Abstract
Seventy-six Pseudomonas aeruginosa isolates recovered from chronically (n = 18) and nonchronically (n = 18) colonized cystic fibrosis (CF) patients (2002 to 2009) were grouped in separate polyclonal populations. International CF epidemic clones were not identified, but the high-risk clone ST274, also found circulating in Spanish hospitals, was present. Persistent isolates were more resistant to antibiotics than nonpersistent isolates.
TEXT
Molecular typing tools applied to Pseudomonas aeruginosa isolates recovered from cystic fibrosis (CF) patients have demonstrated not only local epidemiological differences (1–4) but also the spread of particular multiresistant clones through different CF units (5–8) and the in vivo evolution of single clones involving multilocus sequence type (MLST) shifts (9). Nevertheless, studies addressing differences in terms of population structure between nonpersistent and persistent isolates from CF patients have not yet been reported. This work was performed to describe the genetic diversity of P. aeruginosa colonizing CF patients from a CF unit in a university hospital in Spain, differentiating between nonpersistent isolates (isolates recovered once or during a <6-month period in consecutive sputum cultures) and persistent isolates (genetically related isolates recovered during a minimum of 6 months) (4, 10). All morphologically different colonies cultured from each sputum sample during the follow-up period were analyzed. When only nonpersistent isolates were recovered, patients were considered nonchronically colonized. On the other hand, patients were considered chronically colonized when persistent isolates were recovered.
Seventy-six P. aeruginosa isolates from 36 CF patients in our CF unit between 2002 and 2009 were included. Patients and isolates were distributed in two groups (Tables 1 and 2). The first one included 18 patients from whom 26 P. aeruginosa nonpersistent isolates were recovered. The second group included 18 chronically colonized P. aeruginosa patients from whom 48 persistent isolates were obtained. In two of these chronically colonized patients, two isolates were recovered on one occasion each, and these isolates were grouped within the nonpersistent isolates (Table 1). The follow-up protocol for CF patients in our institution includes 4 to 5 microbiological sputum analyses per year, with additional sputum analysis during exacerbations or admittance to the hospital. The first and the subsequent isolates that were indistinguishable or highly related according to standard pulsed-field electrophoresis (PFGE) typing criteria were considered a single lineage (11), and only one isolate was included for further studies.
Table 1.
Distribution of the 26 nonpersistent clones in 20 CF patients with (C2 and C16) and without (P1 to P18) chronic P. aeruginosa bronchopulmonary colonization
| Patient | Clone found in patients by yr |
|||||||
|---|---|---|---|---|---|---|---|---|
| 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | |
| P1 | ST170 | |||||||
| P2 | ST606 | |||||||
| P3 | ST274 | ST270 | ||||||
| P4 | ST460 | |||||||
| P5 | ST485 | |||||||
| P6 | ST395 | |||||||
| P7 | ST641 | |||||||
| P8 | ST274 | |||||||
| P9 | ST592 | |||||||
| P10 | ST253 | |||||||
| P11 | ST508 | ST499 | ST540 | |||||
| P12 | ST312 | |||||||
| P13 | ST539 | |||||||
| P14 | ST274 | ST312 | ||||||
| P15 | ST620 | ST17 | ST360 | |||||
| P16 | ST274 | ST980 | ||||||
| P17 | ST809 | |||||||
| P18 | ST809 | ST395 | ||||||
| C2 | ST1045 | |||||||
| C16 | ST27 | |||||||
Table 2.
Distribution of the persistent P. aeruginosa clones in CF patients (n = 18)b
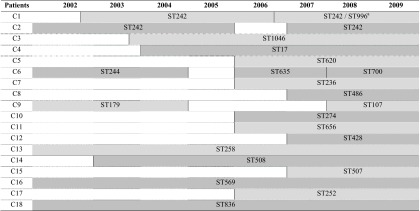
a ST996 is a single-locus variant of ST242 (see the text and reference 9 for further explanation).
ST persistence is represented by a shaded background. Vertical lines indicate initiation or termination of an ST chronic colonization.
Isolates were initially typed by PFGE using SpeI enzyme and interpreted by standard criteria (9, 11). One isolate per pulsotype was subsequently typed by the MLST scheme developed by Keith Jolley (University of Oxford; http://pubmlst.org/paeruginosa) and analyzed by using minimum spanning tree (MST) analysis (http://goeburst.phyloviz.net/). Bayesian phylogenetic trees (BAPS) based on concatenated nucleotidic sequences from the same fragments used in the MLST approach were also performed (12). MLST nucleotide sequences from representative multidrug-resistant clones circulating in 16 Spanish hospitals (1), as well as the Dutch sequence type (ST) ST406 and ST497, Liverpool ST146, and Manchester ST148 international CF epidemic clones (5–8), were included in the analysis. Antibiotic susceptibility was determined by standard broth microdilution and interpreted with CLSI criteria (13). Statistical differences in antimicrobial resistance were explored by chi-square test and Fisher's exact test.
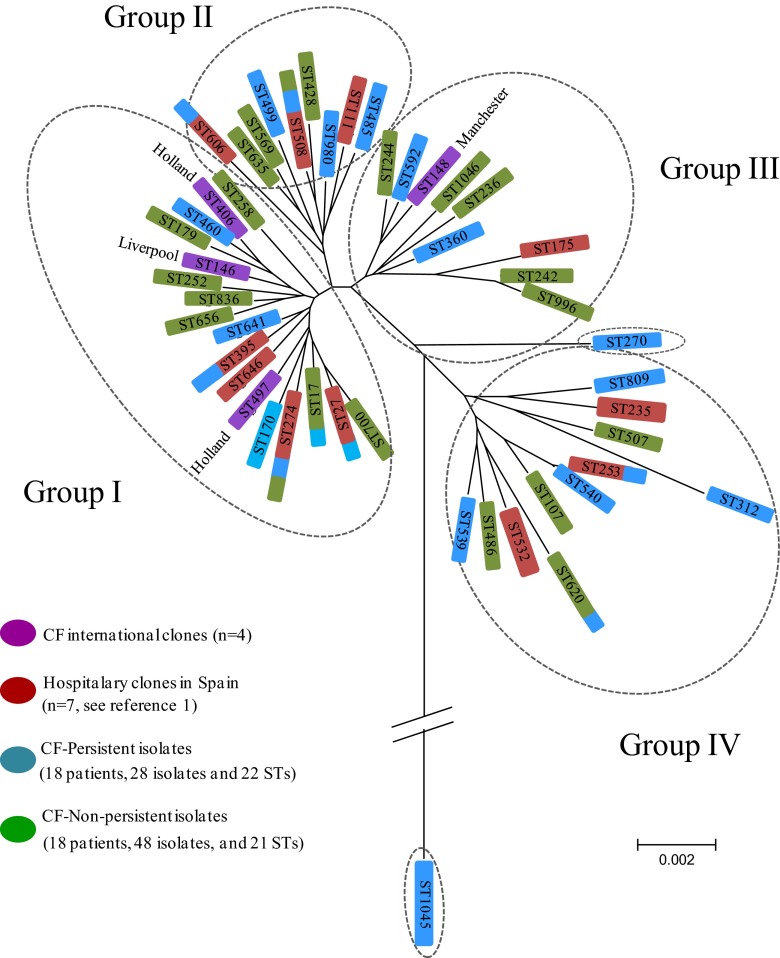
Within the 28 nonpersistent isolates (26 from nonchronically colonized patients and 2 from chronically colonized patients) (Table 1), 22 STs were identified; 20 of them were unrelated, and the other two corresponded to single-locus variants (SLV) (ST253 and ST540) (Fig. 1). STs representing more than one isolate were ST274 (n = 4), ST312 (n = 2), ST395 (n = 2), and ST809 (n = 2). The scarce genetic relationship between these STs suggested that clones involved in primocolonization without persistence corresponded to nonrelated acquisition events (Fig. 1 and 2). The PFGE patterns confirmed this hypothesis. Six of the 22 primocolonizer STs were previously described in Spanish hospitals (ST27, ST253, ST274, ST395, ST508, and ST606) (1), suggesting potential acquisition of these clones from those circulating within the Spanish health care system.
Fig 1.
Minimum spanning tree (MST) of nonpersistent and persistent CF clones compared with international CF clones and multiresistant clones recovered from Spanish hospitals.
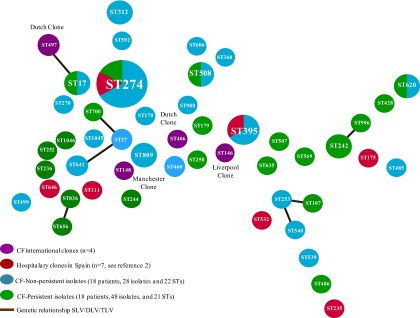
Fig 2.
Phylogenetic BAPS analysis of the 7 MLST concatenated alleles from nonpersistent and persistent CF clones recovered in our CF unit compared with international CF clones and multiresistant clones recovered from Spanish hospitals.
On the other hand, the 48 persistent isolates recovered from 18 chronically colonized patients were grouped in 21 STs (Fig. 2). During the follow-up period of these chronically colonized patients, 15 of them presented a single ST, an ST replacement was detected during chronic colonization in two patients (C6 and C9), and finally an MLST shift was described in patient C1 (9).
ST17, ST274, ST508, and ST620 were detected, causing both nonchronic and chronic colonization. PFGE analysis showed an identical pattern for isolates within ST17 and ST274 clones that pointed out the possibility of patient-to-patient transmission or exposition to a common source. The same statement could be applied to ST242 isolates from two chronically colonized patients sharing an undistinguishable PFGE pattern. However, isolates belonging to the other STs (ST312, ST395, and ST809 in nonchronically colonized patients and ST508 and ST620 in both chronically and nonchronically colonized patients) exhibited different PFGE patterns. Three of the 21 persistent clones had previously been found circulating in hospitalized patients in Spain (ST244, ST274, and ST508) (1).
It is of note that the international CF epidemic clones identified in other countries were not represented within our chronically colonized CF patients, even though the ST17 lineage corresponded to a double-locus variant of the Dutch ST497 CF epidemic clone (Fig. 1). Nevertheless, ST274, found in CF patients from other European countries and Australia (MLST database), was detected among our isolates. ST274 was also identified within multidrug-resistant isolates circulating in Spain (1, 14).
Analysis of the antimicrobial susceptibility results showed that nonpersistent isolates exhibited lower resistance rates than persistent isolates, especially for tobramycin (0% versus 30%; P = 0.015), piperacillin-tazobactam (7% versus 35%; P = 0.03), imipenem (14% versus 30%; P = 0.2), and ciprofloxacin (7% versus 14%; P = 0.5), whereas both groups of isolates presented the same resistance rates for colistin (7%), ceftazidime (14%), and amikacin (35%).
During the first stages of bronchopulmonary P. aeruginosa colonization in CF patients, different nonrelated clones can access this niche, whereas in the advanced stages, the number of clones decreases but the respiratory tract is chronically colonized (15). Subsequently to an initial adaptation period, critical mutations in chromosomal genes and loss of virulence seem to be necessary for lung persistence (16). Independent phylogenetic analysis of the seven MLST loci demonstrated a high genetic diversity for the guaA allele, whereas the more conserved loci were aroE and nuoD genes (see Fig. S1 to S7 in the supplemental material). The mutL locus, which is implicated in the mismatch repair system, is usually altered in the CF isolates, with the direct consequence of hypermutability (17) and the consequent limitation for the application of the MLST technique in CF isolates (9).
In our collection, 4 different main groups (I to IV) were identified using the BAPS analysis without particular distribution (Fig. 2). International CF epidemic clones grouped with our nonpersistent and persistent colonizers in groups I and III. ST1045, a new lineage described in this study, showed remarkable genetic differences, both in the concatenated and independent allele analyses. This fact could be attributed to an independent evolution event or to the acquisition of alien genetic material.
In summary, this is the first description of CF P. aeruginosa isolates representing nonpersistent and persistent clones recovered from a Spanish CF unit. BAPS analysis depicts a deeper picture of the P. aeruginosa population structure from our CF patients. Unlike other CF units, we could not demonstrate the spread of previously identified international CF epidemic clones. Nevertheless, one of the most prevalent STs was ST274, previously described in another Spanish CF unit, which is a multidrug-resistant clone circulating in Spain (1, 18). Our CF unit follows the segregation measures internationally recommended (19), even though patient-to-patient transmission events were suspected in three patients. In addition, persistent clones showed higher resistance rates than nonpersistent ones, particularly for tobramycin.
Supplementary Material
ACKNOWLEDGMENTS
A.F.-O. and B.R. received a Rio Hortega contract (reference no. CM08/166 and 11/181) from the Instituto Carlos III (Ministry of Economy and Competitiveness), Spain. This study was supported by Instituto de Salud Carlos III (Ministry of Economy and Competitiveness) (reference no. PI12/00734) and cofinanced by European Development Regional Fund “A Way to Achieve Europe,” ERDF, Spanish Network for the Research in Infectious Diseases (REIPI RD12/0015).
The authors declare no conflicts of interests with the content of the manuscript.
Footnotes
Published ahead of print 12 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00802-13.
REFERENCES
- 1. García-Castillo M, del Campo R, Morosini MI, Riera E, Cabot G, Willems R, van Mansfeld R, Oliver A, Cantón R. 2011. Wide dispersion of ST175 clone despite high genetic diversity of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical strains in 16 Spanish hospitals. J. Clin. Microbiol. 49:2905–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Selezska K, Kazmierczak M, Müsken M, Garbe J, Schobert M, Häussler S, Wiehlmann L, Rohde C, Sikorski J. 2012. Pseudomonas aeruginosa population structure revisited under environmental focus: impact of water quality and phage pressure. Environ. Microbiol. 14:1952–1967 [DOI] [PubMed] [Google Scholar]
- 3. Waters V, Zlosnik JE, Yau YC, Speert DP, Aaron SD, Guttman DS. 2012. Comparison of three typing methods for Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 31:3341–3350 [DOI] [PubMed] [Google Scholar]
- 4. Tramper-Stranders GA, van der Ent CK, Molin S, Yang L, Hansen SK, Rau MH, Ciofu O, Johansen HK, Wolfs TF. 2012. Initial Pseudomonas aeruginosa infection in patients with cystic fibrosis: characteristics of eradicated and persistent isolates. Clin. Microbiol. Infect. 18:567–574 [DOI] [PubMed] [Google Scholar]
- 5. van Mansfeld R, Jongerden I, Bootsma M, Buiting A, Bonten M, Willems R. 2010. The population genetics of Pseudomonas aeruginosa isolates from different patient populations exhibits high-level host specificity. PLoS One 5:e13482. 10.1371/journal.pone.0013482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng K, Smyth RL, Govan JR, Doherty C, Winstanley C, Denning N, Heaf DP, van Saene H, Hart CA. 1996. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 348:639–642 [DOI] [PubMed] [Google Scholar]
- 7. Johansen HK, Moskowitz SM, Ciofu O, Pressler T, Høiby N. 2008. Spread of colistin resistant non-mucoid Pseudomonas aeruginosa among chronically infected Danish cystic fibrosis patients. J. Cyst. Fibros. 7:391–397 [DOI] [PubMed] [Google Scholar]
- 8. Jones AM, Govan JR, Doherty CJ, Dodd ME, Isalska BJ, Stanbridge TN, Webb AK. 2001. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet 358:557–558 [DOI] [PubMed] [Google Scholar]
- 9. García-Castillo M, Máiz L, Morosini MI, Rodríguez-Baños M, Suarez L, Fernández-Olmos A, Baquero F, Cantón R, del Campo R. 2012. Emergence of an mutL mutation causing multilocus sequence typing–pulsed-field gel electrophoresis discrepancy among Pseudomonas aeruginosa isolates from a cystic fibrosis patient. J. Clin. Microbiol. 50:1777–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cantón R, Cobos N, de Gracia J, Baquero F, Honorato J, Gartner S, Alvarez A, Salcedo A, Oliver A, García-Quetglas E; Spanish Consensus Group for Antimicrobial Therapy in the Cystic Fibrosis Patient 2005. Antimicrobial therapy for pulmonary pathogenic colonization and infection by Pseudomonas aeruginosa in cystic fibrosis patients. Clin. Microbiol. Infect. 11:690–703 [DOI] [PubMed] [Google Scholar]
- 11. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng L, Connor TR, Aanensen DM, Spratt BG, Corander J. 2011. Bayesian semi-supervised classification of bacterial samples using MLST databases. BMC Bioinformatics 12:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. M100-S22 CLSI, Wayne, PA [Google Scholar]
- 14. Cabot G, Ocampo-Sosa AA, Domínguez MA, Gago JF, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A; Spanish Network for Research in Infectious Diseases (REIPI) 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob. Agents Chemother. 56:6349–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 10:841–851 [DOI] [PubMed] [Google Scholar]
- 16. Lorè NI, Cigana C, De Fino I, Riva C, Juhas M, Schwager S, Eberl L, Bragonzi A. 2012. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS One 7:e35648. 10.1371/journal.pone.0035648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oliver A, Mena A. 2010. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin. Microbiol. Infect. 16:798–808 [DOI] [PubMed] [Google Scholar]
- 18. Rojo-Molinero E, López-Causapé C, Cabot G, Mulet X, Mena A, Pérez JL, Oliver A. 2012. Clonal epidemiology and resistance evolution in Pseudomonas aeruginosa strains colonising the respiratory tract of cystic fibrosis patients from the Balearic Islands. Clin. Microbiol. Infect. 18(Suppl S3):57521958149 [Google Scholar]
- 19. Kerem E, Conway S, Elborn S, Heijerman H; Consensus Committee 2005. Standards of care for patients with cystic fibrosis: a European consensus. J. Cyst. Fibros. 4:7–26 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.