Abstract
Lyme disease is a multisystemic disorder caused by Borrelia burgdorferi infection. Upon infection, some B. burgdorferi genes are upregulated, including members of the microbial surface components recognizing adhesive matrix molecule (MSCRAMM) protein family, which facilitate B. burgdorferi adherence to extracellular matrix components of the host. Comparative genome analysis has revealed a new family of B. burgdorferi proteins containing the von Willebrand factor A (vWFA) domain. In the present study, we characterized the expression and membrane association of the vWFA domain-containing protein BB0172 by using in vitro transcription/translation systems in the presence of microsomal membranes and with detergent phase separation assays. Our results showed evidence of BB0172 localization in the outer membrane, the orientation of the vWFA domain to the extracellular environment, and its function as a metal ion-dependent integrin-binding protein. This is the first report of a borrelial adhesin with a metal ion-dependent adhesion site (MIDAS) motif that is similar to those observed in eukaryotic integrins and has a similar function.
INTRODUCTION
Lyme disease is a multisystemic disorder that leads to arthritis in 60% of cases, carditis in 10% of untreated adults, and other neurological symptoms. The causative agent of Lyme disease is the spirochetal pathogen Borrelia burgdorferi, which is transmitted to humans through the bite of an infected Ixodes sp. tick (1). There is currently very little information available on the tissue-specific host-pathogen interactions that lead to pathological manifestations of B. burgdorferi infection. This pathogen's ability to colonize mammals is dependent on its capacity to rapidly alter gene expression in response to highly disparate environmental signals following transmission from infected ticks (2). The open reading frames that are upregulated upon infection include members of the microbial surface components recognizing adhesive matrix molecules (MSCRAMM) protein family, and they facilitate the adherence of B. burgdorferi to extracellular matrix (ECM) components of the host (3).
Comparative genome analysis has also identified a family of von Willebrand factor A (vWFA) domain-containing proteins in B. burgdorferi, including BB0172, BB0173, BB0175, and BB0325 (4–6). The vWFA domains present in ECM proteins and on eukaryotic cells are involved in cell adhesion and protein-protein interactions; they play key roles in the adhesion of platelets to areas of vascular damage by binding to glycoproteins on the platelet surface, to exposed ECM components (5, 7, 8), and to metalloproteases (ADAMTS 13) (7–12). Therefore, the vWFA domain-containing borrelial proteins might be involved in the adhesion of B. burgdorferi to eukaryotic cells, ECM components, and activated platelets, and they may thus play a role in the virulence mechanisms of B. burgdorferi. In humans, absence of vWF results in severe bleeding disorders due to the removal of blood clotting factor VIII from the circulation (12–15). vWF-binding proteins (vWFbp) have been identified in several bacterial species, such as Helicobacter pylori and Staphylococcus aureus (16–19), and these secreted or surface-exposed proteins are involved in the binding of these pathogens to ECM components, platelets, and endothelial cells, thus playing an important role in pathogen colonization and dissemination in the mammalian host. In silico sequence analysis has shown that B. burgdorferi vWFA domain-containing proteins have a sequence domain (DXSXS) that is very similar to the metal ion-dependent adhesion site (MIDAS) found in integrins (20, 21). These proteins also show similarity to the Plasmodium spp. extracellular adhesion molecule TRAP and the LFA-1 integrin (Fig. 1) (6, 22).
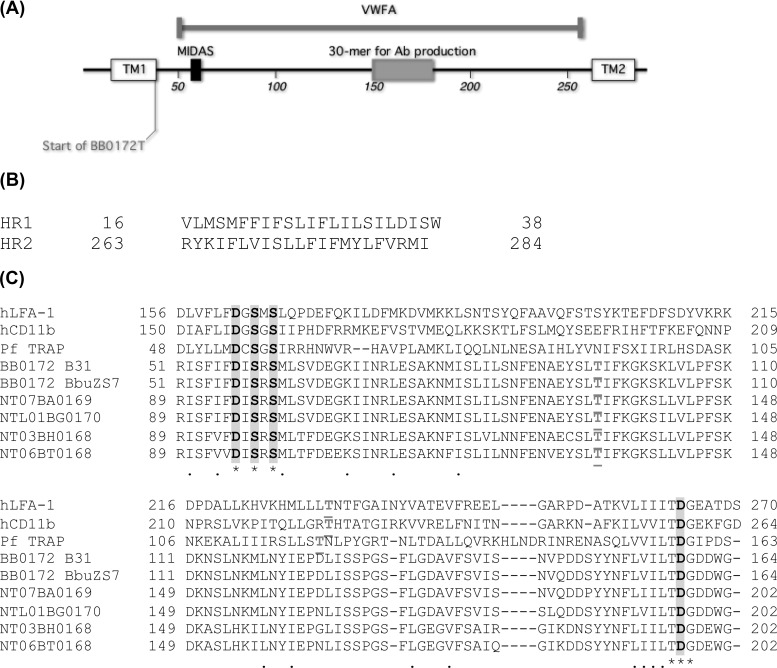
Fig 1.
In silico analysis of BB0172. (A) Schematic representation of BB0172. TM1 and TM2, transmembrane domains 1 (amino acids 17 to 35) and 2 (amino acids 264 to 281), respectively. The vWFA domain includes amino acids 51 to 256. The recombinant BB0172 protein (BB0172T) amino acid sequence spans residues 38 to 291. The anti-BB0172pep antibody (Ab) was generated against a 30-mer peptide, from amino acids 150 to 180. The MIDAS motif is located at residues 57 to 61. (B) Hydrophobic amino acid sequences (HRs) were cloned to study their insertion into membranes. HR1, amino acids 16 to 38, HR2, amino acids 263 to 284. (C) Clustal W (v1.83) alignment of BB0172 of B. burgdorferi B31 (in bold) and ZS7 strains to its homologues in Borrelia garinii (NTL01BG0170), Borrelia afzelii (NTL07BA0169), and the relapsing fever species Borrelia hermsii (NT03BH0168) and Borrelia tunicate (NT06BT0168), as well as to the human adhesins LFA-1 and CD11b and the Plasmodium falciparum membrane protein TRAP. Bold text with gray shadowing indicates the MIDAS domain (DXSXS) and the aspartic acid (D) necessary to complete the metal binding (20, 21). The threonine (T) that is highlighted in gray and underlined is also involved in the MIDAS motif function; it is present in a different location in the bacterial species compared to eukaryotic counterparts. MacVector version 12.6 (MacVector, Inc.) was used for sequence analyses and production of schematics.
The Lyme disease agent binds to a variety of ECM components and integrins, which are metal ion-dependent heterodimeric receptors that mediate cell-to-cell and cell-to-ECM interactions (23). The present study used a well-established in vitro model to investigate the localization and function of the vWFA domain-containing BB0172 protein of B. burgdorferi and to determine its function in adherence to different tissues during infection. Our findings established the topology of the BB0172 protein in biological membranes and its adherence to different ECM components and integrins, emphasizing the complexity of host-pathogen interactions in Lyme disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Borrelia burgdorferi B31 isolate A3 (24) was used throughout this study. To mimic the temperature and pH conditions during the transition of this bacterium from the unfed to the fed tick, the strain was grown in BSK-II medium pH 7.6 complemented with 6% inactivated normal rabbit serum at room temperature (RT) until reaching a density of 107 cells/ml. Then an aliquot of this culture was transferred to BSK-II medium (pH 6.8) and incubated at 37°C with 1% CO2 until reaching a density of 5 × 107 cells/ml (25). Escherichia coli OneShot Top10 cells (Invitrogen, CA) were used for all cloning steps, and Rosetta-gami(DE3)pLysS cells (Novagen, Madison, WI) were used for BB0172 recombinant protein expression. All E. coli strains were grown in LB (Difco) broth with the appropriate antibiotics.
RNA and genomic DNA purification for detecting bb0172 transcripts by PCR.
RNA was extracted as previously described (25, 26). Briefly, B. burgdorferi cultures were grown to a density of 2 × 107 to 3 × 107 spirochetes/ml under the shifting conditions outlined above. RNA was extracted by resuspending the bacterial pellets with 0.2 ml RNA-Bee (Tel-Test, Inc., Friendswood, TX) for every 106 cells. Following extraction with chloroform, RNA was precipitated with isopropanol, washed with 75% ethanol, air dried, and resuspended in RNase-free water. To remove contaminating DNA, the RNA was treated twice with DNase I at 37°C for 45 min. Then, the total RNA was quantified spectrophotometrically and reverse transcribed to cDNA by using TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA). From B. burgdorferi cultures growing under tick-feeding conditions (pH 6.8, 37°C) or regular growing conditions (pH 7.6, 32°C), genomic DNA was obtained by general phenol-chloroform extraction.
RNA, cDNA, and genomic DNA (positive control) samples from each growing condition were used to detect when bb0172 was expressed. A 500-bp fragment of bb0172 was amplified using primers BB0172cDNA-F (B. burgdorferi nucleotides 174705 to 174728) and BB0172cDNA-R (B. burgdorferi nucleotides 174225 to 174249) (Table 1). Primers specific to the flaB, ospC, and p66 genes were also included as controls for the temperature and pH shift (Table 1) (27, 28). PCR products were separated on 0.8% agarose gels and imaged using the Bio-Rad Gel Doc XR system.
Table 1.
Oligonucleotide primers used in this study
| Primer pair | RSa | Sequence (5′→3′)b | Application |
|---|---|---|---|
| BB0172cDNA-F | GCTTGATTTTTTTAATTTTATCC | Amplification of bb0172 500-bp fragment from B. burgdorferi cDNA | |
| BB0172cDNA-R | CGGGATTACTCCCGCCAATCCCAA | ||
| FlaBcDNA-F | AACACACCAGCATCACTTTCAGG | Amplification of flab 234-bp fragment from B. burgdorferi cDNA | |
| FlaBcDNA-R | GAGAATTAACTCCGCCTTGAGAAGG | ||
| P66cDNA-F | CAAAAAAGAAACACCCTCAGATCC | Amplification of p66 683-bp fragment from B. burgdorferi cDNA | |
| P66cDNA-R | CCTGTTTTTAAATAAATTTTTGTAGCATC | ||
| OspCcDNA-F | TATTAATGACTTTATTTTTATTTATATCT | Amplification of ospC 593-bp fragment from B. burgdorferi cDNA | |
| OspCcDNA-R | TTGATTTTAATTAAGGTTTTTTTGG | ||
| BB0172TF | NdeI | ACGCCATATGTGGGGGCAAAGAGCTGTTGAGGA | Amplification of bb0172 for cloning into pCR2.1 and pET23a for protein expression and purification |
| BB0172TR | XhoI | ACGCCTCGAGCTAAAAAGTTTCGTCCCA | |
| BB0172TM1-F | SpeI | ACGCACTAGTGGAGGACCAGGAGTTCTAATGTCAATGTTT | Amplification of the first bb0172 hydrophobic region in frame with Lep sequence |
| BB0172TM1-R | KpnI | ACGCGGTACCCCTCCTGGTCCCCAAGAAATATCCAAAAT | |
| BB0172TM2-F | SpeI | ACGCACTAGTGGAGGACCAGGAAGATATAAAATTTTTTTG | Amplification of the second bb0172 hydrophobic region in frame with Lep sequence |
| BB0172TM2-R | KpnI | ACGCGGTACCCCTCCTGGTCCTATCATCCTGACAAACAA | |
| BB0172-T | ACAGATGGCGATGATTGGGGGGAA | Sequencing | |
| SP6 | ATTTAGGTGACACTATAG | Sequencing | |
| BB0172 SDM MIDAS-1F | GAGAATTTCTTTTATTTTTGCTATTTCTCGTAGCATG | Site-directed mutagenesis of the first amino acid of the MIDAS motif, DXSXS→AXSXS | |
| BB0172 SDM MIDAS-1R | CATGCTACGAGAAATAGCAAAAATAAAAGAAATTCTC | ||
| BB0172 SDM MIDAS-2F | GAGAATTTCTTTTATTTTTGATATTGCTCGTAGCATGTTAAG | Site-directed mutagenesis of the second amino acid of the MIDAS motif, DXSXS→DXAXS | |
| BB0172 SDM MIDAS-2R | CTTAACATGCTACGAGCAATATCAAAAATAAAAGAAATTCTC | ||
| BB0172 SDM MIDAS-3F | GAGAATTTCTTTTATTTTTGATATTTCTCGTGCTATGTTAAGTGTAGATG | Site-directed mutagenesis of the third amino acid of the MIDAS motif, DXSXS→DXSXA | |
| BB0172 SDM MIDAS-3R | CATCTACACTTAACATAGCACGAGAAATATCAAAAATAAAAGAAATTCTC | ||
| BB0172 SDM MIDAS-3X-F | GAGAATTTCTTTTATTTTTGCTATTGCTCGTGCTATGTTAAGTGTAGATG | Site-directed mutagenesis of the three amino acids of the MIDAS motif, DXSXS→AXAXA | |
| BB0172 SDM MIDAS-3X-R | CATCTACACTTAACATAGCACGAGCAATAGCAAAAATAAAAGAAATTCT |
Restriction sites (RS), when present, are indicated by bold text in the sequence.
Site-directed mutations are underlined in the sequence.
Computer-assisted analysis of BB0172 transmembrane (TM) regions.
Membrane-spanning regions in the BB0172 sequence were predicted by using six of the most commonly used prediction methods available online: DAS-TMfilter (29; http://mendel.imp.univie.ac.at/sat/DAS/DAS.html), ΔG Prediction (30; http://dgpred.cbr.su.se/), MEMSAT3 (31; http://bioinf.cs.ucl.ac.uk/psipred/submit), OCTOPUS (32; http://octopus.cbr.su.se/index.php), SOSUI (33; http://bp.nuap.nagoya-u.ac.jp/sosui/sosui_submit.html), and TMHMM (34; http://www.cbs.dtu.dk/services/TMHMM/). The potential presence of a putative signal sequence was also investigated using the SignalP 4.0 server (35; http://www.cbs.dtu.dk/services/SignalP/). All user-adjustable parameters were left at their default values.
Cloning of transmembrane domain regions.
To study the insertion of BB0172 hydrophobic regions into membranes, the segments to be tested (Fig. 1A and B) were engineered into the luminal P2 domain of the integral membrane protein Lep from E. coli (leader peptidase), where it was flanked by two N-glycosylation sites that were used as reporters (see Fig. 5, top). To further clone these regions, the two BB0172 putative TM domains were PCR amplified from total genomic DNA obtained from the B31A3 Borrelia strain by using the primers described in Table 1. PCR product size was verified on 2% agarose gels, and then amplicons were cleaned using the Wizard SV gel and PCR cleanup system (Promega, Madison, WI), following the manufacturer's recommendations. The PCR products were then double digested using SpeI/KpnI enzymes (NEB, Ipswich, MA) and cloned into pGEM-Lep as previously described (36–38). Ligation reaction mixtures were precipitated overnight and electroporated into TOP10 cells. Positive clones were selected on ampicillin plates (100 μg/ml) and verified by sequencing (Eton Biosciences, San Diego, CA). Clones showing the hydrophobic TMs in frame with Lep were used for the in vitro transcription-translation experiments.
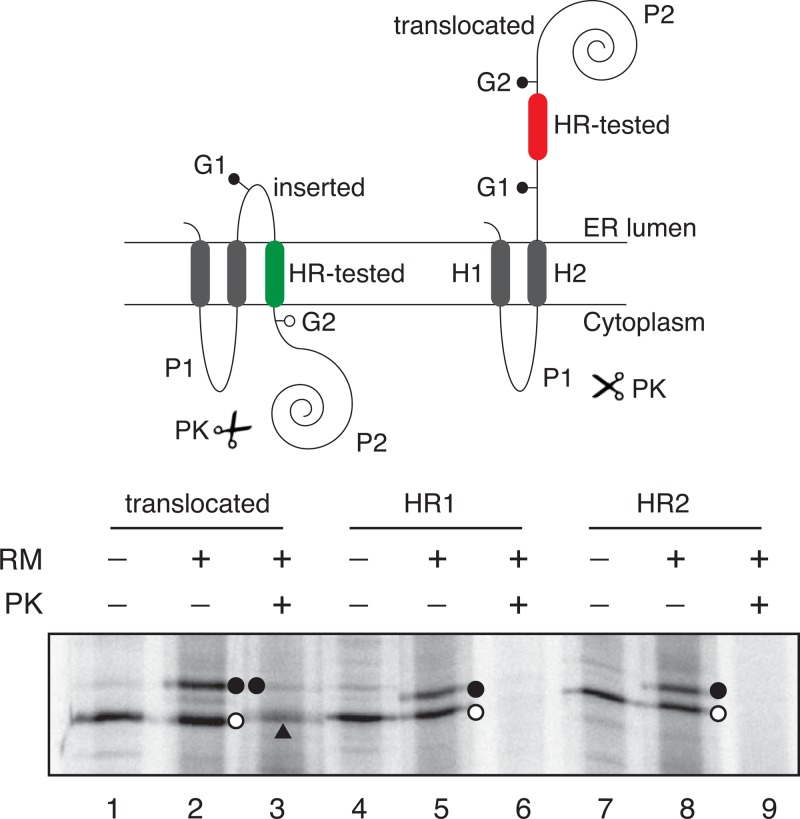
Fig 5.
Hydrophobic regions of the BB0172 insert into microsomal membranes. (Top) Schematic representation of the Lep construct used to report insertion into the endoplasmic reticulum (ER) membrane of BB0172 hydrophobic regions. The TM segment under investigation (HR-tested) was inserted into the P2 domain of Lep, flanked by two artificial glycosylation acceptor sites (G1 and G2). Recognition of the tested sequence as a TM domain by the translocon machinery results in the location of only G1 in the luminal side of the ER membrane, preventing G2 glycosylation (left). The Lep chimera is doubly glycosylated when the sequence being tested is translocated into the lumen of the microsomes (right). (Bottom) In vitro translation of different Lep constructs containing BB0172 HR1 (residues 16 to 38; lanes 4 to 6) and HR2 (residues 263 to 284, lanes 7 to 9) sequences in the presence of membranes. A control construct was used to verify sequence translocation (lanes 1 to 3). Constructs were transcribed and translated in the presence or absence of rough microsomal membranes (RM) and proteinase K (PK), as indicated. Bands of nonglycosylated protein are indicated by a white dot; singly and doubly glycosylated proteins are indicated by one and two black dots, respectively. The arrowhead identifies protected a double-glycosylated P2 domain after PK treatment.
In vitro transcription-translation.
In vitro translation of in vitro-transcribed mRNA was performed in the presence of reticulocyte lysate, [35S]Met-Cys, and dog pancreas microsomes, as described previously (39). Lep constructs with hydrophobic region (HR)-tested segments from the BB0172 sequence (residues 16 to 38 and 263 to 284) were transcribed and translated as previously reported (36). After translation, membranes were collected by ultracentrifugation and analyzed by sodium-dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gels were visualized on a Fuji FLA3000 PhosphorImager with ImageGauge software.
The proteinase K protection assay was performed as previously described (40). Briefly, the translation mixture was supplemented with 1 μl CaCl2 (50 mM) and 1 μl proteinase K (4 mg/ml) and then digested for 40 min on ice. The reaction was stopped by adding 1 mM phenylmethylsulfonyl fluoride (PMSF), followed by SDS-PAGE analysis. This process is shown schematically in Fig. 5, below.
Expression and purification of BB0172T.
For this experiment, a truncated BB0172 (BB0172T) protein was purified to avoid insertion of the protein into E. coli membranes and to ensure purification of BB0172 as a soluble protein. Total genomic DNA obtained from the B31A3 strain (Table 2) was used as a template to PCR amplify bb0172 from nucleotides 150 to 873 (amino acids 50 to 290), using the primers listed in Table 1. The amplicons were cloned into the pCR2.1-TOPO vector (Invitrogen), transformed into E. coli TOP 10 cells, and subjected to blue/white colony screening in the presence of ampicillin (100 mg/ml) and kanamycin (50 mg/ml). The insert was digested with NdeI/XhoI and ligated into the pET23a expression vector. The ligated products were electrotransformed into E. coli TOP10 cells and screened for the presence of the insert by restriction enzyme digestion. The junctions of plasmids containing inserts of the expected sizes were sequenced and used to transform the E. coli expression host. Recombinant BB0172 (rBB0172) with a C-terminal 6×His tag was overexpressed by inducing the E. coli strain containing pET23a-bb0172T with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h. The bacterial pellets were disrupted by using a French press and denaturing lysis buffer (8 M urea, 20 mM imidazole; pH 7.4). The supernatants were collected, clarified by centrifugation, and subjected to affinity purification with a His60 Ni Superflow resin (Clontech, Mountain View, CA), following the manufacturer's instructions. The bound 6×His-tagged proteins were eluted as 0.5-ml fractions with elution buffer (8 M urea, 300 mM imidazole; pH 7.4) and then analyzed by SDS–12.5% PAGE. Select fractions with the largest concentrations of eluted proteins were further purified using dialysis against 50 mM sodium phosphate and 300 mM NaCl (pH 7.4; Slide-A Lyze G2 dialysis cassette; Thermo Scientific). After dialysis, Amicon centrifugal filters (Millipore) were used to concentrate the proteins. A 27.5-kDa protein was purified to homogeneity (data not shown), quantified in a bicinchoninic acid (BCA; Thermo Scientific, Inc.) assay, and stored at −80°C until further use.
Table 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Source |
|---|---|---|
| Borrelia burgdorferi B31A3 | cp9−, wild type | Rocky Mountain Labs (1) |
| E. coli strains | ||
| OneShot Top10 | Cloning host; F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL(Strr) endA1 nupG | Invitrogen |
| Rosetta-gami(DE3)pLysS | Expression host; Δ(ara leu)7697 ΔlacX74 ΔphoA PvuII phoR araD139 ahpC galE galK rpsL DE3 F′ [lac+ lacIq pro] gor522::Tn10 trxB pLysS RARE2 (Camr Kanr Strr Tetr) | Novagen |
| Plasmids | ||
| pGEM-Lep | Cloning host, containing E. coli Lep, leader peptidase (Ampr) | 82 |
| pLE102 | pGEM-1(bb0172TM1-Lep) | This study |
| pLE103 | pGEM-1(bb0172TM2-Lep) | This study |
| pLE136 | pET23a(bb017250–290) | This study |
| pECW1 | pET23a (bb017250–290-AXSXS) | This study |
| pECW2 | pET23a (bb017250–290-DXAXS) | This study |
| pECW3 | pET23a (bb017250–290-DXSXA) | This study |
| pECW3X | pET23a (bb017250–290-AXAXA) | This study |
Site-directed mutagenesis of DXSXS metal-binding domain.
BB0172 contains a MIDAS motif (Fig. 1C) comprising amino acids 57 to 61 (DXSXS). To determine which amino acids are essential for maintaining this protein's function, we performed site-directed mutagenesis to change the motif-relevant amino acids to alanine, creating the mutants D57A, S59A, and S61A and the triple mutant (21). For this process, we used the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies), following the manufacturer's recommendations. Primers (Table 1) were designed as recommended by the manufacturer and using their primer design tool for site-directed mutagenesis. Positive colonies were screened by sequencing using the universal T7 promoter and terminator primers (Eton Biosciences, Ltd., San Diego, CA). Mutant MIDAS motif clones were stored at −80°C for further use. For their expression and purification, 2 μl of purified plasmid for each mutation was electroporated into Rosetta-gami E. coli strains (Novartis). Further binding assays were performed with all isolated mutants, as described below.
Generation of specific BB0172 antibodies.
To generate antibodies specific to the extracellular domain of BB0172, we design a 30-mer-long peptide corresponding to amino acids 150 to 180 (Fig. 1A) with the sequence YNFLVILTDG DDWGENNYYR FSKFVNNLKL conjugated to KLH (Peptide 2.0, Chantilly, VA). The synthetic peptide was dissolved in 10% dimethyl sulfoxide until reaching homogeneity and administered to three BALB/c mice at a dose of 50 μg/mouse in conjugation with TiterMax Gold (Sigma-Aldrich, St. Louis, MO) on days 0, 14, and 21. At 28 days, the mice were euthanized and exsanguinated. Serum was collected and used in the following immunoblot and binding assays; this specific antiserum is henceforth referred to as anti-BB0172pep. Specific serum to the whole BB0172 protein was obtained following the same immunization protocol and using 50 μg of rBB0172T per mouse, but we detected no BB0172 in the B. burgdorferi whole-cell lysates or from purified rBB0172T. Therefore, we used the anti-BB0172pep developed in this study.
Triton X-114 partitioning of B. burgdorferi outer membrane proteins.
To evaluate the localization of BB0172 in B. burgdorferi, we separated the outer membrane proteins (OMPs) from the protoplasmic cylinders (PCs) by Triton X-114 partitioning as previously described (40–43). Briefly, B. burgdorferi was grown at RT and pH 7.6 and then shifted to 37°C, pH 6.8 and grown to a density of 5 × 107 spirochetes/ml. Cells were washed in phosphate-buffered saline (PBS), resuspended in PBS containing 1% Triton X-114, and incubated overnight at 4°C with gentle rocking to ensure separation of the outer membrane proteins without disruption of the protoplasmic cylinders. After the overnight partitioning, PCs were pelleted by centrifugation at 15,000 × g for 15 min at 4°C. Supernatants containing OMPs were incubated at 37°C for 15 min in the presence of 2% Triton X-114. The OMP-containing organic phase was separated by centrifugation at 2,000 × g for 10 min at RT and then washed three times with PBS. Finally the OMPs were precipitated out of the organic phase by adding 10× ice-cold acetone at −20°C for 2 h, followed by centrifugation at 15,000 × g for 30 min at 4°C. PCs, the aqueous phase, and the detergent phase containing the OMPs from different growing conditions were separated in SDS–12% PAGE gels and silver stained (Silver Staining Plus; Bio-Rad Laboratories, Inc., Hercules, CA), following the manufacturer's recommendations, or transferred to polyvinylidene difluoride (PVDF) membranes to determine the BB0172 localization by immunoblot assay as described below. OMPs OspC and P66, the periplasmic protein superoxide dismutase A (SodA), and the cytosolic proteins carbon storage regulator A (CsrA) and the Borrelia oxidative stress regulator (BosR) were used as controls.
SDS-PAGE gels and immunoblot analysis.
Borrelia burgdorferi whole-cell lysates (prepared from cultures grown at RT and pH 7.6, then shifted to pH 6.8 and 37°C) were treated with proteinase K (20 μg/ml) and separated by SDS–12.5% PAGE. The separated proteins were either visualized by Coomassie brilliant blue staining or transferred onto a PVDF membrane (Hybond-P; GE Healthcare, Piscataway, NJ) and subjected to immunoblot analysis. The PVDF membranes were blocked overnight at 4°C in Tris-buffered saline containing 0.2% Tween 20 (TBS-T) and 10% skim milk. After blocking, membranes were probed with mouse anti-BB0172pep. Polyclonal mouse anti-P66 and anti-OspC (25) sera were used as controls for proteinase K activity on outer membrane proteins and controls for the fractioning of outer membrane proteins in the Triton-X114 partitioning assay. Polyclonal mouse anti-SodA and rabbit anti-BosR sera were used as controls of cytoplasm-located proteins (44). The blots were developed following incubation with appropriate dilutions of horseradish peroxidase (HRP)-conjugated secondary antibodies and using enhanced chemiluminescence Western blotting reagents (GE Healthcare, Piscataway, NJ) as previously described (25, 26, 44, 45).
Binding assays.
To understand the possible capability of BB0172 to bind to ECMs, we purified a truncated BB0172 protein lacking the N terminus and the first TM segment (BB0172T) (Fig. 1), and we analyzed its binding to collagen type I, collagen type IV, laminin, and human fibronectin (BD BioCoat, Bedford, MA). All incubations were performed using HBS buffer (HEPES-buffered saline) containing 0.25 mM Ca2+, 1 mM Mn2+, 1 mM Mg2+, and 1% bovine serum albumin (BSA) (46, 47). After blocking plates with 3% BSA in HBS containing 1% Tween 20 (HBS-T), we added 500 ng/well of rBB0172T or rBBK32 (positive binding control) to the plates in quadruplicate wells and incubated them at RT for 1 h (47–49). The plates were washed three times with HBS-T to remove unbound BB0172T protein. Then, plates were incubated with a 1:1,000 dilution of mouse-anti-rBB0172pep or 1:2,000 dilution of the mouse anti-rBBK32 antibody for 1 h at RT, followed by a 1-hour incubation with a 1:5,000 dilution of anti-mouse HRP-conjugated antibody (GE Healthcare, Piscataway, NJ). Plates were washed and incubated with o-phenyldiamine dihydrochloride (OPD) tablets (Thermo Scientific), and the absorbency at 450 nm was measured by using a FLUOstar Omega plate reader (BMG LabTech, Cary, NC). A blank control (BK) was included in quadruplicate in all plates.
To evaluate the binding of rBB0172T to a wider range of ECMs and integrins, and to prevent any interference of the anti-rBB0172 antibody with binding capabilities to the substrates, 96-well flat-bottom plates (Nunc) were coated with 500 ng/well of each of the recombinant proteins: rBB0172, rBBK32, and rP66. All incubations used HBS buffer containing 0.25 mM Ca2+, 1 mM Mn2+, and 1 mM Mg2+ (46, 47). After coating overnight at 4°C in carbonate buffer, pH 9.2, plates were washed and blocked using HBS-T with 3% BSA buffer for 2 h at RT. Blocked plates were washed with HBS-T and incubated for 1 h at RT with 500 ng/well of each of the ECM components (human plasma fibronectin, human tissue fibronectin, aggrecan, decorin, vitronectin, elastin, and fibrinogen; Sigma-Aldrich, Co., LLC) and integrins (α1β1, α3β1, αVβ3, αVβ5, and α5β1; EMD Millipore Corporation) (46, 47). After washing, plates were incubated for 1 h at RT with each respective anti-ECM component or anti-integrin antibody. The plates were washed again, and a secondary HRP-conjugated antibody was applied for 1 h at RT, followed by development with OPD tablets (Thermo Scientific) and measurement of the A450 as described above. Wells without ECM components or integrins were used as blanks. The effects of the mutations in the MIDAS motif of BB0172 were also evaluated by analyzing their binding to α3β1 integrin following the same protocol described above.
Statistical analysis.
Two-way analysis of variance was used to determine significant differences in the binding of BB0172T to the different integrins and extracellular matrix components, along with the Sidak multiple comparison test with a 95% confidence interval. Analyses were performed using Prism 6.0d (GraphPad Software, Inc.).
RESULTS
B. burgdorferi expression and localization of BB0172 protein.
We found that the bb0172 gene was not expressed when B. burgdorferi was grown at RT and pH 7.6, while an mRNA product was detected when cultures were shifted from RT/pH 7.6 to either 37°C/pH 6.8 or 32°C/pH 7.6 (Fig. 2, compare lanes 1 and 3 in A to lanes 1 and 4 in B). Evaluation of flaB, p66, and ospC gene expression at the mRNA level showed the presence of flaB and p66 under both the RT/pH 7.6 and the temperature-pH (T/pH)-shifting conditions, while ospC was expressed only under T/pH-shifting conditions (Fig. 2A). These results suggested that bb0172 gene expression is selectively triggered by a change in temperature. Therefore, in subsequent studies, we adopted the temperature and pH shift strategy to induce BB0172 expression on borrelial cells and to detect the protein.
Fig 2.
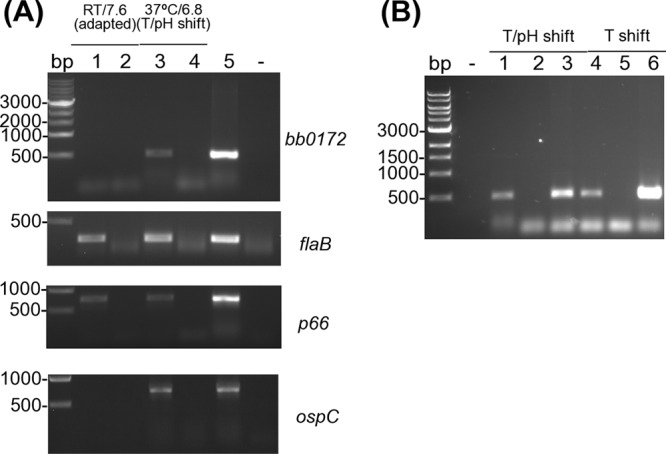
BB0172 expression upon temperature shift. B. burgdorferi B31A3 strain was grown at RT/pH 7.6 and shifted to 37°C/pH 6.8 (T/pH shift) or to 32°C/pH 7.6 (T shift). The purified mRNA was reverse transcribed to cDNA, and PCR was performed to detect bb0172, flaB, p66, and ospC. Water was used as a negative control (-). (A) RNA samples from all purifications were checked for DNA contamination and applied in lanes 2 and 4. Genomic DNA was included as a positive control in lane 5. cDNA results are shown in lanes 1 and 3. (B) cDNA samples of cultures growing after T/pH shift or T shift are represented in lanes 1 and 4, RNA is in lanes 2 and 5, and genomic DNA is in lanes 3 and 6. The DNA ladder is shown on the left side of each panel, with the sizes indicated in base pairs.
We next evaluated the cellular localization of the BB0172 protein in B. burgdorferi based on proteinase K sensitivity and Triton X-114 partitioning of OMPs, followed by immunoblot analysis (Fig. 3). The BB0172 protein was only present in cultures that were shifted to 37°C/pH 6.8, not in RT cultures (Fig. 3B, lane 1). This result correlates with the RNA data, supporting the hypothesis that this protein is expressed during the change from RT conditions to warmer conditions (32°C to 37°C). Furthermore, the protein was not observed after proteinase K treatment, suggesting that the vWFA domain of this protein (against which the antibodies were generated) is accessible from outside the cell.
Fig 3.
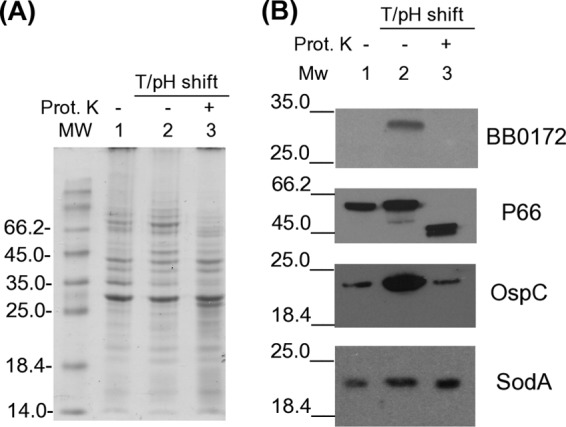
BB0172 localizes on the B. burgdorferi surface. B. burgdorferi B31A3 was cultured at RT/pH 7.6 (lane 1), shifted to 37°C/pH 6.8 (lane 2), and shifted to 37°C/pH 6.8 and treated with proteinase K (lane 3). Whole-cell lysates of all treatments were separated on a 12% SDS-PAGE gel and stained with Coomassie blue (A) or transferred to PVDF membranes and probed with mouse anti-BB0172pep followed by anti-mouse HRP-conjugated antibody (B). OspC (outer membrane lipoprotein), SodA (intracellular), and P66 (outer membrane integral protein) were used as controls for the proteinase K treatment. T/pH shift denotes the shift from RT/pH 7.6 to 37°C/pH 6.8.
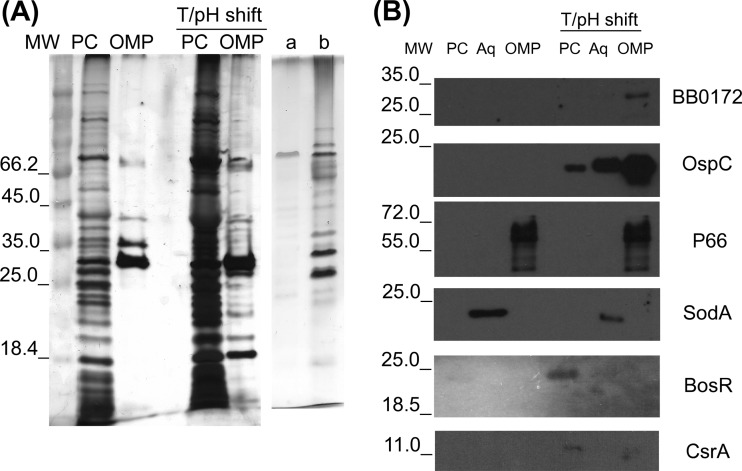
We also evaluated the presence of BB0172 in the outer membrane fraction of B. burgdorferi by using Triton X-114 partitioning of OMPs after shifting the cultures from RT/pH 7.6 to 37°C/pH 6.8. BB0172 was only detected in the OMP fraction of the shifted B. burgdorferi cultures, not in the PC or OMP fractions extracted from cultures grown at RT/pH 7.6 (Fig. 4B). In addition, when B. burgdorferi was grown at 37°C/pH 6.8, the borrelial outer membrane lipoprotein OspC was detected at larger amounts in the OMP fraction than in the PC fraction. Very small amounts of OspC were detected in the OMP fraction isolated from cells growing at RT/pH 7.6, and it was only detectable after overexposing the immunoblot (data not shown). Moreover, SodA was only detected in the Borrelia aqueous phase, regardless of the growing conditions used, while BosR was only detected in the PC fraction from cells growing at 37°C/pH 7.6. Altogether, these results confirmed the presence of the BB0172 protein in the outer membrane of B. burgdorferi, with its vWFA domain oriented toward the extracellular milieu.
Fig 4.
B. burgdorferi BB0172 anchors to the outer membrane. The B. burgdorferi B31A3 strain was cultured at RT/pH 7.6 and shifted to 37°C/pH 6.8, and the PCs and OMPs were separated by Triton X-114 partitioning. PC and OPM fractions from all treatments were separated on 12% SDS-PAGE gels and stained with Silver Staining Plus (Bio-Rad) (A) or transferred to PVDF membranes and probed with mouse anti-BB0172pep followed by anti-mouse HRP-conjugated antibody (B). Serum specific for OspC (outer membrane lipoprotein), SodA (periplasmic), BosR (cytoplasmic), and CsrA (cytoplasmic) were used as a controls. T/pH shift denotes the shift from RT/pH 7.6 to 37°C/pH 6.8. MW, molecular weight marker; lane 1, PCs from cultures at RT/pH 7.6; lane 2, OMPs from cultures at RT/pH 7.6; lane 3, PCs from cultures at 37°C/pH 6.8; lane 4, OMPs from cultures at 37°C/pH 6.8; lane a, aqueous phase from cultures at RT/pH 7.6; lane b, aqueous phase from cultures at 37°C/pH 6.8.
BB0172 is anchored to biological membranes through two transmembrane segments.
The BB0172 amino acid sequence was parsed to test the performance of several commonly used algorithms for predicting TM regions of integral membrane proteins. We submitted the BB0172 sequence to the most current online versions of six widely used prediction methods: DAS-TMfilter (29), ΔG Predictor (30), MEMSAT3 (31), OCTOPUS (32), SOSUI (33), and TMHMM (34). The predicted outcomes significantly coincided across the different methods used, with all algorithms predicting the presence of two TM domains at similar positions in the protein sequence (Table 3). It has been established that the reliability of a given prediction is very high when many different methods agree (40). Additionally, the SignalP 4.0 (35) results predicted that it is unlikely that BB0172 contains a signal sequence.
Table 3.
In silico analysis of the BB0172 amino acid sequence
| Algorithm | Membrane protein? | Resulting no. of TM segments (starting amino acids/ending amino acids) |
|---|---|---|
| DAS-TMfilter | Yes | 2 (17/35 and 264/281) |
| ΔG Prediction | Yes | 2 (16/38 and 263/284) |
| MEMSAT3 | Yes | 2 (21/39 and 264/283) |
| OCTOPUS | Yes | 2 (17/38 and 263/283) |
| SOSUI | Yes | 4 (16/38, 177/198, 219/241, and 258/280)a |
| TMHMM | Yes | 2 (20/39 and 266/284) |
Bold numbers denote “primary” helices.
The membrane insertion of the two BB0172 hydrophobic regions was investigated by using an in vitro experimental system that accurately reports the integration of TM helices into microsomal membranes (Fig. 5, top) (37, 50). The translation of the chimeric constructs harboring the predicted BB0172 TM regions as HR-tested sequences mainly resulted in single glycosylated forms (Fig. 5, lanes 4 to 9), suggesting membrane integration of these two regions. Lanes 1 to 3 in Fig. 5 show the control construct (38, 40, 51) with a previously tested computer-designed translocated (nonintegrated) sequence. Proteinase K treatment of these samples rendered complete loss of detectable fragments in the case of the BB0172 tested sequences, while a clear protected band was observed in the translocated control construct (Fig. 5, compare lane 3 to lanes 6 and 9), confirming membrane insertion of the BB0172 hydrophobic regions. Although these in vitro experiments are not entirely representative of in vivo conditions, the results strongly suggest that the translocon machinery can recognize the assayed regions, ultimately ensuring their proper integration into the intended target membrane.
Binding of BB0172 to human ECM components and integrins.
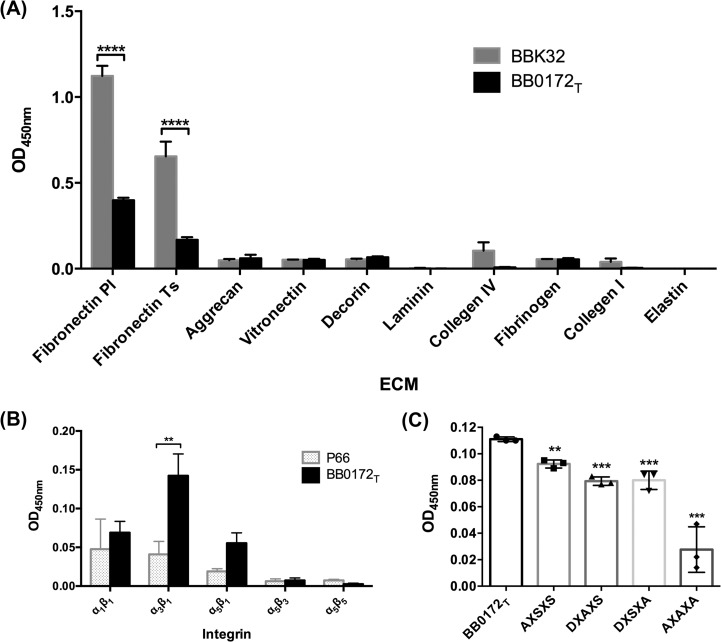
After characterizing the orientation in the membrane of the BB0172 vWFA domain, the recombinant BB0172 protein was used to perform a number of binding assays to different human ECM components and integrins. Recombinant expression of full-length BB0172 protein in Escherichia coli cells was not detected on Coomassie-stained gels or Western blots (data not shown). Since it was previously reported that the first TM segment can prevent translation of membrane proteins in E. coli cells (52), we then expressed a truncated version of the BB0172 protein lacking the N terminus and the first TM segment (BB0172T) (Fig. 1). This strategy enabled expression and purification of the truncated BB0172 as a recombinant protein in sufficient concentrations for further characterization. The first set of binding experiments showed that the BB0172 protein was slightly bound to human plasma fibronectin in the presence of the metals Ca2+, Mn2+, and Mg2+; binding to other ECM components was not observed (Fig. 6A). The binding of the BB0172 protein to human plasma fibronectin was very weak and significantly lower than that observed for BBK32, an extensively characterized plasma fibronectin-binding protein of B. burgdorferi (48, 53–55). We next evaluated the binding capability of the BB0172 protein to integrins. The BB0172 protein significantly bound to α3β1 integrin in the presence of divalent cations, while P66 (a β3 integrin-binding protein from B. burgdorferi) showed very low binding (Fig. 6B). This result was consistently observed, suggesting that the BB0172 protein could be a α3β1 integrin-binding protein.
Fig 6.
BB0172 is an integrin-binding protein. (A) Purified BB0172T was incubated with different ECM components: aggrecan, collagen I and IV, decorin, elastin, laminin, fibrinogen, human plasma fibronectin, human tissue fibronectin, and vitronectin. Recombinant BBK32 was used as a borrelial positive-control protein for the binding to human plasma fibronectin. (B) The binding of BB0172 to different integrins by using HBS buffer containing Ca2+, Mg2+, and Mn2+. Recombinant P66 was used as a positive control for the binding to human integrins. BB0172 bound more strongly to α3β1 integrin compared with the P66 binding. (C) Purified recombinant BB0172 proteins containing mutations in the MIDAS motif (AXSXS, DXAX, DXSXA, and AXAXA) were incubated with α3β1 integrin. BB0172T corresponds to the wild-type sequence. Each mutation of the MIDAS motif altered efficacy of the binding of BB0172 to α3β1 integrin. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with the control protein). All binding assays entailed four replicates for each protein in each ECM component or integrin.
Ligand binding to integrins is usually dependent upon divalent cations. The presence of a MIDAS motif in the BB0172 amino acid sequence (Fig. 1C) suggests that this region may accomplish this function by adopting a specific fold. To investigate whether this region of BB0172 is functionally equivalent to a MIDAS domain, we performed an alanine-scanning analysis by replacing critical residues of the MIDAS motif (DXSXS) (Table 2) with alanine through site-directed mutagenesis. Each single-point mutant strain (AXSXS, DXAXS, and DXSXA) plus a triple mutant strain harboring all three replacements (AXAXA) was used in a α3β1 integrin-binding assay. The results showed that all three individual mutations significantly reduced the binding of BB0172 to α3β1 integrin compared to the wild-type sequence (Fig. 6C), suggesting that all three amino acid residues are important for the coordination of the metal required to maintain the function of the BB0172 protein. As expected, the triple mutation induced the highest reduction of binding, indicating that these amino acids play a critical role in maintaining the coordination of metals in the BB0172 protein.
DISCUSSION
vWFA domain-containing proteins are found in a broad array of systems, from integrins in mammalian organisms to bacteria (5, 13, 56–58). To be functional, these proteins must exist on the cell surface to interact with their receptors. The chromosome of Borrelia burgdorferi encodes a series of vWFA domain-containing proteins (BB0172, BB0173, BB0175, and BB0325). Of all these hypothetical proteins, in silico analysis only predicts those encoded by the bb0172 and bb0173 genes to be membrane proteins. The main objectives of the present work were to determine the BB0172 protein expression pattern, to demonstrate its membrane association, and to characterize its binding to potential host components. Furthermore, we evaluated the cellular localization of the BB0172 vWFA domain by using proteinase K treatment and Triton X-114 partitioning, together with immunoblot analyses. Altogether, the results showed that BB0172 is a helical membrane protein with two TM segments expanding from amino acids 16 to 38 and 263 to 284, suggesting a specific membrane disposition (Fig. 7). Immunoblot analysis with proteinase K-treated whole cells consistently revealed that the vWFA domain of BB0172 was not visible due to its degradation during treatment, indicating that the vWFA domain was exposed to the outer cell surface. Triton X-114 partitioning experiments also demonstrated the localization of BB0172 in the outer membrane of B. burgdorferi.
Fig 7.
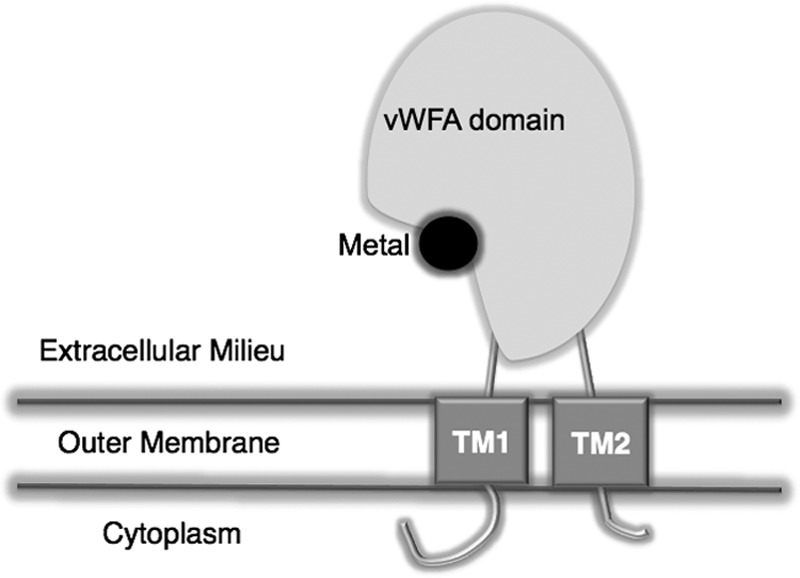
Schematic representation of B. burgdorferi BB0172 topology. We propose that BB0172 is anchored to the borrelial outer membrane through two TM segments located at the N terminus and C terminus of the protein and that the vWFA domain will be exposed to the exterior of the cell. The MIDAS motif is also present in the external segment of this protein, actively participating in the adhesion of BB0172 to α3β1 integrin.
This is the first study demonstrating that a vWFA domain-containing borrelial protein is anchored to the cell surface through two TM helices. We could not obtain the full-length protein by in vitro translation techniques, probably due to differences in the codon usage between B. burgdorferi and the rabbit system utilized in these experiments. For instance, the codons for Arg (AGA), Gln (CAA), Ile (AUU), Leu (UUA) Pro (CCU), Ser (UCU), and Thr (ACU) that are preferentially used by B. burgdorferi are poorly used in rabbit (Oryctolagus cuniculus). In addition, previous experiments conducted in our laboratory showed that some borrelial proteins are only recombinantly expressed in E. coli strains (Rosetta strain from Novagen) that are engineered to recognize rare codons for Leu and Ile (data not shown). Due to these limitations, here we evaluated short polypeptide regions by using in vitro translocation/insertion of putative TM domains in the presence of canine pancreatic microsomes and rabbit reticulocytes. While the hydrophobic regions tested here came from prokaryotic proteins, the high degree of sequence conservation in the translocon components between prokaryotes and eukaryotes (59) suggests that general conclusions can be drawn based on studies using the microsomal assay system and that the present results are relevant to the study of the BB0172 surface-exposed regions of B. burgdorferi. This contention is further supported by previous observations of prokaryotic sequences based on measurements obtained using this system (60). Based on our present results, this strategy appears to be a valuable tool for studying complex membrane proteins from prokaryotic organisms that are difficult to culture in vitro but that have a sequenced genome. Further modifications of the current system utilizing a synthetic bb0172 gene recoded with a more appropriate codon combination are in progress.
In our study, both bb0172 RNA and protein were only detected when the pathogenic B. burgdorferi B31A3 strain was shifted from RT/pH 7.6 to 37°C/pH 6.8 or 32°C/pH 7.6. Additionally, bb0172 expression was inhibited when cells were adapted to any of the studied temperature/pH combinations. These results suggest that BB0172 is only present under the conditions found during the temperature shift that is part of the transition of B. burgdorferi from the unfed tick to the fed tick, indicating that BB0172 might be of great importance during the migration of this pathogen from the tick to the mammalian host. It should be noted that a recent proteomic analysis of B. burgdorferi in response to culture condition changes detected two BB0172-derived peptides at RT/pH 7.6 but not at 34°C/pH 6.6 (61). This discrepancy with the present data could have arisen from the differences in culture shifting conditions between studies and from the low total number of spectral counts found with the proteomic approach, which indicates a very low abundance of BB0172 under all the conditions studied. Taking all these observations together, we hypothesize that BB0172 plays an important role as an adhesin during the first hours postinfection. Therefore, we performed a number of binding assays to ECM components as well as to several commercially available integrins. Our findings suggested that the BB0172 protein is a metal-dependent adhesin to the human α3β1 integrin.
Integrins are heterodimeric receptors that mediate cell-to-cell and cell-to-ECM interactions (23, 62). Pathogenic B. burgdorferi can target integrins and ECM components to local sites of damage in host cells to promote bacterial dissemination (23, 63). For instance, the outer membrane protein P66 interacts with integrin αIIbβ3 on the surface of activated platelets, as well as with integrin αvβ3 on endothelial cells (47, 64, 65). Similarly, B. burgdorferi infection induces the expression of selected integrins in the C3H/HeN mouse model, which can influence the severity of Lyme disease-derived carditis and arthritis (66, 67). Moreover, different B. burgdorferi proteins can bind to specific ECM components; for example, the BBK32 protein interacts with fibronectin (48, 53–55), and decorin-binding proteins A and B (DbpA and DbpB) interact with decorin and chondroitin (68–71). However, analysis of borrelial mutants deficient in BBK32 or DbpA/B did not show complete abrogation of the binding to extracellular products, suggesting that this binding might be mediated by unknown functionally and structurally related determinants (49).
Behera and collaborators (72) previously described BBB07, an integrin α3β1-binding protein of B. burgdorferi. They showed that the RGD-dependent binding of the BBB07 protein to integrins specifically stimulated the generation of proinflammatory cytokines in primary human chondrocyte cultures. In the present study, we observed a similar level of binding between the BB0172 protein and the same α3β1 integrin, but in an RGD-independent manner, since BB0172 lacks this motif. Integrin α3β1 binds to collagen I and IV, laminin, fibronectin, and nidogen, and in some cell types to thrombospondin 1 (73–76). Integrin α3β1 is also critical for the formation of kidneys and epidermis, and also in wound healing, and it is significantly expressed by neurons in the brain (73–76).
Interestingly, we found that BB0172 has a MIDAS sequence motif at residues 57 to 61 (DXSXS), which is highly conserved among Borrelia species. Mutations of these amino acid residues reduced recombinant BB0172 binding to integrin α3β1, with the strongest impact seen with the triple mutant (AXAXA), which reduced the binding to near background levels. We hypothesized that these mutations reduced the ability of BB0172 to coordinate metals, thus diminishing capability of the protein to bind integrin α3β1. Consequently, we speculate that the vWFA domain of BB0172 includes a MIDAS domain similar to that found in the I-domain of integrins (77–81), which is essential to maintain the protein's function. Further studies are in progress with the aim of deleting bb0172 from B. burgdorferi and complementing it with its native form and the mutated versions. We propose that this mutation might reduce the ability of B. burgdorferi to bind to tissues where integrin α3β1 is highly expressed, e.g., the basolateral membrane in epidermis, glomerular podocytes in the kidney, and collecting tubes and neurons in the brain (74).
In summary, the results of the present study prove that BB0172 is a surface-exposed membrane protein that displays a significant capacity to bind integrin α3β1. Furthermore, the in vitro transcription/translation strategy utilized here could be useful for further studies of the helical membrane proteins of fastidiously growing bacteria. The technique developed in this paper was demonstrated to correlate well with studies of whole-protein localization on the borrelial surface, validating the potential use of this technology with other hypothetical membrane proteins of B. burgdorferi. Finally, this is the first report of a metal-dependent borrelial adhesin with an active MIDAS motif that is essential for maintaining protein function.
ACKNOWLEDGMENTS
This work was supported by an American Heart Association postdoctoral fellowship (AHA-0825175F), a Scientist Developing Grant (11SDG4990006), a Texas A&M International Research Travel Award Grant (IRTAG) (to M.D.E.-G.), the Texas A&M University Undergraduate Scholar Program, the Undergraduate Biology and Mathematics Program (to E.C.W.), and the Spanish Ministry of Science and Innovation (BFU2009-08401 and BFU2012-38482) to I.M. S.T. received a predoctoral fellowship from the University of Valencia (V Segles program).
We thank Patricia Rosa at Rocky Mountain Labs for kindly providing B. burgdorferi B31A3 strain and J. Seshu at the University of Texas San Antonio for providing the plasmids used for expression of the BB0172 recombinant protein.
Footnotes
Published ahead of print 17 May 2013
REFERENCES
- 1.Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 10:87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuels DS. 2011. Gene regulation in Borrelia burgdorferi. Annu. Rev. Microbiol. 65:479–499 [DOI] [PubMed] [Google Scholar]
- 3.Kenedy MR, Lenhart TR, Akins DR. 2012. The role of Borrelia burgdorferi outer surface proteins. FEMS Immunol. Med. Microbiol. 66:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV. 1999. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289:729–745 [DOI] [PubMed] [Google Scholar]
- 5.Whittaker CA, Hynes RO. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13:3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian G, Koonin EV, Aravind L. 2000. Comparative genome analysis of the pathogenic spirochetes Borrelia burgdorferi and Treponema pallidum. Infect. Immun. 68:1633–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishio K, Anderson PJ, Zheng XL, Sadler JE. 2004. Binding of platelet glycoprotein Ibα to von Willebrand factor domain A1 stimulates the cleavage of the adjacent domain A2 by ADAMTS13. Proc. Natl. Acad. Sci. U. S. A. 101:10578–10583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter S, Clark IM, Kevorkian L, Edwards DR. 2005. The ADAMTS metalloproteinases. Biochem. J. 386:15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng X, Nishio K, Majerus EM, Sadler JE. 2003. Cleavage of von Willebrand factor requires the spacer domain of the metalloprotease ADAMTS13. J. Biol. Chem. 278:30136–30141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Majerus EM, Sadler JE. 2002. ADAMTS13 and TTP. Curr. Opin. Hematol. 9:389–394 [DOI] [PubMed] [Google Scholar]
- 11.Xie L, Chesterman CN, Hogg PJ. 2001. Control of von Willebrand factor multimer size by thrombospondin-1. J. Exp. Med. 193:1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pimanda JE, Ganderton T, Maekawa A, Yap CL, Lawler J, Kershaw G, Chesterman CN, Hogg PJ. 2004. Role of thrombospondin-1 in control of von Willebrand factor multimer size in mice. J. Biol. Chem. 279:21439–21448 [DOI] [PubMed] [Google Scholar]
- 13.Bernardo A, Ball C, Nolasco L, Choi H, Moake JL, Dong JF. 2005. Platelets adhered to endothelial cell-bound ultra-large von Willebrand factor strings support leukocyte tethering and rolling under high shear stress. J. Thromb. Haemost. 3:562–570 [DOI] [PubMed] [Google Scholar]
- 14.Castaman G, Federici AB, Rodeghiero F, Mannucci PM. 2003. Von Willebrand's disease in the year 2003: towards the complete identification of gene defects for correct diagnosis and treatment. Haematologicaae 88:94–108 [PubMed] [Google Scholar]
- 15.Sadler JE. 2005. von Willebrand factor: two sides of a coin. J. Thromb. Haemost. 3:1702–1709 [DOI] [PubMed] [Google Scholar]
- 16.Bjerketorp J, Jacobsson K, Frykberg L. 2004. The von Willebrand factor-binding protein (vWbp) of Staphylococcus aureus is a coagulase. FEMS Microbiol. Lett. 234:309–314 [DOI] [PubMed] [Google Scholar]
- 17.Byrne MF, Kerrigan SW, Corcoran PA, Atherton JC, Murray FE, Fitzgerald DJ, Cox DM. 2003. Helicobacter pylori binds von Willebrand factor and interacts with GPIb to induce platelet aggregation. Gastroenterology 124:1846–1854 [DOI] [PubMed] [Google Scholar]
- 18.Grewal PK, Uchiyama S, Ditto D, Varki N, Le DT, Nizet V, Marth JD. 2008. The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat. Med. 14:648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson M, Bjerketorp J, Wiebensjo A, Ljungh A, Frykberg L, Guss B. 2004. A von Willebrand factor-binding protein from Staphylococcus lugdunensis. FEMS Microbiol. Lett. 234:155–161 [DOI] [PubMed] [Google Scholar]
- 20.Loftus JC. 1990. A β3 integrin mutation abolishes ligand binding and alters divalent cation-dependent conformation. Science 249:915–918 [DOI] [PubMed] [Google Scholar]
- 21.Lee JO, Rieu P, Arnaout MA, Liddington R. 1995. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18). Cell 80:631–638 [DOI] [PubMed] [Google Scholar]
- 22.Gross DM, Forsthuber T, Tary-Lehmann M, Etling C, Ito K, Nagy ZA, Field JA, Steere AC, Huber BT. 1998. Identification of LFA-1 as a candidate autoantigen in treatment-resistant Lyme arthritis. Science 281:703–706 [DOI] [PubMed] [Google Scholar]
- 23.Coburn J, Fischer JR, Leong JM. 2005. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol. Microbiol. 57:1182–1195 [DOI] [PubMed] [Google Scholar]
- 24.Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, Bono JL, Akins DR, Radolf JD, Schwan TG, Rosa P. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karna SL, Sanjuan E, Esteve-Gassent MD, Miller CL, Maruskova M, Seshu J. 2011. CsrA modulates levels of lipoproteins and key regulators of gene expression critical for pathogenic mechanisms of Borrelia burgdorferi. Infect. Immun. 79:732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanjuan E, Esteve-Gassent MD, Maruskova M, Seshu J. 2009. Overexpression of CsrA (BB0184) alters the morphology and antigen profiles of Borrelia burgdorferi. Infect. Immun. 77:5149–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunikis J, Garpmo U, Tsao J, Berglund J, Fish D, Barbour AG. 2004. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150:1741–1755 [DOI] [PubMed] [Google Scholar]
- 28.Jaulhac B, Heller R, Limbach FX, Hansmann Y, Lipsker D, Monteil H, Sibilia J, Piemont Y. 2000. Direct molecular typing of Borrelia burgdorferi sensu lato species in synovial samples from patients with lyme arthritis. J. Clin. Microbiol. 38:1895–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cserzo M, Eisenhaber F, Eisenhaber B, Simon I. 2002. On filtering false positive transmembrane protein predictions. Protein Eng. 15:745–752 [DOI] [PubMed] [Google Scholar]
- 30.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030 [DOI] [PubMed] [Google Scholar]
- 31.Nugent T, Jones DT. 2009. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics 10:159. 10.1186/1471-2105-10-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viklund H, Elofsson A. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24:1662–1668 [DOI] [PubMed] [Google Scholar]
- 33.Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 34.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 35.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Gil L, Johnson AE, Mingarro I. 2010. Membrane insertion and biogenesis of the Turnip crinkle virus p9 movement protein. J. Virol. 84:5520–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamborero S, Vilar M, Martinez-Gil L, Johnson AE, Mingarro I. 2011. Membrane insertion and topology of the translocating chain-associating membrane protein (TRAM). J. Mol. Biol. 406:571–582 [DOI] [PubMed] [Google Scholar]
- 38.Saaf A, Wallin E, von Heijne G. 1998. Stop-transfer function of pseudo-random amino acid segments during translocation across prokaryotic and eukaryotic membranes. Eur. J. Biochem. 251:821–829 [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Gil L, Bano-Polo M, Redondo N, Sanchez-Martinez S, Nieva JL, Carrasco L, Mingarro I. 2011. Membrane integration of poliovirus 2B viroporin. J. Virol. 85:11315–11324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Gil L, Sauri A, Vilar M, Pallas V, Mingarro I. 2007. Membrane insertion and topology of the p7B movement protein of Melon necrotic spot virus (MNSV). Virology 367:348–357 [DOI] [PubMed] [Google Scholar]
- 41.Brooks CS, Vuppala SR, Jett AM, Akins DR. 2006. Identification of Borrelia burgdorferi outer surface proteins. Infect. Immun. 74:296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryksin AV, Tomova A, Godfrey HP, Cabello FC. 2010. BmpA is a surface-exposed outer-membrane protein of Borrelia burgdorferi. FEMS Microbiol. Lett. 309:77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skare JT, Shang ES, Foley DM, Blanco DR, Champion CI, Mirzabekov T, Sokolov Y, Kagan BL, Miller JN, Lovett MA. 1995. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J. Clin. Invest. 96:2380–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esteve-Gassent MD, Elliott NL, Seshu J. 2009. sodA is essential for virulence of Borrelia burgdorferi in the murine model of Lyme disease. Mol. Microbiol. 71:594–612 [DOI] [PubMed] [Google Scholar]
- 45.Raju BV, Esteve-Gassent MD, Karna SL, Miller CL, Van Laar TA, Seshu J. 2011. Oligopeptide permease A5 modulates vertebrate host-specific adaptation of Borrelia burgdorferi. Infect. Immun. 79:3407–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonara S, Ristow L, Coburn J. 2011. Adhesion mechanisms of Borrelia burgdorferi. Adv. Exp. Med. Biol. 715:35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coburn J, Chege W, Magoun L, Bodary SC, Leong JM. 1999. Characterization of a candidate Borrelia burgdorferi β3-chain integrin ligand identified using a phage display library. Mol. Microbiol. 34:926–940 [DOI] [PubMed] [Google Scholar]
- 48.Li X, Liu X, Beck DS, Kantor FS, Fikrig E. 2006. Borrelia burgdorferi lacking BBK32, a fibronectin-binding protein, retains full pathogenicity. Infect. Immun. 74:3305–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, Hook M, Skare JT. 2006. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol. Microbiol. 59:1591–1601 [DOI] [PubMed] [Google Scholar]
- 50.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. 2005. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433:377–381 [DOI] [PubMed] [Google Scholar]
- 51.Bano-Polo M, Baldin F, Tamborero S, Marti-Renom MA, Mingarro I. 2011. N-glycosylation efficiency is determined by the distance to the C-terminus and the amino acid preceding an Asn-Ser-Thr sequon. Protein Sci. 20:179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiefer H, Krieger J, Olszewski JD, von Heijne G, Prestwich GD, Breer H. 1996. Expression of an olfactory receptor in E. coli: purification, reconstitution, and ligand binding. Biochemistry 35:16077–16084 [DOI] [PubMed] [Google Scholar]
- 53.Kim JH, Singvall J, Schwarz-Linek U, Johnson BJ, Potts JR, Hook M. 2004. BBK32, a fibronectin binding MSCRAMM from Borrelia burgdorferi, contains a disordered region that undergoes a conformational change on ligand binding. J. Biol. Chem. 279:41706–41714 [DOI] [PubMed] [Google Scholar]
- 54.Probert WS, Kim JH, Hook M, Johnson BJ. 2001. Mapping the ligand-binding region of Borrelia burgdorferi fibronectin-binding protein BBK32. Infect. Immun. 69:4129–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raibaud S, Schwarz-Linek U, Kim JH, Jenkins HT, Baines ER, Gurusiddappa Hook S M, Potts JR. 2005. Borrelia burgdorferi binds fibronectin through a tandem beta-zipper, a common mechanism of fibronectin binding in staphylococci, streptococci, and spirochetes. J. Biol. Chem. 280:18803–18809 [DOI] [PubMed] [Google Scholar]
- 56.Emsley J, Cruz M, Handin R, Liddington R. 1998. Crystal structure of the von Willebrand factor A1 domain and implications for the binding of platelet glycoprotein Ib. J. Biol. Chem. 273:10396–10401 [DOI] [PubMed] [Google Scholar]
- 57.Pinto AF, Terra RM, Guimaraes JA, Fox JW. 2007. Mapping von Willebrand factor A domain binding sites on a snake venom metalloproteinase cysteine-rich domain. Arch. Biochem. Biophys. 457:41–46 [DOI] [PubMed] [Google Scholar]
- 58.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. 2000. Ligand binding to integrins. J. Biol. Chem. 275:21785–21788 [DOI] [PubMed] [Google Scholar]
- 59.Martinez-Gil L, Sauri A, Marti-Renom MA, Mingarro I. 2011. Membrane protein integration into the endoplasmic reticulum. FEBS J. 278:3846–3858 [DOI] [PubMed] [Google Scholar]
- 60.Hedin LE, Ojemalm K, Bernsel A, Hennerdal A, Illergard K, Enquist K, Kauko A, Cristobal S, von Heijne G, Lerch-Bader M, Nilsson I, Elofsson A. 2010. Membrane insertion of marginally hydrophobic transmembrane helices depends on sequence context. J. Mol. Biol. 396:221–229 [DOI] [PubMed] [Google Scholar]
- 61.Angel TE, Luft BJ, Yang X, Nicora CD, Camp DG, Jacobs JM, Smith RD. 2010. Proteome analysis of Borrelia burgdorferi response to environmental change. PLoS One 5:e13800. 10.1371/journal.pone.0013800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell ID, Humphries MJ. 2011. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 3:pii a004994. 10.1101/cshperspect.a004994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fikrig E, Narasimhan S. 2006. Borrelia burgdorferi: traveling incognito? Microbes Infect. 8:1390–1399 [DOI] [PubMed] [Google Scholar]
- 64.Coburn J. 2001. Adhesion mechanisms of the Lyme disease spirochete, Borrelia burgdorferi. Curr. Drug Targets Infect. Disord. 1:171–179 [DOI] [PubMed] [Google Scholar]
- 65.Coburn J, Cugini C. 2003. Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease agent, Borrelia burgdorferi, to integrin αvβ3. Proc. Natl. Acad. Sci. U. S. A. 100:7301–7306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glasner J, Blum H, Wehner V, Stilz HU, Humphries JD, Curley GP, Mould AP, Humphries MJ, Hallmann R, Rollinghoff M, Gessner A. 2005. A small molecule alpha 4 beta 1 antagonist prevents development of murine Lyme arthritis without affecting protective immunity. J. Immunol. 175:4724–4734 [DOI] [PubMed] [Google Scholar]
- 67.Guerau-de-Arellano M, Alroy J, Huber BT. 2005. β2 integrins control the severity of murine Lyme carditis. Infect. Immun. 73:3242–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown EL, Guo BP, O'Neal P, Hook M. 1999. Adherence of Borrelia burgdorferi. Identification of critical lysine residues in DbpA required for decorin binding. J. Biol. Chem. 274:26272–26278 [DOI] [PubMed] [Google Scholar]
- 69.Fischer JR, Parveen N, Magoun L, Leong JM. 2003. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. U. S. A. 100:7307–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo BP, Brown EL, Dorward DW, Rosenberg LC, Hook M. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30:711–723 [DOI] [PubMed] [Google Scholar]
- 71.Pikas DS, Brown EL, Gurusiddappa S, Lee LY, Xu Y, Hook M. 2003. Decorin-binding sites in the adhesin DbpA from Borrelia burgdorferi: a synthetic peptide approach. J. Biol. Chem. 278:30920–30926 [DOI] [PubMed] [Google Scholar]
- 72.Behera AK, Durand E, Cugini C, Antonara S, Bourassa L, Hildebrand E, Hu LT, Coburn J. 2008. Borrelia burgdorferi BBB07 interaction with integrin α3β1 stimulates production of pro-inflammatory mediators in primary human chondrocytes. Cell. Microbiol. 10:320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kreidberg JA, Symons JM. 2000. Integrins in kidney development, function, and disease. Am. J. Physiol. Renal Physiol. 279:F233–F242 [DOI] [PubMed] [Google Scholar]
- 74.Kreidberg JA. 2000. Functions of α3β1 integrin. Curr. Opin. Cell Biol. 12:548–553 [DOI] [PubMed] [Google Scholar]
- 75.Dulabon L, Olson EC, Taglienti MG, Eisenhuth S, McGrath B, Walsh CA, Kreidberg JA, Anton ES. 2000. Reelin binds α3β1 integrin and inhibits neuronal migration. Neuron 27:33–44 [DOI] [PubMed] [Google Scholar]
- 76.DiPersio CM, van der Neut R, Georges-Labouesse E, Kreidberg JA, Sonnenberg A, Hynes RO. 2000. α3β1 and α6β4 integrin receptors for laminin-5 are not essential for epidermal morphogenesis and homeostasis during skin development. J. Cell Sci. 113:3051–3062 [DOI] [PubMed] [Google Scholar]
- 77.Arnaout MA, Goodman SL, Xiong JP. 2007. Structure and mechanics of integrin-based cell adhesion. Curr. Opin. Cell Biol. 19:495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Askari JA, Buckley PA, Mould AP, Humphries MJ. 2009. Linking integrin conformation to function. J. Cell Sci. 122:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruoslahti E. 1991. Integrins. J. Clin. Invest. 87:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takada Y, Ye X, Simon S. 2007. The integrins. Genome Biol. 8:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takagi J. 2007. Structural basis for ligand recognition by integrins. Curr. Opin. Cell Biol. 19:557–564 [DOI] [PubMed] [Google Scholar]
- 82.Vilar M, Sauri A, Monne M, Marcos JF, von Heijne G, Perez-Paya E, Mingarro I. 2002. Insertion and topology of a plant viral movement protein in the endoplasmic reticulum membrane. J. Biol. Chem. 277:23447–23452 [DOI] [PubMed] [Google Scholar]