Abstract
We evaluated the effect of storage at 2 to 8°C on the stability of human genomic and human papillomavirus (HPV) DNA stored in BD SurePath and Hologic PreservCyt liquid-based cytology media. DNA retained the ability to be extracted and PCR amplified for more than 2.5 years in both medium types. Prior inability to detect DNA in archived specimens may have been due to failure of the extraction method to isolate DNA from fixed cells.
TEXT
Liquid-based cytology (LBC) media (BD SurePath [Becton, Dickinson and Company] and Hologic PreservCyt [Hologic Inc.]) are routinely used to prepare liquid Pap preparations for cervical cancer screening. In the United States, molecular human papillomavirus (HPV) testing out of the LBC vial is recommended (i) as a triage test for determining whether a woman with atypical squamous cells of undetermined significance (ASC-US) needs immediate colposcopic evaluation and (ii) as an adjunct to cervical cytology, “cotesting,” for cervical cancer screening of women of age 30 years and older. In addition, there is a growing interest in the use of LBC media for primary HPV screening in which positive specimens could be reflexed to cytology, HPV type-specific genotyping, or another triage method, although molecular tests have yet to be approved for this intended use (1, 2). LBC media were originally developed and approved for the preservation and preparation of cells for Pap smear evaluation (3, 4). BD SurePath and Hologic PreservCyt media are both alcohol-based preservatives and are designed to fix cellular and subcellular components and aid in the cytological evaluation of the obtained smears. Fixative solutions can prove problematic for downstream molecular extraction methods because of the potential to inhibit cellular lysis, because of the potential to interfere with the proteolytic enzymes used in this process, or due to cross-linking of nucleic acids and proteins (5, 6). Some prior studies have reported poor recovery of nucleic acids from SurePath and PreservCyt media and inferred that the DNA had degraded over time during storage (7–10). SurePath medium contains a low concentration of formalin, which is known to cross-link nucleic acids and protein (11, 12). Formalin treatment results in reversible cross-linking of proteins to DNA, which can render it refractory to magnetic- or silica-based purification and subsequent downstream nucleic acid biochemistry, such as PCR (13). Cross-linking can be removed using proteinase K digestion and/or heat (14–16).
We have developed a simple, one-step chemical lysis method for use with the BD hrHPV-GT assay that can process 0.5 ml of either LBC specimen without prior cell harvesting or the use of lytic enzymes. The method uses a combination of heat and chemicals to lyse LBC-preserved cells directly in the media. It also efficiently reverses cross-linking effects and allows biologically active DNA to be extracted directly from the resulting lysate (17–19). The objective of the current study was to examine the effect of long-term storage of LBC specimens at 2 to 8°C on the ability to detect both human genomic and HPV target DNA and to determine if there was any degradation of the nucleic acid material over this time period using an extraction method specifically designed for LBC media.
Forty-five BD SurePath-preserved specimens and 45 Hologic PreservCyt-preserved specimens were selected from a database of previously tested samples with the BD hrHPV-GT assay. The samples were originally collected from cytology labs as deidentified residual vial specimens after the liquid-based cytology slides had been prepared. Thus, the samples were originally exposed to ambient storage temperatures (15 to 30°C) in accordance with the respective manufacturers' recommendations during collection, transit to the cytology laboratory, and final transfer of the residual material to long-term storage at 2 to 8°C. Upon receipt, approximately 16% of the SurePath specimens and 31% of the PreservCyt specimens were beyond their recommended storage dating, and at the time of first testing, all samples had expired. The original testing was performed in the fall of 2009, and each specimen tested was positive for at least one HPV type. The second test was performed in the spring of 2012, at which time the mean age of both SurePath and PreservCyt specimens was 2.6 years. All 90 samples were collected from different individual patients but were purposely selected so as to provide a balanced representation of high-risk types detected by the assay for both medium types (Table 1). One SurePath sample was removed from the study due to a labeling error. A subset of samples from the same collection was also analyzed by endpoint PCR for detection of the human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene.
Table 1.
2 by 2 comparison results for primary and repeat testing
| Medium and analytea | nb | No. of specimens with each result |
Total no. with a positive result | No. matched | % agreement | 95% CIc | |||
|---|---|---|---|---|---|---|---|---|---|
| Pos/Pos | Pos/Neg | Neg/Pos | Neg/Neg | ||||||
| PreservCyt | |||||||||
| HPV16 | 45 | 7 | 0 | 1 | 37 | 8 | 7 | 87.50 | 47.35, 99.68 |
| HPV18 | 45 | 1 | 0 | 0 | 44 | 1 | 1 | 100.00 | 5.00, 100.00 |
| HPV31 | 45 | 4 | 0 | 0 | 41 | 4 | 4 | 100.00 | 47.29, 100.00 |
| HPV39_68_35 | 45 | 9 | 1 | 1 | 34 | 11 | 9 | 81.82 | 48.22, 97.72 |
| HPV45 | 45 | 1 | 0 | 0 | 44 | 1 | 1 | 100.00 | 5.00, 100.00 |
| HPV51 | 45 | 5 | 0 | 0 | 40 | 5 | 5 | 100.00 | 54.93, 100.00 |
| HPV52 | 45 | 10 | 1 | 0 | 34 | 11 | 10 | 90.91 | 58.72, 99.77 |
| HPV59_56_66_33_58 | 45 | 17 | 1 | 1 | 26 | 19 | 17 | 89.47 | 66.86, 98.70 |
| G1.HBB | 45 | 45 | 0 | 0 | 0 | 45 | 45 | 100.00 | 93.56, 100.00 |
| G2.HBB | 45 | 45 | 0 | 0 | 0 | 45 | 45 | 100.00 | 93.56, 100.00 |
| G3.HBB | 45 | 45 | 0 | 0 | 0 | 45 | 45 | 100.00 | 93.56, 100.00 |
| HPV total | 360 | 54 | 3 | 3 | 300 | 60 | 54 | 90.00 | 79.49, 96.24 |
| HBB total | 135 | 135 | 0 | 0 | 0 | 135 | 135 | 100.00 | 97.81, 100.00 |
| Grand total | 495 | 189 | 3 | 3 | 300 | 195 | 189 | 96.92 | 93.42, 98.86 |
| SurePath | |||||||||
| HPV16 | 44 | 8 | 2 | 0 | 34 | 10 | 8 | 80.00 | 44.39, 97.48 |
| HPV18 | 44 | 2 | 0 | 0 | 42 | 2 | 2 | 100.00 | 22.36, 100.00 |
| HPV31 | 44 | 4 | 0 | 0 | 40 | 4 | 4 | 100.00 | 47.29, 100.00 |
| HPV39_68_35 | 44 | 9 | 0 | 0 | 35 | 9 | 9 | 100.00 | 71.69, 100.00 |
| HPV45 | 44 | 3 | 1 | 0 | 40 | 4 | 3 | 75.00 | 19.41, 99.37 |
| HPV51 | 44 | 2 | 1 | 1 | 40 | 4 | 2 | 50.00 | 6.76, 93.24 |
| HPV52 | 44 | 4 | 3 | 1 | 36 | 8 | 4 | 50.00 | 15.70, 84.30 |
| HPV59_56_66_33_58 | 44 | 12 | 1 | 2 | 29 | 15 | 12 | 80.00 | 51.91, 95.67 |
| G1.HBB | 44 | 44 | 0 | 0 | 0 | 44 | 44 | 100.00 | 93.42, 100.00 |
| G2.HBB | 44 | 44 | 0 | 0 | 0 | 44 | 44 | 100.00 | 93.42, 100.00 |
| G3.HBB | 44 | 44 | 0 | 0 | 0 | 44 | 44 | 100.00 | 93.42, 100.00 |
| HPV total | 352 | 44 | 8 | 4 | 296 | 56 | 44 | 78.57 | 65.56, 88.41 |
| HBB total | 132 | 132 | 0 | 0 | 0 | 132 | 132 | 100.00 | 97.76, 100.00 |
| Grand total | 484 | 176 | 8 | 4 | 296 | 188 | 176 | 93.62 | 89.12, 96.66 |
HBB, human beta-globin internal control, detected in each of the three assay wells, G1 to G3.
Number of analytes tested in primary and repeat tests.
95% CI, 95% confidence interval.
HPV testing was performed as described previously (18). DNA amplification and detection were performed simultaneously using real-time PCR detection, and the signal output from each of the four optical channels was read independently in each of the three assay wells using standard fluorescent dyes. HPV-positive specimens were identified using a cycle threshold (CT) algorithm method. Identical primer and probe sequences were used for both the initial and second tests except for a single base replacement in the reverse primer for HPV type 31 (HPV31). The optical channels (dyes) on which HPV type 59 and the paired types HPV33_58 are detected were also swapped between the first (2009; assay version 1.0) and second (2012; assay version 2.0) tests. The results from both of these channels (which include HPV types 33_58, 56_66, and 59) were combined in the data analysis to account for this change. The human beta-globin (HBB) gene was detected on one channel as an internal sample processing control. Confirmatory endpoint PCR was performed using a human GAPDH primer set (G3PDH For/Rev primers; Integrated DNA Technologies Inc.) to amplify a 452-bp fragment using a Stratagene Mx3005P thermocycler (Agilent Technologies) with a 15-min 95°C enzyme activation step followed by 40 cycles of 96°C for 0.5 min and 61°C for 1 min.
Cycle threshold scores for both the beta-globin and HPV targets were analyzed using Minitab statistical software (Minitab Inc.). Clinical positives were identified using a previously established cutoff for CIN2+ histology endpoints (17–19). Analysis of variance was examined using a 2-proportion test and analysis of variance (ANOVA). Comparing the first and second tests and applying the clinical cutoff, the total agreement for both the beta-globin internal control and HPV results across all samples for all optical channel outputs of the assay was 98.2% (n = 979). There was no statistical difference between the reproducibilities of the BD SurePath and PreservCyt specimens across all possible test results (BD SurePath overall agreement = 97.5% [n = 484]; PreservCyt overall agreement = 98.8% [n = 495]; P = 0.141). The agreement for the beta-globin internal control (tested in triplicate [3-well assay] for each of the two specimen runs) was 100% for both sample types. The positive agreement for all HPV genotypes (both single and multiple infections) was 78.57% for BD SurePath and 90.0% for PreservCyt (Table 1). This difference was not statistically significant (P = 0.089). Analysis of the individual discordant results in Table 1 suggests that this was likely due to sampling variation, with all of the samples remaining analytically positive with CT scores close to the clinical cutoff. There were equal numbers (3) of positive/negative and negative/positive results among the PreservCyt specimens and 8 positive/negative and 4 negative/positive results among the SurePath specimens, suggesting a CT score distribution on either side of the clinical cutoff. All of the discordant CT scores were close to the clinical cutoff and further suggest that even weakly positive samples were not adversely affected by long-term storage and can fall on either side of the clinical cutoff when retested. This is not unexpected since LBC is known to be a heterogeneous specimen type due to the exfoliation (scraping) method used to collect the cells and the fixing of cell clumps by the LBC fixative solution, which, in particular, can lead to variation in the number of diseased (infected) cells from one sampling to another (20, 21). It is also noteworthy that there was 100% detection of the human beta-globin target in both tests, since this target is present in both diseased and normal tissue. Looking at the data set as a whole, the average CT scores (including discordant samples) from the first and second tests across all of the assay channels show a high degree of concordance (Fig. 1).
Fig 1.
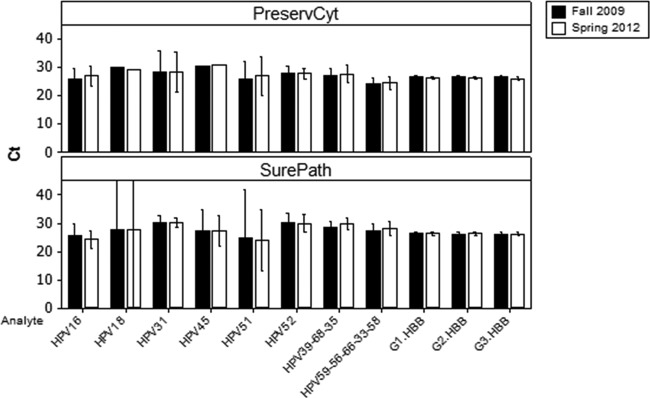
Clinical positive specimen results for PreservCyt- and SurePath-stored specimens. Bar charts represent the average CT scores for the HPV types and the beta-globin (HBB) internal control in each of the three assay wells (G1, G2, G3), tested in the fall of 2009 and the spring of 2012. Results from individual patients were combined by HPV type for both SurePath and PreservCyt media. Bars represent 95% confidence intervals. The large confidence intervals observed for the SurePath HPV18 and HPV51 specimens are due to the low sample number (Table 1).
The DNA fragments detected in the current study were small (75 to 137 bp). In order to determine if only smaller fragments were able to be amplified due to DNA degradation during storage, we employed primers to the human GAPDH gene that amplify a fragment 452 bp in length. This amplicon length also represents the upper size limit of fragments routinely used to detect HPV. The products of an endpoint PCR were readily visualized on an agarose gel using DNA extracted from >4-year-old BD SurePath and >5-year-old Hologic PreservCyt specimens (Fig. 2). In addition, we were routinely able to amplify a 3.5-kb HPV fragment from both fresh and aged (>3-year-old) BD SurePath and Hologic PreservCyt specimens using a commercially available extraction kit (NucleoSpin Tissue Kit, Clontech Laboratories Inc.) which utilizes a high-heat and proteinase K extraction method (data not shown). This suggests that there is no significant DNA degradation of fragments of this size over this time period.
Fig 2.
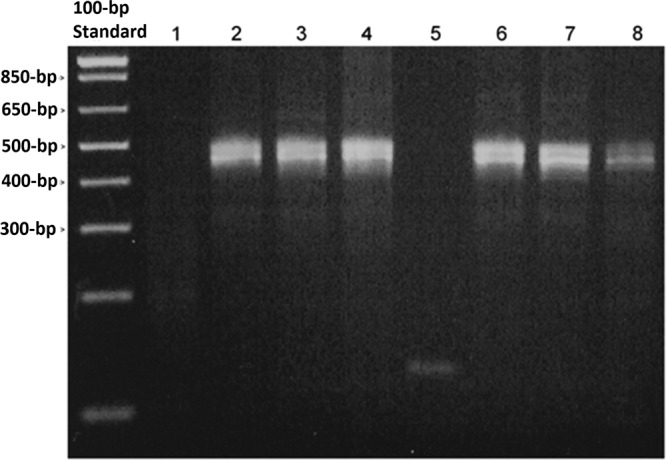
PCR amplification from aged LBC specimens using human GAPDH primers. Ethidium bromide-stained agarose gel. Lane 1, SurePath medium; lanes 2 to 4, >4-year-old residual SurePath specimens; lane 5, PreservCyt medium; lanes 6 to 8, >5-year-old residual PreservCyt specimens. Note that the amplification of more than one genomic product of approximately 452 bp is expected due to the presence of GAPDH pseudogene copies in the human genome.
The BD hrHPV-GT assay provided nearly identical results when repeat aliquots were tested approximately 2.5 years apart from both BD SurePath and Hologic PreservCyt specimens stored at 2 to 8°C. Both human genomic and HPV viral DNA targets were stable over this time period. The long-term stability of human genomic and HPV DNA in PreservCyt medium has been previously studied by Castle et al. (7, 10). These authors investigated the ability to detect HPV DNA (by hybrid capture 2 detection; Qiagen) and human beta-globin DNA (by endpoint PCR and agarose gel electrophoresis of DNA fragments of 268, 610, and 1,327 bp) in PreservCyt specimens stored at ambient temperature over an 8-year period. Hybrid capture 2 detection was found to be unaffected by storage over this period whereas detection of DNA by PCR was inconsistent, and approximately 15% of all specimens were not positive for any size of PCR fragment after 5 years. Larger fragments were also less likely to be detected over time. This is in contrast to the findings in this study in which a wide variety of HPV targets were consistently detected in 2- to 3-year-old specimens and a 452-bp fragment was recovered from >5-year-old PreservCyt specimens (Fig. 1 and 2). Negri et al. also studied the effects of long-term storage at ambient temperature of PreservCyt specimens on the hybrid capture 2 assay signal and found that, while the samples remained positive, there was an almost-5-fold reduction in relative light units (RLUs) over a mean time period of 31.3 months (9). Both of the above-cited studies concluded that there was a time-dependent degradation of DNA over the period of the study. Another possibility, not considered by the authors, is that the DNA became more refractory to extraction over time as a result of the long-term storage in the fixative solution. Long-term storage in alcohol-containing fixative will likely result in progressive dehydration of the cells and reduce the solubility of proteins, which may render them more difficult to lyse and/or reduce access to the DNA during extraction. In addition, cross-linking of DNA can occur in fixative solutions, such as SurePath, which contains a low concentration of formalin. The effect of formalin on DNA and protein has been widely studied and is also frequently exploited as a tool to study protein-DNA interactions (12, 22, 23). Formalin-induced cross-linking can be reversed by using heat, proteinase K, or a combination of both (5, 14, 15). The BD hrHPV-GT assay uses a high-heat extraction step with a proprietary buffer that efficiently lyses cells and reverses cross-linking. It has also been shown to be effective on formalin-fixed paraffin-embedded (FFPE) tissue samples (F. Castro, J. Koshiol, M. Gillison, W. Quint, L. Vaughan, C. Wheeler, and N. Wentzensen, unpublished data). There have been a number of other reports of RNA and DNA “degradation” in samples stored in LBC media (7–9). However, none of the studies provided any direct visualization of the degraded nucleic acid and simply inferred the degradation due to the absence of or reduction in assay signal or failure to recover the material. Cross-linking of nucleic acid to protein is known to result in DNA-protein complexes that disrupt normal DNA function and inhibit its ability to be bound using standard silica- or magnetic-based purification methods (13, 24). The data presented here and elsewhere support the view that DNA can be efficiently recovered from both LBC- and FFPE-preserved specimens (5, 15, 25). Similar findings have also been reported for RNA extraction methods in which high-quality RNA has been isolated from LBC and FFPE specimens following high temperature and/or proteinase K digestion prior to the RNA purification step (25, 26). Prior reports of variable success in RNA and DNA purification from BD SurePath- and Hologic PreservCyt-preserved specimens over shorter storage periods may also be explained by the ability of the extraction method to reverse fixative-induced cross-linking or other deleterious effects on the cells and their nucleic acid that render it refractory to lysis and extraction. Extraction methods that utilize high temperature and/or proteinase K digestion for extended periods reported higher success rates in DNA detection and the ability to detect longer fragments (5, 16). This suggests that prior reports of nucleic acid degradation following storage in LBC media need to be reevaluated and may have been limited by the extraction and amplification methods used rather than degradation of the target nucleic acid. We cannot exclude the possibility that the ambient storage temperature used by prior reports versus the refrigerated storage temperature used here accounts for the observed differences in stability. However, we consider this unlikely given that the samples in this study also experienced room temperature storage and almost half of them had aged beyond their recommended expiration date prior to initial refrigerated storage and testing in our laboratory. FFPE specimens are also routinely stored at room temperature for extended periods, and nucleic acids can be successfully isolated from these specimens using optimized extraction procedures (16, 26).
In summary, the results of this study suggest that both BD SurePath- and Hologic PreservCyt-preserved cells are stable for extended periods when stored at 2 to 8°C and that human genomic and viral DNAs are stably preserved and can be reliably detected using PCR amplification using the BD hrHPV-GT assay extraction and purification procedure. Further studies are required to understand the effects of long-term ambient temperature storage on assay performance. These results have important implications for nucleic acid testing of LBC specimens from bio-bank collections because access to archived material is critical for epidemiological and other long-term retrospective studies (27).
ACKNOWLEDGMENTS
This article is dedicated to the memory of Jeff Peck (14 May 1986 to 11 January 2011).
All of the contributing authors are employees of Becton, Dickinson and Company, and this work was funded by Becton Dickinson.
Footnotes
Published ahead of print 15 May 2013
REFERENCES
- 1. Saslow D, Solomon D, Lawson HW, Killackey M, Kulasingam SL, Cain J, Garcia FA, Moriarty AT, Waxman AG, Wilbur DC, Wentzensen N, Downs LS, Jr, Spitzer M, Moscicki AB, Franco EL, Stoler MH, Schiffman M, Castle PE, Myers ER, American Cancer Society, American Society for Colposcopy and Cervical Pathology, American Society for Clinical Pathology 2012. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. Am. J. Clin. Pathol. 137:516–542 [DOI] [PubMed] [Google Scholar]
- 2. Rijkaart DC, Berkhof J, van Kemenade FJ, Coupe VM, Hesselink AT, Rozendaal L, Heideman DA, Verheijen RH, Bulk S, Verweij WM, Snijders PJ, Meijer CJ. 2012. Evaluation of 14 triage strategies for HPV DNA-positive women in population-based cervical screening. Int. J. Cancer 130:602–610 [DOI] [PubMed] [Google Scholar]
- 3. Becton Dickinson 2011. BD SurePath collection product insert (for use with the PREPSTAIN system). REF 490522. Becton Dickinson, Franklin Lakes, NJ [Google Scholar]
- 4. Hologic Inc 2011. The ThinPrep® 2000 system instructions for use. MAN-02060-002 Rev. 001. Hologic Inc, Bedford, MA [Google Scholar]
- 5. Steinau M, Patel SS, Unger ER. 2011. Efficient DNA extraction for HPV genotyping in formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 13:377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keegan H, Boland C, Malkin A, Griffin M, Ryan F, Lambkin H. 2005. Comparison of DNA extraction from cervical cells collected in PreservCyt solution for the amplification of Chlamydia trachomatis. Cytopathology 16:82–87 [DOI] [PubMed] [Google Scholar]
- 7. Castle PE, Solomon D, Hildesheim A, Herrero R, Bratti MC, Sherman ME, Rodriguez AC, Alfaro M, Hutchinson ML, Dunn ST, Kuypers J, Schiffman M. 2003. Stability of archived liquid-based cervical cytologic specimens. Cancer 99:89–96 [DOI] [PubMed] [Google Scholar]
- 8. Powell N, Smith K, Fiander A. 2006. Recovery of human papillomavirus nucleic acids from liquid-based cytology media. J. Virol. Methods 137:58–62 [DOI] [PubMed] [Google Scholar]
- 9. Negri G, Rigo B, Vittadello F, Egarter-Vigl E, Mian C. 2004. Human papillomavirus typing with hybrid capture II on archived liquid-based cytologic specimens: is HPV typing always reproducible? Am. J. Clin. Pathol. 122:90–93 [DOI] [PubMed] [Google Scholar]
- 10. Castle PE, Hildesheim A, Schiffman M, Gaydos CA, Cullen A, Herrero R, Bratti MC, Freer E. 2003. Stability of archived liquid-based cytologic specimens. Cancer 99:320–322 [DOI] [PubMed] [Google Scholar]
- 11. Becton Dickinson 2011. BD SurePath™ preservative fluid material safety data sheet. Catalog number 490527. Becton Dickinson, Franklin Lakes, NJ [Google Scholar]
- 12. Dryer RL, Covey LR. 2006. Use of chromatin immunoprecipitation (ChIP) to detect transcription factor binding to highly homologous promoters in chromatin isolated from unstimulated and activated primary human B cells. Biol. Proced. Online 8:44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koon HE, Loreille OM, Covington AD, Christensen AF, Parsons TJ, Collins MJ. 2008. Diagnosing post-mortem treatments which inhibit DNA amplification from US MIAs buried at the Punchbowl. Forensic Sci. Int. 178:171–177 [DOI] [PubMed] [Google Scholar]
- 14. Campos PF, Gilbert TM. 2012. DNA extraction from formalin-fixed material. Methods Mol. Biol. 840:81–85 [DOI] [PubMed] [Google Scholar]
- 15. Duval K, Aubin RA, Elliott J, Gorn-Hondermann I, Birnboim HC, Jonker D, Fourney RM, Fregeau CJ. 2010. Optimized manual and automated recovery of amplifiable DNA from tissues preserved in buffered formalin and alcohol-based fixative. Forensic Sci. Int. Genet. 4:80–88 [DOI] [PubMed] [Google Scholar]
- 16. Kotorashvili A, Ramnauth A, Liu C, Lin J, Ye K, Kim R, Hazan R, Rohan T, Fineberg S, Loudig O. 2012. Effective DNA/RNA co-extraction for analysis of microRNAs, mRNAs, and genomic DNA from formalin-fixed paraffin-embedded specimens. PLoS One 7:e34683. 10.1371/journal.pone.0034683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cuzick J, Cadman L, Mesher D, Austin J, Ashdown-Barr L, Ho L, Terry G, Liddle S, Wright C, Lyons D, Szarewski A. 2013. Comparing the performance of six human papillomavirus tests in a screening population. Br. J. Cancer 108:908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castle PE, Gutierrez EC, Leitch SV, Maus CE, McMillian RA, Nussbaumer WA, Vaughan LM, Wheeler CM, Gravitt PE, Schiffman M. 2011. Evaluation of a new DNA test for detection of carcinogenic human papillomavirus. J. Clin. Microbiol. 49:3029–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Szarewski A, Mesher D, Cadman L, Austin J, Ashdown-Barr L, Ho L, Terry G, Liddle S, Young M, Stoler M, McCarthy J, Wright C, Bergeron C, Soutter WP, Lyons D, Cuzick J. 2012. Comparison of seven tests for high-grade cervical intraepithelial neoplasia in women with abnormal smears: the Predictors 2 study. J. Clin. Microbiol. 50:1867–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siebers AG, van der Laak JA, Huberts-Manders R, Vedder JE, Bulten J. 19 June 2012. Accurate assessment of cell density in low cellular liquid-based cervical cytology. Cytopathology 10.1111/j.1365-2303.2012.00990.x [DOI] [PubMed] [Google Scholar]
- 21. Chen G, Kobayashi L, Nazarenko I. 2007. Effect of sample aliquot size on the limit of detection and reproducibility of clinical assays. Clin. Chem. 53:1962–1965 [DOI] [PubMed] [Google Scholar]
- 22. Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, Ball LM, Swenberg JA. 2010. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 132:3388–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mostegl MM, Richter B, Dinhopl N, Weissenbock H. 2011. Influence of prolonged formalin fixation of tissue samples on the sensitivity of chromogenic in situ hybridization. J. Vet. Diagn. Invest. 23:1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nocker A, Camper AK. 2006. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl. Environ. Microbiol. 72:1997–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tarkowski TA, Rajeevan MS, Lee DR, Unger ER. 2001. Improved detection of viral RNA isolated from liquid-based cytology samples. Mol. Diagn. 6:125–130 [DOI] [PubMed] [Google Scholar]
- 26. Joseph A, Gnanapragasam VJ. 2011. Laser-capture microdissection and transcriptional profiling in archival FFPE tissue in prostate cancer. Methods Mol. Biol. 755:291–300 [DOI] [PubMed] [Google Scholar]
- 27. Arbyn M, Andersson K, Bergeron C, Bogers JP, von Knebel-Doebertitz M, Dillner J. 2011. Cervical cytology biobanks as a resource for molecular epidemiology. Methods Mol. Biol. 675:279–298 [DOI] [PubMed] [Google Scholar]


