Abstract
High concentrations of free metal ions in the environment can be detrimental to bacterial survival. However, bacteria utilize strategies, including the activation of stress response pathways and immobilizing chemical elements on their surface, to limit this toxicity. In this study, we characterized LA4131, the HtpX-like M48 metalloprotease from Leptospira interrogans, with a putative role in bacterial stress response and membrane homeostasis. Growth of the la4131 transposon mutant strain (L522) in 360 μM FeSO4 (10-fold the normal in vitro concentration) resulted in the production of an amorphous iron precipitate. Atomic force microscopy and transmission electron microscopy analysis of the strain demonstrated that precipitate production was associated with the generation and release of outer membrane vesicles (OMVs) from the leptospiral surface. Transcriptional studies indicated that inactivation of la4131 resulted in altered expression of a subset of metal toxicity and stress response genes. Combining these findings, this report describes OMV production in response to environmental stressors and associates OMV production with the in vitro activity of an HtpX-like metalloprotease.
INTRODUCTION
Iron is a highly abundant, redox-reactive metal. The redox chemistry of iron allows it to readily transition between Fe (II) and Fe (III) states. This property, combined with its abundance, makes iron a key metal in environmental microbe-metal interactions (1). In order to manage their environment, bacteria have developed the capacity to associate with and precipitate metal ions from aqueous sources (2, 3). Extracellular biogenic iron oxides are amorphous iron precipitates closely associated with cell walls and exopolymers of bacteria and are the most common extracellular metal species affiliated with microorganisms (4, 5). The precipitation of iron on the surface of bacteria resulting in the formation of amorphous iron oxides occurs by both active and passive processes (5).
Active formation of iron precipitates can result from metabolic processes, whereby metabolic byproducts oxidize the metal ions, causing the formation of amorphous minerals (6). An example of active metal ion precipitation is the selective regulation of iron oxyhydroxide formation on the surface of the neutrophilic, Fe (II)-oxidizing bacteria of the genus Leptothrix (6, 7). Although the metabolic pathways involved in this process have not been fully characterized, regulation of iron oxidation by Leptothrix may enhance metabolic energy generation under certain environmental conditions (5).
Passive iron oxide formation is the result of structures at the bacterial surface facilitating the accumulation, nucleation, and precipitation of metal ions (5). Bacterial cell surfaces most commonly have an overall net negative charge resulting from the presence of cell wall components such as polysaccharides (5). Electrostatic interaction between positively charged metal ions and the negatively charged bacterial surface can result in metal adsorption (3, 5). Nucleation and precipitation of adsorbed Fe (III) occur when localized concentrations are sufficiently high at surface reactive sites (5). Precipitates become stabilized at the bacterial surface and serve as sites for further metal aggregation (3). Under extreme conditions, visible flocs of precipitated iron oxyhydroxide can be formed, leading to destabilized outer membranes (OM) and outer membrane vesicle (OMV) formation (2).
Bacteria do not gain any metabolic advantage from passive nucleation events. Instead, it is thought that passive iron oxide formation may represent a survival mechanism to prevent cell death by decreasing the concentration of iron in solution to nontoxic levels (5). Thus, passive nucleation, metal precipitation, and OMV formation represent a putative stress-response mechanism for heavy metal resistance (3).
Outer membrane vesicles are membranous structures derived from the OMs of Gram-negative bacteria. Vesiculation is a ubiquitous process, occurring in both pathogenic and saprophytic organisms during normal growth (8–11). OMVs are primarily composed of soluble periplasmic proteins encased within an OM sheath (8). Roles for OMVs in bacterial pathogenesis have been previously described (12–17). However, functions associated with bacterial survival remain unclear. To date, only a role in environmental stress response has been demonstrated.
Bacterial stress responses can be defined as “a cascade of alterations in gene expression and protein activity for the purpose of surviving extreme and rapidly changing and potentially damaging conditions” (18). The σE proteolysis pathway (19) and Cpx two-component signal transduction system (20) are involved in envelope maintenance, adaptation, and protection in response to environmental stress (21–25). Both pathways regulate the expression and constitutive degradation of misfolded proteins within the cell periplasm. At high temperatures, the protease DegP is transcriptionally regulated by both systems to prevent the accumulation of toxic products (10, 26). Studies have demonstrated induction of OMV production, under high temperatures, in response to degP mutation and associated accumulation of misfolded proteins within the periplasm (8, 10, 26, 27). Multiple proteases, other than DegP, have been identified as key components in both Cpx and σE pathways. Transcription of proteases is regulated by a wide range of environmental stimuli, suggesting, in turn, that OMV production may be stimulated under a range of different conditions.
The Escherichia coli metalloprotease HtpX degrades accumulated and misfolded protein products in both the σE and Cpx pathways (21, 28, 29). Orthologs of HtpX are present in nearly all bacteria. Mutational inactivation of HtpX causes increased thermal sensitivity, growth retardation, abnormal protein translocation, accumulation of misfolded products, and altered surface adhesiveness, cellular morphology, and surface antigen expression (28–31). This suggests that HtpX plays a central role in maintaining the various functions of the outer membrane.
In this paper, we present a phenotypic analysis of a strain of Leptospira interrogans in which the gene encoding LA4131, an OM-associated HtpX-like metalloprotease, has been insertionally inactivated. In particular, we show that this strain is defective in the normal active processes used by Leptospira to manage high concentrations of extracelluar soluble iron. Mass spectrometry (MS), atomic force microscopy (AFM), and transmission electron microscopy (TEM) analysis of the mutant demonstrated the production of an amorphous iron precipitate associated with the generation and release of OMVs from the leptospiral surface. Combined, these data indicate that passive nucleation stress-response processes are induced in direct response to inactivation of a HtpX-like metalloprotease. This report extends the repertoire of functions maintained by HtpX and adds to an existing body of work that shows that HtpX metalloproteases are essential for the normal function of the bacterial outer membrane.
MATERIALS AND METHODS
Bioinformatics analysis.
The nucleotide and deduced amino acid sequences of la4131 were obtained from GenBank (NP_714311) (Table 1). Identification of conserved domains was conducted with RPS-BLAST (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). A cellular location for LA4131 was predicted using a combination of pSORTB (32, 33), SignalP (34), LipoP (35), SpLip (36), and SecretomeP (37, 38). Protein membrane topology was predicted using ConPredII (39).
Table 1.
Database sources for comparative sequence analysis
| Gene | Species/strain | GenBank accession no. | Reference |
|---|---|---|---|
| la4131 | L. interrogans sv Lai | AAN51329 | 48 |
| htpX | E. coli | AAA62779 | 62 |
| lic13293 | L. interrogans sv Copenhageni | AE016823 | 47 |
| lman3580 | L. interrogans sv Manilae | AHPU00000000 | 77 |
| lip0012 | L. interrogans sv Pomona | NZ_AFLT02000043 | |
| L. biflexa sv Patoc | CP000777 | 78 | |
| L. borgpetersenii sv Hardjobovis | CP000348 | 66 |
Bacterial strains and culture conditions.
Random transposon mutagenesis of L. interrogans serovar (sv) Lai strain L56601 (designated L521 in this study) was described previously (40). Disruption of la4131 in la4131 mutant clone L56 (designated L522 in this study) was confirmed by direct sequencing from genomic DNA as described by Murray et al. (41). Leptospires were routinely cultured at 30°C in EMJH medium (Becton, Dickinson and Co., Sparks, MD) with kanamycin (50 μg/ml) where appropriate. Based on the observation of phenotypic variation in particular lots of medium, lots 701688 and 8170035 were selected for iron phenotype experiments. Supplementation with 360 μM FeSO4 was used, where specified, for the cultivation of strains during iron precipitation investigations.
ICP-MS.
Inductively coupled plasma mass spectrometry (ICP-MS) is a technique for quantification of analytes to subnanogram concentrations. Isotopic fingerprints display all constituent elemental isotopes at their respective atomic masses; each peak height represents the quantitative number (counts) of ions detected for each isotope (42). Four stable isotopes of iron (Fe54, Fe56, Fe57, and Fe58) (43) can be resolved by ICP-MS. Three replicate 50-ml cultures of L521 and L522 were cultivated at 30°C in EMJH medium with or without iron supplementation until a density of 1 × 109 cells/ml was reached. The cultures were pelleted and washed twice at 8,500 × g for 15 min and resuspended in 10 ml of EMJH base solution (Becton, Dickinson and Co., Sparks, MD). The washed cell pellets were resuspended in 3 ml of 1% nitric acid to dissolve organic and inorganic cellular material. Inductively coupled plasma mass spectroscopy was conducted at the Monash University School of Geosciences (Clayton, Victoria, Australia) (44).
Atomic force microscopy (AFM).
Cultures of L. interrogans were grown to a density of 5 × 108 cells/ml in 5 ml of EMJH medium with or without iron supplementation. One milliliter of culture was removed and centrifuged at 5,000 × g for 15 min. The culture supernatant was removed and the pellet carefully resuspended in 100 μl sterile deionized H2O. The culture was pelleted at 5,000 × g for 15 min, and the cells were resuspended in sterile deionized H2O to a final density of 1 × 107 cells/ml. A 10-μl aliquot of the diluted cell suspension was air dried onto heat-treated glass microscope slides under aseptic conditions.
Surface characterization was undertaken in air with a PicoPlus atomic force microscope (AFM) interfaced with a Picoscan 3000 controller (Molecular Imaging Inc.). A silicon cantilever (Ultrasharp, NSC15/AIBS; MikroMasch) was used with a typical spring constant of 40 N/m in tapping and phase-contrast modes. The resulting images were analyzed using WS×M 4.0 Develop 11.6 software. AFM imaging was performed twice on independently prepared samples.
Transmission electron microscopy (TEM).
Cultures of L. interrogans were grown in EMJH medium with or without iron supplementation, pelleted, and resuspended in sterile deionized H2O to a final density of 1 × 107 cells/ml. Paired samples of cells were fixed onto Formvar/carbon-coated grids for 10 min and washed for 5 min with sterile deionized water. One grid from each pair was negatively stained with 2% phosphotungstic acid (pH 7.0) for 30 s to serve as a stained-image control. Samples were examined with a Hitachi H7500 electron microscope (Hitachi High Technologies America, Pleasanton, CA).
Microarray analysis.
Three biological replicate RNA samples from L. interrogans sv Lai (L521) and the L. interrogans sv Lai la4131 mutant (L522) were grown in 100 ml EMJH medium with or without iron supplementation. Cells were harvested at a density of between 4 × 108 and 7 × 108 cells/ml. RNA purification, reverse transcription, microarray construction, cDNA hybridization, and array image analysis were conducted as described previously (45). All conditions tested were cross-compared.
RESULTS
LA4131 is a putative outer membrane M48 metalloprotease.
The la4131 gene of L. interrogans sv Lai strain L55601 consists of 1,977 nucleotides, encoding a protein of 659 amino acids. The distribution of la4131 in the genus Leptospira is unusual, with no ortholog identified in the genome of the bovine pathogen L. borgpetersenii sv Hardjobovis L550 strain (GenBank accession no. CP000348). However, a protein sharing 28% similarity was identified in the saprophyte L. biflexa sv Patoc Patoc1 strain (GenBank accession no. CP000777). Orthologs were also identified in isolates from the pathogenic species L. interrogans, L. kirschneri, L. kmetyi, and L. noguchii as well as intermediate species L. inadai, L. broomii, and L. licerasiae (47–49).
RPS-BLAST analysis indicated that LA4131 contains an M48 superfamily domain (pfam01435) between residues 75 and 264; this M48 domain has a conserved zinc-binding motif, HELSH, between residues 143 and 147. These features are consistent with the identification of LA4131 as a putative bacterial metalloprotease. Analysis of the peptide sequence using SpLip, LipoP, and SignalP predicted that LA4131 has a leader sequence with a signal peptidase 1 cleavage site between residues 22 and 23. Thus, LA4131 is predicted to be localized in the OM. Beta strands and coiled regions were predicted at the C-terminal end of the protein after residue 530, further supporting an OM localization (50).
Inactivation of la4131 results in growth medium-dependent iron precipitate production.
Strain L522 was derived from L. interrogans sv Lai strain L56601 (40). The transposon inserted at nucleotide 178 of the coding region of la4131. Quantitative reverse transcription-PCR (qRT-PCR) comparisons between the parent L521 strain and mutant L522 measured an 8-fold reduction (P < 0.01) in transcript levels after the point of transposon insertion in la4131 (data not shown).
No difference between the parent and mutant strains in growth rates was observed after routine culture in EMJH medium. However, with EMJH medium containing a high level of iron (360 μM FeSO4), phenotypic differences between L522 and the parental strain (L521) were observed; an orange precipitate formed in cultures of the la4131 mutant (L522) but not in those of the parent (L521). The formation of this precipitate was associated with an increase in cell pellet size; when 1 ml of culture (standardized for cell density) was centrifuged, the cell pellet sizes ranged from 100 μl to 250 μl. The wild-type (WT) strain consistently produced a pellet of 100 μl (Fig. 1A). We were alerted to the phenotype of L522 during routine culture in EMJH medium, with some cultures forming the orange precipitate. In EMJH medium not producing a phenotypic difference, such as lot 8170035, supplementing the medium with a final concentration of 360 μM FeSO4 induced an identical phenotypic difference. Precipitate production by the WT strain was not observed under the conditions tested. We note that iron concentration is not reported as part of the manufacturer's specifications sheet for EMJH medium.
Fig 1.
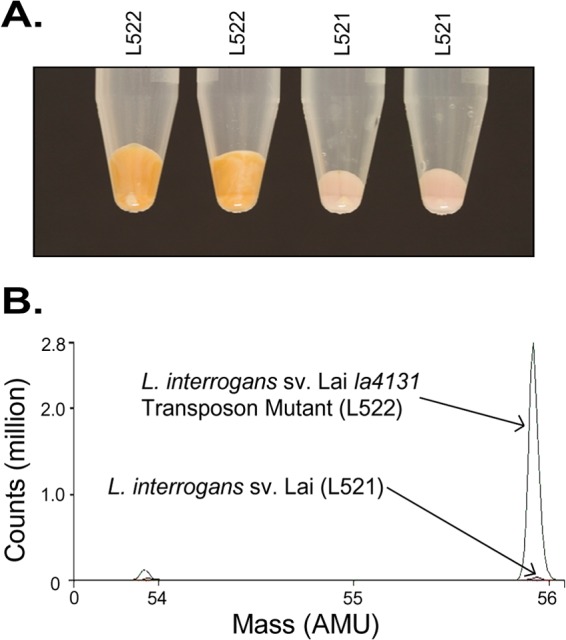
Iron-associated phenotypic difference between the L. interrogans sv Lai la4131 mutant (L522) and the L. interrogans sv Lai (L521) parent strain after culture in EMJH lot 701688. Cell pellets were prepared from 5-ml cultures of L521 and L522 pelleted at 4,000 × g for 5 min. (A) Overlaid mass spectra generated using inductively coupled plasma mass spectrometry (ICP-MS) conducted on pelleted samples of L522 and L521. (B) The spectra in the region of 53.6 to 56.1 atomic mass units (AMU) are shown.
ICP-MS was used to examine the metal and trace element isotope composition of cell pellets from cultures in which the phenotypic difference was observed. Figure 1B shows the ICP-MS spectra between 53.6 and 56.1 atomic mass units (AMU). Differences were observed in isotope Fe56, with a final ion count of >2,760,000 for L522 in comparison to <50,000 measured for the parent strain. The results indicated that the precipitate produced by the mutant contained a higher concentration of iron than the parent strain precipitate. Across the spectrum, no other difference in metal ion profiles was observed.
la4131 mutation causes surface association of precipitated iron and vesicle formation.
Iron acquisition by bacterial cells is an active process mediated by proteins in the OM. Iron precipitate production by the la4131 mutant may affect, or be the result of, alterations in OM architecture. To determine if L522 displayed altered surface characteristics, two imaging techniques, atomic force microscopy (AFM) and transmission electron microscopy (TEM), were employed.
AFM produces topographical images of bacterial cell surface structures. Highly viscous materials, such as outer membrane vesicles (OMVs), are identified as bright or illuminated features by phase-contrast AFM (51). By AFM, cells of the parental L521 strain, cultured under conditions of high or normal concentrations of iron, exhibited typical leptospiral features, such as helical morphology, periplasmic flagella, and OMVs (Fig. 2A and B). Vesicles were primarily associated with the periphery of the cell. Characterization of membranous features as OMVs was based on morphological comparison to surface structures previously observed in leptospiral TEM images (52, 53) (Fig. 2C). The mutant L522 strain cultured with normal iron concentrations displayed features similar to those observed for strain L521. In contrast, substantial differences in cell surface characteristics were observed for L522 cells cultured under conditions of high iron concentrations (Fig. 2D and E); these were associated with an array of extracellular materials (Fig. 2E). In comparison to the parent strain (Fig. 2C), L522 cells had an illuminated surface indicative of increased surface viscosity consistent with excessive membrane blebbing (Fig. 2F). Thus, the absence of a functional la4131 gene caused an overall increase in surface viscosity and accumulation of precipitated material on the cell periphery.
Fig 2.
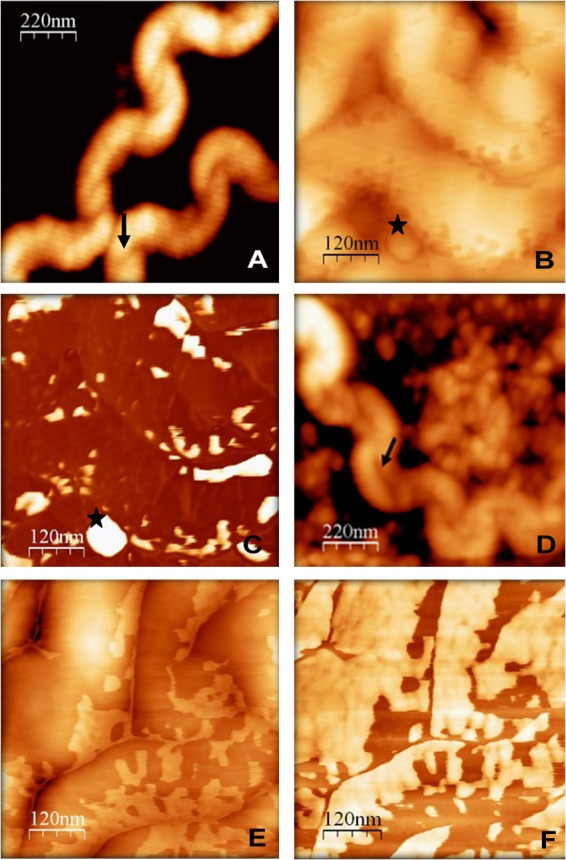
Micrographs generated using atomic force microscopy (AFM) showing the surface characteristics of L. interrogans sv Lai strain L521 (A, B, and C) and L. interrogans sv Lai la4131 mutant L522 (D, E, and F). Bacteria were grown in BD EMJH lot 8170035 supplemented with 360 μM FeSO4. Cultures of the la4131 mutant (D) show the presence of cell-associated material. Images were acquired in tapping (A, B, D, and E) or phase-contrast (C and F) modes. Increased viscoelasticity is shown as light coloration on phase-contrast images. Cultures of the la4131 mutant showed overall increased surface viscosity. Stars (★) indicate outer membrane vesicles. Arrows (↑) indicate flagella. The fine white lines that occur in the AFM images are image-processing artifacts due to the line-by-line leveling process.
TEM images of unstained L521 cells showed no electron-dense material associated with the cell irrespective of the growth medium (Fig. 3A and B). In contrast, electron-dense material was observed associated with the L522 mutant cultured under conditions of high and normal iron concentrations (Fig. 3C and D). At normal EMJH concentrations of FeSO4 (36 μM), electron-dense material was closely associated at the junction between aggregated cells (Fig. 3C). Iron precipitates were not observed on the surface of singular separate leptospires. At high iron concentrations, there was a large increase in the amount of electron-dense material in L522 cultures (Fig. 3D). Precipitates were associated with both single and aggregated L522 cells. The production and release of OMVs from the cell surface were also increased under high iron conditions. The majority of electron-dense material was associated with the L522 cell periphery and the released OMVs. However, a proportion of the precipitate was clumped in the extracellular milieu and no longer associated with the cells, consistent with the hypothesis that mutation of la4131 increased iron association and passive precipitation at the surface of L522.
Fig 3.
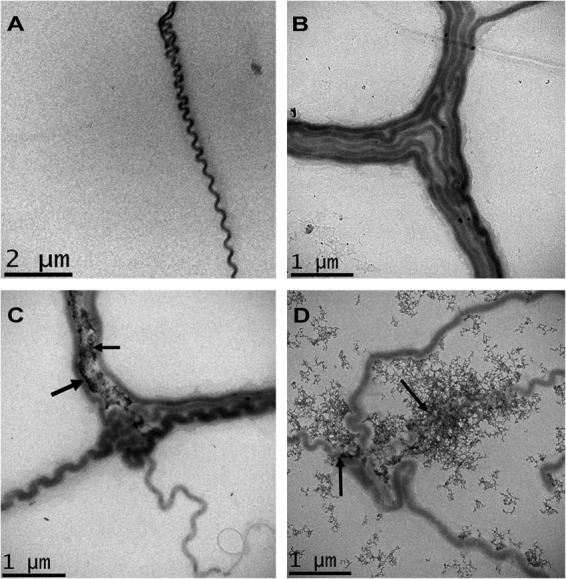
Transmission electron microscopy of L. interrogans strain L521 (A and B) and L. interrogans sv Lai la4131 mutant L522 (C and D) cultured in EMJH medium or EMJH medium supplemented with 360 μM FeSO4. The bacteria were grown in BD EMJH lot 8170035 (A and C) and BD EMJH lot 8170035 supplemented with 360 μM FeSO4 (B and D). Arrows indicate accumulated electron-dense material.
Effect of la4131 mutation on global gene regulation.
The effect of la4131 mutation was assessed by microarray. Mutation of la4131 combined with growth at normal iron levels resulted in the differential expression of 11 genes (Table 2) in comparison to L521 cultured under the same conditions. Of these, the protein products of only three genes, namely, GTP pyrophosphokinase SpoT (LA3085), leptospiral endostatin-like protein LenD (LA1433; binds to laminin and fibronectin of host organisms [54]), and a methyl-accepting chemotaxis protein (MCP) (SPN3218), had known or predicted functions.
Table 2.
Comparison of global gene expression characteristics of strains L521 and L522
| Strain comparison and locus tag | Fold differencea | Gene | Gene product | Locationb | COGc |
|---|---|---|---|---|---|
| L522 vs L521—both normal iron | |||||
| LA2066 | −6.5 | Hypothetical protein | |||
| LA3085 | −6.3 | spoT | GTP pyrophosphokinase | Cyt | T, K |
| LA2065 | −3.9 | Hypothetical protein | Cyt | ||
| LA2013 | −3.8 | Hypothetical protein | |||
| LA2020 | −3.0 | Hypothetical protein | |||
| LA1762 | −2.3 | Hypothetical lipoprotein | OM | ||
| LA1433 | −2.2 | lenD | Leptospiral endostatin-like protein, LenD | OM | |
| LA1761 | −2.0 | Hypothetical lipoprotein | |||
| LA3145 | +2.0 | Hypothetical protein | S | ||
| LA0598 | +2.1 | Hypothetical protein | S | ||
| SPN3218 | +2.3 | Methyl-accepting chemotaxis protein | CM | N, T | |
| L522 vs L521—both high iron | |||||
| LA2065 | −5.5 | Hypothetical protein | Cyt | ||
| LA2066 | −5.4 | Hypothetical protein | |||
| LA1844 | −2.1 | Hypothetical protein | |||
| LA0189 | +2.1 | Mu-like bacteriophage protein | Cyt | R | |
| LA0802 | +2.1 | TPR repeat-containing protein | N, U | ||
| LA3145 | +2.2 | Hypothetical protein | S | ||
| LA3926 | +2.3 | AcrB family efflux pump | CM | V | |
| LA1369 | +2.4 | Hypothetical protein | |||
| LA2110 | +2.4 | bolA | Putative transcriptional regulator, BolA | Cyt | T |
| LA3018 | +2.5 | Hypothetical protein | CM | R | |
| LA1879 | +2.5 | clpB | Endopeptidase ClpB | Cyt | O |
| LA0593 | +4.0 | copZ | Copper chaperone | Cyt | P |
| LA1124 | +6.1 | lnt-1 | Apolipoprotein N-acyltransferase | CM | M |
| L522 normal iron vs L522 high iron | |||||
| LA0634 | −5.7 | Putative permease | CM | E, P | |
| LA3781 | −2.7 | Hypothetical lipoprotein | |||
| LA3780 | −2.5 | Hypothetical lipoprotein | |||
| LA4128 | −2.5 | Hypothetical protein | |||
| LA3778 | −2.3 | ligB | LigB lipoprotein | N | |
| LA1072 | −2.3 | d-Alanine d-alanine ligase | H, J | ||
| LA1027 | −2.1 | Sphingomyelinase C precursor | OM | R | |
| LIC12947 | −2.1 | Putative endoflagellum-like protein | N | ||
| LA2200 | −2.1 | Putative amidase | M | ||
| LB105 | −2.0 | Putative methyltransferase | |||
| LB107 | −2.0 | Putative ferredoxin | C | ||
| LA0863 | +2.0 | Putative acetolactate synthase | E, H | ||
| LA0212 | +2.2 | Hypothetical protein | |||
| LA_SPN1928 | +2.4 | Hypothetical lipoprotein | |||
| LA0593 | +2.7 | copZ | Copper chaperone | Cyt | P |
| LA3018 | +2.8 | Hypothetical protein | CM | R | |
| LA0980 | +3.5 | thiC | Thiamine biosynthesis protein, ThiC | Cyt | H |
| LA3017 | +3.8 | Hypothetical lipoprotein | S | ||
| LA3016 | +4.1 | Hypothetical protein | |||
| L521 normal iron vs L521 high iron | |||||
| LA2200 | −3.1 | Putative amidase | M | ||
| LA0101 | −2.8 | Hypothetical protein | |||
| LA3781 | −2.5 | Hypothetical lipoprotein | |||
| LA3778 | −2.5 | ligB | LigB lipoprotein | N | |
| LA0815 | −2.3 | Signal transduction histidine kinase | CM | T | |
| LA0100 | −2.2 | Hypothetical protein | CM | ||
| LA1027 | −2.1 | Sphingomyelinase C precursor | OM | R | |
| LA_SPN1928 | +2.1 | Hypothetical lipoprotein | |||
| LA1468 | +2.5 | Hypothetical lipoprotein | CM |
Differentially expressed genes have at least a 2-fold difference in gene expression at a confidence level of 95%.
Cellular localization predicted using PSORTB. Localization abbreviations: Cyt, cytoplasmic; CM, cytoplasmic membrane; Per, periplasmic; OM, outer membrane; Ext, extracellular.
Cluster of orthologous group (COG) categories (79). COG categories were as follows: cellular processing and signaling (COGs D, Y, V, M, T, N, Z, W, U, and O), metabolism (COGs C, G, E, F, H, I, P, and Q), information storage (COGs J, A, K, L, and B), and processing or poorly characterized (COGs R and S).
Comparison of the mutant strain, L522, to the parent strain, L521, in the presence of increased FeSO4 identified 13 differentially expressed genes. Of these genes, the protein products of five were orthologous to those of known bacterial cellular stress response genes (Table 2) (55–59). Comparison of levels of gene expression in the L522 mutant grown under normal and high iron conditions identified 19 differentially expressed genes; 10 of these genes had annotated functions (Table 2). The same comparison performed with the parental L521 strain showed reduced expression for la3778, la1027, and la2200; these same three genes followed the same pattern of expression in the L522 mutant strain. These genes encode potential virulence factors, LigB (la3778), sphinomyelinase C (la1027), and a putative amidase (la2200). Thus, the differential expression of these genes is related to iron concentration and is independent of the la4131 mutation.
DISCUSSION
This study has investigated a transposon mutant of a putative HtpX-like metalloprotease, LA4131, and examined the phenotypic alterations associated with in vitro iron concentrations. Sequence similarity analysis demonstrated that LA4131 is most related to the M48 family of metalloproteases. M48 metalloproteases share similarity with heat shock proteins, such as E. coli HtpX and Saccharomyces cerevisiae Ste24p. These heat shock proteins respond to changes in temperature, exposure to UV irradiation, bacteriophage infection, and the presence of accumulated misfolded proteins (60–65). The proposed role of M48 metalloproteases is in the degradation of misfolded intracellular proteins (21, 28, 31, 62).
Genes encoding protein orthologs of the HtpX-like metalloprotease LA4131 were identified within the available sequenced genomes of pathogenic leptospires, with the exception of the bovine pathogen L. borgpetersenii. It has been suggested that L. borgpetersenii is undergoing a process of genome reduction (66) centered on the inactivation of genes associated with environmental sensing, metabolite transport, and utilization. It can thus be hypothesized that the absence of a LA4131 ortholog, a protein with a putative role in environmental stress response, is a direct result of these processes. The ortholog identified within the saprophyte L. biflexa shares 28% similarity with LA4131. However, based on protein topology, the saprophytic protein is more similar to classical HtpX metalloprotease than that of LA4131, suggesting that LA4131 may play a unique but related function within pathogenic leptospires.
The N-terminal signal peptide is essential for protein translocation and localization of the E. coli M48 metalloprotease, HtpX, to the inner membrane (IM) (21). Characterized HtpX proteins have two to four transmembrane helices that aid integration into the IM after protein translocation (21, 22, 28). Despite regions of local similarity to the E. coli HtpX, it is apparent that the leptospiral LA4131 has a different membrane topology. LA4131 contains an N-terminal signal peptide, but the presence of small hydrophobic residues preceding the predicted signal peptidase I cleavage site indicates that this protein may be translocated across the IM by SecII-dependent processes and targeted to the OM. Supporting this notion, and in contrast to the E. coli HtpX, LA4131 has no predicted transmembrane helices but rather predicted C-terminal β-structures, consistent with localization in the OM. The OM localization is further supported by the MudPit analysis by Lo et al. (67) that found LA4131 located exclusively in the TX-114 extracted OM fractions from L. interrogans. Combined, our data suggest that L4131 is different from other M48 metalloproteases in that LA4131 is the first to have a predicted OM localization.
Generally, M48 metalloproteases have roles in the maintenance of membrane homeostasis, specifically in the removal of misfolded, nonfunctional proteins. Thus, phenotypic changes are likely to be indirect and due to interruption of normal membrane operation. Under high iron conditions, mutational inactivation of la4131 resulted in a phenotype where precipitated iron was associated with the bacterial surface along with the production of OM blebs and the subsequent release of outer membrane vesicles (OMVs). Thus, it is hypothesized that membrane-associated proteins, used in the management of excessive environmental iron concentrations, were not able to function normally in the LA4131 mutant due to the absence of a functional M48 metalloprotease to enable the appropriate reconfiguration of the membrane, resulting in OMV formation. This leads to the suggestion that M48 metalloproteases may play a role in facilitating the rapid retooling of membranes to suit prevailing environmental conditions.
The control of the intracellular iron concentration at below toxic levels is essential for life. The phenotype observed in the mutant is indicative of the mechanism used to maintain intracellular iron below toxic concentrations. In the LA4131 mutant, under high iron conditions, an alternative passive mechanism for management of intracellular iron concentrations is activated.
Examination of the passive management of iron concentration observed in the LA4131 mutant suggested that the destabilization of the OM and the sloughing of OMVs are likely to be part of a more generalized mechanism used by bacteria to manage intracellular toxicity. Vesiculation can function as part of cellular stress response system by aiding bacterial survival through specific enrichment and release of toxic compounds from the cell (2, 9, 68–70). However, the mechanism of OMV formation and release is not well characterized. McBroom and Kuehn (10) investigated defined mutants of E. coli Cpx and σE stress response pathway proteins and found a positive correlation between vesicle production, intracellular concentration of accumulated misfolded proteins, and the impairment of stress response mechanisms. Our report furthers this work and is the first to demonstrate a correlation between vesicle production and accumulation of surface iron due to the impairment of the activity of the LA4131 metalloprotease.
Transcription analysis allowed us to hypothesize a molecular basis for the phenotype observed in the LA4131 mutant. The reduced expression of the transcriptional regulator spoT (la3085), which is upregulated under conditions of iron limitation (71), is consistent with an oversupply of iron. There is enhanced transcription of cell homeostasis and stress response genes, such as that encoding the methyl-accepting chemotaxis protein (MCP), SPN3218. Expression of MCPs enables bacteria to detect extracellular repellents or attractants, alter motility gene expression, and thus move toward a more favorable environment (72), such as in this case, perhaps facilitating migration toward a lower iron concentration.
The enhanced expression of membrane oxidative stress response genes bolA and clpB (58, 73) in conjunction with lnt-1 may directly enhance OMV formation (10). The production of vesicles would result in an overall reduction of the surface iron concentration (9), which in turn would alleviate some of the associated intracellular iron toxicity. Furthermore, upregulation of the expression of copZ and thiC with a concomitant reduction in la0634, encoding a putative metal transport protein, may aid in further limiting the heavy metal toxicity and promote cell survival (74–76).
LA4131 is a member of the HtpX metalloprotease family of stress response proteins. We suggest that this protein has a cellular location in the OM that sets it apart from other members of this family of proteins, although its cellular role is likely to be similar to that of other M48 metalloproteases. This investigation of LA4131 under high iron conditions highlights the complexity of bacterial stress response pathways and their ability to compensate for loss of the activity of one of their component proteins in order to enable survival.
ACKNOWLEDGMENTS
This work was supported by the National Health and Medical Research Council and the Australian Research Council, Canberra, Australia, and in part by the Commonwealth of Australia under the International Science Linkages program and is related to the Australian-European integrated FP6 CHARPAN and FP7 BISNES projects.
Footnotes
Published ahead of print 24 May 2013
REFERENCES
- 1. Lovely DR. 2000. Fe(III) and Mn(IV) reduction, p 3–30 Lovely DR. (ed), Environmental microbe-metal interactions. ASM Press, Washington, DC [Google Scholar]
- 2. Beveridge TJ. 1989. Role of cellular design in bacterial metal accumulation and mineralization. Annu. Rev. Microbiol. 43:147–171 [DOI] [PubMed] [Google Scholar]
- 3. Southam G. 2000. Bacterial surface-mediated mineral formation, p 257–276 Lovely DR. (ed), Environmental microbe-metal interactions. ASM Press, Washington DC [Google Scholar]
- 4. Chatellier X, Fortin D, West MM, Leppard GG, Ferris FG. 2001. Effect of the presence of bacterial surfaces during the synthesis of Fe oxides by oxidation of ferrous ions. Eur. J. Mineral. 13:705–714 [Google Scholar]
- 5. Fortin D, Langley S. 2005. Formation and occurrence of biogenic iron-rich minerals. Earth Sci. Rev. 72:1–19 [Google Scholar]
- 6. Frankel RB, Bazylinski DA. 2003. Biologically induced mineralization by bacteria. Rev. Mineral Geochem. 54:95–114 [Google Scholar]
- 7. Suzuki T, Hashimoto H, Ishihara H, Kasai T, Kunoh H, Takada J. 2011. Structural and spatial associations between Fe, O, and C in the network structure of the Leptothrix ochracea sheath surface. Appl. Environ. Microbiol. 77:7873–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwechheimer C, Sullivan CJ, Kuehn MJ. 2013. Envelope control of outer membrane vesicle production in gram-negative bacteria. Biochemistry 52:3031–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macdonald IA, Kuehn MJ. 2012. Offense and defense: microbial membrane vesicles play both ways. Res. Microbiol. 163:607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63:545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188:5385–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, Wai SN. 2008. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 70:100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berlanda Scorza F, Doro F, Rodriguez-Ortega MJ, Stella M, Liberatori S, Taddei AR, Serino L, Gomes Moriel D, Nesta B, Fontana MR, Spagnuolo A, Pizza M, Norais N, Grandi G. 2008. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DeltatolR IHE3034 mutant. Mol. Cell. Proteomics 7:473–485 [DOI] [PubMed] [Google Scholar]
- 14. Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, Park KS, Lee JO, Kim YK, Kwon KH, Kim KP, Gho YS. 2007. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7:3143–3153 [DOI] [PubMed] [Google Scholar]
- 15. Iwami J, Murakami Y, Nagano K, Nakamura H, Yoshimura F. 2007. Further evidence that major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis stabilize bacterial cells. Oral Microbiol. Immunol. 22:356–360 [DOI] [PubMed] [Google Scholar]
- 16. Button JE, Silhavy TJ, Ruiz N. 2007. A suppressor of cell death caused by the loss of sigmaE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J. Bacteriol. 189:1523–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alaniz RC, Deatherage BL, Lara JC, Cookson BT. 2007. Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. J. Immunol. 179:7692–7701 [DOI] [PubMed] [Google Scholar]
- 18. Giuliodori AM, Gualerzi CO, Soto S, Vila J, Tavio MM. 2007. Review on bacterial stress topics. Ann. N. Y. Acad. Sci. 1113:95–104 [DOI] [PubMed] [Google Scholar]
- 19. Barchinger SE, Ades SE. 2013. Regulated proteolysis: control of the Escherichia coli sigma(E)-dependent cell envelope stress response. Subcell. Biochem. 66:129–160 [DOI] [PubMed] [Google Scholar]
- 20. Dorel C, Lejeune P, Rodrigue A. 2006. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res. Microbiol. 157:306–314 [DOI] [PubMed] [Google Scholar]
- 21. Shimohata N, Chiba S, Saikawa N, Ito K, Akiyama Y. 2002. The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7:653–662 [DOI] [PubMed] [Google Scholar]
- 22. Huang Y, Zhang B, Dong K, Zhang X, Hou L, Wang T, Chen N, Chen S. 2007. Up-regulation of yggG promotes the survival of Escherichia coli cells containing Era-1 mutant protein. FEMS Microbiol. Lett. 275:8–15 [DOI] [PubMed] [Google Scholar]
- 23. Lüdke A, Kramer R, Burkovski A, Schluesener D, Poetsch A. 2007. A proteomic study of Corynebacterium glutamicum AAA+ protease FtsH. BMC Microbiol. 7:6. 10.1186/1471-2180-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanehara K, Ito K, Akiyama Y. 2002. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 16:2147–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 16:2156–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McMahon KJ, Castelli ME, Garcia Vescovi E, Feldman MF. 2012. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J. Bacteriol. 194:3241–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spiess C, Beil A, Ehrmann M. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339–347 [DOI] [PubMed] [Google Scholar]
- 28. Sakoh M, Ito K, Akiyama Y. 2005. Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli. J. Biol. Chem. 280:33305–33310 [DOI] [PubMed] [Google Scholar]
- 29. Akiyama Y. 2009. Quality control of cytoplasmic membrane proteins in Escherichia coli. J. Biochem. 146:449–454 [DOI] [PubMed] [Google Scholar]
- 30. Marciniak BC, Trip H, Fusetti F, Kuipers OP. 2012. Regulation of ykrL (htpX) by Rok and YkrK, a novel type of regulator in Bacillus subtilis. J. Bacteriol. 194:2837–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vickerman MM, Mather NM, Minick PE, Edwards CA. 2002. Initial characterization of the Streptococcus gordonii htpX gene. Oral Microbiol. Immunol. 17:22–31 [DOI] [PubMed] [Google Scholar]
- 32. Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617–623 [DOI] [PubMed] [Google Scholar]
- 33. Rey S, Acab M, Gardy JL, Laird MR, deFays K, Lambert C, Brinkman FS. 2005. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 33:D164–D168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 35. Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Setubal JC, Reis M, Matsunaga J, Haake DA. 2006. Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. 2004. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17:349–356 [DOI] [PubMed] [Google Scholar]
- 38. Tjalsma H. 2007. Feature-based reappraisal of the Bacillus subtilis exoproteome. Proteomics 7:73–81 [DOI] [PubMed] [Google Scholar]
- 39. Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. 2004. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32:W390–W393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bourhy P, Louvel H, Saint Girons I, Picardeau M. 2005. Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J. Bacteriol. 187:3255–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murray GL, Ellis KM, Lo M, Adler B. 2008. Leptospira interrogans requires a functional heme oxygenase to scavenge iron from hemoglobin. Microbes Infect. 10:791–797 [DOI] [PubMed] [Google Scholar]
- 42. PerkinElmer 2011. The 30-minute guide to ICP-MS. http://www.perkinelmer.com/CMSResources/Images/44-74849tch_icpmsthirtyminuteguide.pdf
- 43. Dauphas N, Rouxel O. 2006. Mass spectrometry and natural variations of iron isotopes. Mass Spectrom. Rev. 25:515–550 [DOI] [PubMed] [Google Scholar]
- 44. Balcia N, Bullenb TD, Witte-Lienc K, Shanksd WC, Motelicae M, Mandernacka KW. 2006. Iron isotope fractionation during microbially stimulated Fe(II) oxidation and Fe(III) precipitation. Geochim. Cosmochim. Acta 70:622–639 [Google Scholar]
- 45. Lo M, Bulach DM, Powell DR, Haake DA, Matsunaga J, Paustian ML, Zuerner RL, Adler B. 2006. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 74:5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reference deleted.
- 47. Nascimento AL, Verjovski-Almeida S, Van Sluys MA, Monteiro-Vitorello CB, Camargo LE, Digiampietri LA, Harstkeerl RA, Ho PL, Marques MV, Oliveira MC, Setubal JC, Haake DA, Martins EA. 2004. Genome features of Leptospira interrogans serovar Copenhageni. Braz. J. Med. Biol. Res. 37:459–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888–893 [DOI] [PubMed] [Google Scholar]
- 49. Ricaldi JN, Fouts DE, Selengut JD, Harkins DM, Patra KP, Moreno A, Lehmann JS, Purushe J, Sanka R, Torres M, Webster NJ, Vinetz JM, Matthias MA. 2012. Whole genome analysis of Leptospira licerasiae provides insight into leptospiral evolution and pathogenicity. PLoS Negl. Trop. Dis. 6:e1853. 10.1371/journal.pntd.0001853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. 2004. A Hidden Markov Model method, capable of predicting and discriminating beta-barrel outer membrane proteins. BMC Bioinformatics 5:29. 10.1186/1471-2105-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stukalov O, Korenevsky A, Beveridge TJ, Dutcher JR. 2008. Use of atomic force microscopy and transmission electron microscopy for correlative studies of bacterial capsules. Appl. Environ. Microbiol. 74:5457–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ritchie AE, Ellinghausen HC. 1965. Electron microscopy of leptospires. I. Anatomical features of Leptospira Pomona. J. Bacteriol. 89:223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anderson DL, Johnson RC. 1968. Electron microscopy of immune disruption of leptospires: action of complement and lysozyme. J. Bacteriol. 95:2293–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stevenson B, Choy HA, Pinne M, Rotondi ML, Miller MC, Demoll E, Kraiczy P, Cooley AE, Creamer TP, Suchard MA, Brissette CA, Verma A, Haake DA. 2007. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS One 2:e1188. 10.1371/journal.pone.0001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Peeters E, Sass A, Mahenthiralingam E, Nelis H, Coenye T. 2010. Transcriptional response of Burkholderia cenocepacia J2315 sessile cells to treatments with high doses of hydrogen peroxide and sodium hypochlorite. BMC Genomics 11:90. 10.1186/1471-2164-11-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45–55 [DOI] [PubMed] [Google Scholar]
- 57. Santos JM, Freire P, Vicente M, Arraiano CM. 1999. The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol. Microbiol. 32:789–798 [DOI] [PubMed] [Google Scholar]
- 58. Lourdault K, Cerqueira GM, Wunder EA, Jr, Picardeau M. 2011. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect. Immun. 79:3711–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Capestany CA, Tribble GD, Maeda K, Demuth DR, Lamont RJ. 2008. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J. Bacteriol. 190:1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Heusipp G, Nelson KM, Schmidt MA, Miller VL. 2004. Regulation of htrA expression in Yersinia enterocolitica. FEMS Microbiol. Lett. 231:227–235 [DOI] [PubMed] [Google Scholar]
- 61. Johnstone DB, Farr SB. 1991. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 10:3897–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kornitzer D, Teff D, Altuvia S, Oppenheim AB. 1991. Isolation, characterization, and sequence of an Escherichia coli heat shock gene, htpX. J. Bacteriol. 173:2944–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mogensen JE, Kleinschmidt JH, Schmidt MA, Otzen DE. 2005. Misfolding of a bacterial autotransporter. Protein Sci. 14:2814–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ohnishi K, Matsumoto H, Takahashi A, Wang X, Ohnishi T. 1996. Heat shock transcription factor, HSF, is activated by ultraviolet irradiation. Photochem. Photobiol. 64:949–952 [PubMed] [Google Scholar]
- 65. Raivio TL, Silhavy TJ. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591–624 [DOI] [PubMed] [Google Scholar]
- 66. Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, Cullen PA, Davis J, Johnson M, Kuczek E, Alt DP, Peterson-Burch B, Coppel RL, Rood JI, Davies JK, Adler B. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lo M, Cordwell SJ, Bulach DM, Adler B. 2009. Comparative transcriptional and translational analysis of leptospiral outer membrane protein expression in response to temperature. PLoS Negl. Trop. Dis. 3:e560. 10.1371/journal.pntd.0000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kobayashi H, Uematsu K, Hirayama H, Horikoshi K. 2000. Novel toluene elimination system in a toluene-tolerant microorganism. J. Bacteriol. 182:6451–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kuehn MJ, Kesty NC. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645–2655 [DOI] [PubMed] [Google Scholar]
- 70. McGrath H, Adler B, Vinh T, Faine S. 1984. Phagocytosis of virulent and avirulent leptospires by guinea-pig and human polymorphonuclear leukocytes in vitro. Pathology 16:243–249 [DOI] [PubMed] [Google Scholar]
- 71. Atkinson GC, Tenson T, Hauryliuk V. 2011. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. 10.1371/journal.pone.0023479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schweinitzer T, Josenhans C. 2010. Bacterial energy taxis: a global strategy? Arch. Microbiol. 192:507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moreira RN, Dressaire C, Domingues S, Arraiano CM. 2011. A new target for an old regulator: H-NS represses transcription of bolA morphogene by direct binding to both promoters. Biochem. Biophys. Res. Commun. 411:50–55 [DOI] [PubMed] [Google Scholar]
- 74. Magnani D, Solioz M. 2005. Copper chaperone cycling and degradation in the regulation of the cop operon of Enterococcus hirae. Biometals 18:407–412 [DOI] [PubMed] [Google Scholar]
- 75. Silver S, Phung LT. 2005. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 32:587–605 [DOI] [PubMed] [Google Scholar]
- 76. Thorgersen MP, Downs DM. 2009. Oxidative stress and disruption of labile iron generate specific auxotrophic requirements in Salmonella enterica. Microbiology 155:295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Murray GL, Morel V, Cerqueira GM, Croda J, Srikram A, Henry R, Ko AI, Dellagostin OA, Bulach DM, Sermswan RW, Adler B, Picardeau M. 2009. Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect. Immun. 77:810–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, Wilson PJ, Creno S, Kuczek ES, Bommezzadri S, Davis JC, McGrath A, Johnson MJ, Boursaux-Eude C, Seemann T, Rouy Z, Coppel RL, Rood JI, Lajus A, Davies JK, Medigue C, Adler B. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607. 10.1371/journal.pone.0001607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]


