Abstract
In the gammaproteobacteria, the FeoA, FeoB, and FeoC proteins constitute the Feo system, which mediates ferrous iron [Fe(II)] import. Of these Feo proteins, FeoB is an inner membrane Fe(II) transporter that is aided by the small protein FeoA. However, the role of another small protein, FeoC, has remained unknown. Here we report that the FeoC protein is necessary for FeoB protein-mediated Fe(II) uptake in Salmonella experiencing low levels of oxygen and iron. The FeoC protein was found to directly bind to the FeoB transporter, leading to high cellular levels of FeoB. Depletion of the FtsH protease enabled high levels of FeoB in the absence of FeoC, suggesting that the FeoC protein protects the FeoB transporter from FtsH-mediated proteolysis. Our present study provides a singular example of bacteria that can control expression of iron uptake systems posttranslationally by employing a small iron transporter-binding protein.
INTRODUCTION
Iron, an essential metal for all forms of life, is present in an oxidized (ferric) or a reduced (ferrous) form depending on the oxidation state. Thus, iron mostly exists in an insoluble ferric or as a soluble ferrous form under aerobic or anaerobic conditions, respectively. To utilize ferric iron [Fe(III)], bacteria synthesize and secrete Fe(III)-specific chelators called siderophores (1). Gram-negative bacteria then uptake Fe(III)-siderophore complexes via outer membrane receptors in a process that requires the TonB-ExbB-ExbD protein complex (1). In contrast, bacteria directly import ferrous iron [Fe(II)] via membrane transporters (1).
In the gammaproteobacteria, the feoA, feoB, and feoC genes are likely to constitute an operon (2). The Escherichia coli feo operon encodes the small (8.4-kDa) protein FeoA, the large (84.5-kDa) protein FeoB, and another small (8.7-kDa) protein, FeoC (2). The FeoB protein is an authentic Fe(II) transporter (3) consisting of an N-terminal cytoplasmic domain and a C-terminal transmembrane domain (2). The FeoB N terminus possesses a eukaryotic G-protein function and hydrolyzes GTP using GTPase activity, which is essential for FeoB to import Fe(II) (4). We recently reported that the FeoA protein interacts with the FeoB transporter and is necessary for FeoB-mediated Fe(II) uptake in Salmonella enterica (5). FeoB-mediated Fe(II) uptake appears to play an important role in Salmonella pathogenesis, as evidenced by the fact that a Salmonella feoB mutant was attenuated for virulence in mice (6).
The deduced amino acid sequence of the FeoC protein possesses a motif resembling DNA-binding domains of transcriptional regulators and a putative binding site for an iron-sulfur (Fe-S) cluster (2). On the basis of these features, the FeoC protein was initially proposed as a Fe-S cluster-dependent transcription factor controlling the feo operon (2); however, a feoC mutation was shown to have no effect on transcription of the feo promoter in Yersinia pestis (7). Nonetheless, evidence suggests that the FeoC protein plays a role as the FeoB transporter, because the FeoC protein and the N-terminal domain of FeoB from Klebsiella pneumoniae were shown to interact with each other (8).
Expression levels of the feoB gene change in response to iron and oxygen, which results from the action of the iron-sensing transcriptional regulator Fur (9) and the oxygen-sensing transcriptional regulator Fnr (10). The Fur protein was shown to repress feoB transcription under iron-replete conditions (3, 11), while the activator Fnr protein promotes feoB transcription under anaerobic conditions (3). In the present study, we report that the FeoC protein is another factor that determines cellular levels of the FeoB Fe(II) transporter in Salmonella enterica. We reveal that the FeoC protein contributes to an increase in FeoB protein levels at the posttranslational level. We demonstrate direct interaction between the FeoC and FeoB proteins and provide evidence that the FeoC protein leads to high cellular levels of FeoB by protecting it from FtsH protease-mediated proteolysis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Salmonella enterica serovar Typhimurium strains were derived from strain 14028s. Phage P22-mediated transductions were performed as described previously (12). S. Typhimurium strains were grown at 37°C in Luria-Bertani (LB) medium. Bacteria were grown in Erlenmeyer flasks with vigorous shaking for the preparation of aerobic cultures or grown in screw-cap tubes fully filled with medium without agitation for the preparation of anaerobic cultures. The iron chelator deferoxamine was used at 0.2 mM to reduce the iron levels in LB medium. When necessary, ampicillin (Ap), chloramphenicol (Cm), and kanamycin (Km) were used at 50 μg/ml, 25 μg/ml, and 50 μg/ml, respectively. For induction of genes from plasmids, arabinose and isopropyl 1-thio-β-d-galactoside (IPTG) were used at various concentrations.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristicsa | Reference or source |
|---|---|---|
| S. enterica serovar Typhimurium | ||
| 14028s | Wild type | 31 |
| JH352 | 14028s Δfur | 11 |
| JH362 | 14028s ΔfeoB | 11 |
| JH363 | 14028s ΔfeoC | This study |
| JH323 | 14028s ΔfeoC::Kmr | This study |
| HK715 | 14028s ΔfeoBC | This study |
| HK708 | 14028s Δfnr | This study |
| HK333 | 14028s ΔftsH::Kmr/plac-FtsH | This study |
| HK334 | 14028s ΔftsH::Kmr ΔfeoC/plac-FtsH | This study |
| DN281 | 14028s ΔclpA::Kmr | This study |
| DN297 | 14028s ΔclpX | This study |
| DN296 | 14028s ΔclpP | This study |
| DN283 | 14028s ΔhslUV::Kmr | This study |
| DN282 | 14028s Δlon::Kmr | This study |
| DN299 | 14028s ΔclpA::Kmr ΔfeoC | This study |
| DN311 | 14028s ΔclpX ΔfeoC | This study |
| DN310 | 14028s ΔclpP ΔfeoC | This study |
| DN301 | 14028s ΔhslUV::Kmr ΔfeoC | This study |
| DN300 | 14028s Δlon::Kmr ΔfeoC | This study |
| E. coli BTH101 | F− cya99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | 19 |
| Plasmids | ||
| pUHE21-2lacIq | Plac reppMBI Apr lacIq | 14 |
| plac-FeoC | pUHE21-2lacIq feoC | This study |
| plac-FtsH | pUHE21-2lacIq ftsH | This study |
| pBAD33 | PBAD pACYC184 ori Cmr | 15 |
| pBAD-FeoB | pBAD33 feoB | This study |
| pHis-parallel1 | PT7 reppMBI Apr | 16 |
| pHis-parallel1-FeoC | pHis-parallel1 feoC | This study |
| pHis-parallel1-Fur | pHis-parallel1 fur | This study |
| pKD3 | repR6Kγ Cmr FRT Kmr FRT | 13 |
| pKD4 | repR6Kγ Apr FRT Kmr FRT | 13 |
| pKD46 | reppSC101(Ts) Apr ParaBAD γ β exo | 13 |
| pKD46-CmR | reppSC101(Ts) Cmr ParaBAD γ β exo | This study |
| pCP20 | reppSC101(Ts) Apr Cmr cI857 λPRflp | 13 |
| pKT25 | Plac p15A ori Kmr | 17 |
| pUT18 | Plac ColEI ori Apr | 17 |
| pT18-FeoC | pUT18 feoC | This study |
| pT25-FeoB | pKT25 feoB | 5 |
| pT25-MgtA | pKT25 mgtA | This study |
| pT18-Zip | pUT18 zip | 19 |
| pT25-Zip | pKT25 zip | 19 |
FRT, Flp recombination target; rep, replicon.
Construction of bacterial strains.
S. Typhimurium strains carrying a gene deletion were constructed using the one-step gene inactivation method (13). For deletion of the feoC and feoBC genes, the Kmr cassette from plasmid pKD4 (13) was amplified using primer pairs DEL-feoC-F/DEL-feoC-R and DEL-feoB-F/DEL-feoC-R, respectively. For deletion of the fnr gene, the Kmr cassette from plasmid pKD4 was amplified using the primer pair DEL-fnr-F/DEL-fnr-R. For deletion of the clpA, clpX, clpP, hslUV, and lon genes, the Kmr cassette from plasmid pKD4 was amplified using the primer pairs DEL-clpA-F/DEL-clpA-R, DEL-clpX-F/DEL-clpX-R, DEL-clpP-F/DEL-clpP-R, DEL-hslUV-F/DEL-hslUV-R, and DEL-lon-F/DEL-lon-R, respectively. The resulting PCR products were integrated into the chromosome of strain 14028s as described previously (13). When necessary, the Kmr cassette was removed using plasmid pCP20 (13). The ftsH gene encoding the FtsH protease was essential in S. Typhimurium. Thus, a chromosomal deletion of the ftsH gene was generated in strains producing the FtsH protease from the plasmid-linked lac promoter as follows. The Kmr cassette from plasmid pKD4 was amplified using the primer pair DEL-ftsH-F/DEL-ftsH-R. The resulting PCR products were integrated into the chromosome of strains expressing the λ Red recombinase and the FtsH protease from pKD46-CmR and plac-FtsH plasmids, respectively. Deletion of the corresponding genes was verified by colony PCR. The sequences of all primers used for strain construction are listed in Table S1 in the supplemental material.
Construction of plasmids.
Plasmid plac-FeoC, expressing the feoC gene from the lac promoter, was constructed. The feoC gene was amplified using the primer pair EX1-feoC-F/EX1-feoC-R and chromosomal DNA from the 14028s strain. The PCR products were purified and introduced between the BamHI and PstI restriction sites of pUHE21-2lacIq (14). To construct plasmid pBAD-FeoB, in which the feoB open reading frame (ORF) is expressed from the PBAD promoter, the feoB ORF was amplified using the primer pair EX2-feoB-F/EX2-feoB-R and chromosomal DNA of strain 14028s. The PCR products were then introduced between the XbaI and PstI restriction sites of pBAD33 (15). Plasmid plac-FtsH expresses the ftsH gene from the lac promoter. For its construction, the ftsH gene was amplified using the primer pair EX-ftsH-F/EX-ftsH-R and chromosomal DNA from the 14028s strain, and the PCR products were introduced between the BamHI and PstI restriction sites of pUHE21-2lacIq. Plasmids pHis-parallel1-FeoC and pHis-parallel1-Fur express the feoC and fur genes, respectively, with nucleotides encoding a His6 tag at their N termini. For construction of these plasmids, the feoC and fur genes were amplified using the primer pairs EX2-feoC-F/EX2-feoC-R and EX-fur-F/EX-fur-R, respectively, with 14028s chromosomal DNA as the template. The resulting PCR products were introduced between the NcoI and NotI restriction sites of the pHis-parallel1 vector (16). For the bacterial two-hybrid assay, plasmids pT18-FeoC and pT25-MgtA were constructed. The feoC and mgtA genes were amplified using the primer pairs TH-feoC-F/TH-feoC-R and TH-mgtA-F/TH-mgtA-R, respectively. The resulting PCR products were introduced between the PstI and BamHI sites of the plasmid vector pUT18 (17) or between the XbaI and BamHI sites of pKT25 (17). Plasmid pKD46-CmR, a derivative of plasmid pKD46 (13), expresses the λ Red recombinase gene from the PBAD promoter. For its construction, the bla gene on pKD46 plasmid was inactivated by restriction enzyme digestion with XmnI as described previously (18). Then, the Cmr cassette from plasmid pKD3 (13) was amplified using the primer pair EX-CmR-F/EX-CmR-R, which was introduced into the XmnI-digested pKD46 via blunt-end DNA ligation. Recombinant plasmid gene sequences were confirmed by nucleotide sequencing. The sequences of all primers used for plasmid construction are listed in Table S1 in the supplemental material.
Immunoblot analysis.
S. Typhimurium strains were grown in LB medium to optical density at 600 nm (OD600) values of approximately 0.5. Equivalent amounts of bacterial cells normalized by OD600 values were removed, washed with phosphate-buffered saline (PBS), suspended in 0.5 ml PBS, and opened by sonication. Whole-cell lysates were resolved on 12% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and analyzed by immunoblotting using anti-FeoB antibodies (5). Blots were developed using anti-rabbit IgG horseradish peroxidase-linked antibodies (GE Healthcare) and an enhanced chemiluminescence (ECL) detection system (GE Healthcare).
RNA isolation and quantitative real-time RT-PCR (qRT-PCR) analysis.
Bacteria were grown in LB medium to an OD600 of 0.5 to 0.6. The culture (0.5 ml) was removed and mixed with 1 ml RNAprotect bacterial reagent (Qiagen), and RNA was isolated using the RNeasy minikit (Qiagen). The RNA sample was treated further with RNase-free DNase (Ambion). cDNA was synthesized using the PrimeScript RT reagent kit (TaKaRa), random primers (Invitrogen), and 0.5 μg of template RNA. Amounts of cDNA were quantified by real-time PCR using SYBR green real-time PCR master mix (Toyobo) with an ABI7300 sequence detection system (Applied Biosystems). Primers used for the detection of cDNA corresponding to the feoB mRNA, feoC mRNA, and gyrB mRNA were Q-feoB-F/Q-feoB-R, Q-feoC-F/Q-feoC-R, and Q-gyrB-F/Q-gyrB-R, respectively. Transcription levels of each gene were calculated from a standard curve obtained by PCR with the same primers and serially diluted genomic DNA. mRNA levels of the feoB and feoC genes were normalized to those of the gyrB gene. The sequences of primers used are listed in Table S1 in the supplemental material.
Bacterial two-hybrid (BACTH) assay.
To assess protein-protein interactions in vivo, a BACTH assay was conducted as described previously (19). The E. coli BTH101 (cya) strain was cotransformed with derivatives of the pUT18 and pKT25 plasmids. The strains were grown overnight at 30°C in LB supplemented with Ap (50 μg/ml), Km (50 μg/ml), and IPTG (1 mM). β-Galactosidase activity was determined in the cultures as described previously (20).
Purification of the His6-FeoC and His6-Fur proteins.
E. coli BL21(DE3) cells harboring pHis-parallel1-FeoC plasmid were grown in LB medium at 37°C. When the culture OD600 value reached approximately 0.5, IPTG (0.5 mM) was added to the culture for induction of His6-FeoC protein production, followed by another 16 h of incubation at 21°C. The His6-FeoC protein was purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. The cell pellet was suspended in cold buffer A [50 mM Tris (pH 8.0), 300 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride (PMSF)] and disrupted by sonication. The cell extract was loaded onto a Ni-NTA column equilibrated with buffer A. After the flowthrough fraction had been discarded, the column was washed with 500 ml washing buffer (buffer A containing 20 mM imidazole), and the adsorbed His6-FeoC protein was eluted with elution buffer (buffer A containing 200 mM imidazole). The His6-Fur protein was purified from E. coli BL21(DE3) cells harboring pHis-parallel1-Fur plasmid by the same method. Finally, the eluted proteins were dialyzed with buffer A containing 10% glycerol and stored at −80°C.
His tag protein pulldown assay.
To assess protein-protein interaction in vitro, a pulldown assay was conducted. Two proteins of interest, one with a His6 tag and another without a His6 tag, were mixed at a 1:1 molar ratio (25 μM each). The protein mixture was incubated at 4°C for 3 h in buffer A [50 mM Tris (pH 8.0), 300 mM NaCl, and 1 mM PMSF] and then loaded onto a column packed with Ni-NTA agarose resin (Qiagen). After the flowthrough fraction had been discarded, the column was washed twice with washing buffer (buffer A containing 20 mM imidazole), and the bound proteins were eluted with elution buffer (buffer A containing 200 mM imidazole). The flowthrough, wash, and elution fractions were boiled in SDS sample loading buffer for 10 min and resolved on 15% SDS-polyacrylamide gels. Protein bands were visualized by silver staining.
Iron uptake assay.
Fe(II) uptake levels in S. Typhimurium strains were determined as follows. Bacterial cells grown in LB to an OD600 of 0.5 were washed twice with N-buffer [N-minimal medium (21), pH 7.0, supplemented with 0.1% Casamino Acids, 38 mM glycerol, 1 mM MgCl2, and 0.1 M sodium ascorbate], suspended in the same buffer, and kept on ice. One ml of the cell suspension at an OD600 of 1.0 was placed at 37°C for 15 min, and the Fe(II) transport assay was started by addition of 0.5 μM 55Fe(II). The 55Fe(II) stock solution was prepared in N-buffer and contained 50 μM 55FeCl3 (PerkinElmer). After 15 min of incubation, bacterial cells were loaded onto a 0.45-μm membrane filter (Whatman) placed on a 1225 sampling manifold (Millipore) and washed twice with 0.1 mM LiCl. The filter with bacterial cells was transferred into a vial containing 2 ml scintillation cocktail fluid. Activity as counts per minute was determined using an LS6500 scintillation counter (Beckman) and converted into picomoles of 55Fe(II) using a standard curve.
RESULTS
The FeoB protein is highly produced when levels of both oxygen and iron are low.
We wanted to determine the amount of Salmonella FeoB Fe(II) transporter produced at different levels of oxygen and iron. For this purpose, we grew Salmonella strains aerobically and anaerobically in LB (i.e., high-iron medium) and in LB supplemented with the iron chelator deferoxamine (i.e., low-iron medium). In the wild-type strain, FeoB protein production was negligible under both aerobic and anaerobic conditions with high iron and weak under aerobic conditions with low iron (Fig. 1A). However, FeoB was highly produced under anaerobic conditions with low iron (Fig. 1A).
Fig 1.
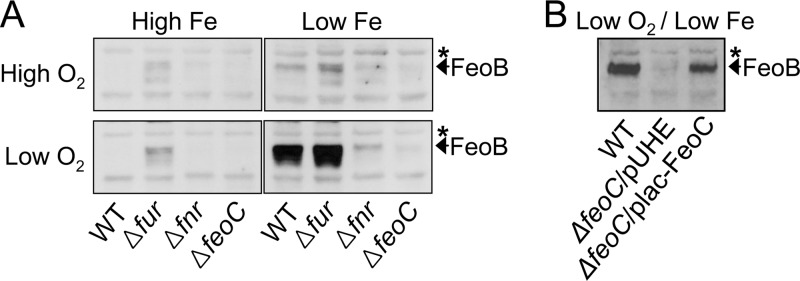
The FeoC protein is necessary for high cellular levels of the FeoB Fe(II) transporter under low-oxygen and -iron conditions. Salmonella strains were grown aerobically (high O2) or anaerobically (low O2) in LB medium (high Fe) or in LB medium supplemented with 0.2 mM deferoxamine (low Fe). By conducting immunoblot analysis, levels of the FeoB protein were determined in wild-type (WT, 14028s), fur deletion (Δfur, JH352), fnr deletion (Δfnr, HK708), and feoC deletion (ΔfeoC, JH363) strains (A) and in the wild-type strain (14028s) and the feoC deletion strain (ΔfeoC, JH363) carrying the empty plasmid vector pUHE21-2lacIq (pUHE) (14) or FeoC-expression plasmid (plac-FeoC) (B). The band indicated with an asterisk corresponds to a protein displaying cross-reactivity against anti-FeoB antibody and serving as an internal loading control.
The iron-sensing Fur regulator represses feoB transcription under iron-replete conditions (3, 11) while the oxygen-sensing Fnr regulator promotes feoB transcription under anaerobic conditions (3). Consistent with these findings, the lack of Fur under anaerobic conditions enhanced FeoB production under high-iron conditions, whereas the lack of Fnr impaired FeoB production under low-iron conditions (Fig. 1A). Taken together, these results suggest that two events that occur upon feoB transcription, the relief of Fur repression and the occurrence of Fnr activation, lead to high-level production of the FeoB transporter when levels of both iron and oxygen are low.
FeoC is necessary for high cellular levels of the FeoB transporter.
Together with the FeoB Fe(II) transporter, the FeoA and FeoC proteins constitute the Feo system in gammaproteobacteria, including Salmonella (2). We explored the possibility that the FeoC protein plays a role in regulating FeoB protein levels. Interestingly, a feoC deletion strain produced very low levels of the FeoB protein under anaerobic conditions with low iron, in contrast to the wild-type strain, which contained high levels of the FeoB protein (Fig. 1A). The defective FeoB production displayed by the feoC deletion mutant was due to the function of FeoC, as evidenced by the fact that ectopic production of the FeoC protein from a plasmid plac-FeoC restored FeoB levels in the feoC deletion mutant (Fig. 1B). Therefore, these results indicate that high cellular levels of the FeoB transporter are dependent on the FeoC protein.
FeoC contributes to FeoB production at the posttranslational level.
We initially explored the possibility that the FeoC protein regulates transcription of the feoB gene. Consistent with FeoB protein levels, the wild-type strain produced the feoB mRNA at high levels under anaerobic conditions with low iron (Fig. 2A). Under the same conditions, the feoC deletion strain was able to express the feoB mRNA slightly more than the wild-type (Fig. 2A), despite the fact that this mutant hardly produced the FeoB protein (Fig. 1A). This result was also in contrast to the finding that an fnr mutant was defective for the expression of feoB mRNA and the production of FeoB protein under the same growth conditions (Fig. 1A and 2A). Thus, unlike the transcriptional regulator Fnr, the FeoC protein did not act on feoB transcription. The FeoC protein also had no effect on the stability of the feoB mRNA. Once RNA synthesis was inhibited with rifampin, the feoB mRNA produced under anaerobic conditions with low iron was degraded at similar rates in both the wild-type and feoC deletion strains (Fig. 2B).
Fig 2.
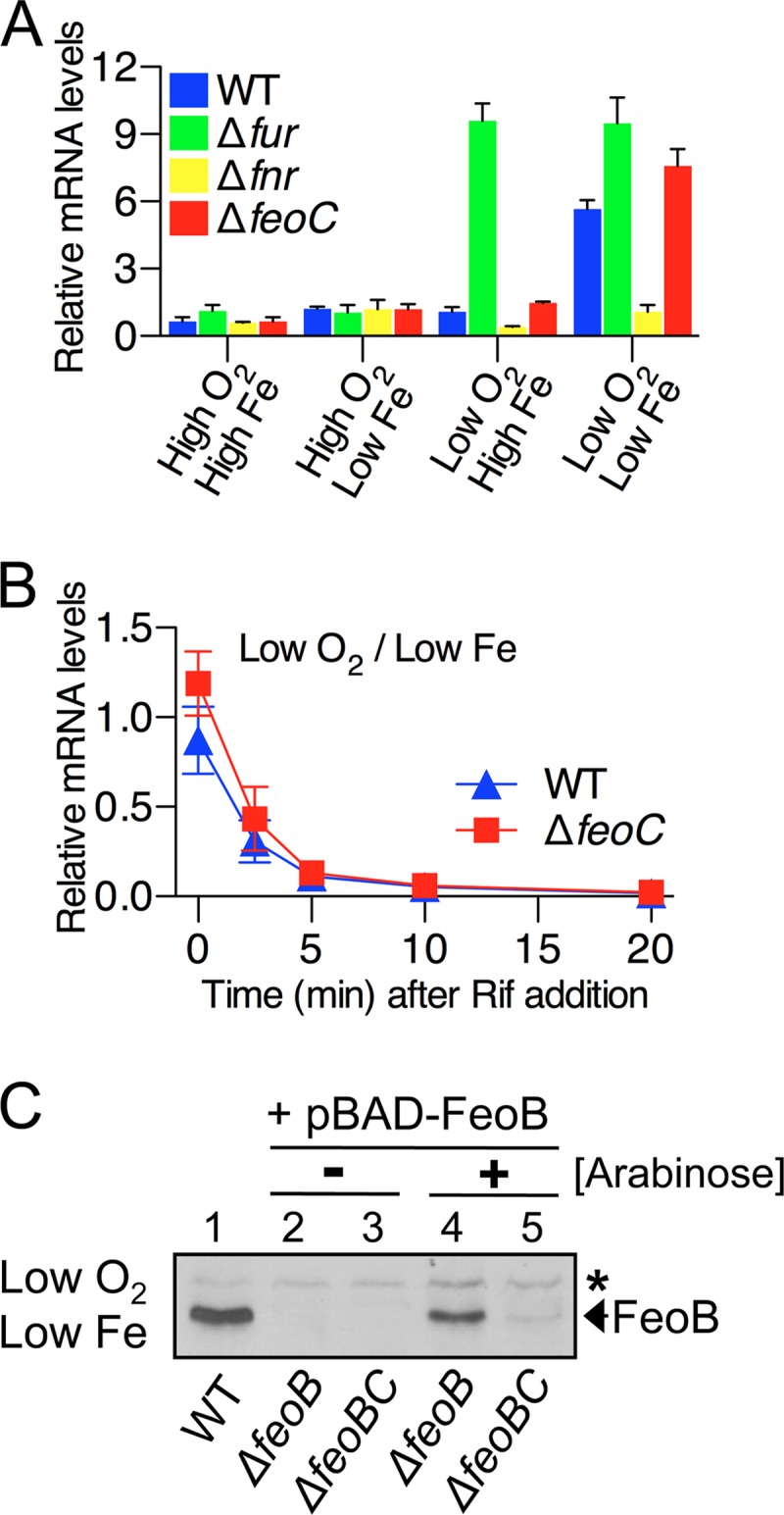
The FeoC protein contributes to cellular FeoB transporter levels at the posttranslational level. Salmonella strains were grown aerobically (high O2) or anaerobically (low O2) in LB (high Fe) or in LB containing 0.2 mM deferoxamine (low Fe). (A) qRT-PCR analysis determined mRNA levels of the feoB gene in wild-type (WT, 14028s), fur deletion (Δfur, JH352), fnr deletion (Δfnr, HK708), and feoC deletion (ΔfeoC, JH363) strains. (B) The stability of the feoB mRNA was examined in the wild-type (WT, 14028s) and feoC deletion (ΔfeoC, JH363) strains. RNA synthesis was inhibited with 200 μg/ml rifampin (Rif). qRT-PCR determined feoB mRNA levels in total RNA isolated at the indicated time points. Values (A and B) are the means and standard deviations (SD) of three independent experiments. (C) Immunoblot analysis determined FeoB protein levels in the wild-type (WT, 14028s), feoB deletion (ΔfeoB, JH362), and feoBC deletion (ΔfeoBC, HK715) strains. The JH362 and HK715 strains harbored the FeoB expression plasmid (pBAD-FeoB) and were grown with (+) or without (−) the inducer arabinose (60 μM). The band indicated with an asterisk corresponds to a protein displaying cross-reactivity against anti-FeoB antibody and serving as an internal loading control.
We then reasoned that if the FeoC protein contributes to FeoB protein levels at the posttranslational level, FeoC-dependent FeoB production would still occur even when the only feoB ORF was expressed from a heterologous promoter. To test this idea, we determined FeoB protein levels in strains deleted for the chromosomal copy of the feoB gene and expressing the feoB ORF from the plasmid-linked PBAD promoter. Under anaerobic conditions with low iron, the FeoB protein was produced at levels similar to the wild-type upon the addition of the inducer arabinose in the feoC+ strain but was hardly produced in the isogenic feoC mutant (Fig. 2C). Taken together, these results indicate that the FeoC protein contributes to FeoB production at the posttranslational level.
The FeoC protein directly interacts with the FeoB transporter.
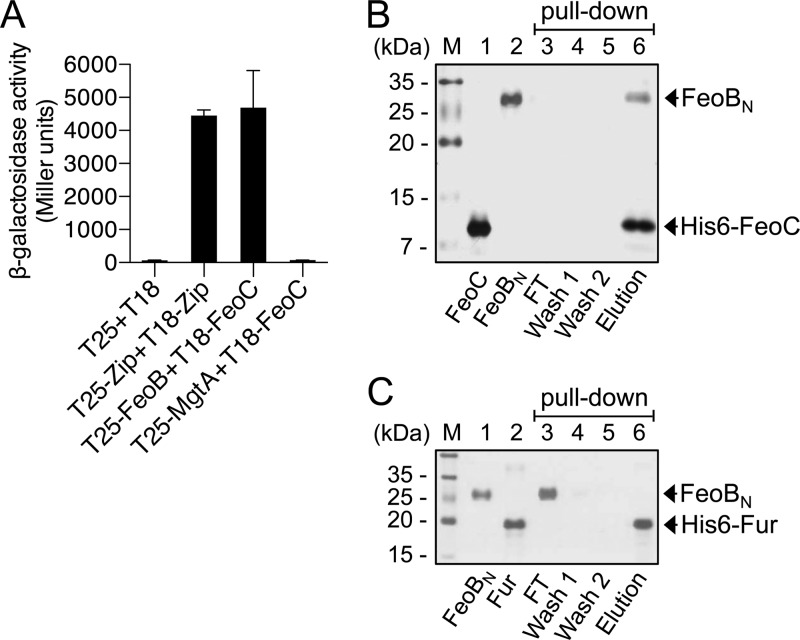
We explored whether the FeoC protein directly acts on the FeoB protein via protein-protein interaction. To assess protein-protein interaction in vivo, a bacterial two-hybrid (BACTH) assay was conducted. We constructed a series of plasmids where proteins are expressed as forms fused to the T18 or T25 domain of Bordetella pertussis adenylate cyclase (19). The cya E. coli strain BTH101 (19) coexpressing the T25-FeoB and T18-FeoC fusion proteins expressed approximately 70-fold higher levels of β-galactosidase than the control strain coproducing the T18 and T25 domains (Fig. 3A). This result indicates that the FeoC and FeoB proteins interact with each other to complement adenylate cyclase activity, which results in cyclic AMP (cAMP)-dependent lacZ expression. In contrast, the FeoC protein did not bind to the Mg2+ transporter MgtA protein (22); therefore, coproduction of the T25-MgtA and T18-FeoC fusion proteins failed to generate β-galactosidase activity relative to the control (Fig. 3A).
Fig 3.
The FeoC protein directly interacts with the FeoB transporter. (A) FeoB-FeoC interaction was assessed using a BACTH system (19). β-Galactosidase activity was determined in cya E. coli (BTH101) strains carrying BACTH plasmids coexpressing T25 and T18, T25-Zip and T18-Zip, T25-FeoB and T18-FeoC, and T25-MgtA and T18-FeoC. β-Galactosidase activity resulting from the coproduction of T25 and T18 or T25-Zip and T18-Zip (19) served as negative and positive controls, respectively. Values are the means and SD of three independent experiments. (B) His tag protein pulldown experiments determined interaction between FeoBN and FeoC. The same amounts of purified FeoBN (without a His tag) and His6-tagged FeoC protein (each at 25 μM) were incubated and subjected to Ni-NTA affinity chromatography. (C) Interaction between purified FeoBN and His6-Fur proteins was assessed as described for panel B. Proteins in the flowthrough (FT), wash (wash 1 and wash 2), and elution fractions were resolved by SDS-PAGE and visualized by silver staining.
To further investigate whether the FeoB-FeoC interaction is direct, we conducted His tag pulldown experiments. The purified N-terminal domain of the FeoB protein (FeoBN) and His-tagged FeoC protein (His6-FeoC) were mixed and subjected to Ni-NTA affinity chromatography. The FeoBN protein was detected together with the His6-FeoC protein in the elution fraction (Fig. 3B), indicating that FeoBN bound to His6-FeoC. In contrast, when the experiment was performed using the His-tagged Fur regulatory protein, the FeoBN protein was mostly detected in the flowthrough fraction but not in the elution fraction, indicating no interaction between these two proteins (Fig. 3C). Taken together, our results demonstrate that the FeoC protein directly and specifically binds to the FeoB transporter.
The FeoC protein prevents FtsH protease-mediated degradation of the FeoB protein.
Based on the findings that the FeoC protein contributes to FeoB protein levels at the posttranslational level and directly interacts with the FeoB transporter, we reasoned that FeoC might function to protect FeoB from proteolytic control mediated by a protease. We further predicted that a lack of such protease activity would recover FeoB protein levels in the feoC mutant.
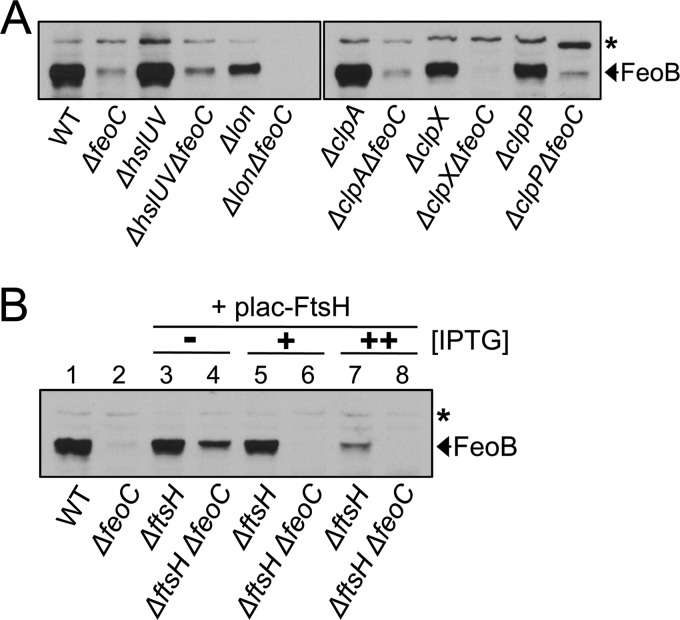
Five proteases—ClpAP, ClpXP, HslUV, Lon, and FtsH—are known to play roles in regulated proteolysis in Gram-negative bacteria (23). We initially examined FeoB levels in a series of feoC deletion mutants carrying an additional deletion of the clpA, clpP, clpX, hslUV, or lon gene. However, all of these mutants produced the FeoB protein at low levels, similar to the feoC deletion mutant (Fig. 4A), indicating that a lack of the ClpAP, ClpXP, HslUV, or Lon protease does not affect FeoB levels in the feoC mutant.
Fig 4.
Depletion of the FtsH protease enables high levels of FeoB in the absence of FeoC. Salmonella strains were grown anaerobically in LB medium with low iron (i.e., LB supplemented with 0.2 mM deferoxamine). Immunoblot analysis determined FeoB protein levels in the wild-type (WT, 14028s), feoC deletion (ΔfeoC, JH363), hslUV deletion (ΔhslUV, DN283), hslUV feoC deletion (ΔhslUV ΔfeoC, DN301), lon deletion (Δlon, DN282), lon feoC deletion (Δlon ΔfeoC, DN300), clpA deletion (ΔclpA, DN281), clpA feoC deletion (ΔclpA ΔfeoC, DN299), clpX deletion (ΔclpX, DN297), clpX feoC deletion (ΔclpX ΔfeoC, DN311), clpP deletion (ΔclpP, DN296), and clpP feoC (ΔclpP ΔfeoC, DN310) strains (A) and wild-type (WT, 14028s) and feoC deletion (ΔfeoC, JH363) strains and ftsH deletion mutants carrying (ΔftsH, HK333) or lacking (ΔftsH ΔfeoC, HK334) the feoC gene (B). Note that ftsH mutants harbored the FtsH expression plasmid (plac-FtsH). ftsH mutants were grown to saturation in the presence of the inducer IPTG (30 μM), and cultures diluted to OD600 values of approximately 0.01 were grown to OD600 values of approximately 0.5 with 30 μM (+) or 60 μM (++) IPTG or without (−) IPTG. The band indicated with an asterisk corresponds to a protein displaying cross-reactivity against anti-FeoB antibody and serving as an internal loading control.
FtsH is a membrane-bound protease that controls membrane protein substrates (24). Thus, we next investigated the role of the FtsH protease in FeoC-dependent FeoB protein levels. Because the ftsH gene is essential in E. coli (25), we anticipated that this might be the case in Salmonella as well. Therefore, we generated an ftsH deletion in a strain expressing the FtsH protease from the plasmid-linked lac promoter. Indeed, the resulting ftsH deletion mutants were able to form colonies on agar plates only when FtsH was induced from the plac-FtsH plasmid by adding IPTG (see Fig. S1 in the supplemental material).
We grew the ftsH mutant to saturation in the presence of IPTG and then diluted the cultures in fresh medium containing 0 to 60 μM IPTG. Interestingly, when grown without IPTG, the ftsH mutant lacking the feoC gene produced the FeoB protein at markedly increased levels compared to the feoC deletion strain (Fig. 4B, lane 4), indicating that FtsH depletion enables FeoB stabilization even in the absence of FeoC. In contrast, FtsH induction by IPTG complemented FtsH depletion, so that the ftsH mutant contained hardly any FeoB protein in the absence of FeoC (Fig. 4B, lanes 6 and 8). Collectively, these results suggest that FeoB transporter levels are negatively controlled by the FtsH protease, where the FeoC protein protects FeoB from FtsH. Regardless of whether FtsH is induced by 30 μM IPTG, the ftsH mutant carrying the feoC gene maintained FeoB protein levels similar to those in the wild-type (Fig. 4B, lanes 3 and 5). In contrast, further FtsH induction by 60 μM IPTG greatly lowered FeoB content in this strain (Fig. 4B, lane 7), suggesting that FtsH overproduction surpasses FeoC protection of FeoB.
Salmonella lacking the feoC gene is impaired for FeoB-mediated Fe(II) uptake.
Because the FeoC protein was necessary for high levels of the FeoB transporter under anaerobic conditions with low iron, we reasoned that the feoC mutant might be defective for FeoB-mediated Fe(II) uptake after growth under such conditions. The wild-type strain was able to import Fe(II) at much higher (approximately 11-fold) levels after growth under anaerobic conditions with low iron than under aerobic conditions with high iron (Fig. 5A). This phenotype required both the FeoB and FeoC proteins, as evidenced by the fact that the feoB and feoC deletion strains imported Fe(II) approximately 6.5- and approximately 4.5-fold less than the wild-type strain, respectively (Fig. 5A). The FeoC protein contributed to Fe(II) uptake in Salmonella via the FeoB transporter as evidenced by the fact that FeoC production from the plac-FeoC plasmid restored Fe(II) uptake in the feoC deletion strain but failed to do so in the feoBC double deletion strain (Fig. 5B). Cumulatively, these results demonstrate that FeoC-dependent stabilization of the FeoB transporter is important for Salmonella to take up Fe(II) under conditions of both oxygen and iron depletion.
Fig 5.
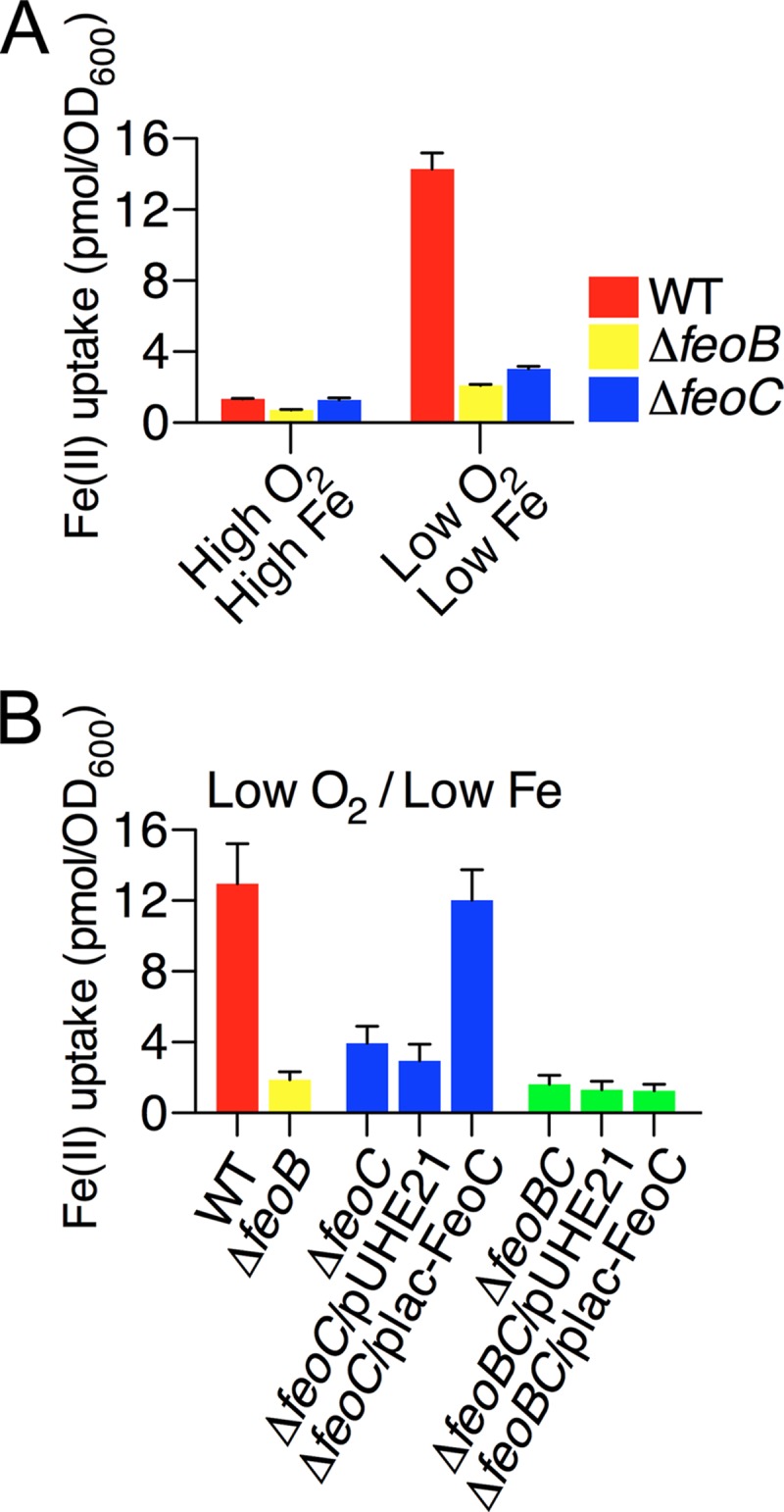
FeoC-dependent FeoB production enables Salmonella to take up Fe(II) under anaerobic conditions with low iron. 55Fe(II) uptake levels were determined in Salmonella strains that were grown aerobically (high O2) or anaerobically (low O2) in LB (high Fe) or LB containing 0.2 mM deferoxamine (low Fe). Bacterial strains used in the experiments were wild-type (WT, 14028s), feoB deletion (ΔfeoB, JH362), and feoC deletion (ΔfeoC, JH363) strains (A) and wild-type (WT, 14028s) and feoB deletion (ΔfeoB, JH362) strains and feoC deletion (ΔfeoC, JH363) and feoBC deletion (ΔfeoBC, HK715) strains carrying the empty plasmid vector pUHE21-2lacIq (pUHE) or FeoC-expression plasmid (plac-FeoC) (B). Values are the means and SD of three independent experiments.
DISCUSSION
Because iron is present mostly in the ferrous form under anaerobic conditions and because production of metal transporters is often induced as levels of their cognate metals are lowered, production of the FeoB Fe(II) transporter should be induced when levels of both oxygen and iron are low. Indeed, the FeoB protein was highly produced in Salmonella only when these two environmental factors (i.e., low oxygen and low iron) were satisfied (Fig. 1A). For such FeoB production, Fur repression should be relieved and Fnr activation should occur on feoB transcription (Fig. 1A and 2A), as previously reported (3, 11). In addition to this regulation, we have now revealed that even under conditions where the feoB mRNA is highly expressed (Fig. 2A), the FeoB transporter cannot be maintained at high levels in the absence of the FeoC protein (Fig. 1).
Based on the notion that FeoC possesses a putative DNA-binding motif and a putative binding site for a Fe-S cluster, this protein was initially proposed as a Fe-S cluster-dependent transcription factor controlling the feo operon (2). However, the FeoC protein does not affect feoB transcription (Fig. 2A) (7). Instead, the FeoC protein regulates FeoB protein levels at the posttranslational level (Fig. 2). The FeoC protein was shown to interact with the FeoB transporter in vivo (Fig. 3A). Moreover, this protein-protein interaction was direct, as evidenced by the fact that the FeoC protein bound to the N-terminal domain of the FeoB protein in vitro, whether these proteins were from Salmonella (Fig. 3B) or from Klebsiella (8).
FtsH is a membrane-bound protease that degrades proteins in the membrane as well as those in the cytoplasm (24). FtsH depletion restored FeoB cellular levels even in the absence of FeoC, while its overproduction impaired FeoB levels even in the presence of FeoC (Fig. 4B), indicating that the FtsH protease negatively controls FeoB levels, and this control is hindered by the FeoC protein. It is known that the FtsH protease initiates proteolysis at a cytoplasmic region of the substrate when degrading a membrane protein (24, 26). Therefore, we propose that FeoC binding to the cytoplasmic N-terminal domain of FeoB protects the FeoB transporter from FtsH-mediated proteolysis initiated at the FeoB N terminus (Fig. 6). Consequently, the action of FeoC on FeoB may stabilize the FeoB transporter under certain circumstances, which enables Salmonella to take up Fe(II) under anaerobic conditions with low iron (Fig. 5 and 6).
Fig 6.
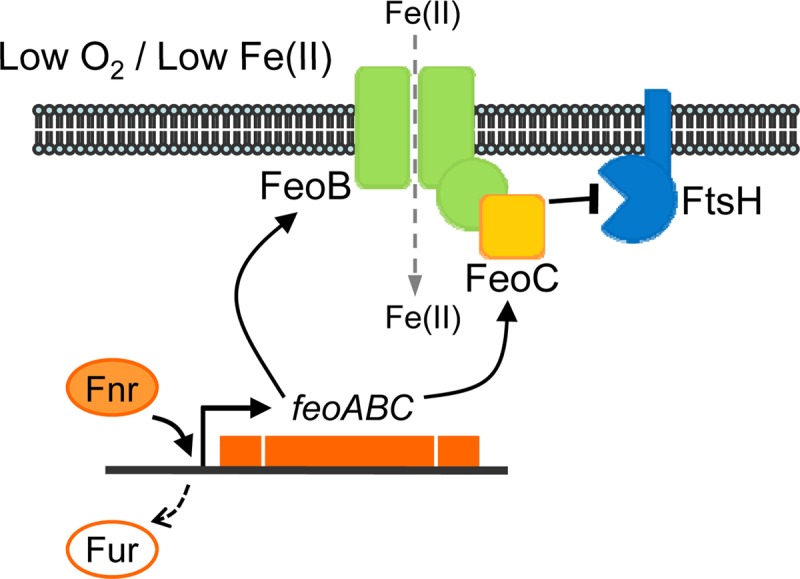
Model illustrating the role of FeoC in FeoB cellular levels. In Salmonella experiencing environmental conditions with low levels of oxygen and iron, Fur repression is relieved on the feo operon while Fnr promotes expression of the feoC mRNA (see Fig. S2 in the supplemental material) together with the feoB mRNA. The FeoB transporter is under proteolytic control by the membrane-bound FtsH protease. The FeoC protein binds to the cytoplasmic N-terminal domain of FeoB, which protects the FeoB transporter from FtsH-mediated proteolysis. Consequently, the FeoC protein leads to high cellular levels of the FeoB transporter, which enables Salmonella to take up Fe(II).
It has been a long-standing notion that bacteria control expression of iron uptake systems at the transcriptional level (27). Likewise, feoB expression is under dual transcriptional control by the iron-sensing Fur and oxygen-sensing Fnr regulators whereby FeoB-mediated Fe(II) uptake is maintained at appropriate levels in response to different levels of iron and oxygen. What then is the physiological relevance of FeoC regulation of FeoB? We note that FeoC could be a Fe-S cluster-binding regulator (2). Fe-S clusters in some regulatory proteins are known to serve as redox sensors or as iron sensors. The Fe-S cluster of Fnr is an example of redox sensors (10). A shift from anaerobic to aerobic conditions oxidizes the Fe-S cluster to cause its disassembly in the Fnr protein, whereby Fnr loses the activity of a transcription factor (10). Moreover, the Fnr regulator that lacks the Fe-S cluster becomes vulnerable to ClpXP protease-mediated proteolysis (28). Although aconitase B (AcnB) is a tricarboxylic acid cycle enzyme carrying a Fe-S cluster, the apo form of AcnB plays a regulatory role by binding to the acnB mRNA (29). AcnB was reported to be demetallated upon iron depletion even under anaerobic conditions, suggesting that the Fe-S cluster of AcnB could serve as an iron sensor (30). Therefore, if the Fe-S cluster of FeoC acts as a redox sensor and/or as an iron sensor, exposure to oxygen or iron might reduce the activity (i.e., FeoB-binding ability) or the levels of FeoC, resulting in rapid elimination of FeoB by the FtsH protease. Given that Salmonella can be exposed to various concentrations of oxygen and iron, this scenario might be particularly important for Salmonella to prevent sudden Fe(II) influx when it experiences a radical environmental change, that is, a growth shift from anaerobic conditions with low iron to aerobic conditions with high iron.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2011-0021585 and 2012R1A2A2A01013521).
Footnotes
Published ahead of print 24 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00343-13.
REFERENCES
- 1. Wandersman C, Delepelaire P. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58: 611– 647 [DOI] [PubMed] [Google Scholar]
- 2. Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo–transport of ferrous iron into bacteria. Biometals 19: 143– 157 [DOI] [PubMed] [Google Scholar]
- 3. Kammler M, Schon C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175: 6212– 6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. U. S. A. 99: 16243– 16248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim H, Lee H, Shin D. 2012. The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem. Biophys. Res. Commun. 423: 733– 738 [DOI] [PubMed] [Google Scholar]
- 6. Boyer E, Bergevin I, Malo D, Gros P, Cellier MF. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70: 6032– 6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fetherston JD, Mier I, Jr, Truszczynska H, Perry RD. 2012. The Yfe and Feo transporters are involved in microaerobic growth and virulence of Yersinia pestis in bubonic plague. Infect. Immun. 80: 3880– 3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hung KW, Tsai JY, Juan TH, Hsu YL, Hsiao CD, Huang TH. 2012. Crystal structure of the Klebsiella pneumoniae NFeoB/FeoC complex and roles of FeoC in regulation of Fe2+ transport by the bacterial Feo system. J. Bacteriol. 194: 6518– 6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Escolar L, Perez-Martin J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181: 6223– 6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bauer CE, Elsen S, Bird TH. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53: 495– 523 [DOI] [PubMed] [Google Scholar]
- 11. Jeon J, Kim H, Yun J, Ryu S, Groisman EA, Shin D. 2008. RstA-promoted expression of the ferrous iron transporter FeoB under iron-replete conditions enhances Fur activity in Salmonella enterica. J. Bacteriol. 190: 7326– 7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis RW, Bolstein D, Roth JR. 1980. Advanced Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640– 6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177: 4364– 4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121– 4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sheffield P, Garrard S, Derewenda Z. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15: 34– 39 [DOI] [PubMed] [Google Scholar]
- 17. Karimova G, Ullmann A, Ladant D. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3: 73– 82 [PubMed] [Google Scholar]
- 18. Doublet B, Douard G, Targant H, Meunier JY, Cloeckaert A. 2008. Antibiotic marker modifications of lambda Red and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J. Microbiol. Methods 75: 359– 361 [DOI] [PubMed] [Google Scholar]
- 19. Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95: 5752– 5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller JH. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 21. Snavely MD, Gravina SA, Cheung TT, Miller CG, Maguire ME. 1991. Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J. Biol. Chem. 266: 824– 829 [PubMed] [Google Scholar]
- 22. Hmiel SP, Snavely MD, Florer JB, Maguire ME, Miller CG. 1989. Magnesium transport in Salmonella typhimurium: genetic characterization and cloning of three magnesium transport loci. J. Bacteriol. 171: 4742– 4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gur E, Biran D, Ron EZ. 2011. Regulated proteolysis in Gram-negative bacteria–how and when? Nat. Rev. Microbiol. 9: 839– 848 [DOI] [PubMed] [Google Scholar]
- 24. Ito K, Akiyama Y. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59: 211– 231 [DOI] [PubMed] [Google Scholar]
- 25. Tomoyasu T, Yuki T, Morimura S, Mori H, Yamanaka K, Niki H, Hiraga S, Ogura T. 1993. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J. Bacteriol. 175: 1344– 1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chiba S, Akiyama Y, Ito K. 2002. Membrane protein degradation by FtsH can be initiated from either end. J. Bacteriol. 184: 4775– 4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27: 215– 237 [DOI] [PubMed] [Google Scholar]
- 28. Mettert EL, Kiley PJ. 2005. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J. Mol. Biol. 354: 220– 232 [DOI] [PubMed] [Google Scholar]
- 29. Tang Y, Guest JR. 1999. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology 145: 3069– 3079 [DOI] [PubMed] [Google Scholar]
- 30. Varghese S, Tang Y, Imlay JA. 2003. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J. Bacteriol. 185: 221– 230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U. S. A. 83: 5189– 5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.