Abstract
Rhizobium leguminosarum bv. trifolii pssA encodes a glucosyl-isoprenylphosphate (IP)-transferase involved in the first step of exopolysaccharide (EPS) synthesis. It was found that the pssA gene is an important target for regulation of this biosynthetic pathway. The data of this study indicate that pssA transcription is a very complex and mainly positively regulated process. A detailed analysis of a 767-bp-long pssA upstream region revealed the presence of several sequence motifs recognized by regulatory proteins that are associated with phosphate-, carbon-, and iron-dependent regulation. In addition, numerous inverted repeats of different lengths have been identified in this region. pssA transcription is directed from two distal P1 and proximal P3 promoters whose sequences demonstrate a significant identity to promoters recognized by RNA polymerase sigma factor σ70. Among rhizobial proteins, RosR seems to be a primary regulator that positively affects pssA expression. This protein binds to RosR box 1 located downstream of the P1 promoter. In addition, phosphate and the carbon source strongly affect pssA transcription. A significantly lower level of pssA expression was observed in both the wild-type strain growing under phosphate-rich conditions and the phoB mutant. In this regulation, the PhoB protein and Pho box 2 located upstream of the P3 promoter were engaged. pssA transcription is also significantly affected by glucose. Transcriptional analysis of a set of pssA-lacZ fusions expressed in Escherichia coli wild-type and cyaA and crp mutants confirmed that cyclic AMP (cAMP) receptor protein (CRP) and two cAMP-CRP boxes located upstream of the P1 are required for this upregulation. Moreover, the production of EPS was totally abolished in R. leguminosarum bv. trifolii mutant strains 4440 and 1012 containing a Tn5 insertion downstream of the P3 promoter and downstream of the P3 −35 hexamer, respectively.
INTRODUCTION
Rhizobium leguminosarum bv. trifolii is a soil bacterium able to induce formation of nitrogen-fixing root nodules on its host plant, clover (Trifolium pratense). The establishment of symbiosis is a complex process involving an exchange of a series of signals between the plant and the bacterium; among them, flavonoids secreted from plant roots and bacterial Nod factors are the best characterized signals (1). In addition, acidic exopolysaccharide (EPS) secreted by rhizobia in large amounts plays a crucial role in the symbiotic interactions with legumes that form indeterminate-type nodules (e.g., Trifolium, Pisum, Vicia, and Medicago) (2). EPS-deficient mutants of R. leguminosarum bv. trifolii induce formation of small, only partially infected nodules on clover plants that are ineffective in nitrogen fixation (3–5). In free-living cells of rhizobia, EPS has several other functions such as nutrient gathering, protection against environmental stresses, attachment to abiotic and biotic surfaces, and biofilm formation, ensuring adaptation of these bacteria to changing environmental conditions (6). EPS produced by R. leguminosarum is a polymer composed of octasaccharide repeating units containing d-glucose, d-glucuronic acid and d-galactose in a molar ratio of 5:2:1, substituted with O-acetyl and pyruvyl groups (2). The assembly of the repeating units is initiated by transfer of glucose-1-phosphate from UDP-glucose to a C55-isoprenylphosphate (IP) carrier located in the inner membrane. This step of EPS synthesis is conducted by a glucosyl-IP-transferase encoded by the pssA gene that represents an individual open reading frame (ORF) and is unlinked to other EPS synthesis genes (4, 7–9). pss genes encoding enzymes involved in the successive steps of EPS synthesis are organized in a large chromosomal EPS cluster I (10). pssDE genes encode a glucuronosyl-(β1-4)-glucosyl transferase catalyzing the second step of the unit synthesis, whereas pssC encoding a glucuronosyl-(β1-4)-glucuronosyl transferase is engaged in the third step of the subunit assembly (11). Mutations in pssA and pssDE genes resulted in failure of EPS synthesis (3–5, 7, 8), whereas the pssC mutant synthesized about 40% of the amount of EPS produced by the wild type (4). Other genes of cluster I are predicted to be involved in the subsequent steps of the unit synthesis (pssGHI coding for putative glycosyl transferases) and EPS modification (pssRMK genes). Recently, Ivashina and coworkers have indicated that pssM encoding a ketal pyruvate transferase is necessary for addition of the pyruvyl group to the subterminal glucose in the EPS repeating unit (12). R. leguminosarum bv. viciae strain VF39 containing a mutation in this gene failed to elicit nitrogen-fixing nodules on peas.
The level of EPS biosynthesis, the polymerization degree, and the type of non-sugar modifications are very important for biological functions of this polymer in the symbiosis. However, until now, very little has been known about EPS synthesis in R. leguminosarum and regulation of expression of pss genes by environmental signals. So far, only a few regulatory genes involved in this process have been described (6); these include rosR, pssB, and exoR located on the chromosome of R. leguminosarum and psrA and psiA genes located on the symbiotic megaplasmids (pSym) of R. leguminosarum bv. phaseoli strains exclusively. Although the mutation in psiA does not affect EPS production, the presence of multiple copies of this gene prevents EPS synthesis. The effect of additional psiA copies was overcome in the presence of extra copies of psrA or pssA, indicating that a balanced number of psiA, psrA, and pssA copies is required for a proper level of EPS production (13–15). Moreover, a negative effect of pssB encoding an inositol monophosphate phosphatase on EPS biosynthesis has been observed (16). In R. leguminosarum, EPS synthesis is probably also regulated by exoR because a mutant in this gene produced more EPS than the wild-type strain (17).
Recently, a role of rosR encoding a transcriptional regulator involved in EPS biosynthesis has been confirmed (18). A mutation in R. leguminosarum bv. trifolii rosR resulted in a 3-fold decrease in EPS synthesis and ineffective symbiosis with clover, whereas multiple copies of this gene caused a 2-fold increase in production of this polymer (5, 18). RosR is a 15.7-kDa protein with a C2H2-type zinc finger motif at its C terminus which binds to a 22-bp-long sequence termed the RosR box. It was confirmed that RosR recognizes and binds specifically to the RosR box motif located in the rosR upstream region and negatively regulates the transcription of its own gene. rosR expression is affected by some environmental signals such as the carbon source, phosphate, and flavonoids (19, 20). The same external factors influence the level of EPS production in R. leguminosarum. It seems possible that transcription of pssA, the key gene in EPS synthesis, is also regulated in response to changing environmental conditions. Until now, there have been only fragmentary data concerning pssA expression. Previously, we have established that clover root exudates and ammonia slightly affect the transcription of this gene (21). In this report, a detailed functional analysis of the pssA upstream region was performed, and the functions of some identified sequence motifs in regulation of transcription of this gene were confirmed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains, plasmids, and oligonucleotide primers used in this study are listed in Table 1. R. leguminosarum and Sinorhizobium meliloti strains were routinely grown in 79CA medium with 1% glycerol as a carbon source (22) and in M1 minimal medium (23) containing 1% glycerol and 2 ml liter−1 vitamin stock solution (24) at 28°C on a rotary shaker (120 rpm). Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37°C (23). To study the effect of phosphate on pssA expression, the strains were cultured in M1 medium buffered with 20 mM morpholinepropanesulfonic acid (MOPS) and supplemented with appropriate concentrations of K2HPO4. To establish the influence of iron on the expression of pssA-lacZ fusions, the rhizobial strains were grown in M1 medium supplemented with 20 μM FeCl3 (high-Fe medium). Low-Fe medium contained 20 μM 2,2′-dipyridyl instead of Fe. To study the effect of the carbon source on pssA transcription, R. leguminosarum and E. coli strains were cultured for 24 h in M1 medium supplemented with 1% glycerol or 1% glucose. When required, antibiotics were used at the following final concentrations: kanamycin, 40 μg ml−1; rifampin, 40 μg ml−1; gentamicin, 10 μg ml−1; streptomycin, 200 μg ml−1; ampicillin, 100 μg ml−1; tetracycline, 10 μg ml−1; and nalidixic acid, 40 μg ml−1.
Table 1.
Bacterial strains, plasmids, and oligonucleotide primers used in this study
| Strain or plasmid | Relevant characteristic(s) or sequencea | Source or reference |
|---|---|---|
| R. leguminosarum bv. trifolii strains | ||
| 24.2 | Wild type, Rifr Nxr | 18 |
| 2472 | 24.2 with Tn5 insertion at bp 151/152 of rosR | 5 |
| 5819 | 24.2 with Tn5 insertion at bp 363/364 of pssA | 5 |
| 1012 | 24.2 with Tn5 insertion at bp −253/−254 of pssA | This work |
| 4440 | 24.2 with Tn5 insertion at bp −77/−78 of pssA | This work |
| R. leguminosarum bv. viciae strains | ||
| J251 | Wild type, 8401(pRL1JI), Smr | 54 |
| J386 | J251 with Tn5 lac insertion in irrA, Smr Gmr Kmr | 55 |
| J397 | J251 with Tn5 insertion in rirA, Smr Kmr | 31 |
| S. meliloti strains | ||
| Rm1021 | SU47 str-21 | 56 |
| RmH406 | ΩphoB3::Tn5-132 | T. M. Finan, McMaster University, Hamilton, Ontario, Canada |
| E. coli strains | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 23 |
| VH1000 | MG1655 derivative, lacZ lacI pyrE+ | 57 |
| VH1000 cyaA | ΔcyaA::Kmr derivative of VH1000 | 57 |
| M182 Δcrp | Δcrp39 Δlac derivative of M182 | 57 |
| S17-1 | 294 derivative; thi RP4-2 (Tc::Mu-Km::Tn7) | 58 |
| Plasmids | ||
| pUC19 | Cloning and sequencing vector, Apr | 23 |
| pMP220 | IncP, mob, promoterless lacZ, Tcr | 59 |
| pBBR1MCS-2 | mob, lacZα, Kmr cloning vector | 60 |
| pSUP202 | pBR325 derivative, mob, Cmr Tcr Apr | 58 |
| pM18 | pUC19 with a 1.7-kb HindIII fragment containing 24.2 pssA | This work |
| pBR12 | pBBR1MCS-2 with a 1.2-kb EcoRI fragment containing 24.2 rosR | This work |
| pM34 | pUC19 containing a 3.4-kb EcoRI fragment with pssA | 5 |
| pMJ440 | pUC19 with a 0.4-kb EcoRI-PstI fragment of the pssA upstream region | This work |
| pMSUP4 | pSUP202 containing a 4.6-kb EcoRI fragment with Tn5 inserted at bp 77/78 upstream of pssA ORF | This work |
| pMSUP10 | pSUP202 containing a 4.6-kb EcoRI fragment with Tn5 inserted at bp 253/254 upstream of the pssA ORF | This work |
| pAX1 | pBBR1MCS-2 with a 0.75-kb EcoRI-XbaI fragment containing the pssA upstream region (bp −750 to bp −2) | This work |
| pAX2 | pBBR1MCS-2 with a 0.53-kb EcoRI-XbaI fragment containing the pssA upstream region (bp −538 to bp −2) | This work |
| pAX3 | pBBR1MCS-2 with a 0.37-kb EcoRI-XbaI fragment containing the pssA upstream region (bp −374 to bp −2) | This work |
| pAX4 | pBBR1MCS-2 with a 0.28-kb EcoRI-XbaI fragment containing the pssA upstream region (bp −284 to bp −2) | This work |
| pAX5 | pBBR1MCS-2 with a 0.13-kb EcoRI-XbaI fragment containing the pssA upstream region (bp −134 to bp −2) | This work |
| pAX6 | pBBR1MCS-2 containing a 1.55-kb EcoRI-XbaI fragment with pssA | This work |
| pAX7 | pBBR1MCS-2 containing a 1-kb EcoRI-XbaI fragment with pssA | This work |
| pPA1 | pMP220 carrying the −750-bp to +152-bp fragment of the pssA regulatory region | This work |
| pPA2 | pMP220 carrying the −538-bp to +152-bp fragment of the pssA regulatory region | This work |
| pPA3 | pMP220 carrying the −374-bp to +152-bp fragment of the pssA regulatory region | This work |
| pPA4 | pMP220 carrying the −284-bp to +152-bp fragment of the pssA regulatory region | This work |
| pPA4-A | pMP220 carrying the −265-bp to +152-bp fragment of the pssA regulatory region | This work |
| pPA4-B | pMP220 carrying the −261-bp to +152-bp fragment of the pssA regulatory region | This work |
| pPA4-C | pMP220 carrying the −284-bp to +152-bp fragment of the pssA regulatory region with a deletion of Pho box 3 (−38 bp to −14 bp sequence) | This work |
| pPA5 | pMP220 carrying the −134-bp to +152-bp fragment of the pssA regulatory region | This work |
| Primers (5′–3′)b | ||
| PA1 | AGATCGTTGAATTCCGGCAAGCG | This work |
| PA2 | TCTTTTGAATTCGCTGCGCGCCAT | This work |
| PA3 | CCCTGTGAATTCCTGCCAAAGGC | This work |
| PA4 | GCGAAAGAATTCCTGCGATGCGA | This work |
| PA4-A | TGCGATGAATTCTTTCTCTGGACAC | This work |
| PA4-B | GCGATAGAATTCTGGACACCATTAC | This work |
| PAC-F | GATGCGGATCCGGCGCCGTTGGT | This work |
| PAC-R | TAACAGGGATCCGGATCTTCAGG | This work |
| PA5 | CAACAGAATTCGATCCTTGAGCATC | This work |
| PAR1 | ATGGCCCTGCAGTTCTTGCCCCA | This work |
| PAR2 | ACCAGATCTAGAAACGGCGCCTG | This work |
| PAR3 | CAGGTTATCTAGAGAGCCCGATG | This work |
| EMSA13 | TGGGTCACGTGTCGAAACGGCACGT | This work |
| EMSA14 | CGCATCGCAGCAAATCTTTCGCATG | This work |
Nxr, nalidixic acid resistance; Rifr, rifampin resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance; Kmr, kanamycin resistance; Smr, streptomycin resistance; Gmr, gentamicin resistance. The sequences for the EcoRI, BamHI, XbaI, and PstI restriction sites are underlined.
Amplification of the pssA promoter region for construction of pPA fusions and EMSAs.
DNA methods and sequence analysis.
Standard techniques were used for plasmid and genomic DNA isolation, restriction enzyme digestion, cloning, and transformation (23). For PCR amplifications, REDTaq Ready PCR Mix (Sigma) and plasmid or genomic DNA isolated from E. coli and R. leguminosarum strains were used as templates. Amplicons and plasmid constructs were sequenced using a BigDye Terminator cycle sequencing kit (Applied Biosystems) and an ABI Prism 310 sequencer. Database searches were done with the BLAST and FASTA programs available from the National Center for Biotechnology Information (Bethesda, MD, USA) and the European Bioinformatic Institute (Hinxton, United Kingdom). Promoter prediction in the sequence upstream of pssA was done using the BDGP Neural Network Promoter Prediction (www.fruitfly.org). The searches for several motifs in the promoter region of this gene were performed with Malign and Fuzznuc programs using CTTGAC-N17/18-CTATAT (S. meliloti promoter consensus), TGAAATCTAGGGGTAGATTTCA (RosR box consensus), CTGTCAT-N4-CTGTCAT (Pho box), TTT-N11-AAA (iron control element [ICE] box), TGA-N9-TCA (iron-responsive operator [IRO] box), and TGTGA-N6-TCACA (cyclic AMP [cAMP]-cAMP receptor protein [CRP] motif) as query sequences (http://www.genebee.msu.su/services/malign; http://emboss.ch.embnet.org/Pise). Inverted repeats (IRs) and RNA secondary structures were predicted using the RNA folding software mfold, version 3.2 (25 [http://www.bioinfo.rpi.edu/applications/mfold]) using the default settings. The data concerning β-galactosidase activities and EPS production were subjected to a linear analysis of variance (ANOVA) model using Microsoft Excel 2000 for Windows. A P value of <0.05 was considered significant.
Construction of plasmids bearing transcriptional pssA-lacZ fusions.
To construct plasmids containing specific regions of the pssA promoter, the broad-host-range plasmid pMP220 carrying a promoterless lacZ gene was used. Based on a 1.7-kb HindIII fragment of plasmid pM18 containing pssA with its long upstream regulatory region, a set of deletion derivatives was generated by PCR amplification using the following primer pairs: PA1/PAR1 (for pPA1 fusion), PA2/PAR1 (pPA2), PA3/PAR1 (pPA3), PA4/PAR1 (pPA4), PA4-A/PAR1 (pPA4-A), PA4-B/PAR1 (pPA4-B), and PA5/PAR1 (pPA5) (Table 1). The PCR products obtained were digested with EcoRI and PstI enzymes, cloned into the corresponding sites in the pUC19 vector, and verified by sequencing. Then, plasmid-derived EcoRI-PstI fragments were cloned between the respective sites of the pMP220, yielding plasmids pPA1 to pPA5 containing individual fragments of the pssA upstream region fused to the lacZ gene. For construction of the pPA4-C fusion (Table 1), two primer pairs, PA4/PAC-R and PAC-F/PAR1, were used, yielding amplicons of 261 bp and 182 bp, respectively. The 261-bp fragment was digested with EcoRI and BamHI enzymes, and the 182-bp fragment was digested with BamHI and PstI enzymes. The EcoRI-BamHI and BamHI-PstI fragments were ligated and cloned into the EcoRI and PstI sites of the pUC19 vector, yielding pMJ440. The insertion of this plasmid was verified by sequencing, digested with EcoRI and PstI enzymes, and cloned into the respective sites of pMP220, resulting in pPA4-C fusion. The constructed plasmids were introduced into E. coli, R. leguminosarum and S. meliloti by electroporation as described earlier (19).
Mutagenesis of the pssA regulatory region.
For mutagenesis of the pssA upstream region, a pM34 plasmid as a target and an EZ::TN<KAN-2> Insertion Kit (Epicentre Technology), which enables generation of random mini-Tn5 transposon insertions into target DNA, were used according to the manufacturer's instruction. Tn5 locations in pM34 derivatives were established by restriction and sequencing analyses. Among these, two plasmids, pMT4 and pMT10, with mini-Tn5 transposons inserted at positions −77/−78 bp and −253/−254 bp of the pssA upstream region, respectively (Fig. 1) (accession number AF316883), were chosen for further studies. Subsequently, 4.6-kb EcoRI inserts of these plasmids were cloned into the appropriate site of pSUP202, resulting in the plasmids pMSUP4 and pMSUP10, respectively. These constructs were introduced into E. coli S17-1 by transformation and then into R. leguminosarum bv. trifolii strain 24.2 via biparental conjugation. Transconjugants were selected on 79CA medium supplemented with kanamycin and rifampin. Two clones, named R. leguminosarum bv. trifolii strains 4440 and 1012, which formed small nonmucoid colonies were isolated as a result of conjugation with the S17-1(pMSUP4) and S17-1(pMSUP10) donors, respectively.
Fig 1.
Nucleotide sequence of the upstream region and 5′ end of the pssA gene of R. leguminosarum bv. trifolii 24.2 (accession numbers AF316883 and AF014791). The amino acid sequence of the PssA N terminus is given in single-letter code. Putative promoter regions are underlined, and −35/−10 motifs of the P1, P2, and P3 promoters are marked by square brackets. Upstream and downstream endpoints of PCR fragments of individual plasmid fusions are marked by angled arrows. RosR boxes, Pho boxes, a ribosome binding site [rbs], and inverted repeats IR1 to IR10 are marked by white boxes. The cAMP-CRP motifs and the ICE and IRO boxes are shaded in black and gray, respectively. Locations of Tn5 insertions in R. leguminosarum bv. trifolii pssA mutants (strains 120, 1012, 4440, and 5819) are marked by gray vertical arrows.
Construction of plasmids containing different fragments of the pssA upstream region.
To construct a set of plasmids containing different fragments of the pssA regulatory region, the following primer pairs were used: PA1/PAR2, PA2/PAR2, PA3/PAR2, PA4/PAR2, and PA5/PAR2 (Table 1). Based on the pM18 plasmid, PCR products 0.75 kb, 0.53 kb, 0.37 kb, 0.28 kb, and 0.13 kb long were obtained. Additionally, 1.55-kb- and 1-kb-long amplicons containing pssA with a full-length upstream region and exclusively the P3 promoter were yielded using the PA1/PAR3 and PA4/PAR3 primers, respectively. All of these PCR products were digested with EcoRI and XbaI enzymes and cloned into respective sites of the pBBR1MCS-2 vector, yielding plasmids pAX1 to pAX7. The constructs obtained were introduced into strain 24.2 by electroporation.
β-Galactosidase assay.
R. leguminosarum, S. meliloti, and E. coli derivatives containing pssA-lacZ fusions were grown for 24 h in 79CA or M1 medium supplemented with tetracycline and, where necessary, appropriate concentrations of FeCl3, 2,2′-dipirydyl, MOPS, or K2HPO4. To study the effect of the carbon source on pssA expression, the bacteria were grown in M1 medium supplemented with 1% glycerol or 1% glucose. The assays for β-galactosidase activity were carried out according to the protocol described by Miller (26). The reported values are given in Miller units and are averages of at least three independent experiments.
Overexpression and purification of RosR protein.
Overproduction and purification of RosR protein under native conditions were done as described earlier (18). Briefly, the E. coli M15(pQE450) strain bearing the expression vector pQE-32 with a 450-bp BamHI-SalI fragment containing the 24.2 rosR gene cloned upstream of a His6 tag sequence was used. The strain was grown in 200 ml of LB medium supplemented with ampicillin and kanamycin at 30°C. After 5 h of growth (optical density at 600 nm [OD600] of ∼0.4), rosR expression was induced by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM, and the induction was conducted for 4 h. Then, cells were harvested by centrifugation, and the pellet was lysed in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 5 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, and 1 mg ml−1 lysozyme) and sonicated. A crude lysate was clarified by centrifugation and applied to a nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen). His6-RosR protein was eluted from the column with 200 mM imidazole, dialyzed two times against a buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 10% vol/vol glycerol) and concentrated using Amicon columns (Millipore). The protein concentration was determined by the Bradford method using a Bio-Rad Protein assay (Bio-Rad). The purity of His6-RosR was confirmed using polyacrylamide gel electrophoresis.
EMSAs.
For electrophoretic mobility shift assays (EMSAs), a 145-bp fragment of the pssA regulatory region matching RosR box 1 was amplified by PCR using plasmid pPA1 as a template and primers EMSA13 and EMSA14 (Table 1). PCR amplification was performed using 100 ng of pPA1 and REDTaq Ready PCR Mix in a final volume of 100 μl. The PCR mixture contained a 0.5 μM concentration of each forward and reverse primer. The temperature cycle used was 94°C for 3 min, followed by 25 cycles of 94°C for 60 s, 54°C for 40 s, and 72°C for 30 s, with a final extension at 72°C for 3 min. The PCR product was purified on columns (A&A Biotechnology), and its concentration and quality were established in agarose gel electrophoresis. Then, the DNA fragment was labeled at the 3′ end with a terminal transferase and digoxigenin (DIG)-11-ddUTP using a DIG Gel Shift Kit (Roche) according to the manufacturer's instruction. The mixture for the EMSA (total volume of 20 μl) contained 20 ng of the DIG-labeled DNA fragment and different amounts of His6-RosR protein in the reaction buffer [20 mM HEPES, pH 7.6, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol (DTT), 0.2%, wt/vol, Tween 20, 30 mM KCl, 5 μg ml−1 poly-l-lysine, and 0.5 μg ml−1 poly(dI-dC)]. After incubation at 20°C for 20 min, the reaction mixtures were loaded onto a 5% nondenaturing gel containing 10% glycerol and resolved in Tris-glycine buffer (25 mM Tris, 190 mM glycine) supplemented with 5 mM MgCl2, 1 mM EDTA, and 0.1 mM DTT at 9 V/cm for 2.5 h. Subsequently, DNA-protein complexes were electrotransferred onto a nylon membrane and visualized by chemiluminescent detection according to the manufacturer's instruction.
EPS isolation and quantification.
For EPS isolation, 10-ml cultures of rhizobial strains were grown in 79CA and M1 media supplemented with 1% glycerol for 2 days at 28°C in a rotary shaker. EPS was precipitated from culture supernatants with 4 volumes of 96% ethanol, collected by centrifugation and, after being redissolved in water, analyzed for carbohydrates according to Loewus (27). The total sugar content was calculated as glucose equivalents.
RESULTS
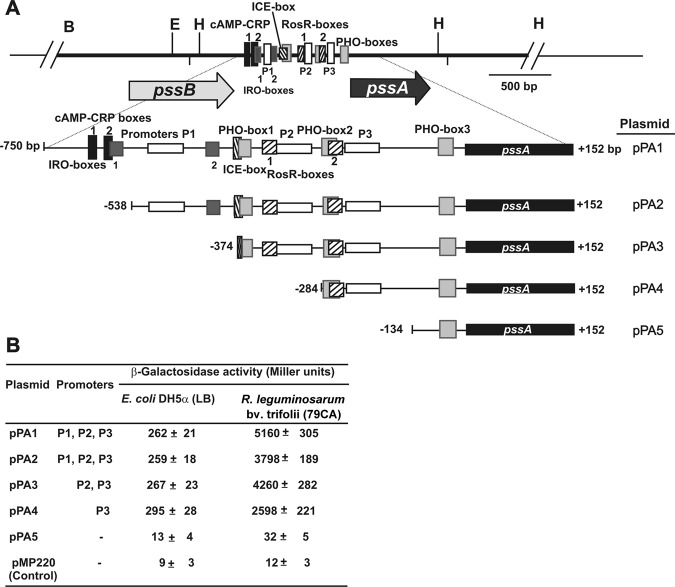
Functional analysis of the pssA promoter region.
Our previous studies have shown that pssA is present in the genomes of strains belonging to all three biovars of R. leguminosarum, and this gene represents an individual open reading frame located downstream of pssB (5, 9). pssA contains a very long upstream region (767 bp), suggesting complex regulation of expression of this gene. Until now, there have been no detailed functional analyses of this sequence and motifs involved in pssA transcription. In this study, a bioinformatic analysis of the pssB-pssA intergenic region has been performed, which revealed the presence of several regulatory elements, including putative promoter motifs and direct and inverted repeats of different lengths (Fig. 1). Upstream of the pssA coding region, three putative promoters, P1, P2, and P3 with a score of 0.71 (a sequence from −511 bp to −452 bp), 0.86 (−354 bp to −305 bp), and 0.79 (−261 bp to −212 bp), respectively, have been found. The above data suggest that these promoters are of a relatively low stringency. Their sequence motifs demonstrated a similarity to eubacterial promoters whose transcription is initiated by the sigma factor σ70 (28). Among these, the distal promoter P1 with the sequence TTGTCA-N17-GACAGT contained a −35 hexamer almost identical to and a −10 hexamer less similar to the consensus sequence ( TTGACA-N17-TATAAT ) with an optimal spacing of 17 nucleotides (nt) (nucleotides identical with the consensus sequence are underlined) (Fig. 1). The second promoter, P2, had a −35 hexamer identical to that of the consensus, but the −10 motif was of a significantly lower sequence identity ( TTGACA-N17-GAAATA) . The proximal promoter P3, similarly to the P1, contained a −35 hexamer with 5 nt identical to the consensus sequence and a −10 hexamer of low sequence identity ( TGGACA-N17-CATTTT ). It is well known that the −35 hexamer is recognized and bound by the σ70 factor of RNA polymerase (RNAP) as the first among promoter motifs (29). The sequences of the −35 motifs of all the three pssA promoters suggested that they might be target sites successively recognized by the σ70 of RNAP. On the other hand, the low homology of the −10 hexamers of the P1, P2, and P3 promoters to the consensus sequence indicated a relatively low stringency of RNAP σ70 binding to these promoter regions. Also, using the promoter consensus CTTGAC-N17/18-CTATAT of S. meliloti, the same fragments of the pssA upstream region have been identified as −35 and −10 motifs of these promoters, but these motifs showed slightly lower identity with the S. meliloti consensus than with the E. coli consensus (30).
To functionally analyze the putative pssA promoters, a set of plasmids (pPA1 to pPA5) containing 5′-end deletion fragments of the promoter region fused to a promoterless lacZ gene were constructed (Fig. 1 and 2). These recombinant plasmids were introduced to E. coli DH5α and R. leguminosarum bv. trifolii 24.2, and β-galactosidase activities were assayed in derivatives of the strains carrying different pssA-lacZ fusions (Fig. 2). In general, very high differences in the levels of pssA transcription were observed in these two genetic backgrounds. We have found low and similar transcription levels for the pPA1 to pPA4 fusions in E. coli. In contrast, very high promoter activity for these fusions in strain 24.2 has been observed, suggesting that some rhizobium-specific activators are engaged in positive regulation of pssA expression. Among them, the highest transcription was observed for the pPA1 fusion, indicating that the full-length upstream region is indispensable for the maximum level of pssA expression. The deletion of the sequence from −750 bp to −538 bp (in pPA2 still containing all three promoters) resulted in a 26.4% decrease in pssA transcription, which suggested a role of some sequence motif(s) located in this region for the optimal expression of pssA (Fig. 2). The subsequent deletion of the sequence from −538 bp to −374 bp (pPA3 fusion) resulted in a slight increase in transcription in comparison to the level of pPA2 transcription. On the other hand, a decrease in pssA transcription to 61% of that in the pPA3 plasmid was observed in the pPA4 plasmid, which does not contain the −374-bp to −284-bp region and the P2 promoter. These data indicate that the P3 promoter is functional in both genetic backgrounds and ensures a basic level of pssA transcription, but the full-length (767-bp) regulatory region is required for very high expression of this gene in R. leguminosarum. The level of β-galactosidase activity in the pPA5 fusion, which was similar to that in the strain containing the pMP220 vector without the pssA regulatory sequence, confirmed that the region downstream of −134 bp did not contain any promoter sequences.
Fig 2.
(A) Physical and genetic map of the R. leguminosarum bv. trifolii 24.2 pssB-pssA region. The light gray and black arrows below the map show the directions of pssB and pssA transcription, respectively. Promoters P1, P2, and P3 are marked by white rectangles. cAMP-CRP boxes, IRO boxes, and Pho boxes are shaded in black, dark gray, and light gray, respectively. ICE box and RosR motifs are left- and right-striped rectangles, respectively. B, BamHI; E, EcoRI; H, HindIII. (B) Transcriptional activity of pssA promoters assayed in E. coli DH5α and 24.2 strains carrying the pMP220 plasmid with different fragments of the pssA upstream region fused with the promoterless lacZ gene. E. coli strains were grown in LB medium, and strain 24.2 derivatives were grown in 79CA medium supplemented with 1% glycerol for 24 h. β-Galactosidase activities shown are means of five independent experiments with standard deviations.
Identification of putative regulatory motifs in the pssA upstream region.
Previously, we have established that EPS production in R. leguminosarum is affected by several environmental factors, including phosphate and the carbon source (19, 20). Also, iron availability influences the synthesis of this polymer, as was evidenced for strain 24.2, which produced about 1.5-fold more EPS in the absence of iron than under iron-rich conditions (2.3 mg and 1.5 mg mg−1 of protein, respectively). Therefore, we decided to establish whether these factors affect EPS production via modulation of pssA transcription. In order to identify motifs potentially involved in regulation of pssA expression, in silico analysis of the upstream region of this gene was performed. As a result, several sequence motifs representing putative target sites for bacterial regulators have been identified. Among them are three motifs sharing significant homology with the Pho box consensus sequence ( CTGTCAT-N4-CTGTCAT ) recognized by PhoB protein, two motifs resembling the E. coli cAMP-CRP consensus sequence (TGTGA-N6-TCGCA), and two motifs with high identity with the RosR box recognized by RosR protein (Fig. 1 and 2). Additionally, two putative IRO boxes and one ICE box showing high homology to motifs recognized by rhizobial proteins RirA (TGA-N9-TCA) and IrrA (TTT-N11-AAA), which are involved in regulation of gene expression in response to high and low iron concentrations, respectively, have been identified (31–33). Moreover, several inverted repeats of different lengths have been found in the pssA upstream region (Fig. 1); among these IR2 and IR9 were the longest, with 15-nt inverted repeats. pssA transcripts generated from the P1 and P3 promoters contained long 5′ untranslated regions, in which a great majority of identified IRs were located. Secondary structure analysis of RNA transcribed from the P1 promoter revealed that the IR2 to IR10 motifs are engaged in formation of several stem structures in the upper part of the transcript, with a total free energy of ΔG = −133.1 kcal/mol. At the 5′ end of this RNA, a very long stem consisting of the IR2, IR3, and IR4 motifs was generated, with a total free energy of ΔG = −34.1 kcal/mol. In addition, the IR8 and IR9 inverted repeats formed a stable structure with an energy of −38.7 kcal/mol. In the case of a shorter transcript generated from the P3 promoter, motifs IR7 to IR10 were involved in stabilizing the secondary structure of its 5′ end (ΔG = −83.7 kcal/mol). The IR10 motif located just downstream of the translation start site formed a stable stem structure (ΔG = −18.6 kcal/mol), which played a significant role in the stability of secondary structures of both pssA transcripts.
The functional analysis of Pho boxes located in the pssA upstream region.
In order to investigate the significance of individual Pho boxes in the regulation of pssA transcription and their response to phosphate availability, fusions of pPA1 to pPA4 were used. Strain 24.2 derivatives bearing respective plasmid fusions were grown in minimal M1 medium containing 0.1 mM phosphate (low Pi) or 15 mM phosphate (high Pi), and β-galactosidase activities were measured (Table 2). In general, this nutrition factor essentially affects pssA transcription. During phosphate starvation, the level of pssA-lacZ transcription was significantly higher than in the presence of a high concentration of phosphate. For pPA1 and pPA2 fusions containing three Pho motifs, the ratios of β-galactosidase activities in strain 24.2 derivatives growing under low-phosphate and high-phosphate (low Pi/high Pi) conditions were 1.7 and 1.64, respectively, confirming the significant role of this environmental signal in regulation of pssA transcription. In the case of the pPA3 which contains Pho box 1 lacking 4 nt at its 5′ terminus, the difference in the transcription levels measured at low and high Pi was slightly lower. For a pPA4 fusion containing Pho boxes 2 and 3, this ratio was very similar to the results obtained for pPA1 and pPA2, indicating that the absence of the distal binding site does not influence the level of pssA expression.
Table 2.
Effect of phosphate on pssA-lacZ transcription in R. leguminosarum bv. trifolii 24.2 and S. meliloti strains
| Plasmid | Pho box genotyped | β-Galactosidase activity by strain and conditiona |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 24.2 (wild type) |
Rm1021 (wild type) |
RmH406 (phoB mutant) |
||||||||
| Low Pib | High Pib | Pi ratioc | Low Pi | High Pi | Pi ratio | Low Pi | High Pi | Pi ratio | ||
| pPA1 | 1, 2, 3 | 8,127 ± 489 | 4,780 ± 386 | 1.70* | 20,720 ± 954 | 16,538 ± 890 | 1.25 | 8,904 ± 664 | 9,659 ± 682 | 0.92 |
| pPA2 | 1, 2, 3 | 6,784 ± 392 | 4,111 ± 312 | 1.64* | 19,182 ± 886 | 15,978 ± 925 | 1.20 | 9,694 ± 564 | 10,467 ± 773 | 0.93 |
| pPA3 | 1†, 2, 3 | 3,867 ± 302 | 2,595 ± 217 | 1.49* | 15,045 ± 790 | 12,470 ± 788 | 1.21 | 8,279 ± 669 | 8,301 ± 532 | 0.99 |
| pPA4 | 2, 3 | 3,120 ± 261 | 1,950 ± 186 | 1.60* | 24,597 ± 1,288 | 20,169 ± 1,176 | 1.22 | 12,402 ± 821 | 12,996 ± 863 | 0.95 |
| pPA4-A | 2†, 3 | 1,574 ± 122 | 1,345 ± 102 | 1.17 | 9,265 ± 757 | 8,659 ± 678 | 1.07 | 6,882 ± 489 | 6,681 ± 512 | 1.03 |
| pPA4-B | 3 | 484 ± 59 | 457 ± 54 | 1.06 | 2,082 ± 189 | 2,169 ± 231 | 0.96 | 1,415 ± 162 | 1,459 ± 128 | 0.97 |
| pPA4-C | 2 | 2,813 ± 211 | 1,888 ± 157 | 1.49* | 19,288 ± 1,136 | 16,209 ± 1,106 | 1.19 | 10,885 ± 926 | 10,672 ± 995 | 1.02 |
The cultures were inoculated to an OD600 of 0.1 and grown for 24 h at 28°C in M1 minimal medium supplemented with different concentrations of phosphate, Tc (10 μg ml−1), and 1% glycerol. Low-phosphate medium additionally contained 20 mM MOPS for pH stabilization. pssA-lacZ transcription was determined from β-galactosidase assays.
Values (Miller unites) are means ± standard deviations of three independent experiments. Low Pi, 0.1 mM phosphate; high Pi, 15 mM phosphate.
Pi ratio, activity at low Pi/activity at high Pi. *, statistically significant difference (P < 0.05) between low- and high-phosphate conditions for the individual fusion assayed.
Numbers refer to Pho boxes (Fig. 1). †, 3′ half.
In order to establish which of Pho boxes 2 and 3 is involved in phosphate-dependent regulation of pssA expression, additional pPA4-A, pPA4-B, and pPA4-C fusions were constructed as pPA4 derivatives. Pho box 2 was located just upstream of P3, whereas Pho box 3 was 195 bp downstream of this promoter (Fig. 1). The pPA4-A and pPA4-B fusions had a progressive deletion of the 5′-end sequence encompassing Pho box 2 (from −284 bp to −265 bp and from −284 bp to −261 bp, respectively), and the pPA4-C had a deletion of the region from −38 bp to −14 bp containing the Pho box 3. pssA-lacZ expression levels in strains 24.2(pPA4-A) and 24.2(pPA4-B) growing under high-phosphate conditions constituted only 69% and 23%, respectively, of the level observed for the strain 24.2 bearing the pPA4 plasmid (Table 2). These data indicated that the sequence located just upstream of the P3 promoter plays a significant role in pssA transcription driven from this promoter. In addition, the pPA4-B fusion has lost its susceptibility to phosphate concentrations. On the other hand, β-galactosidase activity of pPA4-C with the Pho box 3 deletion was similar to that observed in pPA4, which excluded a role of this motif in phosphate-dependent regulation of pssA expression.
In conclusion, phosphate concentration influences the level of pssA transcription in R. leguminosarum, and Pho box 2 located just upstream of the P3 promoter is responsible for this effect.
Further studies of Pho-box-mediated activation of pssA transcription were performed on S. meliloti wild-type and phoB mutant strains bearing plasmids pPA1 to pPA4. Generally, the expression of pPA1 to pPA4 fusions was almost 2-fold higher in the wild-type Rm1021 strain than in the RmH406 mutant growing under low-phosphate conditions (Table 2). In Rm1021, slightly lower low Pi/high Pi ratios for the fusions analyzed (range, 1.25 to 1.2) were detected than in strain 24.2, but in both strains the same tendency was observed. Similarly, in the case of strain Rm1021, Pho box 2 located in the pPA4 and pPA4-C fusions turned out to be essential, whereas Pho box 3 (the pPA4-B plasmid) was not significant for this regulatory effect. In contrast to the wild-type strain, the level of LacZ expression in the phoB mutant carrying all four fusions, pPA1 to pPA4, was similar under both low- and high-phosphate conditions (the low Pi/high Pi ratios ranged from 0.92 to 1.03), confirming that the functional PhoB protein is required for modulation of pssA transcription.
Role of cAMP-CRP binding sites in pssA transcription.
Apart from the regulatory elements described above, two motifs resembling the E. coli cAMP-CRP consensus sequence have been identified in the pssA regulatory region. The two putative cAMP-CRP binding sites were located upstream of the distal P1 promoter at positions −719 bp to −704 bp and −608 bp to −593 bp, respectively. cAMP-CRP box 1 ( TGTAA-N6-GCTCA ) contained both first and second pentamers almost identical to those of the E. coli consensus TGTGA-N6-TCACA (Fig. 1). cAMP-CRP box 2 (TGTGA-N6-TCGCG) shared even higher identity, with the first pentamer identical to this consensus sequence and the second pentamer highly conserved.
To examine the function of the cAMP-CRP binding sites in pssA transcription and their response to the carbon source, strain 24.2 derivatives bearing different pPA fusions were grown in the presence of glycerol or glucose as sole carbon sources, and β-galactosidase activity was measured (Table 3). Among the four pssA-lacZ fusions tested, a substantial difference in the expression levels between these carbon sources was observed only for pPA1 containing the entire upstream region with both cAMP-CRP binding sites 1 and 2. The ratio of β-galactosidase activity with Gly/activity with Glc (Gly/Glc ratio) for strain 24.2(pPA1) was 2, indicating a significant role of these two motifs in pssA transcription. Deletion of both cAMP-CRP sites (pPA2 fusion) rendered the P1 promoter unresponsive to glucose, which was confirmed by the decrease in the Gly/Glc ratio to 0.91. Further deletions of the pssA regulatory region (pPA3 and pPA4) did not change this effect (the Gly/Glc ratios were 1.06 and 1.03, respectively) (Table 3).
Table 3.
Effect of carbon source on pssA-lacZ transcription in R. leguminosarum bv. trifolii 24.2 and E. coli strains
| Plasmid | CRP box genotyped | β-Galactosidase activity by strain and conditiona |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24.2 (wild type) |
VH1000 (wild type) |
VH1000 (cyaA Kmr) |
M182 (Δcrp) |
||||||||||
| +Glyb | +Glcb | Gly/Glcc | +Gly | +Glc | Gly/Glc | +Gly | +Glc | Gly/Glc | +Gly | +Glc | Gly/Glc | ||
| pPA1 | 1, 2 | 4,156 ± 342 | 2,078 ± 168 | 2.0* | 116 ± 28 | 86 ± 16 | 1.37* | 83 ± 19 | 86 ± 32 | 0.96 | 106 ± 25 | 98 ± 33 | 1.08 |
| pPA2 | 4,206 ± 332 | 4,621 ± 332 | 0.91 | 86 ± 19 | 95 ± 17 | 0.91 | 102 ± 26 | 96 ± 29 | 1.06 | 134 ± 20 | 127 ± 28 | 1.06 | |
| pPA3 | 2,982 ± 226 | 2,813 ± 208 | 1.06 | 96 ± 22 | 97 ± 23 | 0.99 | 85 ± 23 | 77 ± 24 | 1.10 | 142 ± 26 | 135 ± 24 | 1.05 | |
| pPA4 | 2,341 ± 198 | 2,273 ± 145 | 1.03 | 94 ± 16 | 89 ± 18 | 1.05 | 81 ± 28 | 75 ± 18 | 1.08 | 141 ± 18 | 136 ± 21 | 1.03 | |
Three cultures per strain and condition were inoculated to an OD600 of 0.08 and grown for 24 h in M1 medium supplemented with Tc and 1% glycerol (+Gly) or 1% glucose (+Glc) and assayed in triplicate.
Values (Miller units) are means ± standard deviations of three independent experiments.
Gly/Glc, activity with Gly/activity with Glc. *, statistically significant difference (P < 0.05) for the individual fusion between glycerol and glucose conditions.
Numbers refer to CRP boxes (Fig. 1).
Since the responses of these regulatory motifs to glucose were similar in R. leguminosarum and E. coli, we further examined cAMP-CRP-mediated activation of pssA transcription in the presence and absence of glucose in the wild-type E. coli and cyaA and Δcrp mutant strains bearing fusions pPA1 to pPA4 (Table 3). The Gly/Glc ratios of β-galactosidase activities in the wild-type strain VH1000 harboring pPA fusions were comparable to those of strain 24.2. In the absence of glucose, significantly higher expression in the VH1000 strain was observed only for pPA1 (ratio of 1.37), whereas loss of responsiveness to glucose was noticed for the remaining pPA2 to pPA4 fusions. No significant differences in pssA-lacZ transcription were observed in either the cyaA or the Δcrp mutant in the presence and absence of glucose (ratio values around 1). All of these data indicate that CRP positively affects pssA transcription and that the cAMP-CRP binding sites located in the upstream region of this gene are engaged in this regulation.
The functional analysis of IRO boxes and the ICE box located in the pssA regulatory region.
In R. leguminosarum, a system responsible for iron homeostasis is mediated by two IrrA and RirA regulatory proteins. IrrA binds to specific conserved motifs (TTT-N11-AAA) termed an iron control element (ICE box) in promoters of iron-regulated genes and represses their transcription under iron limitation. In contrast, RirA recognizes iron-responsive operators (IRO box) of the TGA-N9-TCA consensus sequence and represses gene expression under Fe-rich conditions (32, 33).
In silico sequence analysis of the pssA upstream region revealed the presence of two IRO motifs highly similar to those recognized by RirA and one ICE motif as a putative target site for IrrA protein (Fig. 1 and 2). Among the identified IRO boxes, the first motif (TGA-N9-TCG) was located 82 bp upstream of the P1 −35 hexamer, whereas the second motif (TGA-N9-TCA) identical to the consensus sequence was located just downstream of the P1 −10 element. The ICE box (TGT-N11-AAA) of the sequence, nearly identical to the consensus, was found between the P1 and the P2 promoters (98 bp downstream of the P1 −10 and 11 bp upstream of the P2 −35 motifs).
In order to confirm the significance of these motifs in iron-dependent regulation of pssA expression, β-galactosidase activity was determined in strain 24.2 harboring pPA1 with the longest promoter region cultured in iron-sufficient and iron-deficient M1 medium. Transcription of this fusion increased 1.3-fold in the Fe+3-depleted M1 medium in comparison to the Fe+3-replete medium (15,244 versus 11,726 Miller units), indicating a positive iron response of pssA expression (data not shown). To investigate the function of individual regulatory motifs, the pPA1 to pPA4 fusions were introduced into R. leguminosarum bv. viciae wild-type strain J251 and its derivatives containing a mutation in the irrA (J386) and rirA (J397) genes. These strains were grown under Fe-replete and Fe-depleted conditions, and then β-galactosidase activity of transcriptional pssA-lacZ fusions was measured. The pPA1 plasmid contained both IRO boxes and the ICE box, pPA2 contained only the second IRO box and the ICE box, and pPA3 contained the ICE box lacking 3 bp at its 5′ terminus. The pPA4 plasmid possessed the pssA regulatory region with no IRO and ICE regulatory motifs. The ratios of β-galactosidase activities in strains growing under Fe-depleted and Fe-replete conditions (Fe−/Fe+ ratio) are shown in Table 4. The values of the Fe−/Fe+ ratios were not high, suggesting that the response of the pssA regulatory region to iron availability was moderate. The highest increase in pssA transcription was found in the J251 and J386 strains with the pPA1 fusion containing the full-length upstream region (1.27- and 1.25-fold, respectively). For pPA2 lacking IRO box 1, the Fe−/Fe+ ratio moderately decreased. In the case of pPA3 and pPA4, similar ratios (nearly 1) were found in all the derivatives of the three strains, indicating that this fragment of the pssA upstream region is not engaged in the regulation. Similarly, Fe−/Fe+ values of about 1 were observed in the rirA mutant. All these data indicate that IRO box 2 and RirA protein might be involved in slight iron-dependent modulation of pssA expression.
Table 4.
Effect of iron on pssA-lacZ transcription in R. leguminosarum strains
| Plasmid | Genotype (box) | β-Galactosidase activity by strain and conditiona |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| J251 (wild type) |
J386 (irrA mutant) |
J397 (rirA mutant) |
||||||||
| Fe−b | Fe+b | Fe−/Fe+c | Fe− | Fe+ | Fe−/Fe+ | Fe− | Fe+ | Fe−/Fe+ | ||
| pPA1 | IRO 1, 2; ICE | 14,009 ± 1,354 | 11,052 ± 1,342 | 1.27 | 16,052 ± 1,785 | 12,812 ± 1,462 | 1.25 | 12,302 ± 1,321 | 12,996 ± 1,431 | 0.95 |
| pPA2 | IRO 2, ICE | 12,064 ± 1,232 | 9,651 ± 1,121 | 1.25 | 13,910 ± 1,473 | 11,402 ± 1,543 | 1.22 | 12,307 ± 1,402 | 11,770 ± 1,251 | 1.04 |
| pPA3 | ICEd | 4,856 ± 609 | 5,371 ± 653 | 0.90 | 5,890 ± 663 | 6,134 ± 787 | 0.96 | 5,834 ± 664 | 5,240 ± 709 | 1.11 |
| pPA4 | 3,885 ± 452 | 4,417 ± 483 | 0.88 | 6,499 ± 786 | 6,511 ± 579 | 0.99 | 4,989 ± 576 | 5,379 ± 772 | 0.93 | |
Three cultures per strain and condition were inoculated to an OD600 of 0.1 and grown for 24 h at 28°C in M1 medium supplemented with Tc and 20 μM FeCl3 (Fe-replete [Fe+] condition) or 20 μM 2,2′-dipyridyl (Fe-depleted [Fe−] condition) and assayed in duplicate.
Values are means ± standard errors of β-galactosidase activities (Miller units) of at least three independent experiments.
Fe−/Fe+, activity under Fe− conditions/activity under Fe+ conditions.
Only the 3′ half.
The role of RosR boxes in pssA transcription.
Among the regulatory elements identified in the pssA upstream region, two motifs highly similar to the RosR box consensus have been found (Fig. 1 and 2). The RosR box is a 22-bp-long sequence containing two 9-bp inverted repeats separated by a 4-bp spacer ( TGAAATCTA-N4-TAGATTTCA ), which is a binding site for RosR, a transcriptional regulator uniquely occurring in rhizobia (18). Both RosR boxes identified were located downstream of the P1 promoter, at position −356 bp to −335 bp and at −277 bp to −251 bp. The first motif overlapped the P2 −35 hexamer, whereas the second one overlapped the P3 −35 element. RosR box 1 ( TAGAAACTT-N4-AAAATTCCA ) contained a more accurate palindrome and higher sequence identity to the consensus than RosR box 2 ( TGCGATATT-N4-TGGACACCA ).
To assess the importance of these motifs in pssA transcription, we introduced the pPA1 to pPA4 fusions to the wild-type 24.2, the rosR mutant (R. leguminosarum bv. trifolii 2472), and the 24.2 strain with additional rosR copies on the pBBR1MCS-2 plasmid (Table 5). The pPA1, pPA2, and pPA3 fusions contained both RosR boxes, whereas pPA4 contained only RosR box 2. In general, a significant decrease in pssA-lacZ transcription was observed for all of the tested fusions (with the exception of pPA4) in the rosR mutant in comparison to the wild-type bacteria. The activities of β-galactosidase for the pPA1, pPA2, and pPA3 plasmids were 1.6-, 2.19-, and 3.38-fold higher, respectively, in the wild-type strain than in the rosR mutant. In the case of pPA4, no significant difference in the level of pssA transcription between these strains was found, indicating that RosR box 2 is not essential for this regulation. On the other hand, the presence of multiple rosR copies resulted in a nearly 2-fold increase in pssA-lacZ expression (pPA1 to pPA3 fusions) in comparison to the wild type. In conclusion, our data indicate that RosR box 1 and RosR protein are responsible for the positive regulation of pssA transcription.
Table 5.
Transcriptional activity of pssA-lacZ fusions in R. leguminosarum bv. trifolii 24.2 wild type and its derivatives
| Plasmid | RosR box genotype | β-Galactosidase activity (Miller units) in:a |
||
|---|---|---|---|---|
| 24.2 (wild type) | 2472 (rosR mutant) | 24.2(pBR12)b | ||
| pPA1 | 1, 2 | 5,215 ± 321 A | 3,259 ± 213 B | 9,397 ± 504 C |
| pPA2 | 1, 2 | 3,892 ± 276 A | 1,776 ± 151 B | 6,872 ± 387 C |
| pPA3 | 1, 2 | 4,331 ± 229 A | 1,281 ± 108 B | 7,659 ± 413 C |
| pPA4 | 2 | 2,311 ± 178 | 2,098 ± 183 | 3,081 ± 253 |
The bacteria were grown in 79CA medium supplemented with 1% glycerol for 24 h. For each strain, β-galactosidase activity was assayed in triplicate. Data shown are the means ± standard deviations. Differences that are statistically significant (P <0.001) for the individual fusion present in strains 24.2, 2472, and 24.2(pBR12) are indicated by A, B, and C.
Strain 24.2(pBR12) carries additional copies of the rosR gene.
Binding of His6-RosR protein to the RosR box sequence.
Previously, we have established that the RosR protein binds effectively to the RosR box in the rosR upstream region and downregulates the transcription of its own gene (18). In this work, the binding capacity of RosR to the pssA regulatory region was studied. In order to confirm the direct involvement of this protein in pssA transcription, we performed EMSAs using an 18-kDa His6-RosR fusion protein purified under native conditions. For this experiment, a PCR-amplified 145-bp-long DNA fragment labeled with digoxigenin (DIG) and encompassing the pssA upstream region (−413 bp to −269 bp) with RosR box 1 was used with increasing concentrations of His6-RosR protein (Fig. 3). The observed retardation was dependent on protein concentration, and 0.5 ng μl−1 of the protein was found to be sufficient to form a pssA DNA-His6-RosR complex. The further increase of the protein content in reaction mixtures (to 8 ng μl−1) resulted in formation of larger amounts of this complex. These data confirm that RosR protein binds effectively to the pssA upstream region from −413 bp to −269 bp containing RosR box 1.
Fig 3.
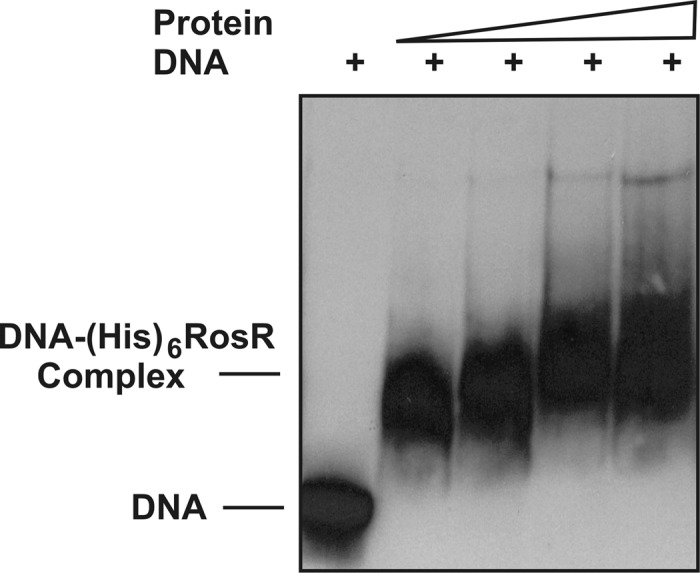
Electrophoretic mobility shift analysis with purified His6-RosR protein of increasing concentration (0, 0.5, 1, 2, and 4 ng μl−1). A digoxigenin-labeled DNA fragment (145 bp) matching the RosR box 1 was added to all reaction mixtures to give a final concentration of 0.2 ng μl−1.
The effect of additional copies of the pssA regulatory region and a mutation in this region on EPS production.
In order to establish whether additional copies of the pssA upstream region influence the level of EPS produced by the R. leguminosarum bv. trifolii 24.2, plasmids containing different fragments of the regulatory region were constructed on a high-copy-number pBBR1MCS vector and introduced into this strain. The presence of the pAX1 plasmid harboring the entire pssA upstream region exerted a strong negative effect on EPS production, decreasing the synthesis of this polymer to 61.5% of the control level (Table 6). Additionally, strain 24.2 derivatives containing pAX2 and pAX3 plasmids produced significantly less EPS than the wild-type strain (66% and 69%, respectively). These data indicate that multiple copies of the pssA regulatory region, present in plasmids pAX1 to pAX3 and encompassing RosR box 1 and other motifs, negatively affected EPS production, probably as a result of binding of RosR and other regulatory proteins. On the other hand, additional pssA copies positively influenced EPS production. Plasmid pAX6 contained the pssA gene with both P1 and P3 promoters, and pAX7 contained the gene preceded by only the P3 promoter. The 24.2 derivatives bearing plasmids pAX6 and pAX7 produced 1.56- and 1.34-fold more EPS than the wild-type strain (Table 6).
Table 6.
The effect of additional copies of the regulatory region and pssA gene on exopolysaccharide production in R. leguminosarum bv. trifolii
| Strain | Additional region of pssA (bp) | EPS concn (mg liter−1 [%])a |
|---|---|---|
| 24.2 (control) | 808 ± 76 (100) | |
| 24.2(pAX1) | −750 to −2 | 497 ± 52 (61.5)* |
| 24.2(pAX2) | −538 to −2 | 533 ± 61 (66)* |
| 24.2(pAX3) | −374 to −2 | 557.5 ± 65 (69)* |
| 24.2(pAX4) | −284 to −2 | 664 ± 63 (82) |
| 24.2(pAX5) | −134 to −2 | 776 ± 84 (96) |
| 24.2(pAX6) | −750 to +815 | 1,260 ± 123 (156)* |
| 24.2(pAX7) | −284 to +815 | 1,083 ± 114 (134)* |
Bacteria were grown in M1 medium supplemented with 1% glycerol, Dilworth's vitamins, and, where needed, kanamycin for 3 days at 28°C. Exopolysaccharide concentrations are expressed as milligrams of glucose equivalents per liter of culture at an OD600 of 1. Average values of three independent experiments are shown. Percentages are relative to the control value, set at 100%. *, statistically significant (P < 0.05) difference compared to the control.
In addition, R. leguminosarum bv. trifolii mutant strains 4440 and 1012 carrying a Tn5 insertion in the pssA upstream region were obtained. Strain 4440 has a mini-Tn5 transposon inserted downstream of P3 (position −77/−78 bp), whereas strain 1012 has the Tn5 located between −35 and −10 hexamers of this promoter (position −253/−254 bp) (Fig. 1). Neither of these mutants produced EPS, confirming the significant role of the P3 promoter in pssA transcription.
DISCUSSION
Previously, it was found that EPS production in R. leguminosarum is affected by such environmental factors as phosphate, ammonium, carbon source, and flavonoids (19, 20). But until now regulatory pathways controlling the expression of pss genes in response to these environmental factors have not been extensively studied (21, 34).
In this work, we performed a detailed analysis of the pssA upstream region to establish environmental signals that influence transcription of this gene. We have found that pssA is an important target for regulation of this biosynthetic pathway. In the pssA upstream region, three putative promoters showing significant similarity to eubacterial promoters recognized by the sigma factor σ70 of RNA polymerase have been identified (28). Among them, the distal promoter P1 and the proximal promoter P3 proved to be responsible for pssA transcription in R. leguminosarum bv. trifolii. Ivashina and coworkers (8) reported that the pss4 gene from R. leguminosarum bv. viciae VF39, which is an ortholog of pssA from R. leguminosarum bv. trifolii 24.2, also possessed three putative promoters. Similarly, S. meliloti exoY, the key gene involved in succinoglycan biosynthesis, was found to be expressed from two distinct promoters (35, 36).
pssA contains a very long (767 bp) regulatory region, in which several inverted repeats (IRs) of different lengths have been identified (Fig. 1). Such long upstream regions have often been described as target sites for the regulation of gene expression (20, 36, 37). Both transcripts generated from the P1 and P3 promoters have long 5′ untranslated regions, and a majority of these IRs proved to be involved in formation of secondary structures of pssA RNAs. Particularly in the case of the longer transcript initiated from the P1 promoter, several stem structures, consisting of the IR2 to IR4, IR7 to IR9, and IR10 motifs, were formed at its 5′ terminus.
In addition, several sequence motifs involved in RosR-dependent and phosphate-, carbon source-, and iron-mediated regulation have been identified in the pssA upstream region. Our results indicate that the transcription of this gene is upregulated by RosR and several nutrition factors as well. These regulatory pathways positively correlate with the levels of EPS production under these environmental conditions (19, 20). RosR protein was evidenced to be essentially involved in this regulation since pssA expression was significantly decreased (1.6-fold) in the rosR mutant but 1.8-fold increased in strain 24.2 carrying additional rosR copies (Table 5). Moreover, it has been confirmed that purified RosR binds effectively to the RosR box 1 sequence located downstream of the distal P1 promoter (Fig. 1 and 3). RosR is the regulatory protein uniquely occurring in rhizobial cells. Previously, we have demonstrated that RosR is a positive regulator of EPS synthesis (18). A mutation in rosR resulted in a 3-fold decrease in EPS synthesis, whereas additional copies of this gene caused a 1.9-fold increase in EPS production. On the other hand, RosR negatively regulates the expression of its own gene by binding to the RosR box sequence located in the rosR upstream region.
Among the environmental factors tested, phosphate and carbon source were found to strongly affect pssA transcription in R. leguminosarum (Table 2 and 3). Under phosphate limitation, pssA expression was 1.7-fold higher than in the presence of a high concentration of this nutrient. Transcriptional analysis of the pPA1 to pPA4 fusions in the S. meliloti wild-type and phoB mutant strains growing under low-Pi conditions indicated significantly lower pssA-lacZ expression in the mutant background, which confirmed the function of PhoB in the upregulation of this gene. The low Pi/high Pi ratios of the tested fusions in both the wild-type strains of R. leguminosarum and S. meliloti showed that Pho box 2 located upstream of the P3 promoter was engaged in the phosphate-dependent modulation of pssA expression (Table 2). In addition, this region (−284 bp to −260 bp) located just upstream of the P3 −35 hexamer proved to be indispensable for the maximal level of pssA transcription driven from this promoter. This finding was in agreement with our previous results indicating that EPS synthesis and biofilm formation in R. leguminosarum were significantly increased under phosphate starvation (20). Similarly, transcription of S. meliloti exoY from the proximal promoter that contained a Pho-box-like sequence in its −35 region was affected by PhoB in a phosphate-dependent manner (36). Moreover, MucR protein that bound to a short DNA region upstream of exoY slightly increased the transcription of this gene (38). In addition, some differences in the levels of pssA-lacZ transcription observed between the Rm1021 and RmH406 strains growing under high-phosphate conditions might suggest an additional phoB-independent effect of phosphate on pssA expression. Similarly, complex phosphate-mediated regulation has been reported for S. meliloti exoY, in which both PhoB-dependent and PhoB-independent regulation was engaged (36).
Phosphate is one of the most essential nutrients for bacteria but is often limited in the soil (39). S. meliloti mutants that are unable to take up phosphate do not fix nitrogen (40), which suggests a substantial role of this factor for effective symbiosis. A two-component regulatory system consisting of a PhoR sensor kinase and a PhoB response regulator is involved in regulation of many rhizobial genes (41–44). PhoB of R. leguminosarum bv. viciae 3841 (accession number RL0547; genome position, 591,764 to 592,447 bp) (45) shows 100% amino acid identity with PhoB of R. leguminosarum bv. trifolii WSM1325, 93% identity with PhoB of S. meliloti, and 54% identity with PhoB of E. coli, which indicates a high sequence conservation within this group of transcriptional regulators (42, 46, 47).
Another nutrient significantly affecting pssA transcription in R. leguminosarum is the type of the carbon source (Table 3). We have found that the presence of glucose caused a 2-fold decrease in pssA expression in strain 24.2 carrying the longest pPA1 fusion, suggesting a role of catabolite repression in the regulation of transcription of this gene. The remaining pPA2 to pPA4 fusions did not show sensitivity to glucose, thus indicating that the pssA regulatory sequence from −750 bp to −539 bp is responsible for this effect (Fig. 1). In this region, two cAMP-CRP motifs for the CRP activator have been identified. Transcriptional analysis of pPA1 to pPA4 fusions in the wild-type E. coli and cyaA and crp mutants confirmed that CRP and cAMP are required for this regulation (Table 3). cAMP-CRP boxes 1 and 2 are located immediately upstream of the P1 promoter and are separated by a 102-bp distance. The location of these motifs suggests that activation of pssA transcription from P1 occurs in a manner similar to an E. coli synergistic class III mechanism, in which two or more CRP dimers or a combination of CRP and other activators is utilized to achieve maximal transcription activation (48, 49). The importance of this regulatory region for pssA expression is additionally confirmed by the phenotype of R. leguminosarum bv. trifolii mutant 120 with a Tn5 transposon insertion at position −666/−667 bp of the pssA upstream region (Fig. 1) (9). This mutant does not produce EPS and elicits nodules on clover plants that are ineffective in nitrogen fixation, similarly to the R. leguminosarum bv. trifolii mutant 5819 with a Tn5 insertion in the pssA coding region (Fig. 1) (5). Previously, we have reported that glucose influenced rosR expression (19). Also in S. meliloti, the production of succinoglycan was found to be affected by catabolite repression (50).
In contrast to the situation in E. coli, elucidation of the physiological roles of CRP and cAMP in rhizobia is difficult since multiple genes encoding transcriptional regulators belonging to the CRP/FNR family and putative adenylate cyclases have been identified in the genomes of these bacteria (45, 51). These CRP/FNR regulators of R. leguminosarum bv. viciae 3841 show 25.1% to 28.9% identity with the CRP of E. coli K-12 (45, 47).
Among the environmental signals tested, iron ions appeared to have only a slight stimulatory effect on pssA expression although we have found that this compound significantly affected EPS production in R. leguminosarum. In the pssA upstream region, the ICE box and two IRO boxes with high sequence identity to consensus sequences recognized by IrrA and RirA regulators, respectively, have been identified (Fig. 1) (32, 33). Our data indicate that RirA and IRO box 2 might be involved in iron-dependent regulation of pssA transcription, but the level of the modulation observed is very low (Table 4). This suggests that genes other than pssA involved in EPS biosynthesis are more efficiently responding targets for RirA and/or IrrA proteins.
The complexity of pssA regulation is additionally confirmed by recent data obtained by Vanderlinde and Yost (52), who have found that the expression of this gene was decreased 1.6-fold in an R. leguminosarum chvG mutant in comparison to its parental strain. This finding suggested a function of the sensor kinase ChvG of a two-component signal transduction system in positive regulation of pssA transcription. In contrast, a small inner membrane-attached protein, PsiA, encoded by a gene located on the symbiotic megaplasmid of R. leguminosarum bv. phaseoli negatively regulates EPS production (14). Since PssA containing a transmembrane domain at its N terminus is also located in the inner membrane, the same subcellular localization of both PsiA and PssA proteins (7, 13–15) suggests that PsiA could function as a posttranslational inhibitor of PssA, which interacts with and inhibits the activity of this enzyme.
In addition, in order to establish a linkage between pssA expression and EPS synthesis in R. leguminosarum, mutations in the pssA regulatory region have been introduced. We have found that a Tn5 insertion within the P3 sequence (mutant strain 1012) exerted an identical phenotypic effect as insertions located downstream of the P3 promoter (mutant 4440) and inside the pssA ORF (mutant 5819) (Fig. 1). None of these mutants produced EPS, confirming that the P3 promoter plays a significant role in pssA expression and, as a result, in EPS synthesis. On the other hand, additional pssA copies significantly enhanced production of this polymer in strain 24.2. Moreover, multiple copies of the pssA regulatory region negatively affected EPS production, most probably by providing additional target sites for binding of regulatory proteins involved in positive regulation of pssA expression. Previously, we have reported that multiple copies of the rosR upstream region containing a RosR box also decreased EPS synthesis (53). These results show that the RosR protein is essential for regulation of expression of both these genes.
In a summary, the data of this study indicate that the regulation of pssA expression is very complex. The high level of pssA transcription in R. leguminosarum is a result of interactions between RosR and other signal-dependent regulatory proteins (PhoB and CRP-like) which positively affect expression of this gene.
ACKNOWLEDGMENTS
We thank Andrew W. B. Johnston and Jonathan Todd (School of Biological Sciences, University of East Anglia, Norwich, United Kingdom) for kindly providing the R. leguminosarum bv. viciae J251, J386, and J397 strains and R. Gourse (via G. Wêgrzyn, Department of Molecular Biology, University of Gdańsk, Gdańsk, Poland) for providing E. coli VH1000, VH1000 cya, and M182 crp strains. Anke Becker (Institute of Biology, University of Freiburg, Freiburg, Germany) is acknowledged for providing S. meliloti Rm1021 wild-type and RmH406 phoB mutant strains. We thank Maria Małek for excellent technical assistance.
This work was partially supported by a grant from the Ministry of Science and Higher Education (number N N303 092234).
Footnotes
Published ahead of print 24 May 2013
REFERENCES
- 1. Cooper JE. 2007. Early interactions between legumes and rhizobia: disclosing complexity in a molecular dialogue. J. Appl. Microbiol. 103: 1355– 1365 [DOI] [PubMed] [Google Scholar]
- 2. Downie JA. 2010. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34: 150– 170 [DOI] [PubMed] [Google Scholar]
- 3. Rolfe BG, Carlson RW, Ridge RW, Dazzo RW, Mateos FB, Pankhurst CE. 1996. Defective infection and nodulation of clovers by exopolysaccharide mutants of Rhizobium leguminosarum bv. trifolii. Aust. J. Plant Physiol. 23: 285– 303 [Google Scholar]
- 4. van Workum WA, Canter Cremers HCJ, Wijfjes AHM, Van der Kolk C, Wijffelman CA, Kijne JW. 1997. Cloning and characterization of four genes of Rhizobium leguminosarum bv. trifolii involved in exopolysaccharide production and nodulation. Mol. Plant Microbe Interact. 10: 290– 301 [DOI] [PubMed] [Google Scholar]
- 5. Janczarek M, Jaroszuk-Ściseł J, Skorupska A. 2009. Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii. Antonie Van Leeuwenhoek 96: 471– 486 [DOI] [PubMed] [Google Scholar]
- 6. Janczarek M. 2011. Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int. J. Mol. Sci. 12: 7898– 7933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borthakur D, Barker RF, Latchford JW, Rossen L, Johnston AWB. 1988. Analysis of pss genes of Rhizobium leguminosarum required for exopolysaccharide synthesis and nodulation of peas: their primary structure and their interaction with psi and other nodulation genes. Mol. Gen. Genet. 213: 155– 162 [DOI] [PubMed] [Google Scholar]
- 8. Ivashina T, Khmelnitsky MI, Shlyapnikov MG, Kanapin AA, Ksenzenko VN. 1994. The pss4 gene from Rhizobium leguminosarum biovar viciae VF39: cloning, sequence and the possible role in polysaccharide production and nodule formation. Gene 150: 111– 116 [DOI] [PubMed] [Google Scholar]
- 9. Janczarek M, Król J, Kutkowska J, Mazur A, Wielbo J, Borucki W, Kopcińska J, Łotocka B, Urbanik-Sypniewska T, Skorupska A. 2001. Mutation in the pssB-pssA intergenic region of Rhizobium leguminosarum bv. trifolii affects the surface polysaccharide synthesis and nitrogen fixation ability. J. Plant Physiol. 158: 1565– 1574 [Google Scholar]
- 10. Król JE, Mazur A, Marczak M, Skorupska A. 2007. Syntenic arrangements of the surface polysaccharide biosynthesis genes in Rhizobium leguminosarum. Genomics 89: 237– 247 [DOI] [PubMed] [Google Scholar]
- 11. Pollock TJ, Workum WA, Thorne L, Mikolajczak MJ, Yamazaki M, Kijne JW, Armentrout RW. 1998. Assignment of biochemical functions to glycosyl transferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J. Bacteriol. 180: 586– 593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ivashina TV, Fedorova EE, Ashina NP, Kalinchuk NA, Druzhinina TN, Shashkov AS, Shibaev VN, Ksenzenko VN. 2010. Mutation in the pssM gene encoding ketal pyruvate transferase leads to disruption of Rhizobium leguminosarum bv. viciae-Pisum sativum symbiosis. J. Appl. Microbiol. 109: 731– 742 [DOI] [PubMed] [Google Scholar]
- 13. Borthakur D, Johnston AWB. 1987. Sequence of psi, a gene of the symbiotic plasmid of Rhizobium phaseoli which inhibits exopolysaccharide synthesis and nodulation and demonstration that its transcription is inhibited by psr, another gene on the symbiotic plasmid. Mol. Gen. Genet. 207: 149– 154 [DOI] [PubMed] [Google Scholar]
- 14. Latchford JW, Borthakur D, Johnston AWB. 1991. The products of Rhizobium genes, psi and pss, which affect exopolysaccharide production, are associated with the bacterial cell surface. Mol. Microbiol. 5: 2107– 2114 [DOI] [PubMed] [Google Scholar]
- 15. Mimmack ML, Hong GF, Johnston AWB. 1994. Sequence and regulation of psrA, a gene on the Sym plasmid of Rhizobium leguminosarum biovar phaseoli which inhibits transcription of the psi genes. Microbiology 140: 455– 461 [DOI] [PubMed] [Google Scholar]
- 16. Janczarek M, Skorupska A. 2001. The Rhizobium leguminosarum bv. trifolii pssB gene product is an inositol monophosphatase that influences exopolysaccharide synthesis. Arch. Microbiol. 175: 143– 151 [DOI] [PubMed] [Google Scholar]
- 17. Reeve WG, Dilworth MJ, Tiwari RP, Glenn AR. 1997. Regulation of exopolysaccharide production in Rhizobium leguminosarum biovar viciae WSM710 involves exoR. Microbiology 143: 1951– 1958 [DOI] [PubMed] [Google Scholar]
- 18. Janczarek M, Skorupska A. 2007. The Rhizobium leguminosarum bv. trifolii RosR: transcriptional regulator involved in exopolysaccharide production. Mol. Plant Microbe Interact. 20: 867– 881 [DOI] [PubMed] [Google Scholar]
- 19. Janczarek M, Skorupska A. 2009. Rhizobium leguminosarum bv. trifolii rosR gene expression is regulated by catabolic repression. FEMS Microbiol. Lett. 291: 112– 119 [DOI] [PubMed] [Google Scholar]
- 20. Janczarek M, Skorupska A. 2011. Modulation of rosR expression and exopolysaccharide production in Rhizobium leguminosarum bv. trifolii by phosphate and clover root exudates. Int. J. Mol. Sci. 12: 4132– 4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Janczarek M, Skorupska A. 2004. Regulation of pssA and pssB gene expression in R. leguminosarum bv. trifolii in response to environmental factors. Antonie Van Leeuwenhoek 85: 217– 227 [DOI] [PubMed] [Google Scholar]
- 22. Vincent JM. 1970. A manual for the practical study of root nodule bacteria. International biological program handbook 15. Blackwell Scientific Publications, Ltd., Oxford, United Kingdom [Google Scholar]
- 23. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Brown CM, Dilworth MJ. 1975. Ammonia assimilation by Rhizobium cultures and bacteroids. J. Gen. Microbiol. 86: 39– 48 [DOI] [PubMed] [Google Scholar]
- 25. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406– 3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Loewus FA. 1952. Improvement in the anthrone method for determination of carbohydrates. Anal. Chem. 24: 219 [Google Scholar]
- 28. Wősten MM. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127– 150 [DOI] [PubMed] [Google Scholar]
- 29. Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2: 57– 65 [DOI] [PubMed] [Google Scholar]
- 30. MacLellan SR, MacLean AM, Finan TM. 2006. Promoter prediction in the rhizobia. Microbiology 152: 1751– 1763 [DOI] [PubMed] [Google Scholar]
- 31. Todd JD, Wexler M, Sawers G, Yeoman KH, Poole PS, Johnston AWB. 2002. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148: 4059– 4071 [DOI] [PubMed] [Google Scholar]
- 32. Todd JD, Sawers G, Rodionov DA, Johnston AW. 2006. The Rhizobium leguminosarum regulator IrrA affects the transcription of a wide range of genes in response to Fe availability. Mol. Genet. Genomics 275: 564– 577 [DOI] [PubMed] [Google Scholar]
- 33. Yeoman KH, Curson AR, Todd JD, Sawers G, Johnston AWB. 2004. Evidence that the Rhizobium regulatory protein RirA binds to cis-acting iron-responsive operators (IROs) at promoters of some Fe-regulated genes. Microbiology 150: 4065– 4074 [DOI] [PubMed] [Google Scholar]
- 34. Wielbo J, Mazur A, Krol JE, Marczak M, Skorupska A. 2004. Environmental modulation of the pssTNOP gene expression in Rhizobium leguminosarum bv. trifolii. Can. J. Microbiol. 50: 1– 11 [DOI] [PubMed] [Google Scholar]
- 35. Cheng HP, Yao SY. 2004. The key Sinorhizobium meliloti succinoglycan biosynthesis gene exoY is expressed from two promoters. FEMS Microbiol. Lett. 231: 131– 136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quester I, Becker A. 2004. Four promoters subject to regulation by ExoR and PhoB direct transcription of the Sinorhizobium meliloti exoYFQ operon involved in the biosynthesis of succinoglycan. J. Mol. Microbiol. Biotechnol. 7: 115– 132 [DOI] [PubMed] [Google Scholar]
- 37. Spinelli SV, Pontel LB, Vescovi EG, Soncini FC. 2008. Regulation of magnesium homeostasis in Salmonella: Mg2+ targets the mgtA transcript for degradation by RNase E. FEMS Microbiol. Lett. 280: 226– 234 [DOI] [PubMed] [Google Scholar]
- 38. Bertram-Drogatz PA, Quester I, Becker A, Pühler A. 1998. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol. Gen. Genet. 257: 433– 441 [DOI] [PubMed] [Google Scholar]
- 39. Bieleski RL. 1973. Phosphate pools, phosphate transport and phosphate availability. Annu. Rev. Plant Physiol. 24: 225– 252 [Google Scholar]
- 40. Bardin S, Dan S, Osteras M, Finan TM. 1996. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol. 178: 4540– 4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bahlawane C, McIntosh M, Krol E, Becker A. 2008. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol. Plant Microbe Interact. 21: 1498– 1509 [DOI] [PubMed] [Google Scholar]
- 42. Rüberg S, Pühler A, Becker A. 1999. Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology 145: 603– 611 [DOI] [PubMed] [Google Scholar]
- 43. Yuan Z, Zaheer R, Morton R, Finan TM. 2006. Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res. 34: 2686– 2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krol E, Becker A. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genomics 272: 1– 17 [DOI] [PubMed] [Google Scholar]
- 45. Young JP, Crossman LC, Johnston AW, Thomson NR, Ghazoui ZF, Hull KH, Wexler M, Curson AR, Todd JD, Poole PS, Mauchline TH, East AK, Quail MA, Churcher C, Arrowsmith C, Cherevach I, Chillingworth T, Clarke K, Cronin A, Davis P, Praser A, Hance Z, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, Whitehead S, Parkhill J. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7: R34. 10.1186/gb-2006-7-4-r34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reeve W, O'Hara G, Chain P, Ardley J, Bräu L, Nandesena K, Tiwari R, Copeland A, Nolan M, Han C, Brettin T, Land M, Ovchinikova G, Ivanova N, Mavromatis K, Markowitz V, Kyrpides N, Melino V, Denton M, Yates R, Howieson J. 2010. Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM1325, an effective microsymbiont of annual Mediterranean clovers. Stand. Genomic Sci. 2:347– 356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, Mori H, Perna NT, Plunkett GIII, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart D, Wanner BL. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot-2005. Nucleic Acids Res. 34: 1– 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293: 199– 213 [DOI] [PubMed] [Google Scholar]
- 49. Beatty CM, Browning DF, Busby SJW, Wolfe AJ. 2003. Cyclic AMP receptor protein-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism. J. Bacteriol. 185: 5148– 5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pinedo CA, Gage DJ. 2009. HPrK regulates succinate-mediated catabolite repression in the gram-negative symbiont Sinorhizobium meliloti. J. Bacteriol. 191: 298– 309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tellez-Sosa J, Soberon N, Vega-Segura A, Torres-Marquez ME, Cevallos MA. 2002. The Rhizobium etli cyaC product: characterization of a novel adenylate cyclase class. J. Bacteriol. 184: 3560– 3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vanderlinde EM, Yost CK. 2012. Mutation of the sensor kinase chvG in Rhizobium leguminosarum negatively impacts cellular metabolism, outer membrane stability and symbiosis. J. Bacteriol. 194: 768– 777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Janczarek M, Kutkowska J, Piersiak T, Skorupska A. 2010. Rhizobium leguminosarum bv. trifolii rosR is required for interaction with clover, biofilm formation and adaptation to the environment. BMC Microbiol. 10: 284. 10.1186/1471-2180-10-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wexler M, Yeoman KH, Stevens JB, de Luca NG, Sawers G, Johnston AW. 2001. The Rhizobium leguminosarum tonB gene is required for the uptake of siderophore and haem as sources of iron. Mol. Microbiol. 41:801– 816 [DOI] [PubMed] [Google Scholar]
- 55. Wexler M, Todd JD, Kolade O, Bellini D, Hemmings AM, Sawers G, Johnston AWB. 2003. Fur is not the global regulator of iron uptake genes in Rhizobium leguminosarum. Microbiology 149: 1357– 1365 [DOI] [PubMed] [Google Scholar]
- 56. Charles TC, Newcomb W, Finan TM. 1991. ndvF, a novel locus located on megaplasmid pRmeSU47b (pExo) of Rhizobium meliloti, is required for normal nodule development. J. Bacteriol. 173: 3981– 3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gaal T, Bartlett MS, Ross W, Turnbough JCL, Gourse RL. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278: 2092– 2097 [DOI] [PubMed] [Google Scholar]
- 58. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Nat. Biotechnol. 1: 784– 791 [Google Scholar]
- 59. Spaink HP, Okker RJH, Wijffelman CA, Pees E, Lugtenberg BJJ. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9: 27– 39 [DOI] [PubMed] [Google Scholar]
- 60. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175– 176 [DOI] [PubMed] [Google Scholar]