Abstract
Pseudomonas aeruginosa chronically infects the lungs of more than 80% of adult patients with cystic fibrosis (CF) and is a major contributor to the progression of disease pathology. P. aeruginosa requires iron for growth and has multiple iron uptake systems that have been studied in bacteria grown in laboratory culture. The purpose of this research was to determine which of these are active during infection in CF. RNA was extracted from 149 sputum samples obtained from 23 CF patients. Reverse transcription–quantitative real-time PCR (RT-qPCR) was used to measure the expression of P. aeruginosa genes encoding transport systems for the siderophores pyoverdine and pyochelin, for heme, and for ferrous ions. Expression of P. aeruginosa genes could be quantified in 89% of the sputum samples. Expression of genes associated with siderophore-mediated iron uptake was detected in most samples but was at low levels in some samples, indicating that other iron uptake mechanisms are active. Expression of genes encoding heme transport systems was also detected in most samples, indicating that heme uptake occurs during infection in CF. feoB expression was detected in all sputum samples, implying an important role for ferrous ion uptake by P. aeruginosa in CF. Our data show that multiple P. aeruginosa iron uptake mechanisms are active in chronic CF infection and that RT-qPCR of RNA extracted from sputum provides a powerful tool for investigating bacterial physiology during infection in CF.
INTRODUCTION
The thick, viscous mucus in the lungs of cystic fibrosis (CF) patients is conducive to infection with bacterial pathogens (1). The lungs of >80% of adult CF patients are chronically infected with Pseudomonas aeruginosa, and infection is associated with poorer clinical scores and rapid decreases in lung function, as well as being a key contributor to the premature mortality of CF patients (2–4). Individuals with CF are intermittently admitted to hospital suffering from exacerbations, which are characterized by pronounced worsening of their lung conditions accompanied by systemic illness with fevers, night sweats, loss of appetite, and weight loss (5), and are treated with intensive antibiotic therapy.
Like other pathogenic bacteria, P. aeruginosa requires iron for numerous proteins, such as cytochromes and catalases. Iron is not freely available to infectious bacteria in the healthy human airway, because it is predominantly assimilated into iron-binding proteins such as transferrin, lactoferrin, and ferritin, but larger amounts of iron are present in CF lungs (6–8). Iron in biological environments is commonly present as ferric (Fe3+) ions, although the reduced pH (9) and the poor availability of O2 in the thick mucus layers of CF airways (10, 11) may mean that significant amounts of ferrous (Fe2+) iron are present.
P. aeruginosa can assimilate Fe3+ ions through multiple pathways, including the siderophore systems pyoverdine (Pvd) and pyochelin (Pch). The bacteria secrete pyoverdine and pyochelin, which chelate Fe3+, enabling it to be internalized through the outer membrane receptors FpvA (for the uptake of ferripyoverdine) (12) and FptA (for the uptake of ferripyochelin) (13). In animal models, pyoverdine is required for P. aeruginosa to cause acute infection (14, 15). The expression of pyoverdine synthesis genes is directed by an alternative sigma factor, PvdS (16, 17). The activity of PvdS is modulated by the antisigma protein FpvR in response to the interaction of ferripyoverdine with FpvA (17). Similarly, the expression of pyochelin synthesis genes is upregulated by the presence of pyochelin in a process involving the transcriptional activator PchR (13, 18). The expression of iron uptake pathways in P. aeruginosa is regulated by the Fur repressor protein, which, in the presence of significant amounts of intracellular Fe2+ ions, binds to the promoters of genes encoding iron uptake systems to repress their activity. Siderophore production is regulated by the binding of Fur-Fe2+ to the promoters of the pvdS and pchR genes, inhibiting their expression and thus inhibiting the synthesis of pyoverdine and pyochelin (19). P. aeruginosa also has multiple pathways for the uptake of exogenous siderophores secreted by other microorganisms (20, 21), and some of these pathways may contribute to iron acquisition in the CF lung (22).
P. aeruginosa can also assimilate iron from hemoglobin and other heme-containing proteins via the heme acquisition (Has) and Pseudomonas heme uptake (Phu) systems (23). In Serratia marcescens, heme is taken up by a hemophore (heme-binding protein) secreted by the bacteria (24). Hemophore-heme complexes are recognized by an outer membrane receptor, HasR, that imports the heme into the periplasm. The sigma factor HasI upregulates hasR gene expression in response to the presence of heme (25). The Has pathway in P. aeruginosa is composed of orthologs of the S. marcescens proteins and is predicted to function in the same way. PhuR is an outer membrane receptor for heme uptake, and PhuS is an intracellular heme-trafficking protein that delivers heme to heme oxygenases for the release of iron (23, 26). The expression of the Has and Phu pathways is repressed by the Fur protein in response to the presence of excess iron (23).
A system for the uptake of ferrous ions, Feo, has been characterized in members of the Enterobacteriaceae and Vibrio cholerae (27–30). FeoB is the major component of this system and acts as a high-affinity transporter of ferrous ions. In P. aeruginosa, an FeoB ortholog is required for the transport of Fe2+ from the periplasm to the cytoplasm during citrate-mediated iron acquisition (31). Regulation of the Feo system by Fur occurs in Escherichia coli and Vibrio cholerae (32) but has not yet been documented in P. aeruginosa, although the expression of feo is increased during growth in an iron-limited medium (33).
Iron acquisition is critical for the growth of bacterial pathogens, and iron withholding, whereby iron is bound to host proteins to reduce its accessibility to infecting bacteria, is an important component of innate immunity (34). CF sputum is a complex mixture that may provide numerous sources of iron, including heme from hemorrhage, plasma leakage, and also potentially epithelial cell iron loss related to the cystic fibrosis transmembrane conductance regulator (CFTR) mutation itself (35). Very little is known about which P. aeruginosa iron uptake systems are active in the lungs of CF patients. Pyoverdine was detected in the sputa of six CF patients, indicating that it plays a role in P. aeruginosa infections (36). However, in a larger study, approximately 33% of P. aeruginosa-containing sputa contained no detectable pyoverdine (37), suggesting that pyoverdine-mediated iron uptake may not always be essential for infection. So far as we are aware, no studies on the roles of other P. aeruginosa iron uptake systems in CF have been reported. In this study, we sought to elucidate the systems used for iron acquisition by P. aeruginosa during CF lung infections.
MATERIALS AND METHODS
Subjects and sampling.
Sputum samples were collected with the approval of the Southern Tasmanian Health and Medical Research Ethics Committee (H9813). Written informed consent was provided by all study participants. Individuals with CF were recruited from outpatient clinics, and inpatients under treatment for acute exacerbations were also recruited. At the time of sputum collection, the clinical status of the patient was assigned a score of 1 (exacerbation with hospital admission), 2 (stable), or 3 (postexacerbation, at the end of antibiotic treatment). For sample set 1, sputum samples were treated with Sputolysin (Calbiochem) for 30 min, as described previously (8), and were stored at −80°C. For sample set 2, sputum was expectorated into 20 ml of RNAlater (Qiagen). A total of 149 sputum samples were collected from 24 CF patients over a period of 117 months. The median patient age was 25 years (interquartile range, 22 to 27 years), and 50% of the patients were male. Amounts of P. aeruginosa in sputum samples were determined by colony counts on Pseudomonas selective agar as described previously (8).
RNA isolation and reverse transcription.
RNA was isolated either from bacteria (0.5 ml) that had been grown aerobically at 37°C to late-log phase (optical density at 600 nm [OD600], 2.2) in King's B medium (38) supplemented as required with FeCl3 or from bacteria that had been grown microaerobically in sealed bottles without shaking in King's B medium supplemented with KNO3 (0.4%) and as required with FeSO4. Samples were pretreated with RNAprotect Bacteria reagent (1 ml) (Qiagen), and RNA was extracted using the RNeasy minikit (Qiagen), including on-column digestion with RNase-free DNase I (Qiagen), and was eluted in RNase-free H2O containing 25 ng · ml−1 tRNA from brewer's yeast (Roche). The quantity and purity of the RNA were measured on a NanoDrop spectrophotometer. Each RNA sample was treated with DNase I (Invitrogen). The DNase was then inactivated by the addition of EDTA to 2.3 mmol and incubation at 65°C for 10 min. Aliquots of as much as 1 μg RNA were reverse transcribed to cDNA by using the Transcriptor First Strand cDNA synthesis kit (Roche) with random hexamer primers according to the manufacturer's instructions. The resulting cDNA was stored at −20°C. To determine whether any contaminating genomic DNA was present, a second aliquot of each RNA was treated with the kit reagents but without the reverse transcriptase.
For sputum sample set 1, sputum samples (0.2 ml) were mixed with an equal volume of Sputolysin (Calbiochem) and were incubated at 37°C for 30 min to reduce viscosity. RNA was then extracted and cDNA prepared in the same way as for laboratory-grown cultures. For sample set 2, as much as 0.15 g of sputum in RNAlater was dried, added to Tri Reagent (Sigma-Aldrich), and homogenized by bead-beating (Mini-BeadBeater; Biospec Products Inc.) for 4 min at a low speed using zirconia/silica beads (0.1 mm; BioSpec Products). The supernatant was collected following centrifugation. RNA was then prepared using the Tri Reagent manufacturer's protocol and was resuspended in Tris-EDTA (TE) buffer (50 μl). Samples of RNA were treated with Turbo DNase (Ambion), and cDNA was made using a SuperScript III First-Strand Synthesis SuperMix kit (Invitrogen).
Reverse transcription–quantitative real-time PCR (RT-qPCR).
Transcripts were quantified with the LightCycler 480 SYBR green I Master kit (Roche) in conjunction with gene-specific primers (see Table S1 in the supplemental material). The high degree of sequence similarity between corresponding genes in different strains of P. aeruginosa (approximately 99.5%) (39) facilitated the development of species-specific primers that will amplify target amplicons from all strains of P. aeruginosa. Reaction mixtures contained SYBR green Master mix (5 μl), 10 pmol of each primer, PCR-grade water (1.8 μl), and cDNA that had been diluted 1:10 in PCR-grade water (3 μl) (Roche). Reaction mixtures for clpX and pvdH contained 7.5 pmol of each primer, since this improved amplification efficiency. Thermocycling was performed on the LightCycler 480 platform (Roche) in 96-well plates and consisted of denaturation at 95°C (10 min); 45 amplification cycles of 98°C (5 s), 58°C (5 s), and 72°C (8 s), with data acquisition at 72°C; and a single cycle of 98°C (30 s) followed by 65°C (5 s) and then ramping to 98°C at 0.11°C/s with continuous acquisition for the collection of melt data.
Data analysis and controls.
RT-qPCR controls included cDNA derived from P. aeruginosa PAO1 cultured in King's B medium, RNA without reverse transcriptase treatment (−RT) as described above, and a water (no-template) control (NTC). Data were discarded if the target product was amplified in the NTC or −RT control run. Technical duplicates of cDNA from sputum samples were used in RT-qPCR experiments and gave high reproducibility with each primer set. Data were analyzed with LightCycler 480 software, version 1.5 (Roche). Melt curve data were used to determine whether only the correct product had been amplified.
The expression of iron metabolism genes was measured relative to the geometric means (√(clpX × oprL)) of clpX and oprL transcripts. For the few reactions in which the amount of transcript was beyond the range of the standard curve, LinRegPCR (40, 41) was used to verify that they amplified with the same efficiency as those within the standard curve range. Amplification data for the PAO1 calibrator cDNA in each LightCycler experiment showed a high level of reproducibility.
Statistical analyses.
Linear regression was used to describe the relationship between the logarithm of gene expression and iron concentration in vitro; P values, derived from a test of the null hypothesis (that the slope of the regression line is zero), indicate the presence or absence of a dose response. Pearson's correlation coefficient was used to measure the association between log gene expression values and the log of P. aeruginosa numbers. Bias-adjusted bootstrap confidence intervals were used to account for the repeated samples from patients.
The generalized estimating equation framework was used to compare gene expression by age, sex, and clinical status and to compare the two sets of data; a linear model was used for the logarithm of the expression levels with an independence working correlation and robust standard errors. The results were transformed back and are presented as relative differences in expression levels. STATA, version 11.0 (StataCorp, 2009), was used for statistical analyses.
RESULTS
Establishment of the RT-qPCR method.
We developed an RT-qPCR approach to measure the transcription of P. aeruginosa iron uptake genes in the CF lung. Our approach was to measure the expression of target genes in sputum samples from CF patients, as a surrogate for expression in the CF lung. We could not carry out an absolute quantification that would indicate the amount of transcript per nanogram of P. aeruginosa cDNA, because the cDNA was synthesized from RNA isolated from a mixture of human cells, P. aeruginosa, and other bacterial species present in sputum. Instead, relative quantification was used, with P. aeruginosa clpX, which encodes a subunit of the cytoplasmic protease ClpXP, and oprL, which encodes an outer membrane protein, as reference genes. No differential expression of clpX and oprL was detected in any of 23 transcriptome studies (42), including 1 in which sputum was added to bacterial growth medium (43). Using in vitro cultures, we confirmed that the expression of these genes is not affected by the amount of iron present (Fig. 1A).
Fig 1.
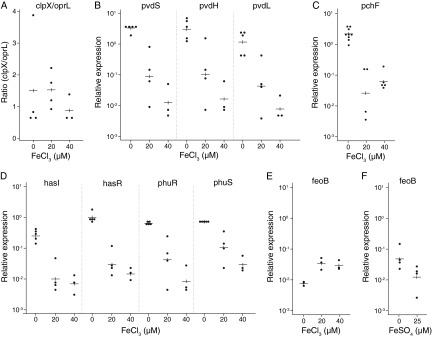
Effects of iron concentration on gene expression during in vitro culture of P. aeruginosa. (A to E) P. aeruginosa PAO1 was grown aerobically in King's B medium containing iron (FeCl3) at the concentrations shown. Gene expression was measured as described in Materials and Methods. The experiment was repeated at least three times for each growth condition, and the value for each replicate, as well as the mean value (horizontal lines), is shown. Linear regression was used to describe the relationship between the logarithm of gene expression and iron concentration; P values, indicating the presence or absence of a dose response, were derived from a test of the null hypothesis, that the slope of the regression line is zero. (A) Ratio of clpX to oprL (the P value from the regression of the log clpX/oprL ratio on iron concentration was 0.45). (B) Variation in the expression of pvd genes with Fe concentration (P, 0.0001 for pvdS, 0.0004 for pvdL, and 0.0004 for pvdH). (C) Variation in pchF expression with Fe concentration (P = 0.0002). (D) Variation in the expression of has and phu genes with Fe concentration (P, 0.0004 for hasI, 0.0001 for hasR, 0.0001 for phuR, and 0.0001 for phuS). (E) Variation in feoB expression with Fe concentration (P = 0.02). (F) Variation in feoB expression with Fe concentration (P = 0.08) for bacteria grown microaerobically with FeSO4 in place of FeCl3.
In clinical and research settings, sputum samples are routinely incubated at 37°C for 30 min in the presence of a denaturant such as Sputolysin to reduce viscosity. Samples prepared in this way (sample set 1) were used to establish the RT-qPCR method for measuring the expression of target genes. Fifty-six samples containing P. aeruginosa were obtained from 23 patients. clpX and oprL transcripts were reproducibly quantified in 53 samples in duplicate RT-qPCR experiments. There was a strong positive correlation between clpX and oprL (r = 0.92 [95% confidence interval, 0.89 to 0.95]) (see Fig. S1A in the supplemental material). There were also positive correlations between the amounts of reference gene transcripts and the number of P. aeruginosa bacteria in each sputum sample (r, 0.39 [95% confidence interval, 0.15 to 0.60] for clpX; r, 0.40 [95% confidence interval, 0.17 to 0.63] for oprL). Collectively, these data validate clpX and oprL as reference genes for the relative quantification of P. aeruginosa transcripts.
Siderophore synthesis genes.
The pyoverdine sigma factor pvdS (16) and two PvdS-dependent biosynthetic genes, pvdH (44) and pvdL (45), were selected as markers of pyoverdine activity because they are conserved across all P. aeruginosa strains (46). All three genes were highly expressed in P. aeruginosa grown in culture medium, and their expression was significantly reduced when iron was added to the growth medium (Fig. 1B), as expected (16, 33).
The activity of the Pvd system during infection of CF airways was investigated by measuring the amounts of pvd transcripts in the 53 cDNA samples derived from CF sputum that contained quantifiable amounts of clpX and oprL transcripts. pvdS, pvdH, and pvdL transcripts were detected and could be quantified for most samples (Table 1, set 1). However, in some samples, secondary amplification products were detected in addition to the expected product, preventing accurate quantification. Such artifacts can occur when the copy number of the target sequence is low (47), due in this case to the use of small sample volumes and the low number of P. aeruginosa bacteria in some samples. It is also possible that RNA from other species impaired specific amplification of the target sequence(s) in these reactions.
Table 1.
Quantification of iron uptake system transcripts in CF sputum samples
| Gene | No. of samples testeda |
% of samples in which transcripts were: |
Median gene expression>c (interquartile range) |
Pd | |||||
|---|---|---|---|---|---|---|---|---|---|
| Detected |
Quantifiedb |
||||||||
| Set 1 | Set 2 | Set 1 | Set 2 | Set 1 | Set 2 | Set 1 | Set 2 | ||
| pvdH | 53 | 79 | 94 | 100 | 89 | 100 | 0.20 (0.11–0.36) | 0.40 (0.22–0.84) | 0.06 |
| pvdL | 53 | 79 | 89 | 100 | 81 | 100 | 0.13 (0.06–0.34) | 0.14 (0.08–0.23) | 0.9 |
| pvdS | 53 | 79 | 98 | 99 | 89 | 99 | 0.23 (0.10–0.43) | 0.46 (0.23–0.72) | 0.1 |
| pchF | 53 | 79 | 85 | 100 | 74 | 99 | 0.03 (0.01–0.08) | 0.19 (0.11–0.49) | 0.002 |
| hasI | 51 | 79 | 94 | 100 | 78 | 95 | 0.40 (0.18–1.22) | 0.08 (0.04–0.17) | <0.0005 |
| hasR | 53 | 79 | 77 | 100 | 53 | 51 | 0.09 (0.03–0.24) | 0.03 (0.01–0.05) | 0.01 |
| phuR | 53 | 79 | 81 | 100 | 57 | 94 | 0.15 (0.05–0.44) | 0.02 (0.01–0.03) | <0.0005 |
| phuS | 51 | 79 | 82 | 100 | 51 | 56 | 0.61 (0.21–1.37) | 0.14 (0.08–0.40) | 0.003 |
| feoB | 52 | 79 | 100 | 100 | 100 | 100 | 1.63 (0.49–6.43) | 0.12 (0.06–0.30) | <0.0005 |
Samples in set 1 (from 23 patients) were processed with Sputolysin for 30 min prior to extraction of RNA. Samples in set 2 (from 10 patients) were added to RNAlater immediately following sputum expectoration.
Data for samples where melt curve analysis showed specific amplification of the target product only.
Relative to the geometric means for clpX and oprL.
For comparison of means of log values between the two sets of data.
There were strong associations between the amounts of pvdS, pvdH, and pvdL transcripts in CF sputum (Table 2; see also Fig. S1 in the supplemental material), demonstrating internal consistency and robustness in the RT-qPCR approach. The concentrations of pyoverdine in these samples were determined previously (37), and the median was 5.02 μM.
Table 2.
Correlation of gene expression in sputum samples
| Gene | Correlation of expressiona (95% confidence limits) with that of: |
|||||||
|---|---|---|---|---|---|---|---|---|
| pvdS | pvdH | pvdL | pchF | hasI | hasR | phuR | phuS | |
| Sample set 1 (Sputolysin treated) | ||||||||
| pvdH | 0.68 (0.46 to 0.82) | |||||||
| pvdL | 0.55 (0.38 to 0.72) | 0.53 (0.27 to 0.72) | ||||||
| pchF | 0.58 (0.28 to 0.75) | 0.53 (0.23 to 0.76) | 0.46 (0.26 to 0.67) | |||||
| hasI | 0.57 (0.28 to 0.86) | 0.40 (0.05 to 0.66) | 0.57 (0.28 to 0.86) | 0.26 (−0.26 to 0.59) | ||||
| hasR | 0.46 (0.18 to 0.73) | 0.14 (−0.16 to 0.33) | 0.13 (−0.18 to 0.49) | 0.21 (−0.42 to 0.62) | 0.81 (0.65 to 0.90) | |||
| phuR | 0.59 (0.36 to 0.77) | 0.45 (0.15 to 0.69) | 0.33 (−0.01 to 0.67) | 0.30 (−0.17 to 0.59) | 0.79 (0.62 to 0.89) | 0.90 (0.79 to 0.96) | ||
| phuS | 0.64 (0.30 to 0.88) | 0.54 (0.10 to 0.80) | 0.42 (−0.01 to 0.71) | 0.08 (−0.37 to 0.52) | 0.81 (0.58 to 0.91) | 0.84 (0.59 to 0.92) | 0.86 (0.72 to 0.93) | |
| feoB | 0.37 (0.06 to 0.62) | 0.42 (−0.02 to 0.71) | 0.61 (0.35 to 0.77) | 0.15 (−0.21 to 0.42) | 0.64 (0.48 to 0.80) | 0.40 (0.04 to 0.68) | 0.60 (0.42 to 0.75) | 0.58 (0.34 to 0.78) |
| Sample set 2 (RNAlater treated) | ||||||||
| pvdH | 0.21 (−0.21 to 0.52) | |||||||
| pvdL | 0.15 (−0.20 to 0.41) | 0.90 (0.77 to 0.95) | ||||||
| pchF | −0.14 (−0.44 to 0.26) | 0.69 (0.42 to 0.87) | 0.75 (0.55 to 0.86) | |||||
| hasI | 0.15 (−0.09 to 0.43) | 0.68 (0.47 to 0.83) | 0.69 (0.53 to 0.82) | 0.50 (0.20 to 0.71) | ||||
| hasR | 0.06 (−0.20 to 0.46) | 0.52 (0.12 to 0.78) | 0.35 (−0.23 to 0.78) | 0.35 (−0.11 to 0.70) | 0.65 (0.36 to 0.79) | |||
| phuR | 0.37 (0.09 to 0.53) | 0.01 (−0.47 to 0.69) | 0.01 (−0.42 to 0.66) | −0.36 (−0.70 to 0.54) | 0.32 (−0.02 to 0.78) | 0.32 (−0.26 to 0.83) | ||
| phuS | 0.07 (−0.39 to 0.53) | 0.58 (0.23 to 0.80) | 0.66 (0.28 to 0.86) | 0.60 (0.13 to 0.87) | 0.79 (0.65 to 0.88) | 0.61 (−0.18 to 0.91) | 0.73 (0.54 to 0.89) | |
| feoB | −0.09 (−0.38 to 0.33) | 0.26 (−0.01 to 0.60) | 0.23 (−0.04 to 0.52) | 0.16 (−0.15 to 0.55) | 0.44 (0.24 to 0.62) | 0.55 (0.34 to 0.72) | 0.47 (0.27 to 0.63) | 0.46 (0.13 to 0.66) |
A value of 1.0 indicates a perfect positive correlation; a value of 0 indicates no correlation; and a value of −1.0 indicates a perfect negative correlation.
The activity of the pyochelin iron uptake system was measured by RT-qPCR of the pyochelin synthetase gene pchF (48). As expected, expression of pchF in vitro was repressed when bacteria were cultured in the presence of Fe3+ (Fig. 1). pchF transcripts were detected in 45/53 CF sputum samples, but only in small amounts (Table 1, set 1). By use of an assay sensitive to 1 μM pyochelin (37), pyochelin was detected in only 6/45 sputum samples tested (data not shown). Collectively, these data indicate that the Pch system is not highly active in our CF patient cohort.
Heme and ferrous iron uptake pathways.
The expression of the Has and Phu heme uptake pathways was examined by RT-qPCR of the Has sigma factor gene hasI and the outer membrane receptor gene hasR and of the Phu outer membrane receptor gene phuR and the cytosolic heme transporter gene phuS. The regulatory effect of iron on has and phu gene expression (23) was confirmed: iron repressed the expression of both systems in laboratory cultures (Fig. 1).
The activities of Has and Phu were measured in sputum samples. Gene transcripts were detected in proportions of samples similar to those for the Pvd and Pch systems; however, a lower proportion could be quantified (Table 1, set 1). Strong correlations in transcript amounts were observed within each system and also between the Has and Phu systems (Table 2; see also Fig. S1 in the supplemental material).
Ferrous ions can be internalized into P. aeruginosa through the inner membrane permease FeoB. The feoB gene was expressed at low levels in aerobically grown cultures, and, for reasons that are not clear, the addition of ferric iron caused an increase in expression (Fig. 1). feoB expression was enhanced by growth under microaerobic conditions, where the addition of iron repressed expression (Fig. 1), as found previously in other species (28, 30). In sputum, feoB transcripts were detected in all 53 samples examined (Table 1, set 1).
Measurement and coordination of gene expression following immediate processing of sputum.
Analysis of the sputum samples described above allowed us to establish our methodology and demonstrate that RT-qPCR can be used to measure Pseudomonas gene expression in sputum. However, P. aeruginosa in sputum typically has a doubling time of 100 to 200 min (49), raising the possibility of changes in transcript amounts during the 30-min Sputolysin treatment. A second set of sputum samples (sample set 2; n = 93), from 9 of the patients who contributed the first sample set and 1 additional patient, was therefore analyzed. These samples were expectorated directly into RNAlater, which inhibits all enzymatic activity, preventing any RNA degradation or changes in gene expression. The expression of P. aeruginosa iron uptake genes in these samples was measured, and valid data were obtained from 79/93 samples (Table 1, set 2). Expression of all the iron uptake genes was detected in these samples, confirming that all of these iron uptake systems are active in CF. In general, median values were higher for siderophore uptake systems (pvdH, pvdS, pchF) and lower for other systems (has, phu, feoB) than those with the Sputolysin-treated samples, and for most of the genes, these differences were significant (P, <0.05 [Table 1]). These differences may have arisen because only a subset of the patients in the first set of samples was represented in the second set and/or because of the differences in sample preparation between the two sample sets.
Data from these samples were analyzed for coordinated expression of different genes. The expression of oprL was very strongly correlated with that of clpX (see Fig. S1 in the supplemental material). As expected, the expression of different genes within each iron uptake system (pvdH and pvdL; hasI and hasR; phuR and phuS) was also coordinated (Table 2; see also Fig. S1). An exception was the lack of correlation between the pyoverdine regulatory gene pvdS and the biosynthetic genes pvdH and pvdL. This may be because the activity of PvdS is posttranslationally regulated, so that its expression need not correlate with the expression of downstream genes (17). The expression of pvdH and pvdL was also strongly correlated with pchF expression, indicating coordinated activity of the pyoverdine and pyochelin siderophore systems. Strikingly, there was a strong positive correlation between feoB expression and the expression of all the has and phu genes (Table 2), something also observed with the first data set.
Correlation of gene expression with clinical parameters.
The data from sample set 2 were analyzed to determine whether there were significant interpatient differences in gene expression and whether Pseudomonas gene expression was correlated with patient age, gender, or clinical status (exacerbation, postexacerbation, or stable). Data from the first set were excluded from these analyses because of the possible confounder effects associated with the sputum-processing method. No significant interpatient differences were identified, and no correlations were observed between gene expression and patient age or gender. The expression of most genes was not significantly different in different clinical states (Table 3). A notable exception is hasR, which showed significantly lower expression in postexacerbation samples than in samples collected either from patients suffering from exacerbations or from stable patients. Three siderophore synthesis genes (pvdH, pvdL, and pchF) showed lower expression in samples from patients with exacerbations than in samples from stable patients, although this difference reached significance only for pvdL. It remains to be determined whether these differences are related to changes in the clinical state of the patient per se or result from altered gene expression associated in some way with antibiotic treatment during exacerbation.
Table 3.
Comparisons of gene expression by clinical state
| Gene | Comparison of the following clinical states: |
|||||
|---|---|---|---|---|---|---|
| Acute vs stable |
Postacute vs acute |
Postacute vs stable |
||||
| Relative expression (95% CI)a | Pb | Relative expression (95% CI) | P | Relative expression (95% CI) | P | |
| pvdS | 1.12 (0.95–1.34) | 0.2 | 0.88 (0.53–1.49) | 0.6 | 0.78 (0.48–1.25) | 0.3 |
| pvdH | 0.78 (0.59–1.03) | 0.08 | 0.72 (0.49–1.06) | 0.1 | 0.92 (0.63–1.36) | 0.7 |
| pvdL | 0.76 (0.62–0.93) | 0.009 | 0.76 (0.54–1.07) | 0.1 | 1.00 (0.66–1.51) | 1 |
| pchF | 0.61 (0.36–1.04) | 0.07 | 0.71 (0.43–1.16) | 0.2 | 1.17 (0.75–1.82) | 0.5 |
| hasI | 0.94 (0.76–1.17) | 0.6 | 0.89 (0.66–1.21) | 0.5 | 0.95 (0.74–1.22) | 0.7 |
| hasR | 0.87 (0.70–1.09) | 0.2 | 0.70 (0.57–0.86) | 0.001 | 0.80 (0.69–0.92) | 0.003 |
| phuR | 1.40 (0.64–3.06) | 0.4 | 0.90 (0.52–1.58) | 0.7 | 0.65 (0.40–1.04) | 0.07 |
| phuS | 0.93 (0.60–1.43) | 0.7 | 0.79 (0.49–1.27) | 0.3 | 0.86 (0.51–1.42) | 0.55 |
| feoB | 1.18 (0.86–1.61) | 0.3 | 1.00 (0.76–1.32) | 1 | 0.85 (0.64–1.13) | 0.26 |
From a regression model estimating the difference between means of the natural logarithms of the gene expression values obtained with sample set 2. CI, confidence interval.
P values are derived from a test of the hypothesis that the difference in means is equal to zero. The P value gives the probability that a ratio this far or farther from 1 would be observed if there were truly no difference in expression levels by clinical state.
DISCUSSION
Numerous studies have investigated P. aeruginosa physiology in vitro, but understanding how the bacteria exist within the CF lung itself, without in vitro growth of the bacteria, has proven much more challenging. One approach has been to use microarray analysis of RNA from P. aeruginosa collected from sputum (50), but this requires the availability of large amounts of sputum and does not lend itself to high-throughput analysis. In this study, we demonstrate the potential of RT-qPCR as a tool for probing bacterial physiology as it occurs in CF. Two different methods were used to extract RNA from sputum, and in each case, gene expression was detected in more than 80% of samples. Posttranscriptional processing and degradation of RNA transcripts can result in different amounts of transcripts for genes within an operon or, potentially, within the transcript for a single gene (51, 52). This may influence the sensitivity of transcript detection and complicates the use of RT-qPCR to compare the exact activities of different genes, although it does not affect the comparison of transcript amounts for each gene between samples. There were strong associations between the expression of genes within each of the pyoverdine and heme (Has and Phu) systems (Table 1), providing added robustness and giving us confidence that our approach was valid.
The expression of siderophore and heme uptake genes was lower in sputum (Table 1) than in P. aeruginosa grown in a standard laboratory culture medium, unless the medium was supplemented with iron (Fig. 1). The expression of these genes is repressed by extracellular iron, which may be present in CF airways as a consequence of alterations in iron trafficking and increased iron loss from epithelial cells in CF (35). Alternatively, the reason for the difference may be that P. aeruginosa grows more rapidly and has a higher requirement for iron in standard laboratory culture.
Research on iron acquisition during infection in CF has focused on pyoverdine. The presence of this siderophore in CF sputum (36, 37) and the expression of pvd genes in most sputum samples (this study) indicate that pyoverdine is an important factor in the survival of P. aeruginosa in CF. However, levels of pyoverdine gene expression were very low in some samples, consistent with the absence of detectable pyoverdine in some sputum samples and the conclusion that pyoverdine is not absolutely required during chronic infection of the CF airway (37). pchF expression was detected in most samples, and pyochelin was detected in some samples, suggesting that pyochelin-mediated iron uptake plays a role in some, but perhaps not all, infections. P. aeruginosa must therefore acquire iron through other pathways as well as (or instead of) via these siderophores. The expression of the has and phu genes in sputa shows that Pseudomonas heme uptake systems are active in CF patients, with heme potentially being supplied by proteins such as hemoglobin, haptoglobin, or myoglobin. The expression of hasR, which is likely to be regulated by the amount of heme available to the bacteria, was notably lower in postexacerbation samples than in samples collected during exacerbation or when patients were stable (Table 3). This suggests that heme may be less readily available to P. aeruginosa following treatment for exacerbations, perhaps reflecting reduced amounts of heme in sputum when patients have recovered following an extensive course of intravenous antibiotics and less airway inflammation is present. P. aeruginosa has a large number of other pathways for the uptake of iron (Fe3+) chelates (20, 21), and it remains to be determined which of these are active during infection in CF.
Expression of feoB, which transports Fe2+ into P. aeruginosa, was detected in all samples in both sample sets (Table 1). feoB expression in vitro was enhanced by growth under microaerobic conditions (Fig. 1), as found previously for Shigella flexneri (53), and feoB expression in CF is consistent with the proposal that low oxygen availability in CF lungs, particularly in biofilms (10, 11), stabilizes the Fe2+ redox state, allowing direct uptake of the iron present in CF sputum. The availability of Fe2+ may also be increased by a lower-than-normal airway pH, which has been reported in CF (9). Additionally, phenazine released by P. aeruginosa can reduce extracellular Fe3+ to Fe2+ (54, 55), potentially providing a substrate for FeoB. The strong correlation between the expression of feoB and that of the has and phu genes (Table 3) may reflect a correlation between the availability of Fe2+ ions and heme (or hemoglobin) in CF sputum. FeoB is required for the utilization of iron taken up as ferric citrate (31), and an alternative explanation for the correlation between feoB expression and the expression of the has and phu genes is that FeoB is also involved in the acquisition of iron that enters P. aeruginosa via the Has or Phu pathway. In general, there were no strong correlations between the expression of siderophore (pvd, pch) genes and that of heme/Fe2+ uptake (has, phu, feoB) genes, suggesting that P. aeruginosa fine-tunes its iron uptake pathways in response to the sources of iron that are immediately available.
In conclusion, this study provides new understanding of how P. aeruginosa acquires iron during the infection of CF airways. It also demonstrates the potential of RT-qPCR in studying the metabolism of infectious bacteria in vivo, an approach that holds considerable promise for the study of other P. aeruginosa pathways implicated in infection in CF. Our data show that iron acquisition by P. aeruginosa is multifaceted and includes heme and Fe2+ uptake pathways that merit greater characterization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karla Mettrick for assistance in establishing the protocol for the extraction and analysis of RNA from sputum and for the design of primers for pvdS and clpX. We also thank Xin Xu for providing expert technical assistance with experiments with P. aeruginosa grown under microaerobic conditions. We are grateful to the anonymous reviewers of a previous version of this manuscript for their helpful comments and suggestions.
This work was supported by a New Zealand Tertiary Education Commission Bright Futures Ph.D. scholarship to A.F.K. and by Australian National Health and Medical Research Council grants 352611 and 544915.
Footnotes
Published ahead of print 20 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00418-13.
REFERENCES
- 1. Harrison F. 2007. Microbial ecology of the cystic fibrosis lung. Microbiology 153:917–923 [DOI] [PubMed] [Google Scholar]
- 2. Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918–951 [DOI] [PubMed] [Google Scholar]
- 3. Kosorok MR, Zeng L, West SE, Rock MJ, Splaingard ML, Laxova A, Green CG, Collins J, Farrell PM. 2001. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr. Pulmonol. 32:277–287 [DOI] [PubMed] [Google Scholar]
- 4. Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, Grimwood K. 2001. Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J. Pediatr. 138:699–704 [DOI] [PubMed] [Google Scholar]
- 5. Stenbit AE, Flume PA. 2011. Pulmonary exacerbations in cystic fibrosis. Curr. Opin. Pulm. Med. 17:442–447 [DOI] [PubMed] [Google Scholar]
- 6. Stites SW, Walters B, O'Brien-Ladner AR, Bailey K, Wesselius LJ. 1998. Increased iron and ferritin content of sputum from patients with cystic fibrosis or chronic bronchitis. Chest 114:814–819 [DOI] [PubMed] [Google Scholar]
- 7. Stites SW, Plautz MW, Bailey K, O'Brien-Ladner AR, Wesselius LJ. 1999. Increased concentrations of iron and isoferritins in the lower respiratory tract of patients with stable cystic fibrosis. Am. J. Respir. Crit. Care Med. 160:796–801 [DOI] [PubMed] [Google Scholar]
- 8. Reid DW, Carroll V, O'May C, Champion A, Kirov SM. 2007. Increased airway iron as a potential factor in the persistence of Pseudomonas aeruginosa infection in cystic fibrosis. Eur. Respir. J. 30:286–292 [DOI] [PubMed] [Google Scholar]
- 9. Tate S, MacGregor G, Davis M, Innes JA, Greening AP. 2002. Airways in cystic fibrosis are acidified: detection by exhaled breath condensate. Thorax 57:926–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock RE, Parsek MR, Noah TL, Boucher RC, Hassett DJ. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593–603 [DOI] [PubMed] [Google Scholar]
- 11. Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schalk IJ. 2008. Metal trafficking via siderophores in Gram-negative bacteria: specificities and characteristics of the pyoverdine pathway. J. Inorg. Biochem. 102:1159–1169 [DOI] [PubMed] [Google Scholar]
- 13. Michel L, Bachelard A, Reimmann C. 2007. Ferripyochelin uptake genes are involved in pyochelin-mediated signalling in Pseudomonas aeruginosa. Microbiology 153:1508–1518 [DOI] [PubMed] [Google Scholar]
- 14. Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takase H, Nitanai H, Hoshino K, Otani T. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunocompromised mice. Infect. Immun. 68:1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cunliffe HE, Merriman TR, Lamont IL. 1995. Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J. Bacteriol. 177:2744–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 99:7072–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heinrichs DE, Poole K. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178:2586–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ochsner UA, Vasil AI, Vasil ML. 1995. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J. Bacteriol. 177:7194–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86:1637–1645 [DOI] [PubMed] [Google Scholar]
- 21. Cornelis P, Bodilis J. 2009. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ. Microbiol. Rep. 1:256–262 [DOI] [PubMed] [Google Scholar]
- 22. Weaver VB, Kolter R. 2004. Burkholderia spp. alter Pseudomonas aeruginosa physiology through iron sequestration. J. Bacteriol. 186:2376–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ochsner A, Johnson Z, Vasil ML. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185–198 [DOI] [PubMed] [Google Scholar]
- 24. Cescau S, Cwerman H, Letoffe S, Delepelaire P, Wandersman C, Biville F. 2007. Heme acquisition by hemophores. Biometals 20:603–613 [DOI] [PubMed] [Google Scholar]
- 25. Rossi MS, Paquelin A, Ghigo JM, Wandersman C. 2003. Haemophore-mediated signal transduction across the bacterial cell envelope in Serratia marcescens: the inducer and the transported substrate are different molecules. Mol. Microbiol. 48:1467–1480 [DOI] [PubMed] [Google Scholar]
- 26. Kaur AP, Lansky IB, Wilks A. 2009. The role of the cytoplasmic heme-binding protein (PhuS) of Pseudomonas aeruginosa in intracellular heme trafficking and iron homeostasis. J. Biol. Chem. 284:56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143–157 [DOI] [PubMed] [Google Scholar]
- 28. Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J. Bacteriol. 188:6515–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wyckoff EE, Boulette ML, Payne SM. 2009. Genetics and environmental regulation of Shigella iron transport systems. Biometals 22:43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fetherston JD, Mier I, Jr, Truszczynska H, Perry RD. 2012. The Yfe and Feo transporters are involved in microaerobic growth and virulence of Yersinia pestis in bubonic plague. Infect. Immun. 80:3880–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marshall B, Stintzi A, Gilmour C, Meyer JM, Poole K. 2009. Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology 155:305–315 [DOI] [PubMed] [Google Scholar]
- 32. Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. 2005. Iron and Fur regulation in Vibrio cholerae and the role of Fur in virulence. Infect. Immun. 73:8167–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277–1287 [DOI] [PubMed] [Google Scholar]
- 34. Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946–953 [DOI] [PubMed] [Google Scholar]
- 35. Moreau-Marquis S, Bomberger JM, Anderson GG, Swiatecka-Urban A, Ye S, O'Toole GA, Stanton BA. 2008. The ΔF508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron bioavailability. Am. J. Physiol. Lung Cell. Mol. Physiol. 295:L25–L37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haas B, Kraut J, Marks J, Zanker SC, Castignetti D. 1991. Siderophore presence in sputa of cystic fibrosis patients. Infect. Immun. 59:3997–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin LW, Reid DW, Sharples KJ, Lamont IL. 2011. Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals 24:1059–1067 [DOI] [PubMed] [Google Scholar]
- 38. King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 39. Spencer DH, Kas A, Smith EE, Raymond CK, Sims EH, Hastings M, Burns JL, Kaul R, Olson MV. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62–66 [DOI] [PubMed] [Google Scholar]
- 41. Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Balasubramanian D, Mathee K. 2009. Comparative transcriptome analyses of Pseudomonas aeruginosa. Hum. Genomics 3:349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Palmer KL, Mashburn LM, Singh PK, Whiteley M. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187:5267–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vandenende CS, Vlasschaert M, Seah SY. 2004. Functional characterization of an aminotransferase required for pyoverdine siderophore biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:5596–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mossialos D, Ochsner U, Baysse C, Chablain P, Pirnay JP, Koedam N, Budzikiewicz H, Fernandez DU, Schafer M, Ravel J, Cornelis P. 2002. Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 45:1673–1685 [DOI] [PubMed] [Google Scholar]
- 46. Lamont IL, Martin LW. 2003. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 149:833–842 [DOI] [PubMed] [Google Scholar]
- 47. Bustin SA. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29:23–39 [DOI] [PubMed] [Google Scholar]
- 48. Reimmann C, Serino L, Beyeler M, Haas D. 1998. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology 144:3135–3148 [DOI] [PubMed] [Google Scholar]
- 49. Yang L, Haagensen JA, Jelsbak L, Johansen HK, Sternberg C, Hoiby N, Molin S. 2008. In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J. Bacteriol. 190:2767–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Son MS, Matthews WJ, Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75:5313–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Belasco JG, Higgins CF. 1988. Mechanisms of mRNA decay in bacteria: a perspective. Gene 72:15–23 [DOI] [PubMed] [Google Scholar]
- 52. Evguenieva-Hackenberg E, Klug G. 2011. New aspects of RNA processing in prokaryotes. Curr. Opin. Microbiol. 14:587–592 [DOI] [PubMed] [Google Scholar]
- 53. Boulette ML, Payne SM. 2007. Anaerobic regulation of Shigella flexneri virulence: ArcA regulates Fur and iron acquisition genes. J. Bacteriol. 189:6957–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. 2011. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J. Bacteriol. 193:3606–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cox CD. 1986. Role of pyocyanin in the acquisition of iron from transferrin. Infect. Immun. 52:263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


