Abstract
We describe an assay which uses broad-spectrum, conserved-site PCR paired with mass spectrometry analysis of amplicons (PCR/electrospray ionization-mass spectrometry [ESI-MS]) to detect and identify diverse bacterial and Candida species in uncultured specimens. The performance of the assay was characterized using whole-blood samples spiked with low titers of 64 bacterial species and 6 Candida species representing the breadth of coverage of the assay. The assay had an average limit of detection of 100 CFU of bacteria or Candida per milliliter of blood, and all species tested yielded limits of detection between 20 and 500 CFU per milliliter. Over 99% of all detections yielded correct identifications, whether they were obtained at concentrations well above the limit of detection or at the lowest detectable concentrations. This study demonstrates the ability of broad-spectrum PCR/ESI-MS assays to detect and identify diverse organisms in complex natural matrices that contain high levels of background DNA.
INTRODUCTION
We describe an assay which uses PCR paired with electrospray ionization-mass spectrometry of amplicons (PCR/ESI-MS) to identify diverse bacteria and Candida species present at low titers in complex, normally sterile biological matrices. This assay is similar to though much more sensitive than previously described PCR/ESI-MS assays designed to identify cultured microorganisms from blood bottles or colonies (1, 2). In PCR/ESI-MS, conserved regions of pathogen genomes are amplified by PCR and subsequently detected using mass spectrometry (1, 3) (Fig. 1). Mass spectrometry signals are translated into amplicon base composition signatures to provide identifying fingerprints of any organisms detected. The assay tested here was designed to identify 611 microbial species through comparison of the detected base composition signatures to reference database signatures. In this study, the general capabilities of this assay were tested using low-titer spikes of phenotypically characterized and culture-quantified microorganisms in blood.
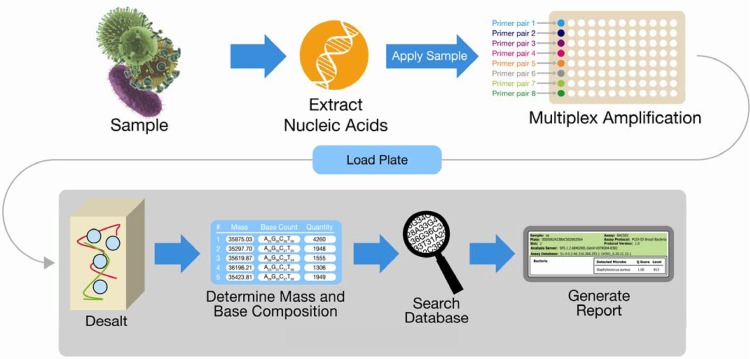
Fig 1.
PCR/ESI-MS system workflow.
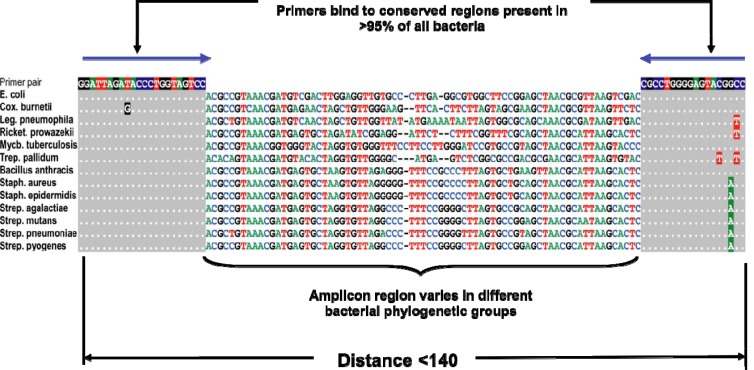
Real-time PCR (4), direct immunofluorescence (5), and serological assays (6) use unique processes and/or reagents for the detection of each targeted organism. Hence, their performance must be independently characterized for each organism. Broad-spectrum techniques, such as PCR/ESI-MS, ribosomal gene sequencing (7), internal transcribed spacer sequencing (8), and multilocus sequence typing (MLST) (9), do not identify organisms with analyte-specific reagents or processes. Instead, they target highly conserved nucleotide sequences shared by many related organisms using conserved-site PCR primers, amplify heterogeneous regions between the primer sites in a nonspecific manner, and derive organism-specific identification data from bioinformatic analysis of the resulting amplicons (Fig. 2). In this sense, broad-spectrum assays can be seen as universal information-based systems that use generic (nonspecific) biochemistry to capture signals from diverse targets and then use information derived from those signals to identify specific organisms.
Fig 2.
An example of broad primer design used in the bacterial assay. E., Escherichia; Cox., Coxiella; Leg., Legionella; Ricket.; Rickettsia; Mycb., Mycobacterium; Trep., Treponema; Staph., Staphylococcus; Strep., Streptococcus.
The nonspecific processes in the PCR/ESI-MS system include nucleic acid extraction, conserved-site PCR, PCR product cleanup, mass spectrometry, and use of base composition determination and signature-matching algorithms to detect and identify all bacterial and Candida species represented in the platform's signature database (3, 10–12). The primers (Table 1) are designed to amplify fragments of highly conserved genes from diverse bacterial and Candida species (Fig. 3). Additional target-specific primers are used to detect common antibiotic resistance genes, though this capability was not addressed in this study. Heterogeneous PCR amplicons from different species can be distinguished by ESI-MS and matched to species-specific database signatures derived from either sequence data or previous analysis of representative strains.
Table 1.
Primers and target organisms
| Primer pair | Wells | Sequence (5′ to 3′) | Target |
|---|---|---|---|
| 346 | A1–A6 | TAGAACACCGATGGCGAAGGC | Eubacteria (16S) |
| TCGTGGACTACCAGGGTATCTA | |||
| 348 | B1–B6 | TTTCGATGCAACGCGAAGAACCT | Eubacteria (16S) |
| TACGAGCTGACGACAGCCATG | |||
| 361 | C1–C6 | TTTAAGTCCCGCAACGAGCGCAA | Eubacteria (16S) |
| TTGACGTCATCCCCACCTTCCTC | |||
| 349 | D1–D6 | TCTGACACCTGCCCGGTGC | Eubacteria (23S) |
| TGACCGTTATAGTTACGGCC | |||
| 3350 | E1–E6 | TCCACACGGTGGTGGTGAAGG | Firmicutes (rplB) |
| TCCAAGCGCAGGTTTACCCCATGG | |||
| 2259 | F1–F6 | TGAACGTGGTCAAATCAAAGTTGGTGAAGA | Staphylococcus (tufB) |
| TGTCACCAGCTTCAGCGTAGTCTAATAA | |||
| 358 | F1–F6 | TCGTGGCGGCGTGGTTATCGA | Enterobacteriaciae (valS) |
| TCGGTACGAACTGGATGTCGCCGTT | |||
| 3346 | G1–G6 | TGAACCACTTGGTTGACGACAAGATGCA | Gammaproteobacteria (rpoB) |
| TCACCGAAACGCTGACCACCGAA | |||
| 3921 | H1–H6 | TCAGTTCGGTGGTCAGCGTTTCGG | Beta-/Gammaproteobacteria (rpoB) |
| TCATCGGACTTCACGGTGAGCATTTC | |||
| 879 | A7–A12 | TCAGGTACTGCTATCCACCCTCAA | mecA methicillin resistance gene |
| TGGATAGACGTCATATGAAGGTGTGCT | |||
| 3767 | B7–B12 | TGGACAAATCGTTGACATACATCGTTG | vanA vancomycin resistance gene |
| TAATAACCCAAAAGGCGGGAGTAGC | |||
| 4675 | B7–B12 | TACACCCGGACGCCTAACAAGGA | KPC carbapenamase gene |
| TGCCCGTTGACGCCCAATCC | |||
| 3768 | C7–C12 | TAGGAAAACGCATGGTCTGCTTGTC | vanB vancomycin resistance gene |
| TGGGAAAGCCACATCAATACGCC | |||
| 3030 | D7–D12 | TGTGAAGCGGCAAAAGCTCAAATTT | Broad fungi (25S) |
| TTCTCACCCTCTGTGACGGCCTGTTCC | |||
| 3031 | E7–E12 | TGGAGTCTAACATCTATGCGAGTGTT | Broad fungi (25S) |
| TCAGCTATGCTCTTACTCAAATCCATC | |||
| 3766 | F7–F12 | TTGTGTAGAATAGGTGGGAGCTTCGGC | Broad fungi (25S) |
| TCTGACAATGTCTTCAACCCGGATC | |||
| 3865 | G7–G12 | TGGTACAGTGGAGTATGCTGTTTAATTGGA | Candida (mitochondrial) |
| TCTGACGACAACAATGTAACGCCTG | |||
| 4437 | H7–H12 | TGACGAGTTCATGAGGGCAGGC | Pumpkin DNA extraction control |
| TCTGGCCTTTCAGCAAGTTTCCAAC |
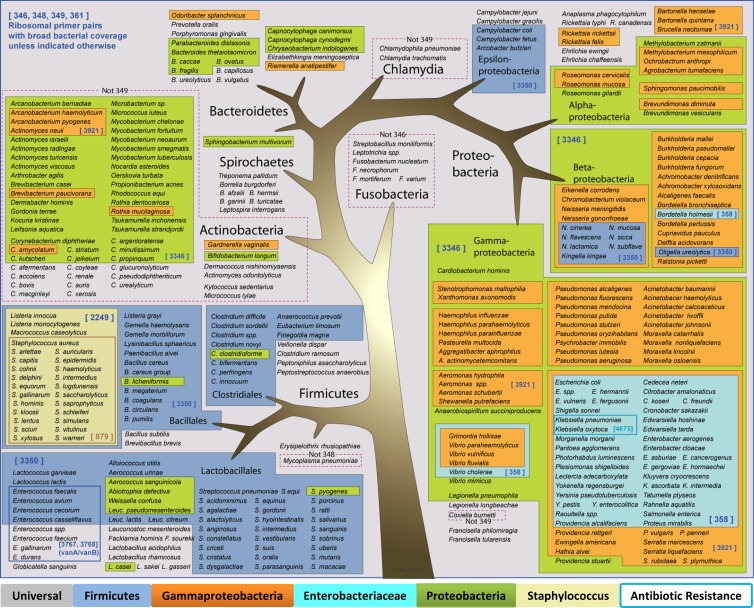
Fig 3.
Primer design and coverage for bacterial detection and identification. Species with abbreviated genus names are grouped with a species where the genus is spelled out.
Each PCR/ESI-MS reaction is calibrated using an internal amplification control which produces an amplicon distinguishable from target amplicons by mass. To maximize sensitivity, the assay described here contained minimal calibrant concentrations, in contrast to previous similar PCR/ESI-MS assays designed to identify cultured organisms (1, 2).
Because the processes and reagents used by the PCR/ESI-MS system to detect diverse biological targets are universal rather than analyte specific, analytical assay characterization data collected using representative species, such as those shown in Table 2, can be used to interpolate general properties of the system, such as sensitivity (limit of detection [LOD]). In this study, the LOD of the BAC Spectrum SF assay was measured for 30 bacterial species and four Candida species chosen on the basis of both clinical relevance and diversity. LODs were initially determined using 5-replicate, 4-fold dilution series of culture-quantified stocks spiked in whole blood and then confirmed to yield at least 95% detection across 20 replicates. The resulting distribution of LOD measurements was analyzed, and a 95% confidence interval was calculated to estimate the mean and range of the LOD for all target species (hereinafter, “assay LOD”). Thirty-six additional species were then tested in duplicate at concentrations near the calculated upper 95% confidence interval limit of the assay LOD in order to verify the predictive nature of the assay LOD range and variance.
Table 2.
Limits of detection for 30 bacterial and 4 Candida species
| Spiked organisma | LOD (CFU/ml) with: |
Detection reported (no. of replicates in which detection was reported) | Taxon | |
|---|---|---|---|---|
| 5 replicates | 20 replicates | |||
| Brucella neotomae | 80 | 320 | Brucella sp. | Alphaproteobacteria |
| Neisseria meningitidis | 5 | 20 | Neisseria meningitidis | Betaproteobacteria |
| Burkholderia cepacia | 0.31 | 20 | Burkholderia cenocepacia/cepacia | Betaproteobacteria |
| Proteus mirabilis | 20 | 20 | Proteus mirabilis | Enterobacteriaceae |
| Proteus vulgaris | 80 | 80 | Proteus vulgaris | Enterobacteriaceae |
| Klebsiella oxytoca | 20 | 20 | Klebsiella oxytoca | Enterobacteriaceae |
| Enterobacter aerogenes | 80 | 320 | Enterobacter aerogenes | Enterobacteriaceae |
| Enterobacter cloacae | 5 | 20 | Enterobacter cloacae complex | Enterobacteriaceae |
| Escherichia coli | 20 | 20 | Escherichia coli | Enterobacteriaceae |
| Shigella sonnei | 80 | 320 | Escherichia coli/Shigella sp. | Enterobacteriaceae |
| Klebsiella pneumoniae | 20 | 20 | Klebsiella pneumoniae | Enterobacteriaceae |
| Salmonella enterica | 80 | 80 | Salmonella enterica | Enterobacteriaceae |
| Serratia marcescens | 20 | 20 | Serratia marcescens | Enterobacteriaceae |
| Stenotrophomonas maltophilia | 320 | 320 | Stenotrophomonas maltophilia | Gammaproteobacteria |
| Acinetobacter baumannii | 5 | 80 | Acinetobacter baumannii | Gammaproteobacteria |
| Pasteurella multocida | 320 | 320 | Pasteurella multocida | Gammaproteobacteria |
| Aeromonas hydrophila | 5 | 80 | Aeromonas hydrophila | Gammaproteobacteria |
| Pseudomonas aeruginosa | 20 | 20 | Pseudomonas aeruginosa | Gammaproteobacteria |
| Arcanobacterium haemolyticum | 80 | 500 | Arcanobacterium haemolyticum | Actinobacteria |
| Corynebacterium diphtheriae | 80 | 80 | Corynebacterium diphtheriae | Actinobacteria |
| Corynebacterium jeikeium | 320 | 320 | Corynebacterium jeikeium | Actinobacteria |
| Micrococcus luteus | 320 | 320 | Micrococcus luteus | Actinobacteria |
| Rothia dentocariosa | 80 | 80 | Rothia dentocariosa | Actinobacteria |
| Listeria monocytogenes | 500 | 500 | Listeria monocytogenes | Firmicutes |
| Enterococcus faecalis | 500 | 500 | Enterococcus faecalis | Enterococcus |
| Enterococcus faecium | 80 | 80 | Enterococcus faecium | Enterococcus |
| Staphylococcus aureus | 80 | 80 | Staphylococcus aureus | Staphylococcus |
| Staphylococcus epidermidis | 320 | 320 | Staphylococcus epidermidis | Staphylococcus |
| Staphylococcus haemolyticus | 320 | 320 | Staphylococcus haemolyticus | Staphylococcus |
| Streptococcus pneumoniae | 20 | 80 | Streptococcus sp. (1) | Streptococcus |
| Viridans/mitis group Streptococcus (6) | Streptococcus | |||
| Streptococcus mitis/pneumoniae (2) | Streptococcus | |||
| Candida glabrata | 80 | 80 | Candida glabrata | Candida |
| Candida tropicalis | 80 | 80 | Candida tropicalis | Candida |
| Candida parapsilosis | 20 | 80 | Candida parapsilosis | Candida |
| Candida albicans | 20 | 20 | Candida albicans | Candida |
All spiked organisms were tested in otherwise negative whole human blood.
Data from all reported detections were analyzed, including data from sporadic detections from samples spiked below the LOD. This was done to ensure that low-titer detections, possibly made on the basis of less information (fewer amplicons) than provided by LOD-level concentrations of the targets, provide accurate identification when an organism is detected.
(Portions of the data presented in this paper were previously presented as a poster at the 22nd European Congress of Clinical Microbiology and Infectious Diseases, London, United Kingdom, 1 April 2012.)
MATERIALS AND METHODS
Growth and quantification of organisms.
The organisms tested in this study were chosen based both on prevalence in clinical laboratory blood culture bottle assays (3) and diversity across the phylogenetic breadth of the assay. Well-characterized bacteria and Candida strains were obtained from ATCC (Manassas, VA) and clinical microbiology laboratories, cultured on appropriate plate media, subcloned to ensure purity, and harvested upon initial appearance of colonies. Fresh colonies were resuspended, dispersed by gentle vortexing, and diluted at various concentrations in sterile, DNA-free media. Dilutions were plated by spreading, and colony counts were used to determine the concentration of the diluted stocks. Live/dead ratio determinations were made on many stocks, though this process was abandoned after it was observed that these generally showed >90% live cells at the time colony counts were performed. Concentration data were used to generate stocks near the estimated LOD of the assay. Final dilutions for purposes of generating spiked samples were made at ratios of approximately 1:10 into whole human blood collected in EDTA blood bags from apparently healthy donors (Biomed, Carlsbad, CA).
Genome preparation and PCR.
The PCR/ESI-MS system used in this study included automated sample lysis, nucleic acid extraction, PCR, PCR cleanup (desalting), mass spectrometry, and bioinformatic data analysis components, as previously described (Ibis Biosciences, Carlsbad, CA) (13, 14). Strict separation of PCR/sample set-up areas and amplification/analysis areas (personnel/equipment movement and airflow) was employed to prevent backflow of amplicons from the thermocycler and mass spectrometer components, according to Clinical and Laboratory Standards Institute (CLSI) guidelines for open PCR processes.
DNA was extracted from specimens of 1 ml of EDTA-treated whole blood spiked with cultured organisms and a nucleic acid extraction control. An amount of 250 μl of 10% bovine serum albumin (BSA) was added as a carrier agent. The initial disruption was performed by mechanical bead-beating with zirconium/yttrium beads in the presence of proteinase K and 20% sodium dodecyl sulfate to provide enzymatic and chemical lysis activities (13, 14).
Following bead beating, samples were briefly incubated, centrifuged, and automatically extracted via a nonspecific magnetic bead capture and washing process as previously described (13, 14), resulting in 280 μl of eluate. An amount of 10 μl of eluate was transferred to each of 16 wells of the PCR plate. Ultimately, 35.7 μl of the original specimen volume were purified and distributed to each PCR well. PCR amplifications were performed using previously described PCR reagents and cycling conditions (15, 16). PCR products were desalted by binding to magnetic microparticles, washing with solutions containing volatile salts and organic solvents, and then eluting with a high-pH buffer solution containing 35% (vol/vol) methanol and 25 mM piperidine-imidazole, as previously described (17).
Amplicon analysis and reporting.
Desalted PCR products were injected into an electrospray ionization-time of flight mass spectrometer (ESI-TOF-MS) (17). Peptide-based mass standards were used to bracket the expected mass-to-charge range and minimize TOF-associated variability. The raw mass-to-charge ratio spectrum from each PCR well was converted computationally to a set of independent mass peaks. Each resulting mass peak was compared to the mean molecular weights of all mathematically possible base compositions. The number of base composition possibilities was defined by a mass error tolerance and by the known information about the primer sets present in each well. The large number of resulting possibilities was reduced by only considering pairs of detected masses which could represent complementary forward- and reverse-strand base compositions (11, 12). Known-organism base composition signatures were then selected from the database based on matches to detected masses across multiple primer pairs (3, 11, 12, 18). The tolerance for matching allowed for multiple known-organism base composition signatures to be associated with a common mass measurement. The organism possibilities identified through the matching process were then tested as competing hypotheses and sorted to yield the most likely match or set of equally likely matches. The identifications were parsed through a series of reporting cutoffs designed to ensure the sufficiency of signal level and resolution for purposes of accurate identification and presented to the user in the form of organism identities.
Large data sets representing known (spiked) positives and presumed negatives in the natural matrix (blood) were generated using the device. Correct detections and identifications were used to positively reinforce specific parameterizations of the analysis model, while apparently spurious detections and misidentifications of apparently real detections were used as negative reinforcers. The resulting models were then tested on sample sets such as the one described in this paper: diverse bacteria spiked above and below the limit of detection to challenge the detection and identification capabilities and define the resulting accuracy of the system.
Database population.
The database used for signature matching was generated through compilation of signatures from two sources. GenBank was searched for close matches to the assay primer pairs. Potentially amplifiable sequences were identified and reduced to base compositions. The resulting signatures were supplemented with empirically observed base compositions captured by direct analysis of type strains with PCR/ESI-MS. Unique strain signatures were included in the database only when information (either a base count or a “no prime” determination based on primer mismatch analysis) was available for all primers in the assay. Nine hundred six complete signatures were included in the database assay version used here (6.24.25.19.1), 611 of which were specifically reportable by the software. The reportable list was selected from the total number of signatures based on clinical relevance (at least one reported instance of association with human disease), with the further inclusion of closely related species for which discrimination from pathogenic relatives is clinically relevant.
PCR/ESI-MS gene targets and selection of primers.
General methods for PCR/ESI-MS using both the current system and earlier PCR/ESI-MS instruments have been described previously (3, 10, 11, 19, 20). The primer pairs used here and their targets are shown in Table 1, and their breadth of coverage is depicted in Figure 3. Nine primer pairs targeted broadly conserved bacterial loci, including both ribosomal genes and other housekeeping genes (the latter provide greater species-level resolution for groups with less variation in ribosomal genes). Four primer pairs targeted common antibiotic resistance cassettes, and four primer pairs targeted fungal ribosomal genes (3). The remaining primer pair amplified an extraction control construct.
Reporting thresholds and cutoffs.
The assay utilized artificial signal thresholds (cutoffs) designed to limit reporting of irreproducible detections. To generate the data shown here, cutoffs were applied to two measurements. The first, termed the “level,” was used to indicate the signal amplitude. This was calculated with reference to an internal calibrant for each primer pair independently and then averaged across the set of amplicons used to define any given detection. The use of this metric in the PCR/ESI-MS system has been described previously (11). The approximate linear range for reporting these levels was between 0.1 and 10 times the levels of calibrants in the assay. In the assay described here, a calibrant was present at 100 copies per well for bacterial ribosomal primers and 20 copies per well for the remaining primers. Calculations of levels from ribosomal primers were adjusted for the multiplicity of ribosomal loci when sufficient data were available. The level was considered a qualitative measure due to the various degrees of mismatch between primers and their targets in different organisms—in cases where primers matched the calibrant significantly better or worse than the target, PCR amplification was expected to be more efficient for one or the other, leading to biases in absolute measurement of concentration. Absolute measurement could also be affected by copy number variation and other factors (21, 22). However, the level does provide a meaningful relative measure of the concentration of any specific target and can be used to set species-specific or general reporting thresholds on the basis of empirical testing.
The second measure to which cutoffs were applied was the quality score (Q-score), a relative measure of the strength of the data supporting identification. The Q-score ranged between 0 (low) and 1 (high), based on parameters including an indicator of how well the hypothesized organisms, as a group, represented the observed data, an indicator of how significant the contribution of a single organism was to the solution, the fraction of missed detections (primers expected to produce a recognizable base composition signature for a detected organism that did not), and the percentage of primers for a detected organism for which no known data existed within the database. Q-score cutoffs were designed to prevent specific identification when the information obtained was not sufficient to confidently resolve an organism's identity. For this study, a Q-score of ≥0.90 was considered a reportable result. In cases where the signatures detected were equally well matched to multiple organisms, the detections were reported as a list of equally possible identities.
Misidentification was also limited through the application of additional organism-specific reporting rules. For most bacterial and Candida species, 50% of the primers expected to generate amplicons were required to yield signatures matching an entry in the database to report detection. For Staphylococcus spp., only two primer pair signature matches were required, and a Staphylococcus-specific primer pair was included to maximize sensitivity and capability to distinguish species. In addition to the primer pair ratio requirement, reporting translations were applied to certain organisms. In cases where multiple species of the same genus could not be resolved when all primers yielded signatures, reporting filters were used to group the unresolvable species into a single category (for example, see Burkholderia cenocepacia/cepacia in Table 2). In the case of Streptococcus spp., three different reporting filters were put in place to generate species, group, or genus level reports on the basis of different proportions of observed/expected primer pair amplifications. Streptococci are less genetically divergent than member species of other genera, and more primer pairs were required to capture species-specific signatures.
Level cutoffs were put in place to prevent contaminants or background noise from generating reported detections. These cutoffs were set on the basis of truth data corresponding to large numbers of contrived or clinical samples in feasibility studies. For the version of the assay used here, all targets required a minimum level of 3 to be reported, in order to limit reporting of suspect detections apparently resulting from artifactual peaks in mass spectra (noise). Higher cutoffs were implemented for specific organisms to limit detections of common contaminants. The highest cutoff was applied to Pseudomonas aeruginosa, a common water contaminant seen in multiple reagents and media; steps taken to limit reagent contamination have since obviated the need for this cutoff. Limits of detection were determined with the cutoffs in place.
The results reported include an identification of bacteria to the genus, group, or species level, Candida to the species level, and the identification of detected antibiotic resistance genes. The assay was tested in a configuration that allowed specific reporting of 601 species of bacteria and 10 species of Candida. This set represents organisms which have been reported as human pathogens and their close relatives. An additional 295 organisms with no recognized association with human disease are reported as “Bacteria detected, not identified.” The inclusion of all organisms for which complete signatures are available allows maximum accuracy in signal sorting and background filtering. Near neighbors of pathogens are included for purposes of accurately assigning single primer pair signatures, as individual primer pair signatures may be shared by multiple detectable organisms. Also, the PCR system is competitive (multiple targets compete with each other for amplification), so it is important to recognize the presence of unrelated commensals and environmental and reagent contaminants that might compete with other targets.
LOD determination and confirmation.
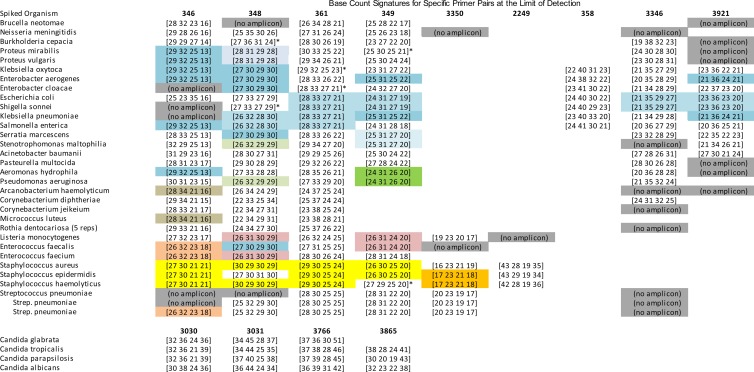
LOD determination and confirmation was performed by testing 30 species of bacteria and four species of Candida (Table 2). The initial LOD determinations were performed using dilution series that began at 320 CFU per ml (CFU/ml) and went down in concentration in 4-fold steps; 5 replicates of each concentration were analyzed. If the initial 320-CFU/ml test failed to yield 5 positive detections, the test was repeated at 500 CFU/ml (this concentration was essentially arbitrary, set at a predefined acceptance limit). The lowest level at which all 5 replicates were detected was then tested across 20 replicates, and this level was considered the confirmed LOD if at least 95% of the replicates were appropriately detected. If this confirmation failed, the experiment was repeated at the next highest concentration. Examples of base composition signatures obtained from LOD-level samples of each tested organism are shown in Figure 4.
Fig 4.
Base composition signatures obtained at the confirmed limit of detection for 30 bacterial species and 4 Candida species. Each row corresponds to the respective row in Table 1. Colors represent signatures shared by more than one of the organisms shown in this figure. Signatures in white boxes are unique among the presented organisms, and gray boxes represent primer pairs which were expected to generate amplicons at higher organism concentrations but did not yield amplicons at the limit of detection. * Secondary organism-associated base compositions were also obtained for these primer pair/organism combinations. These usually represent within-organism polymorphisms among multiple copies of ribosomal genes. Only primary signatures contributing to the final computed (bioinformatically matched) identifications are shown.
Derivation of the assay LOD.
Confirmed LODs from the 30 representative bacterial species were statistically analyzed using a base 4 log-transformed t test analysis to generate a logarithmic mean and 95% confidence interval. A base 4 exponential model was chosen because both PCR and microbial growth are logarithmic processes and because 4-fold steps were used to determine and confirm the LODs. Four-fold steps were chosen as the basis for LOD determination because this step level was shown, over the course of many experiments, to be the lowest step level that yielded robust and repeatable LOD measurements (for example, in experiments with 2-fold steps, the LOD measurements varied by one step in either direction).
Confirmation of the assay LOD.
As the observed distribution of 34 LODs was not log-normal, the statistical model was not assumed to be perfectly predictive. The derived assay LOD, with a logarithmic mean of 100 CFU/ml and a 95% confidence interval of 6 to 600 CFU/ml, was challenged by testing a new set of 34 bacterial species and two Candida species (Table 3), all different than those tested during the initial LOD analysis. These organisms were tested in replicates of 5 at 500 CFU/ml, near the upper 95% confidence interval boundary of the assay LOD. All species were quantified and prepared as spiked samples in EDTA blood, in the same manner as the species tested in primary LOD experiments. This test addressed the specific hypothesis derived from the assay LOD study, that 95% of previously untested bacterial species should exhibit an LOD of less than or equal to 600 CFU/ml.
Table 3.
Confirmatory testing of 34 bacterial and 2 Candida species near the upper 95% confidence interval boundary of the calculated assay LOD
| Spiked organisma | Detection reported (no. of replicates in which detection was reported) | Taxon |
|---|---|---|
| Bartonella henselae | Bartonella henselae | Alphaproteobacteria |
| Bartonella quintana | Bartonella quintana (6) | Alphaproteobacteria |
| Bartonella tribocorum (1) | Alphaproteobacteria | |
| Bordetella pertussis | Bordetella pertussis | Betaproteobacteria |
| Eikenella corrodens | Eikenella corrodens | Betaproteobacteria |
| Ralstonia pickettii | Ralstonia pickettii | Betaproteobacteria |
| Morganella morganii | Morganella morganii | Enterobacteriaceae |
| Citrobacter freundii | Citrobacter freundii | Enterobacteriaceae |
| Providencia stuartii | Providencia stuartii | Enterobacteriaceae |
| Yersinia pseudotuberculosis | Yersinia pseudotuberculosis | Enterobacteriaceae |
| Moraxella catarrhalis | Moraxella catarrhalis/nonliquefaciens | Gammaproteobacteria |
| Acinetobacter lwoffii | Acinetobacter lwoffii | Gammaproteobacteria |
| Haemophilus influenzae | Haemophilus influenzae | Gammaproteobacteria |
| Legionella pneumophila | Legionella pneumophila | Gammaproteobacteria |
| Vibrio parahaemolyticus | Vibrio parahaemolyticus | Gammaproteobacteria |
| Campylobacter coli | Campylobacter coli/jejuni | Epsilonproteobacteria |
| Campylobacter jejuni | Campylobacter coli/jejuni | Epsilonproteobacteria |
| Bacteroides fragilis | Bacteroides fragilis | Bacteroides |
| Fusobacterium nucleatum | Fusobacterium nucleatum | Fusobacterium |
| Gardnerella vaginalis | Gardnerella vaginalis | Actinobacteria |
| Microbacterium spp. | Microbacterium sp. | Actinobacteria |
| Mycobacterium chelonae | Mycobacterium chelonae | Actinobacteria |
| Nocardia asteroids | Nocardia asteroides/farcinica | Actinobacteria |
| Propionibacterium acnes | Propionibacterium acnes | Actinobacteria |
| Bacillus cereus | Bacillus cereus group | Firmicutes |
| Clostridium difficile | Clostridium difficile | Firmicutes |
| Clostridium perfringens | Clostridium perfringens | Firmicutes |
| Lactobacillus acidophilus | Lactobacillus acidophilus | Firmicutes |
| Veillonella dispar | Veillonella dispar/parvula | Firmicutes |
| Staphylococcus saprophyticus | Staphylococcus saprophyticus | Staphylococcus |
| Staphylococcus lugdunensis | Staphylococcus lugdunensis | Staphylococcus |
| Streptococcus mutans | Streptococcus mutans | Streptococcus |
| Streptococcus agalactiae | Streptococcus agalactiae | Streptococcus |
| Streptococcus pyogenes | Streptococcus pyogenes | Streptococcus |
| Streptococcus gordonii | Streptococcus gordonii (3) | Streptococcus |
| Viridans/mitis group Streptococcus (3) | Streptococcus | |
| Streptococcus sp. (1) | Streptococcus | |
| Candida guilliermondii | Candida guilliermondii | Candida |
| Candida krusei | Candida krusei | Candida |
All organisms were tested at 500 CFU/ml in 5 replicates in whole blood, except as noted (in two cases, unexpected results led to further replicate analysis).
RESULTS AND DISCUSSION
The assay described here was designed to detect and identify diverse bacterial species and Candida species at low titers in complex biological matrices. The limits of detection for 30 representative bacteria and four Candida species were evaluated in whole blood (Table 2). The distribution of these LODs was analyzed statistically, and the general assay LOD was determined to be 102 with a 95% confidence interval of 6 × 100 to 6 × 102 (6 to 600). The derived assay LOD of 6 to 600 CFU/ml was then challenged by the further testing of an additional 34 bacterial species and two Candida species at 500 CFU/ml with at least 5 replicates (Table 3). All but one were correctly detected and identified in all replicates. The exception was Bartonella quintana, which was identified correctly in six of seven replicates and incorrectly identified as Bartonella tribocorum in one replicate due to the failure of a single primer pair responsible for discrimination of these two species. This confirmed that 95% of potential analytes for which the assay database contained appropriate signatures could be detected and identified within the 95% confidence intervals of the interpolated general assay LOD.
At lower titers, PCR/ESI-MS can generate partial signatures; that is, some primers may not generate amplicons, but the assay software only requires that at least 50% of the expected amplicons be detected and matched to a reference database signature to report an identification. Since identifications based on partial signatures have the potential to generate ambiguous or incorrect identifications, we analyzed all detections made below the limit of detection and enumerated the cases in which these identifications yielded correct identifications at the species level, correct but ambiguous identifications at the genus or group level, or incorrect identifications. Many organisms were sporadically detected below the limit of detection at least once. In most cases, the reported identifications were species specific and identical to what was reported at the limit of detection. In the cases of Micrococcus luteus, Listeria monocytogenes, and Enterococcus faecium, below-LOD detections were occasionally reported as ambiguous at the species level. These ambiguous detections were reported as lists of possible species identities (“Micrococcus luteus; Micrococcus lylae,” “Listeria monocytogenes; Listeria innocua,” and “Enterococcus faecium; Enterococcus malodoratus/raffinosus”). In these cases, amplicons were not detected for the primers capable of differentiating the listed species, and therefore, the detected signatures were equally well matched to multiple database signatures and the ambiguous reporting was appropriate and reflected the resolution of the detection. All of these identifications were accurate as reported. One below-LOD spiked sample of Rothia dentocariosa resulted in the report of a viridans/mitis group Streptococcus, but this appeared to be the result of contamination rather than misidentification. The detected signature was distinct from that of Rothia and a perfect match to Streptococcus and was made at a spike level of 1/16 the LOD of Rothia, where a negative result was expected. Sporadic detections of coagulase-negative staphylococci (CONS) and non-pneumoniae streptococci are occasionally made in unspiked aliquots of human blood collected from apparently healthy donors, likely derived from venipuncture contamination or other environmental sources. Similar contaminations, especially of CONS, are routinely observed in blood bottle culture systems (23). Reporting of such detections would be limited in any future clinical application by imposing level cutoffs to discriminate between clinically relevant infections and background environmental contamination.
In summary, the PCR/ESI-MS assay tested here is capable of detecting and identifying diverse bacterial and Candida species in whole blood. The limit of detection of the assay varied from 20 to 500 CFU/ml for tested bacterial species capable of being culture quantified and was estimated to be 100 CFU/ml (95% CI = 6 to 600 CFU/ml) for all detectable microorganisms (the 611 species for which the assay reference database contains an appropriate signature). This general limit of detection was confirmed by demonstrating that 33 of 34 additional tested bacterial species were detected and correctly identified at a concentration of 500 CFU/ml, while the remaining species was detected and correctly identified in six of seven replicates. The sensitivity was similar for Candida species. The assay is capable of detecting both culturable and unculturable organisms and can generate information much more quickly than culture (in less than one laboratory shift) and, thus, could be a valuable tool in the microbiology laboratory for broad-spectrum detection of bacteria and Candida.
Current and future studies will address the potential clinical utility of the assay through analysis of normally sterile tissue and fluid specimens collected from patients with suspected infections. Samples will be collected from patients with meningitis, sepsis, pneumonia, orthopedic infections, and others. In these studies, PCR-ESI/MS data will be compared to standard-of-care clinical microbiology results from culture and/or analyte-specific molecular methods to determine the accuracy of identification by the method. Both standard-of-care and PCR-ESI/MS results will be compared to nonmicrobiological clinical findings to define the relative value of the two systems for the diagnosis and detection of etiologically relevant organisms.
ACKNOWLEDGMENTS
All authors except one (C.J.H.) are full-time employees of Ibis Biosciences and Abbott, the businesses that developed and market the PCR/ESI-MS system and the assay described in this paper. Human blood used as a matrix was obtained from a qualified contract research organization vendor (Biomed, Carlsbad, CA).
Footnotes
Published ahead of print 12 June 2013
REFERENCES
- 1. Baldwin CD, Howe GB, Sampath R, Blyn LB, Matthews H, Harpin V, Hall TA, Drader JJ, Hofstadler SA, Eshoo MW, Rudnick K, Studarus K, Moore D, Abbott S, Janda JM, Whitehouse CA. 2009. Usefulness of multilocus polymerase chain reaction followed by electrospray ionization mass spectrometry to identify a diverse panel of bacterial isolates. Diagn. Microbiol. Infect. Dis. 63:403–408 [DOI] [PubMed] [Google Scholar]
- 2. Bhatia NS, Farrell JJ, Sampath R, Ranken R, Rounds MA, Ecker DJ, Bonomo RA. 2012. Identification of Streptococcus intermedius central nervous system infection using PCR and electrospray ionization mass spectrometry. J. Clin. Microbiol. 50:4160–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ecker DJ, Sampath R, Li H, Massire C, Matthews HE, Toleno D, Hall TA, Blyn LB, Eshoo MW, Ranken R, Hofstadler SA, Tang YW. 2010. New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev. Mol. Diagn. 10:399–415 [DOI] [PubMed] [Google Scholar]
- 4. Heid CA, Stevens J, Livak KJ, Williams PM. 1996. Real time quantitative PCR. Genome Res. 6:986–994 [DOI] [PubMed] [Google Scholar]
- 5. Swierkosz EM, Flanders R, Melvin L, Miller JD, Kline MW. 1989. Evaluation of the Abbott TESTPACK RSV enzyme immunoassay for detection of respiratory syncytial virus in nasopharyngeal swab specimens. J. Clin. Microbiol. 27:1151–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Engvall E. 2010. The ELISA, enzyme-linked immunosorbent assay. Clin. Chem. 56:319–320 [DOI] [PubMed] [Google Scholar]
- 7. Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908–934 [DOI] [PubMed] [Google Scholar]
- 8. Leaw SN, Chang HC, Sun HF, Barton R, Bouchara J-P, Chang TC. 2006. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J. Clin. Microbiol. 44:693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ecker DJ, Sampath R, Massire C, Blyn LB, Hall TA, Eshoo MW, Hofstadler SA. 2008. Ibis T5000: a universal biosensor approach for microbiology. Nature Rev. Microbiol. 6:553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hofstadler SA, Sampath R, Blyn LB, Eshoo MW, Hall TA, Jiang Y, Drader JJ, Hannis JC, Sannes-Lowery KA, Cummins LL, Libby B, Walcott DJ, Schink A, Massire C, Ranken R, Gutierrez J, Manalili S, Ivy C, Melton R, Levene H, Barrett-Wilt G, Li F, Zapp V, White N, Samant V, McNeil JA, Knize D, Robbins D, Rudnick K, Desai A, Moradi E, Ecker DJ. 2005. TIGER: the universal biosensor. Int. J. Mass Spectrom. 242:23–41 [Google Scholar]
- 12. Ecker DJ, Drader JJ, Gutierrez J, Gutierrez A, Hannis JC, Schink A, Sampath R, Blyn LB, Eshoo MW, Hall TA, Tobarmosquera M, Jiang Y, Sannes-Lowery KA, Cummins LL, Libby B, Walcott DJ, Massire C, Ranken R, Manalili S, Ivy C, Melton R, Levene H, Harpin V, Li F, White N, Pear M, Ecker JA, Samant V, Knize D, Robbins D, Rudnick K, Hajjar F, Hofstadler SA. 2006. The Ibis T5000 universal biosensor: an automated platform for pathogen identification and strain typing. J. Assoc. Lab. Autom. 11:341–351 [Google Scholar]
- 13. Shin JH, Ranken R, Sefers SE, Lovari R, Quinn CD, Meng S, Carolan HE, Toleno D, Li H, Lee JN, Stratton CW, Massire C, Tang YW. 2013. Detection, identification, and distribution of fungi in bronchoalveolar lavage specimens by use of multilocus PCR coupled with electrospray ionization/mass spectrometry. J. Clin. Microbiol. 51:136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eshoo MW, Crowder CC, Rebman AW, Rounds MA, Matthews HE, Picuri JM, Soloski MJ, Ecker DJ, Schutzer SE, Aucott JN. 2012. Direct molecular detection and genotyping of Borrelia burgdorferi from whole blood of patients with early Lyme disease. PLoS One 7:e36825. 10.1371/journal.pone.0036825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eshoo MW, Crowder CD, Li H, Matthews HE, Meng S, Sefers SE, Sampath R, Stratton CW, Blyn LB, Ecker DJ, Tang YW. 2010. Detection and identification of Ehrlichia species in blood by use of PCR and electrospray ionization mass spectrometry. J. Clin. Microbiol. 48:472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crowder CD, Matthews HE, Schutzer S, Rounds MA, Luft BJ, Nolte O, Campbell SR, Phillipson CA, Li F, Sampath R, Ecker DJ, Eshoo MW. 2010. Genotypic variation and mixtures of Lyme Borrelia in Ixodes ticks from North America and Europe. PLoS One 5:e10650. 10.1371/journal.pone.0010650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sampath R, Mulholland N, Blyn LB, Massire C, Whitehouse CA, Waybright N, Harter C, Bogan J, Miranda MS, Smith D, Baldwin C, Wolcott M, Norwood D, Kreft R, Frinder M, Lovari R, Yasuda I, Matthews H, Toleno D, Housley R, Duncan D, Li F, Warren R, Eshoo MW, Hall TA, Hofstadler SA, Ecker DJ. 2012. Comprehensive biothreat cluster identification by PCR/electrospray-ionization mass spectrometry. PLoS One 7:e36528. 10.1371/journal.pone.0036528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu Z, Hall TA, Frinder M, Walsh TJ, Hayden RT. 2012. Evaluation of repetitive sequence PCR and PCR-mass spectrometry for the identification of clinically relevant Candida species. Med. Mycol. 50:259–265 [DOI] [PubMed] [Google Scholar]
- 19. Hannis JC, Manalili SM, Hall TA, Ranken R, White N, Sampath R, Blyn LB, Ecker DJ, Mandrell RE, Fagerquist CK, Bates AH, Miller WG, Hofstadler SA. 2008. High-resolution genotyping of Campylobacter species by use of PCR and high-throughput mass spectrometry. J. Clin. Microbiol. 46:1220–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ecker JA, Massire C, Hall TA, Ranken R, Pennella TTD, Ivy CA, Blyn LB, Hofstadler SA, Endy TP, Scott PT, Lindler L, Hamilton T, Gaddy C, Snow K, Pe M, Fishbain J, Craft D, Deye G, Riddell S, Milstrey E, Petruccelli B, Brisse S, Harpin V, Schink A, Ecker DJ, Sampath R, Eshoo MW. 2006. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 44:2921–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klappenbach JA, Dunbar JM, Schmidt TM. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148(Pt 1):257–266 [DOI] [PubMed] [Google Scholar]
- 23. Hall KK, Lyman JA. 2006. Updated review of blood culture contamination. Clin. Microbiol. Rev. 19:788–802 [DOI] [PMC free article] [PubMed] [Google Scholar]