Abstract
Bacterial pathogens impose a heavy health burden worldwide. In the new era of high-throughput sequencing and online bioinformatics, real-time genome typing of infecting agents, and in particular those with potential severe clinical outcomes, holds promise for guiding clinical care to limit the detrimental effects of infections and to prevent potential local or global outbreaks. Here, we sequenced and compared 85 isolates of Streptococcus suis, a zoonotic human and swine pathogen, wherein we analyzed 32 recognized serotypes and 75 sequence types representing the diversity of the species and the human clinical isolates with high public health significance. We found that 1,077 of the 2,469 genes are shared by all isolates. Excluding 201 common but mobile genes, 876 genes were defined as the minimum core genome (MCG) of the species. Of 190,894 single-nucleotide polymorphisms (SNPs) identified, 58,501 were located in the MCG genes and were referred to as MCG SNPs. A population structure analysis of these MCG SNPs classified the 85 isolates into seven MCG groups, of which MCG group 1 includes all isolates from human infections and outbreaks. Our MCG typing system for S. suis provided a clear separation of groups containing human-associated isolates from those containing animal-associated isolates. It also separated the group containing outbreak isolates, including those causing life-threatening streptococcal toxic shock-like syndrome, from sporadic or less severe meningitis or bacteremia-only isolates. The typing system facilitates the application of genome data to the fields of clinical medicine and epidemiology and to the surveillance of S. suis. The MCG groups may also be used as the taxonomical units of S. suis to define bacterial subpopulations with the potential to cause severe clinical infections and large-scale outbreaks.
INTRODUCTION
The rapid and accurate classification of bacterial isolates is the most important task of medical microbiology, especially in situations where infectious disease outbreaks pose threats of national or global spread. The classification system of family to species in bacterial taxonomy has remained static, with species being the lowest level of classification used during the past 2 centuries. This classification system using species as the basic unit is well suited to higher organisms, as species defines the biological boundary of sexual reproduction. However, in bacteria, the species definition has long been hotly debated (1, 2). In the clinical care of patients, it is far more relevant to classify bacteria to a level that reveals the mode of pathogenesis and the potential of the strain to cause severe disease (3) so that appropriate clinical care can be rendered. In traditional clinical microbiology, much effort has been devoted to finding phenotypic or genetic traits so as to identify clinically or epidemiologically important pathogens. This goal has not been fully achieved using current methodologies, including the most widely used typing methods, such as multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and multilocus variable-number tandem-repeat analysis (4, 5).
In the coming era of an anticipated wide use of high-throughput and high-coverage sequencing in translational medicine, it is possible to use whole-genome sequence (WGS) data for identification and classification of organisms (6, 7). WGS, in theory, might provide information for diagnosis, clinical care, epidemiological investigation, intervention, and prevention, as well as for vaccine development (8). Ideally, it should be accomplished in a couple of hours to make a real-time diagnosis for clinical management and to provide early warnings and detection of outbreaks.
In this study, we developed a whole-genome sequence-based typing schema to identify and type Streptococcus suis strains. We demonstrate that this novel approach can be an alternative genotyping method for typing bacterial pathogens. S. suis is a swine pathogen posing a serious threat to the pork industry, and is a zoonotic pathogen that causes streptococcal toxic shock-like syndrome in humans with a high mortality rate (4, 9, 10). S. suis has caused severe meningitis in southeast Asia and some European countries (11) and caused two of the largest outbreaks in China in 1998 and 2005 (4, 9, 10, 12–14). In North America, however, there have been few human infections and no deaths, suggesting that some S. suis populations are more pathogenic to humans than others. The differences in disease incidence and severity have been attributed partly to strain differences. S. suis strains have been recognized to have different levels of pathogenicity. Those having caused severe outbreaks or sporadic invasive human infections are treated as highly pathogenic (12, 15). The method we developed here can provide not only the taxonomic identification of S. suis strains, but it can also indicate the pathogenic or epidemic potential of a given strain. The approach used in this study may be applied to other pathogens.
MATERIALS AND METHODS
Bacterial isolates.
We selected 72 isolates from 117 isolates that were previously typed using MLST. Together with 13 available completed genomes (11, 12, 15, 16–18), a total of 85 strains were used for this study. These 85 isolates included all 32 serotypes of reference strains. Serotypes 32 to 34 previously termed S. suis were excluded because they are now classified as another species (19). The 85 isolates include 75 sequence types (STs) and the six ST complexes that are most frequently isolated from animal and human infections; seven are from human infections and three are outbreak-associated (Table 1). The STs represent the diversity of the species, as shown by the ST distribution on the minimum spanning tree (MST) of the 368 known STs in the S. suis MLST database (see Fig. S1 in the supplemental material).
Table 1.
Characteristics of S. suis isolates sequenced in this study
| Isolate | Isolate origin |
Molecular or serological type |
Genome sequencing results |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Countrya | Sourceb | Year | MCGc | Serotyping | PFGE | MLST | No. of reads | Genome length (Mbp) | Coverage (%) | |
| 07SC3 | CHN | DP | 2007 | 1 | 2 | 47 | 7 | 9,975,410 | 2.06 | 97.10 |
| YS1 | CHN | HP | 2011 | 1 | 2 | 49 | 1 | 12,199,108 | 2.03 | 96.90 |
| YS6 | CHN | HP | 2011 | 1 | 2 | 50 | 1 | 11,929,132 | 2.04 | 97.00 |
| R735 | NED | DP | 1980s–1990s | 1 | 2 | 46 | 1 | 8,287,492 | 1.95 | 96.10 |
| YS12 | CHN | HP | 2011 | 1 | 7 | 41 | 17 | 13,225,862 | 2.08 | 91.80 |
| 11611 | GER | NAd | NA | 1 | 14 | 48 | 11 | 9,411,164 | 2.04 | 97.00 |
| 13730 | NED | H | 1980s–1990s | 1 | 14 | 45 | 6 | 10,631,608 | 1.96 | 96.50 |
| S15 | NED | DP | 1980s–1990s | 1 | 15 | 3 | 81 | 11,647,164 | 2.02 | 91.80 |
| RC1 | CHN | HP | 2005 | 2 | 8 | 20 | 308 | 8,300,186 | 2.2 | 89.50 |
| 14636 | NED | DP | 1980s–1990s | 2 | 8 | 22 | 87 | 13,124,752 | 2.08 | 89.30 |
| YS14 | CHN | HP | 2011 | 2 | 8 | 21 | 308 | 9,825,644 | 2.17 | 89.10 |
| 6407 | NED | DP | 1980s–1990s | 3 | 4 | 64 | 54 | 12,478,050 | 2.25 | 89.30 |
| 11538 | NED | DP | 1980s–1990s | 3 | 5 | 65 | 53 | 13,733,866 | 2.17 | 89.20 |
| 93A | CAN | HP | 1980s–1990s | 3 | 17 | 5 | 76 | 14,003,966 | 2.33 | 87.10 |
| NT77 | CAN | HP | 1980s–1990s | 3 | 18 | 69 | 79 | 10,760,626 | 2.32 | 87.00 |
| 42A | CAN | HP | 1980s–1990s | 3 | 19 | 19 | 76 | 12,338,610 | 2.29 | 87.40 |
| 89-2479 | CAN | DP | 1980s–1990s | 3 | 23 | 57 | 80 | 11,345,410 | 2.17 | 88.70 |
| 89-1591 | CAN | DP | 1980s–1990s | 4 | 2 | 66 | 25 | 11,960,982 | 2.15 | 87.10 |
| YS19 | CHN | HP | 2011 | 4 | 2 | 54 | 28 | 12,476,772 | 2.11 | 87.00 |
| 4961 | NED | DP | 1980s–1990s | 4 | 3 | 40 | 35 | 10,310,970 | 2.02 | 85.90 |
| 8074 | DEN | DP | 1980s–1990s | 4 | 7 | 63 | 29 | 8,715,032 | 2.06 | 86.00 |
| YS4 | CHN | HP | 2011 | 4 | 1/2 | 55 | 28 | 9,939,394 | 2.11 | 87.10 |
| YS66 | CHN | HP | 2011 | 4 | 7 | 67 | 32 | 10,211,192 | 2.06 | 85.40 |
| YS53 | CHN | HP | 2011 | 5 | 11 | 35 | 318 | 10,785,148 | 2.21 | 84.20 |
| YS10 | CHN | HP | 2011 | 5 | NA | 32 | 306 | 10,514,108 | 2.22 | 85.10 |
| YS31 | CHN | HP | 2011 | 5 | NA | 1 | 313 | 12,456,376 | 2.23 | 84.60 |
| YS44 | CHN | HP | 2011 | 5 | NA | 34 | 318 | 10,644,018 | 2.21 | 84.30 |
| YS59 | CHN | HP | 2011 | 5 | NA | 33 | 313 | 11,647,296 | 2.2 | 84.40 |
| 161_00P5 | AR | NA | NA | 6 | NA | 23 | NA | 9,099,510 | 2.27 | 77.30 |
| YS21 | CHN | HP | 2011 | 6 | NA | 28 | 310 | 10,086,924 | 2.47 | 79.10 |
| 10581 | DEN | DP | 1980s–1990s | 6 | 13 | 43 | 71 | 11,974,692 | 2.48 | 76.30 |
| YS23 | CHN | HP | 2011 | 6 | NA | 71 | 328 | 12,435,058 | 2.35 | 76.60 |
| YS7 | CHN | HP | 2011 | 6 | NA | 68 | 305 | 10,243,932 | 2.35 | 77.40 |
| YS50 | CHN | HP | 2011 | 6 | 29 | 30 | 330 | 10,451,798 | 2.42 | 78.10 |
| YS27 | CHN | HP | 2011 | 6 | 27 | 26 | 312 | 12,018,336 | 2.43 | 78.70 |
| YS54 | CHN | HP | 2011 | 6 | 29 | 2 | 319 | 11,656,110 | 2.32 | 77.60 |
| 14A | CAN | HP | 1980s–1990s | 6 | 21 | 39 | NA | 10,794,868 | 2.35 | 77.80 |
| YS46 | CHN | HP | 2011 | 6 | NA | 17 | NA | 11,115,414 | 2.43 | 78.90 |
| 88-5299A | CAN | DP | 1980s–1990s | 6 | 24 | 15 | 68 | 9,500,742 | 2.34 | 74.40 |
| 89-5259 | CAN | DP | 1980s–1990s | 6 | 27 | 16 | 72 | 11,393,632 | 2.46 | 78.20 |
| 92-1191 | CAN | DP | 1980s–1990s | 6 | 29 | 14 | 92 | 12,982,848 | 2.36 | 76.60 |
| YS77 | CHN | HP | 2011 | 6 | 29 | 37 | 334 | 9,124,374 | 2.26 | 77.60 |
| YS57 | CHN | HP | 2011 | 6 | NA | 10 | 320 | 11,585,314 | 2.52 | 79.50 |
| YS49 | CHN | HP | 2011 | 6 | NA | 27 | 329 | 11,128,288 | 2.41 | 78.20 |
| YS72 | CHN | HP | 2011 | 6 | NA | 70 | 332 | 9,402,972 | 2.36 | 77.80 |
| YS74 | CHN | HP | 2011 | 6 | NA | 29 | 333 | 11,254,086 | 2.47 | 78.00 |
| 5428 | DEN | DP | 1980s–1990s | UG | 1 | 13 | 10,405,408 | 1.99 | 88.10 | |
| YS16 | CHN | HP | 2011 | 7 | NA | 4 | 309 | 9,878,654 | 2.33 | 80.30 |
| 2524 | NED | DP | 1980s–1990s | 7 | 6 | 52 | 55 | 10,020,074 | 1.88 | 83.40 |
| 22083 | DEN | DP | 1980s–1990s | 7 | 9 | 58 | 82 | 11,732,812 | 2.03 | 79.00 |
| 4417 | DEN | DP | 1980s–1990s | 7 | 10 | 38 | 78 | 9,168,540 | 2.22 | 83.70 |
| YS24 | CHN | HP | 2011 | 7 | 10 | 36 | 311 | 12,459,076 | 2.11 | 83.80 |
| 12814 | DEN | DP | 1980s–1990s | UG | 11 | 8 | 91 | 10,722,342 | 2.07 | 84.20 |
| 8830 | DEN | DP | 1980s–1990s | UG | 12 | 51 | 93 | 12,649,154 | 2.2 | 85.30 |
| YS35 | CHN | HP | 2011 | 7 | NA | 6 | 315 | 11,343,994 | 2.32 | 81.30 |
| 2726 | NED | DP | 1980s–1990s | 7 | 16 | 62 | 73 | 11,549,464 | 2.22 | 84.00 |
| 86-5192 | CAN | DC | 1980s–1990s | 7 | 20 | 13 | NA | 11,496,440 | 2.11 | 60.20 |
| 88-1861 | CAN | DP | 1980s–1990s | 7 | 22 | 24 | NA | 13,397,520 | 2.28 | 61.40 |
| 89-3576-3 | CAN | DP | 1980s–1990s | 7 | 25 | 18 | 69 | 12,291,892 | 2.13 | 83.50 |
| 89-4109-1 | CAN | DP | 1980s–1990s | 7 | 26 | 9 | NA | 9,868,296 | 2.18 | 59.40 |
| 89-590 | CAN | DP | 1980s–1990s | UG | 28 | 60 | 75 | 8,352,086 | 2.16 | 86.80 |
| 92-1400 | CAN | DP | 1980s–1990s | 7 | 30 | 44 | 77 | 10,417,216 | 2.31 | 84.20 |
| 92-4172 | CAN | DC | 1980s–1990s | 7 | 31 | 25 | 70 | 11,390,990 | 2.31 | 80.70 |
| 2651 | NED | DP | 1980s–1990s | UG | 1/2 | 59 | 56 | 11,352,306 | 1.98 | 89.10 |
| YS56 | CHN | HP | 2011 | 7 | NA | 56 | 331 | 10,028,326 | 2.23 | 74.90 |
| YS43 | CHN | HP | 2011 | 7 | NA | 42 | 317 | 8,593,284 | 2.05 | 84.20 |
| YS67 | CHN | HP | 2011 | UG | NA | 12 | 321 | 10,163,838 | 2.18 | 84.60 |
| YS34 | CHN | HP | 2011 | 7 | NA | 11 | 314 | 8,203,060 | 2.38 | 81.50 |
| YS39 | CHN | HP | 2011 | 7 | NA | 61 | 316 | 11,305,136 | 2.24 | 83.80 |
| YS3 | CHN | HP | 2011 | UG | NA | 53 | 304 | 12,544,766 | 2.17 | 82.80 |
| YS17 | CHN | HP | 2011 | 7 | NA | 7 | 327 | 9,649,640 | 2.34 | 82.20 |
| YS64 | CHN | HP | 2011 | UG | NA | 31 | NA | 9,800,376 | 2.07 | 84.20 |
CHN, China; NED, Netherlands; GER, Germany; CAN, Canada; AR, Argentina; DEN, Denmark.
DP, diseased pig; DC, diseased calf; HP, healthy pig; H, patient.
UG, ungrouped.
NA, not applicable.
Genome sequencing and core genome analysis.
The 72 isolates were sequenced using Illumina sequencing by constructing two paired-end (PE) libraries with average insertion lengths of 500 bp and 2,000 bp. Sequences were generated with an Illumina GA IIx (Illumina, San Diego, CA) and assembled into contigs and scaffolds using SOAPdenovo (release 1.04) (20). Low-quality reads were removed if the quality scores of ≥3 consecutive bases were ≤Q20.
Genes were predicted using Glimmer (21). Gene orthologs were determined using OrthoMCL (22). A matrix describing the genome gene content was constructed using OrthoMCL, with a BLAST E value cutoff of 1e−5 and an inflation parameter of 1.5. Genes that were included in all isolates were considered to be the core genome genes. A simulation of the number of core genes was calculated using the median value of each set of randomly selected genomes. The regression analysis of the number of isolates against their shared genes was performed by fitting a double exponential decay function, as previously reported (23): Nc = ϴ + k1 × exp(m1 × Ng) + k2 × exp(m2 × Ng), where Nc is the number of core genes, Ng is the number of genomes, and ϴ, k1, m1, k2, and m2 are free parameters where the model was determined by a weighted least-squares regression analysis. The best fit was obtained with an R2 value of 0.999 for a ϴ value (the asymptotic core genome size) of 980 ± 4, k1 of 382.8 ± 1.87, m1 of −0.016 ± 0.00041, k2 of 247.1 ± 4.3, and m2 of −0.176 ± 0.0051.
Mobile genes were excluded from the core genome based on the pathogenicity island (PAI), plasmid, insertion sequence (IS), transposon, and phage databases from EBI, GenBank, and PAIDB (24).
SNP detection.
We examined single-nucleotide polymorphisms (SNPs) through pairwise comparisons of S. suis genomes using SOAPsnp (25) and MUMmer (26). For isolates with complete genome sequences, SNP selection was performed using the NUCmer program in the MUMmer package (26). For SNP detection in the draft genomes, reads with low quality (>3 consecutive bases with a quality score of ≤Q20) were removed before SNP calling. SNPs were called if they met the following criteria using SOAPsnp (25): (i) each SNP site was covered with ≥20 reads, (ii) ≥5 bp was the distance between two SNP sites, (iii) the SNP was not located in a repeat region, and (iv) the prior probability of heterozygous SNPs is ≤0.1%.
Analysis of recombination and removal of recombinant SNPs.
Gene segments with recombination in the 85 isolates were identified using the method described by Feng et al. (27). In the method we used, if the segments between two adjacent SNPs are defined as ISSs (inter-SNP segments), the distribution of ISSs around the genome is expected to follow an exponential distribution if all the observed SNPs are due to mutations that occur as a Poisson process. However, ISSs brought in by recombination events will disturb this distribution and form anomalous clusters of ISSs, which have shorter distances between SNPs due to imported segments carrying more SNPs. Therefore, the overall distribution observed with these ISSs will have an excess of short ISSs due to recombination and will not follow an exponential distribution. This excess of short ISSs may be removed to fit an exponential distribution if most parts of the genome have not been involved in recombination. A progressive exclusion of the short ISSs will allow one to find a cutoff value to fit an exponential distribution and to identify and remove ISSs due to recombination.
Since there was a large amount of recombination in S. suis (28), we did not remove the whole gene where recombination was detected; instead, only relevant portions of the recombined regions were removed. As such, the phylogenetic content is more likely to reflect the evolutionary history of vertical descent in populations and their true relationships.
Population structure and phylogenetic analyses.
The program Structure 2.2 (29) was used to analyze the SNPs in the 85 isolates at the genome level, assuming one to 15 populations for five iterations each, using the admixture model and uncorrelated allele frequencies. A burn-in of 50,000 replications was discarded, and 150,000 additional replications were analyzed. The burn-in period was sufficient to stabilize log-likelihood values. Each value of K is based on the run with the highest likelihood value. Likewise, a standard measure of genetic distance, Fst, was calculated from the Structure 2.2 run with the highest likelihood value for K at 7 (see Table S4 in the supplemental material) and showed the divergence of each population from the estimated ancestral allele frequencies. An admixture model and independent allele frequency were used for Structure analysis. The assignment of an isolate to a subpopulation was based on the largest percentage of ancestry contained in an isolate.
A phylogenetic tree based on the core genome SNPs was constructed using the neighbor-joining or minimum evolution algorithms in MEGA (30). Bootstraps were performed with 1,000 replicates. Streptococcus pneumoniae R1 was used as an out-group. The program MEGA was also used to calculate the p-distance (p = nd/n) within and between population groups, where nd is the number of sites with difference and n is the total number of sites.
Online whole-genome typing tool.
Fastaq format data using Sanger quality from the Illumina platform were used in our online tools. The uploaded new genome was mapped to the genome of the reference strain S. suis GZ1. All low-quality reads that contained >3 consecutive bases with a quality score of <Q20 were removed. SNP detection criteria used were the same as for the SNP-calling method described above. Only those SNPs located in the 553 group-specific SNP sites were kept for group identification. We assigned an isolate to a particular minimum core genome (MCG) group based on the highest-percentage match (calculated as [number of matched SNPs/total number of group-specific SNPs] × 100) to one of the seven groups.
Nucleotide sequence accession numbers.
The sequences of the 72 representative strains of S. suis were deposited in the GenBank/EMBL/DDBJ database under accession numbers PRJNA171409 to PRJNA171425, PRJNA171428 to PRJNA171478, and PRJNA171480 to PRJNA171483.
RESULTS
Identification of the minimum core genome content of S. suis.
Using Illumina high-throughput sequencing, we sequenced 72 representative isolates of S. suis. Thirteen completed genomes (those of S. suis strains 05ZYH33, P1/7, ST3, BM407, SC84, 98HAH33, JS14, SS12, D12, D9, GZ1, ST1, and A7) were also included in the analysis. These isolates covered all 32 serotypes recently defined for S. suis, and 75 sequence types (STs) refer to MLST data, representing the diversity of the species and the isolates with clinical and public health significance. 16S rRNA gene sequences and biochemical analyses showed that strains from serotypes 32 and 34 were from Streptococcus orisratti and serotype 33 was from a novel species (31). Thus, serotypes 32 to 34 were excluded from this study.
A total of 224 gigabases of 90-bp paired-end sequence data were obtained (Table 1), with approximately 11 million reads and >300× coverage (maximum 572×) per isolate. The sequences for each isolate were de novo assembled, producing 82 contigs and 60 scaffolds on average. The number of genes per isolate ranged from 1,816 to 2,469 (see Table S1 in the supplemental material). Comparisons of these genes among the 85 genomes showed that 1,077 genes were shared by all the isolates.
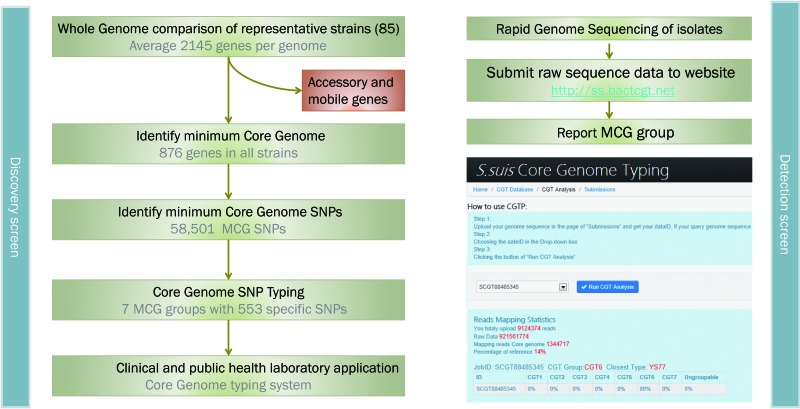
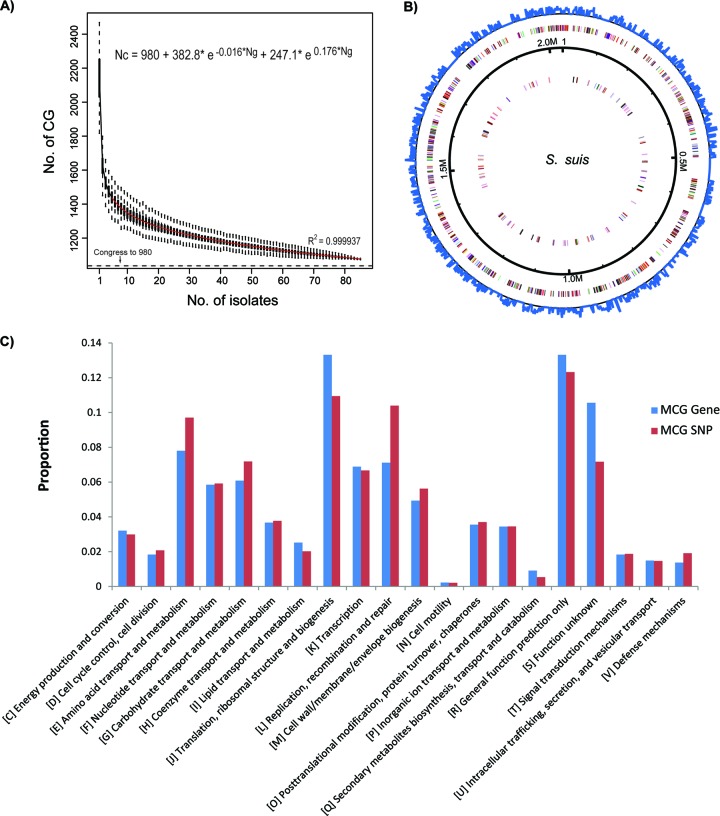
Additionally, 201 of the 1,077 genes were further excluded from analysis, including 120 mobile genes or transposons and 81 genes with high nucleotide diversity. The 81 genes excluded were previously identified as noncore genes (28), and we found that they have an average of 0.11 SNPs/bp, which is much higher than the average of the core genes (0.068), suggesting that this high diversity is due to recombination. Finally, we derived 876 minimum core genome (MCG) genes of S. suis, accounting for approximately 40.8% of the genome (Fig. 1). The theoretical minimal number of core genes was calculated to be 980 by fitting a double exponential decay function based on the 85 genome sequences (Fig. 2A).
Fig 1.
Flowchart to determine minimum core genome (MCG) genes, MCG SNPs, and application of the MCG sequence typing method.
Fig 2.
(A) Descending trend of core genome size of S. suis with the increasing number of isolates. The number, n, of shared genes is plotted as the number of strains sequentially added. For each n, circles are 8!/[(n − 1) × (8 − n)!] values obtained for the different strain combinations. Squares are the averages of such values. (B) Distribution of MCG genes in the reference genome of S. suis GZ1. Outer circle, MCG SNP density; colored circles, coding sequences (CDSs) of core genes shown in a pair of concentric rings representing coding strands; black solid circle, mobile genes. (C) The proportions of MCG genes and MCG SNPs in 20 functional categories.
These 876 MCG genes were randomly distributed across the genome (Fig. 2B), 13.3% of which are in the cluster of orthologous genes (COG) functional category of translation, ribosomal structure, and biogenesis (Fig. 2C; see also Fig. S2 in the supplemental material); these are essential for the survival of an organism. In contrast, only 4.1% of the accessory genome genes fall into this category (see Fig. S2 in the supplemental material). It should be mentioned that three of the seven genes in the MLST scheme of S. suis (4) do not belong to the MCG, namely, cpn (60-kDa chaperonin), dpr (defective proboscis extension response), and gki (glucokinase). gki was found to be missing in S. suis strain 92-1191, whereas cpn and dpr share high nucleotide similarities with a gene on a plasmid and a gene on a pathogenicity island, respectively, and were identified as mobile elements in this study.
Identification of the minimum core genome SNPs.
By comparison to the reference genome of strain GZ1 (12), we found 229,618 SNPs among the 85 isolates, of which 87,646 SNPs were located in the MCG genes. Excluding the 29,145 SNPs in the recombination regions, there were 58,501 MCG SNP sites. The MCG SNPs are found in the genome with a Poisson distribution (see Fig. S3 in the supplemental material). Genes with the highest frequency of MCG SNPs were located in two categories, namely, translation, ribosomal structure, and biogenesis and predicted general function (Fig. 2C). The average density of the SNPs among the 85 isolates is ∼70 SNPs/kb. MCG genes with <60 SNPs/kb are those in the COG category translation, ribosomal structure, and biogenesis (see Fig. S3 and S4 in the supplemental material). Among the 50 genes with the lowest SNP densities, eight encode ribosomal proteins. In contrast, of the 50 genes with the highest SNP densities, five are associated with amino acid transport and metabolism (see Tables S2 and S3 in the supplemental material).
Definition of the minimum core genome group based on population structure.
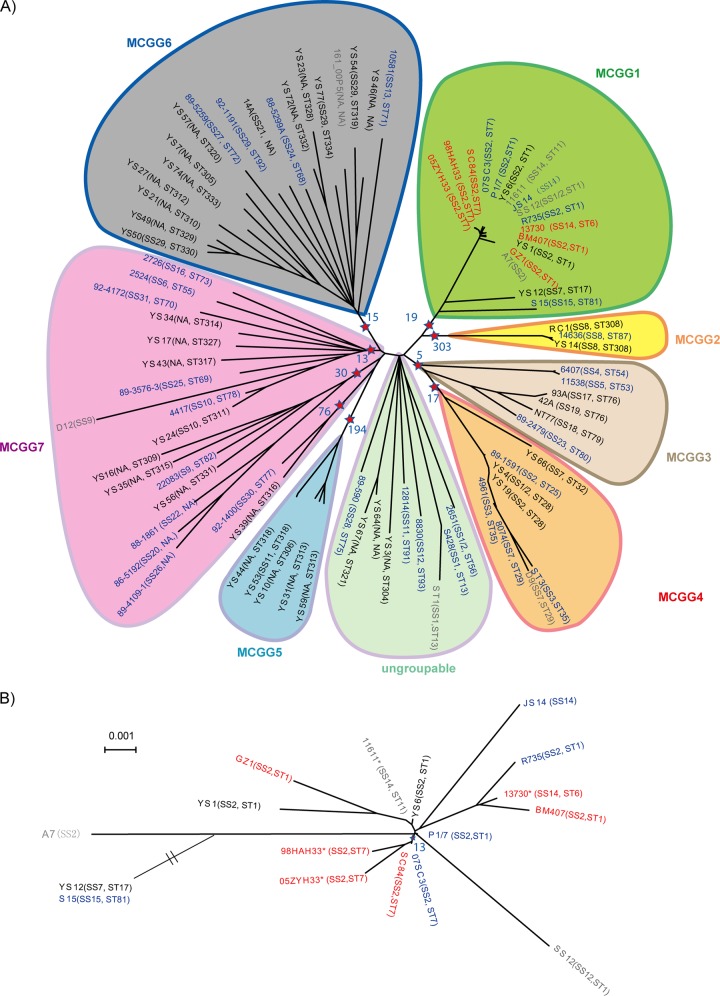
We used the Bayesian statistics tool Structure (32) to show the population structure of S. suis and to establish population genetics-based subdivisions of the species for strain identification and typing. By testing subdivisions of the 85 S. suis isolates into between two and 15 subpopulations, we found that the optimal number of subpopulations was seven, with 17, 3, 6, 8, 5, 18, and 28 isolates being assigned to subpopulations 1 to 7, respectively (Fig. 3; see also Fig. S5 in the supplemental material). These subpopulations were defined as MCG groups. For all groups except MCG group 7, each has isolates with ancestral genes from other subpopulations (Fig. 3).
Fig 3.
Phylogenetic trees and MCG groups of all strains. (A) Phylogenetic tree of 85 strains and MCG groups. The pentagrams and their corresponding blue numbers represent the number of SNPs used for group typing for each MCG group. (B) The phylogenetic tree of MCG group 1 strains. In both panels, the serotypes and MLST are represented in brackets. Strains from patients, diseased pigs or calves, and healthy pigs are shown in red, blue, and black, respectively. The strains in gray are from published data.
Phylogenetic analysis was also performed using both the neighbor-joining algorithm and the minimum evolution algorithm (33, 34). The clustering of the isolates was largely consistent with the Structure analysis (Fig. 3). We computed the genetic distances for all of the combinations of within- and between-group comparisons where the within-group distance is 1.3 to 47 times smaller than the between-group distance, except for MCG group 7. The smallest within-group distance was 0.003 for MCG group 2, which is 50 times less than the distances between MCG group 2 and the other MCG groups. The distance data further support the Structure divisions (see Table S6 in the supplemental material). Since the seven subpopulations were robustly derived from the Structure analysis using core genome variation, these subpopulations have taxonomic significance below the species level. We propose that these Structure-based subpopulations be called MCG groups and be treated as taxonomic units. Thus, S. suis is divided into seven MCG groups, with a small number of ungrouped isolates.
Six of the seven MCG groups can be clearly identified using group-specific MCG SNPs. Among the 58,501 MCG SNPs, 553 SNPs were found to be group specific (Fig. 3). There were 19 MCG group 1-specific SNPs that separate that group from the others. Similarly, there were 303, 5, 17, 194, and 15 SNPs specific for MCG groups 2 to 6, respectively. There were no group-specific SNPs found for MCG group 7, within which the three lineages contain 76, 30, and 13 lineage-specific SNPs, respectively, and a minimum of one SNP from each lineage is required for lineage assignment. All group-specific or lineage-specific SNPs have no reverse or parallel mutations.
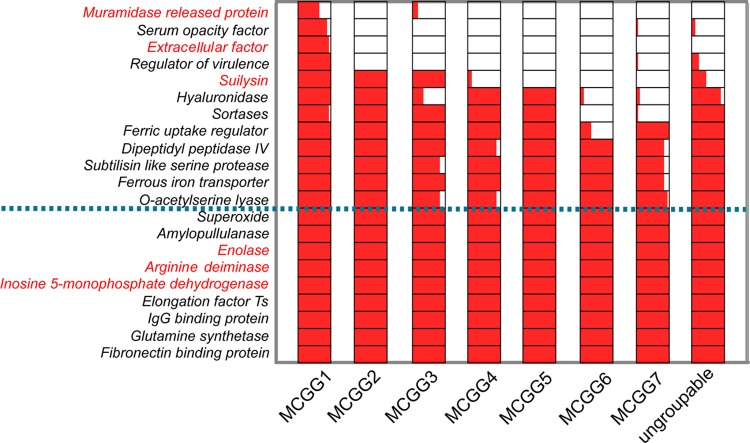
MCG groups and public health significance.
We analyzed the distribution of the 21 known putative virulence genes among the MCG groups, in which nine exist as core genome genes (Fig. 4). MCG group 1 includes all the highly virulent isolates of ST1 and the epidemic isolates of ST7, and it also contains the greatest number of virulent genes (Fig. 4). The previously identified “intermediate virulent” STs, such as ST25 and ST28 (35), were all located in MCG group 4, which carries fewer numbers of virulent genes than MCG group 1. MCG group 6 carries the lowest number of virulence genes. Interestingly, MCG group 6 was found by phylogenetic analysis to diverge the earliest, as it is closest to the out-group S. pneumoniae. Therefore, S. suis may have progressively gained additional virulence genes during its evolution to becoming a human pathogen. Clearly, all isolates that caused severe human infections, death, and outbreaks fell into MCG group 1.
Fig 4.
Distribution of known putative virulence genes in the seven core genome groups. The bar within a group shows the percentage of strains in that MCG group carrying a given virulence gene. Six genes encoding immune proteins are marked in red (46–49). The horizontal dotted blue line demarcates the virulence genes that are present in all isolates. The encoded protein names are shown on the left.
Online bioinformatics tool for real-time clinical and outbreak investigations using minimum core genome typing.
The rapid identification of a given MCG group, such as MCG group 1, which has the capacity to cause countrywide or global outbreaks (4, 12, 13), would be useful for clinical and public health laboratories. To meet this demand, we developed a Web-based S. suis minimum core genome typing system (http://ss.bactcgt.net). This system uses whole-genome sequencing data to identify the minimum core genome type in a very short period of time. The raw reads from Illumina genome sequencing from the test isolate are mapped to the reference genome (that of S. suis GZ1) in the database to identify SNPs in the 58,501 MCG SNP sites, and group assignment is then performed based on 553 group-specific SNPs stored in the database. The output includes the probability (calculated as the number of matched sites/total group-specific sites) of assignment to each of the seven defined groups and the number of SNP differences to the closest reference core genome. In our simulation test, using 10 million reads of 90 bp, which is ∼200-fold coverage of the S. suis GZ1 genome as input, the examination took ∼1 h, demonstrating the rapidity of the computation. We also simulated the number of times for coverage required to perform a reliable assignment. We found that 1×, 5×, 10×, 15×, 20×, 50×, and 100× coverage of the S. suis genome can identify 0%, 35%, 85%, 100%, 100%, 100%, and 100% of the MCG group-specific SNPs, respectively. Therefore, 20× coverage was sufficient for accurate group assignment and identification to the closest reference strain. With multiplexing in current next-generation sequencing (NGS) platforms, the low coverage required can substantially reduce the MCG sequence typing cost.
DISCUSSION
In this study, we developed a novel approach for defining the minimum core genome of S. suis strains for taxonomical classification. This approach is also likely to be useful for other pathogenic bacterial species. The minimum core genome refers to genes that are shared by all strains of a given species (36, 37). We further reduced the minimum core genome genes to exclude mobile elements even if they were present in all strains, since these genes carry mixed phylogenetic signals. Additionally, SNP sites with a high frequency of recombination were removed from the core genome genes to increase the precision of the assignment of the isolates to a subpopulation for taxonomic classification. Using this approach, we found that S. suis has a core genome of 876 genes out of an average of 2,140 genes per isolate and 58,501 MCG SNPs out of a total of 190,894 SNPs.
Population genetic analysis of the MCG SNPs from 85 isolates divided the S. suis population into seven core genome groups. These seven groups were the optimal divisions after trials of two to 15 divisions with 150,000 iterations in the Structure runs. Phylogenetic analysis revealed six of the seven groups to be monophyletic. Further, the within-group distance range of 0.003 to 0.087 is much smaller than the between-group distance range of 0.115 to 0.172 (see Table S6 in the supplemental material), indicating considerable divergence between MCG groups. Thus, these MCG groups may be considered to be robust taxonomic units within S. suis.
In this study, we used Structure analysis to establish the MCG groups. Bacterial species vary in their capacities to recombine among members of the species, which leads to different levels of clonality and affects the inference of phylogeny in the strains. The population genetics approach we used to classify the isolates into subpopulations takes recombination into account, and thus it provides a more-robust means to identify subpopulations than do phylogenetic approaches that group isolates into phylogenetic clusters.
In our study, subpopulation 7 did not group together, as one cluster with seven isolates grouped away from the majority of the isolates. These isolates possibly have had a significant recombinant history that was not identified by our recombination detection method and cannot be reliably allocated into a subpopulation by Structure; therefore, they were assigned to group 7 (see Fig. S6 in the supplemental material). This phenomenon has been previously reported for Salmonella (38). Thus, these isolates were removed from subpopulation 7 and were referred to as ungroupable. The remaining group 7 isolates formed three lineages, with two lineages closest to group 5 on the neighbor-joining tree. Using S. pneumoniae strain R6 as an out-group, group 6 is the earliest diverged group, while group 1 diverged most recently. Group 3 shared the most recent common ancestry with group 4 and clustered into one lineage (see Fig. S5 in the supplemental material).
The differences in the clustering of the strains observed might be due to the difference in the methods and algorithms used. The neighbor-joining and minimum evolution methods use assumptions and clustering algorithms that are different from those used by Structure. The reliability of phylogenetic inference is adversely affected by the frequency of recombination that obscures phylogenetic signals. Structure analysis was applied to MLST data in a range of species and has shown that even a frequently recombining species, such as Helicobacter pylori, can be divided into subpopulations (39, 40). We previously used Structure to divide S. suis into five groups using the MLST of seven genes (28). MCG typing further refines the groupings and also identifies multiple SNPs for reliable group assignment. We advocate that this population genetics approach be applied to the development of MCG typing in other species. MCG groups established in this way are expected to stand the test of time, similar to the divisions of many genera and species.
The seven MCG groups of S. suis can be identified using a minimum of nine SNPs. These SNPs can be used as markers for typing using subgenome sequencing methods, such as real-time PCR (41), providing an interim means of MCG typing in laboratories where NGS is not available during this transitional phase to NGS-based genome sequence typing.
These taxonomical MCG groupings may also have public health significance that cannot be shown by routine species identification and serotyping. Of the seven MCG groups of S. suis, only MCG group 1 contains isolates from human infections and outbreaks (4, 12, 13, 15). MCG grouping provides a clear separation of human-related from non-human-related infection isolates. The MCG SNPs can further provide a separation of outbreak-related from non-outbreak-related isolates, since outbreaks have been restricted to certain STs as based on MLST analysis (4, 12, 15). The outbreak STs tested in this study were all within MCG group 1. Although more isolates and STs will need to be tested, we expect that typing based on the MCG group 1 SNPs to identify strains with severe disease or outbreak potential will provide relevant clinical or public health information that can influence patient care and public health decisions.
Moreover, the full set of MCG SNPs may be useful for tracking the epidemic clones, e.g., ST7. Investigations of outbreaks and interhost transmission using genome sequence data have found SNPs to be very useful markers (42, 43). In a number of pathogens, evolutionary timelines ranging from decades to thousands of years were successfully estimated based on SNPs with given molecular clocks (44–47). Therefore, the use of MCG SNPs may allow the tracking of clones in the time frames of years and decades, providing a more reliable marker for epidemiology.
A caveat of this study is that the number of strains sequenced is still small, and the strains selected may not cover the full diversity of the species. However, our isolates represented the most-frequently isolated STs from animal and human infections with S. suis. Our selection of isolates was based on the MLST diversity and representation on the MST of seven MLST loci (see Fig. S1 in the supplemental material). There are 368 STs in the S. suis MLST database, 150 of which are grouped into 22 clonal complexes. The 85 isolates studied represent 75 of the 368 STs and 11 of the 22 clonal complexes. The geographic representation is also rather limited, with isolates being from only six countries, including three European countries (Netherlands, Denmark, and Germany), two countries of the Americas (Canada and Argentina), and one Asian country (China). Based on the MST of all STs (see Fig. S1 in the supplemental material), the Chinese isolates were widely distributed on the tree, suggesting that at least S. suis diversity in China reflected the overall species diversity with no evidence of geographical restriction. Nevertheless, additional diversity outside the MCG groups may be discovered in the future, and thus additional MCG groups may need to be defined accordingly. MCG group 7, which does not form a monophyletic group, may also require revision once more isolates from this group are typed.
Two major challenges in the use of genome sequencing in clinical microbiology laboratories are the speed of sequence data processing and the accuracy and uniform reporting of results. Our implementation of a Web-based online typing system (http://ss.bactcgt.net) that accepts raw sequence data and returns a report of the minimum core genome group assigned and the closest relative of the test strain in the database within 2 h demonstrates that this approach is computationally highly feasible. The use of core genome data and the recognition of core genome groups can standardize the reporting of typing results to ensure uniformity.
With the rapid reduction in genome sequencing cost and the wide deployment of next-generation sequencing instruments in clinical microbiology laboratories, genome sequencing may soon become a routine diagnostic tool. The approach we developed can be applied to many bacterial species. For example, in Neisseria meningitidis, a subpopulation referred to as the ST4821 complex causes about 300 human infections annually in China (48, 49), and a genome sequencing-based scheme would be highly beneficial. Our core genome sequence typing system provides a timely implementation of strain identification and classification using high-throughput NGS genome data, facilitating the application of genome data to the fields of clinical medicine, epidemiology, and surveillance. Our study marks a significant step in translating bacterial genome sequence data into clinical diagnosis and public health information.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by a China-Canada collaboration grant of the Chinese National Natural Science Foundation (no. 812111251, 81290340, and 81290345) and National Key Program of Mega Infectious Diseases grant no. 2013ZX10004-221.
We thank Yan Wang for technical assistance.
The authors declare no conflicts of interest.
Footnotes
Published ahead of print 29 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00535-13.
REFERENCES
- 1. Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL, Swings J. 2005. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733–739 [DOI] [PubMed] [Google Scholar]
- 2. Lan R, Reeves PR. 2001. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 9:419–424 [DOI] [PubMed] [Google Scholar]
- 3. Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW. 2012. Transforming clinical microbiology with bacterial genome sequencing. Nat. Rev. Genet. 13:601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ye C, Zhu X, Jing H, Du H, Segura M, Zheng H, Kan B, Wang L, Bai X, Zhou Y, Cui Z, Zhang S, Jin D, Sun N, Luo X, Zhang J, Gong Z, Wang X, Wang L, Sun H, Li Z, Sun Q, Liu H, Dong B, Ke C, Yuan H, Wang H, Tian K, Wang Y, Gottschalk M, Xu J. 2006. Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg. Infect. Dis. 12:1203–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, Whatmore AM. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40:3671–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pallen MJ, Loman NJ, Penn CW. 2010. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr. Opin. Microbiol. 13:625–631 [DOI] [PubMed] [Google Scholar]
- 7. Dunne WM, Jr, Westblade LF, Ford B. 2012. Next-generation and whole-genome sequencing in the diagnostic clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 31:1719–1726 [DOI] [PubMed] [Google Scholar]
- 8. Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, Chen W, Pu F, Peng Y, Li J, Xi F, Li S, Li Y, Zhang Z, Yang X, Zhao M, Wang P, Guan Y, Cen Z, Zhao X, Christner M, Kobbe R, Loos S, Oh J, Yang L, Danchin A, Gao GF, Song Y, Yang H, Wang J, Xu J, Pallen MJ, Wang J, Aepfelbacher M, Yang R, E. coli O104:H4 Genome Analysis Crowd-Sourcing Consortium 2011. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 365:718–724 [DOI] [PubMed] [Google Scholar]
- 9. Feng Y, Zhang H, Ma Y, Gao GF. 2010. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol. 18:124–131 [DOI] [PubMed] [Google Scholar]
- 10. Tang J, Wang C, Feng Y, Yang W, Song H, Chen Z, Yu H, Pan X, Zhou X, Wang H, Wu B, Wang H, Zhao H, Lin Y, Yue J, Wu Z, He X, Gao F, Khan AH, Wang J, Zhao GP, Wang Y, Wang X, Chen Z, Gao GF. 2006. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3:e30151. 10.1371/journal.pmed.0030151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mai NT, Hoa NT, Nga TV, Linh LD, Chau TT, Sinh DX, Phu NH, Chuong LV, Diep TS, Campbell J, Nghi HD, Minh TN, Chau NV, de Jong MD, Chinh NT, Hien TT, Farrar J, Schultsz C. 2008. Streptococcus suis meningitis in adults in Vietnam. Clin. Infect. Dis. 46:659–667 [DOI] [PubMed] [Google Scholar]
- 12. Ye C, Zheng H, Zhang J, Jing H, Wang L, Xiong Y, Wang W, Zhou Z, Sun Q, Luo X, Du H, Gottschalk M, Xu J. 2009. Clinical, experimental, and genomic differences between intermediately pathogenic, highly pathogenic, and epidemic Streptococcus suis. J. Infect. Dis. 199:97–107 [DOI] [PubMed] [Google Scholar]
- 13. Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, Wang S, Liu L, Zu R, Luo L, Xiang N, Liu H, Liu X, Shu Y, Lee SS, Chuang SK, Wang Y, Xu J, Yang W, Streptococcus suis Study Groups 2006. Human Streptococcus suis infection outbreak, Sichuan, China. Emerg. Infect. Dis. 12:914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gottschalk M, Xu J, Calzas C, Segura M. 2010. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 5:371–391 [DOI] [PubMed] [Google Scholar]
- 15. Holden MTG, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail MA, Price C, Rabbinowitsch E, Sharp S, Croucher NJ, Chieu TB, Mai NTH, Diep TS, Chinh NT, Kehoe M, Leigh JA, Ward PN, Dowson CG, Whatmore AM, Chanter N, Iversen P, Gottschalk M, Slater JD, Smith HE, Spratt BG, Xu J, Ye C, Bentley S, Barrell BG, Schultsz C, Maskell DJ, Parkhill J. 2009. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One 4:e6072. 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu P, Yang M, Zhang A, Wu J, Chen B, Hua Y, Yu J, Chen H, Xiao J, Jin M. 2011. Comparative genomics study of multi-drug-resistance mechanisms in the antibiotic-resistant Streptococcus suis R61 strain. PLoS One 6:e24988. 10.1371/journal.pone.0024988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu P, Yang M, Zhang A, Wu J, Chen B, Hua Y, Yu J, Xiao J, Jin M. 2011. Complete genome sequence of Streptococcus suis serotype 14 strain JS14. J. Bacteriol. 193:2375–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang A, Yang M, Hu P, Wu J, Chen B, Hua Y, Yu J, Chen H, Xiao J, Jin M. 2011. Comparative genomic analysis of Streptococcus suis reveals significant genomic diversity among different serotypes. BMC Genomics 12:523. 10.1186/1471-2164-12-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chatellier S, Harel J, Zhang Y, Gottschalk M, Higgins R, Devriese LA, Brousseau R. 1998. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int. J. Syst. Bacteriol. 48(Pt 2):581–589 [DOI] [PubMed] [Google Scholar]
- 20. Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J. 2009. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20:265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bottacini F, Medini D, Pavesi A, Turroni F, Foroni E, Riley D, Giubellini V, Tettelin H, van Sinderen D, Ventura M. 2010. Comparative genomics of the genus Bifidobacterium. Microbiology 156(Pt 11):3243–3254 [DOI] [PubMed] [Google Scholar]
- 24. Yoon SH, Park YK, Lee S, Choi D, Oh TK, Hur CG, Kim JF. 2007. Towards pathogenomics: a web-based resource for pathogenicity islands. Nucleic Acids Res. 35:D395–D400. 10.1093/nar/gkl790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, Wang J. 2009. SNP detection for massively parallel whole-genome resequencing. Genome Res. 19:1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kurtz S, Phillippy A, Delcher A, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng L, Reeves PR, Lan R, Ren Y, Gao C, Zhou Z, Cheng J, Wang W, Wang J, Qian W, Li D, Wang L. 2008. A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS One 3:e4053. 10.1371/journal.pone.0004053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng X, Zheng H, Lan R, Ye C, Wang Y, Zhang J, Jing H, Chen C, Segura M, Gottschalk M, Xu J. 2011. Identification of genes and genomic islands correlated with high pathogenicity in Streptococcus suis using whole genome tilling microarrays. PLoS One 6:e17987. 10.1371/journal.pone.0017987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 31. Hill JE, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM, Goh SH. 2005. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet. Microbiol. 107:63–69 [DOI] [PubMed] [Google Scholar]
- 32. Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rzhetsky A, Nei M. 1993. Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol. Biol. Evol. 10:1073–1095 [DOI] [PubMed] [Google Scholar]
- 34. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 35. Fittipaldi N, Xu J, Lacouture S, Tharavichitkul P, Osaki M, Sekizaki T, Takamatsu D, Gottschalk M. 2011. Lineage and virulence of Streptococcus suis serotype 2 isolates from North America. Emerg. Infect. Dis. 17:2239–2244. 10.3201/eid1712.110609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lan R, Reeves PR. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396–401 [DOI] [PubMed] [Google Scholar]
- 37. Dunning Hotopp JC, Grifantini R, Kumar N, Tzeng YL, Fouts D, Frigimelica E, Draghi M, Giuliani MM, Rappuoli R, Stephens DS, Grandi G, Tettelin H. 2006. Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology 152(Pt 12):3733–3749 [DOI] [PubMed] [Google Scholar]
- 38. Didelot X, Bowden R, Street T, Golubchik T, Spencer C, McVean G, Sangal V, Anjum MF, Achtman M, Falush D, Donnelly P. 2011. Recombination and population structure in Salmonella enterica. PLoS Genet. 7:e2191. 10.1371/journal.pgen.1002191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445:915–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hazbón MH, Alland D. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 42:1236–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Mladonicky JM, Somsel P, Rudrik JT, Dietrich SE, Zhang W, Swaminathan B, Alland D, Whittam TS. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, Cui Y, Thomson NR, Jombart T, Leblois R, Lichtner P, Rahalison L, Petersen JM, Balloux F, Keim P, Wirth T, Ravel J, Yang R, Carniel E, Achtman M. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42:1140–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JL, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reeves PR, Liu B, Zhou Z, Li D, Guo D, Ren Y, Clabots C, Lan R, Johnson JR, Wang L. 2011. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS One 6:e26907. 10.1371/journal.pone.0026907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shao Z, Li W, Ren J, Liang X, Xu L, Diao B, Li M, Lu M, Ren H, Cui Z, Zhu B, Dai Z, Zhang L, Chen X, Kan B, Xu J. 2006. Identification of a new Neisseria meningitidis serogroup C clone from Anhui province, China. Lancet 367:419–423 [DOI] [PubMed] [Google Scholar]
- 49. Zhou H, Gao Y, Xu L, Li M, Li Q, Li Y, Liang X, Luo H, Kan B, Xu J, Shao Z. 2012. Distribution of serogroups and sequence types in disease-associated and carrier strains of Neisseria meningitidis isolated in China between 2003 and 2008. Epidemiol. Infect. 140:1296–1303 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.