Abstract
The putative citrate metabolic pathway in Lactobacillus casei ATCC 334 consists of the transporter CitH, a proton symporter of the citrate-divalent metal ion family of transporters CitMHS, citrate lyase, and the membrane-bound oxaloacetate decarboxylase complex OAD-ABDH. Resting cells of Lactobacillus casei ATCC 334 metabolized citrate in complex with Ca2+ and not as free citrate or the Mg2+-citrate complex, thereby identifying Ca2+-citrate as the substrate of the transporter CitH. The pathway was induced in the presence of Ca2+ and citrate during growth and repressed by the presence of glucose and of galactose, most likely by a carbon catabolite repression mechanism. The end products of Ca2+-citrate metabolism by resting cells of Lb. casei were pyruvate, acetate, and acetoin, demonstrating the activity of the membrane-bound oxaloacetate decarboxylase complex OAD-ABDH. Following pyruvate, the pathway splits into two branches. One branch is the classical citrate fermentation pathway producing acetoin by α-acetolactate synthase and α-acetolactate decarboxylase. The other branch yields acetate, for which the route is still obscure. Ca2+-citrate metabolism in a modified MRS medium lacking a carbohydrate did not significantly affect the growth characteristics, and generation of metabolic energy in the form of proton motive force (PMF) was not observed in resting cells. In contrast, carbohydrate/Ca2+-citrate cometabolism resulted in a higher biomass yield in batch culture. However, also with these cells, no generation of PMF was associated with Ca2+-citrate metabolism. It is concluded that citrate metabolism in Lb. casei is beneficial when it counteracts acidification by carbohydrate metabolism in later growth stages.
INTRODUCTION
Lactobacillus represents a large genus of Gram-positive bacteria that contains over 100 species (1). Among these, Lactobacillus casei is a facultative heterofermentative lactic acid bacterium (LAB) that can be isolated from diverse environments. It is an important nonstarter lactic acid bacterium commonly present in cheese and dairy products. The most important energy and carbon source for LAB in general is lactose, which is abundantly present in milk, but few LAB have in addition the capacity to metabolize citrate, which is present in cheese curd at a concentration of approximately 10 mmol · kg−1.
Citrate fermentation by bacteria proceeds via two main routes that are catalyzed by a variety of enzymes and transporters. The genes encoding the citrate lyase complex and accessory proteins are diagnostic for citrate fermentation by bacteria. Citrate lyase catalyzes the common, first metabolic step in the pathways, i.e., the splitting of internalized citrate in oxaloacetate and acetate. The two main pathways diverge at oxaloacetate yielding succinate or pyruvate. The route to succinate via malate and fumarate is found in many lactobacilli but also in the gammaproteobacterium Escherichia coli (2). The pathway is characterized by a transporter that takes up citrate in exchange for the end product succinate (precursor-product exchange). The transporter is a member of the divalent anion:Na+ symporter (DASS) family of secondary transporters (3, 4). The second pathway involves decarboxylation of oxaloacetate, yielding pyruvate, which is added to the central pyruvate pool in the glycolytic pathway during carbohydrate/citrate cometabolism. In LAB, redundant pyruvate in the pool is converted to the flavor compound acetoin. In different bacteria, the pathway up to pyruvate is catalyzed by different combinations of a transporter responsible for the uptake of citrate and an enzyme (complex) that decarboxylates oxaloacetate, which has consequences predominantly for the energetics of the pathways. In Gram-negative bacteria like Klebsiella pneumoniae, citrate is taken up by an Na+-dependent symporter, termed CitS, and oxaloacetate is decarboxylated by a membrane-bound complex, termed OAD, that conserves the free energy released in the reaction by pumping out Na+ ions (5, 6). Subsequently, pyruvate is converted to acetate via the acetate kinase route, which yields 1 ATP per molecule of pyruvate. The free-energy yield is high enough to allow the organism to grow on citrate as the sole energy source (7, 8). In LAB like Lactococcus lactis and Leuconostoc mesenteroides, a cytoplasmic oxaloacetate decarboxylase, termed CitM, which does not conserve the free energy released in the reaction (9), is combined with a transporter, termed CitP, that functions as a citrate/lactate exchanger during carbohydrate/citrate cometabolism (10–12). In spite of the different transport modes, CitP and CitS are homologous proteins that belong to the 2-hydroxycarboxylate transporter (2HCT) family. The pathway in LAB conserves metabolic energy in the form of an electrochemical gradient of protons across the membrane (proton motive force [PMF]) through exchange of divalent citrate with monovalent lactate, which results in a membrane potential of physiological polarity (i.e., inside negative), and through alkalinization of the cytoplasm by the decarboxylation reactions, which result in the generation of a pH gradient (inside alkaline). The pathway functions as an indirect proton pump (10) and, as such, adds to metabolic energy generation or is involved in acid stress resistance (11–16). Recently, in Lb. casei and a few other members of the Lactobacillales, a third combination of a transporter and a decarboxylase was described, which putatively is responsible for citrate fermentation by these bacteria (17). The citrate cluster on the genome of these organisms identified by the citrate lyase genes contains the genes encoding the major subunits of the OAD complex found in Gram-negative bacteria and a citrate transporter, termed CitH, that is a member of the CitMHS family, members of which have been shown to transport citrate in complex with divalent metal ions, more specifically Mg2+ and Ca2+ (3, 18–20).
Here, we characterize the citrate metabolic pathway in Lb. casei with respect to substrate specificity, expression of the pathway, repression by carbohydrates, contribution to growth, and energetics of the pathway.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
Lactobacillus casei ATCC 334 was grown overnight at 37°C in modified MRS broth medium (21) (mMRS; original composition without glucose, ammonium citrate, or Tween 80) supplemented with 30 mM sodium citrate and 10 mM chloride calcium (mMRSCitCa), 0.5% (wt/vol) glucose (mMRSGlc), or galactose (mMRSGal) as indicated. The medium was adjusted to pH 6.0. Cells were grown in 50-ml screw-cap tubes without agitation and at 37°C during approximately 15 h. Growth was monitored by measuring the optical density at a wavelength of 660 nm (OD660). Cells were harvested at the mid-exponential growth phase, when the optical density was 0.6 (or otherwise, when indicated) by spinning for 10 min at 3,000 rpm. Cells were washed two times with 50 mM potassium phosphate (KPi) pH 5.8 buffer and, finally, resuspended in the same buffer at 4°C.
Citrate consumption by resting cells.
Resting cells resuspended at an OD660 of 1.5 in 50 mM potassium phosphate (pH 5.8) buffer were incubated at 37°C for 10 min without agitation. The assay was performed in a total volume of 1.5 ml. At time zero (t = 0), citrate and calcium chloride (Ca2+) were added at concentrations of 2 mM. Samples of 100 μl were taken every 5 or 10 min and immediately centrifuged for 0.5 min at maximum speed in a tabletop centrifuge. The supernatant was stored on ice until further analysis by enzymatic assays and/or high-performance liquid chromatography (HPLC). Measurements of the concentrations of citrate, oxaloacetate, and pyruvate were in good agreement between the two methods. Each experiment was done at least in triplicate, and the averages and standard deviations of three independent experiments were determined.
Enzymatic assays.
Citrate, oxaloacetate, and pyruvate were measured as described before (10), using the commercially available enzymes citrate lyase (CL), l-malate dehydrogenase (L-MDH), and l-lactate dehydrogenase (L-LDH). Briefly, an aliquot of 30 μl of the sample was added to 50 mM glycyl-glycine (pH 7.8) buffer containing NADH and L-MDH. Oxaloacetate in the sample is converted to l-malate at the expense of NADH. Subsequently, pyruvate in the same sample was measured by addition of L-LDH, which results in the conversion of pyruvate to l-lactate at the expense of NADH. Subsequent addition of citrate lyase converts citrate in the sample to oxaloacetate (and pyruvate), resulting in an additional decrease in the NADH concentration equivalent to the citrate concentration present in the samples. The assay was performed in 96-well microtiter plates. The decrease in NADH concentration was measured spectroscopically at 340 nm.
HPLC analysis.
Products of citrate metabolism were determined as described before (10). An aliquot of 10 μl of the supernatant was loaded on an Aminex HPX-87H anion-exchange column (Bio-Rad Laboratories, Inc., Richmond, CA) operated at 30°C in isocratic mode using 5 mM H2SO4 as the mobile phase and a flow rate of 0.8 ml/min. Quantification was based on calibration curves generated by injecting standards of known concentrations.
Colorimetric determination of acetoin and diacetyl.
Acetoin and diacetyl were measured according to the method described in reference 22. Acetoin and diacetyl react with guanide groups of creatine in alkaline medium to give a pink color. A volume of 30 μl of the supernatant was mixed with 10 μl of 0.5% (wt/vol) creatine, 10 μl of 5% (wt/vol) α-naphthol freshly dissolved in 2.5 M NaOH, and 150 μl of water. Color development was measured at OD530 after 5 min and after 40 min of incubation in the dark at room temperature to determine the amounts of diacetyl and acetoin, respectively. The amount of acetoin was calculated from a calibration curve of known concentrations. Only a trace amount of diacetyl was found in the supernatants, indicating negligible spontaneous nonenzymatic oxidation of α-acetolactate to diacetyl.
Measurement of internal pH (ΔpH) and membrane potential (Δψ).
The components of the proton motive force were measured as described before (10). To measure cytoplasmic pH, resting cells resuspended to high density (typically containing 50 mg/ml of protein) in 50 mM KPi pH 5.8 buffer were loaded with 2′,7′-bis-(2-carboxyethyl)-5 (and 6)-carboxyfluorescein (BCECF). Fluorescence measurements were performed in 1-cm cuvettes containing 50 mM KPi pH 5.8 buffer equilibrated at 37°C and cells loaded with BCECF. The cuvette was stirred with a magnetic stirring bar. Fluorescence was measured using excitation and emission wavelengths of 502 and 525 nm, respectively, with slit widths of 4 and 16 nm, respectively. The fluorescence signal was sampled every second. Opening of the measurement compartment caused loss of data during the first 5 to 6 s after an addition to the cuvette was made. The cytoplasmic pH was calculated as described previously (23).
Membrane potential was measured qualitatively with the fluorescent probe 3,3′-dipropylthiocarbocyanine iodide (DiSC3) (24). A decrease in fluorescence intensity correlates with an increase in electrical potential across the membrane. DiSC3 was added from a stock solution to a final concentration of 2 μM to the quartz cuvette containing the cells in 50 mM KPi pH 5.8 buffer containing 2 mM CaCl2. The presence of Ca2+ was required during the preincubation step to obtain a stable DiSC3 fluorescent signal. The system was left to equilibrate for 5 min at 37°C. Fluorescence measurements were performed using excitation and emission wavelengths of 500 and 705 nm, respectively, and slit widths of 8 nm.
Chemicals.
l-Lactate dehydrogenase (L-LDH), l-malate dehydrogenase (L-MDH), and citrate lyase (CL) were obtained from Roche Applied Science. BCECF [2′,7′-bis-(2-carboxyethyl)-5 (and 6)-carboxyfluorescein] (acid form) and DiSC3 (3,3′-dipropylthiocarbocyanine iodide) probes were obtained from Invitrogen Molecular Probes.
RESULTS
Specificity and induction of the citrate metabolic pathway of Lb. casei ATCC 334.
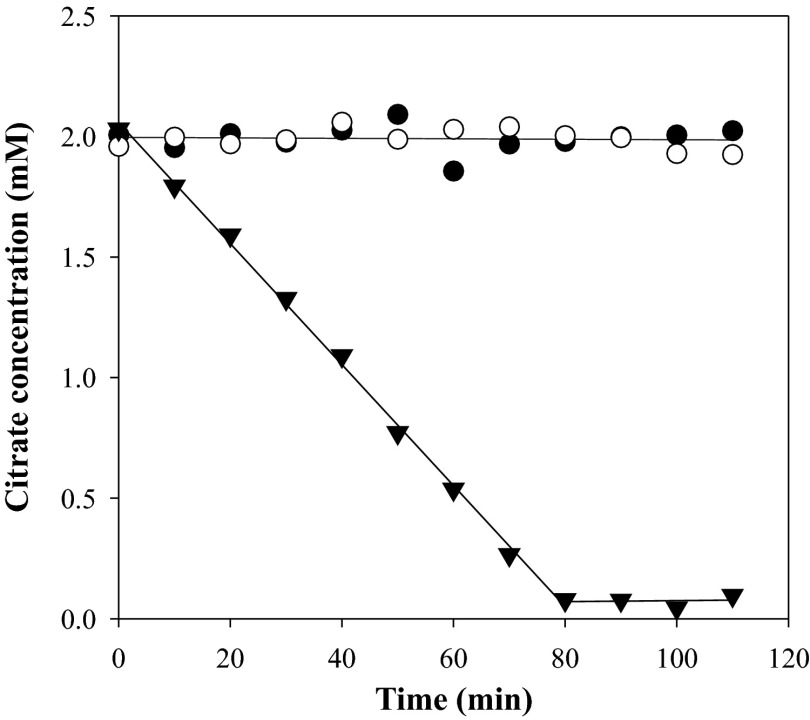
Resting cells of Lb. casei ATCC 334 grown in modified MRS medium (mMRS; see Materials and Methods) supplemented with 30 mM citrate and 10 mM Ca2+ (mMRSCitCa) did not consume citrate for at least 110 min when incubated in 50 mM KPi pH 5.8 buffer containing 2 mM citrate (Fig. 1, open circles). In contrast, complete consumption of citrate was observed in 80 min when in addition 2 mM Ca2+ was present (inverted triangles), strongly suggesting that the product of the citH transporter gene found in the citrate gene cluster on the genome of Lb. casei ATCC 334 recognizes the Ca2+-citrate complex. Specificity for the divalent Ca2+ cation in the metabolized complex was demonstrated by substituting Ca2+ for Mg2+, which resulted in the complete lack of breakdown of citrate (filled circles). In addition, metabolism in the presence of Ca2+ was largely inhibited by a 2 mM concentration of the Ca2+-chelating agent EGTA (data not shown). The breakdown of the Ca2+-citrate complex appeared to be a zero order process down to concentrations of around 100 μM, suggesting that the pathway has a very high affinity for the complex, in the μM range.
Fig 1.
Citrate consumption by resting cells of Lb. casei ATCC 334. Washed cells grown in mMRSCitCa medium were incubated in 50 mM KPi buffer pH 5.8 at an OD660 of 1.5 with 2 mM citrate without further additions (open circles) or in the presence of 2 mM Mg2+ (filled circles) or 2 mM Ca2+ (filled triangles). Citrate in the supernatant was measured enzymatically every 10 min.
The rate of citrate breakdown in the presence of Ca2+ by resting cells grown in the presence of Ca2+ and citrate was 28 ± 2 μM · min−1 (Fig. 1 and 2A). The rate of consumption was reduced to marginal levels when the cells were grown in the absence of Ca2+ and citrate, indicating that constitutive levels of expression of the complete pathway are very low and that the pathway is induced in the presence of Ca2+ and citrate in the medium. Similar to what was observed above, the cells grown in the absence of Ca2+ and citrate did not consume free citrate or citrate in complex with Mg2+ (not shown). Induction of the Ca2+-citrate pathway was enhanced somewhat when only Ca2+ was added to the medium at the same concentration, while citrate alone resulted in a much stronger induction, but not as much as in the presence of both (Fig. 2A). Again, the cells grown in the presence of citrate alone did not consume free citrate or Mg2+-citrate, suggesting the absence of transporters specific for these species (not shown). No special precautions were made in the latter experiments to deplete the experimental system from residual Ca2+, and it is tentatively concluded that the Ca2+-citrate complex is the main inducing agent, which is also in line with the high affinity of the pathway for the complex. Ca2+ contamination of media would explain the observed citrate consumption during growth in the absence of added Ca2+ reported in previous studies (25–27).
Fig 2.
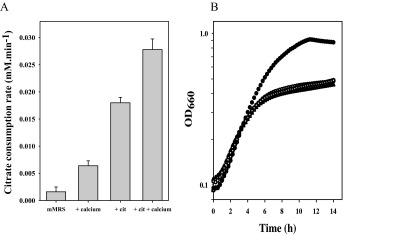
Induction of the citrate catabolic pathway of Lb. casei ATCC 334. (A) Citrate consumption rate of resting cells grown in batches in mMRS medium supplemented with 10 mM calcium chloride and/or 30 mM sodium citrate as indicated. Cells were harvested at late exponential growth (OD660 = 0.6). (B) Growth curves of Lb. casei ATCC 334 in microtiter plates in mMRS medium without further additions (open circles) or in the presence of 10 mM Ca2+ (filled squares), 30 mM citrate (filled triangles), both calcium and citrate (open squares), or both calcium and 10 mM glucose (filled circles). Means of three independent experiments are shown.
In spite of the significantly different citrate metabolic activities, growth of Lb. casei was hardly affected under the different growth conditions. In mMRS medium, containing no carbohydrate as a carbon or energy source, Lb. casei grew at a growth rate of 0.11 h−1 to a modest biomass yield (Fig. 2B). In the presence of glucose, both growth rate and yield improved significantly, demonstrating that in the absence of a carbohydrate growth is carbon/energy limited. The limitation could not be overcome by citrate metabolism. Maximal induction of the citrate metabolic pathway by including both Ca2+ and citrate in the mMRS medium did not improve the growth rate or the biomass yield. Citrate metabolism does not seem to be beneficial for the organism under these growth conditions.
Citrate metabolic pathway in Lb. casei ATCC 334.
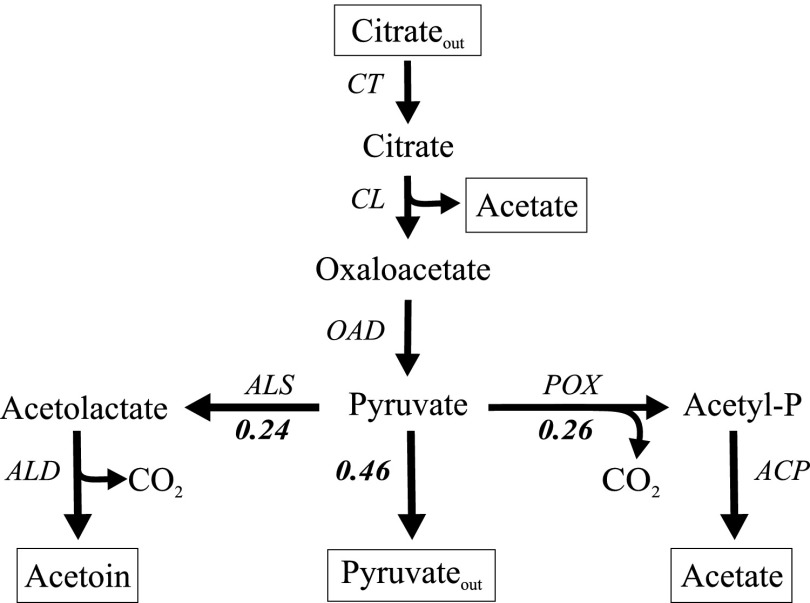
Metabolism of Ca2+-citrate in resting cells of Lb. casei ATCC 334 produced pyruvate, acetate, and acetoin. Following complete consumption of 2 mM citrate, the supernatant contained 0.92 mM pyruvate, 2.51 mM acetate, and 0.24 mM acetoin (Table 1). No oxaloacetate, lactate, formate, ethanol, or α-acetolactate was detected. Since more acetate was produced than expected from the citrate lyase reaction (2 mM), the product profile suggested that following the formation of cytoplasmic pyruvate by the OAD complex, the flux was distributed over the acetoin and acetate pathways and the exit of pyruvate out of the cell (Fig. 3). Clearly, the rate limitations were in the steps following pyruvate. The fluxes to the end products acetoin and acetate were more or less the same, taking into account that 2 mol of pyruvate are required for the production of 1 mol of acetoin. With a little less than half of the pyruvate produced in the cells excreted into the medium, the carbon yield was 96% ± 15%.
Table 1.
Product profile of citrate metabolism by resting cells of Lactobacillus casei ATCC 334
| Metabolite | Concna (mM) |
|---|---|
| Citrate | —b |
| Pyruvate | 0.92 ± 0.13 |
| Acetate | 2.51 ± 0.07 |
| Acetoin | 0.24 ± 0.08 |
| α-Acetolactate | — |
| Formate | — |
| Ethanol | — |
| Lactate | — |
Concentration in supernatant after incubation at an OD660 of 1.5 for 110 min at 30°C in 50 mM KPi pH 5.8 buffer containing initially 2 mM citrate.
—, undetectable.
Fig 3.
Citrate metabolic pathway in resting cells of Lb. casei ATCC 334. The metabolic scheme is based on the product profile obtained when resting cells of Lb. casei were allowed to convert 2 mM citrate in the presence of Ca2+ (Table 1). Indicated are the relative fluxes through the different parts of the scheme following pyruvate. Note the formation of acetate from citrate in the citrate lyase reaction. CT, citrate transporter; CL, citrate lyase; OAD, membrane-bound oxaloacetate decarboxylase; ALS, α-acetolactate synthase; ALD, α-acetolactate decarboxylase; ACP, acetylphosphatase; POX, pyruvate oxidase.
The product profile of citrate metabolism by Lb. casei ATCC 334 was similar to the profile obtained with L. lactis IL1403(pFL3) presented before (28). A comparative analysis of the genomes of both species suggested the genes that constitute the pathways in both organisms (Table 2). The conversion of citrate to pyruvate is done by different sets of enzymes in the two organisms. While in L. lactis citrate is taken up by the citrate/lactate exchanger CitP and, subsequently, degraded by the citrate lyase complex and the soluble oxaloacetate decarboxylase CitM, in Lb. casei citrate is taken up as Ca2+-citrate by CitH, and after splitting by citrate lyase (CL), oxaloacetate is decarboxylated by a membrane-bound OAD complex. The conversion of pyruvate to the flavor compound acetoin requires the presence of α-acetolactate synthase (ALS) and α-acetolactate decarboxylase (ALD), which are found in both organisms. Remarkably, the genes encoding enzymes for the further conversion of acetoin into the flavor compounds diacetyl and butanediol were not found in the genome of Lb. casei ATCC 334. Since no formate or ethanol was detected, the enzymes pyruvate-formate lyase (PFL) and pyruvate dehydrogenase (PDH) are not likely to be involved in the conversion of pyruvate under the experimental conditions, even though the encoding genes are present in the genomes. Rather, pyruvate is converted to acetyl-P by pyruvate oxidase (POX) in a reaction with molecular oxygen yielding hydrogen peroxide and CO2. Subsequently, acetyl-P may be converted to acetate by acetate kinase yielding ATP or be hydrolyzed by acylphosphate phosphohydrolase without conserving the free energy (10). The proposed metabolic scheme for citrate metabolism in Lb. casei is depicted in Fig. 3.
Table 2.
Comparative analysis of the genomes of Lactobacillus casei ATCC 334 and Lactococcus lactis IL1403(pFL3) identifying genes (putatively) involved in citrate metabolism by resting cells
| Protein | Function | Gene |
|
|---|---|---|---|
| Lb. casei ATCC 334 | L. lactis IL1403(pFL3) | ||
| Citrate metabolism | |||
| CitH | M2+-citrate transporter | LSEI1865 | xa |
| CitP | Citrate/lactate exchanger | x | citPb |
| CitC | Citrate lyase γ subunit | LSEI1860 | citC |
| CitD | Citrate lyase γ subunit | LSEI1859 | citD |
| CitE | Citrate lyase γ subunit | LSEI1858 | citE |
| CitF | Citrate lyase accessory protein | LSEI1857 | citF |
| CitG | Citrate lyase accessory protein | LSEI1853 | citG |
| CitX | Citrate lyase accessory protein | citX | —c |
| OadA | Oxaloacetate decarboxylase subunit α | LSEI1855 | x |
| OadB | Oxaloacetate decarboxylase subunit β | LSEI1862 | x |
| OadD | Oxaloacetate decarboxylase subunit δ | LSEI1863 | x |
| OadH | Oxaloacetate decarboxylase subunit ζ | LSEI1864 | x |
| CitM | Oxaloacetate decarboxylase | x | mae |
| Pyruvate metabolism | |||
| ALS | Acetolactate synthase | LSEI1841 | als |
| ALD | Acetolactate decarboxylase | LSEI1840 | aldC |
| POX | Pyruvate oxidase | LSEI2143 LSEI0433 LSEI1784 | poxL |
| ACK | Acetate kinase | LSEI0166 | ackA2 |
| APH | Acyl-P phosphohydrolase | LSEI1676 | yfjC |
| PDHA | Acetoin/pyruvate dehydrogenase complex, E1 component, alpha subunit | LSEI1445 | pdhA |
| PDHB | Acetoin/pyruvate dehydrogenase complex, E1 component, beta subunit | LSEI1444 | pdhB |
| PDHC | Acetoin/pyruvate dehydrogenase complex, E2 component, dihydrolipoamide succinyltransferase | LSEI1443 | pdhC |
| PDHD | Acetoin/pyruvate dehydrogenase complex, E3 component, dihydrolipoamide dehydrogenase | LSEI1446 | pdhD |
| PTA | phosphotransacetylase | LSEI1448 LSEI0996 | eutD |
x, not found.
Introduced on plasmid pFL3.
In L. lactis, citX is part of citG.
Carbohydrate/Ca2+-citrate cometabolism by Lb. casei ATCC 334.
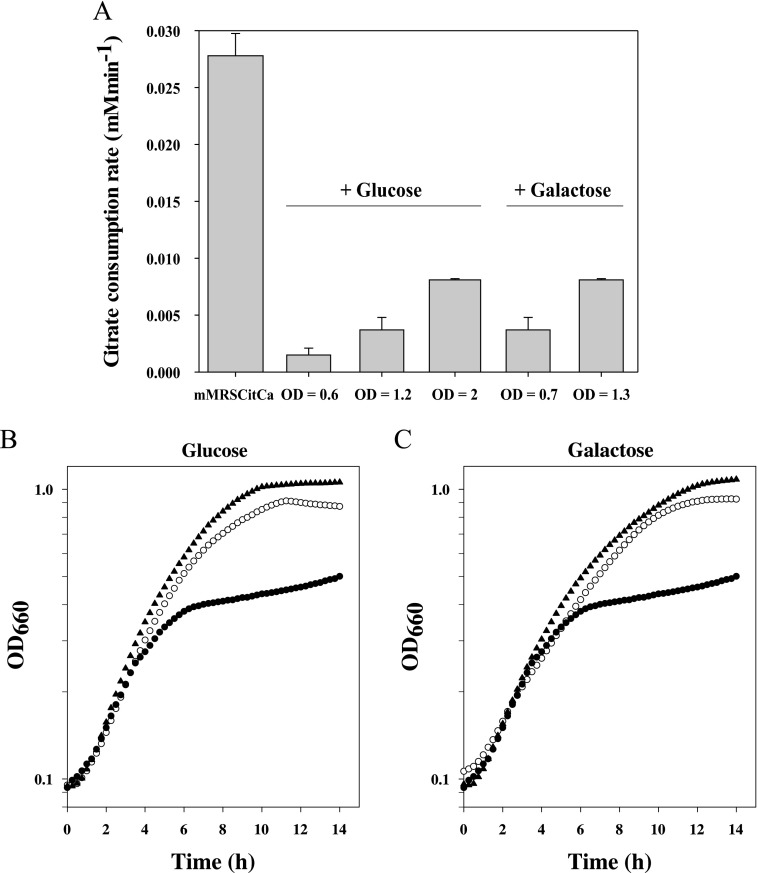
The Ca2+-citrate breakdown rate of resting cells was largely independent of the growth phase of the culture from which the cells were taken. Resting cells grown in mMRS supplemented with Ca2+ and citrate consumed citrate at the same rate irrespective of the cell density of the culture at which they were harvested (Fig. 4A). The rate dropped dramatically, by a factor of 20, when in addition glucose was included in the growth medium (Fig. 4A), strongly suggesting that expression of the citrate metabolic pathway in Lb. casei is, at least in part, under the control of carbon catabolite repression (CCR). In line with this interpretation, repression was partly relieved when the cells were allowed to grow to higher densities. Expression of the pathway was similarly repressed by galactose, but less effectively. At low cell densities, the activity of the pathway was down by a factor of 7.
Fig 4.
Carbohydrate/Ca2+-citrate cometabolism. (A) Citrate consumption rates of resting cells of Lb. casei ATCC 334 grown in mMRSCitCa without further additions and with the addition of glucose and galactose as indicated. The cells were harvested at the indicated OD660. The average values and standard deviations of three independent experiments are shown. (B, C) Growth curves of Lb. casei ATCC 334 during glucose/Ca2+-citrate (B) and galactose/Ca2+-citrate (C) cometabolism. The curves represent growth in mMRS medium with 10 mM calcium chloride (filled circles) supplemented with 28 mM carbohydrate (open circles) and with 28 mM carbohydrate and 30 mM citrate (filled triangles).
Growth of Lb. casei in mMRS (supplemented with Ca2+) was significantly enhanced when glucose was included in the growth medium as a carbon/energy source (Fig. 4B). Most prominent was the significantly higher cell density obtained at the end of growth. When in addition to glucose and Ca2+, citrate was included, additional improvement of growth was observed, especially in the later growth phases, yielding even higher biomass. The results with galactose as the carbohydrate were similar. Galactose in the medium resulted in higher growth yields that were even higher during galactose/Ca2+-citrate cometabolism (Fig. 4C). It follows that, in contrast to the observed lack of growth enhancement by Ca2+-citrate metabolism in mMRS medium (Fig. 2B), Ca2+-citrate metabolism is beneficial during carbohydrate/Ca2+-citrate cometabolism.
Energetics of carbohydrate and citrate metabolism in Lb. casei ATCC 334.
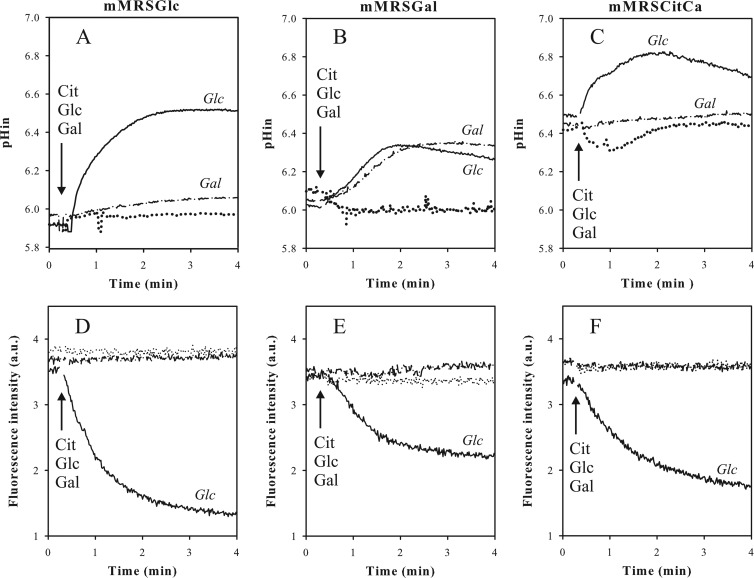
The two components of the proton electrochemical gradient (PMF), i.e., the transmembrane pH gradient (ΔpH) and the membrane potential (Δψ), generated by glucose, galactose, and Ca2+-citrate metabolism in resting cells of Lb. casei were measured using the fluorescent probes BCECF (23) and DiSC3 (24), respectively (Fig. 5). The cytoplasmic pH of cells grown in mMRS supplemented with glucose and resuspended in 50 mM KPi pH 5.8 buffer was slightly more alkaline than the external pH (∼0.1 to 0.2 units). Addition of 5 mM glucose resulted in rapid alkalinization of the cytoplasm resulting in a pH gradient of 0.7 units (Fig. 5A). In a parallel experiment using the same cells and conditions, quenching of the fluorescence of the DisC3 probe upon addition of glucose was indicative of the formation of membrane potential (Fig. 5D). PMF generation by glucose follows from ATP production in glycolysis, which subsequently is used to pump out protons by F1Fo-ATPase. Cells grown in mMRS supplemented with galactose alkalinized the cytoplasm upon metabolizing galactose, but the rate was lower than observed with glucose and the final extent was limited to 0.5 units (Fig. 5B). In addition, no membrane potential was generated (Fig. 5E). Galactose is metabolized in the Leloir pathway (29–31), which produces ATP less efficiently. A proton motive force consisting of ΔpH and Δψ was generated in the galactose-grown cells by glucose, even though the extent of both components was smaller than in glucose-grown cells (Fig. 5B and E). It follows that the glycolytic pathway is expressed constitutively. In contrast, galactose did not generate PMF in glucose-grown cells, indicating that the galactose phosphotransferase system (PTS) and the Leloir pathway require the presence of galactose in the medium for expression, as was reported before (30–32). In line with the lack of expression of the citrate metabolic pathway in glucose- and galactose-grown cells, no effect on cytoplasmic pH and membrane potential was observed upon addition of Ca2+-citrate.
Fig 5.
Energetics of carbohydrate and Ca2+-citrate metabolism in Lb. casei. Lb. casei ATCC 334 was grown in mMRSGlc (A, D), mMRSGal (B, E), and mMRSCitCa (C, F). The cells were harvested at an OD660 of 0.6 and loaded with BCECF for internal pH (pHin) measurements (A, B, and C) and incubated with the DiSC3 fluorescent probe for measurement of the membrane potential (D, E, and F). For cytoplasmic pH measurements, 5 mM glucose, 5 mM galactose, or 2 mM citrate plus 2 mM Ca2+ was added at the time point indicated by the arrow. For membrane potential measurements, the cells were incubated with 2 mM Ca2+ to obtain a stable DiSC3 fluorescent signal before glucose, galactose, and citrate were added (arrow) at the same concentrations. a.u., arbitrary units.
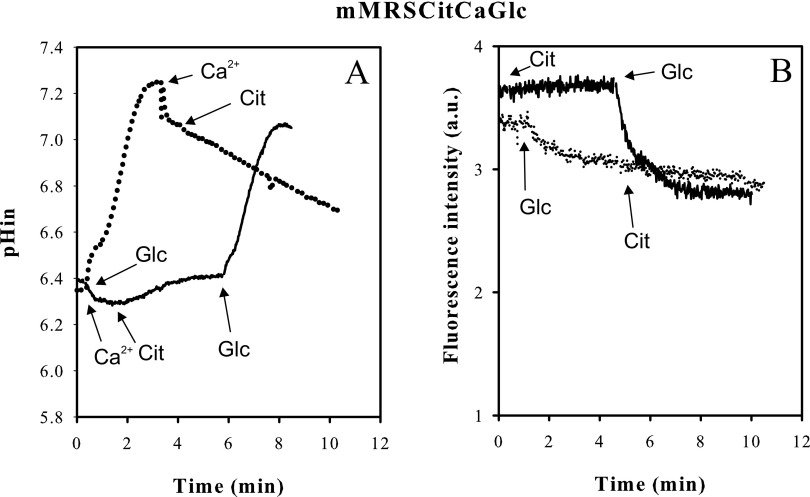
Lb. casei grown in mMRS supplemented with Ca2+ and citrate reproducibly revealed a cytoplasmic pH that was 0.5 to 0.6 units more alkaline than the external pH when the cells were resuspended in 50 mM KPi pH 5.8 buffer (Fig. 5C). Addition of glucose to the cells resulted in the generation of additional ΔpH and Δψ, very similar to what was observed in galactose-grown cells, demonstrating that the cells were healthy in this respect (Fig. 5C and F). Addition of Ca2+-citrate to these cells in which the Ca2+-citrate metabolic pathway is fully induced (Fig. 1) did not result in alkalinization of the cytoplasm or in generation of membrane potential. This suggests that the Ca2+-citrate metabolic pathway as a whole does not produce a net proton electrochemical gradient. Addition of citrate before Ca2+ initially resulted in acidification of the cytoplasm of about 0.1 pH unit, which was reversed when Ca2+ was added subsequently (Fig. 5C). The same effect was observed when Ca2+ was added before citrate (Fig. 6A). The phenomenon was not observed in galactose- and glucose-grown cells and therefore must be associated with growth in the presence of Ca2+ and citrate. A PMF was also not generated by Ca2+-citrate metabolism when Lb. casei was grown in mMRSCitCa medium containing in addition glucose (Fig. 6) or galactose (not shown), i.e., growth conditions under which a growth advantage was observed due to the presence of Ca2+-citrate (Fig. 4).
Fig 6.
Energetics of glucose/Ca2+-citrate cometabolism in Lb. casei. Cells of Lb. casei ATCC 334 grown in mMRSCitCa containing 0.5% (wt/vol) glucose and harvested at an OD660 of 2.1 were loaded with BCECF for internal pH (pHin) measurements (A) and incubated with the DiSC3 fluorescent probe for measurement of the membrane potential (B). (A) Resting cells were incubated in 50 mM KPi pH 5.8 buffer. The dotted line indicates that 5 mM glucose, 2 mM Ca2+ and 2 mM citrate were added subsequently (downward-pointing arrows). The solid line indicates that 2 mM Ca2+, 2 mM citrate, and 5 mM glucose were added subsequently (upward-pointing arrows). (B) Resting cells were incubated in 50 mM KPi pH 5.8 buffer containing 2 mM Ca2+. The dotted line indicates that 5 mM glucose and 2 mM citrate were added subsequently (upward-pointing arrows). The solid line indicates that 2 mM citrate and 5 mM glucose were added (downward-pointing arrows).
DISCUSSION
Lactobacillus species Lb. plantarum, Lb. helveticus, and Lb. rhamnosus have been shown to convert citrate to succinate (2, 33, 34), while several other species of the same genus were shown not to be able to catabolize citrate (35). The conversion of citrate to succinate follows the reductive tricarboxylic acid (TCA) pathway and is typified by the presence of a transporter, termed CitT, that catalyzes the uptake of citrate in exchange with the end product succinate. Atypically, Lb. casei, while metabolizing citrate (26, 36), did not produce succinate from citrate under a variety of growth conditions (27). In line with these observations, the genome of Lb. casei ATCC 334 (37) does not encode a CitT homologue; rather, the citrate cluster identified by the subunits of the citrate lyase complex contains a transporter gene, termed citH, that encodes a member of the CitMHS family (TC 2.A.11 [38]). Members of this family are believed to transport citrate in complex with divalent metal ions (17). The first characterized members were two transporters of Bacillus subtilis, termed CitM and CitH (18, 39). CitM is the major citrate uptake system during growth of B. subtilis on citrate and transports the complex of citrate and Mg2+ (40). The studies demonstrated that the transporters discriminated between the metal ions in the complex with citrate. While the CitM transporter showed activity with Mg2+, Ni2+, Co2+, Zn2+, and Mn2+, CitH transported citrate in complex with Ca2+, Ba2+, and Sr2+. The difference in specificity seemed to correlate with the atomic radii of the ions ranging in size between 65 and 80 pm in the former group and between 99 and 134 pm in the latter. A recent, more detailed study of the specificity of another member of the family, CitH of Enterococcus faecalis, further strengthened the correlation (19). Here, we demonstrate that citrate in complex with Ca2+ is the substrate of the citrate catabolic pathway in Lb. casei ATCC 334. Phylogenetically, the CitH transporter of Lb. casei is close to CitH of E. faecalis (19), which is in agreement with the substrate specificity. Both transport Ca2+-citrate and not Mg2+-citrate. A member of the CitMHS family from Streptococcus mutans is exceptional; it has been reported to transport citrate in complex with trivalent Fe3+ (41).
The product profile of citrate breakdown by resting cells of Lb. casei revealed three major fluxes: (i) excretion of pyruvate, (ii) production of acetoin, and (iii) production of acetate in addition to acetate formed by citrate lyase. The same product profile for nongrowing cells was reported before for Lb. casei strain ATCC 393 (25). The three end products strongly suggested that following the splitting of internalized citrate into acetate and oxaloacetate by citrate lyase, the latter is decarboxylated, yielding pyruvate, most likely involving the membrane-bound OAD complex encoded by the Lb. casei ATCC 334 genome (Fig. 3). Since most of the pyruvate formed was excreted from the cells, there appeared to be a kinetic limitation in the conversion of internal pyruvate into other metabolites. The conversion to acetoin demonstrates that the genes encoding α-acetolactate synthase and α-acetolactate decarboxylase carried in the genome (Table 2) represent an active C4 pathway in Lb. casei ATCC 334. Further metabolism of acetoin to 2,3-butanediol was not observed, which is in line with the absence of the encoding gene on the chromosome. The amount of acetate formed in excess of the equimolar amount formed by citrate lyase most likely also originates from pyruvate, for which a number of possibilities have been suggested (10, 27), involving the pyruvate dehydrogenase (PDH), pyruvate oxidase (POX), and/or pyruvate-formate lyase (PFL) enzymes, the genes of which are all present in the Lb. casei ATCC 334 genome (Table 2). All three routes proceed by the formation of acetyl-P, either directly (POX) or through acetyl coenzyme A (acetyl-CoA), which is then converted to acetyl-P by phosphotransacetylase (PTA), also encoded by the Lb. casei ATCC 334 genome. Most compatible with the present results and experimental conditions would be the conversion by POX. PFL produces formate, which was not detected, and the enzyme is believed to be active only under strict anaerobic conditions (42). The oxidative decarboxylation catalyzed by PDH produces redox equivalents in the form of NADH, which in the normal pathway catalyzed by PDH are used to produce acetaldehyde and ethanol, neither of which was detected. Moreover, the redox equivalents would most likely result in the reduction of (part of) the excess of pyruvate in the cytoplasm by lactate dehydrogenase (LDH), but no lactate was detected either. This leaves, by exclusion, POX, which produces acetyl-P and hydrogen peroxide from pyruvate and molecular oxygen, but the exact nature of the conversion requires further investigation. In the final step, acetyl-P would be converted to acetate, a reaction that is coupled to ATP formation when catalyzed by acetate kinase, which is encoded by the Lb. casei ATCC 334 genome. ATP produced in this reaction would result in proton motive force generation by the action of F1Fo-ATPase, as was observed with ATP produced from glucose and galactose breakdown (Fig. 5). However, no PMF was observed, strongly suggesting that the reaction is catalyzed by the energy-dissipating acylphosphatase enzyme, also encoded by the genome (Table 2).
The product profile of citrate catabolism by resting cells of Lb. casei ATCC 334 was remarkably similar to the profile obtained with L. lactis IL1403/pFL3 (10). The enzyme complement available for pyruvate metabolism is similar in the two organisms but differs considerably for the uptake and conversion of citrate to pyruvate (Table 2). In L. lactis, the transporter CitP takes up citrate in exchange with lactate or any other available intermediate of the pathway (10, 13, 43). Oxaloacetate is decarboxylated by a cytoplasmic decarboxylase not related to the membrane-bound OAD complex. The different transporters used in the two organisms explain the most prominent difference between the two product profiles. L. lactis excretes, in addition to acetoin, the intermediate of the C4 pathway α-acetolactate. The latter is a substrate of CitP of L. lactis and not of CitH of Lb. casei. CitP catalyzes the citrate/α-acetolactate exchange (43). In spite of identical metabolic pathways for citrate breakdown, there is an important difference in the energy-conserving properties of the pathways in the two organisms (see below).
In L. lactis, decarboxylation of oxaloacetate by the cytoplasmic decarboxylase drives indirectly the electrogenic exchange of divalent citrate and monovalent lactate catalyzed by the transporter CitP, which together with proton consumption in the decarboxylation reactions results in the generation of proton motive force when resting cells convert citrate to acetoin (11–13, 43). In Lb. casei, no proton motive force was generated under similar conditions (Fig. 5 and 6). Apparently, there is no net free-energy conservation in the pathway as a whole. Several energy-transducing steps may be discriminated in the pathway, including uptake of the Ca2+-citrate complex, which has been shown to be coupled to the uptake of protons in the case of homologous transporters (3, 19, 39), excretion of Ca2+ by an unknown transporter that is likely to couple the flux to an antiport with protons, and the decarboxylation of oxaloacetate by the membrane-bound OAD that in the Gram-negative counterparts functions as a Na+ pump (6). A very recent study of the OAD complex of the closely related Gram-positive bacterium Enterococcus faecalis showed that the soluble OAD-A subunit that presumably couples the decarboxylation reaction to ion pumping by the membrane-bound OAD-B subunit was found for a significant part in the cytoplasm, which potentially could have a negative effect on the free-energy conservation of the reaction (44). At any rate, the net result of the charge and proton movements across the membrane is zero, and the pathway does not seem to contribute to the metabolic energy pool in Lb. casei ATCC 334.
K. pneumoniae ferments citrate and can use it as an energy and carbon source for growth (45). Citrate is taken up by CitS, a sodium ion motive force-driven symporter, and oxaloacetate is decarboxylated by membrane-bound OAD that pumps out Na+ ions. Metabolic energy for growth is produced mostly by the ability of K. pneumoniae to produce ATP in the acetate kinase pathway, yielding 1 ATP/citrate (46). Acetylphosphate produced from pyruvate is converted to acetate, while the free energy released is conserved in ATP. A number of studies have suggested that Lb. casei is able to derive metabolic energy from citrate metabolism, and energy-producing pathways have been proposed based on the genome sequence and metabolic flux analyses (26, 27, 36). ATP production by acetate kinase plays a pivotal role in these models. In this study, it is demonstrated that resting cells of Lb. casei grown under conditions of maximal citrate utilization do not produce ATP from the conversion of acetyl-P to acetate. Rather, it is concluded that free energy is lost by the action of acetylphosphatase, a conclusion that was also reached in the case of L. lactis IL1403/pFL3. Growth experiments of Lb. casei on complex modified MRS medium without added carbohydrate, conditions under which growth was clearly limited by lack of energy/carbon, supported this conclusion. A high citrate metabolic rate during growth had no effect on growth performance whatsoever (Fig. 2), suggesting no contribution to metabolic energy for growth. During carbohydrate/citrate cometabolism, exponential growth was prolonged, resulting in a higher final biomass. Since also in these cells no metabolic energy production was associated with citrate metabolism (Fig. 6), the beneficial effect is most likely due to the overall alkalinizing effect of citrate breakdown that counteracts the growth-inhibiting acidification by carbohydrate metabolism. Carbon catabolite repression of the citrate pathway when carbohydrate is still high ensures that the pathway becomes active when needed the most, at the final stages of growth.
ACKNOWLEDGMENTS
We thank Jeff Broadbent of Utah State University for the gift of Lb. casei ATCC 334.
This work was supported by the European Community's Seventh Framework Programme (grant agreement no. 211441-BIAMFOOD) and by grants from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) and CONICET (PICT2010-1828 and PIP/2012). P.M. was supported by a Eurotango fellowship 2011 (ERASMUS MUNDUS group). P.M. is a fellow of CONICET, and S.A. and C.M. are Career Investigators of the same institution.
Footnotes
Published ahead of print 24 May 2013
REFERENCES
- 1. Dworkin M, Falkow S. 2006. The prokaryotes: a handbook on the biology of bacteria, 3rd ed Springer, New York, NY [Google Scholar]
- 2. Dudley EG, Steele JL. 2005. Succinate production and citrate catabolism by Cheddar cheese nonstarter lactobacilli. J. Appl. Microbiol. 98:14–23 [DOI] [PubMed] [Google Scholar]
- 3. Krom B, Warner J, Konings W, Lolkema J. 2003. Transporters involved in uptake of di- and tricarboxylates in Bacillus subtilis. Antonie Van Leeuwenhoek 84:69–80 [DOI] [PubMed] [Google Scholar]
- 4. Saier MH, Goldman SR, Maile RR, Moreno MS, Weyler W, Yang N, Paulsen IT. 2002. Transport capabilities encoded within the Bacillus subtilis genome. J. Mol. Microbiol. Biotechnol. 4:37–67 [PubMed] [Google Scholar]
- 5. van der Rest ME, Siewe RM, Abee T, Schwarz E, Oesterhelt D, Konings WN. 1992. Nucleotide sequence and functional properties of a sodium-dependent citrate transport system from Klebsiella pneumoniae. J. Biol. Chem. 267:8971–8976 [PubMed] [Google Scholar]
- 6. Dimroth P. 1982. The generation of an electrochemical gradient of sodium ions upon decarboxylation of oxaloacetate by the membrane-bound and Na+-activated oxaloacetate decarboxylase from Klebsiella aerogenes. Eur. J. Biochem. 121:443–449 [DOI] [PubMed] [Google Scholar]
- 7. Dimroth P, Thomer A. 1989. A primary respiratory Na+ pump of an anaerobic bacterium: the Na+-dependent NADH:quinone oxidoreductase of Klebsiella pneumoniae. Arch. Microbiol. 151:439–444 [DOI] [PubMed] [Google Scholar]
- 8. Pfenninger-Li XD, Dimroth P. 1992. NADH formation by Na+-coupled reversed electron transfer in Klebsiella pneumoniae. Mol. Microbiol. 6:1943–1948 [DOI] [PubMed] [Google Scholar]
- 9. Sender P, Martin M, Peiru S, Magni C. 2004. Characterization of an oxaloacetate decarboxylase that belongs to the malic enzyme family. FEBS Lett. 570:217–222 [DOI] [PubMed] [Google Scholar]
- 10. Pudlik AM, Lolkema JS. 2011. Citrate uptake in exchange with intermediates in the citrate metabolic pathway in Lactococcus lactis IL1403. J. Bacteriol. 193:706–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marty-Teysset C, Posthuma C, Lolkema JS, Schmitt P, Divies C, Konings WN. 1996. Proton motive force generation by citrolactic fermentation in Leuconostoc mesenteroides. J. Bacteriol. 178:2178–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Magni C, de Mendoza D, Konings WN, Lolkema JS. 1999. Mechanism of citrate metabolism in Lactococcus lactis: resistance against lactate toxicity at low pH. J. Bacteriol. 181:1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bandell M, Lhotte ME, Marty-Teysset C, Veyrat A, Prévost H, Dartois V, Diviès C, Konings WN, Lolkema JS. 1998. Mechanism of the citrate transporters in carbohydrate and citrate cometabolism in Lactococcus and Leuconostoc species. Appl. Environ. Microbiol. 64:1594–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marty-Teysset C, Lolkema JS, Schmitt P, Divies C, Konings WN. 1995. Membrane potential-generating transport of citrate and malate catalyzed by CitP of Leuconostoc mesenteroides. J. Biol. Chem. 270:25370–25376 [DOI] [PubMed] [Google Scholar]
- 15. Lolkema JS, Poolman B, Konings WN. 1995. Role of scalar protons in metabolic energy generation in lactic acid bacteria. J. Bioenerg. Biomembr. 27:467–473 [DOI] [PubMed] [Google Scholar]
- 16. Lolkema JS, Poolman B, Konings WN. 1996. Secondary transporters and metabolic energy generation in bacteria, p 229–260 In Konings WN, Kaback HR, Lolkema JS. (ed), Handbook of biological physics, vol 2 North-Holland, Amsterdam, Netherlands [Google Scholar]
- 17. Sobczak I, Lolkema J. 2005. The 2-hydroxycarboxylate transporter family: physiology, structure, and mechanism. Microbiol. Mol. Biol. Rev. 69:665–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krom BP, Warner JB, Konings WN, Lolkema JS. 2000. Complementary metal ion specificity of the metal-citrate transporters CitM and CitH of Bacillus subtilis. J. Bacteriol. 182:6374–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blancato VS, Magni C, Lolkema JS. 2006. Functional characterization and Me2+ ion specificity of a Ca2+-citrate transporter from Enterococcus faecalis. FEBS J. 273:5121–5130 [DOI] [PubMed] [Google Scholar]
- 20. Blancato VS, Repizo GD, Suárez CA, Magni C. 2008. Transcriptional regulation of the citrate gene cluster of Enterococcus faecalis involves the GntR family transcriptional activator CitO. J. Bacteriol. 190:7419–7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23:130–135 [Google Scholar]
- 22. Westerfield WW. 1945. A colorimetric determination of blood acetoin. J. Biol. Chem. 161:495–502 [PubMed] [Google Scholar]
- 23. Molenaar D, Abee T, Konings WN. 1991. Continuous measurement of the cytoplasmic pH in Lactococcus lactis with a fluorescent pH indicator. Biochim. Biophys. Acta 1115:75–83 [DOI] [PubMed] [Google Scholar]
- 24. Šíp M, Heøman P, Plášek J, Hrouda V. 1990. Transmembrane potential measurement with carbocyanine dye diS-C3-(5): fast fluorescence decay studies. J. Photochem. Photobiol. B 4:321–328 [DOI] [PubMed] [Google Scholar]
- 25. Palles T, Beresford T, Condon SM, Cogan TM. 1998. Citrate metabolism in Lactobacillus casei and Lactobacillus plantarum. J. Appl. Microbiol. 85:147–154 [Google Scholar]
- 26. Díaz-Muñiz I, Steele J. 2006. Conditions required for citrate utilization during growth of Lactobacillus casei ATCC 334 in chemically defined medium and Cheddar cheese extract. Antonie Van Leeuwenhoek 90:233–243 [DOI] [PubMed] [Google Scholar]
- 27. Díaz-Muñiz I, Banavara DS, Budinich MF, Rankin SA, Dudley EG, Steele JL. 2006. Lactobacillus casei metabolic potential to utilize citrate as an energy source in ripening cheese: a bioinformatics approach. J. Appl. Microbiol. 101:872–882 [DOI] [PubMed] [Google Scholar]
- 28. Pudlik AM, Lolkema JS. 2011. Mechanism of citrate metabolism by an oxaloacetate decarboxylase-deficient mutant of Lactococcus lactis IL1403. J. Bacteriol. 193:4049–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bettenbrock K. 1997. Molekulare Untersuchung eines PTS-abhängigen und eines Leloir-Abbauweges für D-Galaktose bei Lactobacillus casei 64H. PhD thesis Universität Osnabrück, Osnabrück, Germany [Google Scholar]
- 30. Chassy BM, Thompson J. 1983. Regulation and characterization of the galactose-phosphoenolpyruvate-dependent phosphotransferase system in Lactobacillus casei. J. Bacteriol. 154:1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bettenbrock K, Alpert C-A. 1998. The gal genes for the Leloir pathway of Lactobacillus casei 64H. Appl. Environ. Microbiol. 64:2013–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bettenbrock K, Siebers U, Ehrenreich P, Alpert C-A. 1999. Lactobacillus casei 64H contains a phosphoenolpyruvate-dependent phosphotransferase system for uptake of galactose, as confirmed by analysis of ptsH and different gal mutants. J. Bacteriol. 181:225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Torino MI, Taranto MP, Font de Valdez G. 2005. Citrate catabolism and production of acetate and succinate by Lactobacillus helveticus ATCC 15807. Appl. Microbiol. Biotechnol. 69:79–85 [DOI] [PubMed] [Google Scholar]
- 34. Morishita T, Yajima M. 1995. Incomplete operation of biosynthetic and bioenergetic functions of the citric acid cycle in multiple auxotrophic lactobacilli. Biosci. Biotechnol. Biochem. 59:251–255 [Google Scholar]
- 35. Williams AG, Withers SE, Banks JM. 2000. Energy sources of non-starter lactic acid bacteria isolated from Cheddar cheese. Int. Dairy J. 10:17–23 [Google Scholar]
- 36. Fryer TF. 1970. Utilization of citrate by lactobacilli isolated from dairy products. J. Dairy Res. 37:9–15 [Google Scholar]
- 37. Klaenhammer T, Altermann E, Arigoni F, Bolotin A, Breidt F, Broadbent J, Cano R, Chaillou S, Deutscher J, Gasson M, van de Guchte M, Guzzo J, Hartke A, Hawkins T, Hols P, Hutkins R, Kleerebezem M, Kok J, Kuipers O, Lubbers M, Maguin E, McKay L, Mills D, Nauta A, Overbeek R, Pel H, Pridmore D, Saier M, van Sinderen D, Sorokin A, Steele J, O'Sullivan D, de Vos W, Weimer B, Zagorec M, Siezen R. 2002. Discovering lactic acid bacteria by genomics. Antonie Van Leeuwenhoek 82:29–58 [DOI] [PubMed] [Google Scholar]
- 38. Saier MH., Jr 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boorsma A, van der Rest ME, Lolkema JS, Konings WN. 1996. Secondary transporters for citrate and the Mg(2+)-citrate complex in Bacillus subtilis are homologous proteins. J. Bacteriol. 178:6216–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warner JB, Krom BP, Magni C, Konings WN, Lolkema JS. 2000. Catabolite repression and induction of the Mg(2+)-citrate transporter CitM of Bacillus subtilis. J. Bacteriol. 182:6099–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Korithoski B, Krastel K, Cvitkovitch DG. 2005. Transport and metabolism of citrate by Streptococcus mutans. J. Bacteriol. 187:4451–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abbe K, Takahashi S, Yamada T. 1982. Involvement of oxygen-sensitive pyruvate formate-lyase in mixed-acid fermentation by Streptococcus mutans under strictly anaerobic conditions. J. Bacteriol. 152:175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pudlik AM, Lolkema JS. 2012. Substrate specificity of the citrate transporter CitP of Lactococcus lactis. J. Bacteriol. 194:3627–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Repizo GD, Blancato VS, Mortera P, Lolkema JS, Magni C. 2013. Biochemical and genetic characterization of the Enterococcus faecalis oxaloacetate decarboxylase complex. Appl. Environ. Microbiol. 79:2882–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schwarz E, Oesterhelt D. 1985. Cloning and expression of Klebsiella pneumoniae genes coding for citrate transport and fermentation. EMBO J. 4:1599–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dimroth P, Jockel P, Schmid M. 2001. Coupling mechanism of the oxaloacetate decarboxylase Na+ pump. Biochim. Biophys. Acta 1505:1–14 [DOI] [PubMed] [Google Scholar]