Abstract
Lactobacilli are important for the maintenance of a healthy ecosystem in the human vagina. Various mechanisms are postulated but so far are poorly substantiated by molecular studies, such as mutant analysis. Bacterial autoaggregation is an interesting phenomenon that can promote adhesion to host cells and displacement of pathogens. In this study, we report on the identification of a human vaginal isolate, Lactobacillus plantarum strain CMPG5300, which shows high autoaggregative and adhesive capacity. To investigate the importance of sortase-dependent proteins (SDPs) in these phenotypes, a gene deletion mutant was constructed for srtA, the gene encoding the housekeeping sortase that covalently anchors these SDPs to the cell surface. This mutant lost the capacity to autoaggregate, showed a decrease in adhesion to vaginal epithelial cells, and lost biofilm-forming capacity under the conditions tested. These results indicate that the housekeeping sortase SrtA of CMPG5300 is a key determinant of the peculiar surface properties of this vaginal Lactobacillus strain.
INTRODUCTION
The human vaginal microbiota is commonly inhabited by lactic-acid-producing bacteria (LAB), with Lactobacillus spp. constituting around 70% of the microbial population (1, 2). They appear to protect the urogenital tract from pathogen invasion and prevent urogenital and sexually transmitted diseases (3, 4). Postulated antipathogenic mechanisms include (i) production of lactic acid, hydrogen peroxide (H2O2), bacteriocins, and biosurfactants that directly kill or inhibit bacterial and viral pathogens (5); (ii) stimulation of host defense mechanisms against pathogens (6); and (iii) formation of microcolonies that adhere to the epithelial cell receptors and form a physical barrier to pathogen adhesion (7; M. Petrova, M. van den Broek, J. Balzarini, J. Vanderleyden, and S. Lebeer, unpublished data). The last can occur by virtue of adhesins on the bacterial surface that compete for receptor sites in the presence of pathogens. Although the health benefits of vaginal lactobacilli are widely recognized, the underlying molecular mechanisms, such as vaginal adaptation factors that allow optimal adaptation of lactobacilli to the niche (8), are not yet well understood. For instance, while the genome sequences of various gastrointestinal Lactobacillus strains, such as Lactobacillus plantarum WCFS1 (9), Lactobacillus acidophilus NCFM (10), Lactobacillus salivarius subsp. salivarius strain UCC118 (11), Lactobacillus gasseri ATCC 33323 (12), and Lactobacillus rhamnosus GG (13), have been published since 2004, the vaginal strains have lagged behind, with the genome sequence of Lactobacillus iners AB-1 being an important breakthrough (14), followed by the recently sequenced Lactobacillus pentosus KCA1 (15).
Various lactobacilli, including some vaginal strains, such as L. gasseri 2459 and Lactobacillus johnsonii CRL 1294, have been shown to form large aggregates among themselves, a phenomenon called autoaggregation (16, 17). Some studies also reveal the nature of the molecules involved in autoaggregation, yet they are restricted to strains isolated from the gastrointestinal tract, such as aggregation-promoting factors in L. acidophilus NCFM (18). In addition, a correlation between autoaggregation in LAB and adhesion to host cells has been suggested for the adhesion of intestinal isolates, such as L. acidophilus M92 (19) and Lactobacillus crispatus M247 (20), to a human colonic cell line (Caco-2). Interestingly, Boris et al. (21) also suggest a correlation between autoaggregation and adhesion of some selected strains of L. gasseri, L. acidophilus, and Lactobacillus jensenii isolated from the vagina to vaginal epithelial cells, but without further documentation of the molecules involved.
An important class of cell wall proteins in lactobacilli is formed by the sortase-dependent proteins (SDPs) (8). These surface proteins possess a conserved carboxy-terminal sorting motif, generally LPXTG, that is recognized by the sortase enzyme, which cleaves between the threonine and the glycine residues and covalently links the C-terminally truncated protein to the peptidoglycan layer. Sortases are composed of different classes of enzymes, with srtA encoding the prototype housekeeping enzyme, sortase A (22). In this study, we report on the highly autoaggregative and adhesive capacity of a human vaginal L. plantarum strain, CMPG5300, a single-colony isolate of L. plantarum LAB 129 isolated from a healthy volunteer. Genetic and functional analyses were undertaken to understand the molecular mechanisms of this behavior in relation to the srtA gene.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All Lactobacillus strains used in this study, including L. plantarum strain CMPG5300 and its mutant derivatives (Table 1), were grown at 37°C without agitation in Man-Rogosa-Sharpe (MRS) medium (Difco). Escherichia coli TOP10, used for cloning, was grown with shaking at 37°C in Luria Bertani (LB) medium. When required, antibiotics were added at the following final concentrations: chloramphenicol, 20 μg/ml for L. plantarum and 10 μg/ml for E. coli; erythromycin, 10 μg/ml for L. plantarum and 250 μg/ml for E. coli; and 100 μg/ml ampicillin for E. coli.
Table 1.
Bacterial strains and plasmids used
| Species or plasmid | Strain/host | Relevant genotype or descriptiona | Reference and/or source |
|---|---|---|---|
| Bacterial strains | |||
| L. plantarum | CMPG5300 | Wild type; human vaginal isolate | Mario Vaneechoutte |
| CMPG5376 | srtA mutant of CMPG5300 | This study | |
| CMPG5378 | CMPG5376 complemented with srtA on plasmid pCMPG5378 | This study | |
| LAB135 | Human vaginal isolate | Mario Vaneechoutte | |
| L. jensenii | 25258 | Wild type; human vaginal isolate (Gasser, Mandel, and Rogosa 1970) | LMG/BCCMb |
| L. rhamnosus | PRB016 | Human vaginal isolate | Mario Vaneechoutte |
| GR-1 (ATCC 55826) | Human vaginal isolate | 58; Chr. Hansen | |
| L. reuteri | RC-14 (ATCC 55845) | Human vaginal isolate | 58; Chr. Hansen |
| L. casei/L. paracasei | PRB005 | Human vaginal isolate | Mario Vaneechoutte |
| PRB015 | Human vaginal isolate | Mario Vaneechoutte | |
| L. crispatus | NCIMB 4505 | Wild type; human vaginal isolate (Moore and Holdeman 1970) | LMG/BCCM |
| E. coli | TOP10F | F′ (lacIq Tnr) mcrA_(mrr hsdRMS-mcrBC) 80LacZ_lacX74 deoR recA1 araD139_(ara leu) | Invitrogen |
| Plasmids | |||
| pNZ5319 | E. coli | pNZ5318 derivative for multiple gene replacements in Gram-positive bacteria; Cmr Eryr | 37 |
| pNZ5348 | E. coli | pGID023 derivative containing cre under the control of the lp_1144 promoter; Eryr | 37 |
| pLAB1301 | E. coli | E. coli-Lactobacillus shuttle vector; Apr Eryr | 36 |
| pCMPG5376 | E. coli | pNZ5319 derivative containing homologous regions up- and downstream of CMPG5300 srtA; Cmr Eryr | This study |
| pCMPG5378 | E. coli | pLAB1301 containing the srtA gene of CMPG5300; Apr Eryr | This study |
Cmr, chloramphenicol resistance; Eryr, erythromycin resistance; Apr, ampicillin resistance.
LMG/BCCM, Laboratory for Microbiology of the Faculty of Sciences of Ghent University/Belgian Co-ordinated Collections of Micro-organisms.
Analysis of autoaggregation.
Analysis of the autoaggregation properties of various vaginal strains was performed as previously reported by Kos et al. (19). Briefly, bacteria were grown at 37°C for 18 h in MRS. The cells were washed twice with phosphate-buffered saline (PBS) (pH 7.2) and suspended in the same buffer to an optical density at 600 nm (OD600) of 0.5. After incubation at room temperature for 5 h, 0.1 ml of the upper suspension was transferred to a new tube, diluted 10 times, and then measured for the absorbance at 600 nm. The autoaggregation percentage is expressed as follows: 1 − (At/A0) × 100, where At represents the absorbance at the 5-h time point (t = 5 h) and A0 the absorbance at t = 0.
Autoaggregation was also investigated by microscopy. The strains were cultured overnight in MRS medium and subsequently visualized with the naked eye and/or viewed under a Zeiss Axio Imager Z1 ×40 phase-contrast microscope equipped with an AxioCam MRm Rev. 3 monochrome digital camera.
Assay of adhesion to VK2/E6E7 cells.
The VK2/E6E7 cell line (ATCC CRL-2616), purchased from the American Type Culture Collection (ATCC) (Rockville, MD), was routinely grown in 75-cm2 tissue culture flasks at 37°C in a 5% CO2, 95% air atmosphere in keratinocyte serum-free medium (Gibco-Invitrogen) supplemented with 0.1 ng/ml epithelial growth factor and 25 μg/ml bovine pituitary extract. The cells were passaged every 3 days (at 70 to 80% confluence) by reseeding with a split ratio of 1:7 in fresh culture medium. For adhesion experiments, cells were grown in 12-well Multiwell Plates (BD Falcon) at a density of 3 × 105 cells/cm2 per well, and 5-day-old monolayer cultures were used.
To assess the capacities of various Lactobacillus strains and the mutant derivatives to adhere to vaginal epithelial cells, adhesion studies were carried out as described previously (23). For adhesion assays, bacterial cells at 1 × 107 CFU/ml were added to tissue culture wells containing fully differentiated VK2/E6E7 cells and allowed to incubate at 37°C for 1 h. The vaginal epithelial cells were then rinsed twice with 1× PBS prewarmed at 37°C. Following washing, a 0.1-ml volume of 1× trypsin-EDTA (Invitrogen)/1× PBS was added to the wells, and the plate was incubated for 10 min at 37°C. Next, a 0.9-ml volume of 1× PBS was added to each well, the cell suspension was mixed, and a set of serial dilutions was prepared and plated out on solid MRS medium. After incubation for 48 h at 37°C, the bacterial colonies were counted. The adhesion ratio, expressed as a percentage, was calculated by comparing the total number of bacterial colonies counted after adhesion to the number of cells in the bacterial suspension prior to addition to the wells. The experiment was performed three times, with each sample tested in triplicate.
Biofilm formation assay.
Biofilm formation was assessed as described previously (24). Briefly, the biofilms were grown for 72 h in MRS medium, and the capacity was estimated by crystal violet staining. The sterile growth medium was also included as a negative control. The experiments were performed three times, with eight technical repeats.
Transmission electron microscopy (TEM).
Bacteria were grown overnight in MRS medium. Uncoated copper grids were used as a probe to adsorb bacterial cells. The grids were placed on a drop of bacterial suspension for 30 s, transferred to drops of 0.25% phosphotungstenic acid (pH 7) for 30 s, and washed three times, followed by draining of the excess liquid. The bacteria were observed with a Philips EM 208S transmission electron microscope at 56 kV. Images were digitalized using an SIS image analysis system.
Identification of the sortase gene (srtA) and the substrates of the srtA-encoded protein.
Genomic DNA of L. plantarum CMPG5300 was isolated using early-exponential-phase culture grown in 200 ml of MRS medium. The cells were pelleted down at 4,000 × g for 10 min and washed twice with Tris-EDTA (TE) buffer. After resuspending the cells thoroughly in 7 ml of TE buffer, the cell suspension was subjected to a brief sonication step (pulse on for 30 s, pulse off for 30 s; 4 min) to break the cell clumps. To lyse the cells, 70 μl lysozyme (final concentration, 2 mg/ml) was added, followed by incubation at 37°C for 15 min. Cell lysis was followed by adding 2 ml of 5 M NaCl and 1 ml of 10% SDS and inverting the sample very gently. RNase A (Sigma) was added at a concentration of 50 μg/ml and incubated at 55°C for 15 min. To remove proteins, 0.15 μg/ml of proteinase K (Sigma) was added, and the mixture was incubated at 55°C for 30 min. Subsequently, 2.7 ml of 5 M Na-perchlorate was added to this mixture. Extraction was done by adding the same volume of chloroform-isoamyl alcohol (24:1) and incubating for 1 h at 4°C (with gentle shaking). The cells were pelleted down at 15,000 × g for 10 min, and the supernatant was collected in a fresh tube. Precipitation of DNA was done by adding 2 volumes of 100% ethanol. The precipitated DNA was pelleted by centrifugation at 9,300 × g for 10 min. The supernatant was discarded, and the DNA pellet was washed twice with freshly prepared 70% ethanol and air dried. The final pellet obtained was dissolved in 400 μl of TE buffer and stored at 4°C till further use.
The genome of CMPG5300 was sequenced using the 454 GS FLX+ sequencing platform (Genomics Core, KU Leuven, Leuven, Belgium), and the reads were assembled into contigs using Roche GS De Novo Assembler software (version 2.5p1). The minimum overlap length was set to 40 nucleotides, with a minimum overlap identity of 90%. Open reading frames were extracted using the getorf program from the EMBOSS suite v. 6.3.1 (25) and manually curated by comparison to the genome of L. plantarum WCFS1 using ACT (26). Putative sortase substrates encoded by L. plantarum CMPG5300 were identified using LocateP (27), which incorporates hidden Markov model model for the identification of LPXTG-type sortase substrates. The amino acid sequences of the 27 identified sortase-dependent substrates for L. plantarum WCFS1 were downloaded from http://www.josboekhorst.nl/cgi-bin/sortase_substrates/index.py (28). The sortase substrates of CMPG5300 were submitted to the SignalP 4.1 server (29) for the prediction of signal peptides (see the supplemental material).
DNA manipulations.
Standard protocols were used for all DNA manipulations, including restriction digestion, ligation, and transformation of E. coli (30). Plasmid DNA was isolated using the QIAprep Spin kit according to the manual (Qiagen). DNA amplification by PCR was performed using Pfx polymerase (Roche) and Taq DNA polymerase (Roche) according to the procedure recommended by the manufacturer. Primers for PCR (see Table S1 in the supplemental material) were synthesized by Integrated DNA Technologies (Coralville, IA). Restriction sites were included at the 5′ ends, when desired, to facilitate cloning. DNA fragments were extracted from 1.0% agarose gels using the Qiagen gel extraction kit according to the manufacturer's instructions.
Optimization of electroporation to L. plantarum CMPG5300.
Various protocols, listed in Table S2 in the supplemental material, were used to obtain the optimum efficiency for electroporation of CMPG5300. To obtain competent cells, precultures of CMPG5300 in MRS medium were used to make serial dilutions (102- to 106-fold) in MRS medium supplemented with 2% glycine. They were inoculated overnight, and 5 ml of the culture with an OD600 of 0.8 to 1.4 (cf., 7 × 107 CFU/ml) was used to further inoculate 100 ml of prewarmed MRS medium containing glycine. The cultures were then grown at 37°C with or without agitation till the OD600 reached 0.4 to 0.6 (∼3 h). Afterward, the cells were pelleted down at 4,000 × g for 10 min and subsequently washed with buffers (ice cold) as follows: set I, 2 washes with distilled water, a 5-min treatment with EDTA, and 2 washes with ice-cold 0.5 M sucrose and 10% glycerol, a combination of the protocols of Berthier et al. (31) and Mason et al. (32) designed by Speer et al. (http://openwetware.org/wiki/Lactobacillus_transformation_(Speer_2012)); set II, 2 washes with 0.5 M sucrose, 7 mM potassium phosphate (pH 7.4), 1 mM MgCl2 (34). Finally, the cells were resuspended in a small volume (∼0.8 ml) of the electroporation buffer, and aliquots of 90 μl were prepared in ice-cold 1.5-ml Eppendorf tubes. A highly concentrated (1 μg in 10 μl) replicating plasmid, pLAB1301 (35), was added to each tube. This mixture was transferred into a precooled 2-mm electroporation cuvette (Eurogentec) and immediately electroporated (Gene Pulser; Bio-Rad Laboratories) using the following settings: peak voltage, 1.7 kV; capacitance, 25 μF; and parallel resistance, 200 Ω/400 Ω. Following this, the cells were regenerated by adding 900 μl of the regeneration medium (MRS medium containing 2 mM CaCl2 and 20 mM MgCl2) and incubated (without agitation) at 37°C for 3 h, after which they were plated out on MRS medium containing 10 μg/ml erythromycin. They were then incubated at 37°C for ∼72 h, followed by colony counting to determine the transformation efficiency. The colonies obtained were also subjected to screening for the presence of the plasmid via colony PCR using the primers pseu 383 and pseu 384 (see Table S1 in the supplemental material), designed for the detection of an ∼600-bp product.
Construction of a srtA mutant by double homologous recombination.
To create a construct to knock out the srtA gene by double homologous recombination, PCR primers were designed to amplify two ca. 1,000-bp-long homologous regions (HR1 and HR2) that flank the 5′ and 3′ ends of the srtA gene (see Table S1 in the supplemental material). Both the PCR fragments were subsequently ligated into the respective multiple-cloning sites (MCSs) upstream and downstream of the chloramphenicol resistance cassette of the pNZ5319 plasmid (36) and transferred to competent E. coli TOP10 cells. The resulting suicide plasmid construct was designated the pCMPG5376 vector and then electroporated into competent cultures of wild-type (WT) CMPG5300. The transformants resulting from double homologous recombination were selected for a chloramphenicol-resistant and erythromycin-sensitive phenotype by replica plating. To excise the chloramphenicol (cam) cassette from the genome of the srtA mutant, the mutant was transformed with a high concentration (∼4 μg) of pNZ5348 DNA (36) using the optimized protocol mentioned above. The Emr colonies obtained after 48 to 72 h of growth were checked for Cre-mediated recombination using a PCR with EryintF and EryintR primers (see Table S1 in the supplemental material), as described previously (36). These colonies were grown for six generations in MRS medium without any antibiotic to cure the pNZ5348 vector and remove the cam cassette. The cells were plated out on MRS medium and MRS medium with chloramphenicol to obtain single-colony isolates for estimating the number of colonies on each plate. A few colonies of the MRS plate were selected and replica plated on MRS medium with 20 μg/ml chloramphenicol to confirm curing of the Cre expression vector. Finally, one colony showing the correct allelic replacement and excision of the cam cassette was selected and designated CMPG5376.
Furthermore, the srtA mutant CMPG5376 was also complemented by electroporation with pCMPG5378, resulting in strain CMPG5378. To achieve this, the srtA gene amplified by primers pro 8177 and pro 8150 (see Table S1 in the supplemental material) was ligated in pLAB1301 (35), resulting in pCMPG5378. Plasmid pCMPG5378 was electroporated in CMPG5376 to yield an erythromycin-sensitive strain, CMPG5378.
Analysis of operon structure with RT-qPCR.
L. plantarum CMPG5300 and its srtA mutant derivative CMPG5376 were grown in MRS medium, and their total RNA was isolated from exponential-phase cultures using the commercially available Promega Benelux BV kit for total RNA isolation. cDNA was made using a reverse transcription system (SuperScript III first-strand synthesis system; Invitrogen), and real-time DNA amplification was done using SYBR green Universal PCR Master Mix (Applied Biosystems). Primers (see Table S1 in the supplemental material) were designed using the Primer Express software v3.0.1 and chemically synthesized by IDT Technologies. Real-time quantitative PCR (RT-qPCR) and data analysis were done using the StepOnePlus Real Time PCR system (Applied Biosystems, Lennik, Belgium).
Nucleotide sequence accession number.
The complete nucleotide sequence of srtA (lab129_01547) and its flanking regions has been submitted to NCBI and is available under accession number KC841273.
RESULTS
Identification of L. plantarum CMPG5300 as a vaginal strain with high autoaggregating, adhering, and biofilm-forming capacity.
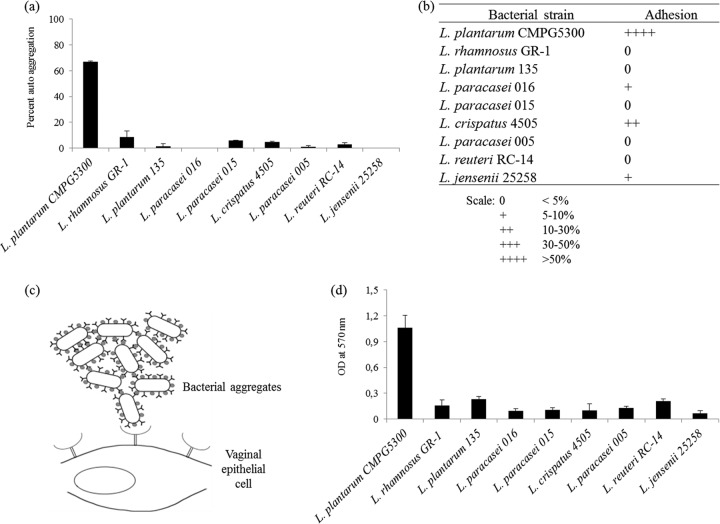
A panel of nine vaginal Lactobacillus isolates (Table 1) was screened for the capacity to autoaggregate. It was observed that most of the strains autoaggregated, although the exact percentages varied a bit with each assay. L. plantarum CMPG5300 displayed the highest percentage of autoaggregation (on average, ∼67%) (Fig. 1a), forming a precipitate and resulting in a clear solution when a suspension of 108 CFU/ml was kept standing for 5 h. In contrast, most of the other strains showed a relatively homogeneous suspension with no distinct upper layer. Subsequently, the capacities of the vaginal strains to adhere to the vaginal epithelial VK2/E6E7 cell line (37) were assessed. It was observed that CMPG5300 also displayed the highest adhesion capacity of the vaginal strains (Fig. 1b). The relatively high adhesion capacity of CMPG5300 (up to 80%) is probably biased by its high autoaggregation capacity, resulting in aggregates of multiple bacteria adhering to single epithelial cells (Fig. 1c), although the bacteria were vigorously vortexed and pipetted up and down immediately before addition to the vaginal cells. Therefore, the adherence data are presented in a relative, semiquantitative way (0 to ++++) (Fig. 1b). In the next step, the abilities of these vaginal strains to form biofilms on polystyrene were assessed. This again revealed that L. plantarum CMPG5300 showed an exceptionally high biofilm-forming capacity compared to other strains, which appeared to form only microcolonies and not confluent biofilms under the tested conditions (Fig. 1d).
Fig 1.
(a) Comparison of autoaggregation abilities of nine vaginal Lactobacillus strains. Overnight cultures of nine Lactobacillus strains were washed and suspended with PBS up to an OD600 of 0.5 and monitored for autoaggregation on the basis of the OD600 after 5 h of incubation. Autoaggregation ability is expressed as percentages. The error bars represent standard deviations of 3 independent experiments. (b) Comparison of the capacities of vaginal lactobacilli to adhere to vaginal epithelial cells. Overnight-grown Lactobacillus cells were coincubated with VK2/E6E7 cells for 1 h. The proportions of adherent bacteria are expressed semiquantitatively as explained at the bottom. (c) Schematic representation of the adhesion of L. plantarum CMPG5300 to VK2/E6E7 cells. The relatively high adhesion percentage of L. plantarum CMPG5300 is probably biased by the fact that multiple aggregates adhere to single host cells. (d) Comparison of the biofilm formation capacities of vaginal lactobacilli. The biofilms formed on polystyrene pegs were quantified after 72 h of incubation in Lactobacillus MRS medium by crystal violet staining. The absorbance values at OD570 represent the biofilm formation capacity.
Optimization of electroporation and genetic engineering of this natural vaginal isolate.
To investigate genetic determinants of the remarkable phenotype of this natural isolate, various genetic tools were optimized. Initially, the genome of a single-colony isolate of CMPG5300 was sequenced as described in Materials and Methods. This revealed the presence of a genome of ca. 3.5 Mbp for CMPG5300 (S. Malik, T. L. A. Verhoeven, B. Renckens, R. J. Siezen, M. Vaneechoutte, J. Vanderleyden, and S. Lebeer, unpublished data). In addition, as CMPG5300 is a highly autoaggregating strain, conditions for making the cells electrocompetent to increase the transformation efficiency were required. For that purpose, pLAB1301 (35), which contains the replicon of a native plasmid isolated from Lactobacillus hilgardii, was used. The parameters tested were different culture conditions, two sets of washing buffers, and two different voltages for electric shock (see Table S2 in the supplemental material). The precultures were made in MRS medium with glycine and ampicillin for cell wall weakening (34) and grown without agitation, while the final cultures were grown with or without shaking. It was observed that the efficiency was higher when L. plantarum strain CMPG5300 was grown in flasks with shaking, permitting better aeration. The efficiency was also enhanced when the sucrose buffer supplemented with salts was used with a voltage of 400 V in 2-mm cuvettes. The electroporation efficiency was expressed as the number of transformants obtained per microgram of DNA (see Table S2 in the supplemental material).
Construction of a srtA knockout mutant.
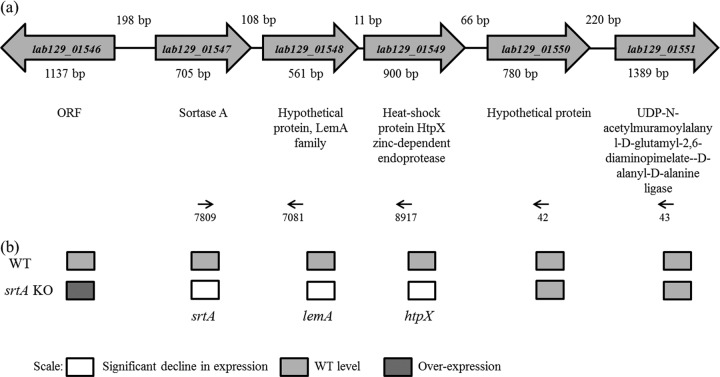
To investigate whether SDPs are involved in the autoaggregating capacity of CMPG5300, the genome of CMPG5300 was screened for the presence of sortase genes. The genome sequence was found to contain only the housekeeping sortase gene, srtA (lab129_01547) (Fig. 2a). In addition, 30 candidate SDPs with an LPXTG motif (see Table S3 in the supplemental material) were found in the draft genome of CMPG5300 (Malik et al., unpublished data), which are putative substrates of this sortase. Among these putative SDPs are seven putative mucus-binding proteins, four putative collagen-binding proteins, and one putative mannose-specific adhesin (see the supplemental material). Analysis of the neighboring genes of srtA suggests that there is no gene encoding an LPXTG motif-containing protein in the neighborhood of the srtA gene (Fig. 2a). Interestingly, qRT-PCR results for the wild type suggest that srtA forms an operon with the downstream genes lemA and htpX (Fig. 2b). To construct a srtA deletion mutant, plasmid CMPG5376 was electroporated with the optimized protocol described above, using the Cre-loxP strategy previously described by Lambert et al. (36), resulting in strain CMPG5376 showing the correct allelic replacement events.
Fig 2.
(a) Genetic organization of the srtA locus in the genome of CMPG5300, along with its flanking genes and their annotations. Reverse transcriptase PCR on isolated total RNA with the indicated primers (arrows) showed a single transcriptomic unit for the genes lab129_01547 to lab129_01549, suggesting that the srtA gene (lab129_01547) is located in an operon with lab129_01548 and with lab129_01549. ORF, open reading frame. (b) Gene expression analysis of the srtA gene and flanking genes in wild-type and mutant strains by qRT-PCR. The shading depicts the levels of gene expression (in a semiquantitative way) of the srtA mutant CMPG5376 compared with the WT strain CMPG5300 as determined by qRT-PCR. More than 2-fold over- or underexpression was considered significant.
Mutant analysis revealed a key role for SrtA in autoaggregation of CMPG5300.
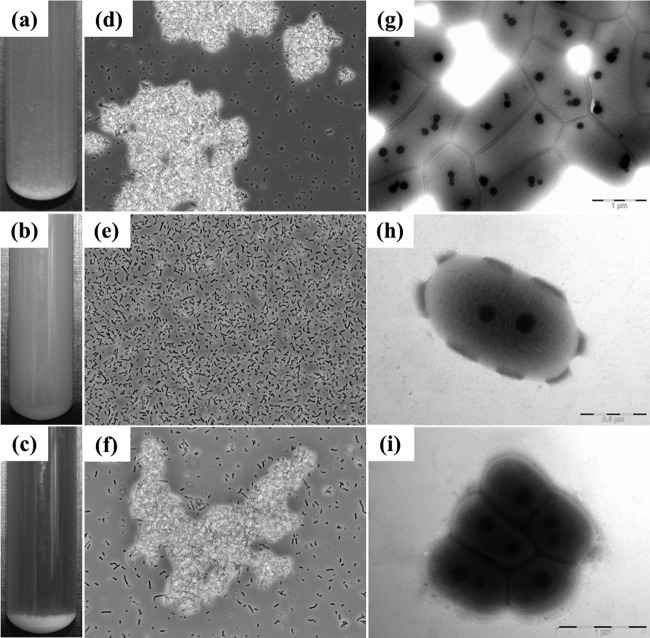
When the srtA mutant CMPG5376 was grown in liquid medium, no difference in the growth rate was observed compared to WT CMPG5300 (data not shown). However, the mutant CMPG5376 clearly did not display cell clumping, as observed for WT CMPG5300 (Fig. 3a and b). Single cells of CMPG5376 were also visualized under the microscope, in contrast to the cell aggregates of the CMPG5300 strain (Fig. 3d and e). In addition, transmission electron microscopy also revealed single cells for the mutant strain CMPG5376 (Fig. 3h), while wild-type CMPG5300 showed clusters of cells through connections to the neighboring cells via their outermost layers (Fig. 3g).
Fig 3.
Phenotypic analysis of the srtA mutant. (a to c) Overnight culture of L. plantarum CMPG5300 (a), its sortase-deficient mutant CMPG5376 (b), and CMPG5378 (complemented strain) (c) in MRS medium. Cell clumps forming a pellet can be seen at the bottom of the tube in the WT and the complemented strain. In contrast, a nearly homogeneous suspension of cells can be seen for the mutant. (d to f) Phase-contrast microscopic images show cell clumps in the case of L. plantarum CMPG5300 (d) and the complemented strain CMPG5378 (f), in contrast to the single cells seen with the srtA mutant CMPG5376 (e). (g to i) TEM images for ultrastructural analysis of the cell morphology of CMPG5300 showing cell clusters (g), a single cell of srtA mutant CMPG5376 (h), and a cell cluster of the complemented strain CMPG5378 (i) grown overnight in MRS medium, washed, and suspended in PBS before fixation for microscopy.
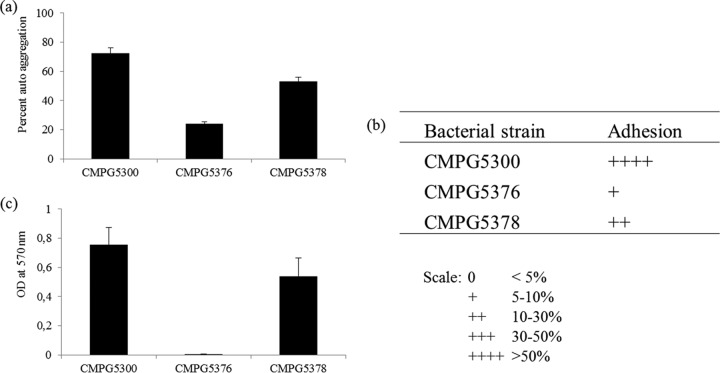
To confirm that the observed phenotype was linked to the srtA mutation, the srtA gene was reintroduced in CMPG5376 using the replicating vector pCMPG5378 (derived from pLAB1301 [35]) for complementation. The complemented strain CMPG5378 grew at the same rate as the WT strain and also exhibited autoaggregation (Fig. 3c). Cell clumps were visualized via phase-contrast (Fig. 3f) and transmission electron (Fig. 3i) microscopy, as seen for CMPG5300. In addition, autoaggregation of CMPG5378 was also quantitatively shown to be near the WT level (Fig. 4a), in contrast to the srtA mutant CMPG5376, which clearly had lost this property.
Fig 4.
(a) Quantitative analysis of autoaggregation of CMPG5300 and its mutant derivatives. The overnight-grown cultures of L. plantarum CMPG5300, the srtA mutant CMPG5376, and the complemented strain CMPG5378 were washed and resuspended to an OD600 of 0.5. The upper suspension was collected and assessed for its OD, and the tubes were left undisturbed for 5 h. Autoaggregation ability is expressed as percentages. The error bars represent standard deviations of 3 independent experiments. (b) Capacities of L. plantarum CMPG5300 and srtA mutant CMPG5376 to adhere to vaginal epithelial cells. A total of 107 CFU/ml of wild-type CMPG5300, the srtA mutant CMPG5376, and the srtA complemented strain CMPG5378 from overnight cultures were coincubated with the VK2/E6E7 cells for 1 h, and thereafter, the proportions of adherent bacteria, expressed as percentages, were determined. The results are presented using a semiquantitative scale as explained at the bottom. (c) Biofilm formation capacities of L. plantarum CMPG5300 and srtA mutant CMPG5376. Biofilm formation by L. plantarum CMPG5300 was compared with that of the srtA mutant CMPG5376 (both grown in MRS medium). The amount of biofilm formed was quantified by crystal violet staining (OD at 570 nm). The error bars represent standard deviations of 8 biological repeats.
Mutant analysis showed the role of SrtA in adhesion of CMPG5300 to vaginal epithelial cells and biofilm formation.
We also investigated the adhesion capacity of the srtA mutant CMPG5376 to VK2/E6E7 cells, which was drastically reduced compared to that of WT CMPG5300 (Fig. 4b). Complementation of the mutant could increase its adhesion percentage 7- to 8-fold, although not completely to the wild-type level. This can be partly explained by the fact that autoaggregation overestimates the exact adhesion percentages (Fig. 1c) and that the copy numbers of the srtA gene are not similar in wild-type (on the chromosome) and complemented (on replicating vector pCMPG5378) strains. However, we cannot exclude polar effects of the srtA mutation on the other genes in its putative operon. qRT-PCR experiments showed that the expression of the neighboring genes is affected in the srtA-deficient mutant (Fig. 2b). In particular, the expression of the upstream gene lab129_01546, encoding an unknown protein, is significantly upregulated in the srtA mutant. Nevertheless, the fact that we could clearly complement adhesion to a major extent compared to the mutant indicates that the srtA mutation is the key factor contributing to the phenotype. In addition, the biofilm formation assay revealed that the srtA mutant CMPG5376 lost the capacity to form a biofilm on a polystyrene surface compared with CMPG5300 (Fig. 4c), while complementation could restore the biofilm-forming ability to nearly the wild-type level.
DISCUSSION
Sortase-dependent cell wall proteins play a crucial role in interactions of Gram-positive bacteria with their environment (38). The vaginal mucosa is colonized by beneficial bacteria, often with lactobacilli dominating the niche (39). In this study, we describe the identification of a vaginal isolate, L. plantarum CMPG5300, with a highly autoaggregative and adhesive phenotype dependent on its srtA gene, encoding the housekeeping sortase A.
To investigate the role of autoaggregation as a possible adaptation factor of this vaginal isolate, genetic tools were first optimized for the construction of knockout mutants of CMPG5300 and the subsequent comparative functional analysis of the wild type and mutant. We showed that the high autoaggregating capacity of CMPG5300 is determined by sortase A, anchoring SDPs covalently to the cell wall, since a srtA mutant (CMPG5376) completely lost its autoaggregative phenotype. Previous studies in other lactobacilli have shown a role for SDPs in autoaggregation but, to the best of our knowledge, not yet for vaginal strains. For instance, Mackenzie et al. (40) have shown that sortase-dependent mucus-binding proteins play a role in autoaggregation of Lactobacillus reuteri strain ATCC 53608, which is intestinal in origin.
Adherence of vaginal lactobacilli to epithelial cells has been suggested to be an important first step in the colonization of mucous membranes and has been shown to prevent colonization by pathogenic microorganisms in vitro (41) and in vivo (42). In the present study, we investigated the adhesion of CMPG5300 and its srtA mutant derivative CMPG5376 to VK2/E6E7 cells (37). The morphological and immunocytochemical characteristics of this cell line closely resemble those of the vaginal epithelium (37), making it an appropriate model system for studying adherence of vaginal microorganisms. The srtA mutant of L. plantarum showed a significant reduction in its ability to adhere to VK2/E6E7 cells. Our results also suggest that autoaggregation and adhesion to vaginal epithelial cells of L. plantarum CMPG5300 are closely linked and that probably multiple aggregates adhere to single host cells. Previous studies have shown a role for SDPs in adhesion of lactobacilli to host cells and intestinal mucus but, to the best of our knowledge, not yet for vaginal epithelial cells. Gene deletion mutant studies have shown a role of SDPs in adhesion for gastrointestinal strains, such as L. plantarum WCFS1 (43) and 299v (44), L. salivarius UCC118 (45), L. johnsonii NCC533 (46), and Lactobacillus casei BL23 (47) to intestinal epithelial cell lines.
Formation of biofilms by Lactobacillus strains is also an interesting phenotype, which has been shown to be related to the expression of potential probiotic properties in some L. reuteri strains (48) and to the prevention of the overgrowth and proliferation of Candida by other lactobacilli (49). In situ formation of loose, nonadherent bacterial biofilms by lactobacilli from vaginal biopsy specimens from women without genital infections was shown by fluorescence in situ hybridization (FISH) analysis by Verstraelen and Swidsinski (50). Only limited studies have yet assessed the potential of vaginal lactobacilli to form biofilms in vitro. These studies indicate that the biofilm-forming capacity of vaginal lactobacilli is strongly influenced by growth media (51, 52), although this ability has not yet been attributed to specific molecules. Interestingly, the observation of this study that the srtA mutant of L. plantarum CMPG5300 lost the capacity to form biofilm on polystyrene indicates that biofilm formation in some vaginal lactobacilli could be by virtue of sortase-dependent targeting of cell wall proteins. In the gastrointestinal isolate L. rhamnosus GG, biofilm formation has also been recently linked to SDPs, i.e., multimeric pilus appendages, termed SpaCBA (53). Moreover, even systemic pathogens, such as Streptococcus mutans (54) and Streptococcus agalactiae (55), appear to mediate biofilm formation via pili. However, the genome sequence of CMPG5300 does not appear to encode pili (Malik et al., unpublished data), as also assessed using the specific bioinformatic tool LOCP to locate pilus operons in Gram-positive bacteria (56). The exact SDPs involved in autoaggregation, adhesion to vaginal epithelial cells, and biofilm formation of CMPG5300 remain to be identified in future studies. Preliminary genome information for strain CMPG5300 indicates that there are at least 30 potential SDPs encoded within the genome (see Table S3 in the supplemental material). Among these putative SDPs are seven putative mucus-binding proteins, four putative collagen-binding proteins, and one putative mannose-specific adhesin, which could be differently expressed on the surface of the mutant strain.
The phenotype of the mutant CMPG5376 could be restored to an autoaggregating, adhesive, and biofilm-forming phenotype during complementation with the srtA gene. However, wild-type percentages were not obtained by the complemented strain compared to the WT, especially for adhesion to epithelial cells. This can be partly explained by the fact that the high adhesion percentage of the wild type is the result of a multiplier effect by aggregates adhering to single vaginal epithelial cells, resulting in the remarkably high adhesion percentages of 80% (Fig. 1c). For comparison, the well-adhering intestinal probiotic strain L. rhamnosus GG adheres at around 10 to 15% to intestinal epithelial cells (53). The fact that the copy numbers of the srtA gene are different in wild-type (on the chromosome) and complemented (on replicating pLAB1301-derived vector) strains probably also has an impact on the adherence percentage, considering the fact that SrtA is estimated to target 30 proteins to the cell wall in L. plantarum CMPG5300. Nevertheless, we also obtained indications that the srtA of CMPG5300 exists in the form of an operon with the two downstream genes lemA and a heat shock protein-encoding gene, htpX, unlike the srtA genes of most of the Gram-positive bacteria, which exist in the form of monocistronic operons (57). As indicated by qRT-PCR experiments, polar effects of the mutation on the downstream genes and even upstream genes cannot be ruled out. However, the fact that we could complement autoaggregation by reintroducing the srtA gene in the mutant and partially complement adhesion and biofilm formation suggest that the polar effects are probably minor and that SrtA is a key factor in these phenotypes. Future studies will be of interest to investigate the operon structure and gene regulation of the srtA gene and its neighboring genes in more detail, especially given the fact that an operon structure for a srtA gene encoding the housekeeping sortase has, to the best of our knowledge, not been documented before.
In addition, future studies will also have to investigate the importance of the high autoaggregating capacity of CMPG5300 upon introduction of the strain into the vaginal niche dominated by other lactobacilli. Recent detailed 16S rRNA gene-sequencing methods suggest that, depending on the dominant species, the vaginal microbiota can be divided into five major “vaginal enterotypes,” with L. crispatus, L. gasseri, L. iners, L. jensenii, or non-Lactobacillus strictly anaerobic bacteria, such as Prevotella, Dialister, and Atopobium, respectively, dominating the niche (2, 58). Furthermore, other Lactobacillus species, such as L. rhamnosus (59), L. plantarum (51), Lactobacillus vaginalis, and L. salivarius (60), can occasionally be detected. Studies with L. rhamnosus GR-1 show that such less common inhabitants can also have significant health effects (61, 62). It is tempting to speculate that CMPG5300 might promote coaggregation of lactobacilli in the vagina, thereby strengthening the barrier against pathogenic bacteria. On the other hand, coaggregation with pathogens, which results in inhibition of pathogen entry, is also plausible, but this requires further documentation.
In conclusion, we have studied the role of srtA in the highly autoaggregating vaginal L. plantarum strain by constructing a srtA gene deletion mutant and assessing its phenotypic effect. This gene was shown to be a key factor involved in autoaggregation, adherence to a human vaginal keratinocyte cell line, and biofilm formation by L. plantarum CMPG5300. To our knowledge, this is the first report that demonstrates such a role of the srtA gene in a vaginal Lactobacillus strain. Considering the potential role of surface localization of SDPs in enhancing bacterial colonization and obstructing pathogen entry, e.g., of HIV in the human vagina, forthcoming studies will be focused on the identification of the substrate(s) imparting these promising properties as a probiotic to the strain.
Supplementary Material
ACKNOWLEDGMENTS
Shweta Malik is supported by a fellowship from Erasmus Mundus External Cooperation Window Lot 13. Work at KU Leuven was supported by BOF program financing (spokesman, Jan Balzarini).
We thank David De Coster from CMPG, KU Leuven, Leuven, Belgium, and Tom Struys from the University of Hasselt, Hasselt, Belgium, for TEM analysis; M. Kleerebezem and Roger S. Bongers from NIZO food research, Ede, The Netherlands, for providing us with the Cre-loxP plasmids; and Bernadet Renckens, Radboud University, Nijmegen, The Netherlands, for in silico analysis of the genome and sortase substrates.
Roland J. Siezen is the chief executive officer (CEO) of Microbial Bioinformatics. There are no patents, products in development, or marketed products to declare. This does not alter the authors' adherence to the ASM policy on sharing data and material.
Footnotes
Published ahead of print 24 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00926-13.
REFERENCES
- 1. Bayo M, Berlanga M, Agut M. 2002. Vaginal microbiota in healthy pregnant women and prenatal screening of group B streptococci (GBS). Int. Microbiol. 5:87–90 [DOI] [PubMed] [Google Scholar]
- 2. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl. 1):4680–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wiesenfeld HC, Hillier SL, Krohn MA, Landers DV, Sweet RL. 2003. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin. Infect. Dis. 36:663–668 [DOI] [PubMed] [Google Scholar]
- 4. Sha BE, Zariffard MR, Wang QJ, Chen HY, Bremer J, Cohen MH, Spear GT. 2005. Female genital-tract HIV load correlates inversely with Lactobacillus species but positively with bacterial vaginosis and Mycoplasma hominis. J. Infect. Dis. 191:25–32 [DOI] [PubMed] [Google Scholar]
- 5. Reid G, Bruce AW. 2003. Urogenital infections in women: can probiotics help? Postgrad. Med. J. 79:428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lebeer S, Vanderleyden J, De Keersmaecker SC. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8:171–184 [DOI] [PubMed] [Google Scholar]
- 7. Reid G, Burton J. 2002. Use of Lactobacillus to prevent infection by pathogenic bacteria. Microbes Infect. 4:319–324 [DOI] [PubMed] [Google Scholar]
- 8. Lebeer S, Vanderleyden J, De Keersmaecker SC. 2008. Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72:728–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U. S. A. 100:1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, Lick S, Hamrick A, Cano R, Klaenhammer TR. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U. S. A. 102:3906–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Claesson MJ, Li Y, Leahy S, Canchaya C, van Pijkeren JP, Cerdeno-Tarraga AM, Parkhill J, Flynn S, O'Sullivan GC, Collins JK, Higgins D, Shanahan F, Fitzgerald GF, van Sinderen D, O'Toole PW. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. U. S. A. 103:6718–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Azcarate-Peril MA, Altermann E, Goh YJ, Tallon R, Sanozky-Dawes RB, Pfeiler EA, O'Flaherty S, Buck BL, Dobson A, Duong T, Miller MJ, Barrangou R, Klaenhammer TR. 2008. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl. Environ. Microbiol. 74:4610–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Macklaim JM, Gloor GB, Anukam KC, Cribby S, Reid G. 2011. At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl. 1):4688–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anukam K, Macklaim JM, Gloor GB, Reid G, Boekhorst J, Renckens B, van Hijum SA, Siezen RJ. 2013. Genome sequence of Lactobacillus pentosus KCA1: vaginal isolate from a healthy premenopausal woman. PLoS One 8:e59239. 10.1371/journal.pone.0059239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boris S, Suarez JE, Barbes C. 1997. Characterization of the aggregation promoting factor from Lactobacillus gasseri, a vaginal isolate. J. Appl. Microbiol. 83:413–420 [DOI] [PubMed] [Google Scholar]
- 17. Juarez Tomas MS, Wiese B, Nader-Macias ME. 2005. Effects of culture conditions on the growth and auto-aggregation ability of vaginal Lactobacillus johnsonii CRL 1294. J. Appl. Microbiol. 99:1383–1391 [DOI] [PubMed] [Google Scholar]
- 18. Goh YJ, Klaenhammer TR. 2010. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 76:5005–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kos B, Suskovic J, Vukovic S, Simpraga M, Frece J, Matosic S. 2003. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 94:981–987 [DOI] [PubMed] [Google Scholar]
- 20. Cesena C, Morelli L, Alander M, Siljander T, Tuomola E, Salminen S, Mattila-Sandholm T, Vilpponen-Salmela T, von Wright A. 2001. Lactobacillus crispatus and its nonaggregating mutant in human colonization trials. J. Dairy Sci. 84:1001–1010 [DOI] [PubMed] [Google Scholar]
- 21. Boris S, Suarez JE, Vazquez F, Barbes C. 1998. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect. Immun. 66:1985–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dieye Y, Oxaran V, Ledue-Clier F, Alkhalaf W, Buist G, Juillard V, Lee CW, Piard JC. 2010. Functionality of sortase A in Lactococcus lactis. Appl. Environ. Microbiol. 76:7332–7337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lebeer S, Verhoeven TL, Francius G, Schoofs G, Lambrichts I, Dufrene Y, Vanderleyden J, De Keersmaecker SC. 2009. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl. Environ. Microbiol. 75:3554–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lebeer S, Verhoeven TL, Perea Velez M, Vanderleyden J, De Keersmaecker SC. 2007. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73:6768–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 26. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 27. Zhou M, Boekhorst J, Francke C, Siezen RJ. 2008. LocateP: genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinformatics 9:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boekhorst J, de Been MW, Kleerebezem M, Siezen RJ. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPXTG-like sorting motifs. J. Bacteriol. 187:4928–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 30. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Berthier F, Zagorec M, Champomier-Verge‘s M, Ehrlich SD, Morel-Deville F. 1996. Efficient transformation of Lactobacillus sake by electroporation. Microbiology 142:1273–1279 [DOI] [PubMed] [Google Scholar]
- 32. Mason CK, Collins MA, Thompson K. 2005. Modified electroporation protocol for lactobacilli isolated from the chicken crop facilitates transformation and the use of a genetic tool. J. Microbiol. Methods 60:353–363 [DOI] [PubMed] [Google Scholar]
- 33. Reference deleted.
- 34. De Keersmaecker SC, Braeken K, Verhoeven TL, Perea Velez M, Lebeer S, Vanderleyden J, Hols P. 2006. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl. Environ. Microbiol. 72:4923–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Josson K, Scheirlinck T, Michiels F, Platteeuw C, Stanssens P, Joos H, Dhaese P, Zabeau M, Mahillon J. 1989. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid 21:9–20 [DOI] [PubMed] [Google Scholar]
- 36. Lambert JM, Bongers RS, Kleerebezem M. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73:1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fichorova RN, Rheinwald JG, Anderson DJ. 1997. Generation of papillomavirus-immortalized cell lines from normal human ectocervical, endocervical, and vaginal epithelium that maintain expression of tissue-specific differentiation proteins. Biol. Reprod. 57:847–855 [DOI] [PubMed] [Google Scholar]
- 38. Remus DM, Bongers RS, Meijerink M, Fusetti F, Poolman B, de Vos P, Wells JM, Kleerebezem M, Bron PA. 2013. Impact of Lactobacillus plantarum sortase on target protein sorting, gastrointestinal persistence, and host immune response modulation. J. Bacteriol. 195:502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang TL, Chang CH, Simpson DA, Xu Q, Martin PK, Lagenaur LA, Schoolnik GK, Ho DD, Hillier SL, Holodniy M, Lewicki JA, Lee PP. 2003. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc. Natl. Acad. Sci. U. S. A. 100:11672–11677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mackenzie DA, Jeffers F, Parker ML, Vibert-Vallet A, Bongaerts RJ, Roos S, Walter J, Juge N. 2010. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology 156:3368–3378 [DOI] [PubMed] [Google Scholar]
- 41. Atassi F, Brassart D, Grob P, Graf F, Servin AL. 2006. Lactobacillus strains isolated from the vaginal microbiota of healthy women inhibit Prevotella bivia and Gardnerella vaginalis in coculture and cell culture. FEMS Immunol. Med. Microbiol. 48:424–432 [DOI] [PubMed] [Google Scholar]
- 42. Anukam KC, Osazuwa E, Osemene GI, Ehigiagbe F, Bruce AW, Reid G. 2006. Clinical study comparing probiotic Lactobacillus GR-1 and RC-14 with metronidazole vaginal gel to treat symptomatic bacterial vaginosis. Microbes Infect. 8:2772–2776 [DOI] [PubMed] [Google Scholar]
- 43. Pretzer G, Snel J, Molenaar D, Wiersma A, Bron PA, Lambert J, de Vos WM, van der Meer R, Smits MA, Kleerebezem M. 2005. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187:6128–6136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gross G, van der Meulen J, Snel J, van der Meer R, Kleerebezem M, Niewold TA, Hulst MM, Smits MA. 2008. Mannose-specific interaction of Lactobacillus plantarum with porcine jejunal epithelium. FEMS Immunol. Med. Microbiol. 54:215–223 [DOI] [PubMed] [Google Scholar]
- 45. van Pijkeren JP, Canchaya C, Ryan KA, Li Y, Claesson MJ, Sheil B, Steidler L, O'Mahony L, Fitzgerald GF, van Sinderen D, O'Toole PW. 2006. Comparative and functional analysis of sortase-dependent proteins in the predicted secretome of Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 72:4143–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Denou E, Pridmore RD, Berger B, Panoff JM, Arigoni F, Brussow H. 2008. Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190:3161–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Munoz-Provencio D, Rodriguez-Diaz J, Collado MC, Langella P, Bermudez-Humaran LG, Monedero V. 2012. Functional analysis of the Lactobacillus casei BL23 sortases. Appl. Environ. Microbiol. 78:8684–8693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jones SE, Versalovic J. 2009. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 9:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strus M, Kucharska A, Kukla G, Brzychczy-Wloch M, Maresz K, Heczko PB. 2005. The in vitro activity of vaginal Lactobacillus with probiotic properties against Candida. Infect. Dis. Obstet. Gynecol. 13:69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Verstraelen H, Swidsinski A. 2013. The biofilm in bacterial vaginosis: implications for epidemiology, diagnosis and treatment. Curr. Opin. Infect. Dis. 26:86–89 [DOI] [PubMed] [Google Scholar]
- 51. Martin R, Soberon N, Vaneechoutte M, Florez AB, Vazquez F, Suarez JE. 2008. Characterization of indigenous vaginal lactobacilli from healthy women as probiotic candidates. Int. Microbiol. 11:261–266 [DOI] [PubMed] [Google Scholar]
- 52. Terraf MC, Juarez Tomas MS, Nader-Macias ME, Silva C. 2012. Screening of biofilm formation by beneficial vaginal lactobacilli and influence of culture media components. J. Appl. Microbiol. 113:1517–1529 [DOI] [PubMed] [Google Scholar]
- 53. Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, Reunanen J, Palva A, Vos WM, Keersmaecker SC, Vanderleyden J. 2012. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 78:185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Levesque CM, Voronejskaia E, Huang YC, Mair RW, Ellen RP, Cvitkovitch DG. 2005. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect. Immun. 73:3773–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Konto-Ghiorghi Y, Mairey E, Mallet A, Dumenil G, Caliot E, Trieu-Cuot P, Dramsi S. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422. 10.1371/journal.ppat.1000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Plyusnin I, Holm L, Kankainen M. 2009. LOCP—locating pilus operons in gram-positive bacteria. Bioinformatics 25:1187–1188 [DOI] [PubMed] [Google Scholar]
- 57. Clancy KW, Melvin JA, McCafferty DG. 2010. Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers 94:385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. 2013. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 9:e1002863. 10.1371/journal.pcbi.1002863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pascual LM, Daniele MB, Ruiz F, Giordano W, Pajaro C, Barberis L. 2008. Lactobacillus rhamnosus L60, a potential probiotic isolated from the human vagina. J. Gen. Appl. Microbiol. 54:141–148 [DOI] [PubMed] [Google Scholar]
- 60. Gustafsson RJ, Ahrne S, Jeppsson B, Benoni C, Olsson C, Stjernquist M, Ohlsson B. 2011. The Lactobacillus flora in vagina and rectum of fertile and postmenopausal healthy Swedish women. BMC Womens Health 11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hummelen R, Changalucha J, Butamanya NL, Cook A, Habbema JD, Reid G. 2010. Lactobacillus rhamnosus GR-1 and L. reuteri RC-14 to prevent or cure bacterial vaginosis among women with HIV. Int. J. Gynaecol. Obstet. 111:245–248 [DOI] [PubMed] [Google Scholar]
- 62. Kohler GA, Assefa S, Reid G. 2012. Probiotic interference of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 with the opportunistic fungal pathogen Candida albicans. Infect. Dis. Obstet. Gynecol. 2012:636474. 10.1155/2012/636474 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.