Abstract
Yeast killer viruses are widely distributed in nature. Several toxins encoded in double-stranded RNA (dsRNA) satellites of the L-A totivirus have been described, including K1, K2, K28, and Klus. The 4.6-kb L-A genome encodes the Gag major structural protein that forms a 39-nm icosahedral virion and Gag-Pol, a minor fusion protein. Gag-Pol has transcriptase and replicase activities responsible for maintenance of L-A (or its satellite RNAs). Recently we reported a new killer toxin, Klus. The L-A virus in Klus strains showed poor hybridization to known L-A probes, suggesting substantial differences in their sequences. Here we report the characterization of this new L-A variant named L-A-lus. At the nucleotide level, L-A and L-A-lus showed only 73% identity, a value that increases to 86% in the amino acid composition of Gag or Gag-Pol. Two regions in their genomes, however, the frameshifting region between Gag and Pol and the encapsidation signal, are 100% identical, implying the importance of these two cis signals in the virus life cycle. L-A-lus shows higher resistance than L-A to growth at high temperature or to in vivo expression of endo- or exonucleases. L-A-lus also has wider helper activity, being able to maintain not only Mlus but also M1 or a satellite RNA of L-A called X. In a screening of 31 wine strains, we found that none of them had L-A; they carried either L-A-lus or a different L-A variant in K2 strains. Our data show that distinct M killer viruses are specifically associated with L-As with different nucleotide compositions, suggesting coevolution.
INTRODUCTION
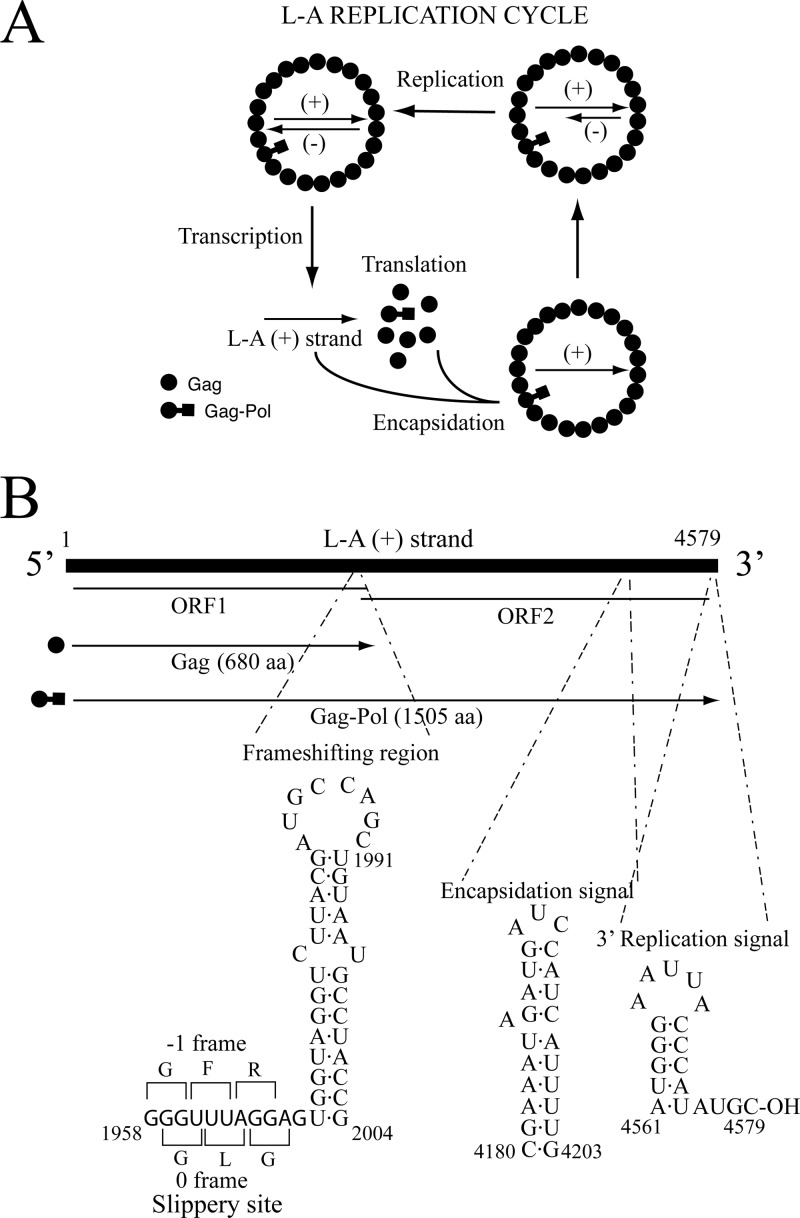
Saccharomyces cerevisiae L-A virus (ScV-L-A) is a cytoplasm-persisting double-stranded RNA (dsRNA) virus of the Totiviridae family. The unsegmented genome (4.6 kb) encodes two virion proteins: a 76-kDa major structural protein, Gag, and a 180-kDa Gag-Pol fusion protein which has two domains, an N-terminal Gag domain and a C-terminal Pol domain (Fig. 1B). Pol has motifs characteristic of viral RNA-dependent RNA polymerases (RdRps) and is translated as a Gag-Pol fusion protein by a −1 ribosomal frameshifting event (1, 2). The viral genome is packed inside 39-nm icosahedral capsids. A total of 60 asymmetric Gag dimers form each capsid, while there are one or two Gag-Pol molecules per capsid (3–5). The L-A replication cycle was well established years ago and is as follows. Inside the virion, the transcriptase activity of Pol conservatively transcribes the dsRNA genome generating L-A (+) strands that are extruded into the cytoplasm. There, they are either translated to produce the two virion proteins aforementioned or else encapsidated to form new virions. Once inside the capsids, Pol replicates the (+) strands to generate the dsRNA viral genome. In the 3′-end region of L-A (+) strands there are two cis-acting signals necessary for packaging and replication (6). Figure 1 summarizes the L-A replication cycle, its genomic organization, and the cis signals. L-A virus, as is typical of fungal viruses, has no extra cellular route of infection. It is transmitted vertically from mother to daughter cells through mitosis or meiosis or horizontally during mating.
Fig 1.
L-A replication cycle, genome organization, and cis signals for frameshifting, encapsidation, and replication. (A) Mature L-A virions carry transcriptase activity. This activity conservatively transcribes the L-A dsRNA genome to produce L-A (+) strands that are extruded from the virions into the cytoplasm. Translation by ribosomes originated two types of products; the first is Gag (closed dots), and, in 1% to 2% of the cases, a −1 frameshifting event occurs to produce the second, a Gag-Pol fusion protein (closed dots-squares). Interaction of Gag-Pol with the (+) strand triggers L-A particle assembly (39). These particles have the same protein composition as mature particles but carry an L-A (+) strand. Then, replicase activity in the virion synthesizes the (−) strand on the (+) strand template, originating the dsRNA genome. (B) L-A genome organization and coding strategy. The two overlapping frames on the (+) strand are indicated. The translation products (Gag and Gag-Pol fusion protein) are shown. The cis signals for frameshifting, encapsidation, and replication on the (+) strand with their secondary structures are displayed at the bottom of the panel. aa, amino acids. (Diagrams adapted from reference 16 with kind permission of Springer Science+Business Media.)
Many yeast strains that carry L-A also harbor smaller satellite dsRNAs generically called M satellites. There are several types of M satellites, and the most thoroughly characterized is M1 (ScV-M1). M1 is separately encapsidated in L-A virions and depends on L-A for its maintenance. Cells carrying both L-A and M1 show a killer phenotype, as M1 encodes the K1 killer toxin, an extracellular protein able to kill nonkiller, sensitive cells. The killer trait was first described in 1963 (7), and its association with two dsRNA species was first reported in 1973 (8). In addition to M1, other toxin-encoding dsRNAs, such as M2, M28, or Mlus, have been described in S. cerevisiae (9–11). Each killer strain harbors only one type of M dsRNA and is immune to the toxin produced but not to other killer toxins. When haploid cells with different killer M dsRNAs mate, the resultant diploids carry only one type of M, a phenomenon known as exclusion. L-A viruses found in natural isolates of K1, K2, or K28 strains are somewhat different, as shown by the limited sequence homology of their genomic RNAs or by differences in the virion major capsid proteins (10, 12, 13). Works in the early 1980s also reported phenotypic variants of L-A affecting the maintenance of K1 and K2 systems designated [EXL], [HOK], and [NEX] (14, 15). For reviews on S. cerevisiae dsRNA viruses and killer toxins, see references 16 and 17 (and references therein). In addition to the M dsRNA-encoded killer toxins, there are other killer toxins in S. cerevisiae named KHR and KHS. They show weak killer activity and are encoded on chromosomal DNA (18, 19).
L-BC is another totivirus (ScV-L-BC), frequently accompanying L-A in S. cerevisiae. L-BC is similar to L-A in terms of genome size and organization (20). It coexists with L-A in several killer or nonkiller strains but has no helper activity for the toxin-producing M dsRNAs.
Several nuclear genes that affect the maintenance of L-A and M1 (or M2) viruses have been described (16). They belong to two categories. Those (more than 30) required for killer activity are known as MAK genes (for maintenance of killer). Mak mutants cannot maintain M1 (or M2); thus, the strains are nonkillers. Only three mak mutants also lose L-A: mak3, mak10, and mak31. MAK3 encodes the catalytic subunit of an N-acetyltransferase that acetylates the N terminus of Gag, a step essential for capsid assembly (21), while MAK10 and MAK31 code for the auxiliary subunits of the enzyme, which form a complex with mak3p (22). Members of the second group of genes (7 are known) have a negative effect on the viruses. Mutations on these genes were initially identified as ski (super killer) mutants with higher levels of toxin production due to increased amounts of M1 (23). Most SKI genes were later shown to be components of the exosome, a complex involved in 3′-to-5′ RNA degradation (24), or modulators of exosome activity. On the other hand, SKI1, also known as XRN1, codes for the main exonuclease involved in the 5′-to-3′ mRNA degradation pathway (25). Overexpression of the 5′ exonuclease XRN1 cures L-A with high frequency, suggesting that it is active on L-A (+) strands when they are outside the virion (26).
Yeast cells carrying killer viruses may have a competitive advantage over cells lacking them, which would explain their positive selection. The overall distribution of the killer character, however, is difficult to establish, as the studies available are partial and difficult to compare. For instance, the K1 killer character is found in S. cerevisiae laboratory strains at a high incidence, probably as a result of inbreeding. Also, killer strains are found among wine, brewing, and baking yeasts (27). Other studies on natural isolates from variable sources report different incidences: i.e., 17% among 154 strains of different yeast genera or the absence of dsRNA-based killer strains (28, 29). Several studies on strains isolated from wine fermentations where K2 strains are the most abundant, and yet are found with variable distribution, have been published (reference 30 and references therein). The K2 toxin, active at low pH (2.9 to 4.9), can be of technological importance for winemaking, as selected K2 killer strains in the must can predominate over indigenous strains.
Recently, the occurrence of killer viruses in S. cerevisiae has been linked to the absence of the RNA interference (RNAi) pathway in this species (31). RNAi is involved in regulating gene expression by silencing specific mRNAs and depends on the activity of two ribonucleases, Dicer and Argonaute. Dicer cleaves dsRNA to small interfering dsRNAs (siRNAs), and Argonaute carries the small dsRNA to the specific mRNA and cleaves it. RNAi is present in animals, plants, and most fungi. RNAi, however, is absent in most budding yeasts, including S. cerevisiae, and yet is present in S. castellii, a closely related yeast. It has been proposed that RNAi loss in S. cerevisiae could have helped cells to acquire and retain the killer system, which, as mentioned before, would represent a selective advantage (32). This hypothesis is supported by the fact that RNAi is absent in species known to carry killer dsRNAs while related species having RNAi lack killer systems (31).
In a previous work, we described a new killer toxin, Klus, in wine yeasts from Spain (11). This new toxin is coded by Mlus dsRNA. Klus strains carry one of four Mlus isotypes (Klus-1 to Klus-4), whose sizes range from 2.1 to 2.3 kb. Klus toxin is active over a wide range of yeasts other than S. cerevisiae, and the ORF for the preprotoxin is ancestrally related to the host chromosomal gene YFR020W (providing for the first time a clue of the origin of a dsRNA-encoded toxin). In that study, we observed that the L-A helper virus for Mlus was somewhat different from L-A in laboratory strains. The aim of the present work is the characterization of that new L-A variant, L-A-lus. Toward this purpose we have (i) cloned and sequenced L-A-lus, (ii) studied the distribution of L-A-lus (or other L-A variants) in a collection of wine yeast strains, (iii) constructed laboratory strains carrying L-A-lus and Mlus or L-A-lus alone to study their properties under controlled laboratory conditions, (iv) analyzed the sensitivity of L-A-lus or L-A to high temperature or in cells that either express the heterologous RNA interference proteins Ago1 and Dcr1 or overexpress the XRN1/SKI1 5′ exonuclease, and (v) analyzed the L-A-lus helper activity over different satellite RNAs that include not only Mlus but also M1 and the L-A deletion mutant X. We also discuss the possible coevolution of different L-A variants and their specific killer toxin-encoding M dsRNAs in wine strains.
MATERIALS AND METHODS
Yeast strains and media.
Table 1 summarizes the strains used in this study. Most were constructed for this work, and others are laboratory strains that have already been described (26, 33). ski2Δ, ski1Δ, and mak10Δ deletion strains, all of them derivatives from strain BY4741, are from the EUROFAN collection (kindly supplied by J. L. Revuelta). Table 2 shows wine strains analyzed for different L-A variants. YPAD medium (1% yeast extract, 2% peptone, 2% glucose, 2% agar, 0.04% adenine) was supplemented with 0.02% uracil. YPG is the same as YPAD except that it contains 3% glycerol (vol/vol) instead of glucose. MB (methylene blue) medium is YPAD buffered at pH 4.7 with sodium citrate and containing 0.003% methylene blue (34). Sporulation medium contains 1% potassium acetate, 0.1% yeast extract, 0.05% glucose, and 2% agar. Synthetic minimal (SD) medium contains 0.67% yeast nitrogen base (without amino acids and with ammonium sulfate; Difco), 2% glucose, and 2% agar. Complete minimal medium is SD medium supplemented with amino acids, uracil, and adenine as described previously (14). For selection of transformants and for genetic experiments, complete minimal medium deprived of tryptophan, histidine, uracil, or leucine (H-trp, H-his, H-ura, or H-leu) was used. When required, Geneticin (Invitrogen) was added to YPAD medium or to SD medium at a final concentration of 300 mg/liter after autoclaving of the medium.
Table 1.
Strains used
| Strain | Genotype and/or description |
|---|---|
| 5X47 | Diploid tester strain sensitive for killer assay |
| 2928 | a ura3 his3 trp1 L-A-o, L-BC, 20S RNA |
| 2927 | a ura3 his3 trp1 ski2-2 L-A-o, L-BC-o, 20S RNA |
| BY4741 | a his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 L-A (low copy number), L-BC |
| 938 | a his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 Genr ski1Δ L-A-o, L-BC |
| 909 | a his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 Genr ski2Δ L-A-o, L-BC-o |
| 2403 | α his4-1, kar1-1 L-A, M1, L-BC |
| 2404 | α his4-1, kar1-1 L-A (high copy number), L-BC |
| 2405 | α his4-1, kar1-1 L-A-o, L-BC |
| 559 | α ura3 trp1 leu2 kar1-1 L-A-o, M1a [pI2L2] |
| 1081 | a ura3 L-A-lus, Mlus, L-BC, 20S RNA, 23S RNAb |
| 1082 | The same as 1081 but without 23S RNA |
| 1083 | Cytoductant of 1081 into 2405; α his4-1 kar1-1 L-A-lus, Mlus, L-BC, 20S RNA, 23S RNA |
| 1084 | Cytoductant of 1082 into 2405; α his4-1 kar1-1 L-A-lus, Mlus, L-BC, 20S RNA |
| 1085 | Cytoductant of 1084 into 909; a his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 Genr ski2Δ L-A-lus, Mlus, L-BC, 20S RNA |
| 1086 | Cytoductant of 559 into 1085; a his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 Genr ski2Δ L-A-lus, M1c, L-BC, 20S RNA |
| 1087 | Cytoductant of 1086 into 2405; α his4-1 kar1-1 L-A-lus, M1, L-BC, 20S RNA |
| 1088 | Cytoductant of 1084 into 938; a his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 Genr ski1Δ L-A-lus, Mlus-od, L-BC, 20S RNA |
| 1089 | Cytoductant of 1088 into 2405; α his4-1 kar1-1 L-A-lus, Mlus-o, L-BC, 20S RNA |
| 1094 | Cytoductant of 1089 into 2927; a ura3 his3 trp1 ski2-2 L-A-lus, L-BC, 20S RNA |
| 1098 | Cytoductant of 1089 into 2928; a ura3 his3 trp1 L-A-lus, L-BC, 20S RNA |
| 1064 | Cytoductant of BY4741 into 2405; α his4-1, kar1-1 L-A (low copy number), L-BC |
| 1127 | Cytoductant of 1064 into 2928; a ura3 his3 trp1 L-A (low copy number), L-BC, 20S RNA |
| TF395 | a ura3 trp1 leu2 L-A-o L-BC |
| 1150 | Strain TF395 that expresses AGO1 and DCR1 genese |
| 1145 | Cytoductant of 1089 into strain 1150; a ura3 trp1 leu2 L-A-lus, L-BC, 20S RNA |
| 1146 | Cytoductant of 1064 into strain 1150; a ura3 trp1 leu2 L-A (low copy number) L-BC |
| 1147 | Cytoductant of 1089 into strain TF395; a ura3 trp1 leu2 L-A-lus, L-BC, 20S RNA |
| 1148 | Cytoductant of 1064 into strain TF395; a ura3 trp1 leu2 L-A (low copy number) L-BC |
| 455 | α prototroph ski2-2, L-A, Xf |
| 1099 | Diploid by mating 1098 and 1064; a/α ura3/+ trp1/+ his3/+ his4-1/+ L-A-lus, L-BC, 20S RNA |
| 1101 | Cytoductant of 455 into 2928 [pI2L2]; a ura3 his3 trp1 L-A-og, X, L-BC, 20S RNA |
| 1113 | Cytoductant of 1101 into 1089; α his4-1 kar1-1 L-A-lus, X, L-BC, 20S RNA |
| 1118 | Cytoductant of 559 into 2928 [pRE1290]; a ura3 his3 trp1 L-A-o, M1h, L-BC, 20S RNA |
M1 viruses are maintained by Gag and Gag-Pol expressed from plasmid [pI2L2] (37).
Spore clone 3 from sporulation of a diploid that carries L-A-lus and Mlus and the Geneticin resistance (Genr) phenotype constructed as described in Results.
In this strain, M1 from strain 559 excluded Mlus initially present in strain 1085.
In the ski1Δ background, Mlus is lost spontaneously and strain 1088 carries only L-A-lus.
AGO1 and DCR1 genes from Saccharomyces castellii were inserted into the strain TF395 genome by transformation with plasmids pRS404-PTEF-Ago1 and pRS405-PTEF-Dcr1 (32).
X is a deletion mutant of L-A of 530 bp from the 5′ and 3′ ends of L-A (6).
In this strain, Gag and Gag-Pol expressed from plasmid pI2L2 exclude L-A originally present in strain 455 (50).
M1 is maintained by the hybrid L-A virions expressed from pRE1290 (see Materials and Methods).
Table 2.
Presence of L-A, M dsRNA satellites, or L-BC in wine yeast strainsa
| Strain | Killer satellite | Helper virus | L-BC | Source |
|---|---|---|---|---|
| CECT1881 | M2b | L-A-lus | − | CECT |
| CECT1887 | Mlusb | L-A-lus | − | CECT |
| DSM-70457 | M-o | L-A-lus | − | DSMZ |
| DSM-70459 | Mlus | L-A-lus | − | DSMZ |
| Uvaferm PM | M2 | L-A-2 | + | Lallemand |
| Lalvin EC1118 | M2 | L-A-2 | + | Lallemand |
| T73 | M2 | L-A-2 | + | Lallemand |
| Fermol Cryoaromae | M2 | L-A-2 | + | AEB Ibérica |
| PB2010 | M2b | L-A-2 | − | AEB Ibérica |
| EX436 | Mlus | L-A-lus | − | Ribera Guadiana (Spain) |
| EX122 | Mlus | L-A-lus | − | Ribera Guadiana (Spain) |
| EX198 | Mlus | L-A-lus | + | Ribera Guadiana (Spain) |
| EX229 | Mlus | L-A-lus | + | Ribera Guadiana (Spain) |
| EX73 | M2 | L-A-2 | + | Toro (Spain) |
| T34 | M-o | L-A-lus | + | Toro (Spain) |
| 5FG-31 | M2b | L-A-2 | − | Toro (Spain) |
| 8F-13 | M2 | L-A-2 | + | Toro (Spain) |
| Ar-12 | Mlusb | L-A-lus | − | Toro (Spain) |
| F5 | M-o | L-A-lus | − | Toro (Spain) |
| 80-P30 | Mlusb | L-A-lus | − | Portugal |
| 47-P07 | M2 | L-A-lus | − | Portugal |
| 96-P45 | M-o | L-A-lus | − | Portugal |
| S4928 | M2 | L-A-2 | + | Rueda (Spain) |
| S3920 | M2 | L-A-2 | + | Rueda (Spain) |
| V4907 | M2 | L-A-2 | + | Rueda (Spain) |
| 58-I | M-o | L-A-lus | − | Galicia (Spain) |
| A143-2 | M2 | L-A-2 | + | Galicia (Spain) |
| A143-6 | M-o | L-A-lus | − | Galicia (Spain) |
| Ca3 | Mlusb | L-A-lus | − | Cádiz (Spain) |
| Ca4 | M2b | L-A-lus | + | Cádiz (Spain) |
| Ca7 | M2 | L-A-2 | − | Cádiz (Spain) |
Strains were kindly provided by A. Palacios and C. Suárez (Lallemand strains), J. M. Álvarez (AEB Ibérica strains), M. Ramírez (Ribera del Guadiana strains), D. Schüller (Portugal strains), M. del Villar (Rueda strains), T. G. Villa (Galicia strains), and J. Cantoral (Cádiz strains). Toro strains were from our laboratory. CECT, Spanish Type Culture Collection; DSM and DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen. The presence of Mlus, M2, or L-BC dsRNAs was determined by Northern blot hybridization. +, presence; −, absence.
These strains are nonkiller, in spite of carrying M dsRNAs.
Killer assay.
Colonies to be tested for killer activity were replica plated onto MB medium previously seeded with a lawn of the sensitive strain 5X47 or appropriate killer strains. MB plates were incubated at 25°C for 2 to 3 days. A clear zone around the colonies indicated killer toxin production.
Transferring the Klus killer trait from a wine strain to a laboratory strain.
Klus-3 strain EX198 was incubated in sporulation medium for 5 days at 28°C and then transferred to YPAD for ascospore germination. An aliquot of this culture was mixed with strain 2928 carrying plasmid pNR41, which confers Geneticin resistance (Genr) (35), and incubated for 1 day on a rich YPAD plate for mating. The plate was then replicated onto SD medium supplemented with Geneticin to select for diploids formed between the Klus ascospore-derived cells (prototrophic and homothallic) and the Geneticin-resistant laboratory strain. Single diploid colonies were tested for Klus activity and then placed on sporulation medium for 5 days. When formation of asci was visible under the optical microscope, a small amount of this sporulating culture was suspended in 200 μl of water and treated with 5 μl glusulase (PerkinElmer Life Sciences, Inc.) for 5 min at 37°C. Afterwards, the sample was diluted with water and plated on H-his medium. A mixture of diploid cells and haploid cells with −ura or −]trp (or both) auxotrophies was able to grow. By successive replica plating onto SD or selective medium, we picked up colonies that were −ura. Only those that were of the a mating type were then used as donors to introduce L-A-lus and Mlus into laboratory strain 2405 by cytoduction (cytoplasmic mixing).
Cytoduction.
Cytoduction (cytoplasmic mixing) is a special kind of mating that allows the transfer of cytoplasmic traits (i.e., killer viruses or mitochondria) from one strain (donor) to another strain (recipient). There is a transient heterocaryon formation without diploidization because one of the strains is a kar1-1 mutant defective in nuclear fusion (36). Previously, the recipient strain was made ρ° by growth on an YPAD plate in the presence of ethidium bromide. The two strains are mixed in water at high density, and one drop of this mixture is placed on an YPAD plate. After mating is allowed for 6 to 8 h at 28°C, cells are streaked for single-colony isolation on plates where the donor strain cannot grow. The resulting colonies (a mixture of diploids, the recipient strain, and the cytoductants) are then replica plated onto YPG (only the respiratory-competent cells grow), SD (identifies a small percentage of diploids that are usually formed, as the kar1-1 mutation is leaky), and finally MB medium to test for the killer phenotype. When the donor strain is nonkiller, cytoductants are selected based on auxotrophies and growth on YPG medium. Further Northern analysis is needed to confirm the transference of the traits under study (i.e., L-A virus alone, which does not confer a killer phenotype).
Plasmids.
We used three types of plasmids in this study: 1, plasmids that express L-A virion proteins (Gag and Gag-Pol), 2, plasmids to synthesize strand-specific RNA riboprobes by runoff transcription with T7 or T3 RNA polymerases, and 3, vectors that carry the AGO1 and DCR1 genes from Saccharomyces castellii. They were used to insert these genes into the S. cerevisiae genome for heterologous expression. We also overexpressed the SKI1 gene.
Plasmid pI2L2 contains the entire L-A sequence and expresses L-A virion proteins under the control of the PGK1 promoter. It carries the TRP1 gene as a selective marker (37). Plasmid pRE1290 is a derivative of pI2L2 that expresses hybrid L-A virions with Gag from L-A and Pol from L-A-lus. It was constructed as follows. First, a NotI site was introduced into pI2L2 after the ORF for Gag by site-directed mutagenesis (SDM) using oligonucleotide RE587. The resulting plasmid was digested with NotI, and the Pol part of L-A (2.5 kb) was replaced by Pol from L-A-lus. The L-A-lus Pol fragment (from nucleotide [nt] 2070 to the end of L-A-lus, nt 4580) was synthesized by reverse transcription-PCR (RT-PCR) from strain EX229 using oligonucleotides NR108 and NR109, digested with NotI, and fused to the C terminus of Gag from L-A. The NotI site at the junction between Gag and Pol was then eliminated by SDM using oligonucleotide RE619.
Plasmids to make L-A-, X-, M1-, Mlus-, or L-BC-specific probes have been described. A summary of the sequences recognized by each probe is as follows. The L-A probes made from plasmids pRE687 and pRE691 recognize the positive strand of L-A from nt 1323 to 1786 and from nt 1783 to 2647, respectively (11, 26). The M1 probe, made from plasmid pRE1119, recognizes the (+) strand of M1 from nt 14 to 500 (38). The X probe was made from plasmid pRE76-2-14 and recognizes the X (+) strand from nt 1 to 530 (6). The Mlus probe, made from plasmid pMlus-11, recognizes the Mlus (−) strand (nt 119 to 935, counting from the 3′ end of Mlus) (11). The L-BC probe, from plasmid pRE442, recognizes the L-BC (+) strand (nt 63 to 502). Plasmid pRE1275 contains the L-A-lus sequence from nt 2070 to 4580 (L-A-lus Pol region) subcloned into the NotI site of the Bluescript-KS+ vector (Stratagene, San Diego, CA). It was used to synthesize an L-A-lus (+) strand-specific probe. Plasmid pRE1280 contains a fragment of 522 bp of M2 cDNA (from nt 332 to 853) obtained by RT-PCR from K2 wine strain A143-2 (Table 2) using oligonucleotides NR112 and NR113. The M2 cDNA fragment was inserted between the BamHI and EcoRI sites of Bluescript KS+ vector and used to synthesize a probe that recognizes the M2 (−) strand.
Plasmids pRS404-PTEF-Ago1 and pRS405-PTEF-Dcr1 were a gift of David Bartel (Addgene plasmid numbers 22313 and 22314, respectively) and express the RNA interference proteins Ago1p and Dcr1p from S. castellii; they contain the TRP1 and LEU2 genes, respectively, as markers. Plasmid pRE914 overexpresses the SKI1 gene under the control of the constitutive PGK1 promoter and carries HIS3 as a selective marker (26).
Total nucleic acid preparation and Northern hybridization.
Strains were grown in YPAD medium or in complete minimal medium deprived of the appropriate amino acid for 2 to 3 days at 28°C. Cells from 1 ml of culture were broken with glass beads by vortex mixing, and nucleic acids were extracted from cell lysates as described previously (33). Total RNAs were separated on 1.3% agarose gels, denatured in the gel, and transferred to neutral nylon membranes (GE Healthcare). Detailed conditions for Northern hybridization were described elsewhere (39). RNAs on the membranes were detected by hybridization with 32P-labeled specific probes made by T3 or T7 runoff transcription from plasmids predigested with appropriate restriction enzymes to make them linear.
cDNA synthesis, cloning, and sequencing of L-A-lus dsRNA.
dsRNAs from strain EX229 (L-A-lus, L-BC, and M-lus) were purified by CF-11 cellulose chromatography as described elsewhere (23). L-A-lus and L-BC were then further separated from Mlus by agarose gel electrophoresis and electroelution. L-A-lus and L-BC cDNA synthesis was carried out using a Universal Riboclone cDNA synthesis system kit from Promega based on the method of Gubler and Hoffman (40), as described previously (11). Without further purification, cDNA fragments corresponding to L-A-lus or L-BC dsRNA were blunt end ligated into the unique SmaI site of Bluescript-KS+ vector and introduced into competent Escherichia coli DH5α cells. Transformants containing inserts were sequenced, and clones corresponding to L-A-lus or to L-BC were distinguished by comparison to the published L-A or L-BC sequences. Using the sequences obtained from 5 L-A-lus random clones and by comparison with standard L-A, we positioned our L-A-lus sequences. Afterwards, we used either of 2 pairs of oligonucleotides (NR88 and NR80 or NR81 and NR89) in order to amplify 2 internal gaps. Finally, the sequences from the ends of L-A-lus were obtained by 3′ rapid amplification of cDNA ends (3′RACE). First, the 3′ ends of the molecule were A-tailed using poly(A) polymerase and the conditions described by the supplier (Epicentre). Conditions for annealing with an oligo(dT) primer (oligonucleotide NR67) and for cDNA synthesis were as described previously (11). For PCR amplification, we used primer NR68 and either primer NR86 or NR87 that annealed near the 5′ or 3′ end of the molecule, respectively. The PCR products were digested with NotI and BamHI, cloned into the Bluescript-KS+ vector digested with the same enzymes, and sequenced.
RT-PCR.
To characterize the type of L-A present in the wine strains of Table 2, we amplified cDNA fragments by RT-PCR using total nucleic acids prepared as mentioned before and two different pairs of oligonucleotides: NR88 and NR80 in the case of L-A-lus and RE548 and RE549 to amplify L-A. Conditions for annealing and first-strand synthesis were as indicated for the 3′RACE protocol, and PCR was done using Go Taq DNA polymerase (Promega). When fragments of larger size were needed for cloning purposes, AccuPrime Taq DNA polymerase High Fidelity (Invitrogen) was used.
Miscellaneous.
DNA manipulations (enzyme digestions and cloning procedures) were done following standard methods according to reference 41. Plasmid DNA for sequencing was obtained and purified using a Wizard Plus SV Minipreps DNA purification system (Promega). DNA fragments were purified with a QIAquick gel extraction kit (Qiagen). Before PCR fragments were sequenced, oligonucleotides present in the samples were removed by Sepharose chromatography using MicroSpin S-400 HR columns (GE Healthcare). Most of the enzymes were purchased from Promega. Synthetic oligonucleotides were purchased from Thermo. Site-directed mutagenesis was done as described previously (6). Antibodies against the Gag protein of L-A virus have been described previously (42). Yeast cells were transformed using lithium acetate to permeabilize the cells (43), and transformants were selected in H-trp, H-ura, or H-his medium. In the case of transformation with integrative plasmids pRS404-PTEF-Ago1 and pRS405-PTEF-Dcr1, the plasmids were digested with HindIII and EcoRI, which cleave in unique sites in the TRP1 and LEU2 genes, respectively. All DNA primers used are listed in Table S6 in the supplemental material. RNA secondary structure predictions were done using the MFOLD program (44).
Nucleotide sequence accession number.
The L-A-lus cDNA nucleotide sequence and the encoded Gag and Gag-Pol proteins appear in NCBI/GenBank under GenBank accession no. JN819511.1.
RESULTS
L-A-lus, a new variant of L-A virus in wine yeast strains.
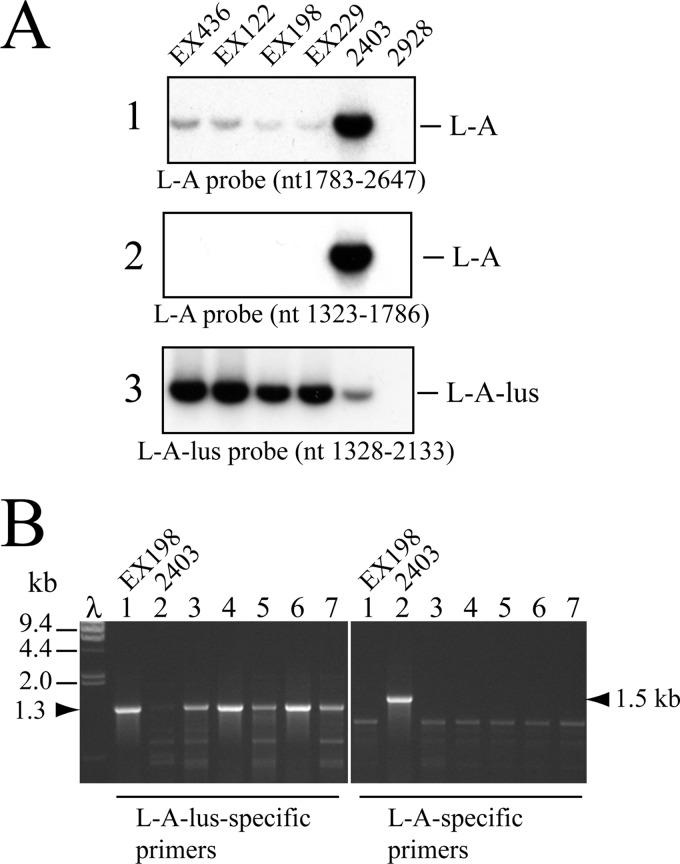
In a previous work, we identified a new killer toxin (Klus) encoded in Mlus dsRNA in wine yeast strains from the Ribera del Guadiana region of Spain (11). Genetic and molecular analysis showed the presence in those strains of a helper virus, L-A, as is the case with other satellite toxin-encoding M dsRNAs. However, Northern blot hybridization with two different L-A probes (whose sequences had been determined years ago) showed poor hybridization to the probes (Fig. 2A), suggesting substantial differences between the nucleotide sequences of the published L-A and the L-A present in yeast strains carrying Mlus. This prompted us to clone and sequence this L-A variant, which we named L-A-lus to distinguish it from standard L-A. We purified L-A-lus dsRNA from strain EX229 by cellulose CF-11 chromatography and used it for RT-PCR as described in Materials and Methods. By random priming, we initially obtained and sequenced 5 independent clones that covered about 50% of the molecule. The sequences of two internal gaps were then resolved by RT-PCR amplification of two fragments that covered the gaps. The sequences at both ends were obtained by 3′-RACE in 2 independent clones in each strand. The entire L-A-lus sequence was thus assembled from 9 clones (GenBank accession no. JN819511.1). Altogether, L-A-lus is 4,580 nt. A ClustalW comparison between L-A-lus and L-A showed 73% identity (see Fig. S1A in the supplemental material). The 27% difference between the two versions of L-A explains our previous hybridization results with two different L-A probes (11) (Fig. 2A). Figure 2A also shows that a probe made from an L-A-lus cDNA clone recognized L-A-lus specifically and L-A only poorly. Certain regions were different enough to allow the design of unique oligonucleotides to amplify specifically L-A or L-A-lus by RT-PCR. Thus, we could examine the presence of one or the other L-A directly in multiple yeast strains isolated from different geographical locations or sources (Table 2).
Fig 2.
Identification of L-A-lus variant in yeast strains by Northern hybridization or RT-PCR. (A) Total RNAs were obtained from strain EX436, EX122, EX198, or EX229 (Klus-1 to Klus-4 isotypes) and from laboratory strain 2403 (standard L-A) or 2928 (L-A-o). Three sets of the samples were separated on an agarose gel, denatured in the gel, blotted onto a nylon membrane, and then hybridized with one of three different probes: probe 1, an L-A-specific probe that recognized nt 1783 to nt 2647; probe 2, a different L-A-specific probe which annealed between nt 1323 and nt 1786; or probe 3, an L-A-lus-specific probe to nt 1328 to nt 2133. Autoradiograms of the membranes are shown. (B) Specific RT-PCR amplification of L-A-lus or L-A. RT-PCRs were performed using total nucleic acids from wine strain Ca7, 47-P07, EC1118, T34, or PB2010 (Table 2 and lanes 3 to 7). For controls, we also amplified RNAs from Klus strain EX198 (L-A-lus, lanes 1) or laboratory strain 2403 (L-A, lanes 2). Reactions were done in parallel using two sets of primers that recognize L-A-lus (left) or L-A (right). λ, lambda DNA digested with HindIII as a size marker. The major RT-PCR products that amplified specifically with the two primers from L-A-lus or from L-A are indicated at both sides of the ethidium bromide-stained gel with the arrowheads.
Analysis of L-A-lus sequence.
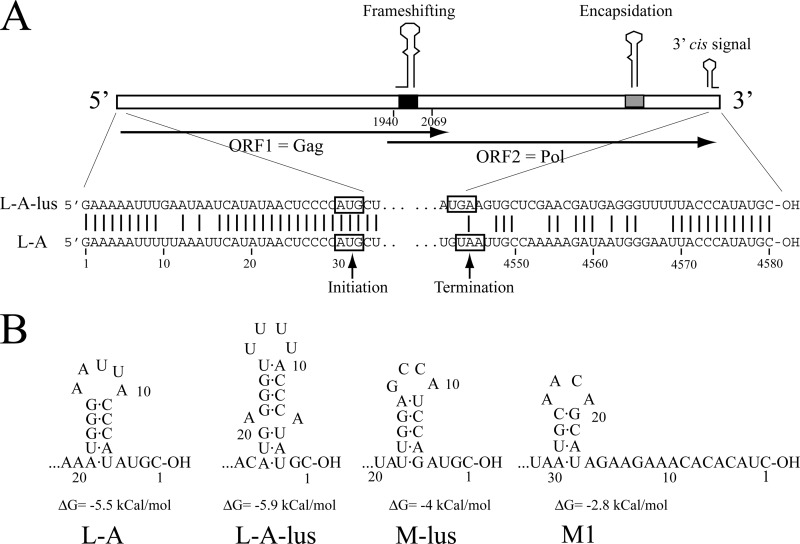
The genomic organization of L-A-lus is the same as that of L-A (Fig. 1B). There are two ORFs. The first ORF starts at nt 30 and extends to nt 2069. The second ORF is from nt 1940 to almost the end of the molecule (nt 4543) and is probably expressed as a fusion protein together with ORF1 by −1 ribosomal frameshifting (Fig. 3A; see also Fig. S1 in the supplemental material). By comparison with the published sequences of Gag and Pol of L-A, we assigned these two ORFs in L-A-lus to the major coat protein of L-A-lus virions (ORF1) and to the fusion Gag-Pol (ORF1-ORF2), respectively (Fig. 3A). A ClustalW comparison at the amino acid level shows that the 73% nucleotide identity between the two L-A genomes rises in the case of proteins to 86% identity (see Fig. S1B in the supplemental material). Polyclonal antibodies against Gag of L-A recognized in a Western blot the major coat protein of L-A-lus. A larger, minor protein, with the size expected for a fusion protein, is also recognized by the antibodies, suggesting that it is the fusion product of ORF1 plus ORF2 (see Fig. S2 in the supplemental material).
Fig 3.
Sequences and secondary structures conserved in the UTRs of L-A-lus and L-A. (A) The diagram shows the genomic organization of L-A-lus. Internal regions indicated as “Frameshifting” (black box) or “Encapsidation” (gray box) correspond to the frameshifting or encapsidation signals depicted in Fig. 1B. The “3′ cis signal” is depicted in panel B. Overlapping arrows show the two ORFs found in the L-A-lus (or L-A) (+) strand. The 5′- and 3′-end sequences of L-A-lus are displayed at the bottom of the panel. For comparison, those of L-A are also shown. Vertical bars indicate identical nucleotides. Initiation and termination codons are boxed. (B) Secondary structures predicted by the MFOLD program at the 3′ end of L-A, L-A-lus, or satellite RNAs that can be maintained by L-A-lus virions are shown. Numbering is from the 3′ ends.
There are two regions in the genomes of L-A and L-A-lus that are 100% identical: (i) a region with a stem-loop structure known to be involved in frameshifting (nt 1969 to nt 2004) adjacent to the slippery site 1958GGGUUUA1964 (Fig. 1B; see also Fig. S1A in the supplemental material) and (ii) a region that contains a 24-nt stem-loop structure responsible for binding to and encapsidation of the L-A (+) strand (nt 4180 to nt 4203) (Fig. 1B; see also Fig. S1A in the supplemental material), indicating the importance of these two regions in the translation and encapsidation steps in the life cycle of either L-A virus (Fig. 1A). We observed only one nucleotide change in the stem-loop secondary structure adjacent to the slippery site in the frameshifting region (at the opening of the loop, U1991 in L-A is C1991 in L-A-lus). This change produces a decrease in the free energy of the structure from ΔG = −15.3 kcal/mol in L-A to ΔG = −17.3 kcal/mol in L-A-lus. We found the same change (U1991C) in the L-A present in strain BY4741 compared to the deposited L-A sequence (R. Esteban, unpublished data). The amount of L-A in BY4741 is much lower than that of L-A in strain 2404. We do not know whether this nucleotide modification in L-A-lus may affect the rate of frameshifting and consequently the ratio between Gag and Gag-Pol, but as described previously (45), changes in the secondary structure affect frameshifting efficiency. The amount of L-A-lus virions in different wine yeast strains or in the laboratory strains constructed in this work is similar to that of L-A in strain BY4741 and much lower than that of L-A in strain 2404 (see Fig. S3 in the supplemental material).
At the 3′ end of L-A (+) strands, a cis signal for replication has been demonstrated (6). The signal is composed of a stem-loop structure and the adjacent 3′-end 4 nt (Fig. 1B and 3B). In the structure, the nucleotide sequence of the loop is important but that of the stem is not. No experimental evidence has been reported for signals at L-A's 5′ end. However, the fact that X, a deletion mutant of L-A, contains only the first 25 nt of L-A's 5′ end and is maintained stably by L-A virions suggests that within that 25-nt sequence reside the cis signals needed for transcription. As is common in other dsRNA viruses, this region is AU rich (Fig. 3A), facilitating the melting of the molecule and the access of Pol to the template strand for conservative transcription. Figure 3A depicts the 5′ and 3′ untranslated terminal regions (UTRs) of L-A-lus and L-A (+) strands. The 5′ UTR in L-A-lus is 29 nt and, like L-A's, is AU rich; both are highly conserved. The 3′ UTR nucleotide sequences are quite different: only the last 12 nt are identical. L-A-lus, however, can be folded into a stem-loop structure that resembles that of L-A (Fig. 3B). We do not know, at present, if this structure constitutes a cis signal for replication similar to that of L-A's.
Because the proteins encoded by L-A or L-A-lus are almost identical (see Fig. S1B in the supplemental material), we would expect the same (or almost the same) tridimensional or spatial organization in the virion and similar mode of actions in the Pol part of the Gag-Pol protein in L-A-lus and L-A. Indeed, the regions that contain the four motifs conserved in RdRps are well conserved (>95% identical) (see Fig. S1B in the supplemental material). Strikingly, we observed a higher variation in one region within Gag around amino acids 160 to 173. We have reported recently the existence of a cap-snatching mechanism in L-A virions (38). The reaction, which transfers cap groups from mRNA to the nascent L-A transcript when extruded from the virions, is carried out by His154 in Gag, which steals cap groups from cellular mRNA and attaches them covalently to His154 (46). Crystallographic studies have shown that His154 in Gag is in the upper rim of a trench where host mRNA can interact through its cap structure (47). Interestingly, comparing these two regions in L-A and L-A-lus, His154 and surrounding amino acids are conserved but a downstream stretch of 14 amino acids shows more variation that the rest of Gag (see Fig. S1B in the supplemental material). This suggests that His154, which is exposed to the outer side of the virion, is not interacting tightly with amino acids in its vicinity. Other parts of Gag are more conserved. A second region with higher variation is an internal fragment of about 43 amino acids in the N-terminal third of Pol (amino acids 731A to 771A in Gag-Pol) that shows a 38% variation; of 43 amino acids, 16 are different. This region is likely to be separating the Gag and Pol domains in the Gag-Pol fusion protein (see Fig. S1B in the supplemental material).
Construction of laboratory strains with L-A-lus and Mlus or with L-A-lus alone.
To go deeper into genetic and biochemical studies with the new L-A-lus variant, we generated laboratory strains with appropriate auxotrophic markers as hosts of L-A-lus alone or L-A-lus and its satellite Mlus. Table 1 shows a list of laboratory strains constructed in this work. We first determined by direct observation under the optical microscope which of the Klus killer strains used in our previous work (11) were able to sporulate. Strain EX198 (Table 2) was selected, and a whole-spore clone population was mated out with strain 2928 expressing the Geneticin-resistant phenotype. The resulting diploids were selected as explained in Materials and Methods. Further sporulation of these diploids produced strains 1081 and 1082 that carry L-A-lus and Mlus. Both were of the a mating type and ura3. Strain 1081 also harbors the narnavirus 23S RNA launched from plasmid pNR41 (35). This narnavirus, which is seldom present in the cytoplasm of strains from the laboratory or nature, was a good marker for further cytoplasmic mixing (cytoduction) experiments. Next, we wanted to generate a strain with appropriate auxotrophic markers with L-A-lus and Mlus to be used for cytoduction. So we chose strain 2405, which is L-A-o and carries the kar1-1 mutation, as the recipient of cytoplasms from strain 1081 or 1082. In this way, we generated strains 1083 and 1084 that carry the kar1-1 mutation and can maintain both viruses stably (and the Klus killer phenotype) after more than 90 generations (three consecutive single-colony isolation experiments). L-A-lus and Mlus in kar1-1 mutants allowed us to introduce the viruses into any strain from the EUROFAN collection (made of derivatives from the BY4741 strain that are a mating type) without nuclear fusion to test the effect of gene deletions on the L-A-lus (or Mlus) copy number.
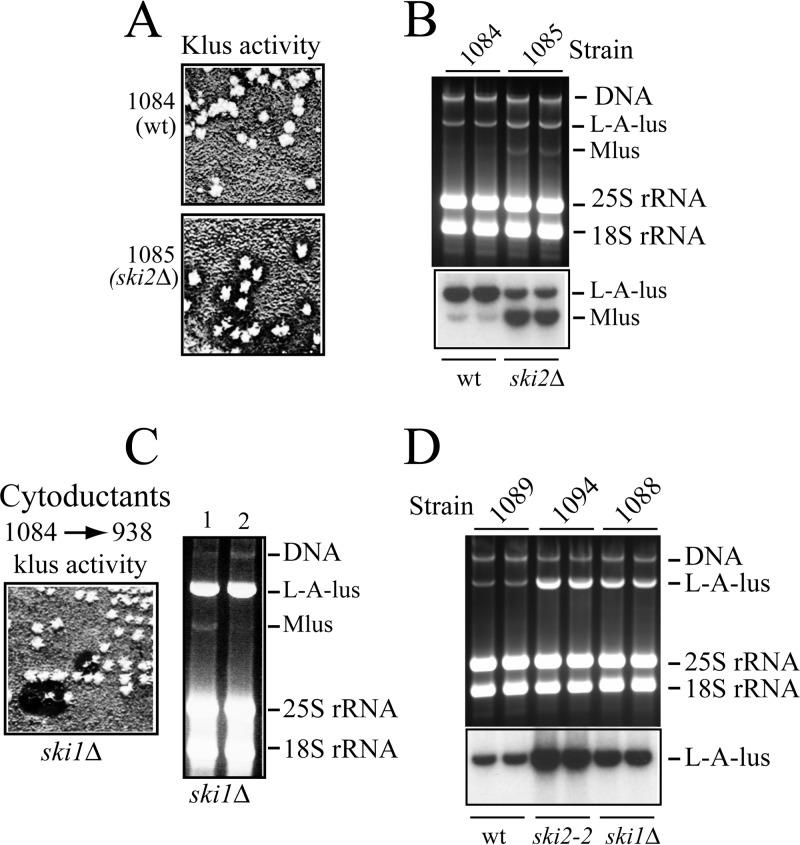
Effect of SKI and MAK genes on L-A-lus.
Because the amounts of L-A in laboratory strains are affected by host mutations such as the ski or mak mutation, we would expect similar behavior for L-A-lus. Thus, we analyzed the amounts of L-A-lus (and Mlus) in the wild type and in the ski1Δ and ski2Δ mutants. Strains 938 and 909 are derivatives of BY4741 that have deleted the SKI1 and SKI2 genes, respectively. We had cured endogenous L-A in these strains by overexpression of the 5′ XRN1/SKI1 exonuclease (strain 938) or by growth at high temperature (strain 909) (26). Thus, strains 938 and 909 were L-A-o. From strain 1084, we cytoduced L-A-lus and Mlus. The resultant ski2Δ cytoductant produced a Klus superkiller phenotype (Fig. 4A), indicating that L-A-lus (and Mlus) are also under the control of the SKI2 gene. Concomitantly, the amount of Mlus in ski2Δ cells is much higher than in the wild-type strain (Fig. 4B). When we introduced L-A-lus and Mlus into the ski1Δ 938 strain, we observed that most cytoductants had lost Mlus spontaneously (Fig. 4C) and that they carried enormous amounts of L-A-lus. This strain (1088) was used as the donor of L-A-lus alone in the next cytoplasmic mixing experiments to generate a set of laboratory strains that carry only L-A-lus. We had interest in analyzing this new variant of L-A in the absence of Mlus. In this way, we generated strain 1089 (Table 1) by introducing L-A-lus into strain 2405. Strain 1089 is the laboratory strain with L-A-lus only that we used for further experiments throughout this work either to introduce it into appropriate strains by cytoduction or to test the stability of L-A-lus virions with respect to growth at high temperature compared to that of L-A virions in an isogenic strain (1064).
Fig 4.
Effect of ski mutations on L-A-lus or Mlus amount and Klus activity. (A) Isolated colonies from strain 1084 (wt) or strain 1085 (ski2Δ) were analyzed for killer activity by replica plating onto an MB plate seeded with the sensitive strain 5X47. Clear big halos surrounding the ski2Δ colonies indicate a Klus superkiller phenotype. (B) RNAs from two colonies of the strains shown in panel A were separated in an agarose gel, blotted onto a nylon membrane, and hybridized with a mixture of L-A-lus- and Mlus-specific probes. The upper panel shows the ethidium bromide-stained gel, and the lower panel shows the autoradiography of the Northern blot analysis. (C) Cytoductants from strain 1084 (L-A-lus and Mlus) into the ski1Δ 938 strain lose Klus activity at high frequency (left panel). The killer assay was done as described for panel A. On the right panel, RNAs prepared from a killer (lane 1) or a nonkiller (lane 2) colony were separated in an agarose gel and visualized by ethidium bromide staining. (D) Strains 1089 (wt), 1094 (ski2-2), and 1088 (ski1Δ) that carry L-A-lus alone (Mlus-o) were grown in YPAD medium in duplicate tubes. Total RNAs were analyzed as described for panel B. The upper panel shows the ethidium bromide-stained gel. The lower panel shows the Northern blot hybridized with an L-A-lus-specific probe.
When L-A-lus alone from strain 1089 was cytoduced into strain 2927 (ski2-2 mutant), the resulting strain (1094) also had enormous amounts of L-A-lus, as happened in the ski1Δ background. Figure 4D shows the amounts of L-A-lus alone (in the absence of Mlus) in a wild-type, ski1Δ mutant, or ski2-2 mutant strain. We observed an increase of about 3- to 5-fold in the mutants, similar to that described for L-A.
We also checked the effect of the mak10 mutation on L-A-lus maintenance. All cytoductants from strain 1084 into the EUROFAN mak10Δ deletion mutant were L-A-lus-o and concomitantly Mlus-o.
Distribution of L-A-lus in nature.
To study which type of L-A was present in wild yeasts, we selected around 30 wine strains from our stock collection (Table 2). Most of them had been analyzed previously by agarose gel electrophoresis, in a screening for the presence of 20S or 23S RNA virus (35). For the specific L-A RT-PCR analysis, we selected only those strains that carried a band of dsRNA with a size compatible with that of L-A or L-BC (4.6 kb). By performing killing assays on MB plates seeded with appropriate sensitive strains, we found that about half of the strains were killers (either K2 or Klus) and the others were nonkillers. Some of the nonkiller strains, however, carried either M2 or Mlus dsRNA (Table 2). None of the strains tested showed the K1 phenotype. The type of L-A present was determined by RT-PCR analysis with two pairs of oligonucleotides: NR88 and NR80, which anneal specifically to L-A-lus and give rise to a 1,270-bp DNA fragment, and RE548 and RE549, which amplify an 1,485-bp fragment from the L-A 5′ end (Fig. 2B). As internal controls, we used RNAs from strain EX198 (for L-A-lus) or 2403 (for L-A) in each amplification experiment (Fig. 2B, lanes 1 and 2). We found that none of the 31 strains analyzed carried L-A. From the previously described Klus strains (Klus-1 to Klus-4) (11), we could amplify an RT-PCR fragment of the expected size, suggesting that they carried the same L-A-lus variant. And indeed, when these RT-PCR fragments were directly sequenced, we confirmed that all of them had the same L-A-lus. A fragment of the same size was also amplified from the remaining strains, though in some cases less efficiently (Fig. 2B shows some of the RT-PCR results). This prompted us to sequence all the PCR products. We found that 13 of the 16 strains that harbor M2 dsRNA in Table 2 carried a different L-A variant with about 75% identity (in the fragment so far sequenced) to the other two L-As (see Fig. S4 in the supplemental material). This new L-A, which we designated L-A-2, was present only in K2 killers. As mentioned in the introduction, a different L-A helper virus in K2 killer strains had already been described (12, 13). However, its nucleotide sequence was never reported. The rest of strains listed in Table 2 (n = 18), including all the strains that harbor Mlus, several nonkiller strains that carry no M dsRNA, and three strains that harbor M2, have L-A-lus. The presence of L-A-lus in M2-containing strains was somewhat unexpected, because we have observed that in haploid laboratory strains, L-A-lus, in fact, cannot maintain M2 dsRNA (N. Rodriguez-Cousiño and R. Esteban, unpublished data). We are in the process of characterizing this association more deeply. In summary, data in Table 2 strongly indicate an association between L-A-lus and Mlus or between L-A-2 and M2, suggesting that each toxin-producing satellite M dsRNA is specifically associated with a distinct L-A helper virus. None of the strains carried either L-A or M1 dsRNA. In our wine strain collection, we did not find any K28 killer strain; thus, we do not know which type of L-A helper virus is accompanying M28 dsRNA. In the original K28 isolate, the L-A present (called L28) also showed certain differences from K1 strains with respect to L-A (10).
L-A-lus excludes L-A.
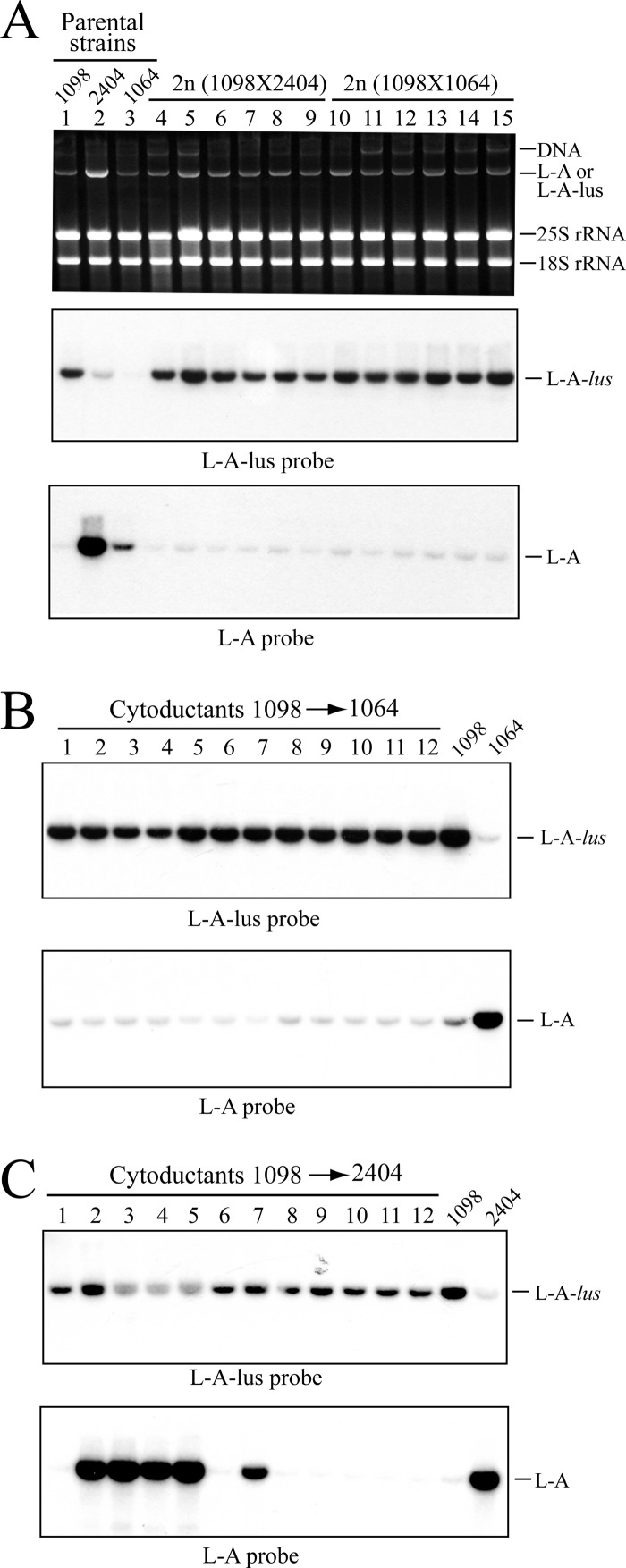
The absence of L-A in killer wine strains from different geographical locations and the prevalence in these natural environments of specific L-A variants (associated with Klus or K2 strains) led us to investigate whether these new L-A variants could work as helper viruses of different satellite killer-producing M dsRNAs under controlled laboratory conditions (i.e., in haploid cells). We focused our attention on L-A-lus and L-A. The amount of L-A in different laboratory strains is variable. Strain 2404 harbors enormous amounts of L-A, while strain BY4741, from which the EUROFAN collection derives, carries an L-A of much reduced copy number (see Fig. S3 in the supplemental material). To generate isogenic strains with different copy numbers of L-A, we constructed strain 1064 by cytoducing the cytoplasm of BY4741 into strain 2405 (isogenic to 2404 but L-A-o). These haploid strains (2404 and 1064; Table 1) were mated out with strain 1098, which carries L-A-lus. Strain 1098 had been obtained by transferring the cytoplasm of 1089 into strain 2928. A total of 12 independent diploid clones, 6 from each cross (1098 × 2404 or 1098 × 1064), were analyzed by Northern hybridization. None of them carried both L-As together, indicating that they are excluded by each other, a phenomenon frequently found and already described for totivitus in the same cell. Unexpectedly, all diploids analyzed carried L-A-lus and none L-A, indicating that L-A-lus was outcompeting L-A (Fig. 5A). This is even more striking in the case of the diploids between strains 2404 and 1098. We estimated that there were at least 5-fold more L-A molecules than L-A-lus molecules in the original zygote after fusing both cytoplasms; nevertheless, in a few generations L-A was completely excluded by L-A-lus. Because L-A-lus in nature is predominantly present in diploid or poliploid yeast cells, we reasoned that perhaps this exclusion was dependent on the ploidy state of the cell. So we carried out cytoplasmic mixing experiments analyzing only haploid cells. Because strains 2404 and 1064 carry the kar1-1 mutation, L-A-lus from strain 1098 was introduced into them by cytoplasmic mixing. A total of 12 cytoductants from each experiment were analyzed by Northern hybridization (Fig. 5B and C). In the case of cytoductants to strain 1064, all them carried L-A-lus (Fig. 5B). In the case of mixed cytoplasms between 2404 and 1098, more than half (seven) carried L-A-lus; three had L-A, and two of them had a mixture of L-A and L-A-lus (Fig. 5C). Further colony isolation from these mixed clones produced colonies either with L-A or with L-A-lus. In conclusion, we have found that, not only in diploid cells but also in haploid cells, L-A and L-A-lus cannot coexist in the same cell and there is an overwhelming predominance of L-A-lus over L-A.
Fig 5.
Exclusion of L-A by L-A-lus. (A) Exclusion in diploids. Strain 1098 carrying L-A-lus was mated either with strain 2404 (L-A, high copy number) or with strain 1064 (L-A, low copy number). Total nucleic acids from parental strains (lanes 1 to 3) and six independent diploid (2n) colonies from cross 1098X2404 (lanes 4 to 9) or from cross 1098X1064 (lanes 10 to 15) were analyzed by agarose gel electrophoresis. The top panel shows the ethidium bromide-stained gel, the middle panel shows a Northern hybridization with an L-A-lus-specific probe, and the lower panel shows the hybridization with an L-A-specific probe. The probes were the same as used for the experiments represented in panels 3 and 1 of Fig. 2A, respectively. (B and C) Exclusion of L-A by L-A-lus in haploid cells. L-A-lus from strain 1098 was introduced into strain 1064 (B) or strain 2404 (C) by cytoduction. Total nucleic acids from 12 independent cytoductants in each strain (panel lanes 1 to 12) together with the respective parental strains were separated in agarose gels and transferred to nylon membranes. The presence of L-A-lus or L-A on the membranes was determined by Northern hybridization using an L-A-lus-specific probe (top panels) or an L-A-specific probe (bottom panels). As shown in the panels in Fig. 2A, there was a minor cross-hybridization between the L-A- or L-A-lus-specific probes and the other type of L-A. This accounts for the weak hybridization signals observed in the lower portions of panels A and B.
L-A and L-A-lus show different sensitivities to growth at high temperature or to different nucleases.
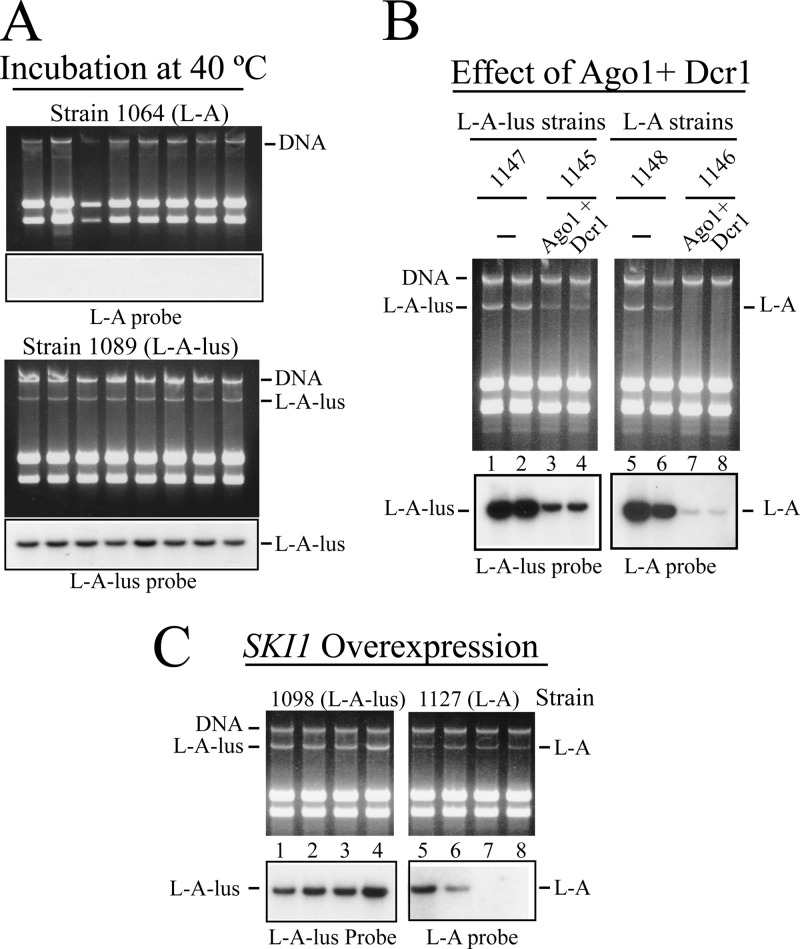
The exclusion of L-A by L-A-lus in diploid or haploid cells (Fig. 5) led us to analyze the behavior of L-A-lus under different conditions known to affect L-A copy number. We selected three conditions: (i) growth at high temperature, (ii) effect of the 5′ SKI1 exonuclease overexpression (26), and (iii) expression of the RNA interfering enzymes Dcr1p and Ago1p from S. castellii (32). (i) First, we tested growth at high temperature (40°C) because this has been the method of choice to eliminate M1 virions (and in most strains, also L-A) (48, 49). Cells from isogenic strain 1064 (L-A) or 1089 (L-A-lus) were spread for single-colony isolation on YPAD plates. After 2 to 3 days, cells from independent colonies were again reisolated on fresh YPAD plates. After 3 consecutive colony isolation experiments, RNAs from 8 independent colonies in each strain were analyzed by Northern hybridization. All colonies from strain 1064 had lost L-A, while all the colonies from strain 1089 maintained similar amounts of L-A-lus (Fig. 6A). Prolonged periods of incubation at this high temperature did not result in L-A-lus-o cells, confirming that L-A-lus is resistant to the treatment. The original Klus strain from which the L-A-lus in strain 1089 derives had been isolated from the Extremadura region (Spain), with an average temperature a few degrees higher than other regions in the country. These conditions may be favorable for L-A-lus maintenance. We also checked the effect of growth at high temperature on L-A-lus curing using strain 1084, which carries Mlus in addition to L-A-lus and thus has a diminished L-A-lus copy number. Again, we did not observe any curing of either L-A-lus or Mlus under these conditions. (ii) We next used two isogenic strains with L-A or L-A-lus to overexpress the 5′ exonuclease SKI1. Strains 1098 and 1127 (Table 1) that carry the his3 mutation were transformed with vector pRE914. This plasmid expresses SKI1 from the constitutive PGK1 promoter and cures L-A with high efficiency (26). Northern hybridization of several transformants of strain 1127 confirmed that L-A could indeed be eliminated in 50% of the colonies analyzed (Fig. 6C, right). In the case of L-A-lus, however, we observed a decrease in the amount of L-A-lus, but nevertheless, after several single-colony isolation rounds, none of the colonies expressing SKI1 were cured of L-A-lus (Fig. 6C, left). (iii) Finally, we tested the effect of Dcr1p and Ago1p endonuclease expression on both types of L-A. It has been published that the genomic insertion and expression of Dcr1p and Ago1p from S. castellii in S. cerevisiae caused the elimination of M1 and L-A dsRNA viruses and so the killer phenotype at high frequency (31). We wondered whether the same was true for L-A-lus. Both genes from plasmids pRS404-PTEF-Ago1 and pRS405-PTEF-Dcr1 (32) were inserted into the genome of strain TF395 that carried no L-A, as described in Materials and Methods, to produce strain 1150 (Table 1). Next, L-A or L-A-lus was introduced by cytoduction from donor strains 1064 or 1089 into strain TF395 or 1150. Figure 6B shows that, in contrast to what was previously described, the expression of Ago1p and Dcr1p could not eliminate either L-A or L-A-lus from the strains expressing the endonucleases. In the case of L-A, there was a big decrease of its copy number due to Dcr1p and Ago1p expression. We again observed that the L-A-lus copy number, though diminished, was much less affected than that of L-A by Dcr1p and Ago1p. We cannot explain the discrepancy between our data and the reported loss of L-A when the RNAi endonucleases are reconstituted in S. cerevisiae cells. Perhaps it might be due to the simultaneous presence of M1 and L-A viruses in those cells. In conclusion, all data presented here confirm that L-A-lus virions are more resistant than those of L-A to any of the traits analyzed and may explain why, in the yeast wine strains of Table 2, isolated from different sources, the L-A from laboratory strains was not found.
Fig 6.
Different sensitivities of L-A and L-A-lus to growth at high temperature or to in vivo nuclease expression. (A) Cells of strain 1064 (L-A, upper panels) or strain 1089 (L-A-lus, lower panels) were streaked for single-colony isolation on an YPAD plate and incubated at 40°C for 3 days. Single colonies were reisolated twice and grown under the same conditions. Then, RNAs from 8 independent colonies from each strain were analyzed in two agarose gels. The upper panels show the ethidium bromide-stained gels. After Northern blotting, RNAs were detected by hybridization with an L-A (strain 1064)- or an L-A-lus (strain 1089)-specific probe. (B) Left panels: L-A-lus-containing strains 1147 and 1145 are isogenic except for the AGO1 and DCR1 genes from S. castellii expressed in strain 1145. RNAs (in duplicate) were prepared after 2 days of growth at 28°C, separated in an agarose gel, and detected by Northern analysis with an L-A-lus-specific probe (lanes 1 to 4). Right panels: strains 1148 and 1146 carrying the L-A virus were analyzed as described for the left panels. Strain 1146 expresses the AGO1 and DCR1 genes from S. castellii. RNAs were analyzed by Northern hybridization with an L-A-specific probe (lanes 5 to 8). (C) Cells of strain 1098 (L-A-lus, left panels) or strain 1127 (L-A, right panels) were transformed with vector pRE914 that overexpresses the SKI1/XRN1 5′ exonuclease. RNAs from 4 transformants in each strain were analyzed by Northern hybridization with probes specific for L-A-lus (strain 1098, lanes 1 to 4) or for L-A (strain 1127, lanes 5 to 8). The upper panels show the ethidium bromide-stained gels and the lower panels the autoradiograms. In lanes 7 and 8 in the agarose gel, there is a band with the same mobility as that corresponding to L-A which is not recognized by the L-A probe (lower panel). It corresponds to L-BC dsRNA present in strain 1127.
Helper activity of L-A-lus for various satellite RNAs.
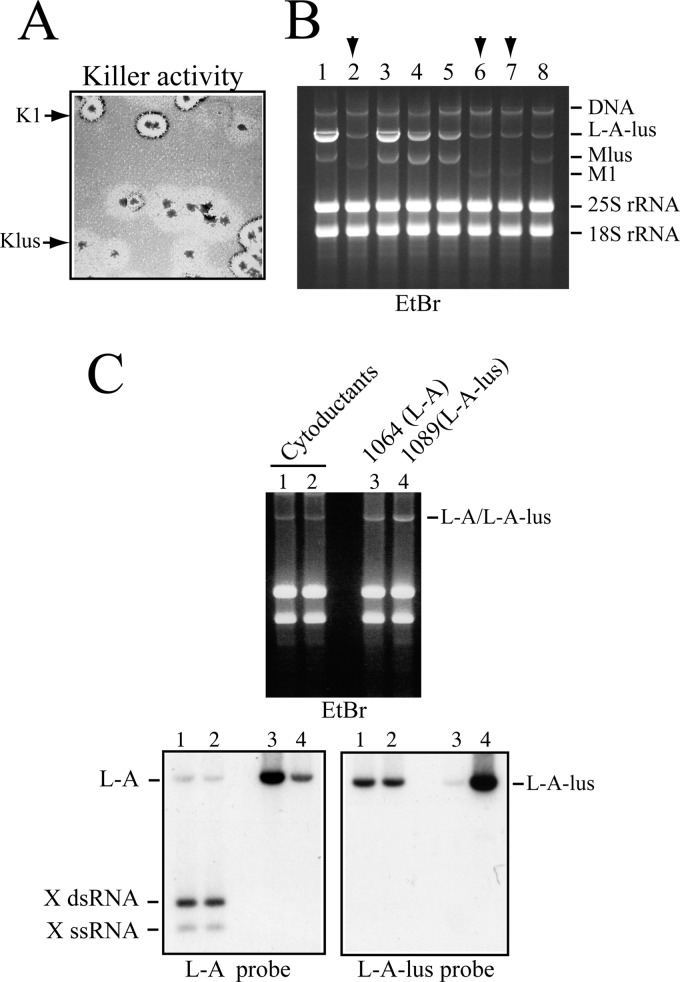
As mentioned above, in wine yeasts L-A-lus was mostly associated with Klus strains (Table 2). Because we did not find any M1-containing strain in that screening, we wondered if L-A-lus could maintain M1. In our laboratory, we have K1 killer strains that carry M1 dsRNA in the absence of L-A helper virus, provided that the L-A coat proteins are expressed from a vector (37). One of those strains (559; Table 1) was used as the donor to cytoduce M1 virus into the 1085 strain that carries L-A-lus and Mlus. Strain 1085 has the SKI2 gene deleted, and it produces on a sensitive lawn a larger Klus killing zone than that seen with the SKI2 wild-type strain (Fig. 4A). Several cytoductants were isolated and analyzed by killer assay, and their RNAs were separated on an agarose gel. Figure 7A shows a killing assay of the original mixture of cytoductants. We can observe two types of killing halos. The clearer ones correspond to K1 killing zones, and the opaque ones correspond to Klus killing (which in general appears not to lyse the cells completely). The ethidium bromide-stained agarose gel shows that the cells carried either M1 or Mlus dsRNA. We could not find any colony with both satellite RNAs together, confirming previous data about exclusion between different satellite toxin-producing M dsRNAs (14). Some of the cytoductants with L-A-lus and M1 were streaked again for single-colony isolation, and we checked their killer activity on MB plates. All of them were stable killers, indicating that L-A-lus can maintain M1. Because this strain (1086) was a ski2Δ mutant, we wondered if the large amount of L-A-lus in this background was necessary for M1 maintenance. To test the behavior of L-A-lus and M1 in a wild-type strain, we introduced the cytoplasm of strain 1086 into strain 2405, which carries no L-A. Strain 1087 now harbors L-A-lus and M1, and after three consecutive single-colony isolations, M1 was maintained stably by L-A-lus. Thus, L-A-lus has helper virus activity for M1 dsRNA. We could not directly address the issue of whether L-A, in turn, could maintain Mlus. As we have shown above, mating an L-A strain to an L-A-lus strain produced exclusion of L-A. However, it has been shown that in a mak10-1 mutant, which cannot support L-A-lus replication, Mlus can be maintained by L-A virions made with Gag and Gag-Pol expressed from the pI2L2 vector (11). Thus, at least under conditions of overexpression of the helper virus coat proteins, that possibility exists.
Fig 7.
Helper activity of L-A-lus virus for various satellite RNAs. (A) Exclusion of M1 and Mlus maintained by L-A-lus. Killer activity of cytoductants from strain 559 (carrying M1 virions supported by vector pI2L2) into the Klus ski2Δ 1085 strain that harbors L-A-lus and Mlus. Two types of killer halos (K1 and Klus) indicated by the arrows are distinguished. (B) Total RNAs from several colonies shown in panel A were analyzed on an agarose gel. The ethidium bromide (EtBr)-stained gel is shown. Colonies carried either Mlus or M1 dsRNA (indicated by the upper arrows). Mlus and M1 show different mobilities (2.3 and 1.8 kb, respectively) on the gel. The amount of L-A-lus was variable, probably as the result of residual activity of vector pI2L2 in the transient heterokaryons, which produced the cytoductants. The presence of vector pI2L2 may increase the L-A-lus copy number. (C) Maintenance of X, a deletion mutant of L-A, by L-A-lus. RNAs from two colonies of strain 1101 (Table 2) that carried L-A-lus, and into which X virions were introduced by cytoduction, were separated in an agarose gel and transferred to a nylon membrane (lanes 1 and 2). In parallel, RNAs from control strains that carry either L-A (1064, lane 3) or L-A-lus (1089, lane 4) were also loaded in the same gel. The upper panel shows the ethidium bromide-stained gel. The lower panels show two Northern blots hybridized with X (left) or L-A-lus (right) probes. Note that the probe that recognized X (the last 530 nt of L-A) also weakly recognized L-A-lus.
L-A-lus can also maintain X dsRNA, a deletion mutant of L-A virus. The X genome is only 530 bp long (about 12% of the parental molecule length) and contains the first 25 nt of L-A 5′ end, and the rest comes from the 3′ end. X contains the transcription, replication, and encapsidation signals of L-A (6). Strain 455, which carries X and its helper virus L-A, was cytoduced into strain 2928 transformed with plasmid pI2L2, which expresses L-A coat proteins. It is known that L-A proteins expressed from a vector exclude the resident L-A virus in the cell (50). In this way, we constructed a strain that carries X maintained by plasmid pI2L2 without its helper virus L-A. This strain (1101) was later used as the donor for cytoduction into strain 1089, which carries L-A-lus. All cytoductants analyzed (strain 1113) could maintain X stably by L-A-lus (Fig. 7C). Even though X carries the replication signals of L-A, there is no exclusion of X by L-A-lus (as we have shown for its parental virus L-A). Probably due to its small size (0.5 kb), X can be replicated at a much higher rate than the helper virus L-A-lus. All these data together reveal that the new L-A-lus variant identified in this work, though specifically associated with Mlus dsRNA in natural strains, is quite versatile in maintaining different types of satellite RNAs under laboratory conditions. Figure 3B shows the 3′-end sequences and the predicted secondary structures of the satellite RNAs that can be maintained by L-A-lus virus.
The Pol domains of L-A and L-A-lus are exchangeable to produce hybrid virions with helper activity over different satellite dsRNAs.
Because of the similarity between Gag and Pol from L-A and L-A-lus, we wondered whether hybrid virions could function as helper viruses, thus supporting different satellite dsRNAs. So we constructed plasmid pRE1290 as described in Materials and Methods in which the Pol part of L-A was substituted by Pol from L-A-lus. Strain 2928 was first transformed with plasmid pRE1290. Next, M1 virions were cytoduced from strain 559 to construct strain 1118. The resultant cytoductants can maintain stably M1 virions (see Fig. S5 in the supplemental material). Also, the hybrid L-A/L-A-lus virions expressed from pRE1290 could maintain stably M2 or Mlus dsRNAs (Rodriguez-Cousiño and Esteban, unpublished). All these data together suggest that the variable amino acid interval of about 45 amino acids that separates the Gag and Pol domains in each helper virus is likely to play a role only in the spatial separation between the domains. Given that the Pol part of Gag-Pol does not interfere in the assembling of Gag and is maintained in the inside part of the virion during the encapsidation process, the amino acid composition of that interval does not seem to be important in the L-A virus cycle. Plasmid pRE1290 expresses enormous amounts of Gag and hybrid Gag-Pol as checked by Western blot analysis. An unlimited supply of these proteins to form virions may account for the apparent lack of specificity that allows encapsidation and replication of the different satellite RNAs. A different situation occurs in nature, where each satellite RNA is specifically associated with a distinct type of helper virus. In the virions produced from a vector, there is no competition with the helper virus and its satellite RNA, because the L-A parental RNA expressed from the vector is not encapsidated to form stable virions.
L-BC virus, another totivirus in the same yeast strains, shows less variation than L-A.
L-BC is another totivitus evolutionarily related to L-A and frequently present in the same yeast strains that carry L-A. In fact, over 50% of the strains analyzed in Table 2 harbor L-BC. L-BC has no helper activity with respect to any killer-producing satellite dsRNA, and its presence is not associated with any particular phenotype. Because we have found differences of around 25% in the nucleotide sequences of L-A viruses isolated from different yeast killer strain populations, we wondered whether the L-BC virus in these strains also showed similar variation. In contrast to the L-A results, however, we found that L-BC isolated from wine strains shows a lesser degree of variation than the L-BC in laboratory strains. Several partial sequences from RT-PCR products amplified by random priming from strain EX229 (or other wine strains) showed about 90% conservation with respect to the published L-BC sequence from a laboratory strain (Rodriguez-Cousiño and Esteban, unpublished).
DISCUSSION
L-A variants in nature.
In this work, we have identified and characterized a new L-A variant in Klus wine strains. This variant, named L-A-lus, shows nucleotides that are 73% identical to those of the L-A virus in K1 laboratory strains and is the helper virus of Mlus dsRNA. The encoded proteins (Gag or Gag-Pol), however, showed a higher degree of conservation (86%), with certain regions that are more than 95% identical (particularly in the central part of Pol in the Gag-Pol fusion protein, where the RdRp consensus motifs reside) (see Fig. S1B in the supplemental material). We have also obtained preliminary data on the existence of another type of L-A helper virus in K2 wine strains, with a similar degree of conservation (about 75%) at the nucleotide level (see Fig. S4 in the supplemental material). This third L-A variant seems to be specifically associated with the M2 toxin-encoding dsRNA in K2 killer strains. Previous works (13, 51) had already mentioned that the L-A in K2 killer strains was different from the L-A in K1 strains on the basis of hybridization experiments. Also, the major coat proteins in purified virions from both types of L-A showed different size and trypsin-derived peptide maps. The authors called these L-As L2A and L1A, respectively. Field et al. (12) had also reported on the basis of T1 fingerprinting analysis and partial sequencing of selected RNA fragments that the L-A in K1 killers and the L-A in K2 killers were different. No further work on the sequence of L-A in K2 killers was reported from that date. Another variant of L-A called L28 has been also reported in the case of K28 strains (10). Again, this variant was not further characterized. Thus, our data on L-A-lus, preliminary data on L-A-2, and data in the literature suggest that there are several types of L-A totiviruses in nature and that each one seems to be associated specifically with a distinct type of killer toxin-encoding M dsRNA. Also, in the course of this work we have observed variation among L-A sequences within the L-A-lus variant (Rodriguez-Cousiño and Esteban, unpublished). In a few of the Mlus or M-o strains in Table 2, we observed various subtypes of L-A-lus. These subtypes (three have been found so far) bore more conservation among them at the nucleotide level (83% to 85% identity) than was seen with the similarities between L-A and L-A-lus (or L-A-2). The nucleotide sequence of one of these L-A-lus variants is identical to that of a small fragment of L-A from a strain isolated in the Hungarian wine region of Tokaj (GenBank accession no. ABO27241). The encoded proteins (Gag or Gag-Pol) in these L-A-lus subtypes are almost (97% to 98%) identical. The variation within L-A-lus nucleotide sequences resembles more that observed in different populations of the L-BC totivirus (see below). We are characterizing these L-A-lus subtypes more deeply to establish an evolutionary tree within L-A totivirus.
Data presented in this report, and work in progress, suggest that each population of L-A virus has evolved to specifically maintain a distinct type of satellite RNA. Satellite RNAs confer to the host different killer phenotypes and probably provide selective advantages in competing with other nonkiller strains in natural environments. In the evolution of RNA viruses, the lack of proofreading activity of their RdRps plays an important role. In each round of replication, new variants can be generated. From them, only those that maintain essential roles in the virus life cycle would remain. In the case of helper viruses of toxin-producing satellite RNAs (such as the L-A totivirus), the toxin produced by the satellite RNA (in environments where different yeast populations coexist) may be important for a certain population to outcompete the others. In this hypothetical situation, the selective pressure presented by the satellite RNA may have played a major role in selecting mainly those L-A variants that were able to maintain the satellite RNA beneficial for the host. The limit of variation in the helper virus nucleotide sequence would be established by the maximum changes in the RNA genome that permit a protein composition in the virion that is not very different from that of the parental virus (we are seeing about 85 to 90% conservation in Gag and Pol) and by the capacity to encapsidate (and replicate) a specific satellite RNA. If changes in the helper virus sequence result in K− cells, they are likely to be eliminated by others' K+ cells. Once different variants of L-A have been established, we find only one type of them in each strain analyzed due to mutual exclusion. Our data show that the L-A in laboratory K1 strains is the weakest of all L-As so far analyzed, based on its sensitivity to different agents (high temperature or nucleases). This may be the reason why we are not finding any K1 strains in our screening but are finding Klus or K2 killers.
In the absence of helper activity for killer toxin production (in other words, of a selective phenotype), a similar type of totivirus, L-BC, in the same strains shows a much lesser degree (about 90% identity) of variation (Rodriguez-Cousiño and Esteban, unpublished), suggesting that L-BC has apparently evolved at much lower rate than L-A. As mentioned before, it also suggests that the toxin-producing satellite RNAs have played an important role in the evolution of their helper viruses. The origin of these toxin-producing RNAs is not clear. Only in the case of Mlus dsRNA, the ORF for the Klus preprotoxin is ancestrally related to a host chromosomal gene (11). K2 killer toxin is frequently found among wine strains, as shown in this study (Table 2) and others (28). A BLAST search of GenBank (February 2013) for putative ORFs with similarity to the K2 preprotoxin produced no results from laboratory or wine strains of Saccharomyces. However, among other yeasts, the ORF DEHA2G11660p from Debaryomyces hansenii CBS 767 shows a significant (24% identity) homology to K2 preprotoxin. Recently, it was reported that Debaryomyces hansenii carries in its genome an inserted copy of L-A virus. Also, a partial sequence of the D. hansenii DB2008 virus is identical to the inserted sequence (52). In both cases, with 44% identity to the L-A or L-A-lus of S. cerevisiae, a close evolutionary relationship is suggested. Thus, it is likely that the K2-producing M2 dsRNA we find in S. cerevisiae today may have an ancestor in the ORF in Debaryomyces that was somehow encapsidated into L-A virions. These virions in Debaryomyces could later on have been transferred horizontally to Saccharomyces and there evolved to produce the L-A-2 we find now widespread in K2 killer wine strains.
Analysis of L-A-lus sequence and comparison to L-A's.
The average 73% identity between L-A and L-A-lus nucleotide sequences does not show any particular bias throughout the 4.6-kb genomes, with the exception of two regions that are 100% identical, (i) the frameshifting region that facilitates the fusion of Gag and Pol and (ii) the encapsidation signal, about 400 nt upstream of the 3′ end of the (+) strand, indicating the importance of these two cis signals in the virus life cycle (see Fig. S1A in the supplemental material). These two signals are indeed conserved in all L-A populations so far analyzed. A pair of oligonucleotides that anneal specifically to the frameshifting or to the encapsidation signal can amplify by RT-PCR a DNA fragment of about 2.2 kb from any L-A dsRNA (L-A, L-A-lus, L-A-2, etc). In fact, we have been using the strict conservation in these two regions as a means of sequencing new L-A variants. With respect to the 5′- or 3′-end region where important cis signals for replication or transcription are, in general, present in RNA viruses, we did not observe such a strict conservation, in particular at the 3′ ends (Fig. 3B). In the last 20 nt of L-A's 3′ end, a cis signal for replication has been analyzed in detail (6). The 3′ end of L-A-lus, however, does not show a high degree of conservation with respect to L-A's (or to several satellite RNAs) (Fig. 3B). It is likely that the secondary or tertiary structure (more than the sequence itself) plays a role in replication in L-A-lus. In the case of the 5′ ends, L-A and L-A-lus are more conserved. Both are also UA rich, in similarity to other dsRNA virus genomes (Fig. 3A).
At the level of the proteins (Gag or Gag-Pol) encoded by L-A or L-A-lus, as mentioned in Results (see also Fig. S1B in the supplemental material), there is 86% conservation. The central part of Gag is quite similar, probably reflecting structural constraints of Gag to interact to another Gag subunit to form the asymmetric Gag dimer present in the icosahedral L-A virion (with a T = 1 triangulation number). There is, however, a striking variation of several amino acids close to H154, an amino acid involved in an enzymatic reaction within a structural protein (cap-snatching). Like His154, the downstream variable amino acids are likely to be facing the outer surface of the virion and they do not seem to be structurally important. A second stretch of variable amino acids is in the N-terminal one-third of Pol within Gag-Pol (see Fig. S1B in the supplemental material) and is likely to be separating the Gag and Pol domains in the fusion protein. In this protein, the Gag part is anchored in the virion interacting with other Gag subunits, while Pol is facing the inner part of the shell. There, it would be engaged in the transcription or replication activities that take place inside the virions on the L-A dsRNA or L-A (+) strand templates, respectively (Fig. 1A). This variable region of about 45 amino acids, rich in hydrophobic amino acids, was also found in the Gag-Pol encoded by L-A-2 (Rodriguez-Cousiño and Esteban, unpublished), suggesting that it is indeed separating the two domains and not interacting tightly with other amino acids from either Gag or Pol. Hybrid virions expressed from vector pRE1290 (with the Pol and Gag domains from different L-A virus) (see Fig. S5 in the supplemental material) support this claim.
The understanding of the complex interactions between an RNA virus and its killer toxin-producing satellite RNAs, or between them and the metabolism of the host where they reside, offers new insights into the factors involved in the colonization of new ecological niches by yeast strains carrying these viruses. Also of interest is the positive role that the presence of these toxin-producing satellite RNAs plays not only in the host to outcompete other nonkiller strains but in the evolution of the helper virus itself, providing a rich variety of viruses ready for the appearance of new killer toxin-producing RNAs.
Supplementary Material
ACKNOWLEDGMENT
This work has been supported by grant BFU2010-15768 from the Spanish Ministry of Education and Science.
Footnotes
Published ahead of print 31 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00500-13.
REFERENCES
- 1. Icho T, Wickner RB. 1989. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J. Biol. Chem. 264:6716–6723 [PubMed] [Google Scholar]
- 2. Dinman JD, Icho T, Wickner RB. 1991. A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. U. S. A. 88:174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheng RH, Caston JR, Wang GJ, Gu F, Smith TJ, Baker TS, Bozarth RF, Trus BL, Cheng N, Wickner RB, Steven AC. 1994. Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric Gag dimers. J. Mol. Biol. 244:255–258 [DOI] [PubMed] [Google Scholar]
- 4. Ribas JC, Wickner RB. 1998. The Gag domain of the Gag-Pol fusion protein directs incorporation into the L-A double-stranded RNA viral particles in Saccharomyces cerevisiae. J. Biol. Chem. 273:9306–9311 [DOI] [PubMed] [Google Scholar]
- 5. Fujimura T, Esteban R. 2000. Recognition of RNA encapsidation signal by the yeast L-A double-stranded RNA virus. J. Biol. Chem. 275:37118–37126 [DOI] [PubMed] [Google Scholar]
- 6. Esteban R, Fujimura T, Wickner RB. 1989. Internal and terminal cis-acting sites are necessary for in vitro replication of the L-A double-stranded RNA virus of yeast. EMBO J. 8:947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bevan EA, Makower M. 1963. The physiological basis of the killer character in yeast, p 202–203 Geerts SJ. (ed), Genetics today, XIth International Congress of Genetics, vol 1 Pergamon Press, Oxford, United Kingdom [Google Scholar]
- 8. Bevan EA, Herring AJ, Mitchell DJ. 1973. Two species of ds RNA in yeast; their preliminary characterization and relationship to the killer character. Nature 245:81–86 [DOI] [PubMed] [Google Scholar]
- 9. Hannig EM, Leibowitz MJ. 1985. Structure and expression of the M2 genomic segment of a type2 killer virus of yeast. Nucleic Acids Res. 13:4379–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmitt MJ, Tipper DJ. 1990. K28, a unique double-stranded RNA killer virus of Saccharomyces cerevisiae. Mol. Cell. Biol. 10:4807–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodríguez-Cousiño N, Maqueda M, Ambrona J, Zamora E, Esteban R, Ramírez M. 2011. A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded RNA virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 77:1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Field LJ, Bobek LA, Brennan VE, Reilly JD, Bruenn JA. 1982. There are at least two yeast viral double-stranded RNAs of the same size: an explanation for viral exclusion. Cell 31:193–200 [DOI] [PubMed] [Google Scholar]
- 13. El-Sherbeini M, Tipper DJ, Mitchell DJ, Bostian KA. 1984. Virus-like particle capsid proteins encoded by different L double-stranded RNAs of Saccharomyces cerevisiae: their roles in maintenance of M double-stranded killer plasmids. Mol. Cell. Biol. 4:2818–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wickner RB. 1980. Plasmids controlling exclusion of the K2 killer double-stranded RNA plasmid of yeast. Cell 21:217–226 [DOI] [PubMed] [Google Scholar]
- 15. Wickner RB. 1983. Killer systems in Saccharomyces cerevisiae: three distinct modes of exclusion of M2 double-stranded RNA by three species of double-stranded RNA, M1, L-A-E, and L-A-HN. Mol. Cell. Biol. 3:654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wickner RB, Bussey H, Fujimura T, Esteban R. 1995. Viral RNA and the killer phenomenon of Saccharomyces, p 211–226 Kück U. (ed), The mycota, vol II. Genetics and biotechnology. Springer Verlag, Berlin, Germany [Google Scholar]
- 17. Schmitt MJ, Breinig F. 2006. Yeast viral killer toxins: lethality and self-protection. Nat. Rev. Microbiol. 4:212–221 [DOI] [PubMed] [Google Scholar]
- 18. Goto K, Fukuda H, Kichise K, Kitano K, Hara S. 1991. Cloning and nucleotide sequence of the KHS killer gene of Saccharomyces cerevisiae. Agric. Biol. Chem. 55:1953–1958 [PubMed] [Google Scholar]
- 19. Goto K, Iwase T, Kichise K, Kitano K, Totuka A, Obata T, Hara S. 1990. Isolation and properties of a chromosome-dependent KHR killer toxin in Saccharomyces cerevisiae. Agric. Biol. Chem. 54:505–509 [PubMed] [Google Scholar]
- 20. Park CM, Lopinski JD, Masuda J, Tzeng TH, Bruen JA. 1996. A second double-stranded RNA virus from yeast. Virology 216:451–454 [DOI] [PubMed] [Google Scholar]
- 21. Tercero JC, Wickner RB. 1992. MAK3 encodes an N acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J. Biol. Chem. 267:20277–20281 [PubMed] [Google Scholar]
- 22. Polevoda B, Sherman F. 2001. NatC N-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p and Mak31p. J. Biol. Chem. 276:20154–20159 [DOI] [PubMed] [Google Scholar]
- 23. Toh-E A, Guerry P, Wickner RB. 1978. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 136:1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tomecki R, Drazkowska K, Dziembowski A. 2010. Mechanisms of RNA degradation by the eukaryotic exosome. Chembiochem 11:938–945 [DOI] [PubMed] [Google Scholar]
- 25. Larimer FW, Hsu CL, Maupin MK, Stevens A. 1992. Characterization of the XRN1 gene encoding a 5′–>3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene 120:51–57 [DOI] [PubMed] [Google Scholar]
- 26. Esteban R, Vega L, Fujimura T. 2008. 20S RNA narnavirus defies the antiviral activity of SKI1/XRN1 in Saccharomyces cerevisiae. J. Biol. Chem. 283:25812–25820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Philliskirk G, Young TW. 1975. The occurrence of killer character in yeasts of various genera. Antonie Van Leeuwenhoek 41:147–151 [DOI] [PubMed] [Google Scholar]
- 28. Stumm C, Hermans JM, Middelbeek EJ, Croes AF, de Vries GJ. 1977. Killer-sensitive relationships in yeasts from natural habitats. Antonie Van Leeuwenhoek 43:125–128 [DOI] [PubMed] [Google Scholar]
- 29. Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. 2005. Yeast prions URE3 and PSI+ are diseases. Proc. Natl. Acad. Sci. U. S. A. 102:10575–10580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maqueda M, Zamora E, Alvarez ML, Ramírez M. 2012. Characterization, ecological distribution, and population dynamics of Saccharomyces sensu stricto killer yeasts in the spontaneous grape must fermentations of southwestern Spain. Appl. Environ. Microbiol. 78:735–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Drinnenberg IA, Fink GR, Bartel DP. 2011. Compatibility with killer explains rise of RNAi-deficient fungi. Science 333:1592. 10.1126/science.1209575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. 2009. RNAi in budding yeast. Science 326:544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramírez-Garrastacho M, Esteban R. 2011. Yeast RNA viruses as indicators of exosome activity: human exosome hCsl4p participates in RNA degradation in Saccharomyces cerevisiae. Yeast 28:821–832 [DOI] [PubMed] [Google Scholar]
- 34. Somers JM, Bevan EA. 1969. The inheritance of the killer character in yeast. Genet. Res. 13:71–83 [DOI] [PubMed] [Google Scholar]
- 35. Esteban R, Rodríguez-Cousiño N. 2008. 23S RNA-derived replicon as a “molecular tag” for monitoring inoculated wine yeast strains. Yeast 25:359–369 [DOI] [PubMed] [Google Scholar]
- 36. Conde J, Fink GR. 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. U. S. A. 73:3651–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wickner RB, Icho T, Fujimura T, Widner WR. 1991. Expression of yeast L-A double-stranded RNA virus proteins produces derepressed replication: a ski- phenocopy. J. Virol. 65:155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fujimura T, Esteban R. 2011. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc. Natl. Acad. Sci. U. S. A. 108:17667–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fujimura T, Esteban R, Esteban LM, Wickner RB. 1990. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell 62:819–828 [DOI] [PubMed] [Google Scholar]
- 40. Gubler U, Hoffman BJ. 1983. A simple and very efficient method for generating cDNA libraries. Gene 25:263–269 [DOI] [PubMed] [Google Scholar]
- 41. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. Fujimura T, Ribas JC, Makhov AM, Wickner RB. 1992. Pol of gag-pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature 359:746–749 [DOI] [PubMed] [Google Scholar]
- 43. Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360 [DOI] [PubMed] [Google Scholar]
- 44. Zuker M, Mathews DH, Turner DH. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p 11–43 Barciszewski J, Clark BFC. (ed), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 45. Dinman JD, Wickner RB. 1992. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 66:3669–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blanc A, Ribas JC, Wickner RB, Sonenberg N. 1994. His-154 is involved in the linkage of the Saccharomyces cerevisiae L-A double-stranded RNA virus Gag protein to the cap structure of mRNAs and is essential for M1 satellite virus expression. Mol. Cell. Biol. 14:2664–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naitow H, Canady MA, Wickner RB, Johnson JE. 2002. L-A dsRNA virus at 3.4 angstroms resolution reveals particle architecture and mRNA decapping mechanism. Nat. Struct. Biol. 9:725–728 [DOI] [PubMed] [Google Scholar]
- 48. Wickner RB. 1974. “Killer character” of Saccharomyces cerevisiae: curing by growth at elevated temperature. J. Bacteriol. 117:1356–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sommer SS, Wickner RB. 1982. Co-curing of plasmids affecting killer double-stranded RNAs of Saccharomyces cerevisiae: [HOK], [NEX], and the abundance of L are related and further evidence that M1 requires L. J. Bacteriol. 150:545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Valle RP, Wickner RB. 1993. Elimination of L-A double-stranded RNA virus of Saccharomyces cerevisiae by expression of gag and gag-pol from an L-A cDNA clone. J. Virol. 67:2764–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tipper DJ, Bostian KA. 1984. Double-stranded ribonucleic acid killer systems in yeasts. Microbiol. Rev. 48:125–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taylor DJ, Bruenn JA. 2009. The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol. 7:88. 10.1186/1741-7007-7-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.