Abstract
Chlamydia pecorum is a significant pathogen of domestic livestock and wildlife. We have developed a C. pecorum-specific multilocus sequence analysis (MLSA) scheme to examine the genetic diversity of and relationships between Australian sheep, cattle, and koala isolates. An MLSA of seven concatenated housekeeping gene fragments was performed using 35 isolates, including 18 livestock isolates (11 Australian sheep, one Australian cow, and six U.S. livestock isolates) and 17 Australian koala isolates. Phylogenetic analyses showed that the koala isolates formed a distinct clade, with limited clustering with C. pecorum isolates from Australian sheep. We identified 11 MLSA sequence types (STs) among Australian C. pecorum isolates, 10 of them novel, with koala and sheep sharing at least one identical ST (designated ST2013Aa). ST23, previously identified in global C. pecorum livestock isolates, was observed here in a subset of Australian bovine and sheep isolates. Most notably, ST23 was found in association with multiple disease states and hosts, providing insights into the transmission of this pathogen between livestock hosts. The complexity of the epidemiology of this disease was further highlighted by the observation that at least two examples of sheep were infected with different C. pecorum STs in the eyes and gastrointestinal tract. We have demonstrated the feasibility of our MLSA scheme for understanding the host relationship that exists between Australian C. pecorum strains and provide the first molecular epidemiological data on infections in Australian livestock hosts.
INTRODUCTION
The obligate intracellular bacterium Chlamydia pecorum is a globally recognized pathogen of livestock and wildlife (1, 2). The best-studied host for C. pecorum infections is the koala (Phascolarctos cinereus) (3), a native Australian arboreal marsupial. Within this host, C. pecorum infections are prevalent and cause acute or chronic keratoconjunctivitis, often leading to blindness (4), and genitourinary tract infections that may lead to infertility (5, 6).
C. pecorum also causes a range of clinically important diseases in economically significant livestock species (cattle, sheep, goats, and pigs) manifesting as encephalomyelitis (7), reduced fertility (8), vaginitis and endometritis, enteric infections, mastitis (9), pneumonia, conjunctivitis, and arthritis (stiff-lamb disease) (10, 11). C. pecorum infections in cattle can be subclinical and asymptomatic but nevertheless exert chronic pathological effects on animal health not seen in herds without infection (12, 13). In affected sheep flocks, C. pecorum infections lead to polyarthritis and conjunctivitis and spread rapidly, leading to increased morbidity and mortality (14). There are also reports implicating C. pecorum in ovine abortions (10). Despite the high prevalence of C. pecorum infections in livestock, there is a lack of information about the genetic diversity of these strains in relation to the anatomical sites that are infected and the diseases observed (15). Virtually nothing is known about the prevalence and epidemiology of infections in Australian livestock, despite the fact that cases of chlamydiosis in sheep and cattle are regularly reported in agriculturally productive areas of central New South Wales (NSW) (14, 16, 17).
The origin of C. pecorum infections in Australian animals is also unclear and of concern, given the potential for C. pecorum “spillover” or “spillback” risk between infected livestock and/or wildlife infections, respectively (18, 19). Soon after C. pecorum was described in the koala (20, 21), Jackson et al. (22) analyzed sequences of the C. pecorum ompA gene and found that (i) koala C. pecorum ompA sequences are highly polymorphic and (ii) some koala C. pecorum ompA sequences cluster more closely to sequences from Australian livestock than to those from other koalas. These earlier observations led to the suggestion that koala C. pecorum infections may have originated from a “recent” cross-host transmission event, possibly from livestock that were introduced to Australia following European settlement in 1788 (3).
The recent completion of the first C. pecorum genome sequence (23) has allowed us to revisit the relationship between C. pecorum infections in koalas and Australian livestock. Using novel molecular markers (tarP in addition to open reading frame 663 [ORF663], incA, and ompA), the findings of Marsh et al. (24) strengthened previous observations that koala C. pecorum strains are genetically diverse (22). In contrast to the previous study, however, the latter study showed that koala C. pecorum strains were phylogenetically distinct from C. pecorum livestock strains (24), although the study was limited by a lack of Australian livestock samples. Use of these polymorphic genes in a previous study, alone or in addition to housekeeping genes as targets of a multivirulence locus-typing scheme (MVLST), allowed for the discrimination of C. pecorum strains from livestock that were isolated from diseased and healthy animals (25). Despite these reports, significant evidence is accumulating to suggest that ompA, which encodes a highly immunogenic surface-exposed chlamydial major outer membrane protein (MOMP), is an unreliable marker for tracing the origin and relationships of other Chlamydia species (26, 27). Due to the numerous schemes presented for the typing of C. pecorum, the need for the establishment of a standardized global epidemiological tracking tool has been evident.
A number of studies have reported the use of multilocus sequence typing (MLST) (28) and multilocus sequence analysis (MLSA) to elucidate intraspecies relationships between members of the genus Chlamydia (29, 30), including analyses of strains from the same species that infect multiple animal hosts (31). These methods utilize the concatenation of four to 12 evolutionarily conserved housekeeping (HK) gene sequence fragments. HK genes are broadly distributed around the chromosome and encode conserved proteins that are not considered to be under diversifying selection (32). Phylogenies based on these loci have been useful in differentiating bacterial pathogens (33–35), including bacteria that are highly recombinogenic and undergo frequent lateral gene transfer (36, 37).
The primary objective of the current study was to develop and apply an MLSA scheme to examine the overall genetic diversity of and relationships between Australian C. pecorum isolates from different geographical origins and hosts. In doing so, we provide the first molecular epidemiological data on this important animal pathogen infecting Australian livestock.
MATERIALS AND METHODS
Chlamydial isolates.
The complete list of isolates from the sequences included for analysis are described in Table S1 in the supplemental material. Five koala isolates from wild koala populations in southeast Queensland (SEQLD), Australia, were previously isolated and described by Marsh and colleagues (24). HK gene fragment sequences for three koala C. pecorum isolates from SEQLD and the sheep C. pecorum polyarthritis type strain IPA were extracted from their draft genomes (A. Polkinghorne, unpublished data). HK gene fragment sequences from other U.S. sporadic bovine encephalomyelitis (SBE), arthritis, conjunctivitis, and healthy fecal C. pecorum isolates, also listed in Table S1, were obtained from the Chlamydiales MLST website (http://pubmlst.org/chlamydiales/) (38). Permission to use these sequences was granted to us by the curator, Yvonne Pannekoek, University of Amsterdam, The Netherlands.
C. pecorum-specific PCR screening of Australian livestock and wild koala samples.
A total of 77 swabs were collected from 40 sheep from various flocks across central New South Wales (NSW), Australia. Swab samples were collected from the eyes, rectum, and vagina by the district veterinarians of the Livestock Health and Pest Authority (LHPA) throughout NSW, Australia, as a part of routine diagnostic testing (see Table S2 in the supplemental material). Two additional sheep with polyarthritis were sacrificed as a part of routine diagnostic testing. Fluid from diseased joints was extracted using a syringe, and swabs were dipped into the synovial fluid prior to processing. DNA extracted from a cultured C. pecorum isolate, originating from a central NSW bovine SBE case, was also used in this study. Additional swab samples were collected from wild Australian koalas from QLD, NSW, Victoria (VIC), and South Australia (SA) as part of ongoing field investigations by veterinary collaborators or for routine diagnostic testing following the presentation of sick animals to various wildlife hospitals. The testing of these swab samples has been considered by the Queensland University of Technology (QUT) Animal Ethics Committee and was approved as tissue use notification number 1100000718.
In a preliminary study to understand the relationships between these C. pecorum isolates, we selected samples from the same animal but different anatomical sites, different individuals from the same population, and/or sympatric populations for the three hosts. The abbreviated names of these C. pecorum-positive samples follow the pattern of the geographical location of the sample/name of animal/site of infection (e.g., Eugowra/Sheep1/Ocular = Eug/Ovi1/Eye). The samples included (i) 11 ovine C. pecorum PCR-positive samples (Eug/Ovi1/Eye, Eug/Ovi1/Rec, For/Ovi5/Eye, Dub/Ovi3/Rec, Mer/Ovi1/Jnt, Mer/Ovi2/Jnt, Nyn/Ovi1/Eye, Nyn/Ovi1/Rec, Nyn/Ovi2/Eye, Nyn/Ovi3/Eye, and Nyn/Ovi4/Eye) from the collection of PCR-positive Australian livestock samples, (ii) C. pecorum PCR-positive swab samples from QUT's extensive collection of Australian koala samples, and (iii) a cell-cultured Australian cow SBE isolate. A list of C. pecorum-positive clinical samples from Australian livestock and 17 koalas detected in this study is included in Table S1 in the supplemental material.
Animal swabs to be screened for the presence of C. pecorum DNA were processed by vortexing and centrifugation (39). For all swabs, DNA was extracted using a QIAamp DNA kit (Qiagen, Doncaster, Victoria, Australia), according to the manufacturer's instructions. DNA purity and yield were determined using a NanoDrop spectrophotometer ND-1000 (Thermo Fisher Scientific, Inc.). Extracted DNA was used as the template for a C. pecorum-specific quantitative real-time PCR for the detection and measurement of the infectious load, targeting a 202-bp region of the C. pecorum 16S rRNA (39). All quantitative PCRs (qPCRs) were performed as previously described (39) on a Rotor-Gene Q (Qiagen). Negative (distilled water [dH2O]) and positive (C. pecorum strain MC/Marsbar) controls were included in each amplification assay.
C. pecorum genes and primers used for MLSA.
The complete list of target genes and primers used in the current MLSA study is found in Table 1. Fragments of seven HK genes (enoA, oppA_3, gidA, hemN, hflX, fumC, and gatA) previously used for the typing of several species in the Chlamydiae phylum by MLST (30) were targeted for MLSA as a part of this study. Initially, we were only able to successfully amplify two HK genes (gatA and enoA) using modified pan-Chlamydiales PCR primers (30) on purified C. pecorum genomic DNA; therefore, we designed new C. pecorum-specific PCR primer pairs for the five remaining HK genes (Table 1). For the genes oppA and fumC, new primer pairs were designed using a combination of an existing modified degenerate primer (30) and a new primer. For the genes hflX, gidA, and hemN, new pairs of primers were used, which were designed based on the C. pecorum E58 bovine genome sequence (23). Using purified C. pecorum genomic DNA as a template, conventional PCR was successfully used to amplify a fragment of each of the HK genes of interest. Additionally, the primers were tested against genomic DNA samples extracted from cultured Australian avian Chlamydia psittaci and koala Chlamydia pneumoniae isolates (data not shown). These C. pecorum-specific HK primers were subsequently used for MLSA of Australian C. pecorum livestock and koala strains.
Table 1.
Genes and primers used in this study
| HK gene | Annotation | Locus tag | Position in the C. pecorum E58 genomea | Primer | Sequence (5′ → 3′) | Annealing temp (°C) | Amplicon size (bp) | Size of sequence analyzed (bp) | Reference or source |
|---|---|---|---|---|---|---|---|---|---|
| gatA | Glutamyl-tRNA amidotransferase subunit A | G5S_0628 | 629005–629429 | MJgatA1F | GCTTTAGAGTTGAGAGAAGCT | 54 | 512 | 425 | 30 |
| MJgatA1R | GATCCTCCTGTATCTGATCC | 54 | 512 | 425 | 30 | ||||
| oppA_3 | Oligonucleotide-binding protein | G5S_0967 | 952191–952671 | MJoppA1F | ATGTGCAAGATCCCAGTGGG | 58 | 605 | 483 | 30 |
| MJoppA1R | GGCGCTACTTGTTATGGG | 58 | 605 | 483 | This study | ||||
| hflX | GTP binding protein | G5S_0597 | 579741–580175 | MJhflX1F | TGAGGAGATCTCTGCATCG | 58 | 607 | 435 | This study |
| MJhflX1R | ATCTTCATGCAAAGCAGCC | 58 | 607 | 435 | This study | ||||
| gidA | Glucose-inhibited division protein A | G5S_0430 | 429868–429395 | MJgidA1F | GCGTCACAACAAAAGAAGGC | 60 | 560 | 474 | This study |
| MJgidA1R | TGACGCTGTATATCACACGG | 60 | 560 | 474 | This study | ||||
| enoA | Enolase | G5S_0242 | 254779–255159 | MJenoA1F | CCTATGATGAACCTTATCAATGG | 58 | 431 | 381 | 30 |
| MJenoA1R | TCTTCCTCCGCTAAGCCATCC | 58 | 431 | 381 | 30 | ||||
| hemN | Oxygen-independent coproporphyrinogen III oxidase | G5S_0144 | 150138–150569 | MJhemN1F | GATCGCGATAGAGATAGACCC | 54 | 634 | 432 | This study |
| MJhemN1R | ATCTTCTCCTGATAGATATCG | 54 | 634 | 432 | This study | ||||
| fumC | Fumarate hydratase class II | G5S_0015 | 18169–17705 | MJfumC1F | TGATTAAGAAATGTGCAGC | 54 | 572 | 465 | This study |
| MJfumC1R | CCTTCAGGTACATTAAGCC | 54 | 572 | 465 | 30 |
GenBank accession number CP002608.
PCR and sequencing.
PCRs for all target gene fragments were prepared to a total reaction mixture volume of 50 μl, including 1× AmpliTaq Gold 360 master mix (Life Technologies, Victoria, Australia), 0.3 μM each forward and reverse primer (Sigma-Aldrich, New South Wales, Australia), and 3 μl DNA template, of an average concentration of 25 ng/μl. All PCRs were performed in an S1000 thermal cycler (Bio-Rad, Singapore). Negative (dH2O) and positive (C. pecorum strain MC/Marsbar) controls were included in each amplification assay. The cycling conditions for all HK genes included an initial denaturation (10 min at 95°C) followed by 40 cycles of denaturation (30 s at 95°C), annealing (30 s at 54°C for fumC, gatA, and hemN; 30 s at 58°C for enoA, hflX, and oppA, and 30 s at 60°C for gidA), and extension (1 min at 72°C), followed by a final extension (7 min at 72°C). Upon amplification, PCR products were detected on a 2% ethidium bromide agarose gel and visualized under an UV transilluminator and purified using a High Pure PCR product purification kit (Roche, New South Wales, Australia). Each PCR product was directly sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Life Technologies, Victoria, Australia) and subsequently was purified according to the manufacturer's instructions. Sequencing was performed at the QUT DNA sequencing facilities using the Applied Biosystems ABI3500 gene analyzer.
Sequence and phylogenetic analysis.
Forward and reverse chromatograms of each sequenced gene were aligned in the Geneious Pro 6.0.4 software package, and a consensus sequence was obtained and trimmed to an appropriate size to correspond with established MLST gene sequence fragments. Concatenation of the seven HK gene fragments was performed in the same order to the established MLST scheme for Chlamydiae (30). Allele numbers for our Australian livestock and koala C. pecorum MLSA data sets were identified (see http://pubmlst.org/chlamydiales/) (38).
Sequence and phylogenetic analyses were performed using the Geneious Pro 6.0.4 software package. Sequences of individual genes and concatenated gene sets were aligned using ClustalW (40). DnaSP 5.0 (41) was used to analyze the level of sequence polymorphisms by determining the number of synonymous (ds) and nonsynonymous (dn) substitutions per site and the average number of nucleotide substitutions per site between the populations (Dxy), with the Jukes-Cantor correction. We also calculated the number of polymorphic (segregating) sites and haplotypes, as well as testing for the minimum number of recombination events (Rm) using the Hudson and Kaplan 1985 algorithm, as implemented in DnaSP 5.0 (41). Best-fit models of nucleotide substitution for constructing phylogenies of our data sets were estimated by considering 11 substitution models using jModelTest v.2.1.1 (42). A phylogenetic tree comprising all C. pecorum strains was constructed based on concatenated MLSA sequences, using the program MrBayes (43) with the HKY85G substitution model, as implemented in Geneious Pro 6.0.4. Run parameters included four Markov chain Monte Carlo (MCMC) chains with a million generations, sampled every 100 generations, and with the first 1,000 trees discarded as burn-in.
Nucleotide sequence accession numbers.
The HK gene sequences from Australian koala, sheep, and cow C. pecorum isolates are available in GenBank (accession numbers KC885978 to KC886180).
RESULTS
C. pecorum in Australian sheep.
In order to assess the presence and prevalence of C. pecorum infections in Australian sheep populations, we screened 77 clinical swabs from a total of 40 sheep with (i) suspected chlamydiosis, (ii) no overt signs of disease, or (iii) presentation of symptoms consistent with other etiological agents from the central NSW region. This analysis revealed 13 C. pecorum-positive sheep from seven out of the nine flocks screened (see Table S2 in the supplemental material). Fifty percent of the PCR-positive swabs were from the eyes of sheep with suspected chlamydial keratoconjunctivitis. In six sheep diagnosed with keratoconjunctivitis from three different flocks (Eugowra, Forbes, and Nyngan), C. pecorum DNA was also detected at the rectal site. Screening of overtly healthy sheep revealed PCR positivity in five animals (38%), including two animals that displayed PCR positivity in ocular swabs, while the remaining three were PCR positive at the rectal sites. Due to the invasive sampling procedures, only two joint samples from sheep with suspected polyarthritis were available for screening in this study, and these were both positive for C. pecorum DNA.
Evaluation and optimization of C. pecorum-specific MLSA PCR assays.
Initially, we were only able to successfully amplify two HK genes (gatA and enoA) using the modified pan-Chlamydiales PCR primers (30) on purified C. pecorum genomic DNA. To resolve these issues, multiple sequence alignments of the seven HK genes from the recently available C. pecorum E58 genome (23), and draft genomes of three koala C. pecorum isolates isolated from SEQLD and the sheep C. pecorum polyarthritis type strain IPA (Polkinghorne, unpublished), as well as other Chlamydia species, were performed, which revealed a number of single nucleotide polymorphisms that may affect the efficiency of PCR amplification using the pan-Chlamydiales degenerate primers. Due to sequence divergence within the Chlamydiaceae family, Zocevic and colleagues (44) were also able to amplify only gatA, enoA, gidA, and hlfX using the pan-Chlamydiales. The newly designed C. pecorum-specific HK gene primers reported here amplified the desired sequences with high efficiency and specificity and were tested against genomic DNA samples extracted from cultured Australian C. psittaci and koala C. pneumoniae isolates (data not shown).
MLSA HK genes are subject to purifying selection.
To understand the relationships and the level of genetic diversity that exists among Australian C. pecorum isolates from livestock and koalas, as well as U.S. strains, an MLSA of concatenated HK gene fragments was performed on 35 isolates, including 18 livestock isolates (11 Australian sheep, one Australian cow, and six U.S. livestock isolates) and 17 Australian koala C. pecorum isolates, detected in a variety of populations across Australia.
Evaluation of the selective pressures on the C. pecorum HK genes was performed by estimating nonsynonymous-to-synonymous-substitution (dn/ds) ratios, where a dn/ds ratio of <1 indicates negative or purifying selection (an excess of synonymous substitutions), a dn/ds ratio of 1 indicates neutral selection, and a dn/ds ratio of >1 indicates positive selection (an excess of nonsynonymous substitutions resulting in an amino acid change) (45). Genetic variability among the sequences at the seven loci of the 35 C. pecorum isolates analyzed in this study was limited (see Table S3 in the supplemental material). The highest number of polymorphic sites and substitutions was limited to three HK genes, gatA, gidA, and enoA. The highest number (n = 3) of nonsynonymous substitutions was observed in gatA. Overall, the number of synonymous substitutions per synonymous site (ds) occurred 3.2 times more than nonsynonymous substitutions per nonsynonymous site (dn). Analysis of the dn/ds ratios, ranging from 0 to 0.237, revealed that all seven loci are under purifying or negative selection, resulting mainly in silent substitutions. Although the level of diversity was limited, we found 26 allelic variants across the seven HK loci, with gatA exhibiting six allelic variants, making it the most diverse locus.
Diversity within livestock and koala C. pecorum populations as assessed by MLSA.
MLSA of Chlamydia HK genes has previously been shown to reflect the level of genetic diversity present across the genome among representative species of Chlamydiae (30, 31, 46). To confirm whether similar observations could be made for C. pecorum, we analyzed the genetic diversity within and between C. pecorum isolates by assessing dn/ds ratios, the number of sequence types (STs), and divergence between different C. pecorum populations based on their respective hosts and/or biogeographical origins.
Koala C. pecorum isolates had the lowest dn/ds ratio among the Australian C. pecorum isolates assessed, with double the number of synonymous substitutions compared to nonsynonymous substitutions (Table 2). Seven previously undescribed and arbitrarily assigned C. pecorum STs (Aa, Ab, Ac, B, C, Ca, and D) were identified in koalas (see Table S4 in the supplemental material). The pool of samples from Australian livestock C. pecorum isolates also had low dn/ds ratios, again with synonymous substitutions in excess. Lesser ST diversity was observed among Australian livestock isolates, with three novel STs (A, E, and F) unique to sheep, one ST (Aa) shared with koalas, and one ST23 previously identified in global C. pecorum livestock isolates (see Table S4 in the supplemental material). Collectively, we have identified 11 STs in Australian C. pecorum isolates, with 10 of them being novel.
Table 2.
C. pecorum population diversity
| C. pecorum population | No. of isolates | Length of sequence (bp) | No. of nonsynonymous substitutions | dna | No. of synonymous substitutions | dsa | dn/ds | No. of sequence types |
|---|---|---|---|---|---|---|---|---|
| Koala | 17 | 3,095 | 3 | 0.00425 | 6 | 0.03271 | 0.130 | 7 |
| Australian livestock | 12 | 3,095 | 3 | 0.00439 | 7 | 0.02413 | 0.182 | 5 |
| All Australia (livestock and koala) | 29 | 3,095 | 5 | 0.00437 | 12 | 0.02793 | 0.156 | 11 |
| U.S. livestock | 6 | 3,095 | 4 | 0.00481 | 10 | 0.04527 | 0.106 | 3 |
| All analyzed | 35 | 3,095 | 6 | 0.00488 | 19 | 0.03051 | 0.160 | 14 |
ds and dn, the average number of synonymous substitutions per synonymous site and nonsynonymous substitutions per nonsynonymous site, respectively (Jukes-Cantor corrected).
Phylogenetic relationships of C. pecorum livestock and marsupial strains assessed by MLSA.
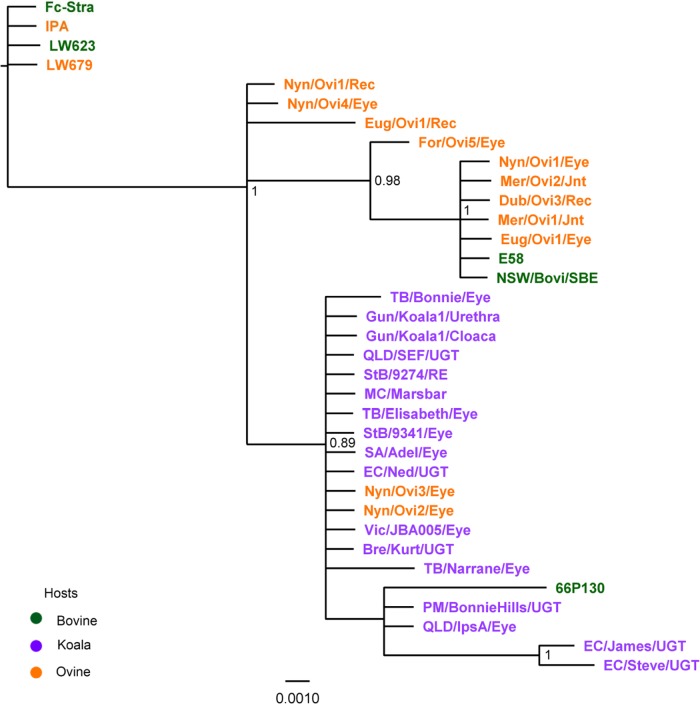
MLSA has been previously used to examine the phylogenetic relationships between closely related species of bacteria (47). In this study, the phylogenetic relationships of each strain were constructed using the aligned concatenated sequences of the seven HK gene fragments for all 35 C. pecorum isolates using Bayesian methods (Fig. 1). Our analysis revealed that C. pecorum isolates separate into three well-supported clades, with an outgroup consisting of four U.S. C. pecorum livestock isolates (Fig. 1). These four isolates had 100% sequence identity among them but differed from the rest of the koala and livestock isolates by an average of 10 nucleotides.
Fig 1.
Bayesian phylogenetic analysis of concatenated sequences of seven HK gene fragments of 35 C. pecorum livestock and koala isolates. Posterior probabilities of >0.85 are displayed on tree nodes. C. pecorum Fc-Stra, IPA, LW623, and LW679 isolates were included as an outgroup. The host origin of each isolate is indicated by the coloring of the isolate name. Rec, rectal; Jnt, joint; RE, right eye; SBE, sporadic bovine encephalomyelitis; UGT, urogenital tract.
A second clade consisted of bovine and ovine C. pecorum isolates from Australia and the United States. Within this clade, we observed a subgroup of six Australian C. pecorum isolates (five sheep and one cow) that have 100% sequence identity to each other, as well as to the U.S. bovine E58 C. pecorum SBE type isolate. This clade also provides several potentially important epidemiological observations regarding the Australian livestock isolates, including the following: (i) isolates found causing SBE in cattle (NSW/Bov/SBE) could also be found in healthy sheep, as well as in association with conjunctivitis or polyarthritis in sheep (Dub/Ovi3/Rec, Eug/Ovi1/Eye, Mer/Ovi1/Jnt), (ii) the same C. pecorum STs could be found circulating within one flock (Mer/Ovi1 and Mer/Ovi2), and (iii) multiple STs could be found within one animal from various anatomical sites (Nyn/Ovi1/Eye and Rec and Eug/Ovi1/Eye and Rec).
All Australian koala C. pecorum isolates grouped together into a third distinct clade. Interestingly, three livestock isolates (U.S. cow 66P130 and Australian sheep Nyn/Ovi2/Eye and Nyn/Ovi3/Eye) also clustered with the koala isolates, sharing 99.8% and 100% sequence similarities, respectively. The identical sheep and koala STs were not sampled from animals in the same region, however. Notably, the latter ovine STs detected on the Nyngan property brought the total of unique STs detected in this single sheep flock to four. As a rule, the diversity of STs detected was more homogenous than that in the previous clade, which contained only C. pecorum isolates from livestock. Nevertheless, an analysis of the diversity of STs within this third clade of koala and livestock C. pecorum isolates showed evidence for (i) C. pecorum STs infecting different anatomical sites within the same koala, as similarly found in STs from the previous livestock clade (e.g., Gun/Koala1/Urethra and Gun/Koala1/Cloaca), (ii) the same STs infecting multiple animals within the same koala population (e.g., StB/9274 and StB/9341), (iii) the presence of multiple C. pecorum STs circulating within a single koala population (e.g., EC/Ned, EC/James, and EC/Steve), and (iv) the presence of a single C. pecorum ST in the koala populations in SA (SA/Adel/Eye), QLD (StB/9341/Eye, Bre/Kurt/UGT, MC/Marsbar, and EC/Ned/UGT), NSW (TB/Elisabeth/Eye), and VIC (Vic/JBA001/Eye).
Divergence between C. pecorum populations assessed in this study.
In our assessment of the phylogenetic relationships between livestock and koala C. pecorum isolates, we observed separation into two distinct clades and a degree of association between the host and isolate. We also assessed the genetic divergence between the koala and livestock C. pecorum populations using these seven concatenated HK gene fragment sequences. The divergence can be assessed by calculating the average number of nucleotide substitutions per site between populations (Dxy) and the number of uniform mutations within specific populations (the fixed differences) and shared polymorphisms between populations.
As observed in Table S5 in the supplemental material, Dxy was very limited and comparable among populations, and there were no fixed differences between populations. Overall, the average number of nucleotide differences between the koala and livestock C. pecorum populations was 7.25, with the highest number of differences observed between U.S. and Australian livestock isolates, which shared 10 polymorphisms. Consistent with phylogenetic observations, the smallest number of differences was noted between Australian koala and livestock, with only two shared polymorphisms.
DISCUSSION
C. pecorum infections continue to cause significant economic losses in livestock, both in Australia and globally (14, 15). C. pecorum infections are also a major contributing factor to the decline of koala populations across Australia (3, 48). Despite this, virtually nothing is known about the epidemiology and genetic diversity of these infections in both livestock and koala hosts. While molecular evidence has previously pointed to a relationship between strains infecting both hosts (24), these relationships had not been subjected to robust phylogenetic analysis using a typing scheme that was demonstrated to reflect the rates of change across the whole chromosome of Chlamydia.
In a pilot study to investigate the overall genetic relationships between C. pecorum isolates infecting koala and livestock hosts, we developed a customized MLSA scheme, based on a previously published MLST scheme for member species of the Chlamydiaceae (30). Sequences of seven genetically stable HK gene fragments were obtained and analyzed from a total of 35 livestock and koala C. pecorum isolates. Although the level of diversity was limited among analyzed C. pecorum strains, we identified 10 novel STs, six found in koala isolates only and three sequence types observed in Australian sheep isolates. For the most part, we observed a distinct phylogenetic separation of koala and livestock isolates, with an observation of only one ST being shared between Australian sheep and koalas.
Based on this analysis and in contrast to previous descriptions of the genetic diversity of this species using the ompA gene (22), C. pecorum appears to harbor limited diversity, at least when HK genes were considered. This observation is supported by low dn/ds ratios across the C. pecorum strains we sampled. No putative recombination events were detected in the seven HK gene fragments, and the number of synonymous substitutions exceeded nonsynonymous substitutions, indicating strong purifying selection. This was expected, as these genes, commonly used for MLST in other bacteria, are widely spaced across the chromosome, are evolutionarily conserved, experience limited or no host immune pressures, and are representative of overall chromosomal change (28, 33). This limited diversity of the C. pecorum species is consistent with the diversity observed in other members of the genus Chlamydia, particularly that observed from MLSA and MLST analysis of the closely related chlamydial species C. pneumoniae (30, 49). The number of synonymous substitutions detected in C. pecorum was comparable to those observed in Chlamydia abortus and C. psittaci (31). Our Australian C. pecorum strains had diversity of 0.4 ST/per strain, which is consistent with observations across the Chlamydiae in general (31, 44).
Koala isolates displayed more diversity of STs, with six unique STs found in this host compared to livestock. This greater ST diversity might reflect the wider geographic range of koala populations sampled here compared to Australian livestock, which were sampled from central NSW only. The greater ST diversity may also reflect the recent diversification of C. pecorum across the geographic range of koalas following its potential introduction from livestock, despite HK genes being under strong purifying selection. Different bacterial populations show different levels of ST diversity per isolate, e.g., 0.92 ST/per strain for Enterococcus faecalis, 0.72 ST/per strain for Neisseria spp., and 0.46 ST/per strain for Staphylococcus aureus, but in general, the values are on the lower end and are comparable to each other (32).
Despite the fact that C. pecorum is a major pathogen of domesticated animals with a worldwide distribution, still little is known about its transmission and the factors associated with C. pecorum infection in these hosts (50). With the establishment of a level of confidence in our C. pecorum MLSA scheme, we then used this scheme to provide insights into the finely detailed molecular epidemiology of C. pecorum infections in Australian livestock and koalas. This analysis of several sheep flocks in central NSW revealed at least four unique STs (see Table S4 in the supplemental material). Among these STs, we identified Australian sheep and cow isolates that were phylogenetically 100% identical to the previously described U.S. bovine SBE type isolate E58 (23). In Australian livestock, this ST, previously described as ST23, could be found in association with (i) ovine conjunctivitis, (ii) ovine polyarthritis, (iii) bovine encephalomyelitis, and, in the present study, (iv) in a single case of clinically healthy ovine fecal shedding. Gastrointestinal strains leading to asymptomatic infection and their fecal shedding may be common in infected Australian sheep flocks. Fecal shedding of C. pecorum by carrier animal hosts has been reported previously (8, 51) and is thought to be the most important mode of transmission (15). Other transmission routes can include sexual and vertical transmission, as C. pecorum isolates were found in the genitourinary tracts of healthy bulls (1) and C. pecorum was detected in infected calves in utero (9). Animals are also susceptible to infection by the fecal-oral route (2). Using our MLSA, we also observed in multiple cases that one sheep host can harbor two distinct STs, with one found in the conjunctiva (Eug/Ovi1/Eye, Nyn/Ovi1/Eye), while another novel ST was detected in the gastrointestinal tract (Eug/Ovi1/Rec, Nyn/Ovi1/Rec). A variety of STs were also present on a flock level. Three distinct STs, 23, 2013A, and 2013Aa, detected in multiple hosts from ocular and rectal sites, were present in a Nyngan sheep flock (see Table S2 in the supplemental material). Infection with these C. pecorum STs in the Nyngan flock could have been from multiple sources, further supporting the possible transmission routes outlined above. The risks of potential transmission of C. pecorum infections between animals are increased with cograzing of sheep and cattle, as in this study we observed the same C. pecorum ST found in both hosts. As the cooccurrence of cattle and sheep in a geographical area is common, this should be a target area for further C. pecorum investigations in order to better understand the epidemiology of the C. pecorum infections.
Using our C. pecorum MLSA scheme, we could also make similarly interesting observations about the epidemiology of C. pecorum infections in koalas. C. pecorum isolates infecting koalas displayed more diversity than those infecting livestock, resulting in seven novel STs being detected (see Table S4 in the supplemental material). The distribution of STs in koala populations was similar to that seen in our sheep epidemiological analysis, including the observation of multiple C. pecorum STs within the same koala population (EC/James, EC/Steve, and EC/Ned), the presence of a single C. pecorum ST in multiple anatomical sites of the same koala (Gun/Koala1/Urethra and Gun/Koala1/Cloaca), and the detection of a single C. pecorum ST within multiple animals in the same population (StB/9341 and SB/9274 koalas). Although koalas were sampled from various geographical locations across Australia, we did not observe biogeographical separation of C. pecorum koala strains using MLSA. Identical STs were observed in koalas originating from different states (SA C. pecorum isolate SA/Adel/Eye was 100% identical to the SEQLD isolates MC/Marsbar, Bre/Kurt/UGT, StB/9274/RE, and StB/9341/RE, as well as the VIC and NSW Vic/Jba001/Eye and TB/Elisabeth/Eye isolates).
The majority of koalas assessed in this study displayed clinical signs of chlamydial disease, with the exception of two koalas (StB/9274 and StB/9341) from a geographically isolated koala population from St. Bees Island, QLD, which were found to be C. pecorum PCR positive. ST2013Aa, observed in these samples (StB/9274/RE and StB/9341/RE), was the same as in the samples derived from animals with keratoconjunctivitis and/or urogenital infection, not allowing for the differentiation of strains based on pathology. Interestingly, this same ST was also observed in C. pecorum isolates from the two sheep samples (Nyn/Ovi2/Eye and Nyn/Ovi3/Eye), which phylogenetically clustered in the koala clade.
The observation of an identical C. pecorum ST type ST2013Aa in two Australian sheep, alongside the clustering of a U.S. bovine ST49 with other koala C. pecorum STs (Fig. 1), provides a potential snapshot of the risk of cross-host transmission between Australian livestock and koalas. Similar observations were reported previously in phylogenetic analyses of koala and livestock C. pecorum using the single highly polymorphic ompA gene (22, 52). In our study, koala isolates formed a single cluster within a larger livestock clade, in contrast to previous observations using novel genetic markers (24), although Australian livestock isolates were not included in the previous study. In agreement with the phylogenetic analyses mentioned above, low population divergence indices (see Table S5 in the supplemental material) also support the possibility that koala C. pecorum isolates may have diverged from livestock isolates, a potential indicator of the origin of C. pecorum infections in koalas. Low population divergence values indicate a potentially very recent evolutionary split between the koala and livestock C. pecorum populations, with no fixed differences and only shared polymorphisms. An increase in shared polymorphisms and reduced fixed differences suggest “recent” active gene flow between the local populations (53), which could explain the observed values in C. pecorum koala and livestock populations that were analyzed here. A better understanding of the origin of C. pecorum in koalas will require a larger cohort of livestock and koala samples from the same geographical area in Australia. A broader assessment of the strains reported in other Australian marsupials (54) and worldwide domesticated and wild ungulates (55, 56) would contribute to our understanding of the genetic diversity of C. pecorum and the origins of these infections. Such an analysis should also include a larger number of samples within each population to build a better picture of the intrapopulation C. pecorum genetic structures, thus improving our understanding of the epidemiology of these infections.
Collectively, our MLSA of seven HK gene fragments of C. pecorum isolates in Australian sheep and cows has provided us with the first molecular epidemiological data on infections in these hosts in Australia. An expansion of cross-sectional studies employing this C. pecorum typing scheme across entire populations of infected animals will be critical for developing effective management strategies for Australian livestock. Beyond this study, our MLSA scheme could be translated into an MLST scheme that is widely used for finely detailed epidemiological studies of other bacteria (32), including Chlamydia trachomatis and C. psittaci (30, 31), further helping us to elucidate the epidemiology and evolution of this widespread and significant pathogen of wild and domesticated animals.
Supplementary Material
ACKNOWLEDGMENTS
We thank District Veterinarians from the Livestock Health and Pest Authority, NSW, Australia, including Susan McClure, Colin Peake, Belinda Edmonstone, Bruce Watt, Jillian Kelly, and Evelyn Walker, for their ongoing collaboration and field sample collections. We also acknowledge the collection and provision of koala swab samples from wild koala populations by Bill Ellis, Centre for Mined Land Rehabilitation, University of Queensland, John Callaghan, Gold Coast City Council, Jon Hanger, Endeavor Veterinary Ecology, David Amos, Gunnedah Veterinary Hospital, Oliver Funnell, Adelaide Hills Animal Hospital, Rod Starr, Tanilba Bay Veterinary Hospital, and Cheyne Flanagan, Port Macquarie Koala Hospital. This study made use of several U.S. C. pecorum MLST sequences available from the Chlamydiales MLST website (http://pubmlst.org/chlamydiales/). These isolates were originally sourced from Bernhard Kaltenboeck, Auburn University, and we thank Yvonne Pannekoek, University of Amsterdam, The Netherlands, for permission to include these sequences for analysis in our study. We thank Charles Wan and Pride Kanyoka for their technical assistance with PCR assays, Eileen Roulis for assistance with figure preparation, and James Marsh and Megan Stride for their helpful advice on the use of the bioinformatics programs.
This work was partially funded by a 2010/2011 Australian Academy of Science Margaret Middleton Fund for Endangered Wildlife Early Career Research award to A.P. and a 2013 Australian Research Council Discovery grant (DP130102066) awarded to P.T. and A.P.
Footnotes
Published ahead of print 5 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00992-13.
REFERENCES
- 1. Mohamad KY, Rodolakis A. 2010. Recent advances in the understanding of Chlamydophila pecorum infections, sixteen years after it was named as the fourth species of the Chlamydiaceae family. Vet. Res. 41:27. 10.1051/vetres/2009075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Longbottom D, Coulter LJ. 2003. Animal chlamydioses and zoonotic implications. J. Comp. Pathol. 128:217–244 [DOI] [PubMed] [Google Scholar]
- 3. Polkinghorne A, Hanger J, Timms P. 2013. Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Vet. Microbiol. pii:S0378-1135(13)00140-5. 10.1016/j.vetmic.2013.02.026 [DOI] [PubMed] [Google Scholar]
- 4. Jackson M, White N, Giffard P, Timms P. 1999. Epizootiology of Chlamydia infections in two free-range koala populations. Vet. Microbiol. 65:255–264 [DOI] [PubMed] [Google Scholar]
- 5. Girjes AA, Hugall A, Graham DM, McCaul TF, Lavin MF. 1993. Comparison of type I and type II Chlamydia psittaci strains infecting koalas (Phascolarctos cinereus). Vet. Microbiol. 37:65–83 [DOI] [PubMed] [Google Scholar]
- 6. McColl KA, Martin RW, Gleeson LJ, Handasyde KA, Lee AK. 1984. Chlamydia infection and infertility in the female koala (Phascolarctos cinereus). Vet. Rec. 115:655. 10.1136/vr.115.25-26.655 [DOI] [PubMed] [Google Scholar]
- 7. McNutt SH, Waller EF. 1940. Sporadic bovine encephalomyelitis (Buss disease). Cornell Vet. 30:437–448 [Google Scholar]
- 8. DeGraves FJ, Gao D, Hehnen HR, Schlapp T, Kaltenboeck B. 2003. Quantitative detection of Chlamydia psittaci and C. pecorum by high-sensitivity real-time PCR reveals high prevalence of vaginal infection in cattle. J. Clin. Microbiol. 41:1726–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reggiardo C, Fuhrmann TJ, Meerdink GL, Bicknell EJ. 1989. Diagnostic features of Chlamydia infection in dairy calves. J. Vet. Diagn. Invest. 1:305–308 [DOI] [PubMed] [Google Scholar]
- 10. Polkinghorne A, Borel N, Becker A, Lu ZH, Zimmermann DR, Brugnera E, Pospischil A, Vaughan L. 2009. Molecular evidence for chlamydial infections in the eyes of sheep. Vet. Microbiol. 135:142–146 [DOI] [PubMed] [Google Scholar]
- 11. Schautteet K, Vanrompay D. 2011. Chlamydiaceae infections in pig. Vet. Res. 42:29. 10.1186/1297-9716-42-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poudel A, Elsasser TH, Rahman KhS, Chowdhury EU, Kaltenboeck B. 2012. Asymptomatic endemic Chlamydia pecorum infections reduce growth rates in calves by up to 48 percent. PLoS One 7:e44961. 10.1371/journal.pone.0044961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jee J, Degraves FJ, Kim T, Kaltenboeck B. 2004. High prevalence of natural Chlamydophila species infection in calves. J. Clin. Microbiol. 42:5664–5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watt B. 2010. Arthritis in sheep, p 109–113 In Proceedings of the 92nd District Veterinarian's Conference District Veterinarian's Association, Dubbo, New South Wales, Australia [Google Scholar]
- 15. Reinhold P, Sachse K, Kaltenboeck B. 2011. Chlamydiaceae in cattle: commensals, trigger organisms, or pathogens? Vet. J. 189:257–267 [DOI] [PubMed] [Google Scholar]
- 16. Pitman B. 2010. A case of sporadic bovine encephalomyelitis (SBE) in Hume LHPA, p 114–115 In Proceedings of the 92nd District Veterinarian's Conference District Veterinarian's Association, Dubbo, New South Wales, Australia [Google Scholar]
- 17. Durham PJ, Paine GD. 1997. Serological survey for antibodies to infectious agents in beef cattle in northern South Australia. Aust. Vet. J. 75:139–140 [DOI] [PubMed] [Google Scholar]
- 18. Daszak P, Cunningham AA, Hyatt AD. 2000. Emerging infectious diseases of wildlife–threats to biodiversity and human health. Science 287:443–449 [DOI] [PubMed] [Google Scholar]
- 19. Daniels PW, Halpin K, Hyatt A, Middleton D. 2007. Infection and disease in reservoir and spillover hosts: determinants of pathogen emergence. Curr. Top. Microbiol. Immunol. 315:113–131 [DOI] [PubMed] [Google Scholar]
- 20. Cockram FA, Jackson AR. 1974. Isolation of a Chlamydia from cases of keratoconjunctivitis in koalas. Aust. Vet. J. 50:82–83 [DOI] [PubMed] [Google Scholar]
- 21. Glassick T, Giffard P, Timms P. 1996. Outer membrane protein 2 gene sequences indicate that Chlamydia pecorum and Chlamydia pneumoniae cause infections in koalas. Syst. Appl. Microbiol. 19:457–464 [Google Scholar]
- 22. Jackson M, Giffard P, Timms P. 1997. Outer membrane protein A gene sequencing demonstrates the polyphyletic nature of koala Chlamydia pecorum isolates. Syst. Appl. Microbiol. 20:187–200 [Google Scholar]
- 23. Mojica S, Huot Creasy H, Daugherty S, Read TD, Kim T, Kaltenboeck B, Bavoil P, Myers GS. 2011. Genome sequence of the obligate intracellular animal pathogen Chlamydia pecorum E58. J. Bacteriol. 193:3690. 10.1128/JB.0054-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marsh J, Kollipara A, Timms P, Polkinghorne A. 2011. Novel molecular markers of Chlamydia pecorum genetic diversity in the koala (Phascolarctos cinereus). BMC Microbiol. 11:77. 10.1186/1471-2180-11-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yousef Mohamad K, Roche SM, Myers G, Bavoil PM, Laroucau K, Magnino S, Laurent S, Rasschaert D, Rodolakis A. 2008. Preliminary phylogenetic identification of virulent Chlamydophila pecorum strains. Infect. Genet. Evol. 8:764–771 [DOI] [PubMed] [Google Scholar]
- 26. Harris SR, Clarke IN, Seth-Smith HMB, Solomon AW, Cutcliffe LT, Marsh P, Skilton RJ, Holland MJ, Mabey D, Peeling RW, Lewis DA, Spratt BG, Unemo M, Persson K, Bjartling C, Brunham R, de Vries HJ, Morré SA, Speksnijder A, Bébéar CM, Clerc M, de Barbeyrac B, Parkhill J, Thomson NR. 2012. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat. Genet. 44:413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brunelle BW, Sensabaugh GF. 2012. Nucleotide and phylogenetic analyses of the Chlamydia trachomatis ompA gene indicates it is a hotspot for mutation. BMC Res. Notes 5:53. 10.1186/1756-0500-5-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klint M, Fuxelius HH, Goldkuhl RR, Skarin H, Rutemark C, Andersson SGE, Persson K, Herrmann B. 2007. High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J. Clin. Microbiol. 45:1410–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pannekoek Y, Morelli G, Kusecek B, Morré S, Ossewaarde J, Langerak A, van der Ende A. 2008. Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol. 8:42. 10.1186/1471-2180-8-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pannekoek Y, Dickx V, Beeckman DSA, Jolley KA, Keijzers WC, Vretou E, Maiden MCJ, Vanrompay D, van der Ende A. 2010. Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS One 5:e14179. 10.1371/journal.pone.0014179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maiden MC. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60:561–588 [DOI] [PubMed] [Google Scholar]
- 33. Feil EJ. 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2:483–495 [DOI] [PubMed] [Google Scholar]
- 34. Abayneh T, Colquhoun DJ, Sørum H. 2012. Multi-locus sequence analysis (MLSA) of Edwardsiella tarda isolates from fish. Vet. Microbiol. 158:367–375 [DOI] [PubMed] [Google Scholar]
- 35. Bell RL, González-Escalona N, Stones R, Brown EW. 2011. Phylogenetic evaluation of the ‘Typhimurium’ complex of Salmonella strains using a seven-gene multi-locus sequence analysis. Infect. Genet. Evol. 11:83–91 [DOI] [PubMed] [Google Scholar]
- 36. Achtman M. 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62:53–70 [DOI] [PubMed] [Google Scholar]
- 37. Papke RT, White E, Reddy P, Weigel G, Kamekura M, Minegishi H, Usami R, Ventosa A. 2011. A multilocus sequence analysis approach to the phylogeny and taxonomy of the Halobacteriales. Int. J. Syst. Evol. Microbiol. 61:2984–2995 [DOI] [PubMed] [Google Scholar]
- 38. Jolley KA, Maiden MCJ. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wan C, Loader J, Hanger J, Beagley KW, Timms P, Polkinghorne A. 2011. Using quantitative polymerase chain reaction to correlate Chlamydia pecorum infectious load with ocular, urinary and reproductive tract disease in the koala (Phascolarctos cinereus). Aust. Vet. J. 89:409–412 [DOI] [PubMed] [Google Scholar]
- 40. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 41. Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452 [DOI] [PubMed] [Google Scholar]
- 42. Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 44. Zocevic A, Vorimore F, Marhold C, Horvatek D, Wang D, Slavec B, Prentza Z, Stavianis G, Prukner-Radovcic E, Dovc A, Siarkou VI, Laroucau K. 2012. Molecular characterization of atypical Chlamydia and evidence of their dissemination in different European and Asian chicken flocks by specific real-time PCR. Environ. Microbiol. 14:2212–2222 [DOI] [PubMed] [Google Scholar]
- 45. Hurst LD. 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486–487 [DOI] [PubMed] [Google Scholar]
- 46. Stephens RS, Myers G, Eppinger M, Bavoil PM. 2009. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol. Med. Microbiol. 55:115–119 [DOI] [PubMed] [Google Scholar]
- 47. Hanage WP, Spratt BG, Turner KME, Fraser C. 2006. Modelling bacterial speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2039–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Timms P. 2005. Chlamydial infection and disease in the koala. Microbiol. Aust. 26:65–68 [Google Scholar]
- 49. Rattei T, Ott S, Gutacker M, Rupp J, Maass M, Schreiber S, Solbach W, Wirth T, Gieffers J. 2007. Genetic diversity of the obligate intracellular bacterium Chlamydophila pneumoniae by genome-wide analysis of single nucleotide polymorphisms: evidence for highly clonal population structure. BMC Genomics 8:355. 10.1186/1471-2164-8-355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Longbottom D. 2004. Chlamydial infections of domestic ruminants and swine: new nomenclature and new knowledge. Vet. J. 168:9–11 [DOI] [PubMed] [Google Scholar]
- 51. Reinhold P, Jaeger J, Liebler-Tenorio E, Berndt A, Bachmann R, Schubert E, Melzer F, Elschner M, Sachse K. 2008. Impact of latent infections with Chlamydophila species in young cattle. Vet. J. 175:202–211 [DOI] [PubMed] [Google Scholar]
- 52. Kaltenboeck B, Kousoulas KG, Storz J. 1993. Structures of and allelic diversity and relationships among the major outer membrane protein (ompA) genes of the four chlamydial species. J. Bacteriol. 175:487–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noor MA, Feder JL. 2006. Speciation genetics: evolving approaches. Nat. Rev. Genet. 7:851–861 [DOI] [PubMed] [Google Scholar]
- 54. Bodetti TJ, Viggers K, Warren K, Swan R, Conaghty S, Sims C, Timms P. 2003. Wide range of Chlamydiales types detected in native Australian mammals. Vet. Microbiol. 96:177–187 [DOI] [PubMed] [Google Scholar]
- 55. Holzwarth N, Pospischil A, Mavrot F, Vilei EM, Hilbe M, Zlinszky K, Regenscheit N, Pewsner M, Thoma R, Borel N. 2011. Occurrence of Chlamydiaceae, Mycoplasma conjunctivae, and pestiviruses in alpine chamois (Rupicapra r. rupicapra) of Grisons, Switzerland. J. Vet. Diagn. Invest. 23:333–337 [DOI] [PubMed] [Google Scholar]
- 56. Pospischil A, Kaiser C, Hofmann-Lehmann R, Lutz H, Hilbe M, Vaughan L, Borel N. 2012. Evidence for Chlamydia in wild mammals of the Serengeti. J. Wildl. Dis. 48:1074–1078 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.