Abstract
Brucella melitensis causes brucellosis, a disease affecting sheep, cattle, and sometimes humans. Attenuated B. melitensis strain M5-90, derived from virulent strain M28, is widely used as a live vaccine in ruminants in China. Genetic differences between the strains may cast light on the mechanism of attenuation. We recently reported the complete genomic sequences of M28 and M5-90. Genome organization is highly conserved between these isolates, and also with virulent strains 16 M and ATCC 23457. Analysis revealed 23 open reading frames (ORFs) with consistent differences between M5-90 and the virulent strains. Notably, the tuf2 gene encoding translation elongation factor EF-Tu from M5-90 contained 50 single nucleotide polymorphisms (SNPs) and 9 gaps (indels) compared to tuf2 of M28 or of the other virulent strains. There were no changes in tuf1. To evaluate the potential role of EF-Tu in pathogenesis, tuf1 and tuf2 mutants of M28 and an M5-90 strain harboring wild-type tuf2 were constructed, and their virulence/attenuation was evaluated in vivo. We report that the tuf2 gene plays an important role in the attenuation of M5-90 virulence.
INTRODUCTION
Brucella melitensis, the first species in the genus Brucella to be described, causes abortion in goats and sheep and Malta fever in humans. Currently, there are nine recognized species of the bacterium Brucella, based on host preferences and phenotypic differences (1). The six classically recognized species of this facultative intracellular parasite are B. melitensis (sheep and goats), B. abortus (cattle), B. suis (pigs, reindeer, and hares), B. canis (dogs), B. ovis (sheep), and B. neotomae (desert wood rats) (2). In humans, B. melitensis is the most highly pathogenic, and infection typically takes place via the consumption of unpasteurized milk and milk products. Attenuated B. melitensis M5-90, derived from virulent strain M28, is widely used as a live vaccine in sheep, goats, and cattle in China (3, 4). However, such strains retain pathogenicity for humans and to date no effective vaccine has been approved for human use. Because of its epidemic potential, the absence of a human vaccine, and the effectiveness of aerosol infection, this agent is classified as a biosafety level 3 pathogen and has been designated a potential bioterrorism agent (5).
The genome of the first Brucella melitensis strain to be sequenced, 16 M, comprises ∼3.3 Mb of unique sequence shared between two circular chromosomes: the larger chromosome (chromosome I) is ∼2.1 Mb, whereas chromosome II is ∼1.2 Mb. Eleven complete genome sequences representing six species of Brucella are currently available, and the genomes of several other Brucella strains/species are in the process of being sequenced (1).
The different genomes are very similar in size and in the numbers of genes and proteins. Sequence comparisons have provided firm evidence of horizontal gene transfer between different Brucella species and strains, despite their predominantly intracellular lifestyle, and phylogenomic analysis of orthologous gene families has demonstrated distinct radiations (6). Importantly, sequence analysis revealed that Brucella species lack classical virulence determinants such as capsules, fimbriae, exotoxins, cytolysins, resistant forms, antigenic variation, plasmids, or lysogenic phages. Compared to other bacterial pathogens, relatively little is known about the factors contributing to Brucella pathogenesis and persistence in the host (7), and previous studies focused primarily on the identification of factors that govern intracellular trafficking and multiplication of the bacterium within its host cell (7).
B. melitensis vaccine smooth strain M5-90 (biovar 1) was generated by serial passage of the virulent B. melitensis M28 ovine isolate through chickens, followed by acriflavine treatment and a further 90 passages in chicken embryo fibroblasts; this isolate was found to be markedly attenuated (8), and its use as a live vaccine is one of the key factors leading to a rapid decline in the incidence of brucellosis in animals and humans in China in the 1970s to 1990s (3, 4).
Genetic differences between virulent and derivative attenuated strains are likely to provide clues to the mechanism(s) of attenuation and therefore assist in the development of new vaccines. We recently reported the complete genome sequences of B. melitensis M28 and M5-90, strains with different virulence backgrounds (9). Comparative sequence analysis reported here revealed 23 open reading frames (ORFs) with consistent differences between the complete genome sequences of the attenuated M5-90 strain versus those of both the parent strain M28 and two other virulent strains, 16 M and ATCC 23457. Notably, there was a consistent difference between the vaccine strain and the three virulent strains in the sequence of the tuf2 gene, encoding translation elongation factor EF-Tu, a highly conserved protein (10). B. melitensis chromosome I harbors two gene copies encoding EF-Tu, termed tuf1 and tuf2. We report that tuf1 sequences are conserved between all complete genome sequences and the tuf1 gene of vaccine strain M5-90 (BM590_A1236) was identical to the equivalent gene (BM28_A1261) of the virulent parent strain M28. In contrast, sequences corresponding to the tuf2 gene of the vaccine strain M5-90 were found to be extensively disrupted and contained multiple SNPs and indels compared to the tuf2 gene (BM28_A1246) of M28. This analysis therefore suggests that disruption of the tuf2 gene could potentially contribute to attenuation of B. melitensis. To address this possibility, we constructed derivatives of virulent strain M28 in which tuf2 was replaced by a selective marker, as well as an M5-90 strain harboring a wild-type tuf2 gene. We report that these modifications significantly modulate the attenuation of B. melitensis M28 pathogenicity in mice, arguing that the tuf2 gene plays an important role in the attenuation of M5-90.
MATERIALS AND METHODS
Ethics statement and animal experimentation.
Care of laboratory animals and animal experimentation were in accordance with the Beijing Administration Guidelines for the Use of Laboratory Animals. All animal studies were approved by the Review Board of Harbin Veterinary Research Institute and by the Animal Care and Use Committee of Heilongjiang Province [SYXK(H)2006-032]. Animal studies with Brucella-infected BALB/c mice were conducted in a biosecurity level 3+ laboratory approved by the Chinese Ministry of Agriculture.
Strain propagation.
B. melitensis strain M28 and the attenuated strain M5-90 (laboratory stock) were grown on tryptic soy broth (TSB) plates containing 1% agar under biosecurity level 3+ conditions (8). After 2 days at 37°C, cells were collected by scraping and were suspended in phosphate-buffered saline (PBS; pH 7.2). CFU were determined by plating on TSB agar and incubating for 3 days at 37°C under 5% CO2.
Genomic comparisons.
B. melitensis M28 and M5-90 complete genome sequences (chromosomes I and II) are deposited in the GenBank database under accession numbers CP002459-CP002460 and CP001851-CP001852, respectively (9). SNP differences between the genomes of M5-90 and the virulent strains M28, 16 M, and ATCC 23457 (11) were identified using pairwise chromosome alignment data generated using the Mauve2.3.0 alignment program (12). Reciprocal pairwise alignments were also performed using the basic local alignment tool (BLAST) version 2.2.21. All identified indels were confirmed by resequencing.
Construction of B. melitensis tuf mutant strains.
Mutant tuf strains of B. melitensis were constructed by separately replacing the tuf1 and tuf2 genes of M28 with a kanamycin resistance marker and flanking homology arms, generating strains M28-tuf1 and M28-tuf2, respectively. In view of flanking sequence homologies, the same construct based on M28 tuf1 and tuf2 flanking sequences permitted targeting the tuf1 and tuf2 genes of M5-90. The M5-90 strain harboring a further disruption of the tuf2 gene is designated M90-tuf2. An M5-90 back mutation strain was reconstituted in which the wild-type tuf2 gene (with the selective marker) was inserted into the tuf2 locus, generating strain M90-rtuf2. Primers for the construction of recombinant plasmids and for the identification of mutant strains are listed in Table S1 in the supplemental material. Mutant strains were characterized by light microscopy after acriflavine or Gram staining and by using the R serum agglutination test (13). The method of Gibby and Gibby (14) was employed to generate proliferation curves for the parental and mutant strains.
In vivo pathogenicity analysis.
BALB/c female mice aged 4 to 5 weeks were purchased from Vital River Laboratories (VRL; Beijing, China). Separate groups of mice (n = 20 in each case) were injected intraperitoneally (i.p.). with 105 CFU of each strain. Mice were sacrificed after 1, 2, 3, and 4 weeks (n = 5 in each group), and spleen weights were measured. Spleen CFU titers of viable B. melitensis were determined by direct plating. Statistical analyses used the Student two-tailed t test; a P value of ≤0.05 was taken to indicate statistical significance.
RESULTS
Global alignment of genome sequences.
The complete genome sequences of B. melitensis M28 and M5-90 predicted 3363 and 3361, respectively, protein-coding genes, 55 tRNA genes, and three rRNA operons in the two genomes. The salient features of the genomes and comparisons with the 16 M and ATCC 23457 genomes are summarized in Table S2 in the supplemental material. Multiple alignments were performed for all four genomes. Global alignment revealed almost perfect colinearity. There were few indels (insertions or deletions) exceeding 30 bp in size. BLAST analysis revealed 54 indels of more than 30 bp in M5-90 relative to 16 M, 9 relative to ATCC 23457, and no indels of >30 bp relative to M28. Overall, the M5-90 genome harbored 382 indels relative to 16 M, 110 relative to ATCC 23457, and 73 relative to M28 (Table 1).
Table 1.
Indels between B. melitensis vaccine strain M5-90 and the virulent strains M28, 16 M, and ATCC 23457a
| M5-90 | M28 |
16 M |
ATCC 23457 |
|||
|---|---|---|---|---|---|---|
| Chr I | Chr II | Chr I | Chr II | Chr I | Chr II | |
| Insertion (+) | 27 | 14 | 121 | 82 | 30 | 24 |
| Deletion (−) | 18 | 14 | 103 | 76 | 36 | 20 |
| Within ORF | 18 | 13 | 106 | 84 | 24 | 19 |
Insertions (+) or deletions (−) relative to M5-90 with a size of ≤30 bp. Chr, chromosome; ORF, open reading frame.
Pyrosequencing technology is unequalled in the accuracy of the sequences generated (15); predominant sequencing errors are single-nucleotide indels rather than substitutions (16). We therefore resequenced and confirmed all 73 indels detected between vaccine strain M5-90 and the parent strain M28. Multiple sequence alignments revealed that 31 indels, within 22 ORFs, were consistently different between vaccine strain M5-90 and the three virulent counterparts. Table 1 lists all confirmed indels within ORFs between M5-90 and the three virulent strains.
Evolutionary analysis.
To determine the evolutionary relationships between the four B. melitensis strains, we based our analysis on pairwise SNP differences between the complete genome sequences of the four strains. The Mauve whole-genome comparison tool was used to identify SNPs within shared sequences of the genomes of B. melitensis strains M5-90, M28, 16 M, and ATCC 23457; these are summarized in Table 2. Comparison of the M5-90 and M28 genomes revealed only 125 SNP differences (0.004%). The SNPs were not equally distributed between the two chromosomes. Whereas SNP differences between M5-90 and 16 M or ATCC 23457 were spread equally across the genome, comparison of M5-90 versus M28 revealed that, of the 125 confirmed SNP differences, 115 (92%) were located on chromosome I, whereas the 10 remaining SNPs (8%) were located on chromosome II; moreover, all chromosome II SNPs were located in intergenic regions. By comparison, there were 372 SNP differences (0.011%) between M5-90 and ATCC 23457 and 2,959 SNPs (0.092%) between M5-90 and 16 M.
Table 2.
Single-nucleotide polymorphisms (SNPs) between B. melitensis vaccine strain M5-90 and virulent strains M28, 16 M, and ATCC 23457
| Virulent strain | No. of SNP differences in virulent strains versus M5-90 |
M5-90 base | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome I |
Chromosome II |
||||||||
| A | C | G | T | A | C | G | T | ||
| M28 | |||||||||
| 6 | 17 | 4 | 0 | 0 | 0 | A | |||
| 8 | 9 | 8 | 0 | 1 | 0 | C | |||
| 14 | 16 | 11 | 2 | 3 | 0 | G | |||
| 4 | 11 | 7 | 1 | 0 | 3 | T | |||
| ATCC 23457 | |||||||||
| 17 | 50 | 8 | 4 | 17 | 1 | A | |||
| 14 | 13 | 26 | 2 | 6 | 12 | C | |||
| 44 | 20 | 18 | 19 | 4 | 4 | G | |||
| 6 | 43 | 11 | 6 | 22 | 4 | T | |||
| 16 M | |||||||||
| 87 | 307 | 46 | 42 | 181 | 18 | A | |||
| 90 | 101 | 338 | 51 | 59 | 173 | C | |||
| 339 | 86 | 94 | 178 | 52 | 48 | G | |||
| 37 | 307 | 108 | 27 | 147 | 43 | T | |||
Virulence-associated differences between the attenuated and virulent strains.
Of the 125 single-nucleotide changes between M5-90 and M28, 28 (22%) were located in intergenic regions and 97 (78%) were located within a total of 35 different ORFs. Comprehensive pairwise and reciprocal comparisons revealed that 23 of the 35 ORFs showed consistent sequence deviations (indels or SNPs) in M5-90 compared to the virulent strains (Table 3). Differences between M5-90, M28, and the other strains and their classification in the COG database are summarized in Table 4.
Table 3.
Indels and SNPs in M5-90 coding sequencesa
| COG category | M28 | M5-90 | M5-90 vs M28 | M5-90 vs 23457 | M5-90 vs 16 M | Function encoded |
|---|---|---|---|---|---|---|
| Not in COG | A0177 | A0176 | IS/PS▽△ | IS/PS▽△ | IS/PS▽△ | Sodium:dicarboxylate symporter, DUF540 |
| A0932 | A0929 | FS▽ | NO▽ | NO▽ | Transposase | |
| A0938 | A0932 | NTΔ | ISΔ | ISΔ | Conserved hypothetical protein | |
| A1528 | NO HIT | FS▽ | FS▽ | FS▽ | Conserved hypothetical protein | |
| B0001 | B0001 | FSΔ | FSΔ | NO | Replication initiation protein RepC | |
| B0564 | NO HIT | FSΔ | FSΔ | FSΔ | Conserved hypothetical protein | |
| B0885 | NO HIT | FSΔ | NOΔ | NOΔ | Hypothetical protein | |
| B0980 | NO HIT | FS▽ | FS▽ | FS▽ | Hypothetical protein | |
| C | A1137 | A1129 | NT▽ | NT▽ | NT▽ | Dehydrogenase, E1 component |
| B0107 | B0107/8 | IS▽ | IS▽ | NO▽ | O-antigen polymerase | |
| B0947 | B0946 | FS▽ | NO▽ | NO▽ | Alanine dehydrogenase | |
| E | A1463 | A1458 | NO1⊙ | NO1⊙ | NO1⊙ | 5,10-Methylenetetrahydrofolate reductase |
| A0904 | A0898 | FS▽ | FS▽ | FS▽ | tRNA (uracil-5-)-methyltransferase Gid | |
| A2028 | A2027 | NT▽ | NT▽ | NT▽ | Alcohol dehydrogenase (acceptor) | |
| B0699 | B0697 | FSΔ | NO | NO | Putative cytosolic protein | |
| H | A0776 | A0775 | YS1⊙ | YS1⊙ | YS1⊙ | Riboflavin biosynthesis protein Ribd |
| I | A1164 | A1156 | FS/IS▽Δ | NO | FS/PS▽Δ | Phosphatidate cytidylyltransferase |
| J | A1103 | A1095 | YS10⊙ | YS10⊙ | YS10⊙ | Queuine tRNA-ribosyltransferase |
| A1261 | A1255/7 | FS▽Δ50⊙ | FS▽Δ50⊙ | FS▽Δ50⊙ | Elongation factor Tu | |
| L | A0145 | NO HIT | FS/PSΔ | FS/PSΔ | FS/PSΔ | Transposase |
| A0989 | NO HIT | FS▽ | FS▽ | FS▽ | IS6501 transposase | |
| A1215 | A1208 | NTΔ | NO | NO | Recombination factor protein RarA | |
| B0522 | B0521/2 | IS▽ | NO▽ | NO▽ | Conserved hypothetical protein | |
| M | A0655 | A0654/5 | ISΔ | ISΔ | ISΔ | Lytic transglycosylase catalytic |
| B0388 | B0386 | NOΔ | NOΔ | NOΔ | LPS biosynthesis protein | |
| B0404 | B0402/3 | IS▽Δ | IS▽Δ | IS▽Δ | Conserved hypothetical protein | |
| O | A0277 | A0274 | NT▽ | NT▽ | NT▽ | Conserved hypothetical protein |
| P | A0508 | A0505 | NO1⊙ | NO1⊙ | NO1⊙ | Chloride channel core protein |
| R | A0422 | A0419 | NT▽ | NT▽ | NT▽ | Methyltransferase |
| B0697 | B0695 | FSΔ | FSΔ | FSΔ | Pyridine nucleotide-disulfide oxidoreductase | |
| B0963 | B0962 | NO▽ | NO▽ | NO▽ | WD-repeat family protein | |
| S | A0683 | A0683 | FS/PS▽ | FS/PS▽ | FS/PS▽ | Conserved hypothetical protein |
| A0936 | A0930 | ISΔ | ISΔ | ISΔ | Conserved hypothetical protein | |
| A1618 | NO HIT | FSΔ | FSΔ | FSΔ | Transcriptional regulator, Cro/cI family | |
| U | B0076 | B0076 | NOΔ | NOΔ | NOΔ | Nodulation protein Nfed, C terminus only |
Abbreviations and symbols: ⊙, single nucleotide substitution; ▽, deletion; Δ, insertion; FS, frameshift; IS, internal stop; NO, no difference; NT, N-terminal truncation; PS, premature stop; vs, versus; YS, SNP resulting in a change in the amino acid net charge; COG, clusters of orthologous groups (see Table 4). Boldface indicates ORFs with consistent differences between the complete genome sequences of the attenuated M5-90 strain versus the virulent strains.
Table 4.
COG-based functional categories of ORFs identified as differences between M5-90 and the virulent B. melitensis strain M28a
| COG category | Function | No. of different ORFs | No. of OCDs |
|---|---|---|---|
| None | Not in COG database | 8 | 5 |
| C | Energy production and conversion | 3 | 1 |
| E | Amino acid transport and metabolism | 4 | 2 |
| G | Carbohydrate transport and metabolism | 1 | 1 |
| H | Coenzyme metabolism | 1 | 1 |
| I | Lipid metabolism | 1 | 0 |
| J | Translation, ribosomal structure, and biogenesis | 1 | 1 |
| L | DNA replication, recombination, and repair | 4 | 2 |
| M | Cell envelope biogenesis, outer membrane | 3 | 2 |
| O | Posttranslational modification, protein turnover, chaperones | 1 | 1 |
| P | Inorganic ion transport and metabolism | 1 | 0 |
| R | General function prediction only | 3 | 3 |
| S | Function unknown | 3 | 3 |
| U | Intracellular trafficking and secretion | 1 | 1 |
| Total | 35 | 23 |
Abbreviations: COG, clusters of orthologous groups (http://www.ncbi.nlm.nih.gov/COG/); OCDs, ORFs with consistent differences.
Previous studies on the attenuated B. abortus S19 strain revealed four ORFs with consistent differences (OCDs) of more than 60 SNP or indel changes (17). These four ORFs are implicated in erythritol uptake or metabolism (BM590_B0823, BM590_B0829, BM590_B0830, and BM590_A0074), and one of them encodes a membrane protein (BM590_A0074). Inspection of the M5-90 sequence revealed that all four ORFs were fully conserved, arguing that the mechanism of attenuation in B. melitensis M5-90 differs from that in B. abortus S19.
For the identification of candidate virulence-associated ORFs, we therefore focused on the 23 chromosome I ORFs consistently differing between M5-90 and the virulent strains, including M28. Nineteen contained conservative substitutions and were not studied further, leaving four ORFs with single-nucleotide indels or SNPs that resulted in net amino acid changes. The queuine tRNA-ribosyltransferase (BM590_A1095) contained multiple alterations, and eight of 11 SNPs caused nonsynonymous codon replacements, although the ORF is conserved in M28. In contrast, the ORFs in M5-90 corresponding to the sodium:dicarboxylate symporter DUF540 (BM590_A0176) and conserved hypothetical protein (BM590_A0683) both contained changes leading to frameshift and premature termination.
This analysis focused our attention on genes encoding the translation elongation factor EF-Tu. B. melitensis contains two genes encoding EF-Tu, designated tuf1 and tuf2. No sequence changes were observed in the tuf1 gene, but remarkably, of the 115 SNPs located on chromosome 1, 50 (43.5%) fell within the region corresponding to the tuf2 coding sequence of M28 (BM28_A1261). Moreover, the M5-90 genomic sequence corresponding to tuf2 contained nine indels that split the tuf2 EF-Tu coding sequence into three adjacent ORFs (BM590_A1255, BM590_A1256, and BM590_A1257). In contrast, all three virulent strains contain an intact tuf2 gene. This observation suggests that disruption of the tuf2 gene in M5-90 may be directly linked to the reduced pathogenicity of this isolate.
Construction of tuf mutants of B. melitensis M28.
To address whether tuf2 disruption is causally linked to attenuation, we constructed deletion mutants of virulent strain M28. In parallel, we reconstituted the wild-type tuf2 gene in the M5-90 strain. We employed a standard method in which a kanamycin resistance cassette flanked by target sequences is electrotransformed into competent cells (18) followed by antibiotic selection for gene replacement by double recombination. Twenty kanamycin-resistant clones were studied in each case; inactivation of each gene was confirmed by PCR, and two M28 strains carrying separate disruptions of the two genes, named M28-tuf1 and M28-tuf2, were selected for study.
The same experiment was performed using the M5-90 strain. As expected, none of the kanamycin-resistant transformants contained a disruption of the tuf1 gene, confirming that tuf2 function is abolished in M5-90 and that EF-Tu is essential for growth. However, kanamycin-resistant derivatives of M5-90 in which the selective marker had inserted into the tuf2 locus (named M90-tuf2) were obtained. In addition, an M5-90 back mutation strain (named M90-rtuf2) in which the wild-type tuf2 gene together with the selective marker is inserted into tuf2 locus was reconstituted; successful reconstitution of tuf2 gene function was confirmed by PCR and sequencing.
The mutant strains were analyzed by light microscopy and the R serum agglutination test. All mutants were smooth B. melitensis strains, as are their parent strains (M28 and M5-90). We then studied their growth properties in vitro. As shown in Fig. 1, the proliferation curves of all strains showed a lag phase of ∼16 h, followed by a logarithmic growth phase, reaching a plateau after ∼40 h. There were no significant differences between the proliferation curves of the parental and mutant bacteria (P > 0.05), demonstrating that separate disruption of either tuf1 or tuf2 of M28 or reconstitution of wild-type tuf2 in M5-90 does not negatively impact upon growth in vitro.
Fig 1.
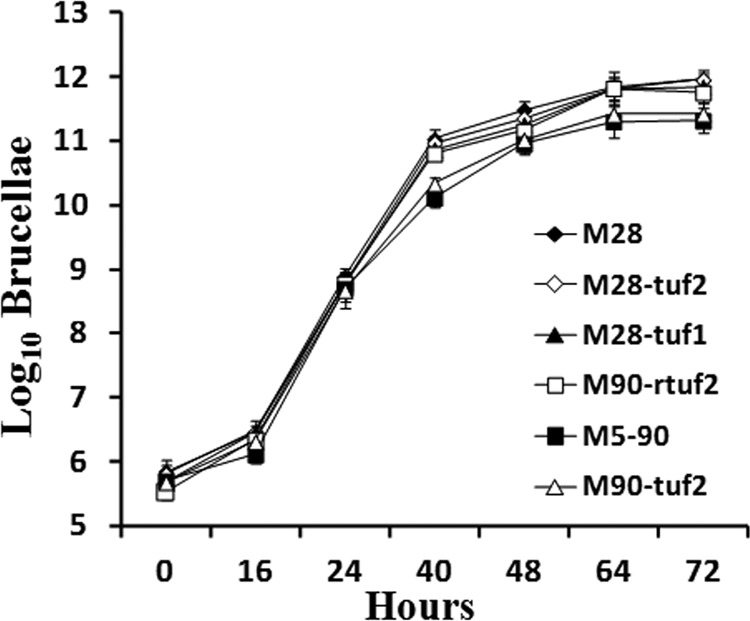
In vitro proliferation curves of M28, M5-90, and mutant strains. Shown are CFU titers plotted against time. Bacteria were grown on TSB medium at 37°C under biosecurity level 3+ conditions; CFU titers were determined by triplicate serial dilution and plating. Values are means, and error bars are standard deviations.
Virulence of tuf mutant strains in mice.
To determine whether tuf1 and/or tuf2 disruption affects attenuation/virulence, separate groups of mice were inoculated (i.p.) with 105 CFU of each of the B. melitensis strains. At different times following challenge, bacterial counts in spleen homogenates were determined by direct plating on agar. As shown in Fig. 2A, all strains showed initial replication in the spleen, with titers elevated at weeks 1 and 2 postinfection (p.i.) and subsequently falling. The attenuated strain M5-90 showed the lowest initial titer and a marked fall in titer between weeks 2 and 4 p.i. that was not seen for the other strains tested. As a live vaccine strain, M5-90 is attenuated but retains low pathogenicity in mice (Fig. 2A); clearing of M5-90 bacteria requires ∼3 months in mice, ∼4 months in goats/sheep, and ∼6 months in cattle (our unpublished data). As expected, further disruption of tuf2 in the M5-90 vaccine strain (M90-tuf2) had no detectable effect on proliferation in vivo versus the attenuated parent strain.
Fig 2.
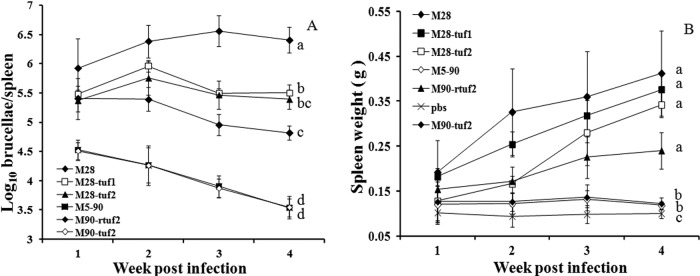
In vivo pathogenicity of M28, M5-90, and mutant strains in BALB/c mice. Different groups of animals were inoculated (i.p.) with 105 CFU of each strain, and spleens were collected at different times p.i. (A) Viable spleen homogenate titers (CFU) as determined by triplicate serial dilution and plating. (B) Mouse spleen weights at different times p.i. All data points are means (n = 5); error bars are standard deviations. Any groups that are not significantly different from one another will have the same lowercase letter on the right of each panel. Different letters indicates significant differences by Student's t test (P < 0.01).
The virulent strain M28 showed a very high initial titer over the course of the experiment. The tuf1 and tuf2 derivatives of M28, however, showed an intermediate level of virulence. Titers were significantly reduced (P < 0.001) and were generally ∼1 log below those achieved by parent strain M28. There was no significant difference between the degree of attenuation of M28-tuf1 and that of M28-tuf2. The back mutant M5-90 strain (M90-rtuf2) showed a level of attenuation that was intermediate between the M28 derivatives and M5-90 strain. Nevertheless, titers remained significantly higher (P < 0.01) than for the attenuated strain M5-90 (Fig. 2A).
A similar profile was seen when another marker of virulence, spleen weight, was compared between groups. As shown in Fig. 2B, spleen weights were largely unchanged in animals inoculated with the attenuated strain M5-90 or its tuf2-disrupted counterpart, whereas a robust increase in spleen weight was recorded in animals inoculated with the virulent strain M28. As before, disruption of either tuf1 or tuf2 in M28 led to a reduction in maximum spleen weight, and the reduction was greater in M28-tuf2 than in M28-tuf1. The reduction of spleen weights between M28 and its tuf derivatives was of high significance (P < 0.01 in both cases) (Fig. 2B), whereas there was no significant difference in spleen weight between the M28-tuf1 and M28-tuf2 strains at 2 to 4 weeks p.i. Moreover, a gradual increase in spleen weight (P < 0.01) was recorded in animals inoculated with the back mutant strain M90-rtuf2 reexpressing wild-type tuf2.
DISCUSSION
B. melitensis M5-90 is an artificially attenuated strain that has been used for decades on a large scale for brucellosis immunization of sheep and goats in China (3). Few studies have addressed the physiological mechanisms of M5-90 attenuation, and the molecular changes underlying the reduced virulence of the vaccine strain are unknown. We therefore determined the complete genome sequences of M5-90 and its parent strain M28. These were comprehensively compared with each other and with the established sequences of two other virulent strains of B. melitensis, 16 M and ATCC 23457. Pairwise and reciprocal comparisons identified 125 SNP and 73 indel differences between M5-90 and M28, and 35 coding sequences were consistently different between the attenuated M5-90 strain and the three virulent strains. Of these, 23 coding sequences were considered to represent candidate attenuation-related genes in view of the nonconservative nature of the changes.
Of these 23 candidates, we focused on the tuf2 gene encoding EF-Tu (BM28_A1261) because this single ORF contained 30% of all changes (SNPs and indels) that differentiate the M5-90 and M28 sequences. In contrast, the related tuf1 gene was fully conserved between M5-90 and M28. We used homologous recombination to construct derivatives of M28 in which the tuf1 and tuf2 genes were disrupted. It was not possible to construct a tuf1 derivative of M5-90 in view of the disruption of the tuf2 ORF in this strain. We also constructed a new back mutant M5-90 strain harboring a wild-type tuf2 gene. Gene disruption had no evident effect on growth in vitro or morphology. The parent strains and their tuf mutant derivatives were inoculated into mice. We report that both the M28-tuf1 and M28-tuf2 strains were significantly attenuated compared to M28 but were still more virulent than M5-90. Furthermore, the virulence of the M90-tuf2 strain harboring a wild-type copy of tuf2 was significantly enhanced versus that of M5-90, and the back mutation restored growth in mice toward M28 levels (Fig. 2). These results indicate that disruption of the tuf2 gene plays an important role in the attenuation of M5-90.
Translation elongation factor EF-Tu is an essential component of the translational machinery. Nevertheless, it may play additional roles. It has recently been demonstrated that EF-Tu is released/secreted from bacterial cells and that it can modulate pathogen-host interactions (19–24). However, the pathways by which EF-Tu protein might interfere with immune regulation in the host cell remain unknown. Studies on host proteins interacting with EF-Tu are likely to provide insights into its role in modulating pathogen-host interactions and Brucella pathogenesis.
Our results argue that disruption of tuf2 is likely to play a role in the attenuation of vaccine strain M5-90. However, tuf2 mutants of the virulent M28 strain, despite demonstrating reduced pathogenicity, remained significantly more pathogenic than the vaccine strain. This indicates that other functions that differ between M28 and M5-90 are likely to synergize with tuf2 disruption in attenuating the pathogenicity of M5-90.
The list of potential attenuation-related genes revealed by complete genome sequencing includes genes involved in DNA/RNA metabolism, amino acid transport and metabolism, and cell envelope biogenesis, as well as outer membrane proteins and several hypothetical proteins. Lipopolysaccharide (LPS) is an important virulence factor for Brucella survival and replication in the host (25), and the genes encoding O-antigen polymerase (BM590_B0107/8) and the LPS biosynthesis protein (BM590_B0386) may also play a role in attenuation/virulence. In particular, our analysis also highlighted queuine tRNA-ribosyltransferase (BM590_A1095), the sodium:dicarboxylate symporter DUF540 (BM590_A0176), and a conserved hypothetical protein (BM590_A0683) as potential attenuation-related genes. Further studies using the approach described here will be required to assess whether these changes contribute to the attenuation of the vaccine strain. We believe that systematic modification of these other functions, in conjunction with tuf2 mutants of M28, and host-response studies will together provide a comprehensive account of the attenuation of the M5-90 vaccine strain.
A systematic review of the literature by Crasta et al. (17) tabulated 236 potential virulence factors and an additional 53 genes previously implicated in Brucella virulence (26–38). Of these 289 potential virulence factors, only BM28_A0655 (encoding a soluble lytic murein transglycosylase), which is involved in amino sugar metabolism and N-glycan biosynthesis, showed consistent differences between M5-90 and the three virulent strains (28). Other prominent virulence factors in Brucella spp., including urease (ure), cytochrome oxidase (cydDCAB), nitric oxide reductase (norD), superoxide dismutase (sod), the type IV secretion system (T4SS), Brucella virulence factor A (bvfA), and a two-component regulatory system (bvrS-bvrR), are all intact in the M5-90 genome (7). Therefore, attenuation of M5-90 does not involve modification of these key functions that govern the ability of Brucella to adapt to the environmental conditions encountered in its intracellular replicative niche, including low levels of nutrients and oxygen, acidic pH, and exposure to reactive oxygen intermediates (39).
The profile of the sequence changes revealed by comparison of the M28 and M5-90 genomes is of interest. M5-90 was derived from M28 by a series of steps, including treatment with acriflavine (acridine orange). Molecules such as acriflavine exert their mutagenic action by intercalating between the bases within DNA, leading to the addition or deletion of single nucleotides. In consequence, acriflavine-induced mutations typically cause frameshifting within ORFs; such mutations give rise to out-of-phase translation and premature termination, leading to radical changes in the function of the encoded polypeptide (40). It is notable that the present study identified a total of 73 indels, of which the large majority (66 indels) are single-nucleotide insertions or deletions that produce frameshift mutations within 27 ORFs, a finding consistent with these mutations having been generated by acriflavine exposure. This contrasts with the situation in B. abortus, where only eight candidate attenuation-related genes in the spontaneously attenuated strain S19 were found to contain frameshifts versus both the 9–941 and 2308 virulent strains (17). Furthermore, of the 45 candidate attenuation-related genes identified in B. abortus S19, none were altered between M5-90 and any one of the three virulent B. melitensis strains M28, 16 M, and ATCC 23457 (17). This suggests that the mechanisms of attenuation in B. melitensis M5-90 and in B. abortus S19 are significantly different and indicates that acriflavine mutagenesis (as in M5-90) may lead to a distinct spectrum of attenuation-related genetic changes.
In conclusion, complete genomic sequencing of M5-90 and M28 has highlighted a series of candidate attenuation-related genes, including tuf2. Inactivation of this gene in virulent strain M28 led to significant attenuation in vivo. Conversely, restoration of wild-type tuf in the attenuated M5-90 strain significantly increased in vivo virulence. These findings argue that disruption of tuf2 in M5-90 contributes to the reduced pathogenicity of the vaccine strain. Future research will need to address the complex role of EF-Tu in the life cycle of Brucella; further studies will also be required to identify other changes that synergize with tuf2 in attenuating the virulence of the vaccine strain.
Supplementary Material
ACKNOWLEDGMENTS
Financial support was provided by the Animal Infectious Disease Control Research Program of the Ministry of Agriculture of China (grants 200803018 and 200903027), the Chinese National S&T plan (grant 2009ZXI0007-214), The Chinese Academy of Agricultural Sciences Basal Research Fund (0302013009), the State Key Laboratory of Veterinary Biotechnology (grant SKLVBF201206), and the Natural Science Foundation of China (grant 31001068). The genomic sequencing and bioinformatics work described here was performed under the auspices of the Shanghai-Ministry Key Laboratory of Disease and Health Genomics, Chinese National Human Genome Center.
Footnotes
Published ahead of print 28 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00224-13.
REFERENCES
- 1. Seleem MN, Boyle SM, Sriranganathan N. 2009. Brucellosis: a re-emerging zoonosis. Vet. Microbiol. 140:392–398 [DOI] [PubMed] [Google Scholar]
- 2. Corbel MJ. 1997. Brucellosis: an overview. Emerg. Infect. Dis. 3:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deqiu S, Donglou X, Jiming Y. 2002. Epidemiology and control of brucellosis in China. Vet. Microbiol. 90:165–182 [DOI] [PubMed] [Google Scholar]
- 4. Cosivi O, Corbel MJ. 1998. WHO consultation on the development of new/improved brucellosis vaccines. 17 December 1997, Geneva, Switzerland. Biologicals 26:361–363 [DOI] [PubMed] [Google Scholar]
- 5. Kaufmann AF, Meltzer MI, Schmid GP. 1997. The economic impact of a bioterrorist attack: are prevention and postattack intervention programs justifiable? Emerg. Infect. Dis. 3:83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wattam AR, Williams KP, Snyder EE, Almeida NF, Jr, Shukla M, Dickerman AW, Crasta OR, Kenyon R, Lu J, Shallom JM, Yoo H, Ficht TA, Tsolis RM, Munk C, Tapia R, Han CS, Detter JC, Bruce D, Brettin TS, Sobral BW, Boyle SM, Setubal JC. 2009. Analysis of ten Brucella genomes reveals evidence for horizontal gene transfer despite a preferred intracellular lifestyle. J. Bacteriol. 191:3569–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seleem MN, Boyle SM, Sriranganathan N. 2008. Brucella: a pathogen without classic virulence genes. Vet. Microbiol. 129:1–14 [DOI] [PubMed] [Google Scholar]
- 8. Research Group of Brucellosis (Harbin Veterinary Research Institute) 1991. Study on the Brucella melitensis strain M5-90 vaccine. Chinese J. Control Endemic Dis. 6:65–68 (In Chinese.) [Google Scholar]
- 9. Wang F, Hu S, Gao Y, Qiao Z, Liu W, Bu Z. 2011. Complete genome sequences of Brucella melitensis strains M28 and M5-90, with different virulence backgrounds. J. Bacteriol. 193:2904–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saxena V, Garg S, Ranjan S, Kochar D, Ranjan A, Das A. 2007. Analysis of elongation factor Tu (tuf A) of apicoplast from Indian Plasmodium vivax isolates. Infect. Genet. Evol. 7:618–626 [DOI] [PubMed] [Google Scholar]
- 11. Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nielsen K. 2002. Diagnosis of brucellosis by serology. Vet. Microbiol. 90:447–459 [DOI] [PubMed] [Google Scholar]
- 14. Gibby IW, Gibby AM. 1964. Population dynamics of Brucella on solid media. Appl. Microbiol. 12:442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Schrijver J, De Leeneer K, Lefever S, Sabbe N, Pattyn F, Van Nieuwerburgh F, Coucke P, Deforce D, Vandesompele J, Bekaert S, Hellemans J, Van Criekinge W. 2010. Analysing 454 amplicon resequencing experiments using the modular and database oriented Variant Identification Pipeline. BMC Bioinformatics 11:269. 10.1186/1471-2105-11-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. 10.1186/gb-2007-8-7-r143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crasta OR, Folkerts O, Fei Z, Mane SP, Evans C, Martino-Catt S, Bricker B, Yu G, Du L, Sobral BW. 2008. Genome sequence of Brucella abortus vaccine strain S19 compared to virulent strains yields candidate virulence genes. PLoS One 3:e2193. 10.1371/journal.pone.0002193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drazek ES, Houng HS, Crawford RM, Hadfield TL, Hoover DL, Warren RL. 1995. Deletion of purE attenuates Brucella melitensis 16M for growth in human monocyte-derived macrophages. Infect. Immun. 63:3297–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shin GW, Nho SW, Park SB, Jang HB, Cha IS, Ha MA, Kim YR, Dalvi RS, Joh SJ, Jung TS. 2009. Comparison of antigenic proteins from Lactococcus garvieae KG- and KG+ strains that are recognized by olive flounder (Paralichthys olivaceus) antibodies. Vet. Microbiol. 139:113–120 [DOI] [PubMed] [Google Scholar]
- 20. Sharma J, Mishra BB, Li Q, Teale JM. 2011. TLR4-dependent activation of inflammatory cytokine response in macrophages by Francisella elongation factor Tu. Cell. Immunol. 269:69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nieves W, Heang J, Asakrah S, Honer zu Bentrup K, Roy CJ, Morici LA. 2010. Immunospecific responses to bacterial elongation factor Tu during Burkholderia infection and immunization. PLoS One 5:e14361. 10.1371/journal.pone.0014361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Araujo FR, Costa CM, Ramos CA, Farias TA, Souza II, Melo ES, Elisei C, Rosinha GM, Soares CO, Fragoso SP, Fonseca AH. 2008. IgG and IgG2 antibodies from cattle naturally infected with Anaplasma marginale recognize the recombinant vaccine candidate antigens VirB9, VirB10, and elongation factor-Tu. Mem. Inst. Oswaldo Cruz 103:186–190 [DOI] [PubMed] [Google Scholar]
- 23. Lopez JE, Palmer GH, Brayton KA, Dark MJ, Leach SE, Brown WC. 2007. Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect. Immun. 75:2333–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Junior DS, Araujo FR, Almeida Junior NF, Adi SS, Cheung LM, Fragoso SP, Ramos CA, Oliveira RH, Santos CS, Bacanelli G, Soares CO, Rosinha GM, Fonseca AH. 2010. Analysis of membrane protein genes in a Brazilian isolate of Anaplasma marginale. Mem. Inst. Oswaldo Cruz 105:843–849 [DOI] [PubMed] [Google Scholar]
- 25. Cardoso PG, Macedo GC, Azevedo V, Oliveira SC. 2006. Brucella spp noncanonical LPS: structure, biosynthesis, and interaction with host immune system. Microb. Cell Fact. 5:13. 10.1186/1475-2859-5-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sangari FJ, Seoane A, Rodriguez MC, Aguero J, Garcia Lobo JM. 2007. Characterization of the urease operon of Brucella abortus and assessment of its role in virulence of the bacterium. Infect. Immun. 75:774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rajashekara G, Covert J, Petersen E, Eskra L, Splitter G. 2008. Genomic island 2 of Brucella melitensis is a major virulence determinant: functional analyses of genomic islands. J. Bacteriol. 190:6243–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu Q, Pei J, Turse C, Ficht TA. 2006. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. BMC Microbiol. 6:102. 10.1186/1471-2180-6-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lavigne JP, O'Callaghan D, Blanc-Potard AB. 2005. Requirement of MgtC for Brucella suis intramacrophage growth: a potential mechanism shared by Salmonella enterica and Mycobacterium tuberculosis for adaptation to a low-Mg2+ environment. Infect. Immun. 73:3160–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loisel-Meyer S, Jimenez de Bagues MP, Basseres E, Dornand J, Kohler S, Liautard JP, Jubier-Maurin V. 2006. Requirement of norD for Brucella suis virulence in a murine model of in vitro and in vivo infection. Infect. Immun. 74:1973–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haine V, Sinon A, Van Steen F, Rousseau S, Dozot M, Lestrate P, Lambert C, Letesson JJ, De Bolle X. 2005. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect. Immun. 73:5578–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hernandez-Castro R, Verdugo-Rodriguez A, Puente JL, Suarez-Guemes F. 2008. The BMEI0216 gene of Brucella melitensis is required for internalization in HeLa cells. Microb. Pathog. 44:28–33 [DOI] [PubMed] [Google Scholar]
- 33. Bandara AB, Schurig GG, Sriranganathan N, Prasad R, Boyle SM. 2009. The putative penicillin-binding proteins 1 and 2 are important for viability, growth and cell morphology of Brucella melitensis. Vet. Microbiol. 133:387–393 [DOI] [PubMed] [Google Scholar]
- 34. Posadas DM, Martin FA, Sabio y Garcia JV, Spera JM, Delpino MV, Baldi P, Campos E, Cravero SL, Zorreguieta A. 2007. The TolC homologue of Brucella suis is involved in resistance to antimicrobial compounds and virulence. Infect. Immun. 75:379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dozot M, Boigegrain RA, Delrue RM, Hallez R, Ouahrani-Bettache S, Danese I, Letesson JJ, De Bolle X, Kohler S. 2006. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell. Microbiol. 8:1791–1802 [DOI] [PubMed] [Google Scholar]
- 36. Delrue RM, Deschamps C, Leonard S, Nijskens C, Danese I, Schaus JM, Bonnot S, Ferooz J, Tibor A, De Bolle X, Letesson JJ. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7:1151–1161 [DOI] [PubMed] [Google Scholar]
- 37. Paulley JT, Anderson ES, Roop RM., II 2007. Brucella abortus requires the heme transporter BhuA for maintenance of chronic infection in BALB/c mice. Infect. Immun. 75:5248–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rambow-Larsen AA, Rajashekara G, Petersen E, Splitter G. 2008. Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J. Bacteriol. 190:3274–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kohler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, Ramuz M, Liautard JP. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. U. S. A. 99:15711–15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nasim A, Brychcy T. 1979. Genetic effects of acridine compounds. Mutat. Res. 65:261–288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


