Abstract
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was used for an extensive identification study of arthroconidial yeasts, using 85 reference strains from the CBS-KNAW yeast collection and 134 clinical isolates collected from medical centers in Qatar, Greece, and Romania. The test set included 72 strains of ascomycetous yeasts (Galactomyces, Geotrichum, Saprochaete, and Magnusiomyces spp.) and 147 strains of basidiomycetous yeasts (Trichosporon and Guehomyces spp.). With minimal preparation time, MALDI-TOF MS proved to be an excellent diagnostic tool that provided reliable identification of most (98%) of the tested strains to the species level, with good discriminatory power. The majority of strains were correctly identified at the species level with good scores (>2.0) and seven of the tested strains with log score values between 1.7 and 2.0. The MALDI-TOF MS results obtained were consistent with validated internal transcribed spacer (ITS) and/or large subunit (LSU) ribosomal DNA sequencing results. Expanding the mass spectrum database by increasing the number of reference strains for closely related species, including those of nonclinical origin, should enhance the usefulness of MALDI-TOF MS-based diagnostic analysis of these arthroconidial fungi in medical and other laboratories.
INTRODUCTION
Species of the genera Trichosporon and Geotrichum occur widely in nature (1), and both are characterized by true hyphae that disarticulate into arthroconidia. Strains of these species are dimorphic and display yeast-like and/or mold-like morphological types of colonies. Trichosporon and Guehomyces spp. are asexually reproducing basidiomycetous fungi. The members of the family Dipodascaceae are ascomycetous yeasts that are able to persist in two reproductive stages, namely, asexual (anamorph) and sexual (teleomorph). These ascomycetous yeasts that form arthroconidia belong to a number of genera; Galactomyces (anamorph Geotrichum) belongs to the same phylogenetic clade as Dipodascus (anamorph Geotrichum) and is a sister genus to Magnusiomyces (anamorph Saprochaete) (Table 1) (2). Species of these arthroconidium-forming genera are important, although rare, human pathogens. In healthy humans, they persist as colonizers of the gastrointestinal tract, the oral cavity and respiratory tract, skin, and hair (3, 4). Their clinical relevance should not be neglected, as bloodstream infections caused by species of Trichosporon and Geotrichum may occur among cancer patients and immunocompromised individuals (5–8). The majority of invasive infections caused by Trichosporon and Geotrichum species are known to be associated with high mortality rates, particularly invasive mycoses that develop in immunocompromised patients (9). In cancer patients, Trichosporon asahii was recognized as one of the important causative agents of catheter-related fungemia (9, 10). Disseminated geotrichosis, although rare in humans, can be fatal in neutropenic patients with acute leukemia (11–14).
Table 1.
Relationships between genus names of anamorphs and teleomorphs of ascomycetous and basidiomycetous arthroconidial yeasts used in this study (2)b
| Teleomorph | Anamorph | Commonly known synonyms |
|---|---|---|
| Galactomyces candidus (2004)a | Geotrichum candidum (1832) | Geotrichum candidum (1809), Oospora lactis (1931), Geotrichum silvicola (2012), Geotrichum bryndzae (2012) |
| Magnusiomyces capitatus (2004)a | Saprochaete capitata (2004) | Trichosporon capitatum (1942), Geotrichum capitatum (1977), Blastoschizomyces capitatus (1985), Dipodascus capitatus (1986) |
| Unknown | Saprochaete suaveolens (2004)a | Oidium suaveolens (1913), Oospora fragrans (1923), Geotrichum fragrans (1964), Geotrichum suaveolens (1966) |
| Unknown | Saprochaete clavata (2004)a | Geotrichum clavatum (1986) |
| Unknown | Guehomyces pullulans (2004)a | Trichosporon pullulans (1942) |
| Unknown | Trichosporon spp. (1890)a |
Currently used names.
Years of publication are indicated in parentheses.
To date, correct identification to the species level of Trichosporon and Geotrichum yeasts recovered from clinical specimens is challenging but remains important because different species respond differently to various antifungal agents (Geotrichum spp. are azole resistant, while Trichosporon spp. are resistant to echinocandins), which affects proper disease treatment and patient care (15). Conclusive identification of pathogens should be fast and reliable to support the appropriate choice and application of antifungal therapy, adjustment of prophylactic treatment, and monitoring of the effects of treatment and the emergence of drug resistance.
In a routine clinical laboratory, identification of arthroconidial yeasts (e.g., Trichosporon and Geotrichum) is performed by investigating macroscopic and microscopic morphological characteristics. When arthroconidia are observed, the urease test is recommended to distinguish between Geotrichum spp. and Trichosporon spp. (16). Especially for uncommon species of Trichosporon and Geotrichum, identification at the species level is difficult to accomplish using commercially available assimilation tests and kits, such as the API 20 C AUX (bioMérieux), ID 32C (bioMérieux), and RapID Yeast Plus (Innovative Diagnostic Systems) systems (15, 17). Use of the automated Vitek 1 and 2 systems (bioMérieux) has increased lately, but the Vitek 2 database includes only three Trichosporon and three Geotrichum species (17–19). Consequently, identifications usually are unreliable (15, 17, 18, 20).
Since the introduction of molecular methods, a plethora of advanced techniques (e.g., PCR-high-resolution melting analysis, real-time PCR, and Luminex xMAP technology) have been used for the identification of fungal pathogens; however, they are not always suitable for routine medical laboratory testing, and their discriminatory power must be increased to identify rarely occurring species (21–23). Currently, the technique used for the identification of uncommon yeasts is sequence analysis of universal bar-coding markers such as internal transcribed spacer (ITS) 1, ITS 2, and large subunit (LSU) D1 and D2 domains of the ribosomal DNA (rDNA) (24).
Recent developments in mass spectrometry-based analysis of pathogens opened new possibilities in diagnostic microbiology and may result in rapid detection and differentiation of pathogenic microorganisms (25). In the present study, the usefulness of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for the identification and differentiation of ascomycetous and basidiomycetous arthroconidial yeasts was investigated. In order to determine the taxonomic resolution of this method, the capabilities and limitations for the identification of closely related species were evaluated.
(This work was presented in part at the 18th Congress of the International Society for Human and Animal Mycology, Berlin, Germany, 11 to 15 June 2012 [26].)
MATERIALS AND METHODS
Strains and culture conditions.
This study included two major representative sets of arthroconidial yeasts. The first set combined type and reference strains from the yeast collection of the CBS Fungal Biodiversity Centre (CBS-KNAW) (Utrecht, the Netherlands) (i.e., the reference CBS set in Table S1 in the supplemental material). This set consisted of 85 strains of medically important ascomycetous and basidiomycetous arthroconidial yeast species, and their current generic names, as used in this study, are listed in Table 1. Strains of ascomycetous arthroconidial yeasts (Geotrichum, Galactomyces, Saprochaete, and Magnusiomyces) were maintained on malt extract agar (MEA) plates (10% sugar solution of malt extract, 2% [wt/vol] agar), while basidiomycetous arthroconidial yeasts (Trichosporon and Guehomyces) were grown on potato dextrose agar (PDA) plates (23% [vol/vol] potato extract, 2% [wt/vol] dextrose, 2% [wt/vol] agar). All strains were cultivated at 30°C for 24 to 48 h. The second set of strains (i.e., the clinical set) consisted of clinical isolates that had been obtained from Qatar, Greece, or Romania. The subset from the Department of Laboratory Medicine and Pathology, Hamad Medical Corporation (Doha, Qatar), included 20 isolates of ascomycetous arthroconidial yeasts and 51 isolates of basidiomycetous arthroconidial yeasts. These isolates were identified previously by conventional methods, including macroscopic and microscopic morphological investigations and the urease test. The automated commercially available Vitek 2 system identified the majority of isolates only to the genus level. Additionally, 27 of 51 isolates of basidiomycetous arthroconidial yeasts were identified previously by molecular methods (15). The remaining isolates were analyzed by ITS and LSU bar-coding during this study. The subset from the Mycology Laboratory, Microbiology Department, Medical School, University of Athens (Athens, Greece), included 4 isolates of Galactomyces spp. and 44 isolates of Trichosporon spp. These isolates were characterized previously to the genus and species levels by sequencing analysis, namely, ITS sequencing for Galactomyces spp. and LSU sequencing (D1/D2 domain analyses; GenBank accession no. JX111912 to JX111953 and KC515399) for Trichosporon spp., according to the protocol reported by Fell et al. (27). Additionally, the T. asahii strains were characterized by sequencing of the rDNA intergenic spacer (IGS) region (GenBank accession no. JX111954 to JX111990 and KC515400), according to a method described previously (28). The last subset of clinical isolates was collected from the Department of Mycology and Mycotoxicology, Ion Ionescu de la Brad University (Iasi, Romania), and included eight isolates of Geotrichum spp. and seven isolates of Trichosporon spp. All of these isolates were identified to the genus level by phenotypic, microscopic, and biochemical methods, using the ID 32C system (bioMérieux, France). Only T. asahii isolates were identified to the species level by ID 32C testing.
MALDI-TOF MS identification and data analyses.
The strains of the reference and clinical sets were subcultured on Sabouraud dextrose agar plates and incubated for 24 h at 30°C. In order to prevent some of the strains from growing into the agar, sterile polycarbonate track etch (PCTE) filtration membranes (diameter, 76 mm; pore size, 0.1 mm; GE Water & Process Technologies) were applied on the agar surface, before inoculation with fungal material. Identification by MALDI-TOF MS was carried out using the formic acid (FA)/ethanol (EtOH) protein extraction method, according to the Bruker Daltonics GmbH protocol (25, 29) with minor modifications, as reported by Cendejas-Bueno et al. (30). Prior to EtOH/FA extraction, 2 loops of cells for ascomycetous arthroconidial yeasts or 3 loops of cells for basidiomycetous arthroconidial yeasts (1-μl sterile inoculation loop; Greiner Bio-One) were collected and resuspended in 300 μl of Milli-Q water. The crucial steps were (i) estimation of the volume of 70% FA in comparison with the size of pellet and (ii) accurate dissolution of the fungal material in FA by vortex-mixing. The optimal volume of FA used ranged between 30 and 50 μl, and an equal volume of acetonitrile (ACN) was added later. One microliter of the crude protein extract was pipetted in duplicate on a 96-spot polished steel target plate (Bruker Daltonics, Bremen, Germany), and 1 μl of bacterial test standard (Bruker Daltonics) solution was spotted twice as a positive control. After air drying, all spots were overlaid with α-cyano-4-hydroxycinnamic acid (HCCA) matrix solution prepared according to the protocol of the manufacturer and, after drying, were analyzed in automatic runs that were operated by FlexControl version 3.3.108.0 (Bruker Daltonics). The strains from the reference and clinical sets were analyzed and identified by MALDI-TOF MS Biotyper RTC software 3.0 (Bruker Daltonics). The identification spectra generated with 240 laser shots for each duplicate of the tested strains were compared with reference main mass spectra (MSP) data selected simultaneously from the Bruker Daltonics (BDAL) database and the CBS-KNAW in-house library. Subsequently, recognition results generated by MALDI-TOF MS (identification spectra versus MSPs) were scored as log score values according to the manufacturer's requirements and were classified as follows: secure genus and species identification, >2.0; secure genus identification, 1.7 to 2.0; no reliable identification (NRI), <1.7. The identification mass spectra obtained during measurements were visualized by Bruker flexAnalysis version 3.3.75.0 software and investigated by ClinProTools version 3.0 software (Bruker Daltonics), as described by Cendejas-Bueno et al. (30). The identification was considered correct if at least one spot from the duplicates gave reliable identification, with a score of >1.7.
CBS-KNAW in-house library.
This research was carried out with two databases, i.e., the original, commercially available Bruker Daltonics (BDAL) database and a CBS-KNAW in-house library. The BDAL database is regularly updated by the manufacturer and currently contains 4,110 MSPs created between 2007 and 2012, including 10 MSPs of Geotrichum spp., 4 MSPs of Magnusiomyces spp., and 11 MSPs of Trichosporon spp.
The CBS-KNAW in-house library of 103 MSPs was created by Bruker Daltonics (Bremen, Germany) for this research project, as indicated in Table S1 in the supplemental material. This library is part of another database that contains a large number of reference spectra (>600 MSPs) generated from different fungal strains, which were obtained from the CBS-KNAW yeast collection and validated previously with ITS and/or LSU sequence data, but these are not yet available in the Bruker Daltonics commercial database. Ethanol extracts of the 103 CBS-KNAW strains of ascomycetous and basidiomycetous arthroconidial yeasts were prepared and sent to Bruker Daltonics for the creation of BDAL database-quality main mass spectra, according to their internal database-creation standard operating procedures. From the protein-rich supernatants that were obtained using the full extraction method (EtOH/FA), 1 μl was pipetted on eight spots on a polished steel target plate and overlaid with HCCA matrix; after air drying, each spot was analyzed three times. The reference MSP was created from at least 20 high-quality spectra generated during these 24 measurements, following the manufacturer's protocol, and was stored in the project database. The library generated by Bruker Daltonics was later uploaded at the MALDI-TOF MS Biotyper 3.0 as the CBS-KNAW in-house library. This library contained 15 MSPs of Galactomyces spp., 2 MSPs of Geotrichum spp. (now reassigned as Galactomyces candidus), 11 MSPs of Magnusiomyces spp., 7 MSPs of Saprochaete spp., 65 MSPs of Trichosporon spp., and 3 MSPs of Guehomyces pullulans (see Table S1 in the supplemental material). The CBS-KNAW in-house library was used in combination with the commercial BDAL database for identification of the target isolates by MALDI-TOF MS. However, it also can be used as an individual source of MSPs.
Sequence analyses.
Genomic DNA was extracted from reference CBS-KNAW strains and clinical isolates grown on MEA (ascomycetous yeasts) or PDA (basidiomycetous yeasts) plates and incubated at 30°C for 48 h. DNA extractions were performed following the phenol-chloroform-isoamyl alcohol protocol according to Bolano et al. (31), with minor modifications as reported by Cendejas-Bueno et al. (30). The primer set V9-F (5′-TGC GTT GAT TAC GTC CCT GC-3′) and LR3-R (5′-GGT CCG TGT TTC AAG AC-3′) was used to obtain the ITS amplicon that was subsequently amplified using the primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′), as reported by White et al. (32). The D1/D2 region was amplified using the primers NL1 (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′) and RLR3R (5′-GGT CCG TGT TTC AAG AC-3′), as suggested by Vilgalys and Hester (33). The PCR and sequencing conditions were reported previously by Cendejas-Bueno et al. (30). Sequence alignments were assembled and edited using SeqMan version 8.0.2 software (DNAStar Inc., Madison, WI) and aligned with MEGA version 5. The sequences were manually corrected. Consensus sequences were compared by the Basic Local Alignment Search Tool (BLAST) with sequences available from the CBS-KNAW in-house library and the GenBank database for correct identification.
RESULTS
MALDI-TOF MS-based identification of reference CBS-KNAW strains.
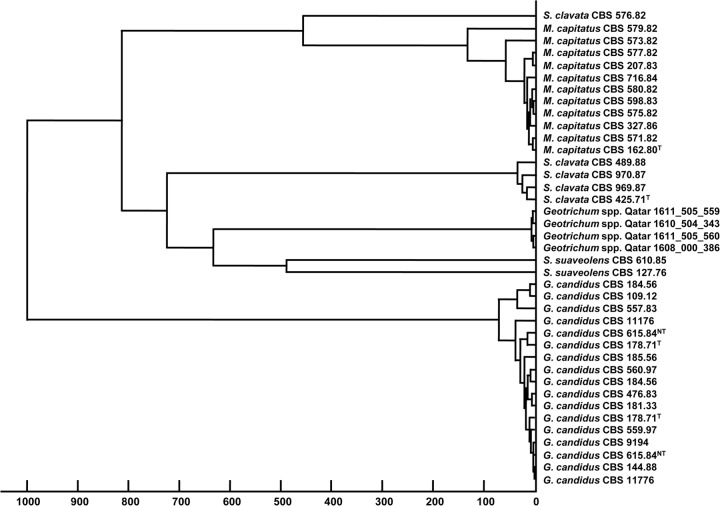
The clinically relevant arthroconidial fungal species from the CBS reference set, represented by 40 strains of ascomycetous yeast, consisted of 2 Geotrichum, 1 Galactomyces, 1 Magnusiomyces, and 2 Saprochaete species together with 45 basidiomycetous yeast strains of 14 Trichosporon and 1 Guehomyces species. This set was used to challenge the CBS-KNAW in-house library and the BDAL database, as the majority of these strains were also used for creation of MSPs (see Table S1 in the supplemental material). All 40 ascomycetous yeast strains were identified correctly at the species level, with 34 strains having log score values above 2.0; five strains had values between 1.7 and 2.0, and a correct identification match at the species level was achieved for one strain of Saprochaete clavata (CBS 758.85) with a NRI score value of <1.7 (Table 2). Strains of Geotrichum silvicola CBS 9194T and Geotrichum bryndzae CBS 11176T were identified by MALDI-TOF MS as Geotrichum candidum/Galactomyces candidus with scores of >2.0. While this research was being conducted, G. silvicola CBS 9194T and G. bryndzae CBS 11176T were reviewed and gained a new taxonomic status, as they were found to be conspecific with G. candidus (teleomorph)/G. candidum (anamorph) (for strain information, see the CBS-KNAW yeast collection at http://www.cbs.knaw.nl/Collections/BioloMICS.aspx?Table=CBS strain database&Rec=5644&Fields=All and http://www.cbs.knaw.nl/Collections/BioloMICS.aspx?Table=CBS strain database&Rec=7759&Fields=All; for the taxonomic name and synonyms, see Mycobank at http://www.mycobank.org/BioloMICS.aspx?Table=Mycobank_Advanced&Rec=431757&Fields=All) (34). The dendrogram grouping MSPs of CBS strains of ascomycetous arthroconidial yeasts (Fig. 1) was used to illustrate the separation of strains at the genus and species levels. It was observed that strains of G. candidus (including G. silvicola and G. bryndzae) formed one cluster, distinct from a cluster containing Magnusiomyces capitatus, Saprochaete suaveolens, and S. clavata. The cluster analysis demonstrated also that S. clavata strain CBS 576.82 deviated from the S. clavata cluster and was found to be more related to the M. capitatus cluster (Fig. 1), although it was correctly identified as S. clavata with a score of >2.0.
Table 2.
Overview of MALDI-TOF MS identification results for arthroconidial yeast species (ascomycetous [Geotrichum-like] or basidiomycetous [Trichosporon-like])
| Log score value ofa: |
No. (%) of identifications for: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CBS-KNAW reference set of ascomycetous yeast spp. (n = 40) |
Set of clinical isolates of ascomycetous yeast spp. (n = 32) |
CBS-KNAW reference set of basidiomycetous yeast spp. (n = 45) |
Set of clinical isolates of basidiomycetous yeast spp. (n = 102) |
||||||
| Spot 1 | Spot 2 | Correct | Other | Correct | Other | Correct | Other | Correct | Other |
| >2.0 | >2.0 | 26 (65) | 0 | 16 (50.0) | 0 | 34 (75.6) | 3 (6.7)b | 53 (52) | 6 (5.8)b |
| >2.0 | 1.7–2.0 | 6 (15) | 0 | 4 (12.5) | 0 | 1 (2.2) | 0 | 11 (10.8) | 1 (1.0)b |
| >2.0 | NRI (<1.7) | 1 (2.5) | 0 | 0 | 0 | 1 (2.2) | 0 | 1 (1.0) | 1 (1.0)b |
| >2.0 | NPF | 1 (2.5) | 0 | 7 (21.9) | 0 | 5 (11.1) | 0 | 28 (27.5) | 0 |
| 1.7–2.0 | 1.7–2.0 | 4 (10) | 0 | 1 (3.1) | 0 | 0 | 1 (2.2)b | 1 (1.0) | 0 |
| 1.7–2.0 | NRI (<1.7) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.7–2.0 | NPF | 1 (2.5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NRI (<1.7) | NRI (<1.7) | 1 (2.5) | 0 | 0 | 4 (12.5)c | 0 | 0 | 0 | 0 |
| NRI (<1.7) | NPF | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NPF | NPF | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 40 (100) | 0 | 28 (87.5) | 4 (12.5)c | 41 (91.1) | 4 (8.9) | 94 (92.2) | 8 (7.8) | |
NRI, no reliable identification; NPF, no peaks found.
Correct identification only at the genus level.
No reliable identification; these isolates may represent a new species.
Fig 1.
Dendrogram of the cluster analysis of 35 MSPs for CBS strains of ascomycetous arthroconidial yeasts used for MALDI-TOF MS general identification and 4 MSPs for Geotrichum-like isolates (1610504343, 1608000386, 1611505559, and 1611505560) from Qatar. The G. silvicola CBS 9194T and G. bryndzae CBS 11176T strains are marked with their new taxonomic designations as G. candidus. Distances are displayed in relative units on the x axis.
MALDI-TOF MS provided correct recognition at the genus level for all 45 CBS strains of basidiomycetous arthroconidial yeasts. Accurate species identification with scores of >2.0 was possible for 41 strains (91.1%) (Table 2). The remaining four strains (8.9%) showed reliable genus identification (three strains with scores of >2.0 and one strain with a score between 1.7 and 2.0), but identification at the species level achieved by MALDI-TOF MS differed from the CBS-KNAW identity. Between the phylogenetically closely related species Trichosporon inkin and Trichosporon ovoides, identification of the tested strains was straightforward with scores of >2.0. A strain of T. ovoides (CBS 5581) was identified as T. inkin with a score between 1.7 and 2.0. At the species level, reliable distinction between T. ovoides and T. inkin was not possible. The results obtained were validated by sequence analysis of the ITS and LSU domains, which confirmed the identity of T. ovoides (1-nucleotide [nt] difference in LSU sequences and 5-nt difference in ITS sequences with T. ovoides and 4-nt difference in LSU sequences and 6-nt difference in ITS sequences with T. inkin). Three strains that were identified with reliable scores of >2.0 revealed the following. Two strains deposited in the CBS collection as T. asahii (CBS 5286 and CBS 7755) were identified as Trichosporon asteroides and occurred in the Trichosporon japonicum/T. asteroides cluster. The molecular data indicated closer relationships of both strains to T. japonicum (no difference in LSU sequences and 0- to 2-nt difference in ITS sequences) than to T. asahii (2-nt difference in LSU sequences and 3- or 4-nt difference in ITS sequences) and thus confirmed the correct identification by MALDI-TOF MS as T. asteroides/T. japonicum.
One strain, CBS 2043T, of Trichosporon dermatis was identified by MALDI-TOF MS as the closely related species Trichosporon mucoides. This result was considered a minor error, as sequencing data revealed that it is difficult to distinguish between these closely related species (1-nt difference in LSU sequences and 4-nt difference in ITS sequences for T. dermatis in comparison with T. mucoides). In the MALDI-TOF MS-based dendrogram of the grouping of MSPs of the CBS strains of basidiomycetous arthroconidial yeasts, diverse clusters occurred (Fig. 2). G. pullulans and Trichosporon species occurred in different clusters with the exception of T. dermatis (CBS 2043T), which clustered with T. mucoides, and T. japonicum, which clustered with T. asteroides (Fig. 2).
Fig 2.
Dendrogram of the cluster analysis of 71 MSPs for CBS strains of basidiomycetous arthroconidial yeasts used for MALDI-TOF MS general identification. Two strains (CBS 5286 and CBS 7755) labeled previously as T. asahii are marked as T. japonicum, thus confirming correct clustering. Distances are displayed in relative units on the x axis.
Identification of clinical isolates by MALDI-TOF MS and ITS/LSU sequencing validation.
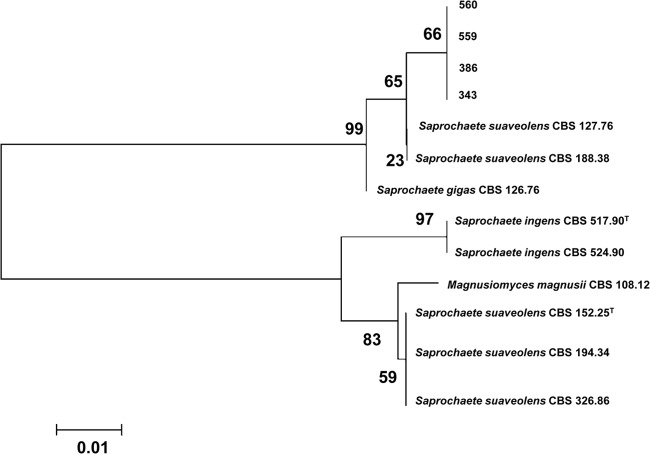
Isolates of ascomycetous and basidiomycetous arthroconidial yeasts from Qatar, Greece, and Romania were used as a test set of clinical isolates. Twenty-eight (87.5%) of 32 clinical ascomycetous arthroconidial yeasts isolates were correctly identified at the species level with log score values above 1.7 (Table 2). Twenty-seven isolates had scores of >2.0 and one isolate had a score of 1.7 to 2.0. Eleven isolates were identified as Geotrichum candidum/Galactomyces candidus and 14 isolates as Magnusiomyces capitatus, but three isolates were identified as Candida krusei/Issatchenkia orientalis with scores of >2.0 (identity confirmed by sequencing of the ITS and LSU regions). The remaining four isolates from the clinical set (12.5%) were not identified reliably either by MALDI-TOF MS or by ITS/LSU domain sequencing (Tables 2 and 3). Three individuals experienced with the MALDI-TOF MS technique independently prepared EtOH/FA extracts and performed MALDI-TOF MS Biotyper 3.0 identification runs (each performed two experiments) and this yielded NRI scores (scores of <1.7), although the spectra were of good quality with appropriate peak intensities (Table 2). The only hit close to the genus level was achieved by MALDI-TOF MS with NRI matching with Geotrichum ingens DSM 70069 (score of 1.429). Molecular data also showed no reliable identification. ITS and LSU region sequencing indicated that these isolates are closely related to Saprochaete suaveolens (99% similarity, based on LSU domain sequencing) or Saprochaete gigas (97% similarity, based on ITS domain sequencing) (Fig. 3 and Table 4). Based on the MALDI-TOF MS dendrogram of MSPs and the phylogenetic ITS tree (Fig. 1 and 3), these four strains may represent a new Geotrichum-like species, which needs further investigation to confirm the final taxonomic identity.
Table 3.
Validation of identification results for arthroconidial yeasts (ascomycetous [Geotrichum-like] or basidiomycetous [Trichosporon-like])
| Set and ID resultsa | No. of isolates for: |
||
|---|---|---|---|
| Routine ID method | ITS/LSU sequencing | MALDI-TOF MS | |
| Reference CBS set of ascomycetous yeast spp. (n = 40) | |||
| Correct genus and species ID | Not performed | 40 | 40 |
| Minor error (correct genus with incorrect species ID) | 0 | 0 | |
| Major error (incorrect genus ID) | 0 | 0 | |
| No ID | 0 | 0 | |
| Clinical set of ascomycetous yeast spp. (n = 32) | |||
| Correct genus and species ID | 5 | 28 | 28 |
| Minor error (correct genus with incorrect species ID) | 22 | 0 | 0 |
| Major error (incorrect genus ID) | 2 Geotrichum candidum isolates were Magnusiomyces; 3 Geotrichum spp. were Candida spp. | 0 | 0 |
| No ID | 0 | 4 (new species) | 4 (new species) |
| Reference CBS set of basidiomycetous yeast spp. (n = 45) | |||
| Correct genus and species ID | Not performed | 45 | 41 |
| Minor error (correct genus with incorrect species ID) | 0 | 4 closely related Trichosporon spp., T. ovoides/T. inkin, T. japonicum/T. asteroides, T. dermatis/T. mucoides | |
| Major error (incorrect genus ID) | 0 | 0 | |
| No ID | 0 | 0 | |
| Clinical set of basidiomycetous yeast spp. (n = 102) | |||
| Correct genus and species ID | 82 | 94 | 94 |
| Minor error (correct genus with incorrect species ID) | 18 | 8 closely related Trichosporon spp., T. ovoides/T. inkin, T. japonicum/T. asteroides, T. asahii/T. faecale | 8 closely related Trichosporon spp., T. ovoides/T. inkin, T. japonicum/T. asteroides, T. asahii/T. faecale |
| Major error (incorrect genus ID) | 2b | 0 | 0 |
| No ID | 0 | 0 | 0 |
ID, identification.
The routine identification test results were incorrect, as MALDI-TOF MS and sequencing data are consistent.
Fig 3.
Phylogenetic tree showing the ITS-based relationships of Geotrichum-like strains (1610504343 [343], 1608000386 [386], 1611505559 [559], and 1611505560 [560]) from Qatar, which likely represent a new species. Note that only the last three digits of the strain code for each Geotrichum-like isolate are used in the figure. Maximum-likelihood analysis of the ITS sequences was used with bootstrap values that are indicated on the branches and that were obtained from 1,000 replicates. Scale bar, 0.01 substitution/site.
Table 4.
Molecular identification of four Geotrichum-like spp. recovered from clinical specimens from Qatar, which may represent a novel taxon
| Strain code | No. of nucleotides in LSU sequence entry | No. of identical nucleotides/total no. of nucleotides in LSU sequence (% identity)a | No. of gaps/total no. of nucleotides | Species of closest hit (BLAST) | No. of nucleotides in ITS sequence entry | No. of identical nucleotides/total no. of nucleotides in ITS sequence (% identity)b | No. of gaps/total no. of nucleotides (%) | Species of closest hit (BLAST) | No. of nucleotides in sequence for closest BLAST hit |
|---|---|---|---|---|---|---|---|---|---|
| 1610504343 | 379 | 376/379 (99) | 0 | S. suaveolens | 532 | 322/331 (97) | 4/331 (1) | S. gigas (CBS 126.67) | 331 |
| 1608000386 | 389 | 384/389 (99) | 1/389 | S. suaveolens | 534 | 322/331 (97) | 4/331 (1) | S. gigas (CBS 126.67) | 331 |
| 1611505559 | 379 | 374/379 (99) | 1/379 | S. suaveolens | 532 | 322/331 (97) | 4/331 (1) | S. gigas (CBS 126.67) | 331 |
| 1611505560 | 389 | 384/389 (99) | 0 | S. suaveolens | 535 | 322/331 (97) | 4/331 (1) | S. gigas (CBS 126.67) | 331 |
The LSU region sequence entries are based on the NL1 primer only.
The ITS sequence entries are based on the ITS1 and ITS4 primers.
For all 102 clinical basidiomycetous arthroconidial yeast isolates, MALDI-TOF MS provided identification at the species level with log score values of >1.7 (Table 2). A total of 101 strains (99.0%) showed identification scores of >2.0, and one isolate (1.0%) had a score between 1.7 and 2.0. Based on the MALDI-TOF MS identification, this set consisted of 10 Trichosporon species, namely, 71 isolates of T. asahii, 12 of T. inkin, 4 of Trichosporon faecale, 4 of T. asteroides, 3 of Trichosporon coremiiforme, 2 of Trichosporon dohaense, and 1 each of T. dermatis, Trichosporon loubierii, Trichosporon moniliiforme, and Trichosporon mycotoxinivorans. However, one strain was identified as Candida ciferrii/Stephanoascus ciferrii with a score of >2.0 (identity was confirmed by sequencing of the ITS and LSU regions). For only 3 of the 102 Trichosporon isolates was species distinction problematic, and that was between T. inkin, T. ovoides, and Trichosporon cutaneum, although identification scores were between 1.7 and 2.0 (Table 2). However, freshly prepared extracts of those three isolates yielded identification as T. inkin with reliable scores of >2.0. ITS and LSU domain sequencing results were concordant with MALDI-TOF MS results as T. inkin, with five nucleotide differences from T. ovoides in the ITS region.
The cumulative percentage of correct scores obtained during successive runs showed that a correct species match with reliable identification results (score ranges of >2.0 and 1.7 to 2.0 were used for calculation of cumulative percentages) was obtained for 78.5% of the tested clinical isolates during the first identification run, which increased to 91.9% after the second run. Occasionally, four runs were needed to achieve identification to the species level for all tested isolates.
The discriminatory power of MALDI-TOF MS was further investigated using 38 isolates of T. asahii that had been characterized previously by sequence analysis of the LSU and intergenic spacer (IGS) rDNA regions. All of these T. asahii isolates were correctly identified by MALDI-TOF MS with scores of >2.0 (Table 2). A Ward clustering dendrogram of the mass spectra (not MSPs) for these T. asahii isolates was created to investigate whether the MALDI-TOF MS technique was able to separate and to identify the known IGS subgroups. Figure S1 in the supplemental material indicates that MALDI-TOF MS separates four clusters that do not represent the same grouping as revealed by IGS typing. It can be observed on the dendrogram that cluster 1 of T. asahii isolates contains a majority of IGS type III strains, cluster 3 contains a majority of IGS type IV strains, and the strains of IGS types V and VII are aligned in cluster 4. Cluster 2, however, includes strains of different IGS types.
MALDI-TOF MS results versus routine identifications applied to the clinical set.
The clinical laboratories from Qatar, Greece, and Romania used different routine identification methods (namely, Vitek 2 analysis, physiological and morphological analyses, ID 32C testing, or LSU or ITS sequencing). These methods, with the exception of rDNA sequencing, allowed correct identification at the species level of only a limited number of isolates (Table 3). Seven of 32 isolates of ascomycetous arthroconidial yeasts (21.9%) were incorrectly identified at the genus and species levels by routine methods. The error rate for conventional identification of Trichosporon was lower, as correct genus identification was not accomplished for only 2 of the 102 isolates (2%). Among clinical isolates of Trichosporon spp., correct identification at the species level was achieved only for T. asahii.
DISCUSSION
Rapid and reliable identification of clinical yeast isolates is crucial for patient care, particularly for critically ill patients, as this allows physicians to start proper treatment. The MALDI-TOF MS Biotyper 3.0 system is an identification tool that proved to deliver fast reliable results in the clinical laboratory (25). To date, identification results obtained by using MALDI-TOF MS for arthroconidial yeasts have not been reported extensively and have been limited to a small number of Geotrichum and Trichosporon spp. (20, 29, 35–37). MALDI-TOF MS has been applied successfully to identify Cryptococcus species, including varieties and hybrids, Candida albicans and non-albicans Candida species, and rare pathogenic yeasts (29, 30, 38–44). Non-Candida species isolates, such as Geotrichum, Galactomyces, Saprochaete, Magnusiomyces, and Trichosporon spp., are underrepresented in the reference library and thus MALDI-TOF MS identification runs were less successful, resulting in low identification rates or failure of reliable analysis (36, 45).
Our data showed a high level of discriminatory power for MALDI-TOF MS, providing reliable identification of both ascomycetous and basidiomycetous arthroconidial yeasts. The EtOH/FA extraction protocol worked well for those yeasts. In general, runs that were performed by the MALDI-TOF MS Biotyper 3.0 to identify strains of arthroconidial yeasts species allowed discrimination among all tested strains and identified most of the strains at the species level (100% of basidiomycetous and 94.7% of ascomycetous arthroconidial yeast strains), with high accuracy. Correct identification was provided not only with log score values of >2.0 but also with scores of >1.7. This observation is in agreement with the results of Van Herendael et al. (46), who considered log score values of >1.7 as the minimal values for secure species identification for yeasts. Importantly, MALDI-TOF MS identification with scores above 2.0 or between 1.7 and 2.0 never gave erroneous identification of the genus. Thus, isolates of ascomycetes were never misidentified as basidiomycetes and vice versa.
Use of the MALDI-TOF MS system resulted in correct recognition of the sister genera Galactomyces, Magnusiomyces, and Saprochaete but also exhibited a high level of discriminatory power between the various Trichosporon species. For instance, T. mycotoxinivorans, a newly recognized pathogen associated with cystic fibrosis (47, 48), was correctly identified. Examination of the relevance, accuracy, and taxonomic resolution of MALDI-TOF MS also was undertaken, to establish whether the identification results obtained were consistent with available molecular data from ITS and LSU bar-coding (15, 24, 27). For most strains of Saprochaete spp., correct identification was achieved with log score values of 1.7 to 2.0. This minor MALDI-TOF MS difficulty in obtaining correct identification was supported by sequencing validation, which also was not straightforward and which revealed the presence of polymorphic sites with double peaks in the raw sequence data. It was recently observed that variable copies of multiple intragenomic rDNAs may occur in the genera Geotrichum, Galactomyces, Saprochaete, and Dipodascus, thus confirming that the ITS region is heterogeneous and ITS-based identification is not readily possible (49). It may be concluded that the taxonomic resolution of MALDI-TOF MS is similar to that of rDNA sequencing. Comparisons of the overall performance of the MALDI-TOF MS Biotyper 3.0 for the identification of arthroconidial yeasts with that of molecular identification through sequence analyses of the ITS regions or the LSU parts of the rDNA as the standard method for yeast identification showed that MALDI-TOF MS provided reliable and highly accurate identification at the genus and species levels, which correlated well with the sequencing data (Table 2).
Some minor limitations of the MALDI-TOF MS Biotyper 3.0 deserve attention. Several strains of Trichosporon species, belonging to T. inkin/T. ovoides and T. japonicum/T. asteroides (all four belonging to the ovoides clade), as well as T. dermatis/T. mucoides (from the cutaneum clade), were not distinguished at the species level by MALDI-TOF MS analysis and usually had log score values of 1.7 to 2.0. For isolates of T. japonicum and T. asteroides, the first best match was always T. asteroides and the second best match was T. japonicum (scores of >2.0). Analogous problems were observed in ITS and LSU sequencing validation, as there were 1- to 4-nt differences in ITS sequences and 1- to 3-nt differences in LSU sequences between T. japonicum and T. asteroides. In fact, T. asteroides and T. japonicum are closely related species (2) and thus the MALDI-TOF MS identification might be considered a minor error. Reliable identification of T. dermatis also might be problematic. It was observed that the MALDI-TOF MS Biotyper 3.0 did not distinguish between the closely related T. dermatis and T. mucoides. All T. dermatis strains tested were initially misidentified as T. mucoides. This situation was observed for CBS strains as well as for the clinical isolates from Greece. Probably this relates to the quality and number of T. dermatis MSPs present in the reference database. After increasing the number of T. dermatis MSPs (manual entries) in the in-house library from 1 to 4, correct identification matches as T. dermatis were made with scores above 2.0. Sugita et al. (28) reported that, for reliable identification and good discrimination between those phylogenetically closely related Trichosporon spp., analysis of the rDNA intergenic spacer (IGS) region should be used instead of ITS sequencing, but this method is not used in routine laboratory testing.
In conclusion, good discriminatory power of the MALDI-TOF MS Biotyper 3.0 depends largely on the number of high-quality MSPs included in the commercial and in-house libraries but also on the sample preparation and the quality of the raw spectra obtained during general identification runs. High-quality real-time mass spectra resulted in increased score values. We found it important to use constant, previously optimized amounts of fresh fungal material to obtain high-quality spectra. As the ascomycetous and basidiomycetous arthroconidial yeasts vary in their growth morphology on Sabouraud's agar (i.e., they can be mold-like or yeast-like), some experience with sample preparation was crucial for obtaining high-quality spectra. During the EtOH/FA extraction procedure, it was important to work with equal, previously optimized volumes of biological sample material and optically adjusted volumes of FA/ACN.
In view of the data presented above, the MALDI-TOF MS Biotyper 3.0 is a sensitive reliable diagnostic system that is able to distinguish between arthroconidial yeast isolates to the genus and species levels with high accuracy comparable to that of ITS and/or LSU sequence analysis. Thus, MALDI-TOF MS is an easy-to-use, high-throughput tool for the identification of these arthroconidial yeasts. In clinical settings, this identification tool should be most useful for identifying these yeasts with minimal effort and with a short turnaround time, which would be beneficial for rapid decision-making regarding treatment and, more generally, optimal management of patients. Further improvements may be possible by increasing the number of MSPs and by adding more environmentally occurring arthroconidial yeast species to the database.
Supplementary Material
ACKNOWLEDGMENTS
This publication was made possible by NPRP grant 5-298-3-086 from the Qatar National Research Fund (a member of the Qatar Foundation) to Teun Boekhout and Saad J. Taj-Aldeen.
Anna Kolecka twice received a travel grant from the Walter Gams nonprofit Foundation for Research in Mycological Taxonomy and Ecology (Studienstiftung Mykologische Systematik und Ökologie). Markus Kostrzewa is an employee of Bruker Daltonics GmbH (Bremen, Germany), the manufacturer of the MALDI-TOF MS Biotyper system used for this study. Bruker Daltonics GmbH did not influence the design of the study. All other authors do not report conflicts of interest. All authors contributed to the content and the writing of the manuscript. The statements herein are solely the responsibility of the authors.
We thank Simone Becker from Bruker Daltonics (Bremen, Germany) for preparation and technical support with the database MSPs. We also appreciate the help of our students Patrick Hoopman and Jonathan Heng with MALDI-TOF MS and Sander van Schaik and Bart Kraak with ITS/LSU sequencing.
Footnotes
Published ahead of print 15 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00470-13.
REFERENCES
- 1. O'Brien M, O'Kiely P, Forristal PD, Fuller HT. 2005. Fungi isolated from contaminated baled grass silage on farms in the Irish Midlands. FEMS Microbiol. Lett. 247:131–135 [DOI] [PubMed] [Google Scholar]
- 2. Kurtzman C, Fell JW, Boekhout T. (ed). 2011. The yeasts: a taxonomic study, 5th ed Elsevier, Amsterdam, Netherlands [Google Scholar]
- 3. Bonifaz A, Vazquez-Gonzalez D, Macias B, Paredes-Farrera F, Hernandez MA, Araiza J, Ponce RM. 2010. Oral geotrichosis: report of 12 cases. J. Oral Sci. 52:477–483 [DOI] [PubMed] [Google Scholar]
- 4. Pottier I, Gente S, Vernoux JP, Gueguen M. 2008. Safety assessment of dairy microorganisms: Geotrichum candidum. Int. J. Food Microbiol. 126:327–332 [DOI] [PubMed] [Google Scholar]
- 5. Chitasombat MN, Kofteridis DP, Jiang Y, Tarrand J, Lewis RE, Kontoyiannis DP. 2012. Rare opportunistic (non-Candida, non-Cryptococcus) yeast bloodstream infections in patients with cancer. J. Infect. 64:68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kustimur S, Kalkanci A, Caglar K, Dizbay M, Aktas F, Sugita T. 2002. Nosocomial fungemia due to Trichosporon asteroides: firstly described bloodstream infection. Diagn. Microbiol. Infect. Dis. 43:167–170 [DOI] [PubMed] [Google Scholar]
- 7. Walsh TJ, Groll A, Hiemenz J, Fleming R, Roilides E, Anaissie E. 2004. Infections due to emerging and uncommon medically important fungal pathogens. Clin. Microbiol. Infect. 10(Suppl 1):48–66 [DOI] [PubMed] [Google Scholar]
- 8. Wolf DG, Falk R, Hacham M, Theelen B, Boekhout T, Scorzetti G, Shapiro M, Block C, Salkin IF, Polacheck I. 2001. Multidrug-resistant Trichosporon asahii infection of nongranulocytopenic patients in three intensive care units. J. Clin. Microbiol. 39:4420–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Girmenia C, Pagano L, Martino B, D'Antonio D, Fanci R, Specchia G, Melillo L, Buelli M, Pizzarelli G, Venditti M, Martino P. 2005. Invasive infections caused by Trichosporon species and Geotrichum capitatum in patients with hematological malignancies: a retrospective multicenter study from Italy and review of the literature. J. Clin. Microbiol. 43:1818–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chagas-Neto TC, Chaves GM, Melo AS, Colombo AL. 2009. Bloodstream infections due to Trichosporon spp.: species distribution, Trichosporon asahii genotypes determined on the basis of ribosomal DNA intergenic spacer 1 sequencing, and antifungal susceptibility testing. J. Clin. Microbiol. 47:1074–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Batlle M, Gimenez M, Sancho JM, Ribera JM. 2004. Disseminated Geotrichum capitatum infection in neutropenic patient with acute leukaemia. Med. Clin. (Barc.) 123:557–558 (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 12. Henrich TJ, Marty FM, Milner DA, Jr, Thorner AR. 2009. Disseminated Geotrichum candidum infection in a patient with relapsed acute myelogenous leukemia following allogeneic stem cell transplantation and review of the literature. Transpl. Infect. Dis. 11:458–462 [DOI] [PubMed] [Google Scholar]
- 13. Mahul P, Piens MA, Guyotat D, Godard J, Archimbaud E, Bui-Xuan B, Motin J. 1989. Disseminated Geotrichum capitatum infection in a patient with acute myeloid leukemia. Mycoses 32:573–577 [DOI] [PubMed] [Google Scholar]
- 14. Pimentel JD, Baker M, Woodgyer AJ, Harris OC. 2005. Fatal disseminated Blastoschizomyces capitatus (Geotrichum capitatum) in a patient with relapse of acute lymphoblastic leukaemia. Pathology 37:319–321 [DOI] [PubMed] [Google Scholar]
- 15. Taj-Aldeen SJ, Al-Ansari N, El Shafei S, Meis JF, Curfs-Breuker I, Theelen B, Boekhout T. 2009. Molecular identification and susceptibility of Trichosporon species isolated from clinical specimens in Qatar: isolation of Trichosporon dohaense Taj-Aldeen, Meis & Boekhout sp. nov. J. Clin. Microbiol. 47:1791–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Howell SA, Hazen KC. 2011. Candida, Cryptococcus, and other yeasts of medical importance, p 1793–1821 In Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW. (ed), Manual of clinical microbiology, 10th ed ASM Press, Washington, DC [Google Scholar]
- 17. Colombo AL, Padovan AC, Chaves GM. 2011. Current knowledge of Trichosporon spp. and trichosporonosis. Clin. Microbiol. Rev. 24:682–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ahmad S, Al-Mahmeed M, Khan ZU. 2005. Characterization of Trichosporon species isolated from clinical specimens in Kuwait. J. Med. Microbiol. 54:639–646 [DOI] [PubMed] [Google Scholar]
- 19. Gunn SR, Reveles XT, Hamlington JD, Sadkowski LC, Johnson-Pais TL, Jorgensen JH. 2006. Use of DNA sequencing analysis to confirm fungemia due to Trichosporon dermatis in a pediatric patient. J. Clin. Microbiol. 44:1175–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iriart X, Lavergne RA, Fillaux J, Valentin A, Magnaval JF, Berry A, Cassaing S. 2012. Routine identification of medical fungi by the new Vitek MS matrix-assisted laser desorption ionization–time of flight system with a new time-effective strategy. J. Clin. Microbiol. 50:2107–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diaz MR, Fell JW. 2005. Use of a suspension array for rapid identification of the varieties and genotypes of the Cryptococcus neoformans species complex. J. Clin. Microbiol. 43:3662–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Landlinger C, Preuner S, Willinger B, Haberpursch B, Racil Z, Mayer J, Lion T. 2009. Species-specific identification of a wide range of clinically relevant fungal pathogens by use of Luminex xMAP technology. J. Clin. Microbiol. 47:1063–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lau A, Stanley K, Sorrell T. 2013. Multiplex-tandem PCR for fungal diagnostics. Methods Mol. Biol. 968:195–201 [DOI] [PubMed] [Google Scholar]
- 24. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. U. S. A. 109:6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. 2011. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17:1359–1365 [DOI] [PubMed] [Google Scholar]
- 26. Kolecka A, Groenewald M, Kostrzewa M, Boekhout T. 2012. The efficient identification of arthroconidial yeasts by MALDI-TOF MS method, abstr P604. Mycoses 55(Suppl 4):285 [Google Scholar]
- 27. Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50:1351–1371 [DOI] [PubMed] [Google Scholar]
- 28. Sugita T, Nakajima M, Ikeda R, Matsushima T, Shinoda T. 2002. Sequence analysis of the ribosomal DNA intergenic spacer 1 regions of Trichosporon species. J. Clin. Microbiol. 40:1826–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG. 2009. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, Cuenca-Estrella M, Gomez-Lopez A, Boekhout T. 2012. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 50:3641–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bolano A, Stinchi S, Preziosi R, Bistoni F, Allegrucci M, Baldelli F, Martini A, Cardinali G. 2001. Rapid methods to extract DNA and RNA from Cryptococcus neoformans. FEMS Yeast Res. 1:221–224 [DOI] [PubMed] [Google Scholar]
- 32. White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, Thomas J, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY [Google Scholar]
- 33. Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Groenewald M, Coutinho T, Smith MT, van der Walt JP. 2012. Species reassignment of Geotrichum bryndzae, Geotrichum phurueaensis, Geotrichum silvicola and Geotrichum vulgare based on phylogenetic analyses and mating compatibility. Int. J. Syst. Evol. Microbiol. 62:3072–3080 [DOI] [PubMed] [Google Scholar]
- 35. Jensen RH, Arendrup MC. 2011. Candida palmioleophila: characterization of a previously overlooked pathogen and its unique susceptibility profile in comparison with five related species. J. Clin. Microbiol. 49:549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lohmann C, Sabou M, Moussaoui W, Prevost G, Delarbre JM, Candolfi E, Gravet A, Letscher-Bru V. 2013. Comparison between the Biflex III-Biotyper and the Axima-SARAMIS systems for yeast identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:1231–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seyfarth F, Wiegand C, Erhard M, Graser Y, Elsner P, Hipler UC. 2012. Identification of yeast isolated from dermatological patients by MALDI-TOF mass spectrometry. Mycoses 55:276–280 [DOI] [PubMed] [Google Scholar]
- 38. Firacative C, Trilles L, Meyer W. 2012. MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS One 7:e37566. 10.1371/journal.pone.0037566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kubesova A, Salplachta J, Horka M, Ruzicka F, Slais K. 2012. Candida “psilosis”: electromigration techniques and MALDI-TOF mass spectrometry for phenotypical discrimination. Analyst 137:1937–1943 [DOI] [PubMed] [Google Scholar]
- 40. McTaggart LR, Lei E, Richardson SE, Hoang L, Fothergill A, Zhang SX. 2011. Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:3050–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pinto A, Halliday C, Zahra M, van Hal S, Olma T, Maszewska K, Iredell JR, Meyer W, Chen SC. 2011. Matrix-assisted laser desorption ionization–time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One 6:e25712. 10.1371/journal.pone.0025712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Posteraro B, Vella A, Cogliati M, De Carolis E, Florio AR, Posteraro P, Sanguinetti M, Tortorano AM. 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based method for discrimination between molecular types of Cryptococcus neoformans and Cryptococcus gattii. J. Clin. Microbiol. 50:2472–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quiles-Melero I, Garcia-Rodriguez J, Gomez-Lopez A, Mingorance J. 2012. Evaluation of matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry for identification of Candida parapsilosis, C. orthopsilosis and C. metapsilosis. Eur. J. Clin. Microbiol. Infect. Dis. 31:67–71 [DOI] [PubMed] [Google Scholar]
- 44. Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buchan BW, Ledeboer NA. 2013. Advances in identification of clinical yeast isolates by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Van Herendael BH, Bruynseels P, Bensaid M, Boekhout T, De Baere T, Surmont I, Mertens AH. 2012. Validation of a modified algorithm for the identification of yeast isolates using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS). Eur. J. Clin. Microbiol. Infect. Dis. 31:841–848 [DOI] [PubMed] [Google Scholar]
- 47. Hickey PW, Sutton DA, Fothergill AW, Rinaldi MG, Wickes BL, Schmidt HJ, Walsh TJ. 2009. Trichosporon mycotoxinivorans, a novel respiratory pathogen in patients with cystic fibrosis. J. Clin. Microbiol. 47:3091–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hirschi S, Letscher-Bru V, Pottecher J, Lannes B, Jeung MY, Degot T, Santelmo N, Sabou AM, Herbrecht R, Kessler R. 2012. Disseminated Trichosporon mycotoxinivorans, Aspergillus fumigatus, and Scedosporium apiospermum coinfection after lung and liver transplantation in a cystic fibrosis patient. J. Clin. Microbiol. 50:4168–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stielow B, Vaas L, Wouters M, Groenewald M. 2012. Taking a closer look into rDNA polymorphism of strains belonging to Geotrichum, Galactomyces and Dipodascus, abstr AP9M. 13th Int. Congr. Yeasts, Madison, WI, 26 to 30 August 2012 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.