Abstract
Acinetobacter baumannii is a major nosocomial pathogen causing infections in critically ill patients. This organism has acquired the propensity to rapidly develop resistance to most antibiotics. At several hospitals within Cape Town, South Africa, tobramycin and colistin are frequently the only therapeutic options. Vitek2 automated susceptibility testing (AST) is used in the clinical laboratory to determine selected susceptibility profiles. The suspicion of a possible AST-related technical error when testing for susceptibility to tobramycin in A. baumannii precipitated this study. Thirty-nine A. baumannii strains isolated from clinical specimens (June to December 2006) were included in this prospective study. Tobramycin susceptibility testing results obtained by AST, disc diffusion, the epsilometer test (Etest), and agar dilution were compared to those for broth microdilution (BMD), the reference method. The tobramycin susceptibility results revealed errors in 25/39 (64%) isolates (10 very major and 15 minor errors) when AST was compared to BMD, 12/39 (31%) (2 very major and 10 minor errors) when Etest was compared to BMD, 16/39 (41%) (3 very major and 13 minor errors) when disc diffusion was compared to BMD, and 21/39 (54%) (10 very major and 11 minor errors) when agar dilution was compared to BMD. Using PCR, we detected aac(3)-IIa, which is associated with tobramycin resistance, in 21/25 of the discrepant isolates. Molecular typing (using pulsed-field gel electrophoresis and repetitive sequence-based PCR [rep-PCR]) showed that these isolates were genetically related. Clinical laboratories that routinely use the Vitek2 system should consider an alternative testing method for determining susceptibility to tobramycin.
INTRODUCTION
Acinetobacter baumannii has gained increased notoriety as a highly resistant nosocomial pathogen globally. This organism has been associated with infections in immunocompromised and debilitated patients, particularly in the intensive care unit (ICU) setting (1, 2).
A. baumannii has the capacity for long-term survival in the hospital environment (3, 4). In addition, its remarkable capacity to acquire resistance has prompted its classification as a high-priority pathogen by the Antimicrobial Availability Task Force of the Infectious Diseases Society of America (5). Pandrug-resistant phenotypes have been isolated in many settings (6–10). In the Western Cape, South Africa, A. baumannii is endemic in many hospital ICUs. The majority of these organisms are multidrug resistant, retaining susceptibility to only tobramycin and colistin. Of the A. baumannii strains isolated from blood cultures during 2006 in the diagnostic laboratory at Groote Schuur Hospital, only 55% and 57% remained susceptible to imipenem and meropenem, respectively (our unpublished data).
The pressure on clinical diagnostic laboratories to produce rapid identification and susceptibility profiles has resulted in increasing use of automated microbiology systems, such as Vitek2 (bioMérieux, Marcy l'Etoile, France). Although there are many advantages to the use of this technology, several studies have indicated inaccurate susceptibility results ranging from false resistance to false susceptibility, especially when testing nonfermenting Gram-negative bacteria such as A. baumannii (11–20). These inaccuracies have a major impact on patient management as they may encourage the use of inactive antimicrobials in critically ill patients. In addition, they promote the use of broader-spectrum antibiotics if narrow-spectrum drugs are falsely reported as resistant.
A discrepancy between tobramycin susceptibility testing using manual methods and the Vitek2 automated susceptibility testing (AST) method alerted the clinical diagnostic laboratory to a possible technical error, thus precipitating this study. We conducted a prospective study to investigate the accuracy of tobramycin susceptibility testing in A. baumannii in comparison to validated susceptibility test methods. In addition, we hypothesized that the resistance was due to the aminoglycoside modifying enzyme, AAC(3)-IIa (aminoglycoside acetyltransferase), as the gene encoding this enzyme was previously isolated from clinical isolates of A. baumannii at our institution (21). The clonal relatedness of these isolates was also investigated.
MATERIALS AND METHODS
Bacterial strains.
A total of 39 nonrecurring clinical isolates of A. baumannii were tested. These isolates were obtained from five hospitals in Cape Town, South Africa (Groote Schuur Hospital, Red Cross War Memorial Children's Hospital, Victoria Hospital, Mowbray Maternity Hospital, and GF Jooste Hospital) over a 7-month period (June 2006 to December 2006). The Groote Schuur (893 beds) and Red Cross hospitals (240 beds) are tertiary academic hospitals with superspecialist services, while the Victoria (158 beds) and GF Jooste hospitals (224 beds) are secondary-level hospitals offering general specialist care. Mowbray hospital (179 beds) is a secondary-level maternity hospital with tertiary level neonatal care.
The isolates were obtained from the following clinical specimens: 14 tracheal aspirates, seven sputum samples, seven pus swabs (including three swabs collected during environmental screening of two different ICUs during an outbreak investigation), four blood cultures, four urine specimens, two tissue cultures, and one fluid culture. The majority of the isolates were obtained from clinical specimens from patients in ICUs at the various hospitals (32/39). Isolates were selected which had zone diameters or MICs (as determined by Vitek2) in each of the different susceptibility categories. Fourteen isolates had MICs close to the susceptible/intermediate breakpoints (as defined by the Clinical and Laboratory Standards Institute [CLSI] [22]). The breakpoints are as follows: for zone diameters, susceptible ≥15 mm, intermediate 13 to 14 mm, and resistant ≤12 mm, and for MICs, susceptible ≤4 μg/ml, intermediate 8 μg/ml, and resistant ≥16 μg/ml (22). Identification and susceptibility testing, with the exception of agar dilution, broth microdilution (BMD), and PCR, were performed on freshly isolated organisms passaged twice on 2% blood agar plates (Greenpoint Media Laboratory, NHLS). The isolates were stored on beads (Viabank VIM tubes, Abtek Biologicals Ltd.) at −70°C while awaiting the remaining susceptibility and molecular testing.
Identification.
All strains were identified by the Vitek2 Gram-negative identification card (bioMérieux, La Balme, France) according to the manufacturer's instructions at 0.6 McFarland (McF). A percent probability greater than 90% was considered an acceptable identification.
Susceptibility test methods.
To ensure that there were no effects of agar pH and cation concentration on the aminoglycoside susceptibility testing, the susceptibility levels of all isolates were tested by disc diffusion (Oxoid, Basingstoke) and Etests (AB Biodisk, Solna, Sweden) using cation-adjusted Mueller-Hinton medium from three commercial manufacturers (bioMérieux [Randburg, South Africa], Bio-Rad Laboratories [Johannesburg, South Africa], and Greenpoint NHLS Media Laboratory [Cape Town, South Africa]) at 0.5 McF per CLSI and manufacturer's guidelines, respectively (23). These tests were performed with inoculum from the same subculture. In addition, automated susceptibility testing using the Vitek2 NO-22 susceptibility card (bioMérieux, La Balme, France) was performed at 0.6 and 1.0 McF, respectively. Although 1.0 McF is not a recommended standard for performing aminoglycoside susceptibility testing, we hypothesized that a heavier inoculum would favor expression of resistance (24). To prevent editing of MICs on the basis of other inferred resistance mechanisms, the advanced expert system (software version 2.01) was not used to analyze the raw antibiotic susceptibility data obtained. To ensure reproducible results, agar dilution and BMD were performed on the isolates in duplicate by two different scientists using cation-adjusted Mueller-Hinton medium at 0.5 McF (25). This testing was performed at the Research and Development Division of bioMérieux (La Balme, France) because the local diagnostic laboratory did not have the experience to carry out this testing. Results were interpreted using CLSI criteria (22). Broth microdilution was used as the reference method because it most closely simulated the methodology used by Vitek2.
Quality control organisms included Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922, as recommended by CLSI and the manufacturer.
Interpretation of susceptibility results.
Discordant results with the reference method (BMD) were classified as very major errors (VMEs), major errors, or minor errors (mEs). The interpretative category errors that were used were defined as follows: VME (isolate resistant by reference method but susceptible by test method), major error (isolate susceptible by reference method but resistant by test method), and mE (difference between reference method and test method differs by 1 interpretative category). For disc diffusion, the mean of the zone diameters obtained from all three media was used in the analysis. For Etests, the mode of the MICs obtained from all three media was used for comparison with BMD. For agar dilution, the single discordant result (isolate 33) was reconciled by choosing the result which most closely matched the results of the reference method. For BMD, discrepant results were reconciled by choosing the result with the higher MIC value. This accords with standard laboratory practice, as theoretically these results would be used to influence the choice of antibiotic in critically ill patients.
PCR for detection of aac(3)-IIa.
Genomic DNA was extracted using the EasyMag (bioMérieux, Durham, North Carolina) per the manufacturer's instruction. Primers (forward [F], CGC GGA AGG CAA TAA C, and reverse [R], GCT TCT CAA GAT AGG TG) described previously by Jacobson et al. were used to amplify a 786-kb fragment of aac(3)-IIa (21). A. baumannii strains designated MOS-1 and MOS-2, which were isolated from a single patient at Groote Schuur Hospital during the early 1980s, were used as positive and negative controls, respectively (21).
The master mix consisted of magnesium chloride (25 mM), 2.5 mM each deoxyribonucleotide triphosphate (dNTP), 20 pmol of each primer, and 2.5 U Taq polymerase in buffer made up to a final volume of 50 μl per reaction. An initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 1 min, 51°C for 45 s, and 72°C for 1 min, and a final extension at 72°C for 5 min was carried out. The amplified fragments were separated on a 2% agarose gel by gel electrophoresis, stained with ethidium bromide, and visualized under UV light. Positive, negative, and extraction controls and a water blank were included for each gel. PCR was repeated for all negative isolates. In addition, a 16s rRNA PCR was performed on all isolates to confirm the integrity of bacterial DNA.
The amplicons from two randomly selected isolates were sequenced using the ABI PRISM BigDye Terminator v3.1 ready reaction cycle sequencing kit (Applied Biosystems) on the ABI genetic analyzer (Applied Biosystems) and compared to available A. baumannii sequences in the National Center for Biotechnology Information (NCBI) GenBank database using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST).
Pulsed-field gel electrophoresis.
The relatedness of the isolates was compared by pulsed-field gel electrophoresis (PFGE) with minor modifications of the previously described technique (21). Following plug preparation and cell lysis, DNA was digested with ApaI (Roche) for 2 h, after which the DNA was separated on a 1.5% agarose gel (Bio-Rad) in 0.5× Tris buffer. Electrophoresis was carried out in a Gene Navigator PFGE machine (Amersham Biosciences AB, Uppsala, Sweden) for 23 h with the pulse time increasing from 5 to 45 s, after which it was stained with ethidium bromide, destained, and photographed using a Fotodyne Inc. UV light box and a Kodak 290 camera. The fingerprint images were analyzed by Gel Compare II software version 4.6 (Applied Maths, Sint-Martens-Latem, Belgium) using a Dice similarity index for cluster analysis and the unweighted pair group average for tree building. Banding patterns were compared with 1.5% optimization and 1.5% band position tolerance (26). All isolates with PFGE banding patterns with >87% similarity were grouped within the same cluster (26).
In addition, repetitive sequence-based PCR (rep-PCR) using DiversiLab (bioMérieux, Marcy l'Etoile, France) was performed by the R&D Division, bioMérieux, La Balme, France.
Ethics.
This research received approval from the University of Cape Town Health Sciences Faculty Research Ethics Committee (REC REF 458/2006).
RESULTS
Characterization of clinical isolates.
All 39 isolates were identified as A. baumannii by the Vitek2 instrument with a high percent probability (99%).
Tobramycin susceptibility and error rates.
The level of agreement between the different methods and the reference method (BMD) varied widely and is shown in Table 1. A significant number of VMEs (10 isolates) and mEs (15 isolates) were observed when Vitek2 AST was compared to BMD. A similar number of VMEs and mEs were detected when agar dilution was compared to BMD. Three VMEs and 13 mEs were noted with disc diffusion testing, and two VMEs and 10 mEs were detected when testing was performed by Etest. Irrespective of the testing method, no major errors were detected (Table 2).
Table 1.
Results from the 39 A. baumannii isolates showing tobramycin susceptibility testing and molecular typing resultsa
| Isolate | Disc diffusion (mm/susceptibility) | Etest (MIC [μg/ml]/susceptibility) | Agar dilution (MIC [μg/ml]/susceptibility) | Vitek 0.6 McF (MIC [μg/ml]/susceptibility)b | Broth microdilution (MIC [μg/ml]/susceptibility) | aac(3)-IIa genec | DiversiLab clone |
|---|---|---|---|---|---|---|---|
| 7 | 0/R | 256/R | 128/R | ≥16/R | >256/R | − | D |
| 40 | 4/R | 8/I | 4/S | 4/S | >256/R | + | C |
| 3 | 3/R | 32/R | 16/R | 8/I | 256/R | − | D |
| 4 | 10/R | 16/R | 8/I | 8/I | 256/R | − | C |
| 27 | 0/R | 64/R | 32/R | ≥16/R | 256/R | − | D |
| 31 | 12/R | 16/R | 16/R | 4/S | 256/R | + | C |
| 18 | 13/I | 16/R | 4/S | 8/I | 128/R | + | C |
| 36 | 13/I | 8/I | 16/R | 4/S | 128/R | + | C |
| 38 | 0/R | 64/R | 16/R | ≥16/R | 128/R | − | A |
| 10 | 13/I | 16/R | 4/S | 8/I | 64/R | + | C |
| 5 | 11/R | 8/I | 8/I | 8/I | 32/R | + | C |
| 6 | 14/I | 4/S | 2/S | 2/S | 32/R | + | C |
| 11 | 13/I | 16/R | 4/S | 2/S | 32/R | − | C |
| 12 | 9/R | 32/R | 8/I | 8/I | 32/R | + | C |
| 13 | 11/R | 32/R | 4/S | 4/S | 32/R | + | C |
| 17 | 13/I | 16/R | 2/S | 4/S | 16/R | + | C |
| 22 | 16/S | 8/I | 2/S | ≤1/S | 16/R | + | C |
| 23 | 21/S | 2/S | ≤0.5/S | ≤1/S | 16/R | − | B |
| 39 | 15/S | 8/I | 2/S | 2/S | 16/R | + | C |
| 8 | 13/I | 8/I | 4/S | 4/S | 8/I | + | C |
| 9 | 15/S | 8/I | 2/S | 2/S | 8/I | + | C |
| 14 | 14/I | 8/I | 4/S | 2/S | 8/I | + | C |
| 15 | 15/S | 8/I | 4/S | 4/S | 8/I | + | C |
| 20 | 15/S | 4/S | 2/S | ≤1/S | 8/I | + | C |
| 26 | 0/R | 64/R | 2/S | ≥16/R | 8/I | + | C |
| 32 | 17/S | 4/S | 2/S | ≤1/S | 8/I | + | C |
| 33 | 10/R | 16/R | 8/I | 4/S | 8/I | + | C |
| 42 | 14/I | 8/I | 4/S | 2/S | 8/I | + | C |
| 29 | 14/I | 8/I | 2/S | 2/S | 4/S | − | C |
| 24 | 22/S | 2/S | ≤0.5/S | ≤1/S | 2/S | − | C |
| 35 | 21/S | 2/S | ≤0.5/S | ≤1/S | 2/S | − | B |
| 2 | 19/S | 2/S | ≤0.5/S | ≤1/S | 1/S | − | B |
| 16 | 23/S | 2/S | ≤0.5/S | ≤1/S | 1/S | − | A |
| 21 | 15/S | 2/S | ≤0.5/S | ≤1/S | 1/S | − | A |
| 28 | 23/S | 1/S | ≤0.5/S | ≤1/S | 1/S | + | A |
| 30 | 20/S | 2/S | ≤0.5/S | ≤1/S | 1/S | − | A |
| 34 | 23/S | 1/S | ≤0.5/S | ≤1/S | 1/S | − | A |
| 37 | 21/S | 2/S | ≤0.5/S | ≤1/S | 1/S | − | B |
| 41 | 21/S | 2/S | ≤0.5/S | ≤1/S | 1/S | − | C |
PCR of aac(C)-IIa and DiversiLab results. S, sensitive; I, intermediate, R resistant.
Shading indicates very major errors; underlining indicates minor errors.
+, aac(3)-IIa gene present; −, aac(3)-IIa gene absent.
Table 2.
Error rates from comparison of the different test methods with BMD for susceptibility testing of the 39 A. baumannii isolates
| Method | % (no.) of: |
Total error rate [% (no.)] | ||
|---|---|---|---|---|
| Very major errors | Major errors | Minor errors | ||
| Disc diffusion | 7.69 (3) | 0 (0) | 33.33 (13) | 41.02 (16) |
| Etest | 5.13 (2) | 0 (0) | 25.64 (10) | 30.77 (12) |
| Agar dilution | 25.64 (10) | 0 (0) | 28.21 (11) | 53.85 (21) |
| Vitek2 | 25.64 (10) | 0 (0) | 38.46 (15) | 64.10 (25) |
There were three discordant results when we used Vitek2 at 1.0 McF versus 0.6 McF. However, the results did not show improvement with a higher inoculum; instead, two additional VMEs would have been detected if 1.0 McF was used for testing.
The specimen types from which A. baumannii was isolated and which gave rise to VMEs included three tracheal aspirates, two sputum samples, two tissue samples, two environmental swabs (from an outbreak investigation), and a blood culture. The minor errors were detected from the following specimen types: seven tracheal aspirates, three urine specimens, two sputum samples, two pus swabs, and one sterile fluid. There was no evidence of an association between specimen type and error rate; however, numbers were small.
Detection of aac(3)-IIa.
Previous research at our institution had shown that the prevalent mechanism of tobramycin resistance among A. baumannii isolates was an aminoglycoside-modifying enzyme encoded by the aac(3)-IIa gene (21). Twenty-one of the 25 isolates that demonstrated discordant results (either sensitive or intermediate by Vitek2 but resistant by BMD) were found to contain the aac(3)-IIa gene (Table 1).
Analysis of the sequencing data of the amplicons from two isolates (17 and 28) showed that they were 100% identical to the sequence of the aac(3)-IIa gene (GenBank accession number M62833).
Relatedness of isolates.
The majority of clinical strains were isolated from various ICUs from four different hospitals. The relatedness of the isolates was determined by rep-PCR using the DiversiLab (bioMérieux) system. The isolates clustered into four clones, with the majority of isolates clustering in cluster C (Table 1). Seventeen of the cluster C isolates were isolated from Groote Schuur Hospital, with four of these isolates from non-ICU wards. Nine isolates from cluster C were isolated from Red Cross Hospital, with a single isolate from Mowbray Maternity Hospital. The results of the DiversiLab analysis confirmed the presence of a predominant clone that was not confined to a single hospital. In addition, isolates 3, 4, and 11 (which lacked the AAC(3′)-IIa enzyme but were resistant to tobramycin) were scattered across clusters C and D, confirming that tobramycin resistance was not linked to a single clone at our hospitals.
PFGE was performed to compare a representative of the two commonest clones (C and A) with the dominant strains that had previously been isolated at our institution (Fig. 1). The MOS-1 and MOS-2 strains were isolated from the same patient during the same period of hospitalization at Groote Schuur Hospital (21). Isolate C2 was highly similar to MOS-1, a tobramycin-resistant strain with the aac(3)-IIa gene, that has been present in Cape Town, South Africa, since the early 1980s (21). Although it was not our intention to do a formal outbreak investigation, 8 of 11 isolates (including 2 environmental swabs) from Red Cross Hospital's ICU belonged to cluster C, confirming a possible outbreak in the unit. A2 represented a strain that was susceptible to tobramycin by all test methods. This strain was not related to cluster C by PFGE, confirming the results obtained by DiversiLab.
Fig 1.
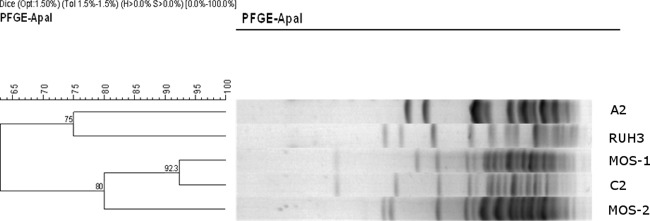
Results of PFGE comparing 2 representative isolates of the most common DiversiLab clones, A and C, and MOS-1. A2, isolate 16 (fully susceptible strain without aac(3)-IIa; DiversiLab clone A); C2, isolate 32 (intermediately susceptible strain with aac(3)-IIa; DiversiLab clone C); RUH3, control strain; MOS-1, clinical strain with aac(3)-IIa; MOS-2, clinical strain without aac(3)-IIa.
DISCUSSION
Inaccuracies with AST have been reported in the literature, particularly among nonfermenters (11–20). Few studies have been performed to evaluate tobramycin susceptibility testing using AST in A. baumannii. A recent study found that up to one-third of A. baumannii isolates (n = 107) tested were incorrectly reported as susceptible to amikacin by the Vitek2 instrument (11). The manufacturers of the Vitek2 and other automated systems suggest either nonreporting or confirmation of the susceptibility result by manual methods in these cases.
After suspecting a possible error in tobramycin susceptibility testing by AST, we determined the susceptibilities of 39 isolates by multiple manual and automated methods. An overall category agreement error rate of <10% was considered an acceptable performance of a susceptibility test method, which included ≤1.5% VMEs and ≤3.0% major errors (27). Our study found a high total error rate of 64% in isolates tested by Vitek2, with 25% of these being VMEs. The use of a higher concentration of inoculum when performing AST did not have an effect on reducing the error rates.
Although CLSI recognizes the agar dilution method as a reference method, a high error rate (54% total errors) was detected when testing tobramycin. Eight out of 10 of the isolates tested by agar dilution with VMEs were confirmed to possess the aac(3)-IIa gene, which confers resistance to tobramycin. There have been no studies reported in the recent literature that have particularly evaluated the accuracy of the agar dilution test method for tobramycin. Further studies to confirm our results are required.
Errors occurred with manual testing methods (disc diffusion and Etests) as well, with the total rates being 41 and 31%, respectively. The Etest had the most acceptable performance (2% VMEs, no major errors). A study at the San Antonio Military Medical Center (San Antonio, TX) revealed errors with manual and automated susceptibility testing of tobramycin in A. baumannii (11). Very major errors were detected in 13.1% of isolates tested by Vitek2, compared to 2.8% VMEs when disc diffusion and Etests were compared to BMD (11). Akers et al. also noted VMEs with tobramycin susceptibility testing using other automated systems (11). We detected a much higher rate of VMEs when testing using Vitek2 (25%) but similar rates when testing using disc diffusion and Etests (3% and 2%, respectively).
Twenty-one of the 25 isolates that demonstrated discordant results (either sensitive or intermediate by Vitek2 but resistant by BMD) in our study were found to contain the aac(3)-IIa gene (Table 1).This confirmed a possible mechanism of resistance to tobramycin. This is not unexpected, as a worldwide study of A. baumannii (which included isolates from South Africa) has shown that another AAC(3) enzyme, AAC(3)-I, was the commonest aminoglycoside-modifying enzyme (AME) present, accounting for resistance to aminoglycosides in 50% (12/24) of Acinetobacter spp. tested (28). Numerous AMEs have been isolated from A. baumannii. The aac(3)-IIa gene confers resistance to gentamicin, tobramycin, dibekacin, netilmicin, and sisomicin. This resistance profile is common among the Enterobacteriaceae (29). The AAC(3)-IIa enzyme is the commonest resistance mechanism in the group exhibiting this resistance profile, accounting for 84.8% of isolates (29). This aminoglycoside resistance profile has been detected in 21.3% of Acinetobacter spp. (29).
Akers et al. concluded that the AME genotype was an inadequate predictor of the aminoglycoside phenotype, suggesting that multiple resistance mechanisms were operating simultaneously (11). This contrasted with our study, which showed a good correlation between the presence of the aac(3)-IIa gene and tobramycin resistance, with the gene being present in 12/19 (63%) isolates with tobramycin MICs of ≥16.
Seven isolates that were resistant to tobramycin by BMD lacked the aac(3)-IIa gene (Table 1). Five of these isolates exhibited tobramycin MICs of ≥128 μg/ml. These isolates likely have other mechanisms of resistance to tobramycin, such as AAC-6′ or ANT-2′′, combinations of AMEs, efflux pumps, or other resistance mechanisms that were not explored further as this was not the focus of our study. Importantly, though, the failure of Vitek2 to detect these resistant isolates suggests a wider failure of the system to detect tobramycin resistance.
Alarmingly, Akers et al. showed that susceptibility to tobramycin by BMD and disc diffusion was retained in the presence of a potentially inactivating AME gene in 16 (18%) isolates due to other AMEs, including combinations such as aph(3′)-Ia and ant(2′′)-Ia, aac(6′)Ih and aph(3′)-Ia (11). We found that just a single isolate that was susceptible by all test methods harbored the aac(3)-IIa gene. This finding may be explained by a so-called cryptic gene, a phenotypically silent DNA sequence. This hypothesis may be supported by similar findings in Salmonella spp., where it has been shown that a 60-kb deletion upstream of an aminoglycoside resistance gene, aac(6′)-Iy, encoding 6′-N-acetyltransferase type I in a strain of Salmonella enterica subsp. enterica serotype Enteritidis, BM4362, resulted in the gene being highly expressed. This gene was silent in the progenitor strain, BM4361 (30). However, as this gene was not the focus of our study, we did not investigate it further.
One hypothesis for the discrepant Vitek2 results may be that the catalytic activity of the AAC(3)-IIa enzyme is too slow to be detected by rapid automated susceptibility test methods, particularly with a poor substrate like tobramycin (31). Whether this has implications on the clinical outcome of patients treated with tobramycin has yet to be determined.
The results of the PFGE suggest that it is unlikely that the tobramycin-resistant strains had recently emerged due to the increased use of tobramycin at our hospitals, as the clone had been present in the hospital since the early 1980s.
Conclusion.
To the best of our knowledge, this is the first study in South Africa that has assessed the ability of Vitek2 to detect A. baumannii isolates with reduced susceptibility to tobramycin. Tobramycin, tigecycline, and colistin are the only antibiotics available in the public health sector in South Africa that remain effective against multidrug-resistant strains of A. baumannii. Thus, accurate susceptibility testing remains critical.
The data from our study confirm the limitations of both automated and manual tobramycin susceptibility test methods. Vitek2 appears to be unreliable for the detection of tobramycin resistance in A. baumannii. It appears that manual methods, such as Etest, may be more reliable for susceptibility testing when tobramycin is considered for use as a potential therapeutic agent.
ACKNOWLEDGMENTS
This work was supported by bioMérieux and OmniMed.
We acknowledge Gilles Zambardi and the Research and Development Division of bioMérieux (La Balme, France) for technical assistance with performing the agar and broth microdilution testing and the DiversiLab rep-PCR.
The authors declare no conflict of interest. The funders had no role in the study design, data analysis, or preparation of the manuscript.
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Gaynes R, Edwards JR; National Nosocomial Infections Surveillance System 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41:848–854 [DOI] [PubMed] [Google Scholar]
- 2. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 3. Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36:1938–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Musa EK, Desai N, Casewell MW. 1990. The survival of Acinetobacter calcoaceticus inoculated on fingertips and on formica. J. Hosp. Infect. 15:219–227 [DOI] [PubMed] [Google Scholar]
- 5. Talbot GH, Bradley J, Edwards JE, Jr, Gilbert D, Scheld M, Bartlett JG. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42:657–668 [DOI] [PubMed] [Google Scholar]
- 6. Apisarnthanarak A, Mundy LM. 2009. Mortality associated with Pandrug-resistant Acinetobacter baumannii infections in Thailand. Am. J. Infect. Control 37:519–520 [DOI] [PubMed] [Google Scholar]
- 7. Bassetti M, Repetto E, Righi E, Boni S, Diverio M, Molinari MP, Mussap M, Artioli S, Ansaldi F, Durando P, Orengo G, Bobbio Pallavicini F, Viscoli C. 2008. Colistin and rifampicin in the treatment of multidrug-resistant Acinetobacter baumannii infections. J. Antimicrob. Chemother. 61:417–420 [DOI] [PubMed] [Google Scholar]
- 8. Cascio A, Conti A, Sinardi L, Iaria C, Angileri FF, Stassi G, David T, Versaci A, Iaria M, David A. 2010. Post-neurosurgical multidrug-resistant Acinetobacter baumannii meningitis treated successfully with intrathecal colistin. A new case and a systematic review of the literature. Int. J. Infect. Dis. 14:e572–e579. 10.1016/j.ijid.2009.06.032 [DOI] [PubMed] [Google Scholar]
- 9. Coelho J, Woodford N, Turton J, Livermore DM. 2004. Multiresistant acinetobacter in the UK: how big a threat? J. Hosp. Infect. 58:167–169 [DOI] [PubMed] [Google Scholar]
- 10. Falagas ME, Rafailidis PI, Matthaiou DK, Virtzili S, Nikita D, Michalopoulos A. 2008. Pan-drug resistant Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumanii infections: characteristics and outcome in a series of 28 patients. Int. J. Antimicrob. Agents 32:450–454 [DOI] [PubMed] [Google Scholar]
- 11. Akers KS, Chaney C, Barsoumian A, Beckius M, Zera W, Yu X, Guymon C, Keen EF, III, Robinson BJ, Mende K, Murray CK. 2010. Aminoglycoside resistance and susceptibility testing errors in Acinetobacter baumannii-calcoaceticus complex. J. Clin. Microbiol. 48:1132–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Silbert S, Pfaller MA, Hollis RJ, Barth AL, Sader HS. 2004. Evaluation of three molecular typing techniques for nonfermentative Gram-negative bacilli. Infect. Control Hosp. Epidemiol. 25:847–851 [DOI] [PubMed] [Google Scholar]
- 13. Snyder JW, Munier GK, Johnson CL. 2008. Direct comparison of the BD phoenix system with the Microscan WalkAway system for the identification and antimicrobial susceptibility testing of Enterobacteriaceae and nonfermentative Gram-negative organisms. J. Clin. Microbiol. 46:2327–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tenover FC, Mizukii TS, Carlson LG. 1990. Evaluation of autoSCAN-W/A automated microbiology system for the identification of non-glucose-fermenting gram-negative bacilli. J. Clin. Microbiol. 28:1628–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kulah C, Aktas E, Comert F, Ozlu N, Akyar I, Ankarali H. 2009. Detecting imipenem resistance in Acinetobacter baumannii by automated systems (BD Phoenix, Microscan WalkAway, Vitek2); high error rates with Microscan WalkAway. BMC Infect. Dis. 9:30. 10.1186/1471-2334-9-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Visser MR, Bogaards L, Rozenber-Arska M, Verhoef J. 1992. Comparison of the autoSCAN W/A and Vitek Automicrobic systems for identification and susceptibility testing of bacteria. Eur. J. Clin. Microbiol. Infect. Dis. 11:979–984 [DOI] [PubMed] [Google Scholar]
- 17. Washington JA. 1993. Rapid antimicrobial susceptibility testing: technical and clinical considerations. Clin. Microbiol. Newsl. 15:153–155 [Google Scholar]
- 18. White RL, Kays MB, Friedrich LV, Brown EW, Koonce JR. 1991. Pseudoresistance of Pseudomonas aeruginosa resulting from degradation of imipenem in an automated susceptibility testing system with predried panels. J. Clin. Microbiol. 29:398–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jung S, Yu JK, Shin SH, Park KG, Jekarl DW, Han K, Park YJ. 2010. False susceptibility to amikacin by the Vitek2 in Acinetobacter baumannii harboring armA. Ann. Clin. Lab. Sci. 40:167–171 [PubMed] [Google Scholar]
- 20. Juretschko S, LaBombardi VL, Learner SA, Schreckenberger PC, and the Pseudomonas Study Group AST 2007. Accuracies of beta-lactam susceptibility test results for Pseudomonas aeruginosa with four automated systems (BD Phoenix, MicroScan WalkAway, Vitek and Vitek2). J. Clin. Microbiol. 45:1339–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacobson RK. 2007. MSc thesis Association of IS1133 with an aminoglycoside resistance gene, aacC2a, in Acinetobacter baumannii isolates. University of Cape Town, Cape Town, South Africa [Google Scholar]
- 22. Clinical and Laboratory Standards Institute 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 23. Clinical Laboratory Standards Institute 2006. Performance standards for antimicrobial disk susceptibility tests, 9th ed Approved standard M2-A9. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 24. Barry J, Brown A, Ensor V, Lakhani U, Petts D, Warren C, Winstanley T. 2003. Comparative evaluation of the Vitek2 Advanced Expert System (AES) in five UK hospitals. J. Antimicrob. Chemother. 51:1191–1202 [DOI] [PubMed] [Google Scholar]
- 25. Clinical and Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 26. Seifert H, Dolzani L, Bressan R, van der Reijden T, van Streijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardisation and interlaboratory reproducibility assessment of pulsed field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clinical and Laboratory Standards Institute 2001. Development of in vitro susceptibility testing criteria and quality control parameters, 2nd ed Approved guideline M23-A2. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 28. Seward RJ, Lambert T, Towner KJ. 1998. Molecular epidemiology of aminoglycoside resistance in Acinetobacter spp. J. Med. Microbiol. 47:455–462 [DOI] [PubMed] [Google Scholar]
- 29. Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Magnet S, Courvalin P, Lambert T. 1999. Activation of the cryptic aac(6′)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J. Bacteriol. 181:6650–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Magnet S, Blanchard JS. 2005. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 105:477–498 [DOI] [PubMed] [Google Scholar]


