]), Promoter 2.0 (http://www.cbs.dtu.dk/services/Promoter), GeneMarkS (version 4.7; Georgia Institute of Technology, Atlanta, GA [http://opal.biology.gatech.edu/GeneMark/genemarks.cgi]), and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) software.
Fermentation conditions.
The inoculums were prepared in 250-ml anaerobic jars containing 200 ml of sterile culture medium supplemented with 10 g/liter yeast extract, 5 g/liter Trypticase soy broth, 2.5 g/liter K2HPO4, and 1.5 g/liter KH2PO4. The jars were sealed with butyl rubber caps, and the cultures were incubated at 30°C for 48 h. Batch fermentations were performed under anaerobic conditions with a nitrogen flux in a 3-liter BIOFLO 115 Bioreactor (Eppendorf, Hamburg, Germany) containing 2 liters of culture medium supplemented with 30 g/liter glycerol. The medium was autoclaved at 121°C for 21 min and then purged with sterile nitrogen gas to remove any traces of oxygen before the inoculation. The temperature and agitation speed were maintained at 30°C and 200 rpm, respectively, and the pH was controlled at 7.0 via automatic addition of Ca(OH)2 (10% [wt/vol]). About 200 ml of seed culture was used as the inoculum.
Analytical methods.
Fifteen-milliliter aliquots were drawn from the fermentor every 12 h, and 3 ml was used to determine the OD600 with a UV mini-1240 spectrophotometer (Shimadzu, Kyoto, Japan) after an appropriate dilution. Dry cell weight (DCW) was calculated from the OD600 according to the following equation: DCW (g/liter) = 0.23 × OD600.
Fifteen-milliliter aliquots were used for the quantification of acetic acid, succinic acid, and PA concentrations. The samples were centrifuged at 7,000 × g for 10 min, and the supernatant was filtered with a 0.22-μm-pore-size-filter membrane (diameter, 25 mm). The sample was analyzed by the use of an Agilent 1200 high-performance liquid chromatography (HPLC) system (Agilent, Santa Clara, CA) equipped with a Zorbax SB-Aq column (Agilent, Santa Clara, CA) (250 by 4.6 mm), a standard G 1329A autosampler (Agilent, Santa Clara, CA), and a G13158 diode array detector (DAD) (Agilent, Santa Clara, CA). Na2HPO4 (0.138 mol/liter) and acetonitrile (1% [vol/vol]), the pH of which was adjusted to 2.0 with phosphoric acid, were used as the mobile phase at a flow rate of 1.0 ml/min. The detection wavelength was 210 nm, and the column temperature was maintained at 35°C. The concentration of the products was calculated by comparing the peak areas with that of the internal standard.
Three-milliliter aliquots were used for analyzing glycerol dehydrogenase activity. The samples were centrifuged at 10,000 × g for 5 min, and then the cell pellets were washed with 0.5 ml of K2HPO4-KH2PO4 buffer (50 mM, pH 7.0) and resuspended in the same buffer. The cell suspension was sonicated to break up the cell walls and centrifuged at 10,000 × g and 4°C for 15 min to remove cell debris. The reaction mixture (1 ml) contained 30 mM ammonium sulfate, 0.2 M glycerol, and 1.2 mM NAD (adjusted to pH 7.0 with 1 M NaOH), and the supernatant of the lysate in 0.1 M potassium carbonate buffer solution (pH 9.8). The absorption intensity was measured by a UV-2450 spectrophotometer (Shimadzu, Kyoto, Japan) at 340 nm and 30°C. One unit of activity was defined as the amount of enzyme required to reduce 1 mmol of substrate per minute under the specified conditions.
Stability of the vector in Propionibacterium.
The transformants were grown in SLB medium supplemented with chloramphenicol (10 μg/ml) for 3 days. The culture was diluted by 50 times with fresh SLB medium and cultured at 30°C for 2 days. To determine the plasmid stability, at least 50 colonies from each tested transformant were transferred to SLB agar plates with or without chloramphenicol after 30 serial transfers. The growth of these colonies was monitored after 7 days of incubation. The plasmids in these colonies were analyzed by colony PCR and sequenced. The percentage of stability was determined as follows: number of colonies grown on the selective medium/number of colonies grown on the nonselective medium × 100%.
Statistical analysis.
All the experiments were independently performed at least three times, and the results were expressed as means ± standard deviations (SD). Statistical analyses were performed using Student's t test. P values of less than 0.05 were considered statistically significant.
Nucleotide sequence accession number.
The obtained nucleotide sequence data have been deposited in the GenBank database under accession number JQ728013.
RESULTS AND DISCUSSION
Comparison of PA production in batch culture by different Propionibacterium species.
The reported species used for PA production include P. freudenreichii (6), P. acidipropionici (5, 7–14), and P. thoenii (15). Among those, P. acidipropionici is the main producer of PA, with a maximal yield of 97 g/liter from a 3-month fed-batch culture of a PA-tolerant P. acidipropionici mutant in a fibrous bed bioreactor (31). Figure 1 shows the time profiles of PA production by batch culture of P. jensenii ATCC 4868. The highest PA production reached 22.06 ± 1.2 g/liter after about 145 h, with a yield of 0.152 g · liter−1 · h−1. Comparisons of levels of PA production in batch culture by different Propionibacterium strains reported in the literature are listed in Table 2. It can be seen that compared to P. acidipropionici and P. thoenii, P. jensenii ATCC 4868 displayed a competitive PA titer and YPA/substrate (the mass ratio of PA to substrate), indicating that P. jensenii ATCC 4868 had a great potential for PA production. To further strengthen the PA synthesis capability of P. jensenii ATCC 4868, it was necessary to develop an engineering tool for the genetic manipulation of P. jensenii ATCC 4868.
Fig 1.
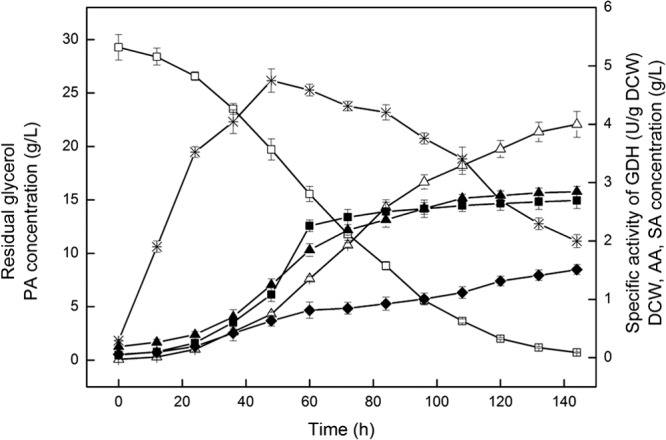
Batch fermentation kinetics of propionic acid production from glycerol with P. jensenii ATCC 4868 at pH 7.0 and 30°C for 144 h. The initial glycerol concentration was 30 g/liter; samples were taken from the fermentor every 12 h. △, propionic acid (PA); □, residual glycerol; ▲, dry cell weight (DCW); ■, acetic acid (AA); ♦, succinic acid (SA); *, specific activity of GDH.
Table 2.
Production of PA in batch fermentation with different strainsa
| Strain | Substrate | PA (g/liter) | YP/A (g/g) | YP/S (g/g) | YPA/substrate (g/g) | Productivity (g liter−1 h−1) | Source or reference |
|---|---|---|---|---|---|---|---|
| P. thoenii | Glycerol | 5 | 0.25 | 0.03 | 15 | ||
| P. acidipropionici ATCC 25562 | Glycerol | 12.7 | 5.73 | 16.53 | 0.635 | 0.42 | 10 |
| P. acidipropionici ATCC 4875 | Lactose | 22 | 2.82 | 2.12 | 0.44 | 0.25 | 9 |
| P. acidipropionici ATCC 4875 adapted mutant | Xylose | 19.5 | 3.45 | 5.45 | 0.33 | 12 | |
| P. acidipropionici ATCC 4965 | Lactate | 15.06 | 2.69 | 0.502 | 7 | ||
| P. acidipropionici ATCC 4875 (ACK-Tet) | Glycerol | 19.3 | 13.4 | 0.55 | 0.026 | 13 | |
| P. acidipropionici CGMCC 1.2230 | Glycerol | 15.72 | 13.1 | 6.81 | 0.79 | 0.13 | 16 |
| P. acidipropionici CGMCC 1.2225 | Glycerol/glucose | 21.9 | 25.17 | 18.4 | 0.572 | 0.152 | 11 |
| P. acidipropionici DSM 4900 | Glycerol | 19.5 | 17.85 | 5.99 | 0.464 | 0.34 | 8 |
| P. acidipropionici ATCC 4875 | Arabinose | 13.8 | 5 | ||||
| P. acidipropionici CGMCC 1.2225 | Glucose | 18.9 | 0.45 | 0.13 | 14 | ||
| P. freudenreichii CCTCC M207015 | Glucose | 14.6 | 4.69 | 14.44 | 0.37 | 0.12 | 6 |
| P. jensenii ATCC 4868 | Glycerol | 22.06 | 8.2 | 14.61 | 0.74 | 0.152 | This study |
| P. jensenii ATCC 4868 (pZGX04-gldA) | Glycerol | 28.23 | 9.08 | 23.52 | 0.94 | 0.2 | This study |
YP/A, the mass ratio of PA to acetic acid; YP/S, the mass ratio of PA to succinic acid.
Plasmid screening and sequencing.
Since the previous attempts to transform plasmids originating from other bacteria into Propionibacterium failed (19), naturally occurring plasmids in Propionibacterium should be used while attempting vector construction. Several plasmids have been screened from dairy Propionibacterium (19, 20, 32, 33).
In this work, nine Propionibacterium strains, representing all four recognized species of dairy Propionibacterium, were used to screen for the presence of endogenous plasmids (Table 1). Only one plasmid, named pZGX01, could be isolated from P. acidipropionici ATCC 4875, while no plasmids were found in the other eight strains tested. The complete nucleotide sequence of pZGX01 was determined. pZGX01 plasmid is 6,868 bp long and, in agreement with other Propionibacterium sequences, displays a GC content of 65%. The sequence analysis of pZGX01 revealed that it carries 11 open reading frames (ORFs, designated orf1 to orf11), where orf1, orf4, and orf5 have opposite orientations with respect to the others. The GC content of most of these ORFs is above 60%, except for orf9 (51.3%) and orf10 (44%).
Prediction of ORF function.
The features of these 11 ORFs are summarized in Table 3. The proteins related to replication are encoded by orf6 and orf7. All the above-mentioned replication proteins show motifs that are typical of theta-replicating plasmids (34). Propionicin SM1, the first bacteriocin encoded by a Propionibacterium plasmid, is related to orf9 and orf10 (35). The derived amino acid sequence of orf10 is believed to originate three transmembrane helices. The gene of orf10, encoding a putative protein of unknown function, also has a low GC content (44%). Interestingly, the three predicted transmembrane helices of orf10 were suggested to be involved in the excretion of propionicin SM1 (21).
Table 3.
Putative ORFs of plasmid pZGX01 from P. acidipropionici ATCC 4875
| ORF | Start position | Stop position | Residue of the protein | Potential function of the putative protein | Highest homology | % amino acid similarity |
|---|---|---|---|---|---|---|
| ORF1 | 623 | 18 | 302 | Double-stranded DNA binding protein | pKNR01 (Rhodococcus opacus) | 52 |
| ORF2 | 1075 | 1308 | 77 | DNA binding domain protein | RHH_1 (Kytococcus sedentarius) | 68 |
| ORF3 | 1292 | 1555 | 87 | DNA binding protein | RelE (Mobiluncus mulieris) | 54 |
| ORF4 | 2258 | 1620 | 212 | Recombinase | ORF3 (Rhodococcus erythropolis) | 34 |
| ORF5 | 2483 | 2316 | 55 | None | None | |
| ORF6 | 2696 | 3604 | 302 | Replicase | repA of pMEC2 (Micrococcus luteus) | 64 |
| ORF7 | 3604 | 3948 | 114 | Replication initiation protein | repB of pRGO1 (Propionibacterium acidipropionici) | 100 |
| ORF8 | 3945 | 4472 | 175 | None | None | |
| ORF9 | 4971 | 5594 | 207 | Propionicin SM1 | PpnA of pLME106 (Propionibacterium jensenii) | 100 |
| ORF10 | 5647 | 5973 | 108 | None | None | |
| ORF11 | 6132 | 6677 | 181 | Resolvase | PinR (Propionibacterium acidipropionici) | 99 |
According to the literature, four plasmids isolated from Propionibacterium have been analyzed (18, 19, 21, 36). Plasmid pPG01 from P. granulosum PF283 has three ORFs, which were predicted to be responsible for DNA transfer and plasmid replication (36). Moreover, plasmid pRGO1 from P. acidipropionici E214 has six ORFs with GC contents ranging from 61.5 to 73.7% and is predicted to encode two replication proteins (i.e., ORF1 and ORF2) (19). Two replication proteins were also predicted for plasmid p545 from P. freudenreichii LMG 16545 (18). Plasmid pLME106 from P. jensenii DF1 contains 10 ORFs; among these ORFs, one was predicted to encode a DNA binding protein, one a DNA invertase, and another one propionicin SM1 (21).
Transcriptional analysis of the ORFs.
Total RNA was extracted from P. acidipropionici ATCC 4875 when the OD600 reached 0.7 in SLB medium. The RT-qPCR results revealed that all 11 of the predicted ORFs could be transcribed to corresponding mRNAs. As shown in Fig. 2, among the 11 ORFs, orf9 and orf10 are characterized by the highest transcription levels. These two genes are predicted to be responsible for the metabolism of propionicin SM1. Additionally, the high expression levels of orf9 and orf10 may benefit the survival of the strain. The transcription levels of orf1, orf2, orf3, orf6, orf7, and orf8 are at intermediate levels, and their encoding proteins seem to be involved in DNA binding and replication, which are important for plasmid maintenance. orf4, orf5, and orf11 have relatively low transcription levels.
Fig 2.
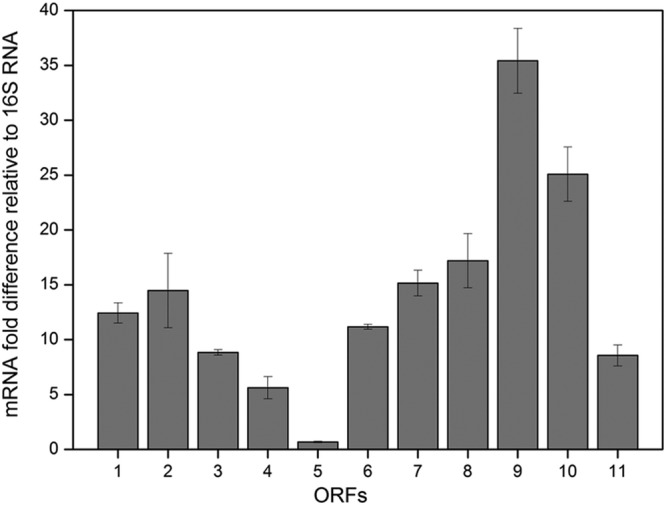
The transcription level relative to 16S RNA of 11 predicted ORFs from pZGX01. The total RNA extracted from cells was reverse transcribed to cDNA. The obtained cDNA was subjected to quantitative PCR, and 16S RNA was set as the guide sample.
Shuttle vector construction and transformation.
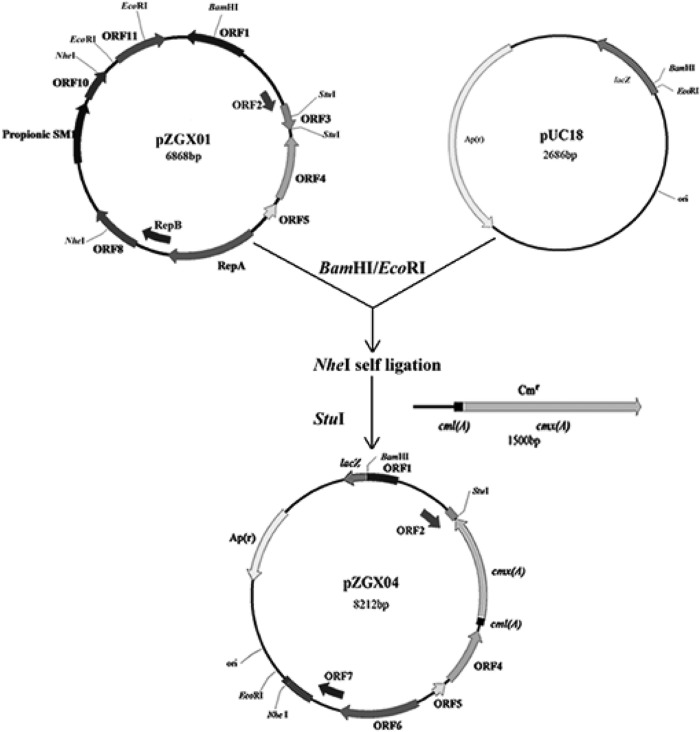
A shuttle vector between Propionibacterium and E. coli was constructed. A large fragment containing orf2 to orf10 was obtained from pZGX01 by digestion with EcoRI and BamHI and ligated to EcoRI-BamHI-digested E. coli pUC18. The resulting pZGX01-1 plasmid was digested with NheI and self-ligated, yielding the pZGX01-2 plasmid. A 1.5-kb PCR product carrying the cmx(A) (chloramphenicol resistance) gene and cml(A) genes was ligated to pZGX01-3, which was obtained by digesting pZGX01-2 with StuI (inside orf2), yielding the shuttle vector pZGX04 (Fig. 3). The GC content of cmx(A) is 63%, which is consistent with the GC content of Propionibacterium. Since it was demonstrated that the expression level of the antibiotic resistance gene with high GC content was good in Propionibacterium (21), this consistency is particularly important.
Fig 3.
Scheme for vector pZGX04 construction. A large fragment containing orf2 to orf10 was obtained from pZGX01 by digestion with EcoRI and BamHI, and the fragment was ligated to EcoRI-BamHI-digested E. coli plasmid pUC18. The resulting plasmid was digested with NheI and self-ligated. Then, the resulting plasmid was digested with StuI (inside orf2) and ligated to a 1.5-kb PCR product carrying the cmx(A) and cml(A) genes, yielding the shuttle vector pZGX04. Cmr, gene encoding chloramphenicol-resistant protein; Apr, gene encoding ampicillin-resistant protein; RepA, gene encoding Rep A protein; RepB, gene encoding Rep B protein; lacZ, β-galactosidase; ori, colE replication origin.
The pZGX04 plasmid cloned in E. coli JM110 was used to transform P. jensenii ATCC 4868. The growth phase for preparation of competent cells, electroporation resistance, voltage, and amount of plasmid DNA have been optimized. It was found that when the competent cells were prepared at a late exponential phase (OD600 = 0.7) and 1.5 μg of pZGX04 was exposed to an electric field strength of 12.5 kV/cm with a resistance of 200 Ω, the transformation efficiency reached a maximum of 30 to 40 CFU/μg DNA.
Transformation host range and stability.
To further examine the transformable capacity of this shuttle vector in other Propionibacterium species, P. acidipropionici ATCC 4875, P. acidipropionici ATCC 4965, P. acidipropionici ATCC 25562, P. jensenii ATCC 4868, P. jensenii ATCC 4870, P. freudenreichii subsp. freudenreichii ATCC 6207, P. freudenreichii subsp. shermanii ATCC 9614, P. thoenii ATCC 4872, and P. thoenii ATCC 4874 (Table 1) have been tested. Interestingly, it was found that strains P. jensenii ATCC 4870 and P. freudenreichii subsp. freudenreichii ATCC 6207 could also be transformed by pZGX04 with transformation efficiencies of 25 to 30 CFU/μg DNA and 30 to 40 CFU/μg DNA, respectively.
The Propionibacterium transformants containing pZGX04 were cultured for 30 generations in medium without chloramphenicol and were then spread on SLB agar plates with and without chloramphenicol. Only a few differences in the number of colonies were observed, indicating that the vector was segregationally stable in all the tested organisms (≤9% loss after 30 generations without selection). Moreover, sequencing of the reisolated vector from P. jensenii also verified that the vector could be replicated in Propionibacterium without reconstruction (data not shown).
Expression of the gldA gene in P. jensenii ATCC 4868.
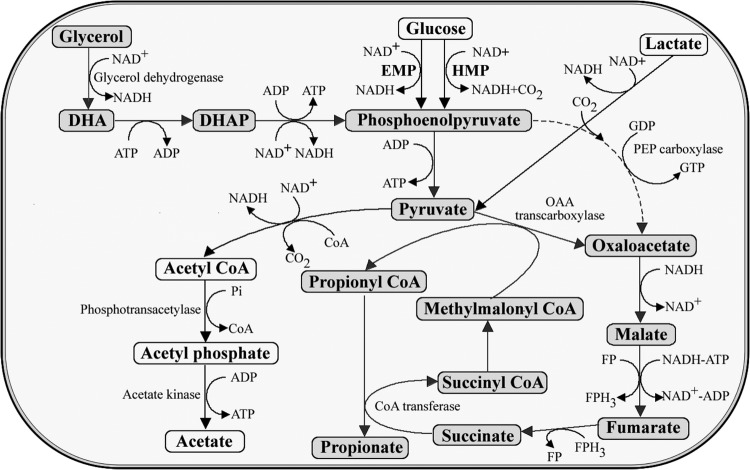
Glycerol dehydrogenase is an enzyme involved in the PA biosynthesis pathway of Propionibacterium and plays an important role during the assimilation of glycerol as the carbon source (Fig. 4). Here, we attempted to express the glycerol dehydrogenase-encoding gene (gldA) from K. pneumoniae within the constructed vector to improve the production of PA by P. jensenii ATCC 4868. The original promoter of pZGX04 was kept in the construction of the expression vector and was used to express gldA. A PCR product of pZGX04 using primers pZGX04-FWD and pZGX04-REV was ligated to another PCR product of the K. pneumoniae subsp. pneumoniae ATCC 12657 genome using primers GLD-FDW and GLD-REV. The resulting vector (named pZGX04-gldA) was transformed into P. jensenii ATCC 4868. The RT-qPCR analysis revealed that the gldA transcription level was 8.07-fold of that of the homologous gene (glpA) in the wild-type P. jensenii ATCC 4868.
Fig 4.
Synthesis pathways for propionic acid from glycerol, glucose, and lactate in Propionibacterium. The biosynthetic pathway from glycerol to propionic acid proceeds from glycerol to dihydroxyacetone (DHA) to dihydroxyacetone phosphate (DHAP) to phosphoenolpyruvate to pyruvate and oxaloacetate and from oxaloacetate to malate, fumarate, succinate, succinyl coenzyme A (CoA), methylmalonyl CoA, propionyl CoA, and propionate (see arrows). Acetate is the main byproduct in propionic acid production. Gray color indicates the biosynthetic pathway from glycerol to propionic acid.
Figure 5 shows the effects of the gldA expression on PA production by P. jensenii ATCC 4868. In batch fermentation, the engineered strain P. jensenii ATCC 4868 (pZGX04-gldA) displays a much higher glycerol dehydrogenase (GDH) activity than the wild-type P. jensenii ATCC 4868 (Fig. 1), and it consumes glycerol more rapidly. Moreover, PA production (amount of PA produced) with the expression of gldA was increased from 22.06 ± 1.2 g/liter to 28.23 ± 1.0 g/liter and the productivity (production per hour) from 0.15 g · liter−1 · h−1 to 0.20 g · liter−1 · h−1. However, the DCW of engineered P. jensenii ATCC 4868 (pZGX04-gldA) decreased, indicating that the plasmid transformation slows cell growth. P. jensenii ATCC 4868 (pZGX04-gldA) also has a higher YP/S (mass ratio of PA to succinic acid), YP/A (mass ratio of PA to acetic acid), and YPA/Glycerol than the wild-type strain.
Fig 5.
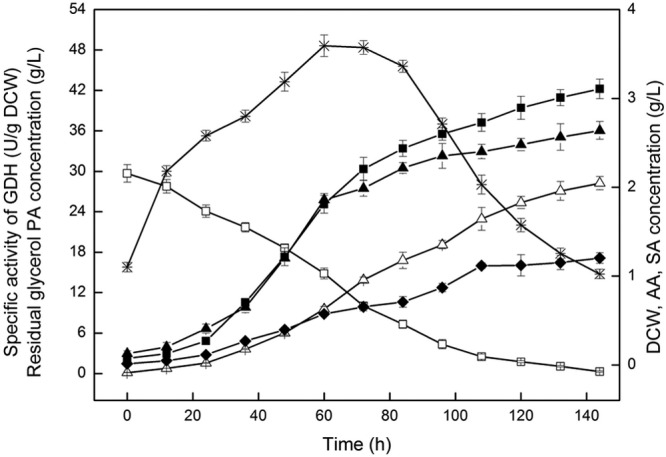
Batch fermentation kinetics of propionic acid production from glycerol with P. jensenii ATCC 4868 (pZGX04-gldA) at pH 7.0 and 30°C for 144 h. Batch fermentations were performed under anaerobic conditions (nitrogen gassing) in a 3-liter Bioreactor containing 2 liters of culture medium supplemented with 30 g/liter glycerol. Samples were taken from the fermentor every 12 h. △, propionic acid (PA); □, residual glycerol; ▲, dry cell weight (DCW); ■, acetic acid (AA); ♦, succinic acid (SA); *, specific activity of GDH.
The results indicate that the expression of gldA enhances the PA yield and reduces the concentration of byproducts (acetic acid and succinic acid) under anaerobic conditions. The expression of the gldA gene increases the transformation rate of glycerol to dihydroxyacetone (DHA) and thus generates more NADH (Fig. 4). Since the synthesis pathway of PA is a NADH consumption process, the accumulation of NADH is considered a favorable event. Although the expression of gldA induced slower cell growth during the batch fermentation, the specific PA synthesis rate results clearly improved. Overall, these findings indicate that gldA expression can noticeably increase the PA titer, productivity, and yield.
Conclusions.
In summary, our results show that the P. jensenii ATCC 4868 strain is a good candidate for PA production. Thus, we developed a shuttle vector for the metabolic engineering of P. jensenii ATCC 4868 to further improve PA production. The constructed vector was successfully transformed into P. jensenii ATCC 4868. The report showing that a vector can be transformed into P. jensenii with a high stability. The constructed shuttle vector was also used to efficiently engineer P. freudenreichii. The glycerol dehydrogenase-encoding gene (gldA) was expressed in P. jensenii ATCC 4868 by the shuttle vector, resulting in a 26.07% increase in the PA production. We believe that the developed shuttle vector could strengthen the engineering capacity of P. jensenii and further improve its production potential by vector-based metabolic engineering strategies.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by National Natural Science Foundation of China (21006040), Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 Project (111-2-06), and 973 Program (2012CB720806).
Footnotes
Published ahead of print 24 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00737-13.
REFERENCES
- 1. Hsu ST, Yang ST. 1991. Propionic acid fermentation of lactose by Propionibacterium acidipropionici: effects of pH. Biotechnol. Bioeng. 38:571–578 [DOI] [PubMed] [Google Scholar]
- 2. Roberto MC, Mayra T. 2002. Production of propionate by fed-batch fermentation of Propionibacterium acidipropionici using mixed feed of lactate and glucose. Biotechnol. Lett. 24:427–431 [Google Scholar]
- 3. Werpy T, Petersen G, Aden A, Bozell J, Holladay J, White J, Manheim A, Elliot D, Lasure L, Jones S, Gerber M, Ibsen K, Lumberg L, Kelley S. 2004. Top value added chemicals from biomass, vol 1—results of screening for potential candidates from sugars and synthesis gas. US Department of Energy, Oak Ridge, TN [Google Scholar]
- 4. Rogers P, Chen J-S, Zidwick MJ. 2006. Organic acid and solvent production. Part II: propionic and butyric acids and ethanol, p 611–671 In Dworkin M. (ed), The prokaryotes: symbiotic associations, biotechnology, applied microbiology 3rd ed, vol 1 Springer, Berlin, Germany [Google Scholar]
- 5. Liu Z, Ma C, Gao C, Xu P. 2012. Efficient utilization of hemicellulose hydrolysate for propionic acid production using Propionibacterium acidipropionici. Bioresour. Technol. 114:711–714 [DOI] [PubMed] [Google Scholar]
- 6. Feng X, Chen F, Xu H, Wu B, Li H, Li S, Ouyang P. 2011. Green and economical production of propionic acid by Propionibacterium freudenreichii CCTCC M207015 in plant fibrous-bed bioreactor. Bioresour. Technol. 102:6141–6146 [DOI] [PubMed] [Google Scholar]
- 7. Coral J, Karp SG, Vandenberghe LPS, Parada JL, Pandey A, Soccol CR. 2008. Batch fermentation model of propionic acid production by Propionibacterium acidipropionici in different carbon sources. Appl. Biochem. Biotechnol. 151:333–341 [DOI] [PubMed] [Google Scholar]
- 8. Dishisha T, Alvarez MT, Kaul RH. 2012. Batch-and continuous propionic acid production from glycerol using free and immobilized cells of Propionibacterium acidipropionici. Bioresour. Technol. 118:553–562 [DOI] [PubMed] [Google Scholar]
- 9. Goswami V, Srivastava AK. 2001. Propionic acid production in an an situ cell retention bioreactor. Appl Microbiol Biotechnol. 56:676–680 [DOI] [PubMed] [Google Scholar]
- 10. Himmi EH, Bories A, Boussaid A, Hassani L. 2000. Propionic acid fermentation of glycerol and glucose by Propionibacterium acidipropionici and Propionibacterium freudenfeichii ssp. shermanii. Appl Microbiol Biotechnol. 53:435–440 [DOI] [PubMed] [Google Scholar]
- 11. Liu Y, Zhang YG, Zhang RB, Zhang F, Zhu J. 2011. Glycerol/glucose co-fermentation: one more proficient process to produce propionic acid by Propionibacterium acidipropionici. Curr. Microbol. 62:152–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suwannakham S, Huang Y, Yang ST. 2006. Construction and characterization of ack knock-out mutants of Propionibacterium acidipropionici for enhanced propionic acid fermentation. Biotechnol. Bioeng. 94:383–395 [DOI] [PubMed] [Google Scholar]
- 13. Zhang A, Yang ST. 2009. Propionic acid production from glycerol by metabolically engineered Propionibacterium acidipropionici. Process Biochem. 44:1346–1351 [Google Scholar]
- 14. Zhu L, Wei P, Cai J, Zhu X, Wang Z, Huang L, Xu Z. 2012. Improving the productivity of propionic acid with FBB-immobilized cells of an adapted acid-tolerant Propionibacterium acidipropionici. Bioresour. Technol. 112:248–253 [DOI] [PubMed] [Google Scholar]
- 15. Boyaval P, Corre C, Madec MN. 1994. Propionic acid production in a membrane bioreactor. Enzyme Microb. Technol. 16:883–886 [Google Scholar]
- 16. Zhu YF, Li JH, Tan M, Liu L, Jiang LL, Sun J, Lee P, Du GC, Chen J. 2010. Optimization and scale-up of propionic acid production by propionic acid-tolerant Propionibacterium acidipropionici with glycerol as the carbon source. Bioresour. Technol. 101:8902–8906 [DOI] [PubMed] [Google Scholar]
- 17. Brede DA, Faye T, Stierli MP, Dasen G, Theiler A, Nes IF, Meile L, Holo H. 2005. Heterologous production of antimicrobial peptides in Propionibacterium freudenreichii. Appl. Environ. Microbiol. 71:8077–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jore JP, van Luijk N, Luiten RG, van der Werf MJ, Pouwels PH. 2001. Efficient transformation system for Propionibacterium freudenreichii based on a novel vector. Appl. Environ. Microbiol. 67:499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kiatpapan P, Hashimoto Y, Nakamura H, Piao YZ, Ono H, Yamashita M, Murooka Y. 2000. Characterization of pRGO1, a plasmid from Propionibacterium acidipropionici, and its use for development of a host-vector system in propionibacteria. Appl. Environ. Microbiol. 66:4688–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rehberger TG, Glatz BA. 1990. Characterization of Propionibacterium plasmids. Appl. Environ. Microbiol. 56:864–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stierli MP. 2002. DNA transformation of propionibacteria based on plasmids pLME106 and pLME108. Ph.D. thesis no. 14468 Swiss Federal Institute of Technology, Zurich, Switzerland [Google Scholar]
- 22. Brede DA, Lothe S, Salehian Z, Faye T, Nes IF. 2007. Identification of the propionicin F bacteriocin immunity gene (pcfI) and the development of a food-grade cloning system for Propionibacterium freudenreichii. Appl. Environ. Microbiol. 73:7542–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheong DE, Lee HI, So JS. 2008. Optimization of electrotransformation conditions for Propionibacterium acnes. J. Microbiol. Methods 72:38–41 [DOI] [PubMed] [Google Scholar]
- 24. Kiatpapan P, Murooka Y. 2001. Construction of an expression vector for propionobacteria and its use in production of 5-aminolevulinic acid by Propionibacterium freudenreichii. Appl. Microbiol. Biotechnol. 56:144–149 [DOI] [PubMed] [Google Scholar]
- 25. Piao YZ, Kiatpapan P, Yamashita M, Murooka Y. 2004. Effects of expression of hemA and hemB genes on production of porphyrin in Propionibacterium freudenreichii. Appl. Environ. Microbiol. 70:7561–7566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiatpapan P, Murooka Y. 2002. Genetic manipulation system in propionibacteria. J. Biosci. Bioeng. 93:1–8 [PubMed] [Google Scholar]
- 27. Ekinci FY, Barefoot SF. 2006. Fed-batch enhancement of jenseniin G, a bacteriocin produced by Propionibacterium thoenii (jensenii) P126. Food Microbiol. 23:325–330 [DOI] [PubMed] [Google Scholar]
- 28. Faye T, Brede DA, Langsrud T, Nes IF, Holo H. 2004. Prevalence of the genes encoding propionicin T1 and protease-activated antimicrobial peptide and their expression in classical propionibacteria. Appl. Environ. Microbiol. 70:2240–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hugenholtz J, Hunik J, Santos H, Smid E. 2002. Nutraceutical production by propionibacteria. Lait 82:103–112 [Google Scholar]
- 30. Vanberg C, Lutnaes BF, Langsrud T, Nes IF, Holo H. 2007. Propionibacterium jensenii produces the polyene pigment granadaene and has hemolytic properties similar to those of Streptococcus agalactiae. Appl. Environ. Microbiol. 73:5501–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang A, Yang ST. 2009b. Engineering Propioniacterium acidipropionici for enhanced propionic acid tolerance and fermentation. Biotechnol. Bioeng. 104:766–773 [DOI] [PubMed] [Google Scholar]
- 32. Fessler DS, Casey MG, Puhan Z. 1999. Propionibacteria flora in Swiss raw milk from lowlands and alps. Lait 79:201–209 [Google Scholar]
- 33. Gautier M, Rouault A, Lemée R. 1995. Electrotransfection of Propionibacterium freudenreichii TL 110. Lett. Appl. Microbiol. 20:125–129 [Google Scholar]
- 34. Ankri S, Bouvier I, Reyes O, Predali F, Leblon G. 1996. A Brevibacterium lines pRBL1 replicon functional in Corynebacterium glutamicum. Plasmid 36:36–41 [DOI] [PubMed] [Google Scholar]
- 35. Miescher S, Stierli MP, Teuber M, Meile L. 2000. Propionicin SM1, a bacteriocin from Propionibacterium jensenii DF1: isolation and characterization of the protein and its gene. Syst. Appl. Microbiol. 23:174–184 [DOI] [PubMed] [Google Scholar]
- 36. Farrar MD, Howson KM, Emmott JE, Bojar RA, Holland KT. 2007. Characterisation of cryptic plasmid pPG01 from Propionibacterium granulosum, the first plasmid to be isolated from a member of the cutaneous propionibacteria. Plasmid 58:68–75 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.