Abstract
Mycobacterium avium subsp. paratuberculosis is the causative agent of Johne's disease (JD) in cattle. M. avium subsp. paratuberculosis infects the gastrointestinal tract of calves, localizing and persisting primarily in the distal ileum. A high percentage of cattle exposed to M. avium subsp. paratuberculosis do not develop JD, but the mechanisms by which they resist infection are not understood. Here, we merge an established in vivo bovine intestinal segment model for M. avium subsp. paratuberculosis infection with bovine-specific peptide kinome arrays as a first step to understanding how infection influences host kinomic responses at the site of infection. Application of peptide arrays to in vivo tissue samples represents a critical and ambitious step in using this technology to understand host-pathogen interactions. Kinome analysis was performed on intestinal samples from 4 ileal segments subdivided into 10 separate compartments (6 M. avium subsp. paratuberculosis-infected compartments and 4 intra-animal controls) using bovine-specific peptide arrays. Kinome data sets clustered into two groups, suggesting unique binary responses to M. avium subsp. paratuberculosis. Similarly, two M. avium subsp. paratuberculosis-specific immune responses, characterized by different antibody, T cell proliferation, and gamma interferon (IFN-γ) responses, were also observed. Interestingly, the kinomic groupings segregated with the immune response groupings. Pathway and gene ontology analyses revealed that differences in innate immune and interleukin signaling and particular differences in the Wnt/β-catenin pathway distinguished the kinomic groupings. Collectively, kinome analysis of tissue samples offers insight into the complex cellular responses induced by M. avium subsp. paratuberculosis in the ileum and provides a novel method to understand mechanisms that alter the balance between cell-mediated and antibody responses to M. avium subsp. paratuberculosis infection.
INTRODUCTION
Johne's disease (JD) of cattle and other ruminants is caused by a chronic enteric infection by Mycobacterium avium subsp. paratuberculosis. JD is characterized by a long asymptomatic latency period during which animals display variable humoral and inflammatory immune responses (1, 2). During the symptomatic phase of infection, there is a progressive inflammatory enteritis, diarrhea, and significant weight loss (3). The infected host sheds M. avium subsp. paratuberculosis throughout the course of infection but especially during the late stages. Shedding occurs primarily in the feces (4) but has also been detected in milk (5). Large quantities of M. avium subsp. paratuberculosis shed by infected cattle survive for extended periods in the environment and persist after high-temperature, short-time pasteurization, raising concerns about contamination of dairy products (6). M. avium subsp. paratuberculosis has been shown to infect primates and has been postulated to be a cause of Crohn's disease in humans (3). The zoonotic potential of this infection and its already devastating impact on cattle and sheep have fueled extensive study into its pathogenesis. A priority is to determine the mechanisms by which M. avium subsp. paratuberculosis subverts the host immune system to establish chronic infection. Understanding these mechanisms may represent a critical step toward the development of either effective vaccines or therapeutics.
Cattle exhibit highly variable responses to M. avium subsp. paratuberculosis infection, and, similar to the well-documented high rates of human resistance to Mycobacterium tuberculosis (7), not all calves exposed to the pathogen develop JD (8). This suggests that genetic and/or environmental factors predispose animals to disease. A recent meta-analysis of two genome-wide association studies revealed multiple loci associated with M. avium subsp. paratuberculosis infection of cattle, indicating that genetic susceptibility to infection is complex, involving 11 different chromosomes (9). Therefore, understanding the regulation of immune responses to this pathogen in an outbred population is a daunting challenge. Furthermore, the eradication of M. avium subsp. paratuberculosis is complicated by its persistence in soil, feed, and water, and as a result, strategies for controlling the spread of infection have focused on herd management. The ability to distinguish animals that effectively control M. avium subsp. paratuberculosis infection from more susceptible animals might provide a way to selectively enhance the health of cattle and decrease the zoonotic threat.
Monitoring global responses at the level of cellular kinase activity (the kinome) is an effective approach to understand complex biology as well as to identify therapeutic targets and biomarkers (10). Many methods have been used to assay kinase activity under different conditions, each with advantages and disadvantages (11). While peptide array approaches that attempt to identify novel phosphorylation sites may lead to false positives, focused arrays that employ a subset of better-characterized phosphorylation sites are powerful tools to profile pathways of interest. Specifically, kinome profiling offers a way to differentiate individuals at a phenotypic level, with the potential to reveal adaptive or maladaptive shifts in host signaling patterns in response to a pathological state such as infection. Previously, using a bovine-specific peptide array developed in our lab, we used kinome analysis to reveal specific mechanisms through which M. avium subsp. paratuberculosis influences the ability of bovine monocytes to respond to gamma interferon (IFN-γ) and Toll-like receptor ligands (12, 13). These investigations highlighted mechanisms employed by the pathogen to alter innate immune responses as well as the power of kinomics to reveal host signaling events following infection.
Here, employing a bovine intestinal segment model developed by our group to restrict M. avium subsp. paratuberculosis infection to specific sites in the intestine (19), we monitored adaptive immune responses in parallel with kinome profiling of ileal tissues from M. avium subsp. paratuberculosis-infected and uninfected intestinal compartments. Kinome profiling of tissues is an ambitious but not unprecedented approach to understand shifts in kinase activities in pathology, and it has provided insights into aberrant signaling in different cancers (14–16). We targeted the early (1-month) phase of M. avium subsp. paratuberculosis infection since antibody responses can be detected in subclinical M. avium subsp. paratuberculosis infections (1). Furthermore, it has been suggested that host responses during the first few weeks after infection may determine whether JD develops (17). We hypothesized that M. avium subsp. paratuberculosis infection for 1 month should provide sufficient time for imprinting the host immune response to M. avium subsp. paratuberculosis and allow us to explore potential relationships between global kinase activity at the site of infection and immune responses occurring in gut-associated lymphoid tissue.
MATERIALS AND METHODS
Calves, surgery, and infection with Mycobacterium avium subsp. paratuberculosis.
All experimental protocols were performed following the guidelines approved by the Canadian Council on Animal Care. Protocols for animal housing, anesthesia, surgery, M. avium subsp. paratuberculosis infection, and postsurgical care were performed as previously described (18, 19). Young calves are most susceptible to JD (4), and four calves that were 2 weeks old were inoculated with M. avium subsp. paratuberculosis in surgically isolated intestinal compartments. Briefly, a 30- to 35-cm segment of intestine was surgically isolated, proximal to the ileocecal fold, and subdivided into three equal compartments using silk ligatures. The distal and middle compartments were injected with 1 × 108 to 3 × 108 CFU of M. avium subsp. paratuberculosis strain K10 (preparation is described below) in a final volume of 5 ml phosphate-buffered saline (PBS). The proximal compartment of the intestinal segment was injected with 5 ml PBS. Postsurgically, calves were treated with 1.1 mg/kg flunixin (Banamine; Schering Plough Canada Inc., Pointe Claire, Quebec, Canada) for 3 days and with 3 to 4 mg/kg enrofloxacin (Baytril; Bayer Inc.) for 5 days. Three uninfected calves of similar age from the same herd were included as negative controls. Calves were maintained on a whole-milk diet for 4 weeks. Blood for the isolation of serum and peripheral blood mononuclear cells (PBMCs) was collected 1 day before animals were euthanized for collection of tissue samples (ileal intestinal compartments and contents, spleen, and mesenteric lymph node [MLN]).
Preparation of M. avium subsp. paratuberculosis inoculum and lysate.
M. avium subsp. paratuberculosis strain K10 was a generous gift from Marcel Behr (McGill University Health Center, Montreal, Quebec, Canada). One loop of K10 culture grown on Middlebrook 7H10 agar (BD Bioscience, Canada) was inoculated into 50 ml of Difco Middlebrook 7H9 broth (BD Bioscience, Canada) and incubated on a rotary shaker for 5 days at 37°C. M. avium subsp. paratuberculosis cultures were centrifuged at 3,000 × g at 4°C for 15 min in a preweighed sterile 50-ml centrifuge tube. The supernatant was discarded, and the bacterial pellet was allowed to dry for 30 min in the inverted tube before obtaining a final weight. The weight of the dry bacterial pellet (tube with pellet minus tube weight) was calculated and total CFU calculated on the basis of previous titration experiments (19). M. avium subsp. paratuberculosis lysate for cell stimulation experiments was prepared from 5-day cultures of M. avium subsp. paratuberculosis grown in Middlebrook 7H9 broth as described previously (19) except that the protein concentration of the lysate was measured using the Bio-Rad Bradford reagent (Bio-Rad Laboratories, Mississauga, Ontario, Canada). Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, Canada) was added to the bacterial lysate (final concentration, 1 mM) before storage at −20°C.
Tissue collection and histology.
Tissues for kinome analysis and pathology were collected from infected and uninfected intestinal compartments immediately after euthanizing the calves. A 4- to 5-cm segment of each infected ileal compartment and the adjacent uninfected ileal compartment, including contents, was collected and fixed in 10% neutral buffered formalin (VWR, Westchester, PA) for histopathological examination. The remaining tissue from each compartment was then opened longitudinally, the contents were removed, and longitudinal strips of intestinal tissue measuring 0.5 cm by 3 cm were collected, placed into cryovials, and snap-frozen in liquid nitrogen prior to storage at −80°C. Histology samples fixed in 10% neutral buffered formalin (NBF) were embedded, sectioned, and acid-fast stained by Prairie Diagnostic Services (Saskatoon, Saskatchewan, Canada). Photos were taken through an Olympus BX41 microscope with a flip-out condenser and a 100× UPlan Fluorite oil immersion lens using a 12 Megapixel Olympus DP71 camera with DP controller acquisition and managing software (Olympus).
Blood was collected from the jugular veins of calves at 4 weeks after M. avium subsp. paratuberculosis infection and 1 day prior to euthanizing. PBMCs were isolated following a previously described protocol (20). Briefly, blood was centrifuged at 1,400 × g for 20 min at room temperature before collecting the buffy coat and resuspending cells in 35 ml Ca2+- and Mg2+-free PBS (PBSA) containing 0.1% EDTA. Cells were layered onto isotonic 54% Percoll (GE Health Bio-Sciences AB, Uppsala, Sweden) and centrifuged at 2,000 × g for 20 min at room temperature. Cells at the Percoll-PBS interphase were collected and washed three times with PBS before resuspending PBMCs at a final concentration of 2 × 106 viable cells/ml in RPMI medium (Invitrogen, Burlington, Ontario, Canada) containing 10% fetal bovine serum (FBS) (Invitrogen) plus antibiotics and antimycotics (Sigma-Aldrich Canada, Oakville, Ontario, Canada). Mesenteric lymph nodes (MLNs) and spleens were collected immediately after euthanizing calves, and tissues were placed in ice-cold Dulbecco modified Eagle medium (DMEM) (Sigma-Aldrich Canada). For lymph nodes, pericapsular fat was removed before the lymph node was cut and immersed in PBSA. The tissue was minced finely with a scalpel blade to release single cells. The cell suspension was passed through a 40-μm nylon cell strainer (Becton Dickson Labware, Franklin Lakes, NJ) and cells washed three times with PBSA containing 0.1% EDTA and one time with only PBSA before being resuspended at a final concentration of 2 × 106 cells/ml in DMEM supplemented with 10% FBS, antibiotics, and antimycotics. Spleens were minced and strained as described above to release and isolate single cells, and the cells were centrifuged at 1,200 rpm for 10 min at 4°C. Medium was poured off and red blood cells lysed by a short treatment with double-distilled water (ddH2O), followed by addition of 1/10 volume of 10× PBSA to restore isotonic pH and three washes in PBSA. Splenocytes were counted on a hemocytometer using trypan blue stain and resuspended to a concentration of 2 × 106 cells/ml in DMEM supplemented with 10% FBS, antibiotics, and antimycotics.
Immune assays.
PBMCs, MLN cells, and splenocytes were cultured in 96-well tissue culture plates (Thermo Fisher Scientific, Rochester, NY) in a final volume of 200 μl DMEM supplemented with 5% FBS, antibiotics, and antimycotics. Separate 96-well plates were set up to allow two separate stimulations, one for measurement of lymphocyte proliferation (2 × 105 cells/well in triplicate) and the other to quantify IFN-γ secretion (5 × 105 cells/well in duplicate). For each lymphoid tissue, cultures were stimulated with either medium alone, 1 μg/ml M. avium subsp. paratuberculosis lysate, or 1 μg/ml concanavalin A (Sigma-Aldrich Canada), and the cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2. IFN-γ secretion into the culture supernatants was measured 48 h after stimulation with a capture enzyme-linked immunosorbent assay (ELISA), as described previously (21). Proliferation assay culture plates were incubated for 5 days. During the last 6 h of culture, 20 μl [3H]thymidine (GE Healthcare Biosciences, Pittsburg, PA) (0.4 μCi per well) was added to each well. Plates were freeze-thawed to lyse cells and harvested using a Microplate cell harvester (model C961961; Perkin-Elmer, Waltham, MA). Radioactivity was detected using a Top Count NXT beta scintillation counter (model C9912V1; Perkin-Elmer, Waltham, MA). Average counts per minute (cpm) were calculated for each set of triplicate cultures, and stimulation indices (SIs) were calculated by dividing average cpm for cells stimulated with M. avium subsp. paratuberculosis lysate by average cpm for cells cultured in medium alone.
Immunoblotting.
Total M. avium subsp. paratuberculosis lysate prepared in PBS containing protease inhibitors as described above was supplemented with concentrated Laemmli SDS-PAGE buffer to 1× with a final concentration of 2.5% β-mercaptoethanol. The lysate was heated for 5 min at 95°C and then cooled to room temperature. Lysate samples (1.25 μg/lane) were separated by SDS-PAGE in a 1.0-mm Tris-glycine minigel and blotted onto a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked with 5% skim milk–Tris-buffered saline (TBS)–0.05% Tween 20. Bovine sera collected prior to and after experimental infection were diluted 1:100 in 5% skim milk–TBS–Tween 20 before being applied to each blot. Blots were washed, and bound antibody was detected with alkaline phosphatase (AP)-conjugated goat anti-bovine IgG(H+L) antibody (KPL Laboratories, Gaithersburg, MD), followed by SigmaFast 5-bromo-4-chloro-3-indolylphosphate (BCIP)–nitroblue tetrazolium (NBT) (Sigma-Aldrich, St. Louis, MO). Blots were dried and scanned with an HP Scanjet G4050 at 600 dpi (grayscale).
Kinome array experiments.
The design, construction, and application of the bovine peptide arrays were carried out essentially as previously described (13, 22, 23). The kinome arrays used were specifically designed for analysis of signaling involved in immune function and were previously applied to bovine monocytes infected with Mycobacterium avium subsp. paratuberculosis (12, 13). Kinome array experiments were all performed within a single assay on the same day to minimize technical and interassay variance. Briefly, ileal intestinal tissue samples flash-frozen and stored at −80°C were crushed in liquid nitrogen using a ceramic mortar and pestle. Pulverized tissue in liquid nitrogen was transferred to a tared sterile 1.5-ml microcentrifuge tube, the liquid nitrogen was allowed to evaporate, and the tissue was weighed. An 80-μl aliquot of ice-cold lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, 1 mM NaF, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mM PMSF, 1 μg/ml pepstatin A, and 2 mM dithiothreitol [DTT] [all products from Sigma-Aldrich unless otherwise indicated]) was added per 15 mg of tissue and vortexed to mix. This homogenate was further diluted 8-fold in ice cold lysis buffer, vortexed thoroughly, and incubated on ice for 20 min to allow adequate lysis. Lysates were spun in a microcentrifuge for 10 min at 4°C. This lysis procedure produced intestinal extracts with protein concentrations of ∼1.0 to 1.5 mg/ml, as determined by the Bio-Rad Bradford method. A 70-μl aliquot of the supernatant was mixed with 10 μl of activation mix (50% glycerol, 500 μM ATP [New England BioLabs, Pickering, Ontario, Canada], 60 mM MgCl2, 0.05% vol/vol Brij 35, 0.25 mg/ml bovine serum albumin [BSA]) and then incubated on the peptide array for 2 h at 37°C. Arrays were then washed with PBS–1% Triton.
Slides were submerged in phospho-specific fluorescent ProQ Diamond phosphoprotein stain (Invitrogen) with agitation for 1 h before washing three times in destain solution containing 20% acetonitrile (EMD Biosciences [VWR distributor], Mississauga, Ontario, Canada) and 50 mM sodium acetate (Sigma) at pH 4.0 for 10 min. A final wash with distilled deionized H2O was done before arrays were air dried for 20 min and centrifuged at 300 × g for 2 min to remove any remaining moisture from the array. Arrays were read using a GenePix Professional 4200A microarray scanner (MDS Analytical Technologies, Toronto, Ontario, Canada) at 532 to 560 nm with a 580-nm filter to detect dye fluorescence. Images were collected using the GenePix 6.0 software (MDS). Spot intensity signals were collected as the mean of pixel intensity using the local feature background intensity calculation with the default scanner saturation level.
Kinome data analysis.
The extent of phosphorylation of each peptide was determined as previously described (24). Briefly, local background intensities were subtracted from foreground intensities, and the resulting measurements were transformed using the variance-stabilizing normalization (VSN) (25) method to bring all the arrays onto the same scale and to eliminate variance-versus-mean dependence. The resulting data set contained the transformed signal intensities associated with each of 300 peptides for the lysates from the different compartments within each animal, excluding the distal infected compartment from animal 1 and the proximal infected compartment from animal 4, which did not yield readable array results. Each array contained three intra-array (technical) replicates for each peptide.
As described previously (24), a chi-square test was used to identify peptides that exhibited significant technical variability, which were then excluded from subsequent analyses. Because a given animal had both treatment and control samples taken from it, the signal intensities for each infected intestinal compartment were subtracted from those of the corresponding uninfected compartment in the same animal. These will be referred to as “biological subtractions.” The resulting values are presented as relative changes in kinase activity within the same animal. This approach minimizes interanimal variability, which can be significant in outbred cattle, and facilitates comparison of kinome responses to infection among animals.
Hierarchical clustering was used to group the samples according to the similarity of their kinome profiles. Euclidean distance was used as the distance metric, while complete linkage was used as the linkage method. A heat map was generated wherein the columns represent samples, the rows represent peptides, and the color of each cell represents the relative level of phosphorylation for a specific peptide in a specific sample. A dendrogram representing the hierarchical clustering results for the samples is shown above the heat map. The heat maps were generated using the R function heatmap.2 from the gplots package.
Using the R function prcomp, principal-component analysis (PCA) was used to reduce the dimensionality of the kinome data. Specifically, the first three principal components (PC1, PC2, and PC3), which explain the greatest amount of variation in the kinome data, were determined. The value of each of these variables was determined for each control-subtracted compartment, and the correlations between each variable and two measures of immune response were determined as described below. The Euclidean distance was also calculated between each pair of biological subtractions. Specifically, let Ai represent the signal intensity of some treatment (e.g., distal ileum) minus the signal intensity of the control for peptide i in the same animal, and let Bi represent a different treatment minus intra-animal control subtraction. The Euclidean distance between these two biological subtractions is then . The relationship between host kinome responses to M. avium subsp. paratuberculosis infection and immune responses of cells draining the site of infection was investigated by plotting principal components (PC1, PC2, and PC3) derived from intra-animal control-subtracted kinome profiles as a function of cellular responses to M. avium subsp. paratuberculosis lysate (IFN-γ secretion in pg/ml) and proliferation as measured by stimulation index (SI). Linear regression analysis was performed with Prism 5 for Mac OSX version 5.0b with default parameters.
Analysis of differentially phosphorylated peptides.
A given peptide was selected for further analysis if two conditions were true: first, the peptide had to be consistently phosphorylated according to the chi-square test for both the treatment and the control conditions; second, the P value resulting from a t test between the transformed treatment intensities and the transformed control intensities had to be less than 0.2. While 0.2 may seem like a liberal threshold, when doing pathway analysis, it is more important to avoid false negatives than to avoid false positives. This is due to the fact that several peptides are involved in the same biological pathway, and we are trying to identify as many of those peptides as possible. Even with a liberal P value threshold, it is unlikely that several peptides from the same biological pathway will be erroneously identified as differentially phosphorylated when that pathway is really not affected by the treatment under investigation; however, it increases the chances that peptides from pathways that really are affected by the treatment will be identified as differentially phosphorylated. An analysis of the impact of different P value thresholds on false-negative probabilities can be found in the supplemental text and in Fig. S1 in the supplemental material.
For peptides meeting the above two conditions, fold change (FC) values were calculated using the formula 2d, where d = averagetreatment − averagecontrol, as previously described (24). For visual interpretation, these peptides were input with their FC values into Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA) to generate top canonical pathways with color-coded measures of relative FC values. Figures for canonical pathways were generated in IPA. Peptide lists were also uploaded to InnateDB (www.innatedb.ca), a publically available analysis resource that predicts biological pathways overrepresented in a data set and assigns a probability value (P) based on the number of proteins present for a particular pathway as well as the degree to which they are differentially expressed or modified relative to a control condition. Pathway analysis was performed in InnateDB using different FC cutoffs (1.0, 1.5, and 2.0), and pathways overrepresented for each cutoff were compiled into lists for each control-subtracted compartment. To identify common pathways overrepresented in infected compartments from the same animal and to compare with infected compartments from other animals, Venn analysis was performed with the pathway names. Common pathways of interest were further investigated by comparing individual players by Venn analysis. To uncover data trends not apparent through pathway overrepresentation analysis, gene ontology analysis (also in InnateDB) was performed by testing different FC cutoffs (1.0, 1.5, and 2.0), compiling lists, and finding common ontologies for responders and nonresponders using Venn analysis. Individual peptides that were significantly differentially phosphorylated in relation to intra-animal controls for all 6 infected compartments were also identified by Venn analysis.
RESULTS
M. avium subsp. paratuberculosis infection of ileal compartments.
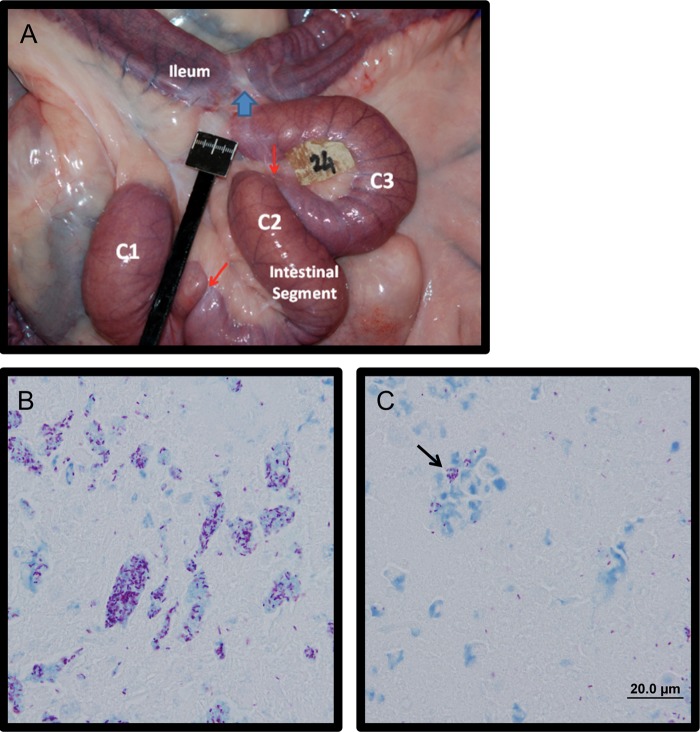
We previously reported that surgically isolated intestinal segments prepared in 2-week-old calves could be stably maintained for up to 11 months (18), and M. avium subsp. paratuberculosis infection remained localized to individual compartments for >9 months (19). Here, isolated segments were divided into three compartments, and the two most distal compartments were each infected with 1 × 108 to 3 × 108 CFU of M. avium subsp. paratuberculosis, while the most proximal compartment was injected with PBS and maintained as an uninfected control. The uninfected compartment provided an intra-animal tissue reference for comparing responses to M. avium subsp. paratuberculosis challenge while controlling for changes that occur when the intestine is surgically isolated (18). This model also facilitated isolation of cells from the specific mesenteric lymph node (MLN) draining the site of infection for assaying immune responses to M. avium subsp. paratuberculosis antigens. A clinical veterinarian observed the animals daily throughout the course of infection, and no significant changes in body temperature, weight, feed intake, or consistency of feces were noted. Gross examination of intestinal segments at the time of collection revealed no gross abnormalities (Fig. 1A). Acid-fast bacilli were frequently observed in the intestinal contents of each infected compartment but not in those of uninfected compartments (Fig. 1B). Furthermore, acid-fast bacilli were also frequently observed within cell remnants present within the intestinal lumen (Fig. 1C, arrow) but were not detected within the mucosa or submucosa of the intestine. The lack of acid-fast bacilli in the intact intestinal tissue at 1 month postinfection mirrored our observation at 9 months postinfection (19). We have previously observed early infiltration of tissues at 1, 3, and 5 days after infection under the same conditions (unpublished observations), but this early infiltration does not necessarily lead to large numbers of detectable acid-fast bacilli in the mucosa at later time points. Still, intestinal tissues within infected compartments were positive for M. avium subsp. paratuberculosis after 9 months of infection as determined by PCR (19), indicating that the host-pathogen interaction is maintained in this infection model for at least 9 months.
Fig 1.
Bovine calf intestines at 1 month after in vivo M. avium subsp. paratuberculosis infection. (A) Gross appearance of a surgically isolated distal intestinal segment in situ at 1 month postinfection. The surgically isolated segment was subdivided into three compartments, C1, C2, and C3, using silk ligatures (indicated by red arrows). The site where intestine proximal and distal to the isolated segment was anastomosed together is indicated with a blue arrow. (B) Ziehl-Neelsen stain of intestinal contents at 1 month after infection, showing diffuse aggregates of acid-fast Mycobacterium avium subsp. paratuberculosis at a magnification of ×100. (C)Acid fast-bacteria observed within cell remnants within the intestinal contents of an infected compartment (black arrow).
Immune responses to M. avium subsp. paratuberculosis infection.
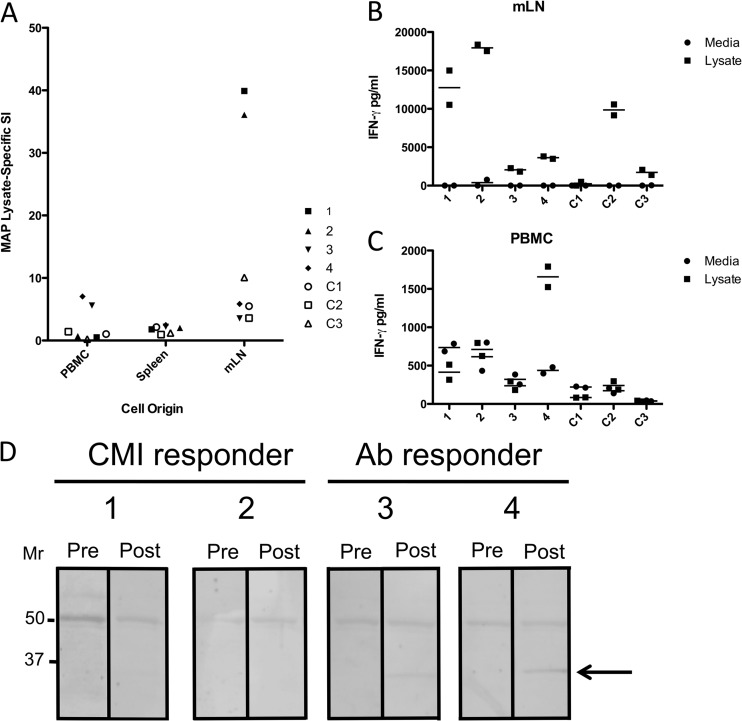
Cells isolated from the blood (PBMCs), spleen, and mesenteric lymph node (MLN) (draining the site of infection) from each calf were incubated with 1 μg/ml M. avium subsp. paratuberculosis lysate, and lymphocyte proliferation (Fig. 2A) and IFN-γ secretion (Fig. 2B and C) were measured. In addition to the young age of the calves when infected, the use of MLN cells specifically draining the site of infection allowed greater confidence in the specificity of the responses observed, as any environmental bacteria in the ingesta encountered during the month by the calves would not be sampled by the lymph node draining the isolated segment (19). PBMCs from animals 3 and 4 responded with significant proliferation responses to lysate (SIs of 5.6 and 7.0, respectively), while PBMCs from animals 1 and 2 did not show significant responses. Splenocytes from all infected animals failed to proliferate in response to lysate, but MLN cells from animals 1 and 2 displayed strong proliferative responses. In contrast, MLN cells from animal 3 and 4 displayed weak proliferative responses. M. avium subsp. paratuberculosis lysate-specific IFN-γ secretion from MLN cells mirrored the results observed for proliferation, where cells from animals 1 and 2 secreted high levels of IFN-γ, while MLN cells from animals 3 and 4 secreted much lower levels (Fig. 2B). Only PBMCs from animal 4 showed significant IFN-γ secretion (Fig. 2C), consistent with the proliferation observed for PBMCs from the same animal (Fig. 2A). We have observed differential immune responses similar to those seen here in 4 other calves at the 1-month time point after intestinal infection of the jejunum (unpublished observations), increasing our confidence that distinct immune responses can be recapitulated in the model used.
Fig 2.
Cell-mediated and antibody immune responses of M. avium subsp. paratuberculosis-infected calves to M. avium subsp. paratuberculosis lysates. (A) Stimulation index (SI) versus cell origin for peripheral blood mononuclear cells, spleens, and mesenteric lymph nodes of four M. avium subsp. paratuberculosis-infected calves (1, 2, 3, and 4) compared to three uninfected control calves (C1, C2, and C3). (B and C) IFN-γ (pg/ml) secreted by MLN cells (B) and PBMCs (C) in response to medium or total M. avium subsp. paratuberculosis lysate from the same calves (separately stimulated duplicate well values and means are shown). (D) Immunoblots of serum collected prior to (Pre) and 1 month after (Post) experimental M. avium subsp. paratuberculosis infection against total M. avium subsp. paratuberculosis lysates. A protein of ∼35 kDa detected with postinfection sera from responder calves 3 and 4 is indicated with an arrow.
To determine if an antibody response to M. avium subsp. paratuberculosis proteins could be detected, we performed immunoblots against lysates using serum samples collected before and 1 month after infection (Fig. 2). Both pre- and postinfection sera reacted with a band near 50 kDa, but sera from two animals reacted with an ∼35-kDa band after infection (animals 3 and 4). PBMCs isolated from the same two animals (3 and 4) showed significant proliferation and IFN-γ responses to M. avium subsp. paratuberculosis lysate (Fig. 2A). Calves that lacked antibodies reactive to the 35-kDa protein at 1 month after infection (animals 1 and 2) (Fig. 2D) showed strong proliferation and IFN-γ responses by MLN cells (Fig. 2A) but not in PBMCs. Therefore, a dichotomy in M. avium subsp. paratuberculosis-specific immune responses was observed when comparing mucosal and systemic responses.
Kinome analysis of M. avium subsp. paratuberculosis-infected ileum.
Tissue samples collected from infected and uninfected compartments were lysed, and the lysates were applied to bovine-specific kinome arrays designed as previously described (22) and processed using established methods (24). Three separate compartments (2 infected and 1 uninfected) were analyzed from each animal (n = 4), and of the 12 compartments assayed, 10 samples provided readable results. The failed samples were from infected compartments of animals 1 and 4 (distal and proximal, respectively). For each of the treatment-control combinations, the majority of the 300 peptides (cardinality range of 280 to 294) exhibited consistent levels of phosphorylation among the technical replicates on the same array for both the treatment array and the control array. Of these peptides, those with a t test P value of less than 0.2 (cardinality range of 112 to 184) were chosen for subsequent analysis to focus on the most significantly altered peptides while retaining many of the target sequences.
Hierarchical clustering and distance calculations.
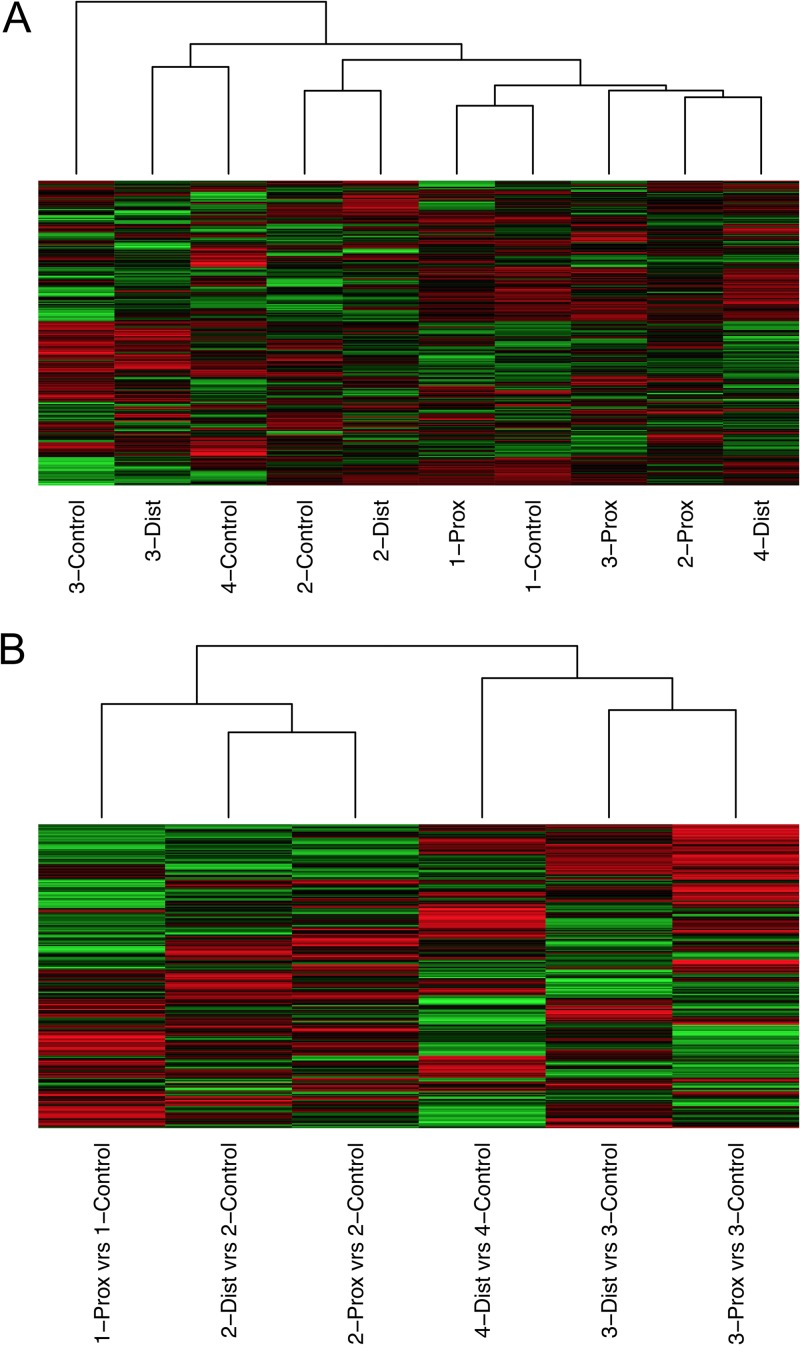
The processed kinome data were subjected to hierarchical clustering using Euclidean distance as the distance metric and complete linkage as the linkage method. Kinome data sets for each intestinal compartment clustered without a clear pattern prior to subtraction of biological controls (Fig. 3A). We have previously observed that individual animals have distinct basal kinase activities and that these distinctions must be accounted for before comparing treatments across animals (12, 13, 23). By considering the response of the treated condition relative to that of the control in the same animal, it was possible to determine and compare responses across animals. When intensity values for uninfected control compartments were subtracted from values for the infected compartments, the resulting data sets clustered perfectly by animal (Fig. 3B). Across the animals, there appeared to be two separate clusters (Fig. 3B). Animals 1 and 2 had the smallest Euclidean distance between the kinome profiles of their control-subtracted compartments, while the distance between those of animals 1 and 3 was the largest (Table 1). These results indicate that animals 1 and 2 shared similar kinomic responses to infection, which were in turn distinct from those shared by animals 3 and 4 (Fig. 3A).
Fig 3.
Kinome analysis of M. avium subsp. paratuberculosis-infected ileal compartments in calves. (A) Hierarchical clustering analysis to ascertain relationships between kinome responses observed in all intestinal compartments analyzed. (B) Hierarchical clustering analysis of the same responses in panel A after subtraction from responses observed for noninfected compartments in the same animal.
Table 1.
Euclidean distances between normalized intensity values for peptides represented on the kinome arraysa
| Compartments |
Euclidean distance | |
|---|---|---|
| 1 | 2 | |
| 2 Prox | 2 Dist | 11.43 |
| 1 Prox | 2 Dist | 14.10 |
| 3 Prox | 3 Dist | 14.42 |
| 1 Prox | 2 Prox | 15.23 |
| 2 Dist | 4 Dist | 15.99 |
| 1 Prox | 3 Dist | 16.54 |
| 3 Prox | 3 Dist | 16.96 |
| 2 Prox | 4 Dist | 17.05 |
| 2 Dist | 3 Dist | 17.78 |
| 3 Dist | 4 Dist | 18.61 |
| 3 Prox | 4 Dist | 18.71 |
| 1 Prox | 4 Dist | 19.59 |
| 2 Dist | 3 Prox | 21.16 |
| 2 Prox | 3 Prox | 21.25 |
| 1 Prox | 3 Prox | 23.03 |
Prox, proximal compartment; Dist, distal compartment. Euclidean distances between control-subtracted compartments were calculated as described in Materials and Methods.
Linear regression analysis of kinome profiles versus cellular responses to M. avium subsp. paratuberculosis lysates.
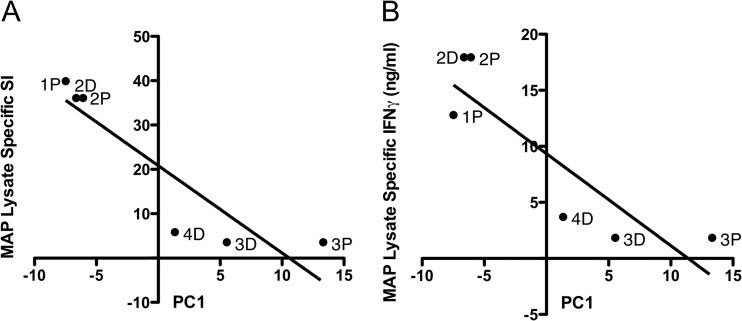
We observed that the animals that clustered together in the hierarchical clustering analysis after subtraction of uninfected control responses (Fig. 3B) also showed similar M. avium subsp. paratuberculosis lysate-specific proliferation, IFN-γ, and antibody responses (Fig. 2). To further examine this relationship, PCA was carried out, allowing us to represent much of the variability in the kinome data by three variables, denoted PC1, PC2, and PC3. PC1 captured a large proportion of the variability (approximately 31%), and the value of PC1 for each control-subtracted compartment was plotted as a function of M. avium subsp. paratuberculosis-lysate specific proliferation (Fig. 4A) and IFN-γ secretion (Fig. 4B) of MLN-derived cells. Linear regression analysis confirmed a significant negative linear correlation between PC1 and both M. avium subsp. paratuberculosis lysate-specific SI (P = 0.014) (Fig. 4A) and IFN-γ secretion (P = 0.023) (Fig. 4B). Neither PC2 nor PC3 showed a significant linear relationship with the MLN cell responses. These results revealed that significant differences in the overall intestinal kinome profiles of infected compartments from animals 1 and 2 or animals 3 and 4 could be correlated with the distinct responses of cells from these animals to M. avium subsp. paratuberculosis lysates.
Fig 4.
Relationships of kinome variability to cell-mediated immune responses. (A) Principal component 1 (PC1) of kinome variability for each compartment in each animal versus stimulation index (P = 0.014; r2 = 0.81) (P, PC1 for proximal infected compartment; D, PC1 for distal infected compartment. (B) PC1 versus IFN-γ secretion (ng/ml) (P = 0.023; r2 = 0.77).
Analysis of kinome array data.
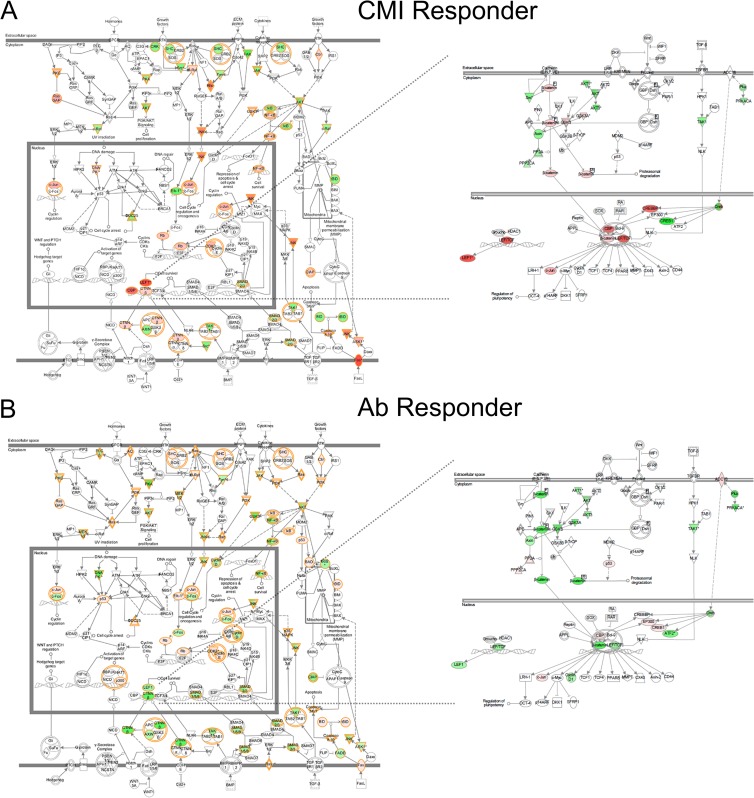
As mentioned above, hierarchical clustering of the kinome data, as well as the Euclidean distance calculations, indicated that the infected compartments from the same animals clustered closest together, followed by a close grouping of animals 1 and 2 (cell-mediated immunity [CMI] responders) distinct from animals 3 and 4 (antibody responders) (Fig. 3B; Table 1). Peptides that were significantly phosphorylated or dephosphorylated relative to the uninfected control (P < 0.2) were compiled for the different groupings (CMI responders versus antibody responders) and compared by Venn analysis (Fig. 5). The greatest similarity was observed between antibody responders (animals 3 and 4), as predicted from the clustering (61 shared significantly altered peptides). Of the 13 peptides that were consistently differentially phosphorylated across animals, 4 belong to a pathway called “inactivation of gsk3 by AKT causes accumulation of β-catenin” (Pathway Interaction Database [PID] BioCarta 4022). To observe trends in the peptides that differed between the groups, data sets (P < 0.2) were uploaded to Ingenuity pathway analysis (IPA) for visual comparisons. IPA determines the top canonical pathways represented within each data set, and it identified “molecular mechanisms of cancer” as the top canonical pathway for all 6 infected compartments analyzed. This allowed for an overall visual comparison of the top pathway for all the data sets in terms of increased or decreased phosphorylation (Fig. 6A and B). It was immediately apparent that animals 1 and 2 showed distinctly higher phosphorylation of many players within this set of pathways than animals 3 and 4, where decreases in phosphorylation predominated for the same targets. Peptides corresponding to the transcription factor LEF-1 and several other players in the Wnt/β-catenin pathway were highly phosphorylated by M. avium subsp. paratuberculosis-infected ileal lysates from animals 1 and 2 relative to uninfected ileal lysates from the same animal. Conversely, the same targets were less phosphorylated by infected intestinal lysates from animals 3 and 4 relative to their intra-animal controls (Fig. 6, right panel, and Table 2). The opposite was true for some of the peptides representing proteins involved in the Wnt/β-catenin pathway, such as ADCY8 and p300 (Table 2).
Fig 5.
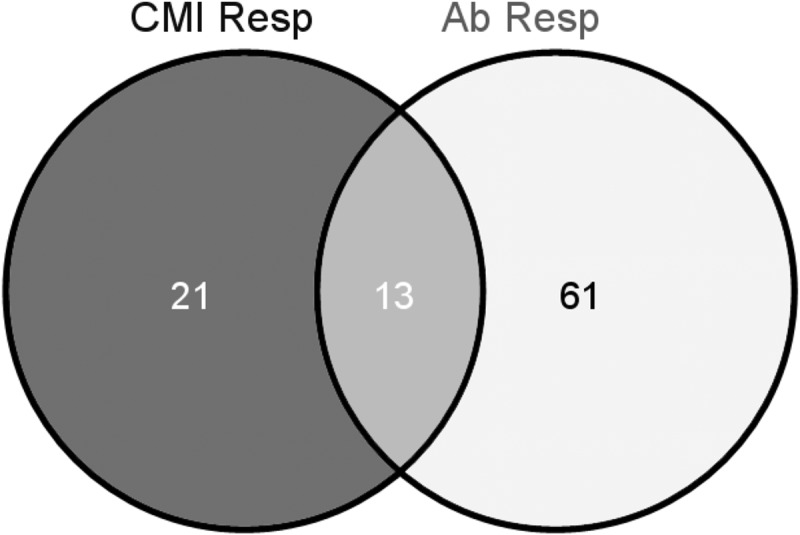
Venn analysis of significantly phosphorylated or dephosphorylated peptides shared between cell-mediated immune responder calves (CMI Resp) and antibody responder calves (Ab Resp).
Fig 6.
Ingenuity pathway analysis (IPA) of kinome profiles, showing top canonical pathway differences between cell-mediated immune responder (CMI Responder) and antibody responder (Ab Responder) calves. The intensity of the color depicts the relative increase (red) or decrease (green) in phosphorylation. (A) Left panel, CMI responder top pathway. Right panel, zoomed-in view of Wnt/β-catenin pathway for the same analysis. (B) Left panel, Ab responder top pathway. Right panel, zoomed-in view of Wnt/β-catenin pathway for the same analysis. Analyses shown are for the animal 2 proximal M. avium subsp. paratuberculosis-infected compartment (CMI responder) and the animal 3 proximal M. avium subsp. paratuberculosis-infected compartment (Ab responder).
Table 2.
Select CREB and Wnt/β-catenin pathway peptide phosphorylation sites differentially phosphorylated by M. avium subsp. paratuberculosis-infected intestinal lysates from CMI responder and antibody responder calvesa
| Compartment | CMI responders |
Antibody responders |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LEF1 (Q9UJU2); T155 or S166 (↑) |
β-Catenin (P35222); Y142, T41/45, or S675 (↑) |
ADCY8 (P40145) Y406 (↑) |
p300 (Q09472); S893 or S1834 (↑) |
PP2CA (P67775); T304/7 (↑) |
||||||
| FC | P | FC | P | FC | P | FC | P | FC | P | |
| 1 Prox | 1.5 | 0.08 | 1.7 | 0.13 | — | — | −1.6 | 0.10 | 1.2 | 0.31 |
| 2 Prox | 5.5 | 0.04 | 1.6 | 0.06 | −1.4 | 0.36 | −1.6 | 0.23 | −1.4 | 0.1 |
| 2 Dist | 3.9 | 0.07 | 1.7 | 0.18 | −1.0 | 0.48 | −1.5 | 0.04 | −1.5 | 0.3 |
| 3 Prox | −2.0 | 0.03 | −3.4 | 0.0004 | 16 | 0.02 | 6.2 | 0.005 | 1.5 | 0.04 |
| 3 Dist | −1.8 | 0.26 | −3.2 | 0.02 | 6.4 | 0.04 | 3.4 | 0.04 | 2.3 | 0.12 |
| 4 Dist | −2.2 | 0.05 | −1.8 | 0.006 | 6.5 | 0.01 | 2.0 | 0.1 | 1.2 | 0.14 |
The fold change (FC) and probability value from a t test between the transformed treatment (infected lysate) intensities and the transformed control (uninfected lysate) intensities for the indicated peptide (Uniprot accession numbers are given in parentheses) and residue(s) are shown. Prox, proximal compartment; Dist, distal compartment. —, inconsistently phosphorylated.
Pathway overrepresentation analysis and gene ontology overrepresentation analysis are often used to focus on specific pathways altered within a homogenous cell population. Here we used these methods to identify trends in the global tissue profile of kinase activity against the array peptides. Significantly increased or decreased phosphorylation of multiple targets within a similar biological process or pathway provides greater confidence in the trends observed but must also be considered with caution because of the averaging of the represented kinases during whole-tissue lysis. We did not find significantly altered common pathways or gene ontologies shared across all 6 infected compartments, but we did identify several pathways and ontologies shared among infected compartments from animals with similar kinome profiles. Animals 1 and 2 exhibited increased phosphorylation of players in the interleukin-1 (IL-1) and transforming growth factor β (TGF-β) signaling through TAK1 pathways, while animals 3 and 4 showed increased phosphorylation of players in the IL-6, natural killer cell-mediated cytotoxicity, and IL-4 signaling pathways (Table 3). Gene ontology overrepresentation analysis revealed significantly increased phosphorylation of players in the innate immune response and decreased phosphorylation of players in peptidyl-tyrosine phosphorylation for lysates of infected compartments from animals 1 and 2 (Table 4). Gene ontologies overrepresented in the arrays exposed to lysates from animals 3 and 4 included increased phosphorylation of players involved in epidermal growth factor (EGF) receptor signaling and decreased phosphorylation of players in the Wnt receptor signaling pathway, matching the observed decrease in phosphorylation of Wnt pathway players by visual assessment of top canonical pathways using IPA.
Table 3.
Pathway overrepresentation analysis of CMI responder and antibody responder calves and associated probabilities of upregulation as determined by InnateDBa
| Compartment | CMI responders |
Antibody responders |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-1 (NETPATH 10429) (↑) |
TGF-β (INOH 10330) (↑) |
IL-6 (NETPATH 10415) (↑) |
NK cell (KEGG 578) (↑) |
IL-4 (NETPATH 10417) (↑) |
|||||||||||
| ↑ | ↓ | P | ↑ | ↓ | P | ↑ | ↓ | P | ↑ | ↓ | P | ↑ | ↓ | P | |
| 1 Prox | 11 | 4 | 0.06 | 6 | 0 | 0.004 | 5 | 5 | 0.66 | 3 | 3 | 0.55 | 6 | 7 | 0.53 |
| 2 Prox | 10 | 4 | 0.05 | 3 | 1 | 0.24 | 7 | 7 | 0.61 | 4 | 5 | 0.66 | 7 | 5 | 0.81 |
| 2 Dist | 7 | 3 | 0.26 | 5 | 1 | 0.08 | 6 | 7 | 0.37 | 2 | 6 | 0.92 | 4 | 4 | 0.47 |
| 3 Prox | 9 | 7 | 0.84 | 3 | 4 | 0.92 | 22 | 9 | 0.04 | 12 | 1 | 0.01 | 13 | 2 | 0.14 |
| 3 Dist | 8 | 7 | 0.89 | 2 | 4 | 0.94 | 17 | 8 | 0.12 | 9 | 5 | 0.30 | 10 | 4 | 0.11 |
| 4 Dist | 9 | 10 | 0.87 | 4 | 5 | 0.79 | 19 | 8 | 0.06 | 13 | 4 | 0.05 | 15 | 5 | 0.08 |
The number of peptides showing increased ↑ or decreased ↓ phosphorylation and associated probabilities of upregulation (↑) for the indicated path (database identification numbers are in parentheses) are shown. Prox, proximal compartment; Dist, distal compartment.
Table 4.
Gene ontology analysis of CMI responders and antibody responders and associated probabilities of up- or downregulation as determined by InnateDBa
| Compartment | CMI responders |
Antibody responders |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Innate immune response (GO:0045087) (↑) |
Peptidyl-tyrosine phosphorylation (GO:0018108) (↓) |
Positive regulation of DNA replication (GO:0045740) (↑) |
Epidermal growth factor receptor signaling (GO:0007173) (↑) |
Wnt receptor signaling pathway (GO:0016055) (↓) |
|||||||||||
| ↑ | ↓ | P | ↑ | ↓ | P | ↑ | ↓ | P | ↑ | ↓ | P | ↑ | ↓ | P | |
| 1 Prox | 17 | 11 | 0.07 | 2 | 4 | 0.37 | — | — | — | 2 | 3 | 0.80 | 0 | 0 | 1 |
| 2 Prox | 23 | 17 | 0.10 | 3 | 8 | 0.08 | 2 | 1 | 0.38 | 3 | 4 | 0.63 | 2 | 2 | 0.53 |
| 2 Dist | 27 | 15 | 0.03 | 3 | 7 | 0.09 | 1 | 0 | 0.53 | 3 | 1 | 0.28 | 1 | 0 | 1 |
| 3 Prox | 33 | 21 | 0.40 | 7 | 3 | 0.79 | 4 | 0 | 0.09 | 9 | 1 | 0.02 | 1 | 4 | 0.07 |
| 3 Dist | 22 | 16 | 0.77 | 7 | 2 | 0.86 | 5 | 0 | 0.05 | 9 | 3 | 0.01 | 1 | 2 | 0.25 |
| 4 Dist | 34 | 23 | 0.32 | 5 | 4 | 0.86 | 4 | 0 | 0.07 | 9 | 2 | 0.09 | 1 | 5 | 0.06 |
The number of peptides and associated probabilities of upregulation (↑) or downregulation (↓) for the indicated ontology (database identification numbers are in parentheses) are shown. Prox, proximal compartment; Dist, distal compartment. —, ontology was not represented in the analysis.
DISCUSSION
We previously used kinome arrays to address how M. avium subsp. paratuberculosis modulates host signaling in isolated infected bovine monocytes (12, 13). Here, we infected surgically isolated intestinal compartments with a controlled dose of M. avium subsp. paratuberculosis and collected intestinal tissue for analysis at 1 month postinfection. Kinome arrays were used to measure global changes in tissue kinase activity relative to intra-animal naive control intestinal compartments. The control-subtracted whole-tissue kinome profiles cluster by animal and then by the type of cell-mediated or antibody responses mounted against M. avium subsp. paratuberculosis lysate. A small number of the proteins represented on the arrays (13) were significantly differentially phosphorylated in the infected compartments compared to uninfected compartments across all 6 biological replicates. However, major differences between pathways, players, and ontologies were observed for animals that showed different immune response profiles, suggesting that global intestinal kinome profiles reflect different host responses following M. avium subsp. paratuberculosis infection.
Kinome analysis of a homogenous cell population, such as M. avium subsp. paratuberculosis-infected monocytes, and kinome profiles of whole-tissue lysates provide distinct information regarding host responses. When studying a homogenous cell population, the kinome arrays can provide insight into specific cell signaling pathways directly altered by M. avium subsp. paratuberculosis infection (12, 13). In contrast, kinome data from tissue lysates reflect the average effective kinase activity within the diverse cell populations sampled, including both direct and indirect effects, and must be understood as a tissue signature rather than representative pathways for a single cell type. Assaying the overall kinase activities within a tissue and subtracting these values from those for an intra-animal control allows for identification of the most significantly altered kinase activities throughout the tissue. The potential exists that these averaged kinase activities will fail to detect important kinase activity occurring in relatively rare cells such as dendritic cells or macrophages that perform critical functions in defining host-pathogen interactions.
Whole-tissue kinase activity measurements in some ways resemble whole-tissue gene expression profiling, where consistent sampling is important (26), and results must be interpreted as overall averages of expression. Tissue gene expression has been useful for comparative studies of cancers (27) that can yield valuable in vivo insights (16). To date, significant success has been achieved for tissue studies focused on kinase activity (28), especially in studies of brain (29, 30), where high numbers of distinct cell types are represented in a given tissue sample.
The kinome profiles generated with whole intestinal samples indicated that M. avium subsp. paratuberculosis infection can result in divergent kinase activity in different calves. The distinct tissue kinase signatures of different animals could be related to differential recruitment of specific immune effector cell populations (19). Differences in kinome profiles are also likely influenced by the unique genotypes of outbred calves, and it is likely that a broader range of kinome profiles will be observed as more animals are studied. Genotypes may confer enhanced susceptibility or resistance to M. avium subsp. paratuberculosis infection (9), but the exact linkages are complex and remain undefined. On the other hand, kinase activity provides a more direct measure of phenotypic responses and may provide more effective parameters to identify cattle that mount protective immune responses to infection. Kinome responses may also uncover links to help better characterize protective genotypes. It is understood that M. avium subsp. paratuberculosis subverts the host immune system by inhibiting phagosome-lysosome fusion in macrophages that take up the bacterium (31). Immune evasion by M. avium subsp. paratuberculosis involves a variety of other mechanisms, including evading cell-mediated immunity through enhancing secretion of suppressor cytokines, activating T-regulatory cells, inhibiting tumor necrosis factor alpha (TNF-α) expression, and inhibiting cytotoxic killing of infected cells (32). One of the antigens recognized by M. avium subsp. paratuberculosis infected cattle is a 35-kDa major membrane protein (MMP) (33, 34) that most likely corresponds to the protein detected with sera from two of the calves in the current study (Fig. 2). Antibodies do not appear to confer protection against progression of the primarily intracellular infection (32). While we cannot predict which early responses to M. avium subsp. paratuberculosis observed at 1 month postinfection would be the most effective for clearance of the pathogen, longer-term studies that correlate early responses with chronic infection or clearance may reveal the most effective early responses. Kinome profiling may be one way to distinguish protective versus nonprotective early responses to M. avium subsp. paratuberculosis infection.
Due to the small sample size, it is difficult to determine the significance of specific differences in kinase activities among the animals exhibiting different immune responses following M. avium subsp. paratuberculosis infection. Further studies with more animals will be required to more confidently determine the kinomic correlates of specific immune responses and more fully define variations that may be influenced by other environmental factors. However, the general increase in phosphorylation of Wnt/β-catenin, IL-1, and TGF-β (through TAK1) pathway proteins in animals that showed strong MLN proliferation and IFN-γ responses to M. avium subsp. paratuberculosis lysate compared to animals that did not show MLN responses to lysates warrants further investigation. Similarly, the increased phosphorylation of IL-6, NK cell, and IL-4 pathway players in animals that showed antibody responses but not MLN proliferation responses to lysate indicates global differences in kinase activity at the site of infection that may reflect either protective or nonprotective responses to M. avium subsp. paratuberculosis infection.
Interestingly, a recent short-term study (0.5, 1, 2, 4, 8, and 12 h) of experimental M. avium subsp. paratuberculosis infection in ileal compartments revealed shifts in gene expression patterns in 4 calves over the course of infection (35). The authors did not comment on differences between calves, perhaps because very-early-stage responses to infection are less variable. However, the shifts they observed included early suppression (at 0.5 and 1 h postinfection) of the Wnt receptor signaling pathway through β-catenin, followed by late-phase activation of the same pathway (at 12 h postinfection) (among several other pathways). They also observed “late-phase” activation of genes implicated in innate immune response gene ontology, which they suggested to be indicative of an effective immune response. Here, we observed increased phosphorylation of innate immune response genes by ileal lysates collected 1 month after infection, most significantly in CMI responder animals, and decreased phosphorylation of Wnt receptor signaling pathway players in animals that failed to mount local M. avium subsp. paratuberculosis-specific CMI but did mount antibody immune responses. These observations seem to suggest a divergence of kinase activity at 1 month postinfection that varied among individual animals. Further studies will be necessary to determine if specific kinase activities may correlate with effective bovine responses to M. avium subsp. paratuberculosis infection and exactly what interval after infection is required before animal-specific differences may be observed. The correlation of specific immune responses against lysates with general kinase activity at the site of infection provides a method to evaluate the effectiveness of host evasion by M. avium subsp. paratuberculosis and may uncover strategies to promote pathogen clearance.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the Saskatchewan Agriculture Development Fund (ADF). Philip Griebel is a holder of a Tier I CRC in Mucosal Immunology, which is funded by the Canadian Institutes of Health Research.
We are grateful to the VIDO Animal Care staff for surgeries, veterinary care, animal handling, and help with sample collection. We thank Donna Dent for help with IFN-γ ELISAs, Chris Stuart from the Western College of Veterinary Medicine for help with microscopy, and Natasa Arsic for help with isolation of splenocytes. We also thank Qingxiang Yan for statistical assistance.
Footnotes
Published ahead of print 28 May 2013
This article is VIDO journal series number 665.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00339-13.
REFERENCES
- 1. Waters WR, Miller JM, Palmer MV, Stabel JR, Jones DE, Koistinen KA, Steadham EM, Hamilton MJ, Davis WC, Bannantine JP. 2003. Early induction of humoral and cellular immune responses during experimental Mycobacterium avium subsp. paratuberculosis infection of calves. Infect. Immun. 71:5130–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu CW, Livesey M, Schmoller SK, Manning EJ, Steinberg H, Davis WC, Hamilton MJ, Talaat AM. 2007. Invasion and persistence of Mycobacterium avium subsp. paratuberculosis during early stages of Johne's disease in calves. Infect. Immun. 75:2110–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sweeney RW, Collins MT, Koets AP, McGuirk SM, Roussel AJ. 2012. Paratuberculosis (Johne's disease) in cattle and other susceptible species. J. Vet. Intern. Med. 26:1239–1250 [DOI] [PubMed] [Google Scholar]
- 4. Marce C, Ezanno P, Seegers H, Pfeiffer DU, Fourichon C. 2011. Within-herd contact structure and transmission of Mycobacterium avium subspecies paratuberculosis in a persistently infected dairy cattle herd. Prev. Vet. Med. 100:116–125 [DOI] [PubMed] [Google Scholar]
- 5. Giese SB, Ahrens P. 2000. Detection of Mycobacterium avium subsp. paratuberculosis in milk from clinically affected cows by PCR and culture. Vet. Microbiol. 77:291–297 [DOI] [PubMed] [Google Scholar]
- 6. Ellingson JL, Anderson JL, Koziczkowski JJ, Radcliff RP, Sloan SJ, Allen SE, Sullivan NM. 2005. Detection of viable Mycobacterium avium subsp. paratuberculosis in retail pasteurized whole milk by two culture methods and PCR. J. Food Prot. 68:966–972 [DOI] [PubMed] [Google Scholar]
- 7. Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. 2011. Innate immune recognition of Mycobacterium tuberculosis. Clin. Dev. Immunol. 2011:405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koets AP, Adugna G, Janss LL, van Weering HJ, Kalis CH, Wentink GH, Rutten VP, Schukken YH. 2000. Genetic variation of susceptibility to Mycobacterium avium subsp. paratuberculosis infection in dairy cattle. J. Dairy Sci. 83:2702–2708 [DOI] [PubMed] [Google Scholar]
- 9. Minozzi G, Williams JL, Stella A, Strozzi F, Luini M, Settles ML, Taylor JF, Whitlock RH, Zanella R, Neibergs HL. 2012. Meta-analysis of two genome-wide association studies of bovine paratuberculosis. PLoS One 7:e32578. 10.1371/journal.pone.0032578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arsenault R, Griebel P, Napper S. 2011. Peptide arrays for kinome analysis: new opportunities and remaining challenges. Proteomics 11:4595–4609 [DOI] [PubMed] [Google Scholar]
- 11. Knight JD, Pawson T, Gingras AC. 2013. Profiling the kinome: Current capabilities and future challenges. J. Proteomics 81:43–55 [DOI] [PubMed] [Google Scholar]
- 12. Arsenault RJ, Li Y, Bell K, Doig K, Potter A, Griebel PJ, Kusalik A, Napper S. 2012. Mycobacterium avium subsp. paratuberculosis inhibits gamma interferon-induced signaling in bovine monocytes: insights into the cellular mechanisms of Johne's disease. Infect. Immun. 80:3039–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arsenault RJ, Li Y, Maattanen P, Scruten E, Doig K, Potter A, Griebel P, Kusalik A, Napper S. 2013. Altered Toll-like receptor 9 signaling in Mycobacterium avium subsp. paratuberculosis-infected bovine monocytes reveals potential therapeutic targets. Infect. Immun. 81:226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kilpinen S, Ojala K, Kallioniemi O. 2010. Analysis of kinase gene expression patterns across 5681 human tissue samples reveals functional genomic taxonomy of the kinome. PLoS One 5:e15068. 10.1371/journal.pone.0015068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hildebrandt MA, Tan W, Tamboli P, Huang M, Ye Y, Lin J, Lee JS, Wood CG, Wu X. 2012. Kinome expression profiling identifies IKBKE as a predictor of overall survival in clear cell renal cell carcinoma patients. Carcinogenesis 33:799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grzmil M, Morin P, Jr, Lino MM, Merlo A, Frank S, Wang Y, Moncayo G, Hemmings BA. 2011. MAP kinase-interacting kinase 1 regulates SMAD2-dependent TGF-beta signaling pathway in human glioblastoma. Cancer Res. 71:2392–2402 [DOI] [PubMed] [Google Scholar]
- 17. Whittington RJ, Begg DJ, de Silva K, Plain KM, Purdie AC. 2012, Comparative immunological and microbiological aspects of paratuberculosis as a model mycobacterial infection. Vet. Immunol. Immunopathol. 148:29–47 [DOI] [PubMed] [Google Scholar]
- 18. Charavaryamath C, Fries P, Gomis S, Bell C, Doig K, Guan LL, Potter A, Napper S, Griebel PJ. 2011. Mucosal changes in a long-term bovine intestinal segment model following removal of ingesta and microflora. Gut Microbes 2:134–144 [DOI] [PubMed] [Google Scholar]
- 19. Charavaryamath C, Gonzalez-Cano P, Fries P, Gomis S, Doig K, Scruten E, Potter A, Napper S, Griebel PJ. 2013. Host Responses to Persistent Mycobacterium avium subspecies paratuberculosis infection in surgically isolated bovine ileal segments. Clin. Vaccine Immunol. 20:156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whale TA, Beskorwayne TK, Babiuk LA, Griebel PJ. 2006. Bovine polymorphonuclear cells passively acquire membrane lipids and integral membrane proteins from apoptotic and necrotic cells. J. Leukoc. Biol. 79:1226–1233 [DOI] [PubMed] [Google Scholar]
- 21. Raggo C, Habermehl M, Babiuk LA, Griebel P. 2000. The in vivo effects of recombinant bovine herpesvirus-1 expressing bovine interferon-gamma. J. Gen. Virol. 81:2665–2673 [DOI] [PubMed] [Google Scholar]
- 22. Jalal S, Arsenault R, Potter AA, Babiuk LA, Griebel PJ, Napper S. 2009. Genome to kinome: species-specific peptide arrays for kinome analysis. Sci. Signal. 2:pl1. 10.1126/scisignal.254pl1 [DOI] [PubMed] [Google Scholar]
- 23. Arsenault RJ, Jalal S, Babiuk LA, Potter A, Griebel PJ, Napper S. 2009. Kinome analysis of Toll-like receptor signaling in bovine monocytes. J. Recept. Signal. Transduct. Res. 29:299–311 [DOI] [PubMed] [Google Scholar]
- 24. Li Y, Arsenault RJ, Trost B, Slind J, Griebel PJ, Napper S, Kusalik A. 2012. A systematic approach for analysis of peptide array kinome data. Sci. Signal. 5:pl2. 10.1126/scisignal.2002429 [DOI] [PubMed] [Google Scholar]
- 25. Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1):S96–S104 [DOI] [PubMed] [Google Scholar]
- 26. Mutch DM, Tordjman J, Pelloux V, Hanczar B, Henegar C, Poitou C, Veyrie N, Zucker JD, Clement K. 2009. Needle and surgical biopsy techniques differentially affect adipose tissue gene expression profiles. Am. J. Clin. Nutr. 89:51–57 [DOI] [PubMed] [Google Scholar]
- 27. Sanz-Pamplona R, Berenguer A, Cordero D, Riccadonna S, Sole X, Crous-Bou M, Guino E, Sanjuan X, Biondo S, Soriano A, Jurman G, Capella G, Furlanello C, Moreno V. 2012. Clinical value of prognosis gene expression signatures in colorectal cancer: a systematic review. PLoS One 7:e48877. 10.1371/journal.pone.0048877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Borst MH, Diks SH, Bolbrinker J, Schellings MW, van Dalen MB, Peppelenbosch MP, Kreutz R, Pinto YM, Navis G, van Goor H. 2007. Profiling of the renal kinome: a novel tool to identify protein kinases involved in angiotensin II-dependent hypertensive renal damage. Am. J. Physiol. Renal Physiol. 293:F428–F437 [DOI] [PubMed] [Google Scholar]
- 29. Sikkema AH, Diks SH, den Dunnen WF, ter Elst A, Scherpen FJ, Hoving EW, Ruijtenbeek R, Boender PJ, de Wijn R, Kamps WA, Peppelenbosch MP, de Bont ES. 2009. Kinome profiling in pediatric brain tumors as a new approach for target discovery. Cancer Res. 69:5987–5995 [DOI] [PubMed] [Google Scholar]
- 30. Hoozemans JJ, Hilhorst R, Ruijtenbeek R, Rozemuller AJ, van der Vies SM. 2012. Protein kinase activity profiling of postmortem human brain tissue. Neurodegener. Dis. 10:46–48 [DOI] [PubMed] [Google Scholar]
- 31. Woo SR, Heintz JA, Albrecht R, Barletta RG, Czuprynski CJ. 2007. Life and death in bovine monocytes: the fate of Mycobacterium avium subsp. paratuberculosis. Microb. Pathog. 43:106–113 [DOI] [PubMed] [Google Scholar]
- 32. Coussens PM. 2004. Model for immune responses to Mycobacterium avium subspecies paratuberculosis in cattle. Infect. Immun. 72:3089–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bannantine JP, Huntley JF, Miltner E, Stabel JR, Bermudez LE. 2003. The Mycobacterium avium subsp. paratuberculosis 35 kDa protein plays a role in invasion of bovine epithelial cells. Microbiology 149:2061–2069 [DOI] [PubMed] [Google Scholar]
- 34. Shin SJ, Chang CF, Chang CD, McDonough SP, Thompson B, Yoo HS, Chang YF. 2005. In vitro cellular immune responses to recombinant antigens of Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 73:5074–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khare S, Lawhon SD, Drake KL, Nunes JE, Figueiredo JF, Rossetti CA, Gull T, Everts RE, Lewin HA, Galindo CL, Garner HR, Adams LG. 2012. Systems biology analysis of gene expression during in vivo Mycobacterium avium paratuberculosis enteric colonization reveals role for immune tolerance. PLoS One 7:e42127. 10.1371/journal.pone.0042127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.