Abstract
In September 2012, the Centers for Disease Control and Prevention (CDC) initiated an outbreak investigation of fungal infections linked to injection of contaminated methylprednisolone acetate (MPA). Between 2 October 2012 and 14 February 2013, the CDC laboratory received 799 fungal isolates or human specimens, including cerebrospinal fluid (CSF), synovial fluid, and abscess tissue, from 469 case patients in 19 states. A novel broad-range PCR assay and DNA sequencing were used to evaluate these specimens. Although Aspergillus fumigatus was recovered from the index case, Exserohilum rostratum was the primary pathogen in this outbreak and was also confirmed from unopened MPA vials. Exserohilum rostratum was detected or confirmed in 191 specimens or isolates from 150 case patients, primarily from Michigan (n = 67 patients), Tennessee (n = 26), Virginia (n = 20), and Indiana (n = 16). Positive specimens from Michigan were primarily abscess tissues, while positive specimens from Tennessee, Virginia, and Indiana were primarily CSF. E. rostratum antifungal susceptibility MIC50 and MIC90 values were determined for voriconazole (1 and 2 μg/ml, respectively), itraconazole (0.5 and 1 μg/ml), posaconazole (0.5 and 1 μg/ml), isavuconazole (4 and 4 μg/ml), and amphotericin B (0.25 and 0.5 μg/ml). Thirteen other mold species were identified among case patients, and four other fungal genera were isolated from the implicated MPA vials. The clinical significance of these other fungal species remains under investigation. The laboratory response provided significant support to case confirmation, enabled linkage between clinical isolates and injected vials of MPA, and described significant features of the fungal agents involved in this large multistate outbreak.
INTRODUCTION
Back and joint injection of glucocorticoids for pain management is widespread, and complications from these injections are uncommonly reported (1, 2). Outbreaks of infection associated with epidural injections are rare, generally manifest as meningitis or epidural abscesses, and are due mainly to bacterial pathogens (1–4). Injection-associated infections due to fungal pathogens are exceedingly rare (5–8).
In September 2012, the Centers for Disease Control and Prevention (CDC) was notified of several cases of meningitis in Tennessee, including one patient with cerebrospinal fluid (CSF) culture positive for Aspergillus fumigatus. Subsequent investigation by a joint team of local, state, and federal health agencies and health care providers revealed a multistate outbreak of fungal infections, primarily Exserohilum rostratum, caused by epidural, paraspinal, and peripheral joint injections of fungus-contaminated lots of methylprednisolone acetate (MPA) produced by the New England Compounding Center (NECC), Framingham, MA (9–14). The majority of early case patients had meningitis or stroke, but later, cases of spinal or paraspinal focal infection at the injection site predominated.
Reliable and well-validated diagnostic assays for these fungal agents largely did not exist prior to this outbreak. When E. rostratum was identified as the primary causative agent, a real-time PCR assay was described (15), and fungal β-d-glucan testing was also investigated as potentially useful for diagnostic purposes (16). During the investigation, the CDC functioned as a reference testing laboratory for patient specimens associated with the outbreak and developed a novel PCR-based detection test with broad-range fungal primers (17). Here, we describe laboratory findings, including results for 150 case patients with specimens positive for E. rostratum, identification of other molds from case patient samples, antifungal susceptibility testing on 50 E. rostratum patients and vial isolates, culture of unopened MPA vials from the three implicated lots, and a correlation of CSF white blood cells (WBC) with the recovery of Exserohilum or its DNA. We also describe the challenges and limitations associated with an outbreak investigation of this magnitude.
MATERIALS AND METHODS
Specimens received.
This report includes specimens received between 2 October 2012 and 14 February 2013. Specimens could be grouped into three broad categories: body fluids, consisting of mostly cerebrospinal fluid (CSF), epidural fluid, and synovial fluid; tissues, primarily from biopsy but also from autopsy; and fungal isolates received from specimens cultured locally. Urine, blood, and serum samples were not included in the analysis. Specimens were shipped to the CDC from state public health laboratories. Fluids were shipped on dry ice. Specimens were processed immediately or stored frozen at −70°C until being processed. Primary bacterial and fungal culture was performed at the submitting institutions, and available fungal isolates were submitted to the CDC for confirmation of identification. Bacterial and fungal isolates recovered from unopened vials of MPA were also submitted for confirmation by the Food and Drug Administration and several state public health laboratories.
Human subjects.
This investigation was considered an emergent public health response and therefore was not subject to review by the CDC's Institutional Review Board.
Case definitions.
A probable case was defined as development of any of the following: meningitis of unknown etiology following epidural or paraspinal injection after 21 May 2012; posterior circulation stroke without a cardioembolic source and without documentation of a normal CSF profile following epidural or paraspinal injection after 21 May 2012; osteomyelitis, abscess, or other infection (e.g., soft tissue infection) of unknown etiology in the spinal or paraspinal structures at or near the site of injection following epidural or paraspinal injection after 21 May 2012; or osteomyelitis or worsening inflammatory arthritis of a peripheral joint (e.g., knee, shoulder, or ankle) of unknown etiology diagnosed following joint injection after 21 May 2012 in a person who received an injection with preservative-free MPA that definitely or likely came from one of the following three lots produced by the New England Compounding Center (NECC): 05212012@68, 06292012@26, 08102012@51. A proven case was defined as a probable case with evidence (by culture, histopathology, or molecular assay) of a fungal pathogen associated with the clinical syndrome.
Isolate processing.
Fungal isolates were identified using polyphasic methods, including morphological analysis and DNA sequencing. Genomic DNA was prepared and used for amplification and sequencing of internal transcribed spacer 1 (ITS1) and ITS2 regions of ribosomal DNA (rDNA) and the D1/D2 regions of 28S rDNA as previously described (18, 19). A portion of the beta-tubulin gene was also used to identify Aspergillus isolates to the species level (18). Identification of other species was made by comparing ITS and D1/D2 identity using the BLAST function in the GenBank and Centraalbureau voor Schimmelcultures (CBS) medical fungi databases (http://www.cbs.knaw.nl/medical/BioloMICS.aspx?searchopt=1; accessed 7 October 2012). We used a 100% cutoff for species identity and 98% for genus identity, although there are no recognized cutoffs for fungal identification (20). Slide culture was performed using Pablum cereal agar (21) or V8 juice agar (22) on dematiaceous isolates and Czapek's agar on presumptive Aspergillus isolates. Morphological identification was made using the methods of de Hoog et al. (23). Morphological and DNA-based identifications were correlated to generate the final identification for each isolate. Bacterial isolates were identified in the CDC Clinical and Environmental Microbiology Laboratory using DNA sequencing and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (24, 25).
Processing of body fluids.
Body fluids were processed by the methodology of Gade et al. (17). Briefly, cells and particulate matter were pelleted by centrifugation and removed. Free DNA was captured using the QIAamp UltraSens virus kit (catalog number 53704; Qiagen, Hilden, Germany) according to the manufacturer's instructions with modifications as described. Fungal DNA was amplified, sequenced, and identified to the species level as described previously (17).
Processing of fresh-frozen and formalin-fixed paraffin-embedded tissue.
Both fresh-frozen and paraffin-embedded tissues were received from a variety of sources, including brain, meninges, epidural tissue, epidural abscess debridement, cyst, and bone. Several types of tissue samples were received: (i) large samples of fresh-frozen intact tissue (≥0.5 cm in diameter), (ii) formalin-fixed paraffin-embedded (FFPE) tissue blocks, (iii) small samples of fresh-frozen intact tissue (≤0.5 cm in diameter), (iv) tissue fragments in sterile saline, from larger tissue biopsy specimens that had been minced in the submitting laboratory. Three different approaches were used for processing tissue specimens. For tissue sample types i and ii, large samples of fresh-frozen tissues as well as formalin-fixed tissues were forwarded to the CDC Infectious Disease Pathology Branch (IDPB) for fixation and immunohistochemical staining (25a). Thin (5-μm) sections were cut from blocks positive by immunohistochemistry and were processed for fungal DNA detection in the CDC Fungus Reference Laboratory as previously described (26). For type iii, small samples of fresh frozen tissues were processed as described previously (17). For type iv, tubes containing tissue fragments sent in sterile saline were centrifuged, and both pellets and supernatants were processed. Pellets containing tissue fragments were processed using the protocol for extracting DNA from fresh tissues (tissue type iii). Supernatants were processed using the body fluid protocol described above.
Susceptibility testing.
Susceptibility testing was performed according to the guidelines of the Clinical and Laboratory Standards Institute document M38-A2 (27). Itraconazole, voriconazole, and posaconazole were part of a custom frozen panel (TREK Diagnostics, Cleveland, OH). Isavuconazole was kindly provided by Astellas Pharma. Isavuconazole powder was diluted in dimethyl sulfoxide (DMSO) and prepared according to M38-A2 (27). The concentration range for each antifungal agent was 0.008 μg/ml to 16 μg/ml. Exserohilum rostratum isolates were grown for 8 to 10 days on V8 juice agar (22) to induce conidial growth. On the sixth day of growth, 0.5 ml of sterile water was added to the tube. The increased humidity increased conidium production, and the water helped dislodge conidia. Prior to inoculation to susceptibility testing plates, the titers of conidia were determined to an optical density at 530 nm (OD530) of 0.25 to 0.3, as suggested for the closely related genera Bipolaris and Alternaria. Plates were incubated at 30°C for 48 h. Endpoints were determined visually, and MICs were read as the lowest drug concentration that prevented any discernible growth for itraconazole, voriconazole, posaconazole, and isavuconazole. Amphotericin B MIC values were determined using Etest strips according to the manufacturer's instructions for testing yeast, since this method has not been approved by the U.S. Food and Drug Administration for testing molds (bioMérieux, Durham, NC). Because of the slow growth of the organism, the Etest could not be read until 72 to 96 h after plating. The MIC value was read at the point of complete inhibition of fungal growth. MIC50, MIC90, and modal (most common) MIC values were calculated. Quality control isolates Candida parapsilosis ATCC 22019, Candida krusei ATCC 6258, Aspergillus fumigatus ATCC MYA 3626, and Paecilomyces variotii ATCC MYA 3630 were tested with each batch of isolates.
Data collection and analysis.
Case patient demographic and clinical data were collected using a standardized case report form developed for the outbreak, and data were entered into a Microsoft Access 2010 database. All analyses were performed using SAS version 9.3 (SAS Institute, Cary, NC). A P value of <0.05 was considered statistically significant. We used a Student t test to examine the association between white blood cell count at initial lumbar puncture and the presence of E. rostratum by either PCR, culture, or histopathology.
RESULTS
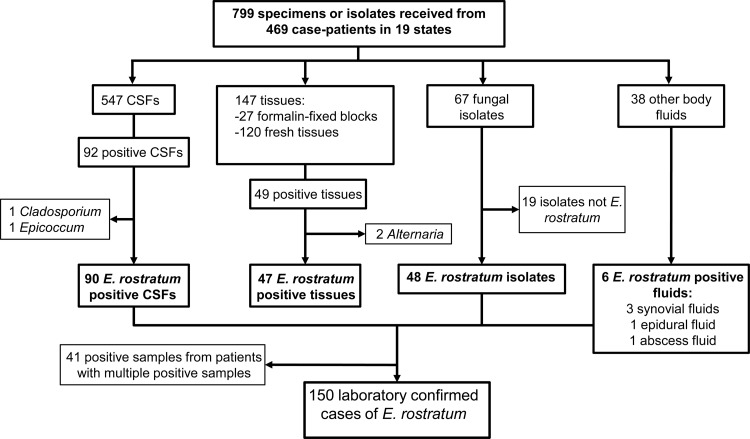
By 14 February 2013, 704 proven or probable cases of fungal infection associated with contaminated steroid injections had been reported to the CDC. Between 2 October and 14 February, 799 specimens from 469 case patients (67% of reported case patients) were received by the CDC Fungus Reference Laboratory (Table 1; Fig. 1). Specimens included 547 CSF samples (68% of specimens received) from 350 case patients, 67 fungal isolates (8% of specimens received) from 64 patients, 120 fresh-frozen tissue samples (15% of specimens received) from 91 patients, 27 FFPE tissue samples (3% of specimens received) from 25 patients, and 38 other body fluid samples, including synovial fluid and epidural fluid samples (5% of specimens received) from 29 patients (Table 1). The types of specimens received changed over the course of the outbreak: during the first 36 days of the investigation, 85% of the samples were CSF samples, 7% were isolates, and 6% were tissue samples. During the next 99 days, the CSF proportion decreased to 47%, while tissue samples increased 6-fold to 33%.
Table 1.
Samples received by state and sample type
| State | No. of case patients with specimens | No. of specimens | No. of specimens by type |
|||
|---|---|---|---|---|---|---|
| CSF | Tissue | Isolate | Othera | |||
| Florida | 23 | 30 | 26 | 2 | 1 | 1 |
| Georgia | 1 | 1 | 1 | |||
| Idaho | 1 | 1 | 1 | |||
| Illinois | 2 | 3 | 3 | |||
| Indiana | 58 | 80 | 62 | 3 | 8 | 7 |
| Maryland | 22 | 37 | 33 | 1 | 3 | |
| Michigan | 142 | 207 | 53 | 109 | 29 | 16 |
| Minnesota | 11 | 18 | 12 | 2 | 2 | 2 |
| North Carolina | 6 | 12 | 7 | 5 | ||
| New Hampshire | 13 | 20 | 12 | 8 | ||
| New Jersey | 42 | 47 | 45 | 2 | ||
| New York | 0 | |||||
| Ohio | 19 | 22 | 19 | 3 | ||
| Pennsylvania | 1 | 1 | 1 | |||
| Rhode Island | 3 | 4 | 3 | 1 | ||
| South Carolina | 1 | 1 | 1 | |||
| Tennessee | 72 | 110 | 78 | 15 | 14 | 3 |
| Texas | 2 | 3 | 3 | |||
| Virginia | 49 | 201 | 186 | 5 | 10 | |
| West Virginia | 1 | 1 | 1 | |||
| Total | 469 | 799 | 547 | 147 | 67 | 38 |
Other specimens included synovial fluids, epidural fluids, and abscess fluids.
Fig 1.
Flow chart of isolate and specimen testing results at the CDC. This flow chart documents the isolate testing at the CDC and includes all of the fungal results.
Of the case patients with submitted specimens, 220 (47%) met the case definition for meningitis, 116 (25%) for meningitis and spinal or paraspinal infection, 110 (23%) for spinal or paraspinal infection only, 21 (4%) for peripheral joint infection only, one for abscess and peripheral joint infection, and one for stroke. The median age of case patients was 64 years (range, 16 to 97 years), 59% were female, and 41% were male. Specimens were received from 19 of the 20 states (95%) that reported cases. Michigan, Tennessee, Indiana, and Virginia reported 72% of the total cases and submitted 75% of total CDC specimens, representing 64% of their case patients.
As of 14 February 2013, fungus had been detected, confirmed, or isolated in 213 specimens from 166 patients from 12 states, which represented 35% of patients from whom specimens were submitted. Exserohilum rostratum was confirmed in one or more specimens from 150 case patients (23% of all case patients), including 90 CSF samples, 48 isolates, 37 fresh-frozen tissue samples, 10 FFPE tissue samples, four synovial fluid samples, one abscess fluid sample, and one epidural fluid sample (Table 2). Nineteen case patients had samples positive for another fungus: these were all isolated from culture, except in the case of two CSF samples in which Cladosporium sp. DNA and Epicoccum sp. DNA were detected by PCR. Several case patient samples showed discordant results, including one where E. rostratum DNA was detected in CSF but Alternaria alternata was isolated from culture of the same CSF, another where E. rostratum DNA was detected in CSF but Cladosporium cladosporioides was isolated in culture of CSF collected on a different date, two patients who each had two different fungal species cultured from CSF on different days, and one patient who had two different fungi cultured from a single sample.
Table 2.
Isolates and specimens positive for E. rostratum
| Sample type | Total no. of specimens submitted | No. of specimens positive for E. rostratum | No. of patients with E. rostratum-positive specimensb |
|---|---|---|---|
| Total | 799 | 191 | 150 |
| CSF | 547 | 90 | 82 |
| Fresh tissue | 120 | 37 | 36 |
| FFPE | 27 | 10 | 9 |
| Isolates | 67 | 48 | 48 |
| Other specimensa | 38 | 6 | 6 |
Other specimens included three synovial fluids, one epidural fluid, and one abscess fluid.
Some case patients had more than one specimen type that was positive.
CSF.
There were 547 CSF samples submitted from 350 case patients. Fungal DNA was detected in 92 CSF samples from 84 case patients. E. rostratum DNA was detected in 90 CSF samples from 82 case patients (23% of the case patients from whom a CSF sample was submitted), Cladosporium DNA in one CSF sample from one case patient, and Epicoccum DNA in one CSF sample from one case patient. These positive CSF samples represented 47% of total positive specimens. E. rostratum DNA was detected in specimens submitted from 12 different states, mostly Virginia, Tennessee, and Michigan, which submitted 22, 21, and 18 positive CSF samples from 18, 20, and 17 case patients, respectively (Table 3). No CSF collected after 13 November was positive for E. rostratum. As a negative control, CSF from 136 patients initially suspected to have fungal meningitis but later shown to have no white cells in their CSF were tested and were all negative by PCR (17).
Table 3.
Isolates and specimens positive for E. rostratum by state and sample type
| State | No. of case patients with specimens | No. of positive case patients (%) | Positive specimens by typea |
|||
|---|---|---|---|---|---|---|
| CSF | Tissue | Isolate | Otherb | |||
| Florida | 23 | 2 (9) | 2 | |||
| Indiana | 58 | 16 (28) | 14 | 1 | 5 | 1 |
| Maryland | 22 | 6 (27) | 5 | 3 | ||
| Michigan | 142 | 67 (47) | 18 | 36 | 25 | 2 |
| Minnesota | 11 | 1 (9) | 1 | |||
| North Carolina | 6 | 2 (33) | 2 | |||
| New Hampshire | 13 | 1 (8) | 1 | |||
| New Jersey | 42 | 4 (10) | 3 | 1 | ||
| Ohio | 19 | 4 (21) | 3 | 3 | ||
| Rhode Island | 3 | 2 (66) | 1 | 1 | ||
| Tennessee | 72 | 26 (36) | 21 | 3 | 10 | 1 |
| Virginia | 49 | 20 (41) | 22 | 1 | 5 | |
| Total | 468c | 151 (32) | 90 | 47 | 48 | 6 |
Some case patients had more than one specimen type that was positive.
Other specimens included three synovial fluids, one epidural fluid, and one abscess fluid.
States with no positive case patients were not included in this table.
Other body fluids.
There were 38 other body fluid samples submitted from 29 patients in seven states, including 25 synovial fluid samples, 11 spinal or paraspinal aspirate samples, and two epidural fluid samples. E. rostratum DNA was detected in 6 fluid samples from six patients in five states, including four synovial fluid samples, one epidural fluid sample, and one spinal or paraspinal aspirate sample.
Tissue.
There were 147 tissue samples, including 27 FFPE tissue samples (type ii), 31 fresh tissue samples that did not arrive in saline (type iii), and 89 tissue fragments received in sterile saline (type iv), from 108 case patients in 10 states (Tables 2 and 3). The majority of tissue samples were received from 84 Michigan case patients. E. rostratum DNA was detected in 47 tissue samples (33%), including 10 FFPE tissue samples, 8 type iii fresh tissue samples, and 29 type iv tissue fragment samples, from 45 case patients in seven states. Fungal elements were identified using histopathology and immunohistochemistry in 21 of the 27 FFPE tissue samples received (Ritter et al., unpublished); PCR testing was performed on all 21, but fungal DNA could not be amplified in 11. In the 29 type iv samples, 26 supernatants and 9 tissue pellets contained detectable Exserohilum DNA, but only six samples had both a positive supernatant and a positive tissue pellet. Alternaria DNA was detected in one saline tissue supernatant and one FFPE specimen from different patients. The FFPE sample was negative for fungus by immunohistochemistry, suggesting possible contamination.
Fungal isolates.
Forty-eight isolates submitted from 48 cases in five states were confirmed as E. rostratum. All of these isolates demonstrated 100% sequence identity at the ITS1 and ITS2 loci and displayed a 100% match to 13 E. rostratum sequences in the GenBank database across the 601 nucleotides sequenced at the ITS1 and ITS2 loci. The D1/D2 locus was less informative. The D1/D2 isolate sequences had 100% identity with the single E. rostratum isolate sequence in the GenBank database; however, in the validated CBS medical fungi database, 11 different species of fungi, including E. rostratum, demonstrated 100% D1/D2 sequence identity to the outbreak E. rostratum sequences. All of the E. rostratum isolates grew well at 37°C and produced conidia within 8 to 12 days on V8 juice agar and within 2 weeks on Pablum agar. Conidia had 6 to 12 distosepta, with an average of 10, and displayed a prominent hilum, making these isolates most similar morphologically to Exserohilum longirostratum, a species that has recently been placed into conspecificity with E. rostratum (28).
Nineteen non-E. rostratum isolates were also received from CSF samples that were cultured in the state of origin (Table 4). Cladosporium cladosporioides was recovered from four case patients in two states. The ITS1 and ITS2 sequences for the four isolates were identical to one another and differed at ITS2 by a single nucleotide from the Cladosporium DNA amplified from the single CSF sample described above. The C. cladosporioides isolates did not grow above 30°C in vitro. The only other species recovered multiple times was Aspergillus terreus, which was recovered from three patients in Indiana, and Alternaria alternata, recovered from two patients in Michigan. Isolates of 10 additional fungal species were submitted from various body sites, each as singlets from a single state. With the exception of the A. alternata and the C. cladosporioides isolates mentioned above, all of the patients with non-E. rostratum isolates had one or more CSF specimens from which fungal DNA was not detected by PCR.
Table 4.
Non-Exserohilum fungal isolates from case patients submitted for confirmation
| Species | No. of isolates | State of origin for isolates |
|---|---|---|
| Alternaria alternataa | 2 | MI |
| Aspergillus fumigatus | 1 | TN |
| Aspergillus terreus | 3 | IN |
| Aspergillus tubingensis | 1 | TN |
| Chaetomium sp. | 1 | MI |
| Cladosporium cladosporioidesb | 4 | VA, MN |
| Cladosporium sp. | 1 | TN |
| Epicoccum nigrum | 1 | VA |
| Paecilomyces niveus | 1 | MI |
| Penicillium paneum | 1 | VA |
| Scopulariopsis brevicaulisc | 1 | MN |
| Stachybotrys chartarumd | 1 | MD |
| Nonspeciated ascomycete | 1 | FL |
CSF with the same collection date was positive for E. rostratum DNA by PCR.
One of these isolates came from a patient with a positive E. rostratum PCR result from a different CSF sample.
This isolate was one colony on a culture plate that also grew C. cladosporioides.
This isolate was recovered as a single colony on one culture plate.
For those patients with an E. rostratum isolate and at least one specimen positive by PCR, the most common combination was an E. rostratum isolate and a positive CSF PCR (n = 16 patients). Eleven of these case patients had an isolate from the same CSF that was positive by PCR, while five had the positive PCR and the positive isolate from different CSF specimens. For tissues, nine patients had both an E. rostratum isolate and a positive PCR from the corresponding tissue. It is unknown whether local cultures from other tissue specimens were negative or whether culture was not attempted.
MPA vials.
Isolates from unopened MPA vials cultured at the Food and Drug Administration or other local institutions were received for culture confirmation (Table 5). Exserohilum rostratum and the basidiomycetous yeast Rhodotorula laryngis were confirmed from lot numbers 08102012@51 and 06292012@26. Rhizopus stolonifer was confirmed in one vial of lot number 08102012@51. E. rostratum, Bacillus subtilis, and Bacillus pumilus were confirmed from one vial of lot number 08122012@51 that was cultured locally in Idaho and submitted for confirmation from the Idaho state public health laboratory. In addition to E. rostratum, the CDC cultured and recovered Cladosporium cladosporioides from one unopened vial of lot number 08122012@51 sent from the Tennessee public health laboratory. Paecilomyces formosus was reconfirmed at the CDC from lot number 05212012@68; 20 vials were cultured and identified at the Wadsworth Center, New York state public health laboratory. The Wadsworth Center also reported to the CDC that Exserohilum DNA was detected in one vial of this lot using a real-time PCR assay (S. Chaturvedi, unpublished data).
Table 5.
Viable bacteria and fungi recovered from unopened MPA vials
| Lot no. | Organisms |
|---|---|
| 05212012@68 | Paecilomyces formosus, Exserohilum rostratuma |
| 06292012@26 | Exserohilum rostratum, Rhodotorula laryngis |
| 08102012@51 | Exserohilum rostratum, Cladosporium cladosporioides, Bacillus subtilis, Bacillus pumilus, Rhodotorula larynges, Rhizopus stolonifer |
E. rostratum DNA was detected in one vial from this lot.
For the five patients from whom Cladosporium cladosporioides was recovered or detected, an attempt was made to correlate the lot number(s) of the vial(s) used for patient injection with the lot number of the culture-positive vial. Three of the patients received an injection from a lot other than the 08122012@51 lot, from which viable Cladosporium was recovered. In the other two cases, the MPA lot number injected was not available. One of these patients received two injections.
Susceptibility testing.
A total of 40 nonduplicative case patient isolates of E. rostratum and 10 additional E. rostratum isolates recovered from MPA vials were tested for antifungal susceptibility to amphotericin B, itraconazole, voriconazole, posaconazole, and isavuconazole (Table 6). Voriconazole had an MIC50 value of 1 μg/ml and an MIC90 value of 2 μg/ml and was bimodal, with modes of 1 to 2 μg/ml. Itraconazole and posaconazole both had an MIC50 value of 0.5 μg/ml, an MIC90 value of 1 μg/ml, and a mode of 0.5 μg/ml. All isolates showed the same result for isavuconazole, with an MIC50, an MIC90, and a mode of 4 μg/ml, except for two, which had an MIC of 2 μg/ml.
Table 6.
MICs for E. rostratum isolates from patients and MPA drug vials
| Antifungal agent (no. of isolates tested) | MIC (μg/ml) at 48 to 72 h |
|||
|---|---|---|---|---|
| Range | 50% | 90% | Mode | |
| Voriconazole (50) | 1–2 | 1 | 2 | 1–2 |
| Itraconazole (50) | 0.25–4 | 0.5 | 1 | 0.5 |
| Posaconazole (50) | 0.25–1 | 0.5 | 1 | 0.5 |
| Isavuconazole (48) | 2–4 | 4 | 4 | 4 |
| Amphotericin B (49) | 0.032–2 | 0.25 | 0.5 | 0.38 |
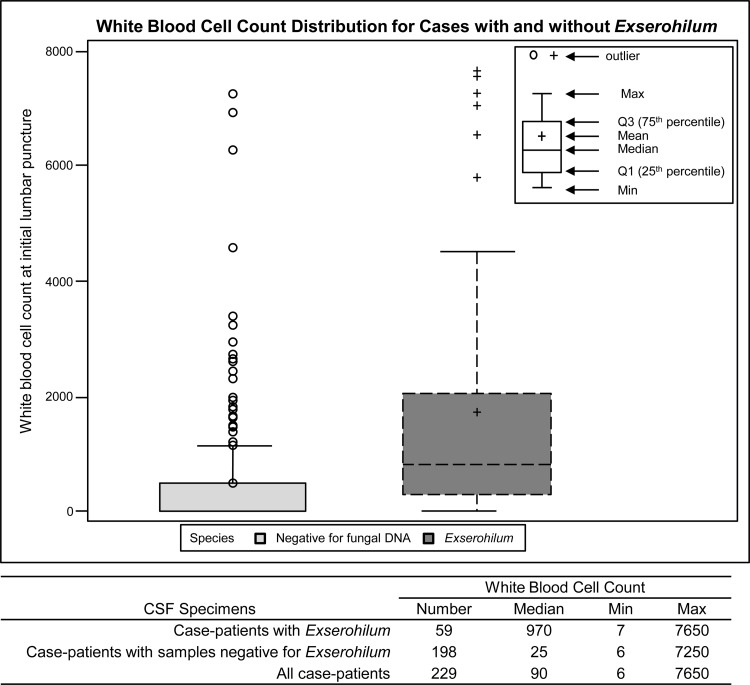
E. rostratum CSF positivity correlated with CSF white blood cell count.
The CSF samples with E. rostratum DNA detected by PCR or isolated in culture had significantly higher WBC counts (median, 970 WBCs) than samples from which fungi could not be detected or isolated (median, 25 WBCs). This difference was statistically significant (P < 0.001) (Fig. 2). Some case patients with specimens negative for fungal DNA had elevated WBC counts; however, 14 of the 18 CSF samples with WBC counts of >1,000 and negative for E. rostratum were collected prior to the date when CDC recommended that CSF to be tested by PCR should be frozen prior to and during shipment. Following the preexisting general laboratory practice, these samples may have been stored and shipped at 4°C or warmer, and fungal DNA may have been degraded.
Fig 2.
Box and whisker plot representing the white blood cell count distribution in CSF specimens of meningitis case patients at initial lumbar puncture, with and without E. rostratum. The box represents the 25th and 75th percentiles of WBC, the bars represent the maximum and minimum observations, the line within the box is the median, and the circles are outliers from the expected mean.
DISCUSSION
The largest health care-associated outbreak of fungal infections reported in the United States involved the nationwide distribution and administration of three lots of a contaminated steroid medication. The manuscript describes the laboratory investigations associated with this large multistate outbreak. The laboratory response was important in order to confirm fungal infection, to identify the species of fungi involved, to link clinical samples to isolates recovered from unopened medication vials, and to examine characteristics of the implicated fungal organisms, such as antifungal susceptibility. The prominent fungal organism identified in patients was the mold Exserohilum rostratum, detected in or recovered in culture from specimens from 150 case patients (32% of case patients with submitted specimens), all associated with injections of the contaminated steroid medication. This organism was also recovered in cultures from unopened vials of two of the three implicated medication lots (13) (Table 5). The second most common fungal species was another environmental mold, Cladosporium cladosporioides, cultured from four patient isolates, detected in one CSF sample, and recovered from one unopened vial of one implicated MPA lot, although not the same lot as those received by some of the patients from whom C. cladosporioides was recovered.
One of the more challenging laboratory questions associated with this outbreak has been attempting to determine whether the non-Exserohilum organisms were also causative agents of injection-associated infections. Thirteen mold species other than Exserohilum were either recovered in culture or detected by PCR from patient samples (Table 4). Furthermore, several nonpathogenic fungi were cultured from a small portion of the approximately 14,000 contaminated vials that were distributed from May to August 2012 (Table 5). It is likely that a variety of fungal organisms may have been injected into patients but not recovered in the culturing of remnant vials that occurred after the outbreak was detected. For example, the outbreak index case died from a stroke, with Aspergillus fumigatus recovered in cultures of CSF and brain tissue (11); however, this organism was never recovered from any vials. It is also possible that “nonpathogenic” fungi may have provoked an inflammatory response when injected into human tissue, thus fulfilling the case definition of MPA injection, consistent symptoms, and CSF pleocytosis (29). On the other hand, environmental molds are well known as a source of laboratory contamination associated with specimen collection or plating. Their recovery from patient samples could have been coincidental and unrelated to this outbreak. It remains difficult to distinguish among these possibilities in interpreting the results related to non-Exserohilum molds. We did find a strong correlation between high numbers of CSF white cells (WBC counts of >5 cells) and detection of Exserohilum either by PCR or in culture (Fig. 2).
The decision early in the investigation to develop a broad-range PCR assay followed by sequencing allowed us to detect and identify any fungal agents present rather than focusing on a single agent. This was important in the early outbreak response, where the finding of Aspergillus fumigatus in the index case suggested that this may have been an outbreak of aspergillosis. Our results also suggest that in this outbreak, much of the fungal DNA was present in a free state unassociated with an intact fungal cell. Prompt freezing of clinical samples and a protocol designed to concentrate viral DNA from fluids allowed us to take advantage of this feature. Many of the clinical samples collected at the beginning of the outbreak response were convenience samples that had been stored for some time and not frozen, which might have contributed to false-negative results due to less-than-optimal specimen handling. Our rate of positive results increased as the outbreak progressed (data not shown), possibly due in part to better quality samples and better attention to shipping and handling conditions.
The receipt of specimens followed the biphasic clinical course reported for this outbreak (T. M. Chiller, M. Roy, D. Nguyen, A. Guh, A. N. Malani, R. Latham, S. Peglow, T. Kerkering, D. Kaufman, J. McFadden, J. Collins, M. Kainer, J. Duwve, D. Trump, C. Blackmore, C. Tan, A. A. Cleveland, T. MacCannell, A. Muehlenbachs, S. Zaki, M. E. Brandt, and J. Jernigan, submitted for publication). At the onset of the outbreak (September 2012), the primary reported syndrome was meningitis (13) and the predominant positive sample was CSF. However, as the outbreak progressed, syndromes such as joint infections, phlegmon, and abscesses became more common and the number of positive tissues had risen dramatically. This result is consistent with the prediction of the decision analysis model developed for this investigation (30), which suggested that the period of greatest risk for developing meningitis was during the first 6 weeks (42 days) after receiving an injection of contaminated MPA. The laboratory findings also track the different patterns of infection that were demonstrated in different states. Most of the samples were submitted from four states (Virginia, Michigan, Tennessee, and Indiana). Most of the tissue samples came from Michigan, while the other three states submitted primarily CSF and other fluid samples.
The azole MIC values for these outbreak isolates are higher than those recently reported for E. rostratum (28), but these drugs were still considered to demonstrate good activity by clinical experts (30). The itraconazole and posaconazole range reported by da Cunha and colleagues (28) was <0.03 to 0.125 μg/ml, while for voriconazole it was slightly higher at <0.03 to 1 μg/ml. Although there are no reported results for isavuconazole against E. rostratum, there are results for the closely related species Curvularia lunata (range, 1 to 4 μg/ml) and Alternaria alternata (range, 0.5 to 2 μg/ml) (31), which are very close to our sole value of 4 μg/ml for E. rostratum for case patient isolates and this drug. Without either susceptibility breakpoints or outcome data, the MIC values are difficult to interpret. Another challenge in this response was the need for conidia to conduct CLSI-standardized susceptibility testing. In the laboratory, Exserohilum sporulated slowly, taking 5 to 8 days to produce conidia, even on plant-based media such as Pablum and V8 juice agars.
There are several limitations to this investigation. As is typical in an urgent outbreak response, some supporting data and specimens are missing or were not collected locally. It is difficult to correlate MIC values with treatment outcome, in part because outcome data are still being collected, patients are still being treated, and the duration of continuing therapy is unknown (30). Because we received samples only from patients who met the outbreak case definition, we did not collect CSF samples from patients not involved in the outbreak to calculate the occurrence of false-positive PCRs. We did, however, find negative PCR results in CSF from 136 patients initially investigated as cases but later found not to be associated with the outbreak (17).
During this complex investigation with over 13,000 potentially infected individuals, the CDC laboratory served as a central reference for molecular detection, confirmation, and susceptibility testing of the primary etiological agent of infection as well as other fungi epidemiologically linked to symptomatic patients. With the assistance of state and local public health laboratories, we were able to correlate the presence of viable Exserohilum rostratum fungi or DNA in unopened MPA vials with 48 patients who met the outbreak case definition and had viable Exserohilum isolated in culture from a compatible body site, 129 patients who met the outbreak case definition and had PCR-based evidence for Exserohilum from a compatible body site, and 27 patients who met the outbreak case definition and showed both a positive culture and positive DNA findings from a compatible body site. Other viable fungi were recovered from contaminated vials, and we assume that they were injected into at least some patients, with unknown results. The outbreak has stimulated discussion of lessons and directions for future research related to these infections (29, 32).
ACKNOWLEDGMENTS
We thank all participants in the Multistate Fungal Infection Outbreak Response Team, especially state and local health department officials, clinicians, and health care professionals who provided clinical samples and data for this study. We thank all members of the CDC Mycotic Diseases Branch and CDC emergency response volunteers for their help with receiving, processing, and cataloging clinical samples, especially Nina Grossman, Joyce Peterson, Carol Bolden, Ngoc Le, Shirley McClinton, Randy Kuykendall, Steve Hurst, Kizee Etienne, Noelle Bessette, Grant Williams, Paris Paul, Sarah Yi, Laura Dickmeyer, Jessica Halpin, Heather O'Connell, and James Graziano of the Bacterial Special Pathogens Branch. We also acknowledge the collaboration of Sherif Zaki and colleagues in the Infectious Diseases Pathology Branch and the staff of the Specimen Management Branch, CDC. We recognize our collaborators at the Food and Drug Administration, Office of Regulatory Affairs, Regional Field Laboratories, especially Michael J. Palmieri, the Wadsworth Center (New York state public health laboratory), the Tennessee state public health laboratory, and the Idaho state public health laboratory for sending vials to be tested or isolates to be confirmed or reconfirmed. We especially thank Russell Zablan, FDA, for critical assistance. We thank Astellas Pharma, Inc., for the kind gift of isavuconazole.
The use of product names in the manuscript does not imply their endorsement by the U.S. Department of Health and Human Services. The findings and conclusions of this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 12 June 2013
REFERENCES
- 1. Benyamin RM, Manchikanti L, Parr AT, Diwan S, Singh V, Falco FJ, Datta S, Abdi S, Hirsch JA. 2012. The effectiveness of lumbar interlaminar epidural injections in managing chronic low back and lower extremity pain. Pain Physician 15:E363–E404 [PubMed] [Google Scholar]
- 2. Staal JB, de Bie RA, de Vet HC, Hildebrandt J, Nelemans P. 2009. Injection therapy for subacute and chronic low back pain: an updated Cochrane review. Spine 34:49–59 [DOI] [PubMed] [Google Scholar]
- 3. Cooper AB, Sharpe MD. 1996. Bacterial meningitis and cauda equina syndrome after epidural steroid injections. Can. J. Anaesth. 43:471–474 [DOI] [PubMed] [Google Scholar]
- 4. Hooten WM, Kinney MO, Huntoon MA. 2004. Epidural abscess and meningitis after epidural corticosteroid injection. Mayo Clin. Proc. 79:682–686 [DOI] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention (CDC) 2002. Exophiala infection from contaminated injectable steroids prepared by a compounding pharmacy—United States, July-November 2002. MMWR Morb. Mortal. Wkly. Rep. 51:1109–1112 [PubMed] [Google Scholar]
- 6. Gunaratne PS, Wijeyaratne CN, Chandrasiri P, Sivakumaran S, Sellahewa K, Perera P, Fernando R, Wanigasinghe J, Jayasinghe S, Ranawala R, Riffsy MT, Seneviratne HR. 2006. An outbreak of Aspergillus meningitis following spinal anaesthesia for caesarean section in Sri Lanka: a post-tsunami effect? Ceylon Med. J. 51:137–142 [DOI] [PubMed] [Google Scholar]
- 7. Gunaratne PS, Wijeyaratne CN, Seneviratne HR. 2007. Aspergillus meningitis in Sri Lanka—a post-tsunami effect? N. Engl. J. Med. 356:754–756 [DOI] [PubMed] [Google Scholar]
- 8. Kolbe AB, McKinney AM, Kendi AT, Misselt D. 2007. Aspergillus meningitis and discitis from low-back procedures in an immunocompetent patient. Acta Radiol. 48:687–689 [DOI] [PubMed] [Google Scholar]
- 9. Bell RW, Dalton JB, McCall CM, Karram S, Pearce DT, Memon W, Lee R, Carroll KC, Lyons JL, Gireesh ED, Trivedi JB, Cettomai D, Smith BR, Chang T, Tochen L, Ratchford JN, Harrison DM, Ostrow LW, Stevens RD, Chen L, Zhang SX. 2013. Iatrogenic Exserohilum infection of the central nervous system: mycological identification and histopathological findings. Mod. Pathol. 26:166–170 [DOI] [PubMed] [Google Scholar]
- 10. Centers for Disease Control and Prevention (CDC) 2012. Multistate outbreak of fungal infection associated with injection of methylprednisolone acetate solution from a single compounding pharmacy—United States, 2012. MMWR Morb. Mortal. Wkly. Rep. 61:839–842 [PubMed] [Google Scholar]
- 11. Kainer MA, Reagan DR, Nguyen DB, Wiese AD, Wise ME, Ward J, Park BJ, Kanago ML, Baumblatt J, Schaefer MK, Berger BE, Marder EP, Min JY, Dunn JR, Smith RM, Dreyzehner J, Jones TF; the Tennessee Fungal Meningitis Investigation Team 2012. Fungal infections associated with contaminated methylprednisolone in Tennessee. N. Engl. J. Med. 367:2194–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pettit AC, Kropski JA, Castilho JL, Schmitz JE, Rauch CA, Mobley BC, Wang XJ, Spires SS, Pugh ME. 2012. The index case for the fungal meningitis outbreak in the United States. N. Engl. J. Med. 367:2119–2125 [DOI] [PubMed] [Google Scholar]
- 13. Smith RM, Schaefer MK, Finks J, Wise M, Kainer MA, Duwve J, Fontaine E, Chu A, Carothers B, Reilly A, Fiedler J, Wiese AD, Feaster C, Gibson L, Purfield A, Cleveland AA, Benedict K, Harris JR, Brandt ME, Blau D, Jernigan J, Weber JT, Park BJ, for the Multistate Fungal Infection Outbreak Response Team 19 December 2012. Fungal meningitis and other infections associated with contaminated methylprednisolone acetate steroid injections from a single compounding pharmacy. N. Engl. J. Med. [Epub ahead of print.] [Google Scholar]
- 14. Kerkering TM, Grifasi ML, Baffoe-Bonnie AW, Bansal E, Garner DC, Smith JA, Demicco DD, Schleupner CJ, Aldoghaither RA, Savaliya VA. 2013. Early clinical observations in prospectively followed patients with fungal meningitis related to contaminated epidural steroid injections. Ann. Intern. Med. 158:154–161 [DOI] [PubMed] [Google Scholar]
- 15. Zhao Y, Petraitiene R, Walsh TJ, Perlin DS. 2013. A real-time PCR assay for rapid detection and quantification of Exserohilum rostratum, a causative pathogen of fungal meningitis associated with injection of contaminated methylprednisolone. J. Clin. Microbiol. 51:1034–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lyons JL, Roos KL, Marr KA, Neumann H, Trivedi JB, Kimbrough DJ, Steiner L, Thakur KT, Harrison DM, Zhang SX. 2013. Cerebrospinal fluid (1,3) β-d-glucan detection as an aid to diagnose iatrogenic fungal meningitis. J. Clin. Microbiol. 51:1285–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gade L, Scheel CM, Pham CD, Lindsley MD, Iqbal N, Cleveland AA, Whitney AM, Lockhart SR, Brandt ME, Litvintseva AP. 2013. Detection of fungal DNA in human body fluids and tissues during a multistate outbreak of fungal meningitis and other infections. Eukaryot. Cell 12:677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balajee SA, Kano R, Baddley JW, Moser SA, Marr KA, Alexander BD, Andes D, Kontoyiannis DP, Perrone G, Peterson S, Brandt ME, Pappas PG, Chiller T. 2009. Molecular identification of Aspergillus species collected for the Transplant-Associated Infection Surveillance Network. J. Clin. Microbiol. 47:3138–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA [Google Scholar]
- 20. Clinical and Laboratory Standards Institute 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. CLSI document MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 21. Padhye AA, Sekhon AS, Carmichael JW. 1973. Ascocarp production by Nannizzia and Arthroderma on keratinous and non-keratinous media. Sabouraudia 11:109–114 [PubMed] [Google Scholar]
- 22. Miller PM. 1955. V-8 juice agar as a general-purpose medium for fungi and bacteria. Phytopathology 45:461–462 [Google Scholar]
- 23. de Hoog GS, Gene H, Figueras MJ. 2001. Atlas of clinical fungi, 2nd edition Kluwer Academic Publishers, Dordrecht, the Netherlands [Google Scholar]
- 24. McCabea KM, Zhanga Y, Huanga B, Wagarb EA, McCabea ERB. 1999. Bacterial species identification after DNA amplification with a universal primer pair. Mol. Gen. Metab. 66:205–211 [DOI] [PubMed] [Google Scholar]
- 25. Senga P, Drancourta M, Gouriet F, La Scola B, Fournier P, Rolain JM, Raoult DD. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 25a. Ritter JM, Muehlenbachs A, Blau DM, Paddock CD, Shieh W, Drew CP, Batten BC, Bartlett JH, Metcalfe MG, Pham CD, Lockhart SR, Patel M, Liu L, Jones TL, Greer PW, Montague JL, White E, Rollin DC, Seales C, Stewart D, Deming MV, Brandt ME, Zaki SR, Exserohilum Infections Working Group A clinicopathologic review of 40 cases of Exserohilum infection caused by injection of contaminated methylprednisolone acetate. Am. J. Pathol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muñoz-Cadavid C, Rudd S, Zaki SR, Patel M, Moser SA, Brandt ME, Gómez BL. 2010. Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: comparison of five tissue DNA extraction methods using panfungal PCR. J. Clin. Microbiol. 48:2147–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard—second edition. CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 28. da Cunha KC, Sutton DA, Gen Jé Capilla J, Cano J, Guarro J. 2012. Molecular identification and in vitro response to antifungal drugs of clinical isolates of Exserohilum. Antimicrob. Agents Chemother. 56:4951–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Casadevall A, Pirofski L-A. 2013. Exserohilum rostratum fungal meningitis associated with methylprednisolone injections. Future Microbiol. 8:135–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pappas PG, Kontoyiannis DP, Perfect JR, Chiller TM. 2013. Real-time treatment guidelines: considerations during the Exserohilum rostratum outbreak in the United States. Antimicrob. Agents Chemother. 57:1573–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. González GM. 2009. In vitro activities of isavuconazole against opportunistic filamentous and dimorphic fungi. Med. Mycol. 47:71–76 [DOI] [PubMed] [Google Scholar]
- 32. Kontoyiannis DP, Perlin DS, Roilides E, Walsh TJ. 6 May 2013. What can we learn and what do we need to know amidst the iatrogenic outbrea of Exserohilum rostratum meningitis? Clin. Infect. Dis. [Epub ahead of print.] 10.1093/cid/cit283 [DOI] [PMC free article] [PubMed] [Google Scholar]