Abstract
A case of Actinomyces hongkongensis pelvic actinomycosis in an adult woman is described. Conventional phenotypic tests failed to identify the Gram-positive bacillus isolated from a fluid aspirate of a pelvic abscess. The bacterium was identified by 16S rRNA gene sequencing and analysis using the SmartGene Integrated Database Network System software.
TEXT
A 47-year-old, gravida 2, para 2, immunocompetent woman presented to the hospital with pelvic pain and fever 13 days after a total abdominal hysterectomy and salpingectomy for menorrhagia and hydrosalpinx. Her medical profile included smoking, perforated gastric ulcer, appendectomy, tubal ligation reversal, and two diagnostic laparoscopies. She had no history of intrauterine contraceptive device (IUCD) use. One gram of intravenous (i.v.) prophylactic cefazolin had been administered before the procedure. The surgery had proceeded without complication (200-ml estimated blood loss), and the patient was discharged on postoperative day 3. On presentation, the patient's white blood cell count was 21.7 × 109 cells/liter (18.6 × 109 neutrophils/liter), the hemoglobin level was 117 g/liter, and the platelet count was 570 × 109 platelets/liter. Electrolyte and creatinine levels were normal. A mass was palpated at the vaginal apex, and computed tomography (CT) showed a bilocular abscess immediately above the vaginal vault (the larger lobule measuring 8.8 by 5.8 cm), with regional inflammation (Fig. 1). The patient received empirical cefazolin and metronidazole, followed by clindamycin and gentamicin. She was then taken to the operating room for incision and drainage via a vaginal approach, but the mass was no longer palpable, and the procedure was aborted. Her antibiotic regimen was changed to ampicillin, gentamicin, and metronidazole, and fluid was drained from the residual mass under CT guidance the following day. Final CT imaging demonstrated a marked decrease in the size of the abscess. The patient improved clinically and was discharged home after 4 days in the hospital. As she had been afebrile for 2 days, no oral antibiotics were prescribed upon discharge. The patient remains free of symptoms 6 years after discharge from the hospital. The conjoint ethics board at the University of Calgary approved this study.
Fig 1.
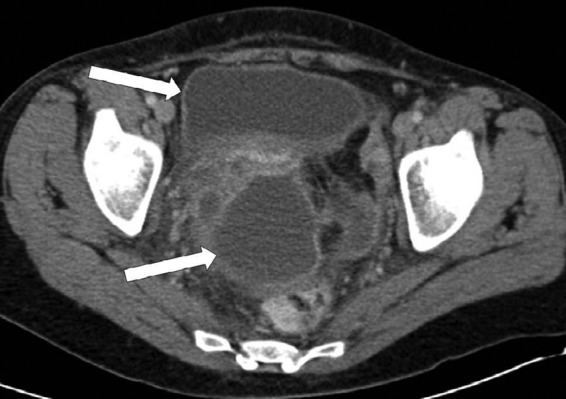
Axial computed tomography scan of the pelvis demonstrating a bilocular perivaginal abscess (arrows).
The fluid aspirate was subcultured on Columbia sheep blood agar (BA), chocolate agar (CHOC), MacConkey agar, and Brucella blood agar (BBA) (PML Microbiologicals, Wilsonville, OR) plates and incubated anaerobically at 35°C for 48 h before examination using an Anoxomat Mark II system (Mark Microbiology, Drachten, Netherlands). Growth occurred on BBA plates as nonhemolytic, pinpoint colonies. Gram staining showed the isolate to be a straight Gram-positive bacillus. Biochemical analysis using the Vitek 2 ANC card (bioMérieux, Laval, Quebec, Canada) provided a low-discrimination organism split between Actinomyces meyeri and Propionibacterium acnes. The isolate was biochemically inert, except for positive arginine dihydrolase, alkaline phosphatase, tyrosine arylamidase, leucine arylamidase, phenylalanine arylamidase, and proline arylamidase reactions. The isolate was nonmotile and did not produce catalase or indole. Metabolic end products were not tested. Vitek matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (bioMérieux) analysis gave a 99% coefficient of variation (CV) for identification as Microbacterium testaceum.
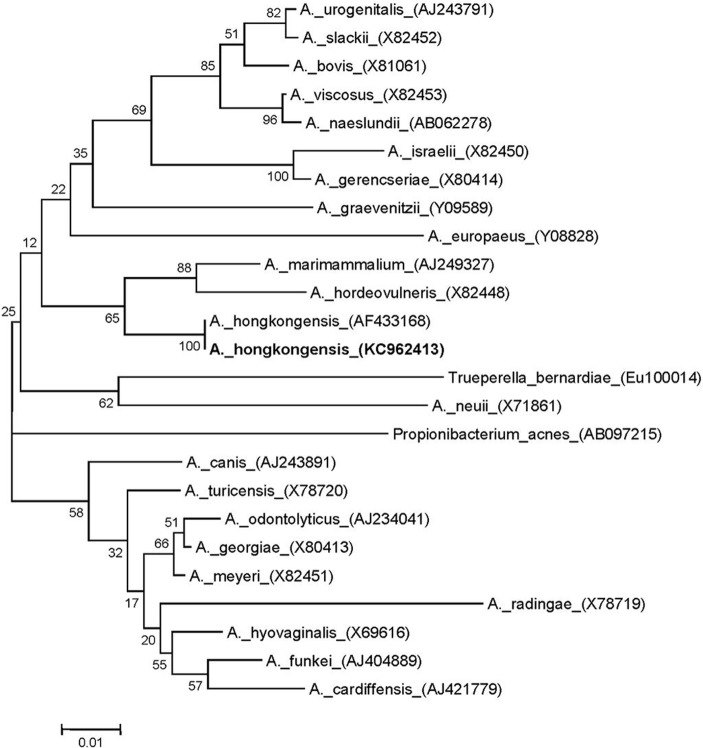
Molecular identification was done by fast partial sequencing of the 16S rRNA gene (523 bp) with MicroSeq 500 kits and an ABI Prism 3130 sequencer (Applied Biosystems, Foster City, CA), using standard methods (1–3). A BLAST search against the SmartGene Integrated Database Network System (IDNS) Bacteria database indicated that the species most closely related to the pelvic isolate was Actinomyces hongkongensis, and the overall identity score was 100%, with no mismatches (4, 5). The references were from a recently described species, Actinomyces hongkongensis (strain HKU8T), within the Actinomycetales order, Actinomycineae suborder, and Actinomyces genus (6), which was recently isolated from another case of pelvic actinomycosis (5). Our isolate also showed good identity (>90%) with other members of the genus Actinomyces (6). The most closely related known species within GenBank was Actinomyces marimammalium, with a sequence identity of 93%, while other Actinomyces species sequences within the IDNS database gave slightly lower identities (87 to 91%) (Fig. 2). Alignment of the partial 16S rRNA sequence from our clinical isolate and A. hongkongensis strain HKU8T showed 100% identity of 16S rRNA sequences using CLUSTALW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) (7). Phylogenetic analysis was done by using MEGA 5.1 software and the neighbor-joining method (2, 8) and gave a bootstrap score of 100 between our pelvic isolate and the sequences from A. hongkongensis strain HKU8T, while other Actinomyces species were related but had much lower bootstrap scores (Fig. 2).
Fig 2.
Neighbor-joining tree of 523 bp of the 16S rRNA gene sequences of the pelvic abscess isolate (GenBank accession no. KC962413) and Actinomyces reference strains. Branch support is recorded at the nodes as a percentage of 1,000 bootstrap iterations. GenBank accession numbers are shown in parentheses.
Antibiotic susceptibility testing using Etest strips (bioMérieux) showed the isolate to be sensitive to penicillin (MIC = 0.2 μg/ml) and clindamycin (MIC < 4 μg/ml) but resistant to metronidazole (MIC > 256 μg/ml).
This report demonstrates that partial or complete sequencing of the 16S rRNA gene is a valuable tool for definitive molecular identification of important clinical isolates that cannot be readily identified by phenotypic methods (1, 3–5). Actinomyces species are primarily anaerobic, filamentous, Gram-positive, opportunistic bacteria that colonize human mucous membranes. There are over 40 currently recognized species of Actinomyces that are morphologically similar and resist identification by traditional phenotypic tests (9). Therefore, sequencing of the 16S rRNA gene has become a useful tool for the identification of members of this genus to the species level, not only to definitively identify clinically relevant isolates but also to improve our understanding of the role of novel Actinomyces spp. in causing specific types of clinical infections (1, 3). In 2003, 16S rRNA sequencing was used to identify a novel species, A. hongkongensis strain HKU8T, isolated from the salpinx of a woman presenting with IUCD-associated pelvic actinomycosis (5). Our isolate demonstrated phenotypic characteristics similar to those previously described for the HKU8T isolate, and fast partial sequencing of the 16S rRNA gene demonstrated complete molecular identity between our strain and HKU8T (GenBank accession no. AF433168) (5). Although Actinomyces israelii has been most frequently described in cases of pelvic actinomycosis (9), many other species have been implicated in human disease (6, 10, 11). Our report is the second to document the isolation of A. hongkongensis and confirms its role in causing pelvic actinomycosis. Due to the rarity of this species, it is currently unknown whether A. hongkongensis has a propensity for localizing to the abdominopelvic region.
We could not determine whether A. hongkongensis was the primary pathogen or part of a polymicrobial flora in this abdominopelvic abscess because the patient received broad-spectrum antibiotics prior to aspiration and culture. Typically, Actinomyces spp. are recovered as a component of mixed bacterial flora in complex abscess formation in various locations. The most commonly affected sites of actinomycosis are cervicofacial (50%), abdominopelvic (20%), and thorax (15%) (9). Abdominopelvic actinomycosis generally occurs in association with abdominal surgery, appendicitis, ruptured viscus, neoplasia, or the prolonged use of an IUCD (12). Progression is usually indolent and leads to nonspecific symptoms, such as intermittent abdominal pain, low-grade fever, and weight loss. While CT imaging can localize the lesion, findings are usually nondiagnostic. For these reasons, abdominopelvic actinomycosis is often diagnosed following laparotomy for a suspected neoplasm. On presentation, the patient described here was thought to have a postoperative bacterial abscess, but actinomycosis was not specifically suspected. The final diagnosis of actinomycosis is usually based upon histological confirmation of actinomycotic sulfur granules or positive cultures, both of which are insensitive tests (9).
The treatment of actinomycosis has traditionally taken the form of extended courses of antibiotics, typically intravenous penicillin G for 2 to 6 weeks followed by oral penicillin V for 6 to 12 months (9). However, with adequate source control, substantially shorter antibiotic regimens have proven effective for abdominopelvic actinomycosis. For example, Atad et al. described a patient cured after only 2 days of effective antibiotics following salpingo-oophorectomy for adnexal actinomycosis (13). The outcome of our patient agrees with this therapeutic strategy and suggests that extended antibiotic courses may not be required. Furthermore, total excision of actinomycotic collections may be unnecessarily invasive if image-guided percutaneous drainage is a viable option. Further studies are required to determine which Actinomyces species and anatomical sites of infection would most benefit from percutaneous drainage and short courses of antibiotics.
In summary, clinical microbiology laboratories should be aware that A. hongkongensis may be a human pathogen in pelvic infections in women. Isolates meeting the preliminary phenotypic characteristics of Actinomyces spp. recovered from pelvic or abdominal abscesses should be referred for definitive identification using partial 16S rRNA gene sequencing. Delineation of the clinical significance and pathogenic potential of A. hongkongensis in humans is dependent upon further isolation from clinical samples and full phenotypic and genotypic characterization.
Nucleotide sequence accession number.
The sequence of the strain described here has been deposited in GenBank under accession number KC962413.
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Clinical and Laboratory Standards Institute 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing. Approved standard MM18-A. CLSI, Wayne, PA [Google Scholar]
- 2. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 3. Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908–934 [DOI] [PubMed] [Google Scholar]
- 4. Simmon KE, Croft AC, Petti CA. 2006. Application of SmartGene IDNS software to partial 16S rRNA gene sequences for a diverse group of bacteria in a clinical laboratory. J. Clin. Microbiol. 44:4400–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woo PC, Fung AM, Lau SK, Teng JL, Wong BH, Wong MK, Hon E, Tang GW, Yuen KY. 2003. Actinomyces hongkongensis sp. nov. a novel Actinomyces species isolated from a patient with pelvic actinomycosis. Syst. Appl. Microbiol. 26:518–522 [DOI] [PubMed] [Google Scholar]
- 6. Sarkonen N, Kononen E, Summanen P, Kononen M, Jousimies-Somer H. 2001. Phenotypic identification of Actinomyces and related species isolated from human sources. J. Clin. Microbiol. 39:3955–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 9. Wong VK, Turmezei TD, Weston VC. 2011. Actinomycosis. BMJ 343:d6099. 10.1136/bmj.d6099 [DOI] [PubMed] [Google Scholar]
- 10. Kayikcioglu F, Akif Akgul M, Haberal A, Faruk Demir O. 2005. Actinomyces infection in female genital tract. Eur. J. Obstet. Gynecol. Reprod. Biol. 118:77–80 [DOI] [PubMed] [Google Scholar]
- 11. Woo PC, Fung AM, Lau SK, Hon E, Yuen KY. 2002. Diagnosis of pelvic actinomycosis by 16S ribosomal RNA gene sequencing and its clinical significance. Diagn. Microbiol. Infect. Dis. 43:113–118 [DOI] [PubMed] [Google Scholar]
- 12. Yeguez JF, Martinez SA, Sands LR, Hellinger MD. 2000. Pelvic actinomycosis presenting as malignant large bowel obstruction: a case report and a review of the literature. Am. Surg. 66:85–90 [PubMed] [Google Scholar]
- 13. Atad J, Hallak M, Sharon A, Kitzes R, Kelner Y, Abramovici H. 1999. Pelvic actinomycosis. Is long-term antibiotic therapy necessary? J. Reprod. Med. 44:939–944 [PubMed] [Google Scholar]