Abstract
Mycoplasma genitalium is a sexually transmitted pathogen associated with several acute and chronic reproductive tract disease syndromes in men and women. To evaluate the suitability of a pig-tailed macaque model of M. genitalium infection, we inoculated a pilot animal with M. genitalium strain G37 in the uterine cervix and in salpingeal pockets generated by transplanting autologous Fallopian tube tissue subcutaneously. Viable organisms were recovered throughout the 8-week experiment in cervicovaginal specimens and up to 2 weeks postinfection in salpingeal pockets. Humoral and cervicovaginal antibodies reacting to MgpB were induced postinoculation and persisted throughout the infection. The immunodominance of the MgpB adhesin and the accumulation of mgpB sequence diversity previously observed in persistent human infections prompted us to evaluate sequence variation in this animal model. We found that after 8 weeks of infection, sequences within mgpB variable region B were replaced by novel sequences generated by reciprocal recombination with an archived variant sequence located elsewhere on the chromosome. In contrast, mgpB region B of the same inoculum propagated for 8 weeks in vitro remained unchanged. Notably, serum IgG reacted strongly with a recombinant protein spanning MgpB region B of the inoculum, while reactivity to a recombinant protein representing the week 8 variant was reduced, suggesting that antibodies were involved in the clearance of bacteria expressing the original infecting sequence. Together these results suggest that the pig-tailed macaque is a suitable model to study M. genitalium pathogenesis, antibody-mediated selection of antigenic variants in vivo, and immune escape.
INTRODUCTION
Mycoplasma genitalium is a recently recognized sexually transmitted pathogen associated with reproductive tract disease in men and women (reviewed in references 1 to 3). Infection with this organism has been associated with urethritis in men (4–6) and cervicitis (7–9), urethritis (10, 11), acute endometritis (12), chronic pelvic inflammatory disease (13), tubal factor infertility (14, 15), and preterm birth (16, 17) in women. When untreated or inappropriately treated, M. genitalium infection can persist for months to years (18–21), suggesting that this organism avoids elimination by the host immune response. Recent studies assessing M. genitalium treatment regimens have found that standard antibiotic therapy fails to cure a significant proportion of infections (reviewed in reference 1), highlighting the importance of a better understanding of M. genitalium pathogenesis so that effective treatment and prevention methods can be devised.
The distinct flask-shaped morphology of M. genitalium cells can be attributed to its complex tip organelle involved in gliding motility (22–25) and adherence to host cells and inanimate surfaces (26–30). Included among the tip organelle-associated proteins are the major adhesin, MgpB (also known as MgPa or P140), and its accessory protein, MgpC (also known as P110), which is required for the stability of MgpB (29). Remarkably, despite having the smallest genome of any fully sequenced bacterium capable of axenic growth (31–33), M. genitalium dedicates 4.7% of its genome to repeat sequences with homology to portions of the mgpB and mgpC genes (31, 34–36). A single locus, the mgpB-mgpC expression site, encodes the full-length MgpB and MgpC proteins; however, sequences homologous to the variable regions of mgpB (regions B, EF, and G) and mgpC (region KLM) are present in nine regions, designated MgPar sequences, distributed throughout the genome (34–36). We (34, 35, 37) and others (38, 39) have shown that the variable regions of mgpB and mgpC recombine with the MgPar sequences, potentially resulting in an almost limitless array of MgpB and MgpC antigenic and phase variants.
The roles of antigenic and phase variation in the pathogenesis and persistence of M. genitalium are unknown but may include differential attachment to (and perhaps invasion of) host cells or avoidance of the host immune response. In support of the latter hypothesis, we and others have observed that serum antibodies to MgpB and MgpC can be detected in infected humans (14, 15, 40, 41) and animal models (42–44) and that sequence variation occurs in vivo in chronically infected men (45; G. E. Wood, S. L. Iverson-Cabral, and P. A. Totten, unpublished results) and women (34, 35, 45). Furthermore, the predominance of MgpB- and MgpC-reactive antibodies in cervicovaginal exudates from M. genitalium-infected women (40) is consistent with their hypothesized role in selection of antigenic variants in the lower genital tract. The potential for gene variation afforded by recombination with the MgPar sequences, the immunodominance of the variable MgpB and MgpC proteins, and the persistent nature of M. genitalium infections has led to the hypothesis that MgpB and MgpC sequence variation provides a mechanism by which this organism avoids immune recognition and clearance from the host, perhaps mediated by specific antibody.
Clearly, an animal model capable of supporting persistent infection is needed to study the development of sequence diversity, antigenic variation, and immune evasion. In the current study, we infected a female pig-tailed macaque (Macaca nemestrina), an animal closely related to humans, with our single-colony-cloned isolate of M. genitalium strain G37 and then monitored the persistence of infection in the cervix and in salpingeal pockets, induction of M. genitalium-reactive antibodies, and the development of mgpB sequence variants. Our results support the utility of this primate model to study persistence, the host immune response, and the role of antigenic variation in evasion of antibodies induced in the host.
(These data were presented, in part, at the 2011 Cold Spring Harbor Meeting on Microbial Pathogenesis & Host Response, Cold Spring Harbor, NY, 13 to 17 September 2011 and the 19th Congress of the International Organization of Mycoplasmology Meeting in Toulouse, France, 15 to 20 July 2012.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Mycoplasma genitalium strains were grown routinely in H broth or on H-agar plates containing 125 U/ml penicillin at 37°C as previously described (46). The pilot primate was inoculated with M. genitalium G37-C, our single-colony filter-cloned isolate of the type strain G37, which contains the mgpB-mgpC expression site and MgPar sequences identical to the fully sequenced genome (35). M. genitalium mutants lacking MgpB (Δmg191), MgpC (Δmg192) (29), or HMW2 (Δmg218 [25]) were kindly provided by Jaume Piñol (Universitat Autonoma de Barcelona, Barcelona, Spain).
Experimental infection of Macaca nemestrina with M. genitalium.
In this pilot study of experimental lower and upper reproductive tract infection, a female pig-tailed macaque (Macaca nemestrina) was inoculated at the cervix and in autotransplanted subcutaneous salpingeal pockets as described previously (47). A single animal was used for this pilot experiment due to the high cost of primate experiments. Prior approval was obtained from the University of Washington Institutional Animal Care and Use Committee and the Washington National Primate Research Center where the animal was housed. To explore upper reproductive tract infection, we used the subcutaneous salpingeal pocket model, as previously described for Chlamydia trachomatis (47). In this model (outlined in Table 1), small segments of the salpingeal fimbriae (∼3 mm) were surgically removed and autotransplanted subcutaneously into pockets made on the anterior abdominal wall. Three weeks after transplantation, the cervix of the primate was inoculated by direct application of 7 × 108 genomes (∼3.3 × 107 color change units/ml) of M. genitalium strain G37-C in 1 ml of phosphate-buffered saline (PBS), and each of 12 salpingeal pockets was injected with 0.2 ml of 3.5 × 109 genomes/ml. Eight pockets were sham inoculated with PBS alone. At the times indicated in Table 1, three inoculated pockets and two control pockets were excised, homogenized in 2 ml mycoplasma transport medium (MTM) (4), and assessed for the presence of M. genitalium DNA by quantitative PCR (qPCR). The presence of viable M. genitalium was measured after 3 weeks incubation in Vero cell cocultures (8.5 ml, containing 100 U/ml penicillin and 50 μg/ml polymyxin B) inoculated with 0.2 ml of homogenate (48) and in H broth (3 ml, containing 125 U/ml penicillin) inoculated with 0.3 ml of homogenate. The antibiotics included in the medium were sufficient to prevent gross bacterial contamination of cultures by normal flora. The H-broth culture was further diluted 10- to 100-fold resulting in three culture tubes containing 10−1, 10−2, and 10−3 dilutions of specimen. Due to the small size of the recovered salpingeal tissue, the entire tissue specimen was homogenized to improve recovery and detection of viable M. genitalium. Vaginal swabs, cervical swabs, and cervical cytobrush (CxCB) specimens were also collected at the time points indicated in Table 1, suspended in 2 ml MTM, and assessed for M. genitalium DNA and viable cells as described above. Eight weeks after inoculation, the animal was treated with azithromycin (14 mg/kg of body weight) for 5 days. Lower genital tract specimens were collected 3 weeks later and analyzed by qPCR and culture to confirm that the infection was cured. Serum samples were collected prior to inoculation and at intervals thereafter until 8 weeks postinoculation; samples were then analyzed by immunoblotting and enzyme-linked immunosorbent assays (ELISAs) for the presence of M. genitalium-reactive antibodies as described below. Cervical cytobrush specimens, collected at 2, 4, and 8 weeks postinoculation, were further analyzed to assess sequence variation in the mgpB gene and MgPar donor sequences during infection (see below).
Table 1.
Detection of M. genitalium in primate specimens by quantitative PCR and culturea
| Specimen type |
M. genitalium detected in specimen by the indicated method at the following time: |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wk 0 |
Wk 1 |
Wk 2 |
Wk 3 |
Wk 4 |
Wk 5 |
Wk 8 |
|||||||||||||||
| qPCRb | H-broth culturec | Vero cocultured | qPCR | H-broth culture | Vero coculture | qPCR | H-broth culture | Vero coculture | qPCR | H-broth culture | Vero coculture | qPCR | H-broth culture | Vero coculture | qPCR | H-broth culture | Vero coculture | qPCR | H-broth culture | Vero coculture | |
| Vaginal swab | − | − | − | + | − | + | + | − | + | − | − | + | + | + | + | − | − | + | − | − | − |
| Cervical swab | − | − | − | + | − | + | + | + | + | + | − | + | + | − | + | − | − | − | − | − | + |
| Cervical cytobrush | ND | ND | ND | ND | ND | ND | + | ND | − | − | − | + | + | − | + | − | − | − | +f | − | + |
| Pocket 1e (PBS) | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| Pocket 2 (PBS) | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| Pocket 3 (Mg) | − | − | − | + | + | + | − | − | − | − | − | − | |||||||||
| Pocket 4 (Mg) | − | − | − | + | + | + | − | − | − | + | − | − | |||||||||
| Pocket 5 (Mg) | + | + | + | + | + | − | − | − | − | − | − | − | |||||||||
Specimens collected at week 11 (three weeks after azithromycin treatment) were negative by qPCR and H-broth culture (data not shown).
M. genitalium genomes in 2.4 μl of primate specimen as determined by quantitative PCR. Samples were considered positive (+) if the number of genomes was more than twice that of preinoculation specimens. −, not detected; ND, not determined.
Growth in 3 ml H broth was considered positive (+) if the color changed from red to orange or yellow, indicating the production of acid, in any of three tubes inoculated with 1:10, 1:100, and 1:1000 dilutions of specimen, and then incubated for 3 weeks. −, not detected; ND, not determined.
Growth in 8.5-ml Vero cocultures was determined by qPCR by detecting M. genitalium genomes in 20 μl of culture supernatant collected 7, 14, and 21 days after inoculation with 0.2 ml of specimen. Positive (+) cultures demonstrated >200-fold increase in M. genitalium genomes. −, not detected; ND, not determined.
A total of 20 abdominal salpingeal pockets were inoculated with PBS or M. genitalium (Mg) as indicated. Five salpingeal pockets were excised at weeks 1, 2, 3, and 4; no pockets were collected for weeks 0, 5, and 8.
The week 8 cervical cytobrush specimen was further analyzed for sequence variants in mgpB region B.
M. genitalium qPCR assays.
Total DNA was isolated from the primate inoculum or directly from primate specimens using the MasterPure DNA purification kit (Epicentre, Madison, WI) according to the manufacturer's instructions. Purified DNA was subjected to quantitative PCR using the LightCycler 480 instrument (Roche Applied Science, Indianapolis, IN) using primers ModMgPa1 and ModMgPa3 (Table 2 and Fig. 1) that were specific for the 5′ conserved region of mgpB, used for detection of M. genitalium in clinical specimens (6, 49). Reaction mixtures consisted of 1.6 μl template DNA (corresponding to 2.4 μl of primate specimen or 20 μl of Vero coculture supernatant concentrated during purification), 0.3 μM each primer, 4 mM MgCl2, 1× FastStart DNA master SYBR green I (Roche Applied Sciences) and water in a total volume of 16 μl. The following PCR conditions were used: 95°C for 10 min followed by 40 cycles, with 1 cycle consisting of 95°C for 10 s, 55°C for 5 s, and 72°C for 10 s with fluorescence acquisition at 65°C. Melting curve analysis and gel electrophoresis confirmed the specificity of the PCR for the 269-bp mgpB product. PCR quantification was performed by comparison to a standard curve composed of serial 10-fold dilutions of M. genitalium whole-cell DNA. The concentration of isolated DNA used to generate the standard curve was determined using the Quant-iT PicoGreen double-stranded DNA (dsDNA) kit (Invitrogen, Grand Island, NY) according to the manufacturer's instructions.
Table 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Region amplified and primer direction | Purpose | Reference |
|---|---|---|---|---|
| ModMgPa1 | TGAAACCTTAACCCCTTGG | Conserved region of mgpB; upstream primer | Quantification of M. genitalium genomes by qPCR | 49 |
| ModMgPa3 | AGGGGTTTTCCATTTTTGC | Conserved region of mgpB; downstream primer | Quantification of M. genitalium genomes by qPCR | 49 |
| 3F | CAAAAATGGAAAACCCCTCAA | Region B of mgpB; upstream primer | Amplification of mgpB region B for assessment of gene variation | 34 |
| 3R | ATCATAGAAACTACCACCGTCG | Region B of mgpB; downstream primer | Amplification of mgpB region B for assessment of gene variation | 34 |
| ModPetF | GTGATGTTGTTAGTGATTGTGTG | Region B of mgpB; upstream primer | Amplification of mgpB region B, second primer pair | This study |
| 1415R | TGGTGGTAAACATCTTAGTAGCAT | Region B of mgpB; downstream primer | Amplification of mgpB region B, second primer pair | This study |
| Par8-F | TCAAAATAGAGTGTTGTGGGTCG | Region B-homologous region of MgPar8; upstream primer | Amplification of MgPar8 of strain G37-C and wk 8 variant for assessment of gene variation | 35 |
| Par8B-R | CGATTCAGGGGAGAAAGTC | Region B-homologous region of MgPar8; downstream primer | Amplification of MgPar8 of strain G37-C and wk 8 variant for assessment of gene variation | This study |
| rMgpB:B-F | GACGACGACAAGATAGGTAAAGTTCCAGTAGAAGTAGTT | Region B of mycobacterial G37-C mgpB inoculum; upstream primer | Construction of plasmids for expression of His-tagged MgpB region B peptides | This study |
| rMgpB:BG37−C-R | GAGGAGAAGCCCGGTGTATGGTTTTCACTGTAGGG | Region B of mycobacterial G37-C mgpB inoculum; downstream primer | Construction of plasmids for expression of His-tagged MgpB region B peptides | This study |
| rMgpB:BWk8-R | GAGGAGAAGCCCGGATCATAGAAACTAACCACCGT | Region B of mgpB week 8 variant; downstream primer | Construction of plasmids for expression of His-tagged MgpB region B peptides | This study |
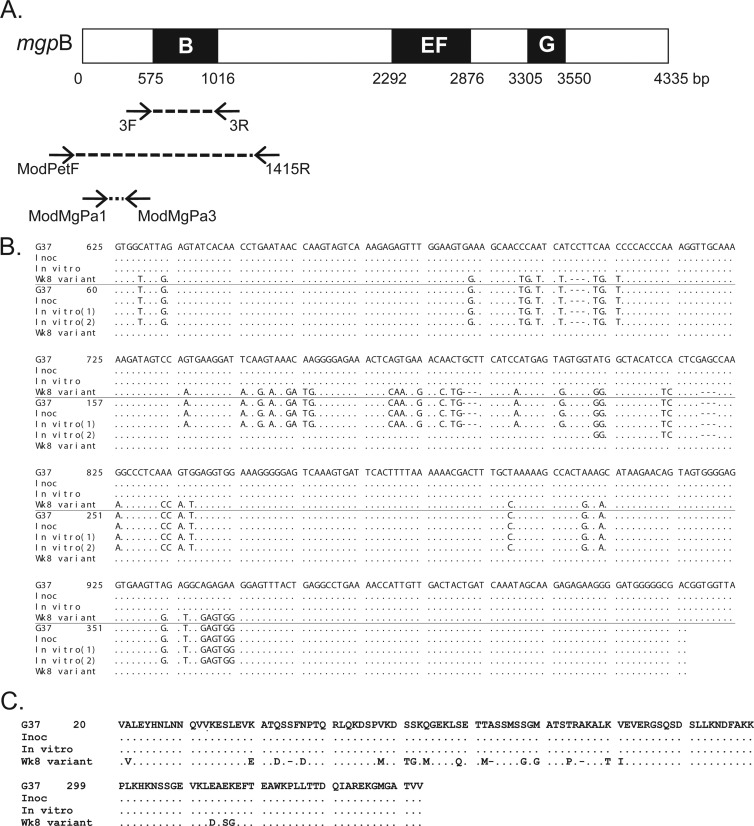
Fig 1.
(A) Schematic of the mgpB expression site of M. genitalium G37-C. Black boxes (B, EF, and G) indicate the variable regions of mgpB, and intervening white areas represent conserved sequences (82). Arrows connected by broken lines show the location and direction of the primers used for PCR amplification. (B) Alignment of mgpB region B and MgPar8 DNA sequences obtained from the primate inoculum (Inoc), bacteria passaged in broth for 8 weeks (In vitro), and the week 8 primate specimen (Wk8 variant) with the published M. genitalium G37 type sequences. In each set of sequences, the top four lines represent mgpB expression site sequences, while the bottom five lines represent MgPar8 sequences. Nucleotides that are identical to the nucleotides in strain G37 are indicated by dots; nucleotides that differ from the G37 type sequence are shown. Deleted nucleotides are indicated by hyphens. Two MgPar8 sequences were identified in the in vitro-propagated control and are indicated as “In vitro (1)” and “In vitro (2)” (see Results). Nucleotides are numbered relative to the first base pair of G37 mgpB or MgPar8. (C) Alignment of the predicted MgpB region B amino acid sequences showing that the primate week 8 DNA sequence predicts an in-frame protein product with variant amino acids indicated by their single-letter abbreviation. Amino acids are numbered according to the start codon of MgpB.
Immunoblot assays.
We prepared M. genitalium G37-C whole-cell lysates from bacteria grown in three 75-cm2 tissue culture flasks in SP-4 broth (50) at 37°C under 5% CO2 until a color change from red to yellow indicated bacterial growth. The culture supernatant was discarded, and the adherent bacteria were washed three times in 1× phosphate-buffered saline, scraped into PBS, and then concentrated by centrifugation into a final volume of 0.5 ml PBS. Because the mgpB and mgpC deletion mutants are nonadherent, bacteria were collected from culture supernatants, then washed, and concentrated in PBS by centrifugation. The bacteria were lysed by adding SDS to a final concentration of 0.1%, and then total protein was quantitated using the bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Rockford, IL). Lysates (8 to 10 μg total protein) were combined with one half volume of SDS sample buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2.3% SDS, 5% 2-mercaptoethanol, 0.05% bromophenol blue), boiled for 5 min, then electrophoresed through an 8% SDS-polyacrylamide gel with a prestained protein size marker (HiMark prestained high-molecular-weight [HMW] protein standard; Invitrogen, Grand Island, New York). Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Invitrogen), and processed as previously described (40) using diluted primate serum (see figure legends) and the following peroxidase-conjugated secondary antibodies (Sigma-Aldrich, St. Louis, MO): goat anti-human IgG (whole molecule), IgA (alpha chain), or IgM (mu chain).
To locate MgpB and MgpC on immunoblots, sera from rabbits immunized with His-tagged recombinant MgpB or MgpC peptide encompassing amino acids 1092 to 1209 or 36 to 308, respectively (40), were combined, diluted 1:10,000, and then detected with a 1:10,000 dilution of peroxidase-conjugated goat anti-rabbit IgG (whole molecule; Sigma-Aldrich).
Analysis of mgpB and MgPar sequences.
DNA from primate specimens and from M. genitalium cultures was isolated using the MasterPure DNA purification kit (Epicentre) according to the manufacturer's instructions. The variable region B of mgpB was amplified in separate experiments using either the 3F/3R (F stands for forward, and R stands for reverse) or ModPetF/1415R primer pair (498 bp or 1,488 bp, respectively; Table 2 and Fig. 1) in a reaction mixture consisting of 4 μl template DNA, 0.25 μM each primer, and 15 μl of Platinum PCR supermix (Invitrogen) using the following conditions: 94°C for 4 min; 35 cycles, with 1 cycle consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min; followed by a final extension step of 7 min at 72°C.
The portion of MgPar8 homologous to mgpB region B was amplified using primers Par8-F and Par8B-R (Table 2) in a reaction mixture consisting of 0.4 μM each primer, 2.3 mM MgCl2, 5 units GoTaq Flexi (Promega, Madison, WI), 1.85 μM each deoxynucleoside triphosphate (dNTP), 1× GoTaq PCR buffer, and 5 μl template DNA. Cycling conditions were as follow: 94°C for 3 min; 35 cycles, with 1 cycle consisting of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min; with a final 10-min extension step at 72°C. Because the Par8B-R primer sequence also occurs within MgPar sequences 1, 2, 5, 7, and 8 and the mgpB expression site (35), the PCR was anchored to MgPar8 using the unique primer Par8-F, thus ensuring amplification of the MgPar8-derived PCR product. Agarose gel electrophoresis and sequencing confirmed that the correct 535-bp product was analyzed.
Amplicons were subsequently cloned into plasmid pCR2.1-TOPO using the TOPO TA cloning kit (Invitrogen). Plasmid DNA was purified, sequenced by the University of Washington Biochemistry Sequencing Facility or by Eurofins MWG Operon (Huntsville, AL), and sequences were aligned with mgpB region B of strain G37-C using the MultAlin program (http://multalin.toulouse.inra.fr/multalin/).
In vitro variation of M. genitalium.
In order to assess the extent of gene variation in the absence of immune selection, the M. genitalium G37-C primate inoculum was cultured in vitro by serial passage in H broth for 8 weeks. H-broth cultures (7 ml) were inoculated with 10 to 25 μl of inoculum in a 25-cm2 tissue culture flask and incubated until a color change indicated growth of M. genitalium (typically 4 to 7 days), after which adherent bacteria were scraped into the culture supernatant. Samples containing 106 to 107 genomes were analyzed for mgpB gene variation as described below and inoculated into a fresh H-broth culture. This procedure was repeated until cultures had been passaged in vitro for 8 weeks.
B-cell epitope predictions.
The variable region B amino acid sequences of the mycobacterial G37-C inoculum and the week 8 variant were analyzed for the presence of B-cell epitopes with the EMBOSS antigenic (http://emboss.bioinformatics.nl/cgi-bin/emboss/antigenic) and BepiPred 1.0 (http://www.cbs.dtu.dk/services/BepiPred/) ([51]) programs using a wide range of epitope lengths and thresholds, respectively. The full-length MgpB protein of strain G37-C was analyzed for predicted conformational B-cell epitopes using the CBTOPE program (http://www.imtech.res.in/raghava/cbtope/ [52]) with a support vector machine threshold of −0.3. The sequence of the full-length week 8 variant MgpB is unknown and consequently was not searched for conformational epitopes.
Cloning and expression of His-tagged proteins.
Recombinant His-tagged MgpB region B peptides representing the wild-type strain G37-C or the week 8 variant sequence were produced as follows. The G37-C sequence (539 bp) was amplified directly from M. genitalium G37-C whole-cell DNA using primers rMgpB:B-F (rMgp stands for recombinant MgpB, the B after the colon stands for region B, and the F after the hyphen stands for forward) and rMgpB:BG37-C-R (BG37-C stands for region B from M. genitalium G37-C, and the R after the hyphen stands for reverse) (Table 2), while the week 8 variant sequence (505 bp) was amplified using primers rMgpB:B-F and rMgpB:BWk8-R (Table 2) from a pCR2.1 clone carrying the week 8 variant mgpB region B sequence. These PCR mixtures contained 10 μl template DNA, 0.2 μM each primer, 2 mM MgCl2, 200 μM each dNTP, 1.25 U native Pfu DNA polymerase (Stratagene, La Jolla, CA), and 1× Pfu buffer in a final volume of 100 μl and used the following conditions: 94°C for 4 min; 35 cycles, with 1 cycle consisting of 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min; followed by a final extension step of 72°C for 10 min. Amplicons were subsequently ligated into the Novagen pET-30 Ek/LIC vector and transformed into Escherichia coli BL21(DE3) (EMD4 Biosciences, Darmstadt, Germany). Candidate plasmids (pET-rMgpB:BG37-C or pET-rMgpB:BWk8) were sequenced to confirm that the correct insert had been cloned.
His-tagged peptides were overexpressed and purified under denaturing conditions as follows. Flasks containing 800 ml LB broth with kanamycin (30 μg/ml) were inoculated with 5 ml of an overnight culture of E. coli BL21(DE3) containing plasmid pET-rMgpB:BG37-C or pET-rMgpB:BWk8 and grown with shaking at 37°C for 2 to 3 h (optical density at 600 nm [OD650] of 0.4 to 0.8). Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM, and expression proceeded for 3 h at which time bacterial cells were collected by centrifugation and frozen. The cell pellets were thawed, resuspended in binding buffer (20 mM Tris [pH 7.9], 500 mM NaCl, 5 mM imidazole, 6 M urea), sonicated to lyse bacteria, and then incubated on ice with shaking for 1 to 2 h. Cellular debris was removed by centrifugation, and the supernatant was combined with nickel-nitrilotriacetic acid (Ni-NTA) agarose and incubated with end-over-end rotation at 4°C overnight to allow protein binding. The resin was washed three times with fresh binding buffer and then washed three more times with wash buffer (20 mM Tris [pH 7.9], 500 mM NaCl, 20 mM imidazole, 6 M urea). Proteins were eluted in 1 ml elution buffer (20 mM Tris [pH 7.9], 500 mM NaCl, 1 M imidazole, 6 M urea) after overnight rotation at 4°C. The elution was repeated with fresh elution buffer for 2 h at 4°C, and the volumes were combined. After buffer exchange to PBS using a PD-10 column (GE Healthcare, Little Chalfont, United Kingdom), the recombinant proteins (rMgpB:BG37-C and rMgpB:BWk8) were quantitated using the BCA protein assay kit (Thermo Scientific).
ELISAs.
Recombinant peptides (rMgpB:BG37-C or rMgpB:BWk8) were diluted in PBS to a final concentration of 5 μg/ml, and then 60 μl of the peptide solution was added to each well in Nunc Maxisorp 96-well flat-bottomed plates (Thermo Scientific) and incubated overnight at 4°C. After removal of the protein solution, 75 μl of blocking/diluent buffer (1× PBS with 10% nonfat milk and 0.1% Tween 20) was added and incubated overnight at 4°C. Wells were washed three times with wash buffer (1× PBS, 0.1% Tween 20) after which primary antibody diluted in blocking buffer (50 μl) was added as indicated in figure legends. After 1 h at 37°C, the wells were washed three times with wash buffer and peroxidase-conjugated secondary antibody (goat anti-human IgG [whole molecule]; Sigma-Aldrich) diluted 1:25,000 was added for another 1 h at 37°C. The secondary antibody solution was removed, the wells were washed five times in wash buffer and then developed using the SureBlue 3,3′,5,5′-tetramethylbenzidine (TMB) microwell peroxidase substrate (KPL, Gaithersburg, MD) for 10 min at which time 1 M HCl was added to stop the reaction, and the OD450 was determined in a microplate reader.
Comparable binding of the two recombinant peptides (rMgpB:BWk8 and rMgpB:BG37-C) to the ELISA plate was confirmed using polyclonal rabbit anti-His antibodies (0.1 mg/ml; Genscript, Piscataway, NJ) diluted 1:500 and detected with goat anti-rabbit IgG peroxidase-conjugated secondary antibody (data not shown).
Nucleotide sequence accession number.
The region B variant sequence identified in the primate at week 8 was deposited in GenBank under accession number JX455755.
RESULTS
Pig-tailed macaque model of persistent M. genitalium infection.
We sought to develop an animal model of persistent infection to study gene variation in M. genitalium in the context of a normal immune response. Female pig-tailed macaques (M. nemestrina) were chosen for this purpose because of the similarities of their reproductive tract anatomy, menstrual cycle, and vaginal flora to those of humans (53–55) and the previous success using this species in models of genital tract infection with other human pathogens, including Chlamydia trachomatis (56–63), Trichomonas vaginalis (64), and simian-human immunodeficiency virus (SHIV) coinfection with C. trachomatis and T. vaginalis (65). Prior to inoculation, the M. genitalium-negative status of a candidate primate was confirmed by culture and qPCR of preinoculation genital tract specimens (Table 1). To explore lower reproductive tract infection, the cervix of this animal was inoculated with M. genitalium strain G37-C (35), and the autologous salpingeal pockets of this animal were inoculated to model upper tract infection (see Materials and Methods). Salpingeal pockets were inoculated in order to assess infection and persistence of M. genitalium at multiple time points in Fallopian tube tissue given the association of this organism with tubal factor infertility (14, 15) and the detection of M. genitalium DNA in salpingitis (66). As outlined in Table 1, the lower genital tract and salpingeal pockets were analyzed at intervals for the presence of M. genitalium DNA by qPCR and for the presence of viable organisms by culture in H broth and in Vero cell cocultures. We detected M. genitalium DNA and viable organisms in the salpingeal pockets for 2 weeks and M. genitalium DNA (but no viable organisms) in a single pocket at week 4. In contrast, viable M. genitalium persisted in the lower reproductive tract for the full 8 weeks of the study, at which time the infection was eradicated with azithromycin. Viable M. genitalium bacteria were more often recovered from the lower reproductive tract specimens (vaginal swab, cervical swab, and cervical cytobrush) in coculture with Vero cells than in broth medium (Table 1), as previously described for recovery of primary cultures from human specimens (48).
Serum antibody response in an M. genitalium-infected primate.
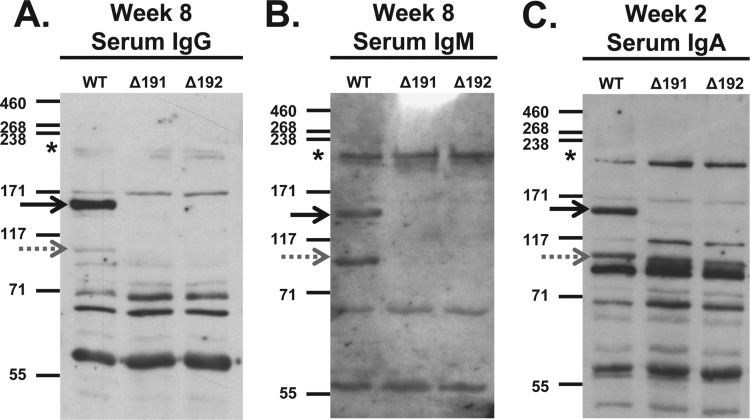
We analyzed serum samples obtained prior to and throughout the course of infection using immunoblotting to determine whether IgG, IgA, and IgM antibodies to M. genitalium antigens were produced during infection. As shown in Fig. 2, IgG, IgM, and IgA antibodies reacting with M. genitalium G37-C proteins were present in serum samples obtained prior to inoculation, indicating reaction of preexisting antibodies with M. genitalium antigens, either due to cross-reaction with other organisms or, conceivably, previous infection with M. genitalium. Laboratory primates can be naturally colonized with various Mycoplasma species (67, 68), antibodies to which are known to cross-react with M. genitalium antigens, including the immunodominant MgpB and MgpC proteins (14, 41, 69). Following inoculation, reactivity of sera to M. genitalium lysates on immunoblots increased in intensity and in the number of protein bands detected. One prominent band of ∼140 kDa, reacting with all three isotypes of primate antibodies, was confirmed to be MgpB by its absence in immunoblots of primate serum with lysates of the M. genitalium G37 mgpB deletion strain, the Δ191 mutant (Fig. 3A, B, and C). Likewise, reaction of primate serum with lysates of G37 Δmg192 mutant, a strain with a deletion in the adjacent mgpC gene, identified an ∼110-kDa protein as MgpC (Fig. 3A, B, and C). Because MgpB and MgpC are reciprocally stabilized, expression of both proteins is reduced when one of the genes is deleted (29) (Fig. 3A, B, and C). Interestingly, IgA reactivity with Δmg192 (and Δmg191) lysates revealed the presence of an unidentified band migrating slightly faster than MgpC that was not visible in the IgG or IgM blots. As previously described (29), the abundance of several proteins is affected when mgpB and mgpC are deleted, including a decrease in MG386 (P200) and an increase in MG305 (DnaK). IgA (but not IgG or IgM) also reacted with an ∼115-kDa protein that was previously noted to increase in abundance when mgpB or mgpC is deleted (29) and that is not visible in wild-type lysates (Fig. 3C).
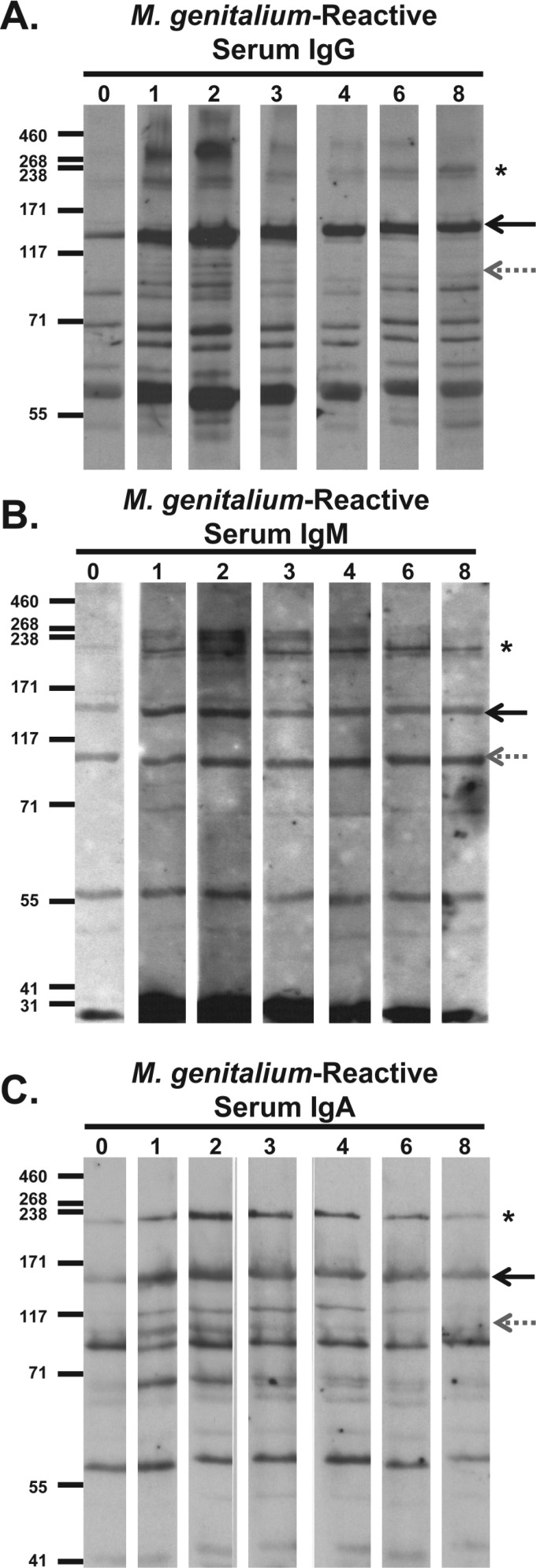
Fig 2.
Immunoblots of primate serum antibody reactivity to M. genitalium whole-cell lysates. Bacterial lysates were electrophoresed through an 8% SDS-polyacrylamide gel, transferred to membranes, and then cut into strips for reaction with primate serum samples collected at weeks 0 to 8 after infection. (A) Serum IgG reactivity. Primate serum samples were diluted 1:5,000, reac-tivity was detected using horseradish peroxidase (HRP)-conjugated goat anti-human IgG secondary antibody diluted 1:10,000, and the membranes were exposed to film for 6 min. (B) Serum IgM reactivity. Primate serum samples were diluted 1:800, reactivity was detected using HRP-conjugated goat anti-human IgM secondary antibody diluted 1:10,000, and then the membranes were exposed to film for 4 min. (C) Serum IgA reactivity. Primate serum samples were diluted 1:200, reactivity was detected using HRP-conjugated goat anti-human IgA secondary antibody diluted 1:8,000, and then the membranes were exposed to film for 13 min. The positions of MgpB (solid black arrows), MgpC (gray dotted arrows), and HMW2 (asterisks) are indicated to the right of the immunoblots. The positions of molecular mass markers (in kDa) are shown to the left of each immunoblot.
Fig 3.
Reactivity of primate serum antibodies to whole-cell lysates of wild-type M. genitalium G37-C (WT) and to mgpB (Δ191) and mgpC (Δ192) deletion mutants. (A) Serum IgG reactivity. Primate serum samples were diluted 1:5,000, reactivity was detected using HRP-conjugated goat anti-human IgG secondary antibody diluted 1:10,000 and then exposed to film for 5 min. (B) Serum IgM reactivity. Primate serum samples were diluted 1:800, and reactivity was detected using HRP-conjugated goat anti-human IgM secondary antibody diluted 1:10,000 and then exposed to film for 4 min. (C) Serum IgA reactivity. Due to the limited volume of week 8 serum samples available, the reactivity of week 2 IgA is shown. Primate serum samples were diluted 1:200, and reactivity was detected using HRP-conjugated goat anti-human IgA secondary antibody diluted 1:8,000 and then exposed to film for 20 min. The positions of MgpB (solid black arrows), MgpC (gray dotted arrows), and HMW2 (asterisks) are indicated. The positions of molecular mass markers (in kDa) are shown to the left of each immunoblot.
All three antibody isotypes (IgG, IgM, and IgA) reacted with a band migrating at approximately 190 kDa (Fig. 2A, B, and C) consistent with the migration of the HMW2 (MG218) protein on SDS-polyacrylamide gels (70) and confirmed as HMW2 by its absence in immunoblots of primate serum to lysates of a M. genitalium G37 Δmg218 deletion mutant (data not shown). Furthermore, we observed an increase in IgG and IgM antibody reactivity to proteins of >200 kDa (Fig. 2A and B) after 2 weeks of infection, perhaps corresponding to P200 (MG386) previously shown to migrate slower than any other M. genitalium protein (24, 29). Reactivity to a 57-kDa band was detected by IgG, IgM, and IgA at all time points (Fig. 2A, B, and C); we speculate that this band corresponds to GroEL, due to the cross-reactivity of the comparable proteins in other bacteria (14), and the observation that nonhuman primates are often colonized with Mycoplasma species.
Presence of M. genitalium-reactive antibodies in the primate genital tract.
To investigate the antibody response to M. genitalium at the site of infection, we analyzed cervical swab specimens for the presence of M. genitalium-reactive IgG, the predominant antibody isotype present in human genital secretions (71, 72) before and after 8 weeks of infection (Fig. 4). Anti-MgpB and anti-MgpC antibodies produced in rabbits (40) identified the locations of MgpB and MgpC on these blots (Fig. 4, left lane). Primate cervical IgG reacted strongly with the ∼140-kDa MgpB protein, but not with the 110-kDa MgpC protein, consistent with previous studies in which 95.7% of M. genitalium-infected women had MgpB-reactive antibodies in their cervical secretions but only 69.6% had MgpC-reactive cervical antibodies (40). A second, less reactive band of approximately 65 kDa reacted with the week 8 cervical antibodies; however, this band also reacted with the week 0 specimen, suggesting that it is unrelated to the current M. genitalium infection (Fig. 4). Interestingly, the pattern of cervical IgG reactivity differed from that of serum IgG (Fig. 2A and 3A) in that no reactivity was observed to the prominent ∼55-kDa band under these conditions, in contrast to its strong reactivity with serum IgG, perhaps suggesting a genital tract-specific IgG response.
Fig 4.
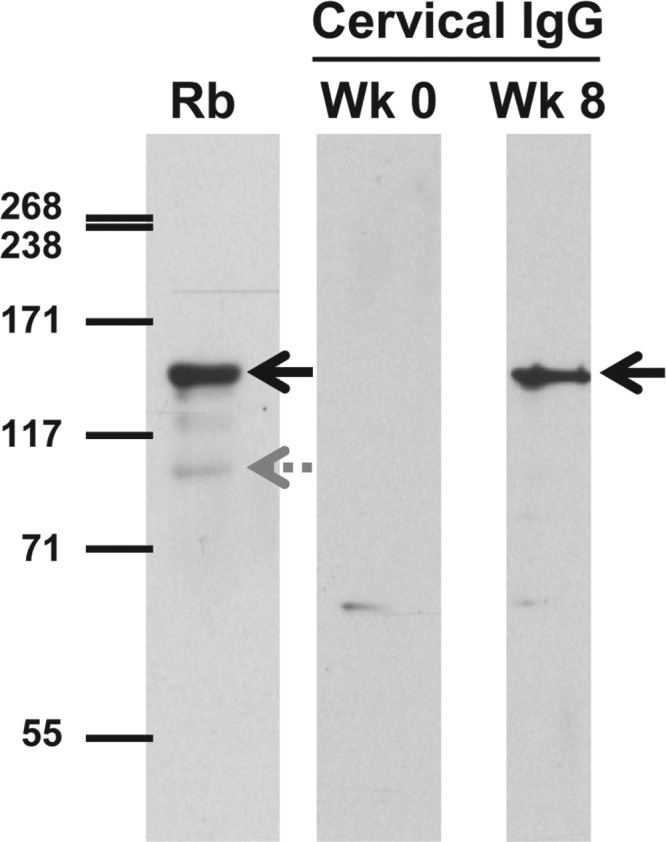
Immunoblot analysis of primate genital tract antibody reactivity to whole-cell lysates of wild-type M. genitalium. Cervical exudates collected prior to inoculation (week 0 [Wk 0]) or after 8 weeks of infection (Wk 8) were diluted 1:50 and reacted with whole-cell lysates of M. genitalium G37-C. To identify the positions of the 140-kDa MgpB and 110-kDa MgpC proteins, bacterial lysates were reacted with antibodies produced in rabbits (Rb), combined, and diluted 1:10,000. HRP-conjugated secondary antibody (anti-human IgG or anti-rabbit IgG) was diluted 1:10,000. After reaction with chemiluminescent reagents, membranes were exposed to film for 5 min or 1 min, respectively. The positions of MgpB (solid black arrows) and MgpC (gray dotted arrow) are indicated.
Variation of mgpB region B sequence during primate infection.
To assess sequence variation in the macaque model, we focused on mgpB region B, previously suggested to be the most variable region of the gene in clinical strains (45) and in persistently infected women (34). Variable region B of mgpB was PCR amplified from the week 8 primate specimen using conserved flanking primers (primers 3F and 3R [Fig. 1A]), and the resulting amplicons were cloned, sequenced, and aligned with region B of the inoculum. All 10 of the cloned amplicons contained an identical variant region B sequence that differed from that of the inoculum (and the published genome sequence of M. genitalium G37) in 56 positions over 316 bp (47 base substitutions and 9 nucleotide deletions [Fig. 1B]). These sequence changes maintain the reading frame of MgpB and predict a protein that differs in 21 amino acids (including three deletions) from the inoculated strain (Fig. 1C). Interestingly, this particular region B sequence has not been previously reported in vitro or in human specimens, consistent with the large number of possible variants predicted for this unique segmental recombination strategy (34). The mgpB region B sequence of the primate inoculum was similarly analyzed, and as expected for this single-colony-cloned strain, all of the 11 plasmid inserts sequenced were identical to the published genome sequence (Fig. 1B). A second PCR using primers ModPetF and 1415R, another primer pair flanking variable region B (Fig. 1A), confirmed these findings: all 25 plasmids cloned from the week 8 specimen contained the identical variant sequence, while all 25 amplicons cloned from the inoculum were identical to the published G37 sequence.
A similar sequence analysis was applied to cervical cytobrush specimens collected at week 2 and week 4 to determine when the new variant sequence emerged. The mgpB region B sequence of the inoculum predominated at both time points: all of the 22 plasmids sequenced from week 2 and 15 of 16 plasmids sequenced from week 4 matched the M. genitalium G37-C sequence (data not shown). Interestingly, a single plasmid sequence from week 4 indicated a novel minority variant population that resulted from recombination with MgPar2 (data not shown). The specimens collected at week 5 were negative by PCR (Table 1), so it was not possible to determine precisely when the week 8 variant arose. We conclude that the week 8 variant sequence arose after 4 weeks of infection.
In order to assess gene variation in the absence of selection in the primate, region B of mgpB was amplified from the primate M. genitalium G37-C inoculum cultured in vitro by serial passage in H broth for 8 weeks. All 25 amplicons sequenced from the in vitro-passaged inoculum were identical to the published G37-C sequence of region B in the mgpB expression site (Fig. 1B). Thus, the variation in the mgpB region B sequence was observed only in M. genitalium present in the genital tract of the primate and not in bacteria cultured in vitro from the same inoculum for a similar length of time.
Sequencing of MgPar8 confirms the reciprocal nature of recombination in vivo.
We have previously shown that mgpB and mgpC sequence variation is extensive in persistently infected women, and using clonal derivatives of the M. genitalium G37 type strain, have demonstrated that this variation is predominantly achieved by reciprocal recombination with the MgPar sequences in vitro (35). Given the complexity of multiple heterologous mgpB, mgpC, and MgPar sequences within individual human specimens, as well as the uncharacterized MgPar donor sequences for the different infecting strains, it has been difficult to determine the recombination partners in vivo (34). Thus, to determine whether the recombination in our persistently infected primate was achieved through reciprocal or unidirectional recombination (gene conversion), we first aligned the novel week 8 variant sequence with the MgPar sequences of M. genitalium G37-C and identified MgPar8 as the most likely recombination partner (Fig. 1B). We also determined that the week 8 mgpB region B variant sequence was identical to the region B-homologous sequences of MgPar8 in the inoculum (34), suggesting that 316 bp of MgPar8 had replaced the region B sequence of the mgpB expression site. To determine whether this recombination event was reciprocal or unidirectional, we PCR amplified the region B-homologous region of MgPar8 from the week 8 primate specimen and then sequenced the cloned amplicons. All of the 18 sequences analyzed from MgPar8 from the week 8 specimen were identical to the region B sequences of the mgpB expression site of the inoculum, confirming the reciprocal nature of this recombination event in vivo (Fig. 1B). In contrast, a similar analysis of the region B-homologous sequences of MgPar8 in the inoculum and in the in vitro-passaged inoculum revealed that all 10 and 9 of 10 cloned amplicons were identical to the published MgPar8 sequence, respectively. One sequence amplified from MgPar8 of the in vitro-passaged inoculum differed from the published MgPar8 sequence over a smaller region (61 bp rather than 316 bp), and thus may represent an independent recombination event [“In vitro(2)” in Fig. 1B]. Given that recombination in M. genitalium is ongoing and frequent (34, 35, 37, 39), it is not surprising to find a small population of other variant sequences present in this in vitro-grown culture, even after it has been cloned from a single colony obtained after filtration.
The observed sequence variations are unlikely to be due to errors introduced during PCR and/or sequencing for the following reasons. (i) Two different PCRs using different primer pairs yielded identical results for the week 8 primate specimen and the inoculum. (ii) The mgpB region B sequence changes were observed only in the week 8 specimen, and none were detected in the inoculum or in the in vitro-passaged bacteria. (iii) All of the observed sequence changes maintained the reading frame of mgpB. (iv) All of the observed changes can be attributed to reciprocal exchange with MgPar8.
In silico identification of predicted B-cell epitopes.
We used in silico analysis to compare predicted B-cell epitopes within variable region B of the inoculum and the week 8 variant. The BepiPred 1.0 program (51), which identifies linear B-cell epitopes, predicts that the majority of the M. genitalium G37-C region B peptide (88 of 133 amino acids) comprises a B-cell epitope when the default threshold of 0.35 (49% sensitivity, 75% specificity) is used (data not shown). When the threshold was increased to 0.9 (25% sensitivity, 91% specificity), four linear epitopes were identified in the G37-C region B sequence including amino acids that differ in the week 8 variant (Fig. 5A, bold amino acids). Interestingly, this analysis predicts the loss of one B-cell epitope and the gain of a new epitope in the variant sequence. In comparison, a similar analysis of the N-terminal 208 amino acids of MgpB predicts that only 9 amino acids are included in predicted epitopes. Conformational B-cell epitopes were predicted for the full-length MgpB protein of strain G37-C using CBTOPE (52) as shown in Fig. 5A (overlines). A similar analysis for the week 8 variant was not performed because the sequences of the other variable regions of mgpB have not been determined. Notably, several conformational and linear epitopes were predicted to include amino acids that vary between the infecting and week 8 variant mgpB region B sequence. While these results represent in silico predictions, they do support the hypothesis that epitopes have changed in response to immune pressure.
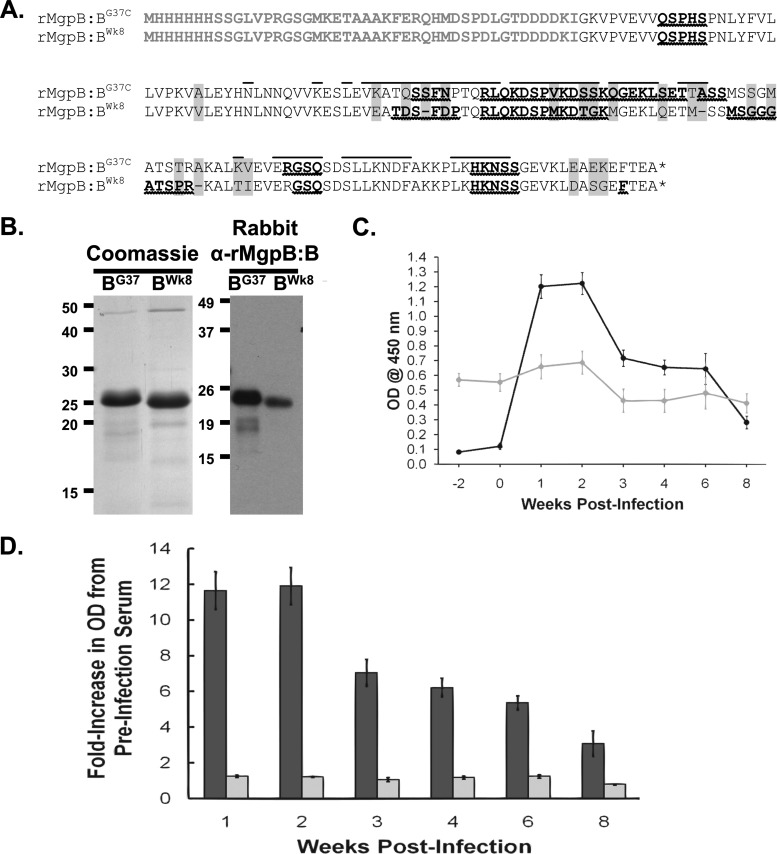
Fig 5.
Analysis of reactivity of primate serum to recombinant wild-type and variant MgpB region B peptides. (A) Amino acid sequences of the wild-type (rMgpB:BG37) and week 8 variant (rMgpB:BWk8) MgpB region B His-tagged recombinant peptides. Plasmid vector-associated amino acids, including the His tag, are shown in gray type. Amino acids that differ between the wild-type and week 8 peptides are indicated by gray background, and deleted amino acids are represented by a dash. The asterisk indicates the TGA codon that encodes stop in E. coli. Predicted linear B-cell epitopes are shown in bold type with wavy underlines; conformational B-cell epitopes for mycobacterial G37-C region B are overlined. His tag and vector-encoded amino acids were omitted from B-cell epitope predictions. (B) (Left) Coomassie blue stain of purified recombinant peptides. (Right) Reactivity of rabbit anti-MgpB region B (α-rMgpB:B) antibodies to purified recombinant peptides. Rabbit serum samples from animals immunized with a recombinant MgpB region B peptide consisting of amino acids 185 to 352 (Iverson-Cabral and Totten, unpublished data) were used at 1:1,000,000 and detected with HRP-conjugated goat anti-rabbit IgG secondary antibody diluted 1:1,000. (C) ELISA reactivity of primate serum to recombinant peptides over the course of infection. Reactivity of serum to rMgpB:BG37 (black circles) increased over time, while reactivity to rMgpB:BWk8 (gray circles) remained unchanged from the high background reactivity observed at weeks −2 and 0 (prior to infection). Primate serum samples were diluted 1:50 and detected with HRP-conjugated goat anti-human IgG secondary antibodies diluted 1:25,000. Data shown are averages of triplicate wells from a typical experiment repeated at least three times, and the error bars show standard errors. A no-antigen control was included for each plate, and this value was subtracted from the data shown. (D) ELISA data presented as fold increase compared to preinoculation values (average of week −2 and week 0). The graph shows the averages of five experiments, each with triplicate wells. Dark bars, wild-type peptide; light bars, week 8 variant peptide. Error bars indicate standard errors. The fold increase for reactivity to the rMgpB:BG37-C peptide was significantly different by Student's t test for weeks 1 through 6 (P < 0.001) and for week 8 (P = 0.0035) compared to preinoculation values.
Reactivity of primate serum to the variant MgpB region B peptide sequence.
The predominance of the novel variant sequence and the complete absence of the original infecting sequence in the week 8 cervical sample suggested that specific MgpB variants are selected in vivo. To explore the reactivity of primate antibodies to this new sequence within the mgpB expression site and to test the epitope predictions experimentally, we cloned, expressed, and purified peptides corresponding to the MgpB region B sequences of the inoculum and the week 8 variant (Fig. 5A). The resulting peptides (encompassing amino acids 185 to 320) are predicted to differ in 21 positions, including 18 amino acid substitutions and 3 deletions (Fig. 5A). Coomassie blue-stained SDS-PAGE analysis (Fig. 5B, left) confirmed that a protein of the correct size had been purified to homogeneity. Antibodies produced in rabbits after immunization with a mycobacterial G37-C region B peptide encompassing amino acids 185 to 352 identical to the inoculum (S. L. Iverson-Cabral and P. A. Totten, unpublished data) reacted with both peptides in immunoblots; however, reactivity with the purified variant peptide was reduced (Fig. 5B, right). To determine whether antibodies induced in the infected primate were less reactive to the variant peptide than with the inoculated sequence, we assessed reactivity of both peptides in ELISAs. As shown in Fig. 5C, there was an increase in reactivity of primate serum to region B of the inoculated protein sequence over time. This increase was not observed when the same serum was reacted with the comparable peptide representing the week 8 variant sequence. Figure 5D shows the ELISA results presented as fold change compared to the preinoculation values (week −2 and week 0 combined) and illustrates the 11- to 12-fold increase in reactivity of the primate serum to the region B peptide of the inoculum, with no significant increase in reactivity to the week 8 variant sequence. These results corroborate the in silico B-cell epitope predictions (Fig. 5A) and are consistent with a model in which antibodies are generated to MgpB region B during infection and in which antibodies reacting to a particular region B select against cells expressing that sequence.
DISCUSSION
With a disease spectrum, including urethritis, mucopurulent cervicitis, and upper genital tract infection, M. genitalium has a clinical significance rivaling that of Chlamydia trachomatis and Neisseria gonorrhoeae. In addition to lower tract infections in men and women—urethritis and cervicitis, this organism has been associated with serious upper tract infections in women—acute endometritis, chronic pelvic inflammatory disease, preterm birth, and possibly salpingitis (4–14, 16, 17, 41). Our previous studies have demonstrated that lower tract infection with M. genitalium can persist for months and even years in infected women (18), possibly increasing the ability of this organism to disseminate to the upper tract to cause upper tract disease and its sequelae. We and others (34–36, 45) have hypothesized that the ability of M. genitalium to persist in vivo is due in part to antigenic variation in the sequences of two immunodominant surface-exposed proteins, accomplished by recombination with archived partial sequences of these genes distributed throughout the chromosome.
In the current study, we sought to develop an animal model that could address these hypotheses. Such a model should allow persistent infection and recovery of viable organisms, development of an immune response similar to that seen in human infection, and allow infection of both the upper and lower genital tracts. The results with a pilot primate support the utility of pig-tailed macaques to serve as an informative animal model for the study of M. genitalium reproductive tract infections. Cervicovaginal infection persisted throughout the experiment (8 weeks) and for 2 to 4 weeks in autologously transplanted salpingeal tissue. Serum and cervicovaginal antibodies that recognize M. genitalium antigens including MgpB were induced, and sequence variation of mgpB, mediated by reciprocal recombination with MgPar sequences, occurred in vivo. Finally, we show that antibodies elicited during infection preferentially recognize the MgpB peptides derived from the infecting strain compared to peptides derived from the predominant variant present after 8 weeks of infection.
To our knowledge, this study is the first description of the use of pig-tailed macaques as a potential animal model in which to study infection and persistence of M. genitalium in the lower genital tract. Previous studies describe genital tract infection established in hormone-treated mice (42, 73–75). Mice have the advantages of small size, low cost, genetic homogeneity, and ready availability and have been used extensively in models of human genital tract pathogens. However, the distant evolutionary relationship between mice and humans and the need for hormone treatment to establish M. genitalium infection in mice highlight the need for additional complementary systems in which to model human genital tract infection. Infection of hamsters has been reported; however, because the respiratory systems of the hamsters were inoculated, the relevance to genital tract infection is unclear (76). Among primates, male chimpanzees and cynomolgous monkeys (Macaca fascicularis) were infected after urethral challenge, but male rhesus monkeys (Macaca mulatta) resisted infection (44, 77, 78). Lower genital tract infection has been established in female chimpanzees, tamarins, squirrel monkeys, marmosets, and baboons (44, 77, 78). In some studies, oviducts were inoculated directly with M. genitalium resulting in salpingitis (74, 77). Experiments with the female pig-tailed macaque, a species with the advantages of small size, a reproductive physiology similar to that of humans, and a greater availability for research purposes, will expand the findings of these animal models. Previous studies determined that the normal vaginal flora cultured from female macaques is similar to bacteria recovered from the human vagina (55). Comparison of the macaque and human microbiomes by molecular methods, which is under way (Dorothy L. Patton, personal communication), will ascertain whether those similarities extend to unculturable organisms.
The salpingeal pocket model, successfully used to model Fallopian tube infection by C. trachomatis (47), may allow studies relevant to tubal factor infertility and salpingitis caused by M. genitalium infection in humans. As we found that viable M. genitalium persisted in salpingeal tissue for only 2 weeks, the pocket model may be best suited to model the early stages of infection. Future experiments will endeavor to optimize this method to improve persistence of M. genitalium in the pocket model given the value of inoculating a single animal with multiple strains and collection of longitudinal samples.
The development of serum antibodies reacting with several M. genitalium antigens including the immunodominant MgpB protein in this macaque is consistent with previously published studies in humans (14, 40, 41) and in animal models (42, 43, 79). The infected primate serum also reacted with MgpC, similar to human infections (40), and HMW2, proteins that, along with MgpB, are components of the tip organelle (25). The observation that our pilot primate exhibited M. genitalium-reactive antibodies prior to infection suggests that other mycoplasma species with cross-reactive antigens may have previously (or currently) colonized this primate. Regardless of the source of these antibodies, they were not effective in preventing the establishment or persistence of infection in our pilot primate. The increase in antibody reactivity to M. genitalium in our pilot animal by the first week postinfection was likely a consequence of multiple subcutaneous pocket inoculations, in addition to the cervical inoculation. The MgpB-specific IgG present in the lower genital tract may be produced locally in response to cervical infection or secreted from plasma (80) as a result of the salpingeal pocket inoculations. Importantly, MgpB-specific antibodies were induced in cervicovaginal secretions consistent with our previously published report documenting the presence of MgpB- and MgpC-specific cervicovaginal antibodies in infected women (40).
The persistence of M. genitalium in the lower genital tract throughout the experiment, despite the presence of MgpB-specific antibodies, prompted the analysis of sequence variation during infection. Because we inoculated this animal with M. genitalium G37-C, our clonal derivative of the G37 type strain, we were able to track sequence variation in mgpB and correlate sequence changes with the appearance of antibodies reacting to the resulting protein. While there are three variable regions (B, EF, and G) within mgpB and one (KLM) within the adjacent mgpC, all of which we predict to vary over the course of an infection, we chose to focus our analysis on mgpB variable region B, as this region is the most immunogenic (Iverson-Cabral and Totten, unpublished) and is most diverse among strains of M. genitalium (45). We found that the predominant sequence of mgpB region B in the mycobacterial G37-C inoculum was replaced by a novel sequence detected in the cervical specimen collected at 8 weeks postinoculation, consistent with our hypothesis of antibody-mediated selection in vivo. This interpretation is supported by the predicted differences in B-cell epitopes between these regions in the inoculated and week 8 variants, the reduced reactivity of the week 8 sequences in sera from rabbits immunized with the inoculated strain, and the induction of antibodies reactive with the inoculated variant, but not to the week 8 variant, in this animal. Further, the selection of variant sequences in vivo, but not in vitro, in our experiment supports our hypothesis. However, we cannot rule out the possibility that unknown differences in replication rates between the in vitro- and in vivo-passaged bacteria affected recombination rates resulting in different sequences predominating after 8 weeks. In addition, it is not known whether the week 8 variant sequence in the primate was present in low numbers in the initial inoculum or whether recombination occurred during genital tract infection. These two possibilities might be differentiated using our anchored PCR method (35, 37) in which one primer anchored in the expression site is paired with a second primer targeting variant sequences in the MgPar sites. This highly sensitive approach produces a PCR product only if the target variant sequence recombines into the expression site.
Although preexisting, presumably cross-reactive antibodies to the week 8 variant in the primate were present prior to inoculation, we failed to detect an induction in antibodies to the peptide that predominated at week 8. Interestingly, the inoculum and week 8 recombinant peptides are 84% identical, indicating that even small sequence changes can affect antibody recognition of these variable regions. Together these observations are consistent with a role for antibodies in selecting against M. genitalium expressing the original sequence and promoting the emergence of antigenic variants that avoid antibody-mediated clearance. In silico methods identified potential linear and conformational B-cell epitopes within variable region B, including amino acids that differ between the inoculum and week 8 variants. Consistent with these computer-based predictions, sequence changes in mgpB region B reduced antibody reactivity, suggesting the occurrence of antibody-mediated selection in vivo. Future experiments measuring the biologic activity of variant-specific antibodies (such as complement-mediated killing, opsonization, and/or inhibition of adherence) would support this hypothesis.
Partial, segmental recombination between the mgpB-mgpC expression site and the MgPar sites (and between the MgPar sites themselves) as documented by our group and others (34, 35, 37, 39, 45), results in an almost limitless possible array of sequence variants. Although we have determined that RecA is required for mgpB and mgpC variation (37) and that recombination between mgpB or mgpC and the MgPar sites is predominantly reciprocal in vitro (35), the nature of recombination in vivo has been controversial. Ma et al. (39, 45) recently suggested that recombination in vivo is predominantly unidirectional (also termed gene conversion) after examining the sequences of the mgpB or mgpC expression site and MgPar sequences in specimens from urethritis patients. Infection of a model animal using a known, clonal population of M. genitalium provided a controlled opportunity to examine the outcome of recombination events in vivo more accurately than in humans in which multiple variants are present in a single specimen. Accordingly, after identifying MgPar8 as the putative donor site in the recombination event that generated the week 8 variant in our primate, we examined the MgPar8 sequence in the same specimen. We found that the original expression site sequence now resides in MgPar8 consistent with reciprocal recombination during infection and supporting our previous conclusions using bacteria cultured in vitro (35). However, we cannot rule out the possibility that DNA repair subsequent to reciprocal recombination might mimic gene conversion or that the balance between DNA repair and recombination might differ between strains or under different inducing conditions. The predominance of reciprocal recombination both in vitro and in vivo is remarkable in that most other bacterial pathogens utilize unidirectional recombination to accomplish antigenic variation (81).
The success of our pilot primate experiment suggests that pig-tailed macaques will be a valuable model animal in which to study M. genitalium pathogenesis and persistence in the context of an active immune response. While the costly nature of experiments involving laboratory primates justified the use of a single animal, this experiment clearly needs to be repeated with other female pig-tailed macaques to ascertain whether the results obtained with this animal generalize to other individual pig-tailed macaques. Future experiments will include cervical infection in the absence of salpingeal pocket inoculations to assess antigenic variation induced by cervical infection alone, localization of M. genitalium with respect to cervical and immune cell types (including intra- and extracellular location), and assessment of potential upper reproductive tract infection and sequelae subsequent to cervical infection. Defining the biological activity of MgpB-specific antibodies is of particular importance in understanding the contribution of sequence variation to persistence, as is the relationship between sequence variation and the configuration of these proteins on the cell surface.
ACKNOWLEDGMENTS
We thank Arturo Centurion and Raul Burgos for helpful discussions.
This work was supported by grants AI082316 and AI074898 from the National Institutes of Health, and funding from the Washington National Primate Research Center and the Department of Medicine, University of Washington. S.L.I.-C. was supported by the University of Washington STD/AIDS research training grants 124 NIH/NIAID T32 AI7140-33 and -34.
Footnotes
Published ahead of print 3 June 2013
REFERENCES
- 1. Manhart LE, Broad JM, Golden MR. 2011. Mycoplasma genitalium: should we treat and how? Clin. Infect. Dis. 53(Suppl 3):S129–S142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McGowin CL, Anderson-Smits C. 2011. Mycoplasma genitalium: an emerging cause of sexually transmitted disease in women. PLoS Pathog. 7:e1001324. 10.1371/journal.ppat.1001324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taylor-Robinson D, Jensen JS. 2011. Mycoplasma genitalium: from chrysalis to multicolored butterfly. Clin. Microbiol. Rev. 24:498–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Horner PJ, Gilroy CB, Thomas BJ, Naidoo RO, Taylor-Robinson D. 1993. Association of Mycoplasma genitalium with acute non-gonococcal urethritis. Lancet 342:582–585 [DOI] [PubMed] [Google Scholar]
- 5. Jensen JS, Orsum R, Dohn B, Uldum S, Worm AM, Lind K. 1993. Mycoplasma genitalium: a cause of male urethritis? Genitourin. Med. 69:265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Totten PA, Schwartz MA, Sjostrom KE, Kenny GE, Handsfield HH, Weiss JB, Whittington WL. 2001. Association of Mycoplasma genitalium with nongonococcal urethritis in heterosexual men. J. Infect. Dis. 183:269–276 [DOI] [PubMed] [Google Scholar]
- 7. Gaydos C, Maldeis NE, Hardick A, Hardick J, Quinn TC. 2009. Mycoplasma genitalium as a contributor to the multiple etiologies of cervicitis in women attending sexually transmitted disease clinics. Sex. Transm. Dis. 36:598–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Manhart LE, Critchlow CW, Holmes KK, Dutro SM, Eschenbach DA, Stevens CE, Totten PA. 2003. Mucopurulent cervicitis and Mycoplasma genitalium. J. Infect. Dis. 187:650–657 [DOI] [PubMed] [Google Scholar]
- 9. Pepin J, Labbe AC, Khonde N, Deslandes S, Alary M, Dzokoto A, Asamoah-Adu C, Meda H, Frost E. 2005. Mycoplasma genitalium: an organism commonly associated with cervicitis among west African sex workers. Sex. Transm. Infect. 81:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anagrius C, Lore B, Jensen JS. 2005. Mycoplasma genitalium: prevalence, clinical significance, and transmission. Sex. Transm. Infect. 81:458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moi H, Reinton N, Moghaddam A. 2009. Mycoplasma genitalium in women with lower genital tract inflammation. Sex. Transm. Infect. 85:10–14 [DOI] [PubMed] [Google Scholar]
- 12. Cohen CR, Manhart LE, Bukusi EA, Astete S, Brunham RC, Holmes KK, Sinei SK, Bwayo JJ, Totten PA. 2002. Association between Mycoplasma genitalium and acute endometritis. Lancet 359:765–766 [DOI] [PubMed] [Google Scholar]
- 13. Haggerty CL, Totten PA, Astete SG, Lee S, Hoferka SL, Kelsey SF, Ness RB. 2008. Failure of cefoxitin and doxycycline to eradicate endometrial Mycoplasma genitalium and the consequence for clinical cure of pelvic inflammatory disease. Sex. Transm. Infect. 84:338–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clausen HF, Fedder J, Drasbek M, Nielsen PK, Toft B, Ingerslev HJ, Birkelund S, Christiansen G. 2001. Serological investigation of Mycoplasma genitalium in infertile women. Hum. Reprod. 16:1866–1874 [DOI] [PubMed] [Google Scholar]
- 15. Svenstrup HF, Fedder J, Kristoffersen SE, Trolle B, Birkelund S, Christiansen G. 2008. Mycoplasma genitalium, Chlamydia trachomatis, and tubal factor infertility–a prospective study. Fertil. Steril. 90:513–520 [DOI] [PubMed] [Google Scholar]
- 16. Edwards RK, Ferguson RJ, Reyes L, Brown M, Theriaque DW, Duff P. 2006. Assessing the relationship between preterm delivery and various microorganisms recovered from the lower genital tract. J. Matern. Fetal Neonatal Med. 19:357–363 [DOI] [PubMed] [Google Scholar]
- 17. Hitti J, Garcia P, Totten P, Paul K, Astete S, Holmes KK. 2010. Correlates of cervical Mycoplasma genitalium and risk of preterm birth among Peruvian women. Sex. Transm. Dis. 37:81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cohen CR, Nosek M, Meier A, Astete SG, Iverson-Cabral S, Mugo NR, Totten PA. 2007. Mycoplasma genitalium infection and persistence in a cohort of female sex workers in Nairobi, Kenya. Sex. Transm. Dis. 34:274–279 [DOI] [PubMed] [Google Scholar]
- 19. Horner P, Thomas B, Gilroy C, Egger M, McClure M, Taylor-Robinson D. 2003. Antibodies to Chlamydia trachomatis heat-shock protein 60 kDa and detection of Mycoplasma genitalium and Ureaplasma urealyticum are associated independently with chronic nongonococcal urethritis. Sex. Transm. Dis. 30:129–133 [DOI] [PubMed] [Google Scholar]
- 20. Taylor-Robinson D, Gilroy CB, Thomas BJ, Hay PE. 2004. Mycoplasma genitalium in chronic non-gonococcal urethritis. Int. J. STD AIDS 15:21–25 [DOI] [PubMed] [Google Scholar]
- 21. Hjorth SV, Bjornelius E, Lidbrink P, Falk L, Dohn B, Berthelsen L, Ma L, Martin DH, Jensen JS. 2006. Sequence-based typing of Mycoplasma genitalium reveals sexual transmission. J. Clin. Microbiol. 44:2078–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burgos R, Pich OQ, Querol E, Pinol J. 2007. Functional analysis of the Mycoplasma genitalium MG312 protein reveals a specific requirement of the MG312 N-terminal domain for gliding motility. J. Bacteriol. 189:7014–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burgos R, Pich OQ, Querol E, Pinol J. 2008. Deletion of the Mycoplasma genitalium MG_217 gene modifies cell gliding behaviour by altering terminal organelle curvature. Mol. Microbiol. 69:1029–1040 [DOI] [PubMed] [Google Scholar]
- 24. Pich OQ, Burgos R, Ferrer-Navarro M, Querol E, Pinol J. 2006. Mycoplasma genitalium mg200 and mg386 genes are involved in gliding motility but not in cytadherence. Mol. Microbiol. 60:1509–1519 [DOI] [PubMed] [Google Scholar]
- 25. Pich OQ, Burgos R, Ferrer-Navarro M, Querol E, Pinol J. 2008. Role of MG218 and MG317 cytoskeletal proteins in terminal organelle organization, gliding motility and cytadherence. Microbiology (Reading, Engl.) 154:3188–3198 [DOI] [PubMed] [Google Scholar]
- 26. Baczynska A, Funch P, Fedder J, Knudsen HJ, Birkelund S, Christiansen G. 2007. Morphology of human Fallopian tubes after infection with Mycoplasma genitalium and Mycoplasma hominis–in vitro organ culture study. Hum. Reprod. 22:968–979 [DOI] [PubMed] [Google Scholar]
- 27. Opitz O, Jacobs E. 1992. Adherence epitopes of Mycoplasma genitalium adhesin. J. Gen. Microbiol. 138:1785–1790 [DOI] [PubMed] [Google Scholar]
- 28. Svenstrup HF, Nielsen PK, Drasbek M, Birkelund S, Christiansen G. 2002. Adhesion and inhibition assay of Mycoplasma genitalium and M. pneumoniae by immunofluorescence microscopy. J. Med. Microbiol. 51:361–373 [DOI] [PubMed] [Google Scholar]
- 29. Burgos R, Pich OQ, Ferrer-Navarro M, Baseman JB, Querol E, Pinol J. 2006. Mycoplasma genitalium P140 and P110 cytadhesins are reciprocally stabilized and required for cell adhesion and terminal-organelle development. J. Bacteriol. 188:8627–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mernaugh GR, Dallo SF, Holt SC, Baseman JB. 1993. Properties of adhering and nonadhering populations of Mycoplasma genitalium. Clin. Infect. Dis. 17(Suppl 1):S69–S78 [DOI] [PubMed] [Google Scholar]
- 31. Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, Fritchman RD, Weidman JF, Small KV, Sandusky M, Fuhrmann J, Nguyen D, Utterback TR, Saudek DM, Phillips CA, Merrick JM, Tomb JF, Dougherty BA, Bott KF, Hu PC, Lucier TS, Peterson SN, Smith HO, Hutchison CA, III, Venter JC. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397–403 [DOI] [PubMed] [Google Scholar]
- 32. Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, III, Smith HO, Venter JC. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U. S. A. 103:425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165–2169 [DOI] [PubMed] [Google Scholar]
- 34. Iverson-Cabral SL, Astete SG, Cohen CR, Rocha EP, Totten PA. 2006. Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences. Infect. Immun. 74:3715–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iverson-Cabral SL, Astete SG, Cohen CR, Totten PA. 2007. mgpB and mgpC sequence diversity in Mycoplasma genitalium is generated by segmental reciprocal recombination with repetitive chromosomal sequences. Mol. Microbiol. 66:55–73 [DOI] [PubMed] [Google Scholar]
- 36. Peterson SN, Bailey CC, Jensen JS, Borre MB, King ES, Bott KF, Hutchison CA., III 1995. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc. Natl. Acad. Sci. U. S. A. 92:11829–11833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burgos R, Wood GE, Young L, Glass JI, Totten PA. 2012. RecA mediates MgpB and MgpC phase and antigenic variation in Mycoplasma genitalium, but plays a minor role in DNA repair. Mol. Microbiol. 85:669–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma L, Jensen JS, Mancuso M, Hamasuna R, Jia Q, McGowin CL, Martin DH. 2012. Variability of trinucleotide tandem repeats in the MgPa operon and its repetitive chromosomal elements in Mycoplasma genitalium. J. Med. Microbiol. 61:191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma L, Jensen JS, Myers L, Burnett J, Welch M, Jia Q, Martin DH. 2007. Mycoplasma genitalium: an efficient strategy to generate genetic variation from a minimal genome. Mol. Microbiol. 66:220–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iverson-Cabral SL, Manhart LE, Totten PA. 2011. Detection of Mycoplasma genitalium-reactive cervicovaginal antibodies among infected women. Clin. Vaccine Immunol. 18:1783–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Svenstrup HF, Jensen JS, Gevaert K, Birkelund S, Christiansen G. 2006. Identification and characterization of immunogenic proteins of Mycoplasma genitalium. Clin. Vaccine Immunol. 13:913–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McGowin CL, Spagnuolo RA, Pyles RB. 2010. Mycoplasma genitalium rapidly disseminates to the upper reproductive tracts and knees of female mice following vaginal inoculation. Infect. Immun. 78:726–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morrison-Plummer J, Jones DH, Daly K, Tully JG, Taylor-Robinson D, Baseman JB. 1987. Molecular characterization of Mycoplasma genitalium species-specific and cross-reactive determinants: identification of an immunodominant protein of M. genitalium. Isr. J. Med. Sci. 23:453–457 [PubMed] [Google Scholar]
- 44. Tully JG, Taylor-Robinson D, Rose DL, Furr PM, Graham CE, Barile MF. 1986. Urogenital challenge of primate species with Mycoplasma genitalium and characteristics of infection induced in chimpanzees. J. Infect. Dis. 153:1046–1054 [DOI] [PubMed] [Google Scholar]
- 45. Ma L, Jensen JS, Mancuso M, Hamasuna R, Jia Q, McGowin CL, Martin DH. 2010. Genetic variation in the complete MgPa operon and its repetitive chromosomal elements in clinical strains of Mycoplasma genitalium. PLoS One 5:e15660. 10.1371/journal.pone.0015660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Phillips E, Nash P. 1985. Culture media, p 1051–1092 In Lennette EH, Balows A, Hausler WJ, Jr, Shadomy HJ. (ed), Manual of clinical microbiology, 4th ed American Society for Microbiology, Washington, DC [Google Scholar]
- 47. Patton DL, Kuo CC, Wang SP, Brenner RM, Sternfeld MD, Morse SA, Barnes RC. 1987. Chlamydial infection of subcutaneous fimbrial transplants in cynomolgus and rhesus monkeys. J. Infect. Dis. 155:229–235 [DOI] [PubMed] [Google Scholar]
- 48. Jensen JS, Hansen HT, Lind K. 1996. Isolation of Mycoplasma genitalium strains from the male urethra. J. Clin. Microbiol. 34:286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dutro SM, Hebb JK, Garin CA, Hughes JP, Kenny GE, Totten PA. 2003. Development and performance of a microwell-plate-based polymerase chain reaction assay for Mycoplasma genitalium. Sex. Transm. Dis. 30:756–763 [DOI] [PubMed] [Google Scholar]
- 50. Tully JG, Rose DL, Whitcomb RF, Wenzel RP. 1979. Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly-modified culture medium. J. Infect. Dis. 139:478–482 [DOI] [PubMed] [Google Scholar]
- 51. Larsen JE, Lund O, Nielsen M. 2006. Improved method for predicting linear B-cell epitopes. Immunome Res. 2:2. 10.1186/1745-7580-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ansari HR, Raghava GP. 2010. Identification of conformational B-cell epitopes in an antigen from its primary sequence. Immunome Res. 6:6. 10.1186/1745-7580-6-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blakley GB, Beamer TW, Dukelow WR. 1981. Characteristics of the menstrual cycle in nonhuman primates. IV. Timed mating in Macaca nemestrina. Lab. Anim. 15:351–353 [DOI] [PubMed] [Google Scholar]
- 54. Lichtenwalner AB, Patton DL, Klebanoff SJ, Headley CM, Hillier SL. 2000. Vaginal myeloperoxidase and flora in the pig-tailed macaque. J. Med. Primatol. 29:36–41 [DOI] [PubMed] [Google Scholar]
- 55. Patton DL, Sweeney YC, Rabe LK, Hillier SL. 1996. The vaginal microflora of pig-tailed macaques and the effects of chlorhexidine and benzalkonium on this ecosystem. Sex. Transm. Dis. 23:489–493 [DOI] [PubMed] [Google Scholar]
- 56. Bell JD, Bergin IL, Schmidt K, Zochowski MK, Aronoff DM, Patton DL. 2011. Nonhuman primate models used to study pelvic inflammatory disease caused by Chlamydia trachomatis. Infect. Dis. Obstet. Gynecol. 2011:675360. 10.1155/2011/675360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Patton DL. 1985. Immunopathology and histopathology of experimental chlamydial salpingitis. Rev. Infect. Dis. 7:746–753 [DOI] [PubMed] [Google Scholar]
- 58. Patton DL, Halbert SA, Kuo CC, Wang SP, Holmes KK. 1983. Host response to primary Chlamydia trachomatis infection of the Fallopian tube in pig-tailed monkeys. Fertil. Steril. 40:829–840 [PubMed] [Google Scholar]
- 59. Patton DL, Wolner-Hanssen P, Cosgrove SJ, Holmes KK. 1990. The effects of Chlamydia trachomatis on the female reproductive tract of the Macaca nemestrina after a single tubal challenge following repeated cervical inoculations. Obstet. Gynecol. 76:643–650 [PubMed] [Google Scholar]
- 60. Peeling RW, Patton DL, Cosgrove Sweeney YT, Cheang MS, Lichtenwalner AB, Brunham RC, Stamm WE. 1999. Antibody response to the chlamydial heat-shock protein 60 in an experimental model of chronic pelvic inflammatory disease in monkeys (Macaca nemestrina). J. Infect. Dis. 180:774–779 [DOI] [PubMed] [Google Scholar]
- 61. Van Voorhis WC, Barrett LK, Sweeney YT, Kuo CC, Patton DL. 1996. Analysis of lymphocyte phenotype and cytokine activity in the inflammatory infiltrates of the upper genital tract of female macaques infected with Chlamydia trachomatis. J. Infect. Dis. 174:647–650 [DOI] [PubMed] [Google Scholar]
- 62. Van Voorhis WC, Barrett LK, Sweeney YT, Kuo CC, Patton DL. 1997. Repeated Chlamydia trachomatis infection of Macaca nemestrina Fallopian tubes produces a Th1-like cytokine response associated with fibrosis and scarring. Infect. Immun. 65:2175–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wolner-Hanssen P, Patton DL, Holmes KK. 1991. Protective immunity in pig-tailed macaques after cervical infection with Chlamydia trachomatis. Sex. Transm. Dis. 18:21–25 [DOI] [PubMed] [Google Scholar]
- 64. Patton DL, Sweeney YT, Agnew KJ, Balkus JE, Rabe LK, Hillier SL. 2006. Development of a nonhuman primate model for Trichomonas vaginalis infection. Sex. Transm. Dis. 33:743–746 [DOI] [PubMed] [Google Scholar]
- 65. Henning T, Fakile Y, Phillips C, Sweeney E, Mitchell J, Patton D, Sturdevant G, Caldwell HD, Secor WE, Papp J, Hendry RM, McNicholl J, Kersh E. 2011. Development of a pigtail macaque model of sexually transmitted infection/HIV coinfection using Chlamydia trachomatis, Trichomonas vaginalis, and SHIV(SF162P3). J. Med. Primatol. 40:214–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cohen CR, Mugo NR, Astete SG, Odondo R, Manhart LE, Kiehlbauch JA, Stamm WE, Waiyaki PG, Totten PA. 2005. Detection of Mycoplasma genitalium in women with laparoscopically diagnosed acute salpingitis. Sex. Transm. Infect. 81:463–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schoeb TR, Dybvig K, Keisley KF, Davidson MK, Davis JK. 1997. Detection of urogenital mycoplasmal infections in primates by use of polymerase chain reaction. Lab. Anim. Sci. 47:468–471 [PubMed] [Google Scholar]
- 68. Somerson NL, Cole BC. 1979. The Mycoplasma flora of human and nonhuman primates, p 191–212 In Tully JG, Whitcomb RF. (ed), The mycoplasmas, vol II Academic Press, New York, NY [Google Scholar]
- 69. Lind K, Lindhardt BO, Schutten HJ, Blom J, Christiansen C. 1984. Serological cross-reactions between Mycoplasma genitalium and Mycoplasma pneumoniae. J. Clin. Microbiol. 20:1036–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dhandayuthapani S, Rasmussen WG, Baseman JB. 1999. Disruption of gene mg218 of Mycoplasma genitalium through homologous recombination leads to an adherence-deficient phenotype. Proc. Natl. Acad. Sci. U. S. A. 96:5227–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kutteh WH, Prince SJ, Hammond KR, Kutteh CC, Mestecky J. 1996. Variations in immunoglobulins and IgA subclasses of human uterine cervical secretions around the time of ovulation. Clin. Exp. Immunol. 104:538–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mestecky J, Alexander RC, Wei Q, Moldoveanu Z. 2011. Methods for evaluation of humoral immune responses in human genital tract secretions. Am. J. Reprod. Immunol. 65:361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Furr PM, Taylor-Robinson D. 1993. Factors influencing the ability of different mycoplasmas to colonize the genital tract of hormone-treated female mice. Int. J. Exp. Pathol. 74:97–101 [PMC free article] [PubMed] [Google Scholar]
- 74. Taylor-Robinson D, Furr PM, Tully JG, Barile MF, Moller BR. 1987. Animal models of Mycoplasma genitalium urogenital infection. Isr. J. Med. Sci. 23:561–564 [PubMed] [Google Scholar]
- 75. Taylor-Robinson D, Horner PJ. 2001. The role of Mycoplasma genitalium in non-gonococcal urethritis. Sex. Transm. Infect. 77:229–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dhandayuthapani S, Blaylock MW, Bebear CM, Rasmussen WG, Baseman JB. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183:5645–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Moller BR, Taylor-Robinson D, Furr PM, Freundt EA. 1985. Acute upper genital-tract disease in female monkeys provoked experimentally by Mycoplasma genitalium. Br. J. Exp. Pathol. 66:417–426 [PMC free article] [PubMed] [Google Scholar]
- 78. Taylor-Robinson D, Tully JG, Barile MF. 1985. Urethral infection in male chimpanzees produced experimentally by Mycoplasma genitalium. Br. J. Exp. Pathol. 66:95–101 [PMC free article] [PubMed] [Google Scholar]
- 79. Morrison-Plummer J, Lazzell A, Baseman JB. 1987. Shared epitopes between Mycoplasma pneumoniae major adhesin protein P1 and a 140-kilodalton protein of Mycoplasma genitalium. Infect. Immun. 55:49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Russell MW, Mestecky J. 2002. Humoral immune responses to microbial infections in the genital tract. Microbes Infect. 4:667–677 [DOI] [PubMed] [Google Scholar]
- 81. Citti C, Nouvel LX, Baranowski E. 2010. Phase and antigenic variation in mycoplasmas. Future Microbiol. 5:1073–1085 [DOI] [PubMed] [Google Scholar]
- 82. Dallo SF, Baseman JB. 1991. Adhesin gene of Mycoplasma genitalium exists as multiple copies. Microb. Pathog. 10:475–480 [DOI] [PubMed] [Google Scholar]