Abstract
The lysogenic state of phage λ is maintained by the CI repressor. CI binds to three operators each in the right operator (OR) and left operator (OL) regions, which lie 2.4 kb apart. At moderate CI levels, the predominant binding pattern is two dimers of CI bound cooperatively at each regulatory region. The resulting tetramers can then interact, forming an octamer and a loop of the intervening DNA. CI is expressed from the PRM promoter, which lies in the OR region and is subjected to multiple regulatory controls. Of these, the most recently discovered is stimulation by loop formation. In this work, we have investigated the mechanism by which looping stimulates PRM. We find that two cis-acting sites lying in the OL region are involved. One site, an UP element, is required for stimulation. Based on the behavior of other promoters with UP elements located upstream of the −35 region, we suggest that a subunit of RNA polymerase (RNAP) bound at PRM binds to the UP element located in the OL region. In addition, adjacent to the UP element lies a binding site for integration host factor (IHF); this site plays a less critical role but is required for stimulation of the weak prm240 allele. A loop with CI at the OL2 and OL3 operators does not stimulate PRM, while one with CI only at OL2 provides some stimulation. We discuss possible mechanisms for stimulation.
INTRODUCTION
The lysogenic state of phage λ is maintained by the CI repressor, which represses two early lytic promoters, thereby preventing initiation of the lytic pattern of gene expression. Expression of the host SOS response disrupts the lysogenic state by promoting proteolytic self-cleavage of CI, inactivating it and leading to expression of the lytic genes. Remarkably, this regulatory state is balanced in such a way that it is almost completely stable under normal growth conditions and almost completely unstable in the face of a sustained SOS response. Hence, the circuitry acts as a switch.
A major part of this behavior results from the interplay of multiple controls that influence the expression of the cI gene. These controls operate on the PRM promoter, which is regulated at the level of transcription initiation by several different regulatory events. First, CI acts as a positive regulator of its own expression. When CI is bound to the OR2 site in the right operator (OR) region (Fig. 1D), it stimulates PRM ∼10-fold (1, 2). Second, occupancy of OR2 is a nonlinear function of CI concentration, arising from two mechanistic features of CI. CI dimerizes rather weakly, so that the concentration of the dimer, the DNA-binding species, increases nonlinearly. In addition, CI binds cooperatively to OR2 and OR1. OR1 is a strong binding site, while OR2 is a weak binding site; cooperativity results in the two sites filling up largely in parallel and with a sigmoid binding curve.
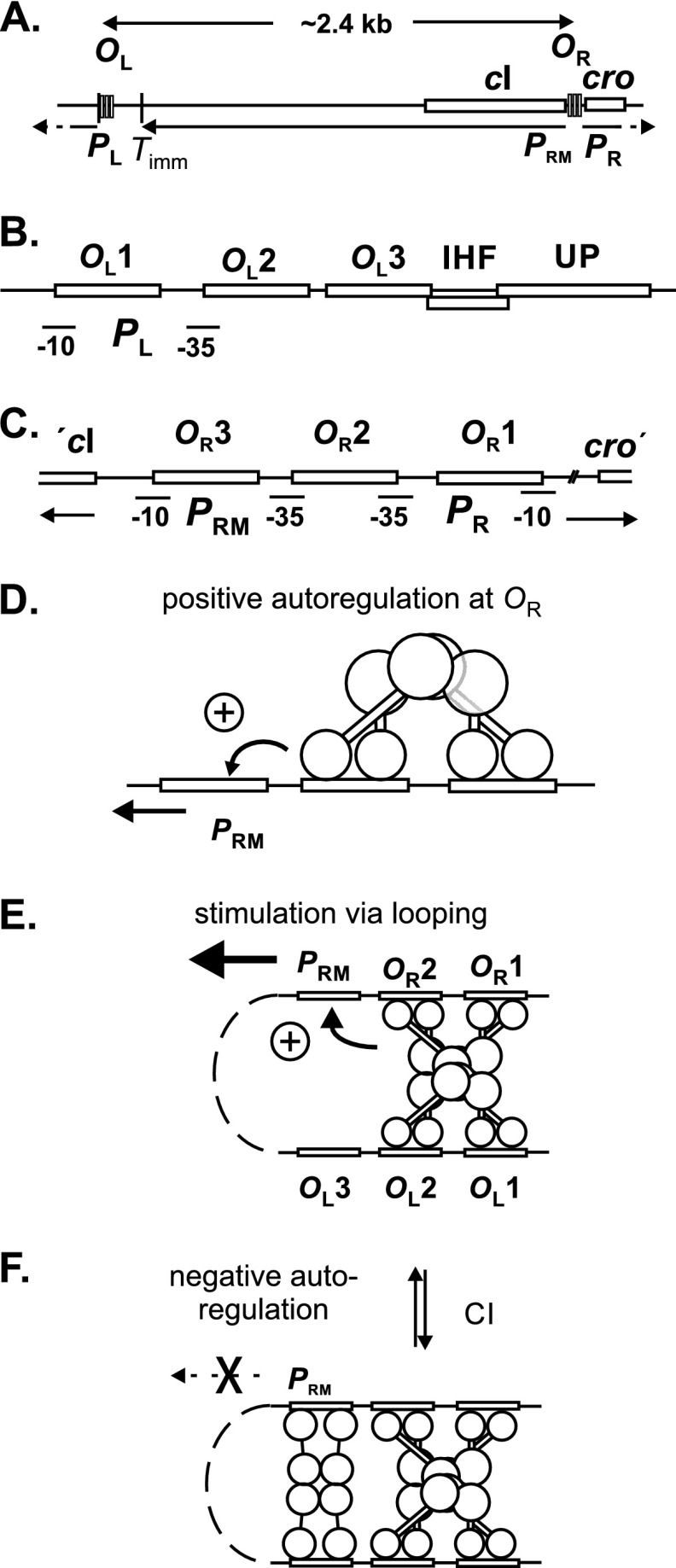
Fig 1.
Maps of λ, functions of CI, and regulation of CI expression. (A) Map of the immunity region of λ. (B) Expanded map, to scale, of the OL region, showing the location of the OL1, OL2, and OL3 sites, to which CI binds, and the location of the UP element and IHF binding site. Another IHF binding site, termed L2, is located close to the Timm site and not shown here. Binding of CI to OL1 and/or OL2 represses the lytic PL promoter. (C) Expanded map of the OR region, to scale, showing the location of the CI binding sites OR1, OR2, and OR3 and the location of the PRM and PR promoters. “ ′cI” and “cro′ ” indicate the start of the cI and cro genes. Binding of CI to OR1 and/or OR2 represses the lytic PR promoter. (D) Positive autoregulation of PRM by CI bound at OR2; CI binds cooperatively to OR1 and OR2. Both subunits of a dimer contact a subunit in the other dimer (27), as schematized. (E, F) Positive and negative autoregulation of PRM by CI-mediated looping. In panel E, an octamer forms by interaction of tetramers at OL and OR, and PRM is further stimulated about 2-fold. In panel F, binding of two more CI dimers leads to dodecamer formation and repression of PRM. The details of protein-protein interactions in looped forms are unclear; likely possibilities for favored forms are shown. Adapted from reference 11 with modifications.
The combination of nonlinear binding to OR2 and positive autoregulation of PRM results in a transition from a low to a high level of PRM expression over a relatively narrow CI concentration range. This is a simple example of emergent behavior, and it was one of the earliest regulatory circuits to be analyzed at a mechanistic level (3, 4). This behavior is largely responsible for the wide range in stability of the lysogenic state; below a given threshold of CI concentration, the protein is no longer expressed at a rate sufficient to counterbalance its depletion by proteolysis.
More recently, two additional levels of control of PRM expression have been recognized (5–8). Both these mechanisms involve a long-range interaction between molecules of CI bound at the OR region and CI bound at the left operator (OL) region, which lie 2.4 kb apart (Fig. 1E and F). Two types of loops have been identified. Interaction between pairs of dimers at OL and at OR form an octamer (Fig. 1E). At higher CI concentrations, additional dimers bind to the remaining operators, forming a dodecamer (Fig. 1F). In the latter form, PRM is repressed, since the OR3 site is occupied, resulting in negative autoregulation (5, 6). Intrinsically, OR3 is a weak binding site, and its occupancy requires dodecamer formation. At the level of CI present in a λ lysogen, this effect represses PRM about 2-fold compared to the level of expression without dodecamer formation (9).
Formation of the octamer leads to another form of positive autoregulation. It provides a further increase of about 2-fold in PRM expression (7, 8). This effect is most readily observed when CI cannot bind to OR3 or to OL3 due to mutations in these sites. It has also been observed in vitro on supercoiled templates (10). In this work, we study the mechanism by which this stimulation occurs.
One possible inroad into analysis of this mechanism is provided by our recent finding (11) that a mutant allele of the PRM promoter, prm240, is stimulated by looping to a far greater extent than is the wild-type (WT) PRM. When cells are grown in minimal medium, prm240 is stimulated about 5-fold by looping, as judged by the behavior of reporter genes. Hence, we have included analysis of prm240 in the present study.
It is plausible that looping-mediated stimulation operates, at least in part, with the involvement of cis-acting sites in the vicinity of the OL operators. We previously found (12) that the Timm terminator (Fig. 1A) and the 160 bp between Timm and OL3 are highly conserved among λ and a set of phages with λ immunity specificity that were isolated from the wild. In λ, previous studies (13–16) show the existence of two sites adjacent to OL3 (Fig. 1B). The first is a binding site for the host protein integration host factor (IHF), lying adjacent to and slightly overlapping OL3. We term this the IHF site for brevity; it is sometimes termed L1 to distinguish it from another IHF binding site (not depicted), termed L2 and lying close to Timm. The second site is an UP element lying further upstream and partially overlapping the IHF site. It is known that the C-terminal domain (CTD) of the α subunit of RNA polymerase (RNAP) can interact with UP elements, increasing the strength of certain promoters (17), and the magnitude of this effect is quite variable. Both the IHF site and the UP element stimulate the lytic PL promoter (13–16), as judged by reporter assays.
Two molecular mechanisms have been proposed (7, 8) to account for looping-mediated stimulation of PRM. It might result from interaction of RNA polymerase bound at PRM with the UP element at OL. This model predicts that removal of the UP element would abolish the stimulatory effect. In this work, we have tested this prediction. Alternatively, looping might somehow lead to conformational changes, either in CI or in RNAP, that make the promoter more active. Here, we have used templates mutated in one or more OL operators to test whether other loops can also stimulate PRM.
We find that the UP element plays an important role in looping-mediated stimulation. Recent evidence from a separate study (9) also supports this conclusion. In addition, we find that other looped forms do not confer stimulation, arguing that looping per se does not suffice to support the stimulatory effect. Finally, we have examined the role of the IHF site and find that it is also important in stimulation, particularly for the mutant prm240 promoter.
MATERIALS AND METHODS
Media, chemicals, and reagents.
LB and M9 minimal media are as described previously (18). LB was supplemented with 0.2% glucose and 1 mM MgSO4 (19); the carbon source for M9 was 0.2% glucose. Restriction enzymes and DNA ligase were from New England BioLabs or Fermentas Inc. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were from GoldBioTech. Ortho-nitrophenyl-β-galactoside and polymyxin B (both used in assay of β-galactosidase) were from Sigma Chemical Co. Pfu Turbo DNA polymerase for site-directed mutagenesis (20) was from Stratagene; GoTaq master mix (Promega) for routine PCR was used as directed by the supplier.
Bacterial, phage, and plasmid strains.
Bacterial strains, with only relevant genotype given, were the following: JL2497, N99 lacZΔM15/F′ lacIq lacZΔM15::Tn9 (2), used as the wild type; JL6142, JL2497 Δ(lacIPOZYA) F− (21); JL6994, JL6142/pJWL615/pJWL486 (22); and JL6995, JL6142/pJWL615/pA3B2 (22). Other bacterial strains are listed in Table S1 in supplemental material. Plasmids were pA3B2, a pACYC184 Cmr derivative encoding a weak lacP::cI fusion (2, 23); pJWL486, a vector control for pA3B2 (2); and pJWL615, a derivative of the Spcr plasmid pGB2, carrying the lacIq gene (2). Construction of phages carrying PRM::lacZ protein fusion reporter genes was as described previously (11) and is detailed in the supplemental material. These phages are derivatives of the Simons vector λRS45 (24), modified as described previously (2); a generic structure of these reporters is shown in Fig. 2A. Regulatory elements downstream (2) and upstream (24) of the lacZ gene were introduced as described. Reporter phages are listed in Table S3 in the supplemental material. Single lysogens of reporter phages were made in strains JL6994 and JL6995; those in JL6994 are indicated in the figures as “no CI.” Construction of other phage and plasmid strains is described in the supplemental material. The OL1-3 allele (this work) changes OL1 to TACCAATGGAGTTGATA , where the underlined bases are C, C, and G, respectively, in OL1+; these positions are important for CI binding (25), so this allele should abolish specific binding to OL1. The OL3-4 allele (6) changes OL3 to TATCACTAGAGTTGGTT (underlined bases are C, G, C, and A in the WT, respectively) and blocks specific CI binding to OL3 (6). The r1 mutation changes OR3 to TATCACACGCAAGGGATA (underlined base is C in the WT) and is proposed (5) to weaken CI binding to OR3 by about 2.9 kcal/mol, based on measurements in the context of OR1 (25).
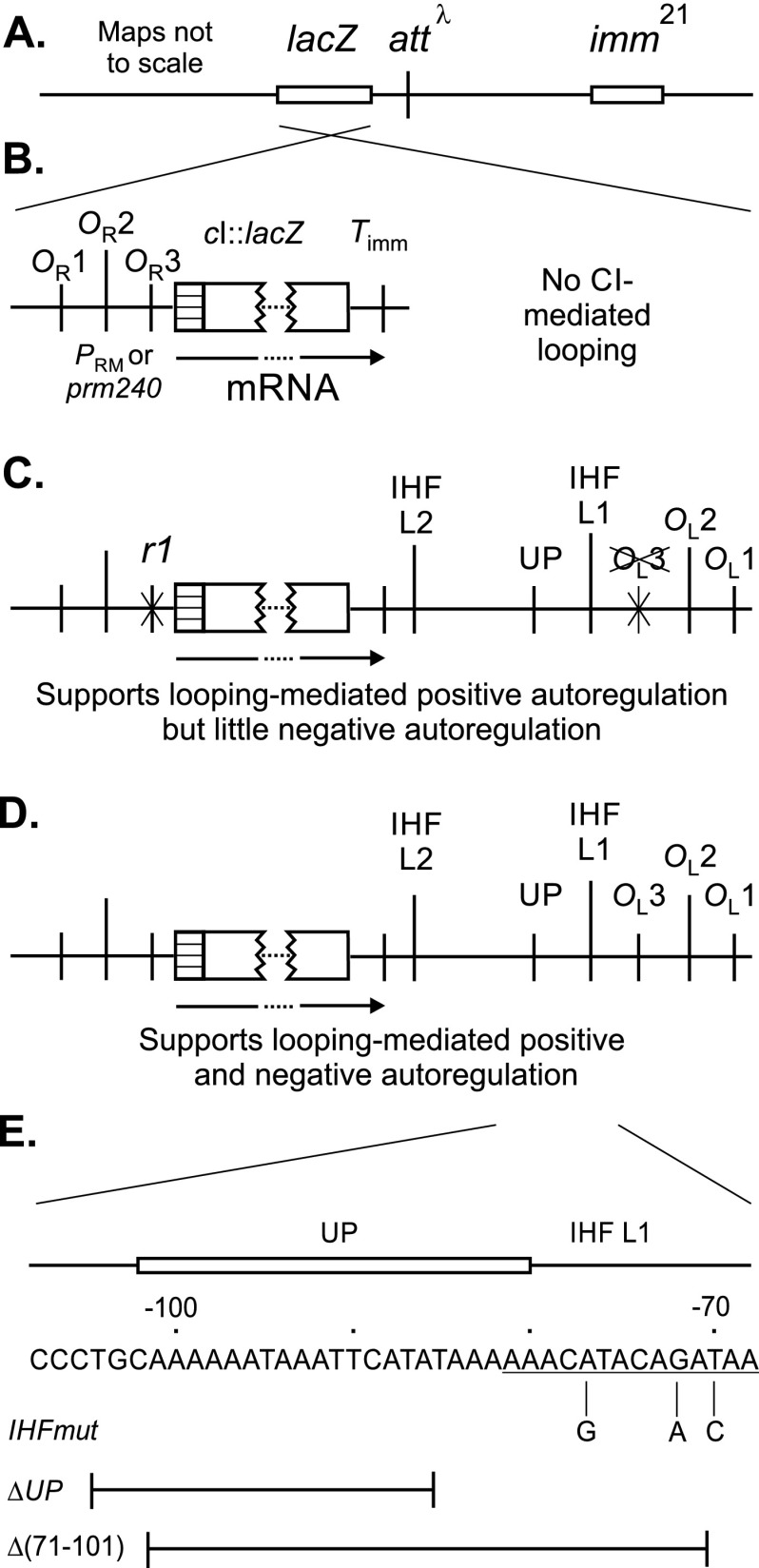
Fig 2.
Structure of reporter constructs and location of mutations. (A) Simplified map of the reporter phages used, which are derived from a Simons vector (24). This vector is a derivative of phage λ with the immunity region of phage 21 (imm21). The lacZ reporter is located in a nonessential region to the left of the λ attachment site (attλ). (B, C, D) Reporter constructs. Maps are not to scale; maps in panels C and D are aligned to the one in panel B. In each case, the mRNA is terminated at Timm, so that levels of expression are directly comparable (8). Some constructs contained the r1 allele in OR3, which weakens CI binding (see the text). The first construct (B) is a control, which cannot loop; the second one (C) cannot confer negative autoregulation, particularly when the r1 allele is present; the third (D) can carry out negative autoregulation. The PRM::cI::lacZ protein fusion joins the first 18 codons of λ cI (hatched box) to codon 9 of lacZ (2) and is termed PRM::lacZ in the text for brevity. (E) Locations of the mutations used to alter the IHF site and/or the UP element are indicated; these were isolated both with OL3-4 and OL+ alleles (see the text). Location of a sequence resembling the IHF consensus site is underlined. The −72 G→A change is expected to weaken PL2. The 17-bp OL3 site starts with the AA bases shown at the right end of the sequence; hence, it overlaps slightly with the IHF site. Adapted from reference 11 with modifications.
Methods.
General phage methodology, including measurement of burst sizes and lysogenization frequencies, phage-by-plasmid crosses, and prophage induction, was as described previously (19). The tests for single lysogens (26) and assay for β-galactosidase activity (11) were as described.
RESULTS
Rationale for experimental approach.
We initially sought to identify cis-acting sites that contribute to the increase in promoter activity conferred by looping, using a classical genetic selection and screen (see the supplemental material). This approach did not yield the desired mutants. Accordingly, we turned to a reverse genetics approach, testing whether site-directed changes in either the IHF binding site or the UP element (Fig. 2) affected the activity of PRM, using the following reporter system (11).
This system (Fig. 2C) has a PRM::cI::lacZ protein fusion, termed PRM::lacZ for brevity, with the OL region lying downstream of the reporter gene. A control (Fig. 2B) has only the Timm terminator, which lies 160 bp upstream of OL3 in λ; the remaining constructs have Timm, followed by λ sequence to and through the OL region, with various alleles of several cis-acting sites. All these constructs make the same mRNA, terminated at Timm, so that expression levels are directly comparable among various mutant derivatives (8). To test the response of these reporters to a range of CI concentrations, cells carried a plasmid with a lacP::cI fusion. This plasmid provides some CI in the absence of IPTG; with increasing amounts of IPTG, cells make progressively more CI. A control strain with no CI lacked this plasmid. As previously noted (11), this system behaves differently in rich medium (LB) and minimal medium (M9), in that much more CI is made in M9 than in LB, both without IPTG and at a given IPTG level. Hence, the shapes of the dose-response curves differ, and these shapes cannot be compared between the two growth media.
In the absence of CI, the unstimulated PRM promoter activity leads to a very low level of expression (1). As the CI level increases, a mixture of different CI-bound template configurations is present, so that at a given CI concentration the observed activity reflects the sum of activities of the various species. In the Timm control, the expression level is dictated by events occurring locally in the OR region. At moderate CI levels, the predominant species is one with CI bound cooperatively to OR1 and OR2, giving the CI-stimulated level of PRM-directed lacZ activity. At higher CI concentrations, CI is expected to bind to OR3, repressing PRM, and lacZ expression would decline, but in our experiments, the CI concentrations are not high enough to have this effect.
When looping is allowed by providing the OL region, additional species can form as CI levels increase. The first is an octamer, in which the predominant occupancy pattern is most likely CI bound to OR1, OR2, OL1, and OL2 (Fig. 1E). This is the species in which additional stimulation of PRM is conferred by looping. At higher CI levels, additional CI dimers bind to OR3 and OL3, forming the dodecamer; the resulting complex has little or no PRM activity (Fig. 1F). We can largely prevent dodecamer formation by blocking CI binding to OR3 and OL3, allowing us to examine the activation curve. At high CI levels, the remaining operators should be saturated with CI, and the expression curve should reach a plateau. However, since octamer formation is apparently only marginally favored, with an estimated change in free energy, ΔG, of −0.5 kcal/mol (6), not all of the template molecules will be looped; hence, the population is a mixture of unlooped and looped species, and the plateau level underestimates the activity of the looped octameric species.
We assessed the effects of changing the cis-acting sites. To remove the UP element, we deleted it by removing residues −86 to −104 relative to the PL start site, giving a deletion we term ΔUP (Fig. 2E). To change the IHF site without changing the relationship between the UP element and the OL operators, we introduced two point mutations at positions known from other studies (14) to weaken IHF binding and function, as well as a third mutation (G−72A) that should weaken a second weak promoter, termed PL2, which lies upstream of the PL promoter (sometimes termed PL1) and is more active in the absence of IHF (15). The result is a triple mutant termed IHFmut. Finally, we deleted both the IHF site and the UP element, removing residues −71 to −101, giving a deletion we term Δ(71-101). Constructs also contained one of several alleles of the OR region, as indicated below.
Effects on activation.
To analyze activation of PRM in the absence of repression, we prevented specific CI binding to OL3 by including the OL3-4 allele (6) and weakened binding to OR3 by including the r1 mutation in OR3 (5). These changes largely prevent dodecamer formation. As CI levels increased, levels of lacZ expression reached a plateau and in most cases declined only slightly as CI levels increased further.
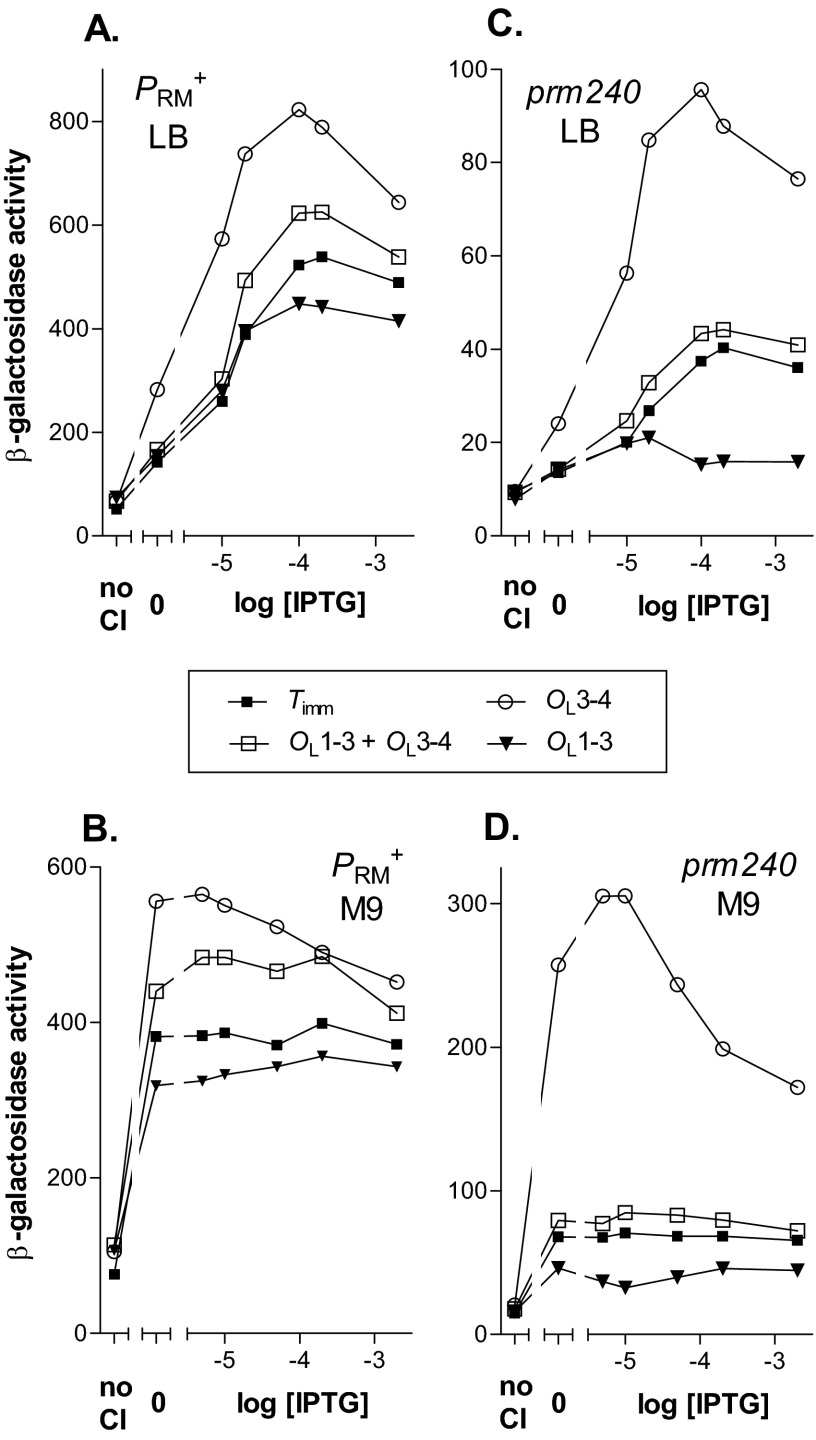
Presence of the OL region stimulated wild-type PRM about 1.6-fold, as judged by comparing the OL3-4 reporter with the Timm control (Fig. 3A), as previously observed in vivo (11), and with a different allele of PRM in vitro (10). Deletion of the UP element abolished this stimulatory effect (Fig. 3A). Mutation of the IHF site weakened but did not abolish the stimulatory effect. Removal of both sites by the Δ(71-101) deletion resulted in mild inhibition of lacZ expression. This result suggests that both sites contribute to activation of PRM at least somewhat independently of one another. We conclude that stimulation of PRM+ by looping requires the UP element and that the IHF site plays a role in this process, but one less significant than the UP element.
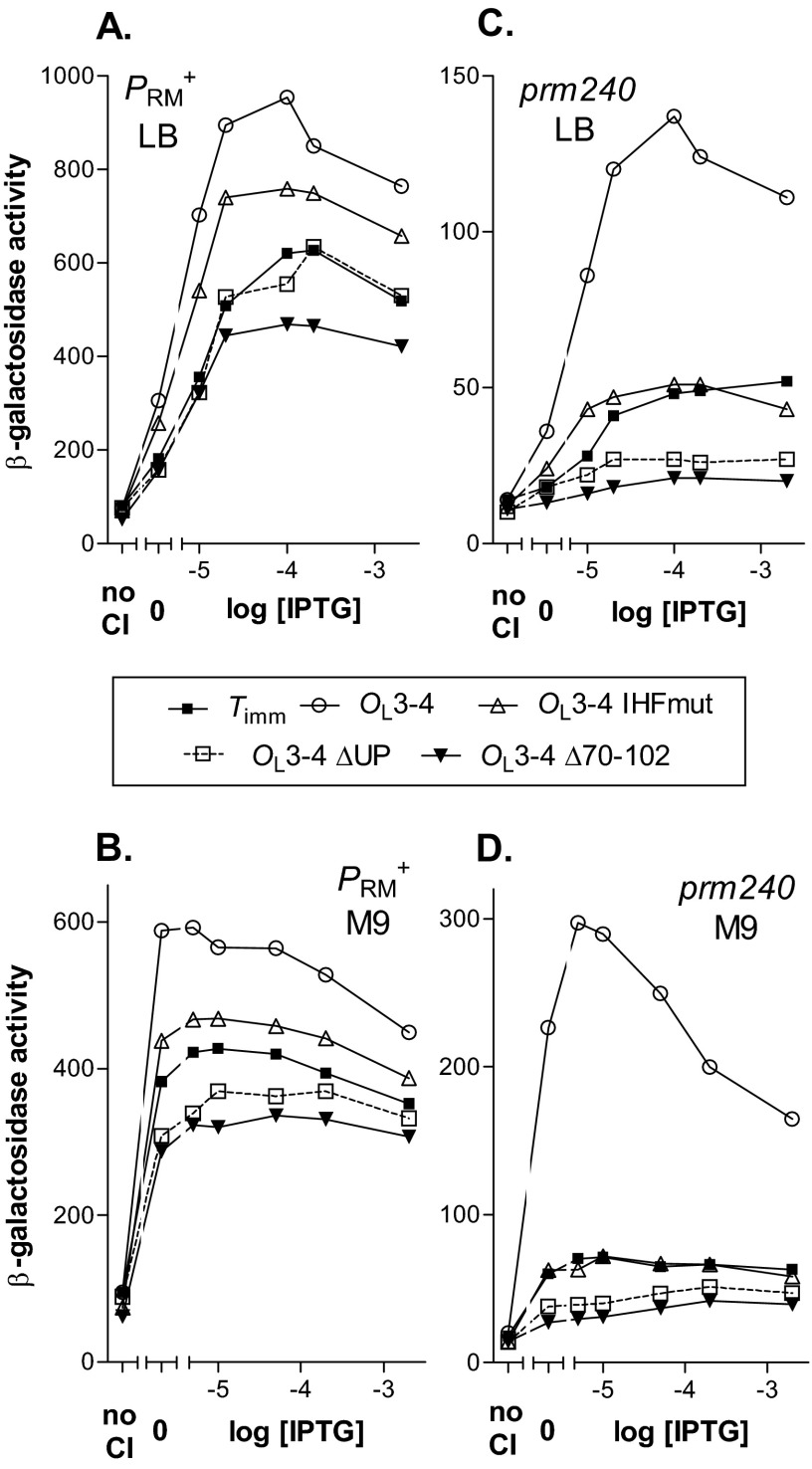
Fig 3.
Effects of IHFmut and ΔUP mutations on activation of PRM+ and prm240. Reporters (Fig. 2) contained the OL3-4 allele and the OR3 r1 mutation to prevent negative autoregulation. Cells were grown in the indicated medium and assayed as described in Materials and Methods. The results of a typical experiment are presented. In each pair of panels, the same strains were grown either in LB at 37°C (A and C) or in M9 at 30°C (B and D). The OL alleles are indicated in the inset. (A and B) PRM+; (C and D) prm240. In each case, we used two strains, one lacking CI (“no CI”), and a second one with a plasmid bearing a weak lacP::cI fusion. This strain was grown without IPTG (“0”) or with the indicated IPTG concentration, affording a range of CI levels. As described previously (11), at 2 mM IPTG, CI levels in the latter strain in LB at 37°C and in M9 at 30°C were about equal to and 3 times, respectively, the level in a lysogen (11). Some CI is made in the absence of IPTG; in LB at 37°C, the CI levels in cells grown at 0.01 and 0.2 mM IPTG were roughly 0.1 and 0.5 times the lysogen level, respectively (11).
We also infer that the reduced expression level seen in the Δ(71-101) deletion is evidence that this template was able to form looped complexes; otherwise, it is difficult to explain why the expression level differs from the unlooped Timm control. We assume, here and below, that all the mutant templates we analyze likewise can assume the looped form, including a few that show an expression level similar to that of the Timm control. Based on this assumption, we discuss in several cases how changes in the template might affect the activity of the looped form. In this case (Fig. 3A), the reduced level suggests that PRM is less active in the looped form than in the unlooped form.
We next tested the effects of the mutations on lacZ expression directed by the weak prm240 promoter in M9 medium at 30°C (Fig. 3D). Under these conditions, the relative effect of looping on lacZ expression is greatest (11). We found, first, that both the IHF site and the UP element were required for stimulation of the mutant promoter by looping. Second, with the ΔUP allele, the observed level of expression was lower than seen in the Timm control. This lower level once again implies that a loop can form on this template and that in this looped form the promoter is less active than in the absence of the loop. Third, the Δ(71-101) deletion gave a value slightly but reproducibly lower than the ΔUP template. Fourth, if we assume that the IHFmut template can form a loop, then loop formation has no effect on the activity of PRM; that is, the IHF site is essential for looping-mediated stimulation under these conditions. Finally, at high CI levels, there was significant repression of the template with the wild-type UP element and IHF sites. We previously interpreted this result to indicate that cooperative interactions can stabilize a dodecamer in which some of the CI dimers are bound nonspecifically (11).
We then tested whether the IHF site was also essential for looping-mediated stimulation of wild-type PRM in M9 at 30°C. As in LB, looping stimulated the OL3-4 reporter about 1.6-fold relative to the Timm control (Fig. 3B). A template with the ΔUP mutation displayed a slight inhibition relative to the Timm construct, and the Δ(71-101) construct showed a somewhat lower activity. The template with IHFmut showed a reduced level of stimulation.
Finally, we analyzed the effect of changing the growth medium from M9 to LB on expression of prm240 (Fig. 3C). As previously observed (11), the mutant promoter was considerably weaker in LB than in M9, and the degree of stimulation by looping was less in LB. As in M9, mutation of the IHF site markedly reduced the stimulatory effect. The contrast with its slight effect on the PRM+ promoter suggests that the IHF site plays a greater role with the weaker prm240 promoter. The ΔUP mutation reduced the level of expression below that seen with the Timm control, and the Δ(71-101) deletion caused an even greater reduction. With the IHF mutation, at low CI levels the expression was higher than in the Timm control. This suggests that presence of the OL operators increases the occupancy of prm240 by RNAP at low CI but that in the looped form the promoter is not stimulated in the absence of the IHF site.
These templates contain the OR3 r1 mutation, which has a slight stimulatory effect on PRM (5) (our data not shown). In other experiments (not shown), templates with OR3+ instead of OR3 r1 showed essentially the same behavior as their r1 counterparts; hence, the behavior described above does not reflect a peculiarity of the r1 mutation.
Taken together, these findings reveal unexpected complexity in the mechanism of looping-mediated stimulation. In most cases, the plateau expression levels from looped constructs differed from those of the Timm control. Interpreting this finding is complicated (see Discussion), but we infer that the activity of the looped form, or the proportion of looped species, is changed in the mutants from the wild-type case. For those cases in which lacZ expression is the same as the control, we assume that the looped form can also occur, and we infer that in these cases, PRM activity is unaffected by looping.
The detailed effects of mutations in the UP element and the IHF site on the activity of the looped form depend on the growth medium and on the identity of the PRM allele. With wild-type PRM, the UP element is required for stimulation; hence, our data support the proposal (7, 8) that the interaction of RNAP bound at PRM with the UP element at OL plays a part in looping-mediated stimulation of PRM. In rich medium, the looped form lacking the UP element had the same activity as the unlooped form, while in minimal medium its activity was somewhat reduced relative to that of the unlooped form. The larger deletion, Δ(71-101), further reduced the activity of the looped form. Mutation of the IHF site resulted in a level of activity that was intermediate between those of the unlooped form and the unmutated looped form. Since deletion of both sites had lower activity than the ΔUP mutation alone, it is plausible that the IHF mutation is not acting indirectly by reducing the effectiveness of the UP element but instead that the IHF site makes a small independent contribution to the mechanism of stimulation.
With the prm240 allele, the ΔUP allele reduced expression below that of the control in both media, and the Δ(71-101) deletion had a somewhat lower level. The IHF mutation reduced expression to the same level as the control; we infer that the looped form has activity roughly equal to that of the unlooped form. Thus, the IHF site appears to play a more important role when PRM is weakened by mutation.
Effects on repression.
The pioneering work of Dodd et al. (6) suggested that loop formation accompanying octamer formation is only marginally favored energetically. In contrast, once the loop has formed, dodecamer formation is much more strongly favored. It is plausible that the energetics of dodecamer formation might be altered in the absence of the IHF site and/or the UP element. We addressed this issue by analyzing reporter constructs that allowed us to assay repression as well as activation of PRM.
We used sets of reporters similar to those described above, except that they were WT at OL3 and OR3, allowing dodecamer formation to occur, and tested WT PRM and prm240 in both LB and M9 minimal medium (Fig. 4). In each case, lacZ expression at low CI concentrations resembled that seen with the templates mutated in OL3 and OR3 (see Fig. 3). Expression rose from a low level to a level resembling that at the plateau seen when repression could not occur (Fig. 3). At higher CI levels, a substantial decline in expression was seen, which was more severe in cells grown in M9, presumably because the CI levels at a given IPTG concentration are higher (11).
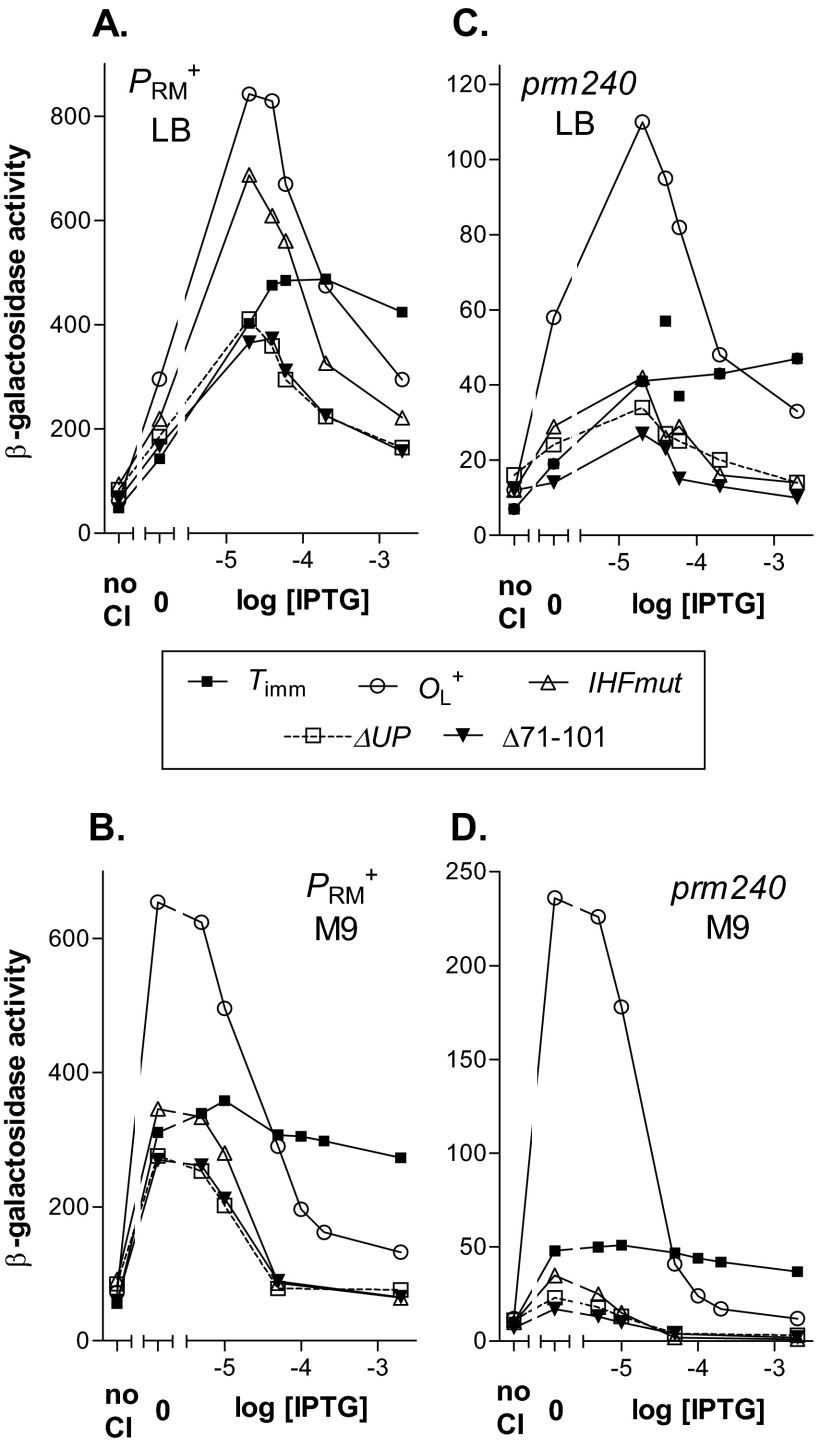
Fig 4.
Effects of IHFmut and ΔUP mutations on repression of PRM+ and prm240. The experiment was as described for Fig. 3, except that templates carried OL3+ and OR+ to allow repression at higher CI levels. (A) PRM+, grown in LB at 37°C; (B) PRM+, grown in M9 at 30°C; (C) prm240, grown in LB at 37°C; (D) prm240, grown in M9 at 30°C.
We interpret this pattern as follows. First, at relatively low CI concentrations, we suggest that the predominant form with templates that allow looping is the octamer, because the highest level observed was similar to that seen in the templates that did not allow repression. As discussed above, with different templates, the octameric looped form had different activities. Second, at higher CI concentrations, the repressed dodecameric form became increasingly predominant, and the overall activity decreased.
These data provide little or no indication that the IHFmut or ΔUP mutations affect the energetics of dodecamer formation. For a given promoter and growth condition, the CI concentration at which repression ensued was about the same with the OL+ template as with the mutant templates. If dodecamer formation were more favored, for instance, one would expect repression to take place at substantially lower IPTG levels. There is a hint of this in the case of prm240 in M9, but the effect is small at best.
Activity of other looped forms.
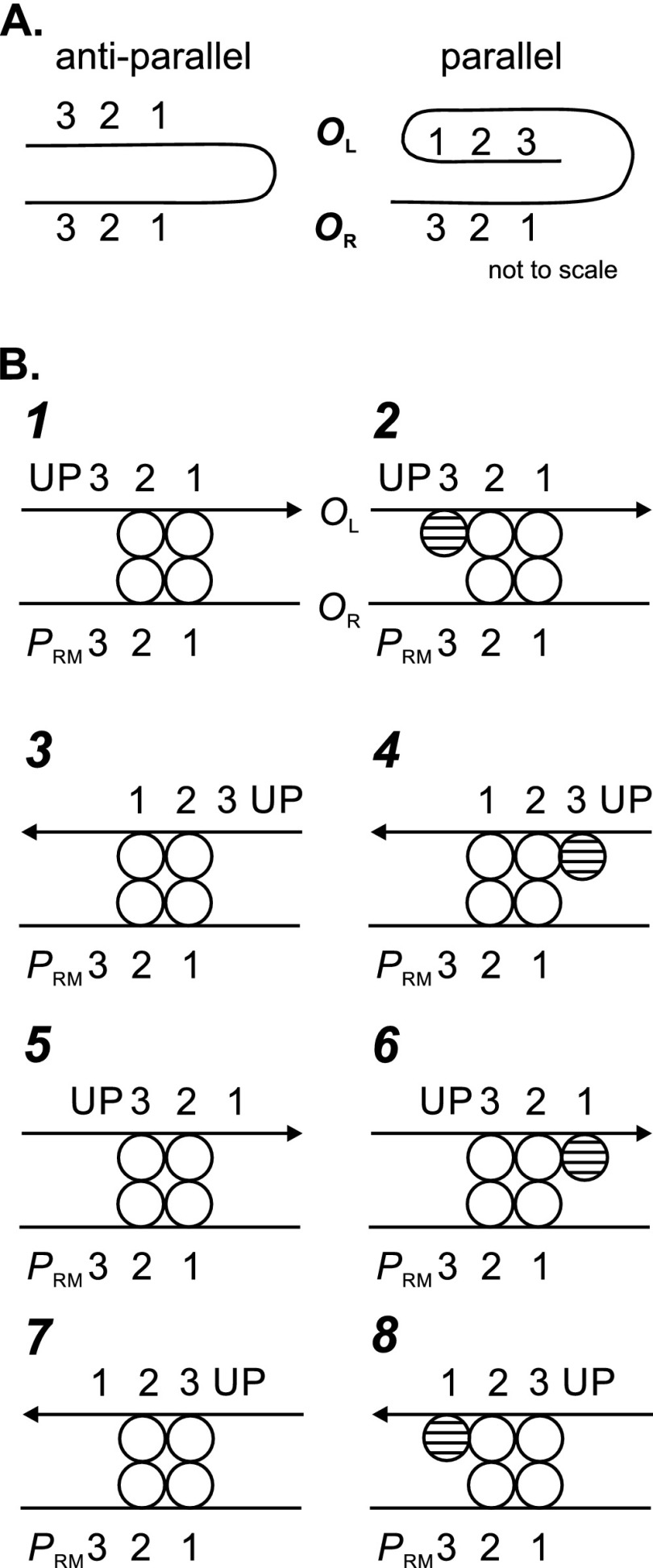
In addition to the complex depicted in Fig. 1E, other looped forms are likely to exist. Since the OL and OR regions are 2.4 kb apart in the λ genome, and DNA is flexible over this distance, the two regions can approach each other in two orientations, termed antiparallel and parallel (Fig. 5A), creating different relationships between the operators at the two regions. Although the spatial relationship between the two duplexes remains unsettled (9, 27) (see Discussion), for simplicity we depict them here as pointing in the same direction.
Fig 5.
Forms of looped complexes. (A) Antiparallel (left) and parallel (right) looped forms. The OR and OL regions approach one another in two different ways. The operators can also be juxtaposed in different registers (e.g., as in panel B). Not to scale. The relative spatial orientation between the two duplexes is not known (see the text), but they are shown as pointing in the same direction for simplicity. (B) Forms of looped complexes. The numbering scheme differs from that in reference 7. CI dimers are depicted as spheres. Forms in the right column are the same as those in the left column, but with a fifth dimer (hatched) bound to the remaining free OL site. Hatched dimers are not part of the octamer. It is not certain that CI binds cooperatively to OL2 and to OL3. These sites are separated by only 3 bp rather than the 6- or 7-bp spacing separating OR2 from OR3 and OR1, respectively, for which cooperativity has been extensively analyzed (33). With the CI repressor of phage HK022, reducing the spacing lowers the cooperativity parameter ω and changes the conformation of CI in the cooperative complex (45, 46). Footprinting data with an intact λ OL region (47) were interpreted as indicating cooperative binding, but this interpretation involved the assumption that the value of ω between OL1 and OL2 is the same as between OL2 and OL3.
Anderson and Yang (7) considered looped forms in which CI is bound to OR1 and OR2 but not to OR3 (since CI bound at OR3 would be expected to repress PRM). These forms are shown in Fig. 5B; in each case, the OR region is shown at the bottom and in the same orientation. Forms 1, 2, 5, and 6 are antiparallel and forms 3, 4, 7, and 8 are parallel. Form 1 juxtaposes the UP element and PRM in this representation, but that depends on the relative disposition of the two duplexes (see Discussion). The forms also differ in whether the CI tetramer at OL lies at OL1 and OL2 or at OL2 and OL3. Those in the right column are the same as those in the left column, but with a fifth dimer (hatched) bound to the remaining free OL site. Anderson and Yang (7) considered several models that varied according to which forms in addition to form 1 are active in transcription. We sought to test the activity of these forms by altering various sites in the OL region. It is not presently possible to control the orientation, limiting the scope of these tests.
If the loop-mediated stimulation of PRM requires only the formation of the loop per se, stimulation should occur with CI bound only to OL2 and OL3 but not to OL1 (7, 8), as in forms 5 and 7. We tested this possibility by mutating OL1, creating the OL1-3 allele (see Materials and Methods), with three changes in positions in OL1 important for CI binding (25). This should allow a possible OL2 - OL3 configuration to form. Higher levels of CI will likely be required for tetramer formation than at OL1 and OL2, since OL2 and OL3 are weak binding sites (25). To prevent CI binding to OR3, templates contained the OR3 r1 mutation. We also tested whether tetramer formation at OL was required for looping-mediated stimulation by using a template with mutations at both OL1 and OL3. Its response to various CI levels might also indicate how much CI is needed to give occupancy of OL2.
When wild-type PRM was assayed in LB (Fig. 6A), the effects of mutations in OL were somewhat surprising. When both OL1 and OL3 were mutated, the level of expression was unexpectedly higher than that of the Timm control at higher CI concentrations. This finding indicates that at least some occupancy of OL2 occurred. It suggests that stimulation mediated by looping could occur when only OL2 is occupied. Alternatively, it is possible that protein-protein interactions stabilized nonspecific CI binding to the altered OL1 site, maintaining some form of the octamer.
Fig 6.
Effects of the OL1 mutation on stimulation of PRM+ and prm240. As described for Fig. 3, cells were grown in two growth media as indicated; growth in LB and M9 was at 37°C and 30°C, respectively. Templates carried various alleles in the OL operators, as indicated; all had the r1 allele in OR3.
When only OL1 was mutated, the lacZ expression level was similar to that of the unlooped Timm control at low CI concentrations and somewhat lower at high CI concentrations. The requirement for higher CI levels to give repression is consistent with the weak binding of CI to OL2 and OL3 (see above). This finding suggests that when both OL2 and OL3 are occupied, looping-mediated stimulation cannot occur; instead, the looped form has a lower level of expression than that of the unlooped form. Its residual activity cannot readily be assessed, since we do not know the fraction of templates that are looped (see Discussion). In addition, the fact that some looping-mediated stimulation is observed for the OL1− OL3− template suggests that inhibition of activity when only OL1 is mutated is due to CI binding to OL3 on that template.
A similar pattern was observed in M9, except that the template retaining only OL2 was nearly as active as the OL3-4 template (Fig. 6B). Again we infer that CI occupancy of OL2 suffices to stimulate activity in the looped form, though it remains possible that cooperative interactions support nonspecific CI binding at OL1. With the prm240 mutant promoter, assayed both in LB and in M9, only a small amount of stimulation was seen with the OL2-only template, and the OL2-3 template exhibited a more marked reduction in activity than did the corresponding PRM+ template (Fig. 6C and D).
The finding that some stimulation occurred with the OL2-only template suggests that a wider variety of looped forms can exist than had been believed. Though it may be structurally implausible, perhaps an octamer could form with a single dimer at OL2 and three dimers at OR1, OR2, and OR3. Presumably, this form would be repressed due to CI binding at OR3. This model predicts that a template with OR3+ and only OL2+ at OL might give less expression than its OR3 r1 counterpart. We repeated the analysis described above on a set of templates with OR3+. Contrary to this prediction, the pattern of expression was essentially the same as that seen with the r1 versions (data not shown). This argues against the possibility that an octamer of the type mentioned can form.
Activity of template with CI bound to OL3.
In looped form 2 (Fig. 5), Anderson and Yang (7) suggested that CI at OL3 would repress PRM due to steric hindrance of RNAP binding at PRM. To test this proposal, we compared expression on OL3+ and OL3-4 templates. In addition, to allow for the possibility that bound IHF could sterically prevent CI from binding to OL3 (see Fig. S1 in the supplemental material), we compared OL3+ and OL3-4 templates in the presence of the IHFmut mutation. All templates also contained the OR3 r1 allele to prevent dodecamer formation or independent CI binding to OR3. If CI binding to OL3 is repressive in the looped configuration, we expected that the level of expression would be reduced, relative to the unlooped Timm control, when CI is bound to OL3. We found instead with both pairs of templates that the level of expression with the OL3+ template was intermediate between those of the Timm control and the corresponding OL3-4 template (Fig. 7). Since the effect was about the same in both pairs of templates, we infer that IHF binding does not have a large effect on CI binding to OL3.
Fig 7.
Effect of CI binding to OL3 on activation of PRM+. Cells were grown and assayed as described for Fig. 3, except that for each panel the averages of two experiments are presented. (A) Growth in LB at 37°C; (B) growth in M9 at 30°C. All templates contained the r1 allele in OR3.
We believe that CI levels in these experiments sufficed to allow CI binding to OL3. Otherwise, the curves for the OL3+ and OL3-4 templates would be the same. In addition, the affinities of CI for OL2 and OL3 are about the same in vitro (25), and our data with the OL1-3 OL3-4 reporter (Fig. 6) showed that OL2 was occupied in this concentration range. We conclude that binding of CI to OL3 does not repress PRM (as long as OR3 is unoccupied) and infer that the level of expression is reduced to some extent relative to that when OL3 is free. Recent structural modeling (9) also suggests that CI binding to OL3 prevents RNAP from contacting the UP element.
We concluded above (“Effects on repression”) that mutating the IHF site had little or no effect on the energetics of dodecamer formation. If IHF bound very tightly to its site, one would expect that IHF binding would compete with CI binding to OL3 and that blocking IHF binding would favor dodecamer formation. We infer that IHF binding to this site is relatively weak, again suggesting that CI can bind to OL3 to some extent in the absence of dodecamer formation.
The finding that CI binding to OL3 is not repressive suggests that there is not a severe clash between CI at OL3 and RNAP at PRM in the looped form. Recent structural modeling (9) is consistent with this conclusion.
Properties of phages carrying IHF or ΔUP mutations.
To analyze the effects of IHF and ΔUP mutations on regulation of CI in the intact phage, we crossed the mutations onto λ, using a newly developed selection for phage recombinants carrying a plasmid-borne OL region (see the text and Fig. S2 in the supplemental material). Both the IHFmut and ΔUP mutants were mildly defective in burst size and lysogenization frequency (see the supplemental material). They formed stable lysogens, but these showed drastic defects in prophage induction (see Fig. S3 in the supplemental material). Several lines of evidence (see the supplemental material) suggest that the defect in prophage induction results, at least in part, from reduced expression of PL, probably through defects in expression of Int and/or Xis proteins. The mutants also showed a growth defect under particular conditions (see the supplemental material), which can also be explained by effects on PL expression. Hence, these mutations have pleiotropic effects on λ gene regulation, and we cannot readily analyze their effects on PRM expression in the context of the intact phage circuitry.
At the outset of Results, we mentioned an unsuccessful selection for mutants defective in looping-mediated stimulation. The properties of the IHFmut or ΔUP mutants would likely prevent their isolation by this approach (see the supplemental material). At the same time, this negative result suggests that there do not exist other cis-acting sites in the OL region that are required for stimulation by looping but not for PL function.
DISCUSSION
Our evidence indicates that the stimulation of PRM by CI-mediated looping requires an UP element located near the CI binding sites in OL and is influenced by an adjacent IHF binding site. We interpret these data to indicate that stimulation requires a relatively specific configuration of the looped complex. We then discuss models for the role of the UP element.
Evidence for a specific activated complex and requirement for UP element.
Two lines of evidence indicate that stimulation of CI-mediated activation occurs in a relatively specific form of the looped complex. First, a template lacking OL1 did not support stimulation (Fig. 6); in addition, this and several other mutant templates gave lower activity than the unlooped Timm control in the presence of CI (Fig. 3, 4, and 6). We infer from the reduced activity that loops are forming on these templates and conclude that loop formation per se is not sufficient for stimulation. Second, the carboxy-terminal domain (CTD) of the RNAP α subunit of RNAP is known to contact the UP element in other promoters (17); for this to occur in the present case, RNAP bound at PRM must lie within reach of the UP element in the OL region.
The αCTD is linked to the α N-terminal domain (αNTD) by a flexible tether; this allows the αCTD to interact with UP elements with a certain amount of variation in the spatial relationship between the promoter and the UP element. Our evidence supports the suggestion of Anderson and Yang (8) that an αCTD contacts the UP element in the OL region and that this somehow stimulates CI-mediated activation of PRM. Evidence from a separate study (9) also supports this conclusion.
In a proposed model for the DNA-bound octamer (27), the two DNA duplexes were held apart by ∼100 Å and pointed in roughly the same direction. With this relative orientation of the DNA duplexes, the antiparallel form (form 1 in Fig. 5B) juxtaposes the UP element and the PRM promoter, while in the parallel form (form 3), these elements are much farther apart, so the αCTD could probably not reach the UP element. It is known that the flexibility and reach of the linker attaching the αCTD to the rest of RNAP are not unlimited (28). In studies of the rrnB P1 promoter, which has a strong UP element lying upstream of the −35 region, moving the UP element relative to the −35 region reduced or abolished stimulation (28). Hence, if the duplexes pointed in the same direction, as suggested (27), our findings would favor a model in which only the antiparallel configuration allows stimulation.
However, recent structural modeling (9) suggests a different relative orientation of the two duplexes. When flexibility is allowed for the DNA and for the hinge region of CI, this modeling suggests, first, that the DNA duplexes are bent and not pointing in the same direction and, second, that the αCTD can contact the UP element in both the antiparallel and parallel configurations (forms 1 and 3 in Fig. 5). These authors concluded that both configurations allow stimulation. They also found experimentally that both orientations of the OL region (located 2.3 kb upstream of the promoter in this case) gave the same degree of stimulation, suggesting either that both orientations allow stimulation and/or that the flexibility of DNA over this distance allows both orientations to occur. More detailed structural information will be needed to resolve this issue.
Mechanism for stimulation by the UP element.
We suggest two nonexclusive mechanisms by which the UP element stimulates CI-mediated activation of PRM in looped complexes. First, the αCTD-UP element interaction may drive the equilibria among various species toward RNAP binding, thereby increasing the proportion of closed complexes at PRM. Second, the αCTD-UP element interaction may lead to conformational changes in the complex which increase the activity of the promoter. We first recall the early steps in transcription initiation and then discuss and evaluate these models in turn.
Transcription initiation proceeds through formation of an open complex, in which the two DNA strands are separated prior to initiation. Its formation can be simplified to a two-step model:
where R, P, RPc, and RPo are free RNAP holoenzyme, free promoter, and closed and open complexes, respectively; KB is the association constant for binding, and kf is the rate constant for the overall isomerization reaction. Values for these parameters can be measured in vitro by kinetic analysis of transcription initiation (29). Changes in either parameter can affect the rate of expression from a promoter (30). For most promoters, including PRM (31, 32), formation of the open complex is rate limiting, and open complexes usually initiate promptly, leading to promoter clearance and accessibility of the promoter to subsequent RNAP molecules. Accordingly, open complexes and later initiating forms are a small fraction of the total templates, and we can ignore them in considering the distribution of species.
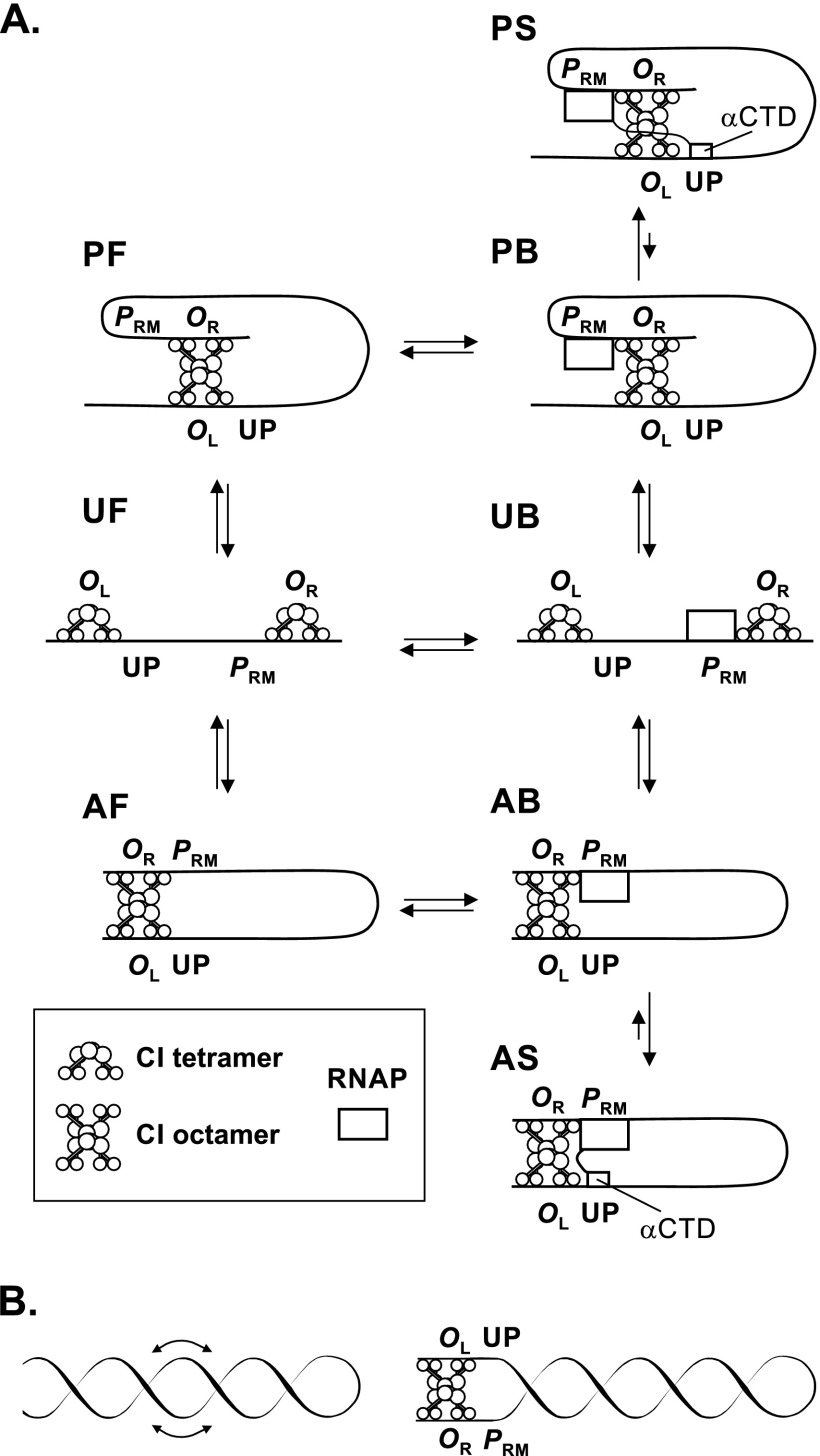
According to the first model for the effect of the UP element, the effective value of KB is increased. The equilibria among the species that can form in our activation experiments (Fig. 3) are depicted in Fig. 8. At the CI levels used in these experiments (Fig. 3), PRM activity reached a plateau value as CI concentrations increased, suggesting that operator occupancy by CI was complete. Even in the wild-type case (with wild-type OL3 and OR3), these are the predominant tetrameric species, since the OR1 - OR2 occupancy pattern is energetically favored (33) and it is likely that the OL1 - OL2 pattern is also favored. Hence, at the moderate CI concentrations at which looping-mediated stimulation occurs, this framework should be applicable to the wild-type as well. The forms include those with RNAP bound (B) at PRM and those free (F) of RNAP, those in the parallel (P) and antiparallel (A) looped configurations, and those that are unlooped (U). In addition, our data support the existence of stimulated forms (S) in which the αCTD contacts the UP element.
Fig 8.
Interconversions of looped complexes and effects of supercoiling. (A) Interconversions among looped complexes. Only complexes with CI tetramers bound at OL1 and OL2 and at OR1 and OR2 are shown. For simplicity, the later stages in open complex formation are not depicted, although clearly these would play a role. In form AB, the αCTD of RNAP can bind to the UP element, giving AS; the chelate effect would likely favor AS in the AB ↔ AS equilibrium. We include a stimulated parallel form, PS, based on recent structural modeling suggesting (9) that the αCTD-UP element interaction can also take place in the parallel form. This work (9) assumes that the equilibrium constant for the PB ↔ PS interconversion is the same as for AB ↔ AS, but direct evidence for this is lacking. (B) Left, idealized structure of a plectonemic superhelix. The line represents a DNA duplex. Negative supercoiling causes the duplex to wrap about itself in a right-handed superhelix, as shown. Helices can readily slide with respect to each other (44), as indicated by the arrows. Right, supercoiled form of the CI-mediated loop. Supercoiling favors the antiparallel configuration, as shown. See the supplemental material for further discussion.
Equilibrium constants for these interconversions are not presently known, but some information is available. The energy change upon looping, termed ΔGoct (6, 9), has a value of about −0.5 kcal/mol, based on fitting reporter data at various CI levels to physicochemical models of the λ circuitry. This value reflects the equilibrium between UF and AF (or between UB and AB) (Fig. 8). At a value of −0.5 kcal/mol, about 70% of the templates would be in the looped form.
The in vivo equilibrium constant is also not known for the interconversion between UF and UB. For the unlooped molecule, KB = [UB]/[RNAP] × [UF]. Although an in vitro value for KB has been measured for unlooped PRM (31, 32), in general the value of KB depends markedly on the ionic conditions (34, 35), and it is unclear how to relate these to in vivo conditions. The in vivo concentration of free RNAP is also not known. Hence, the fraction of time that PRM is occupied by RNAP cannot be calculated.
If we make the reasonable assumption, however, that this fraction is less than half, a simple and straightforward mechanism follows for the action of the UP element in looping-mediated stimulation. If, as seems likely, the UP element-αCTD interaction stabilizes AS relative to AB, it would drive these coupled equilibria toward AS. This would increase the fraction of templates on which RNAP is bound, thereby increasing the overall activity of the promoter. A quantitative analysis of this model, which we term the “KB model,” is presented in the supplemental material. If PS is as active as AS, the same argument would apply to the parallel forms.
In this model, no other changes would need to occur in the activity of the promoter to account for the stimulatory effect. There is a limit on the degree to which PRM activity could be stimulated by this mechanism, since the fraction of templates in the closed complex cannot exceed 1.0. This model predicts that in an in vitro system (10), the apparent degree of stimulation would be decreased with increasing RNAP concentrations and under conditions that favor RNAP binding to PRM (34, 35).
This model is also consistent with the greater degree of stimulation observed with the mutant prm240 promoter (Fig. 3) (11). We have suggested (11) that prm240 reduces the value of KB, since it changes the same base in PRM as does prm116, which has this effect (32). If this is the case, the proposed mechanism would allow a greater degree of stimulation, because the fraction of the UB form on the unlooped template would be substantially lower than in the WT case.
It is plausible that the degree to which the αCTD-UP element interaction is favored would be different in certain cases due to steric effects. For instance, preventing IHF binding, or occupancy of OL3 by CI, might weaken this interaction, thereby reducing stimulation by this mechanism, as observed in these two cases (Fig. 3 and 6). In addition, the equilibrium between PS and PB might differ from that between AS and AB.
A second, nonexclusive model for the effect of the UP element is that conformational changes in the PRM-CI-RNAP complex lead to an increase in promoter activity. Ample precedent exists for such a model. The interaction of an UP element with αCTD can stimulate either KB or kf; the two published studies, with different promoters, concluded that it probably acts at both steps (36, 37). In addition, the αCTD can contact DNA nonspecifically upstream of the −35 region. At several promoters, this interaction somehow stimulates a step subsequent to RNAP binding (38, 39).
In the case of PRM, several examples (with unlooped templates) are compatible with effects arising from subtle conformational changes. First, when CI can bind only to OR2 but not to OR1 or to OR3, the degree of stimulation of PRM in vivo is only 5-fold instead of 10-fold (1). Second, the RNAP σ70 subunit contacts CI, and this contact increases kf (31, 32, 40) in vitro; when this contact is altered by a mutation in σ70, the interaction with WT CI increases KB but not kf (41). Finally, certain PRM mutations appear to weaken cooperative CI binding between OR1 and OR2 in vitro, an effect interpreted as resulting from a conformational change in RNAP (42).
With the mutant prm240 promoter, we saw differences from PRM+ in its responses to changes in growth media and in the cis-acting sites in the OL region, notably with templates carrying only a functional OL2 at OL or the IHFmut allele. Possibly, prm240 is stimulated, at least in part, by a mechanism different from that operating on PRM+. We speculate (see the supplemental material) that a higher proportion of prm240 templates are in the antiparallel form than in the wild-type case; such an effect could contribute to the greater degree of stimulation observed with prm240 than with PRM+, if AS has higher activity than PS. Finally, conformational changes such as those just cited may also affect the properties of RNAP-CI interactions at the mutant promoter or in the interaction with the UP element; for instance, the interaction with the UP element may be more favorable with prm240 than with WT PRM (see the text and Fig. S5 in the supplemental material).
It was previously suggested (10) that looping might stimulate PRM expression simply by increasing the occupancy of OR2. However, this model predicts that increasing the level of CI with the unlooped template would afford the same level of expression as with the looped one, contrary to our data and those of Lewis et al. (10).
Effects of mutational changes on expression.
With many mutant templates, we saw decreases in the level of lacZ expression, and in some cases the activity was lower than that of the unlooped Timm control (Fig. 3, 4, 6, and 7). Decreases could in principle arise from several sources. These include changes in the value of ΔGoct, or in the activity of AS, or in the proportion of AS on templates allowing AS formation. If PS cannot form, or its formation is less favored than that of AS, or it has lower activity than AS, then an increase in the proportion of parallel forms would also reduce the overall activity. A combination of all these changes is also possible. Our data do not permit us to infer which of these possible changes occur in the various mutants we analyzed. However, decreases below the level of the unlooped control imply that some of the looped species (Fig. 5) are repressed, partially or completely, relative to the unlooped control.
Role of IHF site.
We found that the IHF binding site adjacent to OL3 plays a role in looping-mediated stimulation of PRM (Fig. 3). With wild-type PRM, mutating this site reduced the level of stimulation; with the prm240 promoter, stimulation was abolished, but the residual activity was somewhat greater than that seen with the ΔUP template. In addition, the Δ(71-101) deletion, which removes both the UP element and the IHF binding site, decreased PRM activity below that of a template containing only the ΔUP deletion. This suggests that the IHF site is not simply enhancing the contribution of the UP element to looping-mediated stimulation of PRM.
IHF bends DNA by roughly 180° (43). Such a bend could place the UP element in a location that is more favorable for interaction with the αCTD; possibly, in the absence of a bend, the αCTD-UP element interaction is less effective. This model is not, however, consistent with the greater effect of the Δ(71-101) deletion.
In studies of looping-mediated stimulation in vitro (10), done in the absence of IHF, looping increased the activity of a mutant PRM allele by a factor of about 1.6. Our findings suggest that adding IHF might increase the degree of stimulation.
Closing remarks.
An additional complexity of this system deserves further investigation. Although the loop is conventionally depicted as a simple structure (Fig. 1 and 8), this segment of DNA is most likely supercoiled a large fraction of the time. Supercoiling causes DNA to form a plectonemic superhelix (Fig. 8B, left), in which a segment of duplex DNA (represented by a single line) forms a loop with the two arms intertwined (44). The two arms can slide or “slither” rapidly with respect to one another (arrows in figure), rapidly juxtaposing a given site on one helix with many sites on the opposite helix, raising their effective local concentration perhaps 100-fold relative to that on relaxed DNA (44). This “slithering” should favor octamer formation (Fig. 8B, right) and formation of the antiparallel form. In the supplemental material, we consider these issues and the likely impact of transcription on superhelix formation and stability. We also discuss how these issues affect interpretation of the present work.
Clearly, much remains to be learned about this relatively simple system. It has long been appreciated that the OR region is densely packed with interdigitated sites (3, 4). Our findings, taken with those of others (13–16), indicate that the OL region is similarly dense in sites and that these sites play multiple roles. Time will tell whether the complexity of the λ regulatory regions is exceptional, or is typical of biological systems and only appears exceptional because of the great depth in which λ has been analyzed.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Gary Gussin for helpful discussions, to Sankar Adhya, Carol Dieckmann, and Gary Gussin for comments on the manuscript, to Anca Segall for plasmids, and to Ian Dodd and Keith Shearwin for communicating unpublished results.
This work was supported by grant GM24178 from the National Institutes of Health.
Footnotes
Published ahead of print 24 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02148-12.
REFERENCES
- 1. Meyer BJ, Maurer R, Ptashne M. 1980. Gene regulation at the right operator (OR) of bacteriophage λ II. OR1, OR2, and OR3: their roles in mediating the effects of repressor and cro. J. Mol. Biol. 139: 163– 194 [DOI] [PubMed] [Google Scholar]
- 2. Michalowski CB, Short MD, Little JW. 2004. Sequence tolerance of the phage λ PRM promoter: implications for evolution of gene regulatory circuitry. J. Bacteriol. 186: 7988– 7999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ptashne M, Jeffrey A, Johnson AD, Maurer R, Meyer BJ, Pabo CO, Roberts TM, Sauer RT. 1980. How the λ repressor and cro work. Cell 19: 1– 11 [DOI] [PubMed] [Google Scholar]
- 4. Ptashne M. 2004. A genetic switch: phage lambda revisited, vol 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 5. Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. 2001. Octamerization of λ CI repressor is needed for effective repression of PRM and efficient switching from lysogeny. Genes Dev. 15: 3013– 3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB. 2004. Cooperativity in long-range gene regulation by the λ CI repressor. Genes Dev. 18: 344– 354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson LM, Yang H. 2008. A simplified model for lysogenic regulation through DNA looping. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2008: 607– 610 [DOI] [PubMed] [Google Scholar]
- 8. Anderson LM, Yang H. 2008. DNA looping can enhance lysogenic CI transcription in phage lambda. Proc. Natl. Acad. Sci. U. S. A. 105: 5827– 5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cui L, Murchland I, Shearwin KE, Dodd IB. 2013. Enhancer-like long-range transcriptional activation by λ CI-mediated DNA looping. Proc. Natl. Acad. Sci. U. S. A. 110: 2922– 2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lewis D, Le P, Zurla C, Finzi L, Adhya S. 2011. Multilevel autoregulation of λ repressor protein CI by DNA looping in vitro. Proc. Natl. Acad. Sci. U. S. A. 108: 14807– 14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Little JW, Michalowski CB. 2010. Stability and instability in the lysogenic state of phage lambda. J. Bacteriol. 192: 6064– 6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Degnan PH, Michalowski CB, Babić AC, Cordes MHJ, Little JW. 2007. Conservation and diversity in the immunity regions of wild phages with the immunity specificity of phage λ. Mol. Microbiol. 64: 232– 244 [DOI] [PubMed] [Google Scholar]
- 13. Giladi H, Koby S, Prag G, Engelhorn M, Geiselmann J, Oppenheim AB. 1998. Participation of IHF and a distant UP element in the stimulation of the phage λ PL promoter. Mol. Microbiol. 30: 443– 451 [DOI] [PubMed] [Google Scholar]
- 14. Giladi H, Murakami K, Ishihama A, Oppenheim AB. 1996. Identification of an UP element within the IHF binding site at the PL1-PL2 tandem promoter of bacteriophage λ. J. Mol. Biol. 260: 484– 491 [DOI] [PubMed] [Google Scholar]
- 15. Giladi H, Koby S, Gottesman ME, Oppenheim AB. 1992. Supercoiling, integration host factor, and a dual promoter system participate in the control of the bacteriophage λ pL promoter. J. Mol. Biol. 224: 937– 948 [DOI] [PubMed] [Google Scholar]
- 16. Giladi H, Gottesman M, Oppenheim AB. 1990. Integration host factor stimulates the phage lambda pL promoter. J. Mol. Biol. 213: 109– 121 [DOI] [PubMed] [Google Scholar]
- 17. Gourse RL, Ross W, Gaal T. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37: 687– 695 [DOI] [PubMed] [Google Scholar]
- 18. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19. Little JW, Shepley DP, Wert DW. 1999. Robustness of a gene regulatory circuit. EMBO J. 18: 4299– 4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giese KC, Michalowski CB, Little JW. 2008. RecA-dependent cleavage of LexA dimers. J. Mol. Biol. 377: 148– 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Atsumi S, Little JW. 2004. Regulatory circuit design and evolution using phage λ. Genes Dev. 18: 2086– 2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Michalowski CB, Little JW. 2005. Positive autoregulation of cI is a dispensable feature of the phage λ gene regulatory circuitry. J. Bacteriol. 187: 6430– 6442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whipple FW, Kuldell NH, Cheatham LA, Hochschild A. 1994. Specificity determinants for the interaction of λ repressor and P22 repressor dimers. Genes Dev. 8: 1212– 1223 [DOI] [PubMed] [Google Scholar]
- 24. Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53: 85– 96 [DOI] [PubMed] [Google Scholar]
- 25. Sarai A, Takeda Y. 1989. λ repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proc. Natl. Acad. Sci. U. S. A. 86: 6513– 6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL., Jr 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22: 5765– 5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stayrook S, Jaru-Ampornpan P, Ni J, Hochschild A, Lewis M. 2008. Crystal structure of the λ repressor and a model for pairwise cooperative operator binding. Nature 452: 1022– 1025 [DOI] [PubMed] [Google Scholar]
- 28. Meng WM, Belyaeva T, Savery NJ, Busby SJW, Ross WE, Gaal T, Gourse RL, Thomas MS. 2001. UP element-dependent transcription at the Escherichia coli rrnB P1 promoter: positional requirements and role of the RNA polymerase α subunit linker. Nucleic Acids Res. 29: 4166– 4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McClure WR. 1980. Rate-limiting steps in RNA chain initiation. Proc. Natl. Acad. Sci. U. S. A. 77: 5634– 5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McClure WR. 1985. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 54: 171– 204 [DOI] [PubMed] [Google Scholar]
- 31. Hawley DK, McClure WR. 1982. Mechanism of activation of transcription initiation from the λ PRM promoter. J. Mol. Biol. 157: 493– 525 [DOI] [PubMed] [Google Scholar]
- 32. Shih MC, Gussin GN. 1983. Mutations affecting two different steps in transcription initiation at the phage λ PRM promoter. Proc. Natl. Acad. Sci. U. S. A. 80: 496– 500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koblan KS, Ackers GK. 1992. Site-specific enthalpic regulation of DNA transcription at bacteriophage λ OR. Biochemistry 31: 57– 65 [DOI] [PubMed] [Google Scholar]
- 34. Roe JH, Record MT., Jr 1985. Regulation of the kinetics of the interaction of Escherichia coli RNA polymerase with the λ PR promoter by salt concentration. Biochemistry 24: 4721– 4726 [DOI] [PubMed] [Google Scholar]
- 35. Leirmo S, Harrison C, Cayley S, Burgess RR, Record MT., Jr 1987. Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry 26: 2095– 2101 [DOI] [PubMed] [Google Scholar]
- 36. Rao L, Ross W, Appleman JA, Gaal T, Leirmo S, Schlax PJ, Record MT, Jr, Gourse RL. 1994. Factor independent activation of rrnB P1: an “extended” promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol. 235: 1421– 1435 [DOI] [PubMed] [Google Scholar]
- 37. Tang Y, Murakami K, Ishihama A, DeHaseth PL. 1996. Upstream interactions at the lambda pRM promoter are sequence nonspecific and activate the promoter to a lesser extent than an introduced UP element of an rRNA promoter. J. Bacteriol. 178: 6945– 6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ross W, Gourse RL. 2005. Sequence-independent upstream DNA-αCTD interactions strongly stimulate Escherichia coli RNA polymerase-lacUV5 promoter association. Proc. Natl. Acad. Sci. U. S. A. 102: 291– 296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davis CA, Capp MW, Record MT, Jr, Saecker RM. 2005. The effects of upstream DNA on open complex formation by Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 102: 285– 290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jain D, Nickels BE, Hochschild A, Darst SA. 2004. Structure of a ternary transcription activation complex. Mol. Cell 13: 45– 53 [DOI] [PubMed] [Google Scholar]
- 41. Li M, McClure WR, Susskind MM. 1997. Changing the mechanism of transcriptional activation by phage lambda repressor. Proc. Natl. Acad. Sci. U. S. A. 94: 3691– 3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hwang JJ, Gussin GN. 1988. Interactions between Escherichia coli RNA polymerase and lambda repressor. Mutations in PRM affect repression of PR. J. Mol. Biol. 200: 735– 739 [DOI] [PubMed] [Google Scholar]
- 43. Rice PA, Yang SW, Mizuuchi K, Nash HA. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87: 1295– 1306 [DOI] [PubMed] [Google Scholar]
- 44. Vologodskii AV, Levene SD, Klenin KV, Frank-Kamenetskii M, Cozzarelli NR. 1992. Conformational and thermodynamic properties of supercoiled DNA. J. Mol. Biol. 227: 1224– 1243 [DOI] [PubMed] [Google Scholar]
- 45. Mao C, Carlson NG, Little JW. 1994. Cooperative DNA-protein interactions: effects of changing the spacing between adjacent binding sites. J. Mol. Biol. 235: 532– 544 [DOI] [PubMed] [Google Scholar]
- 46. Liu Z, Little JW. 1998. The spacing between binding sites controls the mode of cooperative DNA-protein interactions: implications for evolution of regulatory circuitry. J. Mol. Biol. 278: 331– 338 [DOI] [PubMed] [Google Scholar]
- 47. Senear DF, Brenowitz M, Shea MA, Ackers GK. 1986. Energetics of cooperative protein-DNA interactions: comparison between quantitative deoxyribonuclease footprint titration and filter binding. Biochemistry 25: 7344– 7354 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.