Abstract
Inappropriate activation of the transcription factors STAT3 and STAT5 has been shown to drive cancer pathogenesis through dysregulation of genes involved in cell survival, growth, and differentiation. Although STAT3 and STAT5 are structurally related, they can have opposite effects on key genes, including BCL6. BCL6, a transcriptional repressor, has been shown to be oncogenic in diffuse large B cell lymphoma. BCL6 also plays an important role in breast cancer pathogenesis, a disease in which STAT3 and STAT5 can be activated individually or concomitantly. To determine the mechanism by which these oncogenic transcription factors regulate BCL6 transcription, we analyzed their effects at the levels of chromatin and gene expression. We found that STAT3 increases expression of BCL6 and enhances recruitment of RNA polymerase II phosphorylated at a site associated with transcriptional initiation. STAT5, in contrast, represses BCL6 expression below basal levels and decreases the association of RNA polymerase II at the gene. Furthermore, the repression mediated by STAT5 is dominant over STAT3-mediated induction. STAT5 exerts this effect by displacing STAT3 from one of the two regulatory regions to which it binds. These findings may underlie the divergent biology of breast cancers containing activated STAT3 alone or in conjunction with activated STAT5.
INTRODUCTION
Signal transducers and activators of transcription (STATs) are latent transcription factors that become activated following tyrosine phosphorylation by receptor and nonreceptor tyrosine kinases. Once phosphorylated, they dimerize into their active forms, translocate to the nucleus, and bind to cognate STAT DNA binding sites, where they modulate transcription of key target genes involved in a variety of cellular processes, such as proliferation, survival, differentiation, and apoptosis. While STATs are often thought of as activators of transcription, we and others have shown that STATs can also repress the expression of some key target genes (1–3). Interestingly, it has been shown that most of the seven STATs bind preferentially to TTCNNNGAA sequences and regulate some of the same genes; however, they are not just redundant transcription factors, as they often regulate many distinct target genes.
Given their regulation of key genes involved in proliferation and protection against apoptosis, it is not surprising that STATs are often found to be aberrantly active in cancer. In fact, both STAT5, which refers to the nearly identical proteins STAT5a and STAT5b, and STAT3 are commonly activated constitutively in breast cancer, as opposed to the transient activation that occurs physiologically. STAT3 is activated in ∼70% of breast tumors and is often associated with aggressive tumors (4, 5). In addition, it has recently been shown that STAT3 activation is associated with CD44+ stem cell-like triple-negative breast tumors (6). Overexpression studies have shown that STAT5 can promote breast tumor formation in mice (7). In addition, increased levels of prolactin, the cytokine that activates STAT5 in breast cells, are associated with increased risk of breast cancer in postmenopausal women, though the tumors are often estrogen receptor (ER)/progesterone receptor (PR)-positive differentiated tumors (8). Furthermore, STAT5 and STAT3 can both be activated in the same tumor, which results in distinct consequences. Tumors that have activation of both STATs are more likely to be ER/PR positive and low grade than tumors that have activation of STAT3 alone (5). Therefore, this suggests that STAT5 may counterbalance the effects of STAT3 in these cells. In fact, it was shown that STAT5 affects the expression of some STAT3 target genes, one of which is BCL6.
The transcriptional repressor BCL6 prevents terminal differentiation of B cells and has been shown to be oncogenic in diffuse large B cell lymphoma. In addition, BCL6 has been shown to prevent terminal breast differentiation in mammary cells and prevents the expression of the STAT5 beta-casein target gene (9). In addition, BCL6 is expressed in high-grade ductal carcinomas (10). Given our emerging understanding of the important role of BCL6 in breast cancer pathogenesis and the distinct effects of the related transcription factors STAT3 and STAT5 in its regulation, we examined the molecular mechanism for these divergent effects.
MATERIALS AND METHODS
Cell lines and stimulations.
SKBR3 cells, kindly provided by Lyndsay Harris (Dana-Farber Cancer Institute), were maintained in RPMI containing 10% fetal calf serum. MDA-MB-468 cells, kindly provided by Myles Brown (Dana-Farber Cancer Institute), and the paired MDA-MB-468 sublines (5) were maintained in 10% Dulbecco's modified Eagle's medium (DMEM). The cells were stimulated with 100 ng/ml prolactin (R&D Systems, Minneapolis, MN, USA) and/or 10 ng/ml leukemia inhibitory factor (LIF) (Calbiochem, Temecula, CA, USA) for 15 min for immunoprecipitations, 90 min for mRNA expression, and 30 min for chromatin immunoprecipitation (ChIP), unless otherwise indicated. Cells were pretreated with depsipeptide (Fujisawa Pharmaceutical Co., Osaka, Japan) or trichostatin A (TSA) (Sigma, St. Louis, MO, USA) for 2 h prior to stimulation with cytokine for mRNA expression and ChIP. Cells were pretreated with 10 μM 3-aminobenzamide (3-aba) (Calbiochem) for 4 h prior to stimulation with cytokine.
Immunoprecipitations, immunoblotting, and nuclear-cytoplasmic fractionation.
Immunoprecipitations and immunoblotting were performed as described previously (11). Cleared lysates were immunoprecipitated with 1 μg anti-STAT5 (sc-835) or 1 μg anti-STAT3 (sc-482) from Santa Cruz Biotechnology (Santa Cruz, CA). Nuclear and cytoplasmic fractionation was performed according to the manufacture's protocol (Active Motif, Carlsbad, CA). Antibodies used for immunoblots included phosphospecific STAT5 (9351 and 9359), phosphospecific STAT3 (9131), and poly(ADP-ribose) polymerase (PARP) (9542) from Cell Signaling; tubulin (T-5168) from Sigma; and BCL6 (sc-858), STAT3 (sc-482), and total STAT5 (sc-835) from Santa Cruz Biotechnology.
mRNA analysis.
RNA was harvested using an RNeasy Minikit from Qiagen (Valencia, CA). cDNA was generated using a TaqMan reverse transcription kit (Applied Biosystems, Foster City, CA). Quantitative PCR (qPCR) was performed in triplicate using SYBR green master mix (Applied Biosystems) on a 7300 or 7500 real-time PCR system (Applied Biosystems). Data are expressed as mean fold change and standard error of the mean (SEM). Primers included BCL6 (CTGCAGATGGAGCATGTTGT and TCTTCACGAGGAGGCTTGAT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (AATCCCATCACCATCTTCCA and TGGACTCCACGACGTACTCA), and SOCS3 (TCAAGACCTTCAGCTCCAAG and TGACGCTGAGCGTGAAGAAG).
Reporter gene assays.
SK-BR-3 cells were transfected with A-Luc or TBR-Luc (see below) luciferase reporter plasmid (2) or the STAT3-responsive reporter M67-Luc (kindly provided by J. Bromberg, Memorial Sloan Kettering, New York, NY). Twenty-four hours after transfection, the cells were stimulated for 6 or 24 h with the indicated cytokines and analyzed as described previously (2).
Chromatin immunoprecipitation.
ChIP was performed essentially as described previously (1). Briefly, cells were fixed in 1% formaldehyde for 10 min, sonicated 5 times for 15 s each time using a Fisher Scientific sonic dismembrator model 500 PDQ on setting 13, and lysates were immunoprecipitated overnight with the indicated antibody: normal rabbit IgG from Caltag (Burlingame, CA); STAT5 (sc-835), STAT3 (sc-482 and sc-7179), RNA polymerase (Pol) II (sc-9001), STAT5a (sc-1081), BCL6 (sc-858 and sc-368), Brca1 (sc-646), Brca2 (sc-8326), CtBP (sc-11390), MTA3 (sc-48799), TGIF1 (sc-9084), p300 (sc-585), STAT1 (sc-346), histone deacetylase 1 (HDAC1) (sc-7872), FoxA1 (sc-9186), and C/EBPβ (sc-150) from Santa Cruz Biotechnology; acetyl-histone 4 (Lys8) (2594) and PARP (9542) from Cell Signaling Technology; phospho-Pol II 5 (ab5131) and phospho-Pol II 2 (ab5095) from abCam (Cambridge, MA); and STAT5b (71-2500) from Zymed/Invitrogen (Carlsbad, CA). Quantitative PCR was performed using primers for region B (CGGCAGCAACAGCAATAATC and GGAGAGCTGACACCAAGTCC), region A (TTCCTGTTACGCCGTCAATG and CGGCAGCTTCCTGGAAAGTT), the control region (CACAGGGACAGGAAGATGGT and GGATGGCATACAACCTCCAA), or rhodopsin (TGGGTGGTGTCATCTGGTAA and GGATGGAATGGATCAGATGG). The results were normalized to the input and expressed relative to binding to the rhodopsin-negative binding region.
RNA interference.
Cells (5 × 105) were reverse transfected using Lipofectamine RNAiMax (Invitrogen) with 10 nM small interfering RNA (siRNA) C/EBPβ (sc-29229), FoxA1 (sc-37930) from Santa Cruz, or control siRNA 3 (D-001210-03; Dharmacon). Seventy-two hours after transfection, the cells were stimulated as described above. For ChIP performed with siRNA targeting FoxA1 (siFoxA1), 7.5 × 106 cells were plated in each 20-cm dish. Reverse transfection was performed for 72 h, after which the cells were stimulated with LIF and ChIP was performed.
Transfections.
SK-BR-3 cells (5 × 104) were transfected with 0.4 μg (1×) or 0.2 μg (0.5×) plncx2, plncx2-STAT5a1*6, and/or plncx2-STAT3C for 30 h. mRNA was isolated, and gene expression was analyzed as described above.
Statistical analyses.
Data are expressed as means and standard deviations or standard errors of the means, as indicated in the figure legends. Paired two-tailed Student t tests were performed for all binary analyses.
RESULTS
STAT3 and STAT5 have opposite effects on BCL6 expression.
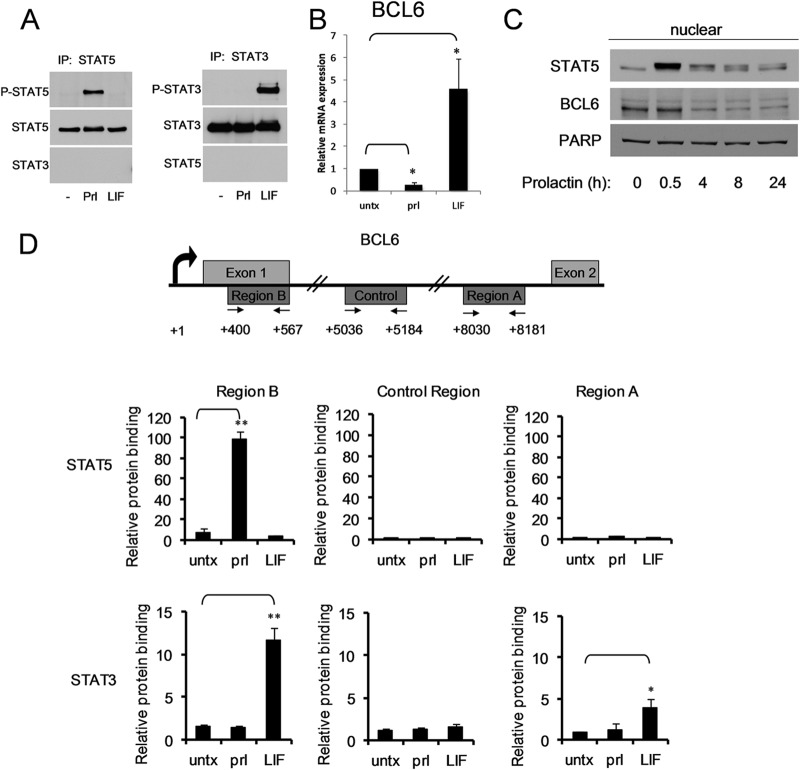
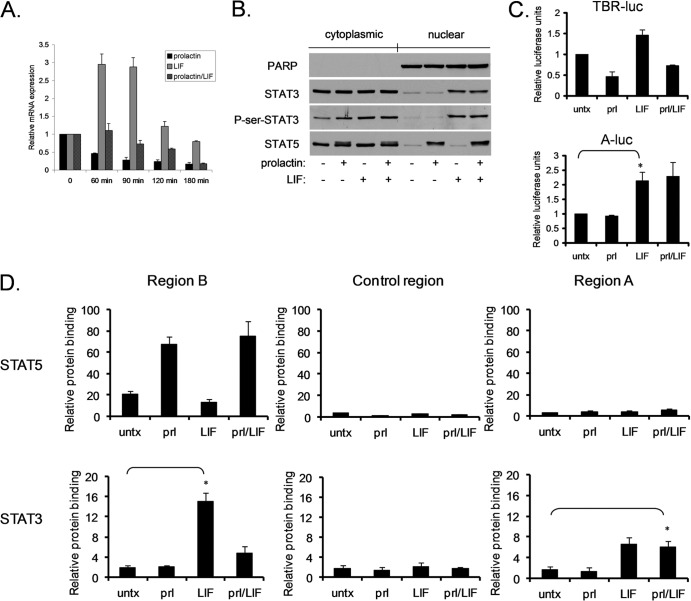
Although STAT5 and STAT3 are highly homologous and bind to overlapping cognate sequences, they play distinct roles in mammary gland biology. To determine the mechanism by which STAT5 and STAT3 exert these disparate effects, we focused on BCL6, which is oppositely regulated by STAT5 and STAT3 in breast cancer cells (5). The SK-BR-3 breast cancer cell line was chosen to analyze BCL6 regulation because neither STAT5 nor STAT3 is constitutively activated in these cells; however, each can be specifically activated by a physiologically relevant cytokine: prolactin activates STAT5 phosphorylation, while LIF activates STAT3 phosphorylation (Fig. 1A). As we had reported previously (5), prolactin stimulation led to a prominent decrease in BCL6 mRNA expression, while LIF stimulation resulted in upregulation of BCL6 mRNA (Fig. 1B). Within 4 h of prolactin stimulation, BCL6 levels in the nucleus decreased by 40%, and this decrease was sustained for 24 h (Fig. 1C). These data confirm that STAT5 and STAT3 oppositely regulate BCL6 expression in SK-BR-3 cells.
Fig 1.
STAT5 and STAT3 oppositely regulate BCL6 expression. (A) Lysates from SK-BR-3 cells that were untreated (−) or stimulated with prolactin (Prl) or LIF were immunoprecipitated (IP) for STAT5 or STAT3 and analyzed by immunoblotting for the phosphorylated form of each STAT. (B) RNA from SK-BR-3 cells stimulated with prolactin or LIF for 90 min was analyzed by quantitative reverse transcription (qRT)-PCR for BCL6 expression normalized to GAPDH (n = 3; *, P < 0.05). (C) Nuclear extracts from SK-BR-3 cells treated with prolactin for the indicated times were analyzed by immunoblotting for BCL6 expression. (D) (Top) Schematic map of the first two exons of the BCL6 gene and the locations of the regions (and primer pairs) analyzed in ChIP experiments. (Bottom) SK-BR-3 cells were stimulated with prolactin or LIF, and ChIP was performed using the indicated antibodies. Binding to the indicated sites was analyzed by qPCR relative to a negative-control binding region in the rhodopsin gene (n = 3; *, P < 0.05; **, P < 0.01). untx, untreated. The error bars indicate standard deviations.
STAT3 and STAT5 show distinct binding to BCL6 regulatory regions.
We previously identified two potential STAT binding sites within the BCL6 gene, region A and region B, and demonstrated that in hematopoietic cells, STAT5 binds to region B, which is located within the first exon of the BCL6 gene (2). Given the opposite biological and transcriptional roles of STAT3 and STAT5, we first wanted to determine their binding preferences in breast cancer cells for these two regulatory regions of BCL6. SK-BR-3 cells were left untreated or were stimulated with prolactin or LIF, and then ChIP was performed with antibodies directed toward STAT5 or STAT3. The ChIP product was analyzed for STAT binding to region A, region B, and a control region located between the two regions that does not contain any putative STAT binding sites. Upon prolactin stimulation, STAT5 bound to region B but not to either the control region or region A (Fig. 1D). Both STAT5a and STAT5b inducibly bound to this site, although the magnitude of STAT5b binding was somewhat greater (see Fig. S1 in the supplemental material). As expected, LIF, which does not lead to STAT5 activation, had no effect on STAT5 binding to these sites. In contrast, following LIF stimulation, STAT3 bound to both region A and region B but not to the control region. These data demonstrate that in the BCL6 gene, STAT5 and STAT3 share a binding site, region B, while STAT3 also has a distinct binding site, region A.
To determine whether similar findings have been observed independently, we analyzed publicly available ChIP-sequencing (ChIP-seq) data sets. These demonstrated that in human cells, STAT5 typically binds to region B (see Fig. S2 in the supplemental material). STAT3 binds to both region A and region B in about half of the human data sets analyzed and to only region B in the other sets (see Fig. S2). Interestingly, in mouse cells, STAT5a bound only to region B in 5 of the 6 data sets analyzed, whereas STAT5b was found only at region B in lymph nodes but at a number of locations, including regions A and B, in mouse liver cells (see Fig. S3 in the supplemental material). Finally, STAT3 typically bound only to region B in the mouse cells analyzed (see Fig S3). These data suggest that region B is the major site of regulation for STATs and region A may function in only a subset of cell types.
The gene-regulatory effects of STAT3 and STAT5 can be recapitulated in reporter constructs.
We next focused on determining the functional effects of STAT5 and STAT3 binding to these regions. We first analyzed the regulation of a luciferase reporter under the control of each of these two regions. Activation of STAT5 by prolactin resulted in downregulation of luciferase when regulated by region B (TBR-Luc), though it had no effect on luciferase regulated by region A (A-Luc) (see Fig. S4 in the supplemental material). In contrast, activation of STAT3 by LIF resulted in upregulation of luciferase when regulated by either region A or region B. Therefore, in these chimeric reporter constructs, STAT3 and STAT5 exert effects that reflect their binding preferences and functional effects on the endogenous BCL6 gene.
STAT3 and STAT5 have distinct effects on recruitment of RNA polymerase II to BCL6.
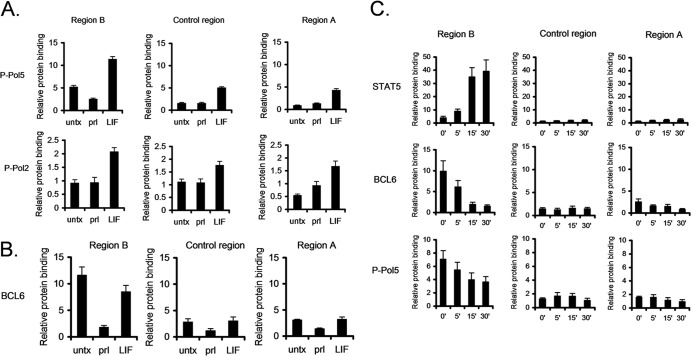
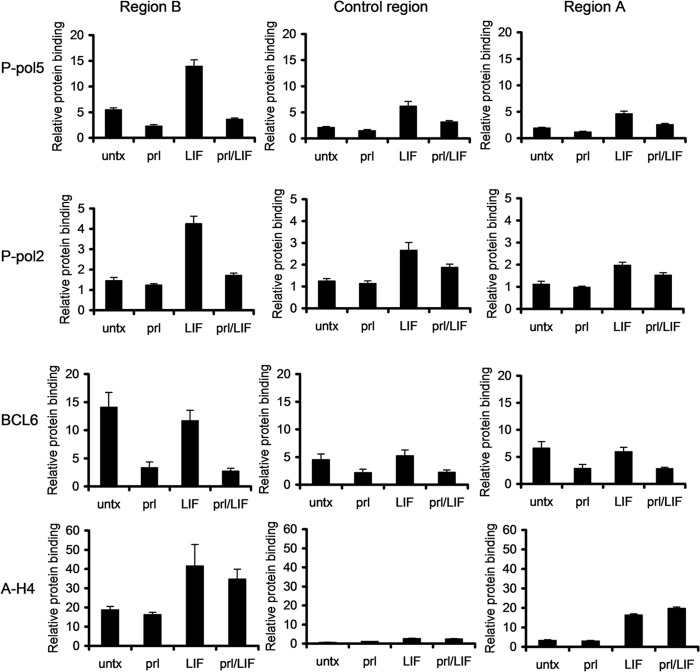
We next focused on the mechanism by which STAT3 and STAT5 mediate these opposite functions. Using ChIP, we first determined the effect of STAT5 binding on recruitment of RNA Pol II. Following prolactin-induced STAT5 binding to region B, RNA Pol II binding was decreased by 40%, suggesting that STAT5 represses BCL6 expression by reducing RNA Pol II recruitment (see Fig. S5 in the supplemental material). To better understand the effects of STAT5 on RNA Pol II recruitment, we performed ChIP using antibodies directed to specific phosphorylated forms of RNA polymerase II. RNA polymerase II is phosphorylated on the 5 position of the carboxy-terminal domain (CTD) repeat (P-Pol5) during the initiation phase, while RNA polymerase II is phosphorylated on the 2 position of the CTD repeat (P-Pol2) during the early stages of elongation. Analysis using antibodies specific to these sites demonstrated that there was a reduction of P-Pol5 upon prolactin stimulation, coincident with STAT5 binding to region B (Fig. 2A); however, prolactin had no effect on P-Pol2 in region B. Therefore, STAT5 binding to region B is coincident with a reduction of RNA polymerase II in the initiation phase.
Fig 2.
STAT5 and STAT3 oppositely modulate RNA Pol II initiation. (A) SK-BR-3 cells stimulated with prolactin or LIF were analyzed by ChIP-qPCR using the indicated antibodies. Binding to the indicated sites was analyzed by qPCR relative to a negative-control binding region in the rhodopsin gene. (B) SK-BR-3 cells were stimulated with prolactin or LIF, and binding of BCL6 was analyzed by ChIP. (C) SK-BR-3 cells were stimulated with prolactin at the indicated time points, and BCL6, STAT5, and P-Pol 5 binding was analyzed by ChIP. The error bars indicate standard errors of the means.
In contrast, LIF stimulation induced recruitment of both P-Pol5 and P-Pol2 in both regions B and A (Fig. 2A), consistent with STAT3 promoting upregulation of BCL6 expression. In addition, LIF stimulation promoted enhanced binding of both P-Pol5 and P-Pol2 in the intervening control region, suggesting that RNA polymerase II tracks from regions B to A to promote gene expression.
STAT5 can displace BCL6 from region B.
BCL6 has been shown to repress its own expression by binding to a region that is located within region B. This BCL6 binding site is distinct from the major STAT5 binding site within this region, although they are in close proximity to each other (2, 12). Therefore, we considered the hypothesis that STAT5 binding altered BCL6 binding to region B. To test this, ChIP was performed using antibodies to BCL6. Upon prolactin-induced STAT5 activation, BCL6 binding was lost at region B (Fig. 2B). Additionally, activation of STAT3 by LIF resulted in a slight reduction of binding of BCL6, but to a much lesser extent than that induced by STAT5 binding (Fig. 2B). This indicated that STAT3 and BCL6 do not appear to be competing for binding to region B. Importantly, BCL6 binding is lost in a time-dependent manner, inversely related to STAT5 binding to region B (Fig. 2C), suggesting that BCL6 and STAT5 compete for binding in this region.
We considered the possibility that the epitope on BCL6 recognized by the antibody is no longer be accessible following STAT5 binding, raising the possibility that BCL6 remains bound to region B. To distinguish these possibilities, we performed ChIP using antibodies to the C terminus of BCL6, whereas our initial ChIPs utilized an antibody directed to the N terminus of BCL6. Similar results were obtained with both antibodies (see Fig. S6 in the supplemental material), suggesting that BCL6 is in fact lost from region B upon STAT5 binding and that this finding does not reflect epitope masking. Furthermore, this loss of BCL6 binding is not due to decreased amounts of BCL6 in the nucleus, since the levels of nuclear BCL6 at this time point (30 min of prolactin stimulation) are similar to that of cells prior to prolactin stimulation (Fig. 1C). Taken together, these data demonstrate that BCL6 itself is not involved in STAT5-mediated repression of BCL6 mRNA expression.
STAT5 binding does not alter histone acetylation.
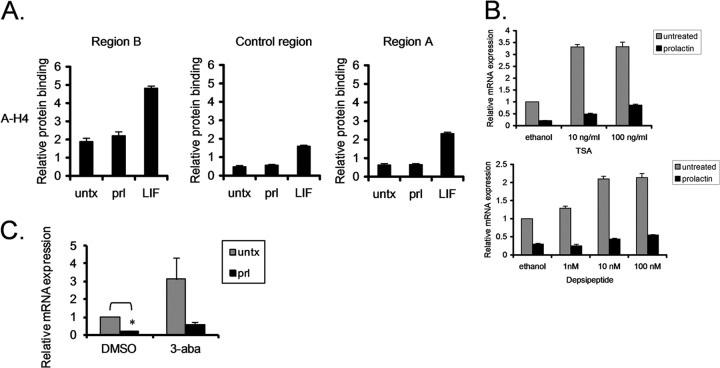
Given that histone acetylation is a known epigenetic mechanism of gene regulation, we analyzed acetylation of histones 3 and 4 in the regulatory regions of BCL6 upon prolactin and LIF stimulation. In region B, prolactin stimulation had no effect on acetylation of histone 4 (Fig. 3A) or histone 3 (data not shown). In contrast, LIF stimulation promoted histone acetylation in both regions B and A, as well as the intervening control region. This demonstrates that histone deacetylation in region B is not necessary for STAT5-mediated repression of BCL6, though increased histone acetylation is correlated with BCL6 upregulation. Consistent with this, HDAC1 is not recruited to region B upon prolactin stimulation (data not shown).
Fig 3.
Histone acetylation and PARP are not involved in STAT5-mediated repression of BCL6. (A) SK-BR-3 cells stimulated with prolactin or LIF were analyzed by ChIP using antibodies directed toward acetylated histone 4 (A-H4). (B) SK-BR-3 cells were pretreated with TSA or depsipeptide for 2 h and stimulated with prolactin for 90 min. BCL6 mRNA expression was analyzed by qRT-PCR relative to GAPDH. (C) SK-BR-3 cells were treated for 4 h with the PARP inhibitor 3-aba prior to prolactin stimulation. BCL6 mRNA expression was analyzed by qPCR relative to GAPDH (n = 2; *, P < 0.05). The error bars indicate standard errors of the means.
Since many transcription factors and other proteins can also be acetylated, we hypothesized that acetylation or deacetylation of these factors may be important for BCL6 gene regulation mediated by STATs. Therefore, we utilized two pharmacologic inhibitors of HDACs to test this hypothesis. We pretreated cells with TSA or depsipeptide, stimulated them with prolactin, and then analyzed BCL6 mRNA expression. Both TSA and depsipeptide resulted in upregulation of BCL6 expression, but neither treatment prevented the downregulation of BCL6 expression induced by prolactin (Fig. 3B). ChIP analysis demonstrated that depsipeptide treatment had no effect on the binding of STAT5 or BCL6 to region B. Depsipeptide by itself promoted transcriptional initiation in region B, as indicated by increased initiation of phosphorylation of RNA polymerase II at this site. This is consistent with the increased BCL6 expression seen with depsipeptide alone. However, STAT5 was still able to repress BCL6 expression, even in the presence of depsipeptide (see Fig. S7 in the supplemental material), providing further evidence that STAT5 does not require deacetylase activity for repression of BCL6 expression.
PARP has been reported to be involved in repression of BCL6 expression (13). Using a specific inhibitor of PARP function (3-aba), we determined that inhibiting PARP activity resulted in upregulation of BCL6 expression. However, STAT5 still repressed expression of BCL6 in the presence of 3-aba (Fig. 3C). In addition, PARP recruitment to region B was not seen upon prolactin stimulation (data not shown). Therefore, this suggests that PARP is not involved in STAT5-mediated repression of BCL6 expression.
It has recently been demonstrated that BCL6 is downregulated in B cells upon NF-κB activation via upregulation of IRF-4 (14). Therefore, we analyzed IRF-4 binding to regions A and B upon prolactin or LIF stimulation; however, we did not detect any IRF-4 binding to either region (data not shown), suggesting that IRF-4 is not involved in STAT-mediated regulation of BCL6.
Taking an alternative approach, we wanted to identify cofactors that were recruited by STAT3 but not by STAT5. Therefore, we analyzed SRC-1 and the related proteins AIB1 and GRIP1, since these cofactors are expressed in breast cancer cells and SRC-1 has been shown to interact with STAT3 (15); however, we did not find significant binding upon stimulation with prolactin or LIF, demonstrating that these cofactors are not involved in prolactin- or LIF-mediated modulation of BCL6 in region A or B (data not shown). Therefore, the specific cofactors recruited by STAT3 to promote upregulation of BCL6 remain unknown.
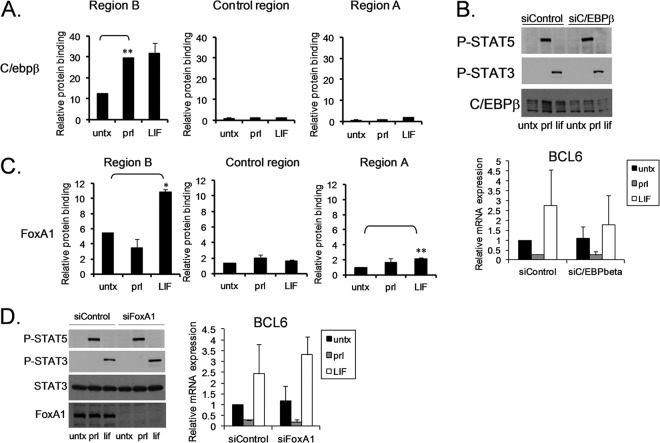
C/EBPβ is a transcription factor involved in mammary gland development. Since STAT5, STAT3, and BCL6 are all involved in mammary gland development, this raises the possibility that C/EBPβ is involved in STAT-mediated regulation of BCL6 expression. ChIP analysis demonstrated that C/EBPβ is bound to region B and that stimulation with either prolactin or LIF resulted in enhanced binding (Fig. 4A). Reducing the levels of C/EBPβ by siRNA resulted in decreased overall expression of BCL6, slightly enhanced repression by STAT5, and reduced upregulation by LIF (Fig. 4B). This suggests that C/EBPβ is involved in regulation of BCL6; however, C/EBPβ is not the factor that promotes differential regulation by STAT3 and STAT5.
Fig 4.
C/EBPβ is coordinately recruited by STAT5 and STAT3 to regulate BCL6 expression, whereas FoxA1 is oppositely recruited. (A) SK-BR-3 cells stimulated with prolactin or LIF were analyzed by ChIP using antibodies directed toward C/EBPβ (n = 2; **, P < 0.01). (B) SK-BR-3 cells were transfected with siC/EBPβ, and STAT phosphorylation (top) and BCL6 mRNA expression (bottom) were analyzed (n = 2). (C) SK-BR-3 cells stimulated with prolactin or LIF were analyzed by ChIP for FoxA1 binding to the indicated regions (n = 2; *, P < 0.05; **, P < 0.01). (D) SK-BR-3 cells transfected with siFoxA1 were analyzed by immunoblotting for the indicated proteins (left) or by qRT-PCR for BCL6 mRNA expression (right) (n = 2). The error bars indicate standard deviations.
FoxA1 is a pioneer factor for estrogen receptor-mediated gene regulation (16). Therefore, we wanted to determine if FoxA1 was involved in STAT-mediated regulation of BCL6. ChIP analysis demonstrated that FoxA1 is present in region B in untreated cells. FoxA1 binding is reduced in cells treated with prolactin and enhanced in cells treated with LIF (Fig. 4C). This suggests that FoxA1 may play an important role in STAT-mediated regulation of BCL6 expression. However, reducing the levels of FoxA1 by siRNA resulted in decreased BCL6 expression, little to no additional effect of prolactin, and enhanced STAT3-mediated gene expression by LIF (Fig. 4D), suggesting that while FoxA1 is important for BCL6 gene expression, it is not the factor involved in differential regulation by STATs. Furthermore, ChIP analysis after reducing FoxA1 by siRNA showed no effect on STAT3 binding (see Fig. S8 in the supplemental material). This is distinct from the functional role of FoxA1 in ER binding (16), confirming that FoxA1 is not the factor that drives the differential effects of STAT3 and STAT5.
STAT5-mediated repression of BCL6 is dominant over STAT3-mediated induction.
We have determined that STAT5 and STAT3 differentially modulate RNA polymerase II recruitment, as well as having different effects on histone acetylation and BCL6 binding in the BCL6 regulatory regions. Having gained an understanding of the differences mediated individually by STAT5 and STAT3 on BCL6, we next determined the effects of concomitant activation of STAT3 and STAT5, as can occur in breast cancer cells. The presence of STAT5 activation supersedes the effects of STAT3 activation in the prognosis of breast cancers, the growth and sensitivity to chemotherapy in breast cancer cell lines, and the regulation of BCL6 expression (5). Therefore, we wanted to determine the mechanism by which STAT5 affects STAT3-mediated regulation of BCL6 expression. Prolactin-induced activation of STAT5 resulted in repression of BCL6 expression to less than 50% of baseline levels as early as 1 h after treatment, and this effect persisted for at least 3 h. LIF-induced activation of STAT3 led to prominent induction of BCL6 mRNA at 1 h, which returned to basal levels by 3 h after stimulation (Fig. 5A). When both STAT5 and STAT3 were activated simultaneously, there was no increase in BCL6 mRNA, and the levels gradually declined to the level induced by prolactin alone (approximately 20% of baseline) by 3 h. This provides further evidence that STAT5 is dominant over STAT3, since STAT5 can repress BCL6 expression even in the presence of activated STAT3.
Fig 5.
STAT5 inhibits STAT3 binding to region B. (A) SK-BR-3 cells were stimulated with prolactin, LIF, or the combination for the indicated times, and BCL6 mRNA expression was analyzed. The error bars indicate standard errors of the means. (B) SK-BR-3 cells were stimulated as described above, and the nuclear and cytoplasmic locations of the indicated proteins were analyzed by immunoblotting. (C) SK-BR-3 cells were transfected with the STAT5/STAT3-responsive region B luciferase construct (TBR-Luc) or with the STAT3-responsive region A luciferase construct (A-Luc) and were then stimulated with prolactin, LIF, or the combination for 24 h and analyzed for luciferase activity (n = 2; *, P < 0.05). (D) Cells stimulated with prolactin, LIF, or the combination were analyzed for STAT5 and STAT3 binding to the indicated regions by ChIP. The error bars (C and D) indicate standard deviations.
We considered the possibility that the dominant effect of STAT5 was due to competition with STAT3 for nuclear transport when both are activated. To test this hypothesis, nuclear and cytoplasmic fractions were isolated following prolactin and LIF stimulation. STAT5 and STAT3 entered the nucleus upon prolactin or LIF stimulation, respectively, and simultaneous activation of both pathways has no effect on nuclear accumulation of either STAT (Fig. 5B). Serine phosphorylation of STAT3 has been found to be induced by prolactin in other cell types (17), and STAT3 serine phosphorylation may affect STAT3 both positively and negatively (18, 19). To determine whether this might be a mechanism by which prolactin modulated STAT3 function, we analyzed the phosphorylation of STAT3 on serine 727 upon treatment with prolactin alone or with LIF. Transcriptionally active nuclear STAT3 shows increased serine phosphorylation upon LIF treatment; however, prolactin alone has no effect on serine-phosphorylated STAT3 (Fig. 5B). In addition, simultaneous prolactin and LIF treatment resulted in no measureable difference from LIF treatment alone. This suggests that prolactin does not directly modulate STAT3 via serine-phosphorylated STAT3. To further assess this mechanism, we analyzed the effects of prolactin treatment on the LIF-induced upregulation of a STAT3-dependent promoter, m67, which is independent of STAT5. Prolactin treatment had no effect on LIF induction of luciferase activity, providing further evidence that prolactin does not inhibit global STAT3 function (see Fig. S9 in the supplemental material).
We next evaluated whether simultaneous activation of STAT5 and STAT3 altered their relative functions in the regulatory regions of BCL6. First, we analyzed luciferase reporter constructs regulated by either region A or region B. Coactivation of STAT5 and STAT3 led to repression of luciferase regulated by region B. In contrast, STAT5 had no effect on STAT3-mediated induction of luciferase activity regulated by region A (Fig. 5C). We next performed ChIP analysis to determine the relative binding of STAT3 and STAT5 at the endogenous BCL6 sites. Similar to the results with the luciferase reporter constructs, STAT5 activation had no effect on STAT3 binding in region A. Furthermore, activation of STAT3 had little effect on STAT5 binding to region B; however, activation of STAT5 reduced STAT3 binding to region B almost back to basal levels (Fig. 5D). To rule out the possibility that coactivation of STAT5 sterically inhibits the ability of the C-terminal antibody to bind to STAT3, ChIP was repeated with an antibody directed to the N terminus of STAT3 (see Fig. S10 in the supplemental material). ChIP performed with this antibody displayed results similar to ChIP data obtained with the C-terminal antibody (Fig. 5D). Taken together, these findings suggest that STAT5 binding to region B prevents STAT3 from binding to this site, thereby allowing STAT5 to have a dominant effect over STAT3 in regulating BCL6 expression.
We next determined the effects of coactivation of STAT5 and STAT3 on RNA polymerase II activity. Activation of STAT3 alone resulted in recruitment of RNA polymerase II phosphorylated on the 5 and 2 positions in regions B and A and the control region of BCL6 (Fig. 6). However, when STAT5 was activated simultaneously with STAT3, recruitment of RNA polymerase II phosphorylated on either site was inhibited in all three regions. This suggests that STAT5 inhibits recruitment of RNA polymerase II to region B, as well as its tracking from region B to region A, thereby preventing BCL6 transcription. It is notable that binding of RNA polymerase II phosphorylated at the 5 position, reflecting transcriptional initiation, is decreased below basal levels when both STATs are activated. This is consistent with STAT5 repressing BCL6 transcription even when STAT3 is activated by preventing the initial binding of RNA polymerase II in region B.
Fig 6.
STAT5 inhibits the effects of STAT3 on regulation of BCL6. SK-BR-3 cells stimulated with prolactin, LIF, or the combination were analyzed by ChIP using the indicated antibodies for binding to regions B and A and the intervening control region. The error bars indicate standard errors of the means.
In contrast to these changes, the binding of a number of other factors is unchanged when both STATs are activated. STAT5 activation alone or in the context of STAT3 activation had no effect on acetylation of histone H4 (Fig. 6). This further suggests that alteration in histone acetylation in region B is not necessary for STAT5-induced repression of BCL6 expression.
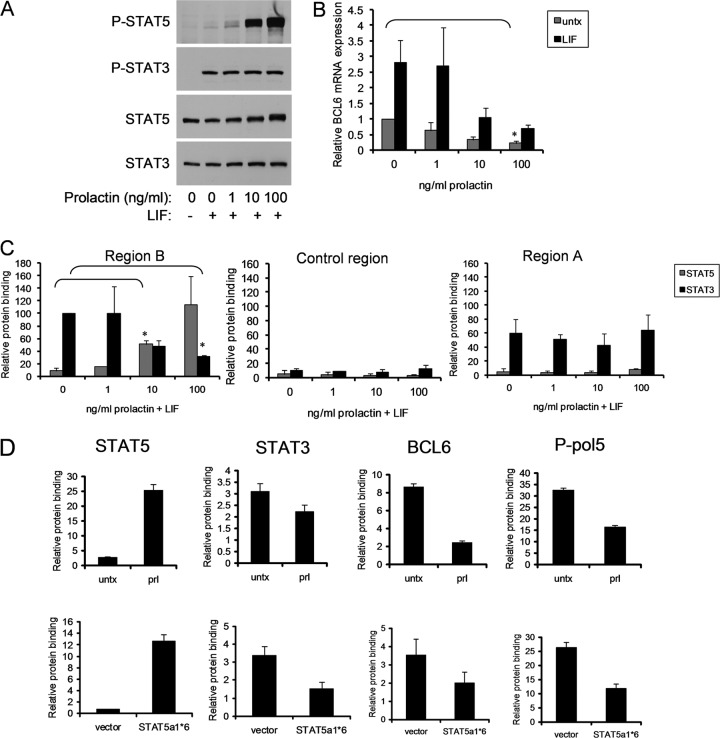
Graded activation of STAT5 displaces STAT3 from region B.
To determine the mechanism by which STAT5 affects STAT3 binding to the BCL6 regulatory regions, we modulated the level of STAT5 activation using increasing doses of prolactin while keeping the level of STAT3 activation constant at the maximally induced level (achieved with 10 ng/ml LIF). Treatment with 1 ng/ml of prolactin resulted in weak induction of STAT5 phosphorylation. Increasing dosing of prolactin resulted in a gradated induction of STAT5 phosphorylation, with 100 ng/ml being maximal (Fig. 7A). A threshold level of STAT5 activation (induced by between 1 and 10 ng/ml of prolactin) was necessary to begin to inhibit STAT3-mediated BCL6 induction (Fig. 7B). As expected at maximal activation, STAT5 repressed BCL6 expression below even basal levels. We then considered the hypothesis that STAT5 could compete with STAT3 for binding at a key regulatory site. Using ChIP, we analyzed STAT3 and STAT5 binding to region B with increasing STAT5 activation. We found that STAT5 was able to bind to region B, and reduce STAT3 binding to the site, in a graded manner roughly paralleling the inhibition of BCL6 mRNA at each level of STAT5 activation (Fig. 7C). This was not a nonspecific effect on STAT3 DNA binding, as STAT3 binding to region A (to which STAT5 does not bind in these cells) was unaffected by prolactin. Taken together, these findings demonstrate that STAT5 outcompetes STAT3 for binding to region B to regulate BCL6 gene expression.
Fig 7.
STAT5 inhibition is dominant over STAT3 activation of BCL6. (A to C) SK-BR-3 cells were stimulated with LIF and increasing doses of prolactin. (A) Phosphorylation of STAT3 and STAT5 was measured by immunoblotting. (B) BCL6 mRNA expression was measured by qRT-PCR (n = 2; *, P < 0.05). (C) STAT5 and STAT3 binding was measured by ChIP. STAT3 binding with LIF only and STAT5 binding with prolactin only in region B were normalized to 100 (n = 2; *, P < 0.05). (D) ChIP was performed for binding of the indicated proteins to region B in MDA-MB-468 cells stimulated with prolactin (top) or the paired MDA-MB-468 sublines stably transfected with constitutively active STAT5a1*6 or empty vector (bottom). The error bars indicate standard deviations.
Constitutively active STAT5 is dominant over constitutively active STAT3 in regulating BCL6 expression.
To determine if the primacy of STAT5 could also be seen in cells that were not stimulated with cytokines, we transfected SK-BR-3 cells with equal concentrations of plasmids expressing the constitutively active forms of STAT5 (STAT5a1*6) and STAT3 (STAT3C) alone and together. As expected, constitutively active STAT5 repressed BCL6 expression, while constitutively active STAT3 upregulated BCL6 expression. Importantly, transfection of equal amounts of each plasmid resulted in repression of BCL6, confirming that the inhibitory effect of STAT5 is dominant over that of STAT3 in regulating BCL6 expression in the absence of cytokine stimulation (see Fig. S11 in the supplemental material). Furthermore, even when only half of the STAT5 plasmid was transfected relative to the constitutively active STAT3 plasmid, BCL6 was maximally repressed, providing further evidence that STAT5 is dominant over STAT3 in regulating BCL6 expression. This was not a global effect, however, as induction of the STAT3 target SOCS3 gene was unaffected by cotransfection with the constitutively active STAT5 plasmid.
STAT5 outcompetes constitutively activated STAT3 in region B.
We have found that in primary human breast cancers, tumors with coactivation of STAT5 and STAT3 have significantly different clinical characteristics than tumors with constitutively activated STAT3 alone. Therefore, we next wanted to determine if STAT5 could exert a repressive effect on BCL6 expression even when STAT3 is constitutively activated in a cell. We utilized MDA-MB-468 breast cancer cells, which contain constitutive activation of STAT3 and in which STAT5 can be activated by prolactin (5). Prolactin stimulation of MDA-MB-468 cells results in downregulation of BCL6 expression (5). Following prolactin stimulation, ChIP analysis demonstrated that STAT5 bound to region B, and STAT3 binding to this site was reduced by 25% (Fig. 7D). In addition, concomitant with STAT5 activation, BCL6 binding was reduced and the initiation of phosphorylation of RNA polymerase II at the site (P-Pol 5) was inhibited, as was seen with the SK-BR-3 cells (Fig. 7D).
To further elucidate the mechanism of regulation of BCL6 in breast cancer, we next evaluated binding when both STAT5 and STAT3 are constitutively activated. While STAT5 activation occurs commonly in breast tumors, alone and in combination with STAT3 activation (5), we have been unable to identify a breast cancer cell line in which STAT5 is constitutively activated (6). Therefore, we engineered paired MDA-MB-468 sublines in which STAT3 is activated in conjunction with STAT5 (having been stably transfected with the constitutively activated STAT5 mutant STAT5a1*6) or in which STAT3 is activated in isolation (having been stably transfected with the corresponding empty vector) (5). In these paired lines, BCL6 expression is downregulated when both STATs are activated compared to STAT3 activation alone (5). ChIP in these paired lines demonstrated that when both STATs are constitutively activated, STAT5 bound preferentially to region B and reduced the initiation of phosphorylation of RNA polymerase II at the site (Fig. 7D). Taken together, these findings demonstrate that the dominant repressive effects of STAT5 over STAT3 on BCL6 expression occur over the full range of physiological activation states of these transcription factors.
DISCUSSION
We have found that STAT5 and STAT3 oppositely regulate BCL6 expression by binding to the same regulatory site, region B, and promoting opposite effects on RNA polymerase II initiation. It was recently demonstrated independently that STAT5 bound to region B in breast cancer cells (20), which is consistent with our data in both hematopoietic and breast cells. In addition, we have found that these STATs compete for binding to this site, and STAT5 can outcompete STAT3, thereby preventing BCL6 gene expression. We have ruled out many plausible mediators, including histone deacetylation, PARP, and Brca1/2. We also showed that other potential cofactors, including C/EBPβ and the pioneer factor FoxA1, do not drive the differential regulation of BCL6 by STAT3 and STAT5. However, the full mechanism by which STAT5 represses BCL6 activity remains unknown.
STAT5-mediated repression of BCL6 has also been demonstrated in mouse liver cells (21, 22); however, the mechanism of repression by STAT5 is currently unknown. It was found that STAT5 bound to region B in both male and female mouse livers when growth hormone was present. Interestingly, STAT5 bound to region A in male livers with low growth hormone levels, but STAT5 binding to region A was not detected in female livers (22). Consistent with the female mouse livers, STAT5 binding was not detected in region A in female breast cancer cell lines when activated by prolactin (Fig. 1 and data not shown). We have also determined that STAT5 does not regulate region A when isolated in a reporter construct and stimulated with prolactin (see Fig. S4 in the supplemental material). In addition, we previously reported that STAT5 does not bind to region A in a variety of hematopoietic cell lines (2). While STAT5 has been shown to bind to region A in other systems (see Fig. S3 in the supplemental material), the role of region A in regulation of BCL6 by STAT5 is currently unknown. However, in the breast cancer cells analyzed, region A does not play a role in STAT5-mediated regulation of BCL6 expression or in the dominance over STAT3, since STAT5 does not affect STAT3 binding to this region. The role of region A in the sex differences seen in mouse livers (22) suggests that region A may play an important role in liver cells. Further analysis of STAT binding to region A in additional cell types may help elucidate the role of region A in regulating BCL6 expression.
Repression of BCL6 by STAT5 has also been demonstrated in TH1 cells after interleukin 2 (IL-2) treatment (23), which is consistent with our previous work demonstrating that IL-2 activation of STAT5 in NK cells represses BCL6 expression (2). It was found that STAT3 binding to the BCL6 gene is reduced and STAT5 binding is increased upon IL-2 stimulation (23). This supports our data in breast cancer cells showing that STAT5 outcompetes STAT3 for binding to the BCL6 gene to regulate BCL6. Interestingly, the authors did not analyze binding to region B but to a site approximately 0.4 kb upstream of region B (23). There is a weak site corresponding roughly to that region that is detected in some of the other human ChIP-seq data (see Fig. S2 in the supplemental material). This may be an additional site that STATs utilize to regulate BCL6 in some cell types. Interestingly, in contrast to TH1 cells, which are regulated by IL-2 and STAT5, follicular helper T cells (TfH cells) are regulated by IL-6 and IL-21, cytokines that activate STAT3. BCL6 has been shown to be upregulated by IL-6 and IL-21 to promote differentiation along the TfH lineage (24). It has also recently been reported that STAT5 negatively regulates TfH cell generation (25). In addition, BCL6 expression is low in these cells when STAT5 is activated (25). The authors suggest that STAT5 promotes upregulation of Blimp-1, which then represses BCL6 expression, though they do not directly show that STAT5 regulates Blimp-1 expression. BCL6 and Blimp-1 reciprocally repress each other; therefore, it is possible that STAT5 represses BCL6 expression, which allows Blimp-1 to be upregulated, thereby preventing TfH generation. This suggests that STAT5 and STAT3 modulation of BCL6 expression may affect helper T cell lineages based upon BCL6 regulation and further supports a role for STAT5 and STAT3 competing to regulate BCL6 expression.
BCL6 and STATs also compete to regulate some of the same genes (20, 21, 26). In liver cells, there are sex-biased genes regulated by STAT5 that affect many cellular processes (22). Interestingly, in male liver cells, BCL6 bound to STAT5 targets that were female biased and thus not expressed in male liver cells (21). This suggests that BCL6 may help to modulate STAT target genes. We have found that STAT5 competes with BCL6 for binding to region B to regulate BCL6 expression (Fig. 2B). BCL6 has been shown to partially repress its own expression (27), leading to a stable equilibrium of expression. However, we demonstrate that STATs do not utilize BCL6 to modulate its expression, since BCL6 binding is reduced when STATs bind (Fig. 2). Importantly, reduction of BCL6 binding to region B upon STAT5 binding is concomitant with decreased RNA polymerase II binding and activity (Fig. 2 and 6; see Fig. S5 in the supplemental material), demonstrating that STAT5 binding promotes greater inhibition of transcription than BCL6 does on its own. As further evidence that STAT binding at this site supersedes the effect of BCL6, STAT3 binding does not greatly reduce BCL6 binding to region B; however, RNA polymerase II is recruited and is activated when STAT3 binds, and transcription of BCL6 increases markedly. Potentially, STAT3 upregulation of BCL6 might be enhanced even further if BCL6 is not bound to region B.
Recent work has found that, like BCL6, IL-17 is oppositely regulated by STAT5 and STAT3 (28). STAT5-mediated repression of IL-17 is associated with recruitment of the transcriptional repressor NCoR2 and a loss of permissive modifications of histone H3 at this locus. This is distinct from the mechanism by which BCL6 is regulated, since histone acetylation is not involved in STAT5-mediated repression of the gene. It was further demonstrated that approximately 300 additional genes in T cells are oppositely regulated by STAT3 and STAT5, though whether this is true in breast cells remains to be seen.
Analysis of STAT target genes in mammary epithelial cells using gene expression microarrays identified the prosaposin gene (Psap) as being upregulated by STAT5 and repressed by STAT3 (29). This provides further evidence that STAT3 and STAT5 can oppositely regulate specific genes and shows that either STAT can mediate upregulation or downregulation in this setting. The mechanism by which Psap is oppositely regulated by these STATs is currently unknown. Further analysis in breast cancer cells by RNA expression studies and ChIP-seq may identify other genes that are oppositely regulated by these STATs, and this may provide insights into the mechanism by which this occurs, as well as its biological significance.
Our work and that of others clearly demonstrates that STAT5 and STAT3 are not redundant transcription factors and in fact can oppositely regulate certain genes. This is important in our understanding of transcriptional regulation, as both of these highly homologous transcription factors are ubiquitously expressed, have similar DNA binding specificities, and are generally thought of only as transcriptional activators. This finding also has implications for the therapeutic use of STAT inhibitors in cancers in which both STATs are activated (30, 31). In the case of breast cancer, STAT5 has been shown to modulate the effects of STAT3, allowing reduced growth and increased sensitivity to cancer therapy (5). Therefore, in this setting, STAT5 inhibitors may not be a useful therapy, since genes that are normally repressed by STAT5, including BCL6, would be upregulated by STAT3, promoting a potentially more aggressive tumor. Both STAT3 and STAT5 can also be activated in leukemias (32, 33), and these STATs have similar opposite effects on the regulation of BCL6 expression in hematopoietic cells (data not shown), though the relative contributions of STAT5 and STAT3 to the biology of these cells are unclear. Understanding the patterns of activation of these STATs in malignant cells, and their effects alone and in combination on gene expression, may have important implications for targeted cancer therapy.
In conclusion, we have demonstrated that STAT5 and STAT3 compete for binding to region B of the BCL6 gene and lead to opposite effects on gene transcription. Taken together with previously published data, it is clear that STAT5 and STAT3 can regulate distinct sets of genes or regulate the same group of genes in either a coordinated or opposing manner. These findings highlight the importance of the genomic context in the intricacies by which transcription factors regulate gene expression.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R01-CA160979, Susan G. Komen for the Cure, a BCRF-AACR grant for Translational Breast Cancer Research, the Brent Leahey Fund, and Friends of the Dana-Farber Cancer Institute.
Footnotes
Published ahead of print 28 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01620-12.
REFERENCES
- 1. Nelson EA, Walker SR, Alvarez JV, Frank DA. 2004. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J. Biol. Chem. 279:54724–54730 [DOI] [PubMed] [Google Scholar]
- 2. Walker SR, Nelson EA, Frank DA. 2007. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene 26:224–233 [DOI] [PubMed] [Google Scholar]
- 3. Horvai AE, Xu L, Korzus E, Brard G, Kalafus D, Mullen TM, Rose DW, Rosenfeld MG, Glass CK. 1997. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc. Natl. Acad. Sci. U. S. A. 94:1074–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alvarez JV, Febbo PG, Ramaswamy S, Loda M, Richardson A, Frank DA. 2005. Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer Res. 65:5054–5062 [DOI] [PubMed] [Google Scholar]
- 5. Walker SR, Nelson EA, Zou L, Chaudhury M, Signoretti S, Richardson A, Frank DA. 2009. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol. Cancer Res. 7:966–976 [DOI] [PubMed] [Google Scholar]
- 6. Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, Wu Z, Gonen M, Mulvey LA, Bessarabova MO, Huh SJ, Silver SJ, Kim SY, Park SY, Lee HE, Anderson KS, Richardson AL, Nikolskaya T, Nikolsky Y, Liu XS, Root DE, Hahn WC, Frank DA, Polyak K. 2011. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24- stem cell-like breast cancer cells in human tumors. J. Clin. Invest. 121:2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iavnilovitch E, Cardiff RD, Groner B, Barash I. 2004. Deregulation of Stat5 expression and activation causes mammary tumors in transgenic mice. Int. J. Cancer 112:607–619 [DOI] [PubMed] [Google Scholar]
- 8. Tworoger SS, Sluss P, Hankinson SE. 2006. Association between plasma prolactin concentrations and risk of breast cancer among predominately premenopausal women. Cancer Res. 66:2476–2482 [DOI] [PubMed] [Google Scholar]
- 9. Logarajah S, Hunter P, Kraman M, Steele D, Lakhani S, Bobrow L, Venkitaraman A, Wagner S. 2003. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene 22:5572–5578 [DOI] [PubMed] [Google Scholar]
- 10. Bos R, van Diest PJ, van der Groep P, Greijer AE, Hermsen MA, Heijnen I, Meijer GA, Baak JP, Pinedo HM, van der Wall E, Shvarts A. 2003. Protein expression of B-cell lymphoma gene 6 (BCL-6) in invasive breast cancer is associated with cyclin D1 and hypoxia-inducible factor-1alpha (HIF-1alpha). Oncogene 22:8948–8951 [DOI] [PubMed] [Google Scholar]
- 11. Nelson EA, Walker SR, Kepich A, Gashin LB, Hideshima T, Ikeda H, Chauhan D, Anderson KC, Frank DA. 2008. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood 112:5095–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X, Li Z, Naganuma A, Ye BH. 2002. Negative autoregulation of BCL-6 is bypassed by genetic alterations in diffuse large B cell lymphomas, Proc. Natl. Acad. Sci. U. S. A. 99:15018–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ambrose HE, Papadopoulou V, Beswick RW, Wagner SD. 2007. Poly-(ADP-ribose) polymerase-1 (Parp-1) binds in a sequence-specific manner at the Bcl-6 locus and contributes to the regulation of Bcl-6 transcription. Oncogene 26:6244–6252 [DOI] [PubMed] [Google Scholar]
- 14. Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, Pernis A, Pasqualucci L, Dalla-Favera R. 2007. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell 12:280–292 [DOI] [PubMed] [Google Scholar]
- 15. Zhao H, Nakajima R, Kunimoto H, Sasaki T, Kojima H, Nakajima K. 2004. Region 752–761 of STAT3 is critical for SRC-1 recruitment and Ser727 phosphorylation. Biochem. Biophys. Res. Commun. 325:541–548 [DOI] [PubMed] [Google Scholar]
- 16. Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- 17. Peters CA, Maizels ET, Robertson MC, Shiu RPC, Soloff MS, Hunzicker-Dunn M. 2000. Induction of relaxin messenger RNA expression in response to prolactin receptor activation requires protein kinase C delta signaling. Mol. Endocrinol. 14:576–590 [DOI] [PubMed] [Google Scholar]
- 18. Wen Z, Zhong Z, Darnell JE., Jr 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241–250 [DOI] [PubMed] [Google Scholar]
- 19. Chung J, Uchida E, Grammer TC, Blenis J. 1997. STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol. Cell. Biol. 17:6508–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, Ryder A, Johnson KJ, Neilson LM, Liu C, Brill KL, Rosenberg AL, Witkiewicz AK, Rui H. 2010. Prolactin inhibits BCL6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res. 70:1711–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y, Laz EV, Waxman DJ. 2012. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol. 32:880–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meyer RD, Laz EV, Su T, Waxman DJ. 2009. Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol. Endocrinol. 23:1914–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oestreich KJ, Mohn SE, Weinmann AS. 2012. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol. 13:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science 325:1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, Watowich SS, Qi H, Dong C, Wang D. 2012. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J. Biol. Chem. 287:11234–11239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fernandez de Mattos S, Essafi A, Soeiro I, Pietersen AM, Birkenkamp KU, Edwards CS, Martino A, Nelson BH, Francis JM, Jones MC, Brosens JJ, Coffer PJ, Lam EW. 2004. FoxO3a and BCR-ABL regulate cyclin D2 transcription through a STAT5/BCL6-dependent mechanism. Mol. Cell. Biol. 24:10058–10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mendez LM, Polo JM, Yu JJ, Krupski M, Ding BB, Melnick A, Ye BH. 2008. CtBP is an essential corepressor for BCL6 autoregulation. Mol. Cell. Biol. 28:2175–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, Kanno Y, O'Shea JJ, Laurence A. 2011. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12:247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clarkson RW, Boland MP, Kritikou EA, Lee JM, Freeman TC, Tiffen PG, Watson CJ. 2006. The genes induced by signal transducer and activators of transcription (STAT)3 and STAT5 in mammary epithelial cells define the roles of these STATs in mammary development. Mol. Endocrinol. 20:675–685 [DOI] [PubMed] [Google Scholar]
- 30. Benekli M, Baumann H, Wetzler M. 2009. Targeting signal transducer and activator of transcription signaling pathway in leukemias. J. Clin. Oncol. 27:4422–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frank DA. 2003. STAT signaling in cancer: insights into pathogenesis and treatment strategies. Cancer Treat. Res. 115:267–291 [DOI] [PubMed] [Google Scholar]
- 32. Lin TS, Mahajan S, Frank DA. 2000. STAT signaling in the pathogenesis and treatment of leukemias. Oncogene 19:2496–2504 [DOI] [PubMed] [Google Scholar]
- 33. Bar-Natan M, Nelson EA, Walker SR, Kuang Y, Distel RJ, Frank DA. 2012. Dual inhibition of Jak2 and STAT5 enhances killing of myeloproliferative neoplasia cells. Leukemia 26:1407–1410 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.