Abstract
tRNA isopentenyltransferases (Tit1) modify tRNA position 37, adjacent to the anticodon, to N6-isopentenyladenosine (i6A37) in all cells, yet the tRNA subsets selected for modification vary among species, and their relevance to phenotypes is unknown. We examined the function of i6A37 in Schizosaccharomyces pombe tit1+ and tit1-Δ cells by using a β-galactosidase codon-swap reporter whose catalytic activity is sensitive to accurate decoding of codon 503. i6A37 increased the activity of tRNACys at a cognate codon and that of tRNATyr at a near-cognate codon, suggesting that i6A37 promotes decoding activity generally and increases fidelity at cognate codons while decreasing fidelity at noncognate codons. S. pombe cells lacking tit1+ exhibit slow growth in glycerol or rapamycin. While existing data link wobble base U34 modifications to translation of functionally related mRNAs, whether this might extend to the anticodon-adjacent position 37 was unknown. Indeed, we found a biased presence of i6A37-cognate codons in high-abundance mRNAs for ribosome subunits and energy metabolism, congruent with the observed phenotypes and the idea that i6A37 promotes translational efficiency. Polysome profiles confirmed the decreased translational efficiency of mRNAs in tit1-Δ cells. Because subsets of i6A37-tRNAs differ among species, as do their cognate codon-sensitive mRNAs, these genomic variables may underlie associated phenotypic differences.
INTRODUCTION
tRNAs contain nucleotide modifications, residing mostly in the tRNA body, that confer a range of activities (reviewed in references 1 and 2). Another class of modifications occurs in the anticodon loop (ACL), mostly at positions 34 and 37, and contributes to tRNA fit in the ribosome decoding center and to codon-anticodon interactions (3–5). Positions 34 and 37 bear the most diverse modifications, with some requiring multiple enzymatic activities (1). Position 34 modifications control the ability of tRNA to wobble to synonymous codons and/or restrict pairing with noncognate codons (5). The TRM9 U34 methyltransferase modification occurs on a subset of tRNAs that sensitize specific mRNAs bearing a high abundance of cognate codons that contribute to a DNA damage response (6). A different U34 modification on different tRNAs sensitizes other mRNAs with cognate codon usage (7).
Position 37 is considered part of the extended anticodon (8) and can carry three types of modification: N6-isopentenyladenosine (i6A), N6-threonylcarbamoyladenosine (t6A) or its cyclized derivative ct6A (in bacteria and archaea) (9), and wybutosine (yW), among which the A37 variety is further modified in some organisms. In bacteria, i6A37 is modified to 2-methylthio-N6-isopentenyladenosine-37 (ms2i6A37) and/or the hydroxyl derivative, ms2i(o)6A37, whereas in eukaryotes it remains i6A37. The i6A37 and (c)t6A37 modifications occur on nonoverlapping sets of tRNAs and function to strengthen the adjacent (position 36), weak anticodon-codon pairs A-U and U-A, respectively, in the ribosome decoding center (5). It was recently reported that Cdkal1, encoded by a risk gene for type 2 diabetes (10), methylthiolates t6A37 to ms2t6A37 on tRNALysUUU, and its deletion leads to type 2 diabetes in mice (11).
Translational misreading can have positive or negative effects on growth (reviewed in reference 12). Misreading is largely affected by competition among tRNAs for ribosome fit and codon match, and depending on the system at hand, rates can vary from 10−3 to 10−5 (13–15). A potential source of differing rates might be the species-specific composition of the tRNA gene complement (tRNAome) (16). Consistent with this possibility are mutants in Maf1, a repressor of tRNA synthesis, that exhibit asymmetric increases in the levels of different tRNAs and increased translational fidelity at UAA and UAG codons (17).
The MiaA gene encodes the Escherichia coli i6A-transferase, and its eukaryotic homologs encode Tit1 in Schizosaccharomyces pombe, MOD5 in Saccharomyces cerevisiae, the life span gene product GRO-1 in Caenorhabditis elegans, and the tumor suppressor TRIT1 in humans (18–23). Curiously, i6A37 occurs on different subsets of tRNAs in different model organisms. In bacteria, ms2i6A37 or ms2i(o)6A37 is found on tRNAs for UNN codons, i.e., for Cys, Leu, Phe, Ser, Trp, and Tyr (2, 24). In eukaryotes, i6A37 is significantly more restricted, as it is found on tRNAs for Ser, Tyr, and sometimes Cys or Trp. tRNACysGCA carries i6A37 in S. cerevisiae but not in S. pombe, whereas tRNATrpCCA carries i6A37 in S. pombe but not in S. cerevisiae (18), and the distribution in humans differs from those in both yeasts (T. N. Lamichhane and R. J. Maraia, unpublished data).
i6A37 does not affect charging, as yeast tRNAs lacking or containing i6A37 are equally aminoacylated (25), consistent with work on E. coli showing that the modification increases the efficiency of ribosome binding but not aminoacylation (reviewed in references 24 and 26). A lack of i6A37 derivatives slows bacterial cell growth (27–29). In the yeasts S. cerevisiae and S. pombe, the defects known to be attributable to a lack of i6A37 are due to decreased activity of mutant tRNAs known as suppressors (18, 19, 25, 30, 31). Suppressor tRNA activity reflects competition with translation termination/release factors, whereas natural tRNAs compete only with each other during normal elongation. In yeast, the absence of i6A37 decreases suppressor tRNA-mediated suppression but has little effect on growth (19, 25, 30).
A crystal structure of the bacterial ribosome-mRNA-tRNAPhe complex shows that the sulfur moiety of ms2i6A37-tRNAPhe makes direct contacts with and stabilizes the codon-anticodon interaction (32). Yet, as noted above, sulfonation of i6A37 does not occur in eukaryotes. To our knowledge, the function of tRNA-i6A37 in normal translation has not been examined in any eukaryotic system. Moreover, while published data link modifications of the wobble base U34 to translation of mRNA subsets related to observable phenotypes, whether or not this would be extendable to the anticodon-adjacent position 37 was unknown.
We provide evidence that i6A37 increases the functional efficiency of tRNAs to which it is added for cognate and near-cognate codons in S. pombe, increasing fidelity at cognate codons while decreasing fidelity at noncognate codons. Depletion of i6A37 by Tit1 deletion appears to reset levels of tRNA competition and misreading, translational efficiency, and translational fidelity. The data indicate that there is a translational deficiency in tit1-Δ cells that leads to rapamycin sensitivity and slow growth in glycerol. Computational searches for biased use of synonymous cognate codons for i6A37-tRNAs identified highly abundant mRNAs for ribosome subunits, translation factors, and enzymes involved in energy metabolism, congruent with observed phenotypes. Because subsets of i6A37-containing tRNAs vary among species, as do mRNAs with cognate codon biases, these genome-specific parameters may help to explain the diverse phenotypes associated with i6A37.
MATERIALS AND METHODS
S. pombe strains used in this study are listed in Table 1. S. pombe cells were seeded to an optical density at 600 nm (OD600) of 0.1 and grown until an OD600 of 0.5, and 10-fold dilutions were plated on appropriate medium. For rapamycin (AG Scientific Inc.) treatment, cells were treated with the indicated concentrations for 2 h or with the drug vehicle (dimethyl sulfoxide [DMSO]; Sigma). For mitochondrial tRNA (mt-tRNA) analysis, cells were grown in Edinburgh minimal medium (EMM) lacking uracil.
Table 1.
Strains used in this study
Plasmids.
lacZ was amplified from E. coli and cloned into pREP4X with a 3×-FLAG tag at the C terminus. The UAC sequence at codon 503 was changed to UGC by site-directed mutagenesis. tRNACys-G37 from S. pombe, including 200 bp upstream, was amplified, cloned into pREP3X, and then subjected to site-directed mutagenesis to create tRNACys-A37. All constructs were confirmed by sequencing.
RNA preparation and Northern blotting.
Cells were grown to an OD600 of 0.5 in appropriate medium. Purified RNA was electrophoresed in Novex 10% Tris-borate-EDTA (TBE)–urea gels, transferred to GeneScreen nylon by use of an Invitrogen iBlot device, and probed with 32P-labeled oligonucleotides as described previously (18).
Protein extraction and Western blotting.
Cells were grown to an OD600 of 0.5, and protein was extracted in lysis buffer (150 mM NaCl, 50 mM Tris-Cl, pH 7.5, 1 mM EDTA, 0.1% NP-40, and 0.1 mM phenylmethylsulfonyl fluoride [PMSF]), using glass beads and a Biospec Mini-Beadbeater 4 times for 30 s each, with 1 min on ice between beatings. Debris was pelleted, and the supernatant was collected, quantified by the Bradford assay, analyzed by 4 to 12% Bis-Tris PAGE (Novex), stained with Ponceau S, rinsed, and transferred to nitrocellulose by use of an Invitrogen iBlot transfer apparatus.
Beta-galactosidase.
tit1+ and tit1-Δ cells were grown to an OD600 of 0.5 and resuspended in 100 mM Tris-HCl, pH 8, 1 mM dithiothreitol (DTT), and 20% glycerol. Total protein was prepared and quantified by the Bio-Rad Bradford assay. Equal amounts of protein were used. ortho-Nitrophenyl-β-galactoside (ONPG) was freshly prepared in Z buffer (60 mM Na2HPO4 · 7H2O, 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO4, 5 mM β-mercaptoethanol, pH 7.0). A solution of 1 mg ONPG was incubated with protein at 37°C. With wild-type lacZ, 100 μg protein was incubated for 10 min; with lacZ encoding a protein with the Y503C mutation (lacZ-Y503C), 1 mg protein was incubated for 16 h. Reactions were stopped with 1 mM sodium carbonate. Absorbance was measured at 420 nm, and lacZ activity was calculated as described previously (Stratagene) (33).
Polysome profiles were obtained from fresh extracts made from yYH1 and yNB5 cells after growth in yeast extract with supplements (YES) medium, using a programmable density gradient fractionation system spectrophotometer (Foxy Jr.; Teledyne Isco, Lincoln, NE) with a gradient master (Biocomp) as described previously (34). Specific mRNAs were analyzed by Northern blotting.
[35S]methionine labeling.
Wild-type and tit1-Δ strains were grown in 5 ml EMM in the presence of DMSO or rapamycin (100 ng/ml). After the OD600 reached 0.5, 150 μCi of [35S]methionine (PerkinElmer) was added, and the cells were pulse labeled for 30 min at 32°C. Equal volumes of ice-cold water were added to each culture, and the cells were harvested. The cells were resuspended in 1 ml ice-cold water. Washed cells were harvested and whole-cell extracts separated by 10% PAGE. Gels were stained with SimplyBlue (Invitrogen, Carlsbad, CA), fixed, dried, and quantified with VisionWorks software to confirm that signals were in the linear range. Quantification of 35S was performed with a model FLA3000 phosphorimager. The ratios of 35S to SimplyBlue stain for 3 independent experiments were averaged and plotted.
Generation of codon-biased gene lists and GO term enrichment.
Based on previously developed methods (6), codon bias was defined utilizing a simple metric of the number of i6A37-tRNA cognate codons versus the total number of synonymous codons. UCU codon or i6A37-tRNA(Ser) cognate codon counts versus total serine codon counts were compiled over all S. pombe mRNAs (GenBank accession no. GCF_000002945.1). Based upon fractional compositions, a Z score was derived for each gene as a metric of individual gene codon bias. Lists were generated based upon gene Z-score sorting. From each list, the top 200 genes were selected and subjected to gene ontology (GO) term enrichment analysis by AmiGO (35).
Nomenclature note.
The S. pombe gene that encodes the tRNA isopentenyltransferase was initially named sin1 (30, 31), but subsequently, another gene was officially named sin1 in the contemporary S. pombe database. Therefore, we had to choose another name.
RESULTS
i6A37 increases translation-decoding activity of tRNATyrGUA.
ACL modifications decrease tRNA rejection by the translating ribosome (36). An in vivo reporter that monitors amino acid misincorporation based on a mutant firefly luciferase with inactivating mutations in the essential active site residue lysine-529 has been used to monitor decoding of specific codons in yeast and bacteria (13, 15). However, lysine is not among the tRNAs that carry i6A37 in S. pombe, which are three tRNASer species (DGA; D = A, C, or U) that decode four of the six Ser codons; tRNATyrGUA, which decodes both Tyr codons; and tRNATrpCCA, which decodes the Trp codon (18). A reporter that is applicable for our purpose is β-galactosidase (lacZ), which requires Tyr at position 503 for enzymatic activity (37, 38). While activity was severely diminished by amino acid substitutions at this position, the resulting LacZ proteins suffered no detectable change in overall structure or stability (37–39). Tyr-503 is encoded by UAC and decoded in S. pombe by tRNATyrGUA containing i6A37 (18). We cloned lacZ into an S. pombe expression vector and mutated the second position (A) of the Tyr-503 UAC codon to G to create the near-cognate Cys codon UGC; the resultant gene is referred to as lacZ-Y503C. Thus, only if tRNATyrGUA misread the UGC 503 codon would lacZ-Y503C activity be observed. Consistent with expected misreading rates, we observed an ∼1,000-fold decrease in lacZ activity due to replacement of the Tyr-503 codon with the UGC Cys codon in S. pombe (not shown) (Fig. 1A). For the present study, we used lacZ-Y503C as an in vivo reporter.
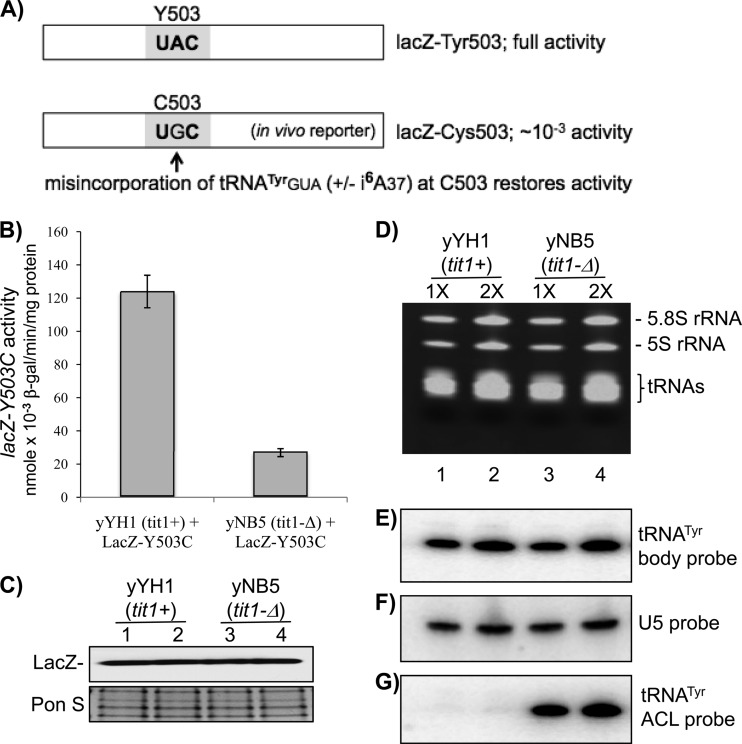
Fig 1.
i6A37 increases competitive decoding activity of tRNATyrGUA at a near-cognate codon. (A) Schematic of experimental design. (B) lacZ activities of extracts from yYH1 (tit1+) and yNB5 (tit1-Δ) cells after transformation with lacZ-Y503C. Error bars reflect the ranges for triplicate experiments. (C) Immunoblot detection of FLAG-tagged LacZ-Y503C protein in extracts used for panel B, using anti-FLAG antibody (upper panel). (Lower panel) Ponceau S (Pon S)-stained gel prior to transfer, as a loading control. (D to G) RNA analysis. (D) Ethidium bromide-stained denaturing polyacrylamide gel used to make a blot that was sequentially probed as indicated to the right of panels E to G (see the text). Total RNAs from yYH1 (tit1+) and yNB5 (tit1-Δ) cells were loaded at two concentrations, i.e., 1× (5 μg) and 2× (10 μg), as indicated above the lanes.
We first used lacZ-Y503C to compare activities in the wild-type tit1+ (yYH1) and tit1-Δ (yNB5) strains, in which tRNATyrGUA is modified with i6A37 and unmodified, respectively (18). S. pombe tRNACysGCA has an encoded G at position 37 and is not a Tit1 substrate in either strain (18), so it was not a variable here. We observed higher lacZ activity in tit1+ cells than in tit1-Δ cells (Fig. 1B). There was no difference in the amounts of LacZ protein accumulated in these cells as monitored by anti-FLAG immunoblotting (duplicate loadings) (Fig. 1C). Thus, i6A37 on tRNATyrGUA caused misreading of the near-cognate Cys codon. We showed previously that i6A37 does not affect levels of tRNASer or tRNATrp in S. pombe (18). We showed this here for tRNATyr in the cells used for lacZ analysis: there was no significant difference in tit1+ and tit1-Δ cells (Fig. 1D to F). Differential i6A37 modification of tRNATyrGUA in the tit1+ and tit1-Δ cells was confirmed using the PHA6 assay (positive hybridization in the absence of i6A37), in which the signal intensity increases as modification decreases (18) (Fig. 1G). The i6A modification decreases hybridization with an oligonucleotide probe because the large, bulky, hydrophobic isopentenyl group on N6 of A interferes with base pairing to T. The data suggest that i6A37-tRNATyrGUA competes ∼4 times better than unmodified tRNATyrGUA for the near-cognate lacZ UGC 503 codon.
This result was counterintuitive to the expectation that the presence of a modification should improve rather than worsen decoding fidelity. The presence of mnm5s2 on wobble base U34 of tRNALys was unexpectedly found to increase third-position codon misreading in bacteria (40). To our knowledge, increased second-position misreading due to the presence of i6A37 had not previously been observed.
The unexpected results with tRNATyrGUA raised the question of how i6A37 would affect the decoding fidelity of a tRNA for its cognate codon. To answer this question, we monitored the degree to which tRNACysGCA in the i6A37-modified versus unmodified state could prevent decoding by tRNATyrGUA by using lacZ-Y503C. While tRNACysGCA from bacteria and eukaryotes, including the yeast S. cerevisiae, contains i6A37, the S. pombe tRNACysGCA does not, because its tRNACysGCA genes encode G rather than A at position 37 (18, 41; http://gtrnadb.ucsc.edu/). We could circumvent this with an S. pombe tRNACysGCA gene whose position 37 was changed from G to A by single nucleotide mutagenesis, comparing the tRNACysGCA-G37 and tRNACysGCA-A37 genes for lacZ-Y503C activity in the tit1+ and tit1-Δ strains.
i6A37 increases tRNACysGCA activity at its cognate codon.
We cloned the S. pombe tRNACysGCA gene into a plasmid. Point mutation of tRNACysGCA G37 to A37 created an A36A37A38 recognition site for Tit1p (18). For plasmid selection, we needed to use another tit1-Δ strain, yTL1, and its tit1+ isogenic control, yAS99 (Fig. 2).
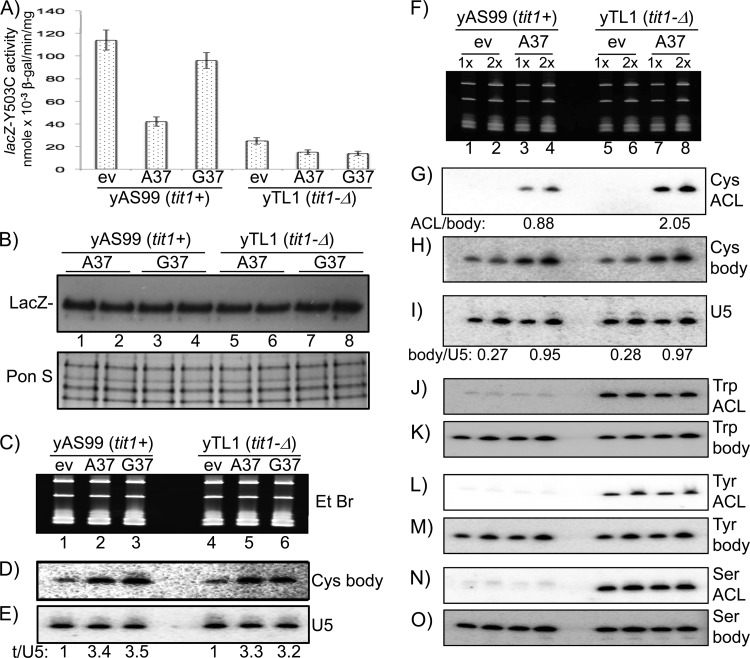
Fig 2.
i6A37 increases tRNACysGCA activity at its cognate codon. (A) lacZ-Y503C activities as shown in Fig. 1, after transformation of yYH1 (tit1+) and yTL1 (tit1-Δ) cells with empty vector (ev) or vector containing the S. pombe tRNACysGCA-G37 or tRNACysGCA-A37 gene, as indicated below the bars. (B) Immunoblot detection of FLAG-tagged LacZ-Y503C protein in extracts used for panel A, using anti-FLAG antibody (upper panel). (Lower panel) Ponceau S-stained gel prior to transfer, as a loading control. (C to E) Analysis of tRNACysGCA-G37 and tRNACysGCA-A37 levels. (C) Ethidium bromide (EtBr)-stained gel used to make a blot that was sequentially probed as indicated to the right of panels D and E for comparison of expression of ectopic tRNACysGCA-G37 and tRNACysGCA-A37 by use of a body probe that has perfect complementarity to both, with U5 RNA as a loading control. Quantitation of U5 and tRNA levels was done by phosphorimager analysis, and the relative ratios are indicated below the lanes. (F to I) Analysis of i6A37 modification of tRNACysGCA-A37. (F) Ethidium bromide-stained gel. (G) Blot probed for i6A37 modification of tRNACysGCA-A37 by use of the PHA6 assay. See the text for interpretation of the quantification shown under the lanes. (H) Blot probed for tRNACysGCA-G37 and tRNACysGCA-A37 by use of a body probe with perfect complementarity to both. (I) Blot probed for U5 snRNA. Quantitation of U5 and tRNA levels in panels H and I was done by phosphorimager analysis, and the relative ratios are indicated below the lanes. (J to O) ACL and body probing for tRNATrp, tRNATyr, and tRNASerAGA on the same blot, as indicated to the right of the panels.
According to tRNA competition, an increase in tRNACysGCA levels would decrease the lacZ-Y503C activity resulting from misreading by tRNATyrGUA-i6A37 in tit1+ cells. While tRNACysGCA-G37 and -A37 decreased lacZ-Y503C activity, the A37 residue did so more than the G37 residue in the tit1+ strain (Fig. 2A). Significantly, in contrast to the case with the tit1+ strain, tRNACysGCA-G37 and -A37 decreased activity to the same extent in tit1-Δ cells, in which neither tRNA contains i6A37 (Fig. 2A and B).
We examined the tRNAs and found similar expression from the ectopic tRNACysGCA-A37 and -G37 genes with a probe complementary to their identical ψ stem-loops (Fig. 2C and D, “body” probe). Normalization to U5 RNA revealed that tRNACysGCA in the transformed cells accumulated to about 3-fold higher levels than those of endogenous tRNACysGCA (Fig. 2E).
We next examined the i6A37 modification status of ectopic tRNACysGCA-A37 in the tit1+ cells by using the PHA6 assay. The ACL A37 probe detected ectopic tRNACysGCA-A37 without cross-reactivity with endogenous tRNACysGCA-G37 (Fig. 2F and G). A detectable signal from tit1+ cells (Fig. 2G, lanes 3 and 4) indicated that the extent of i6A37 modification of ectopic tRNACysGCA-A37 was incomplete, consistent with partial modification due to overexpression. Quantification after normalization to the body probe (Fig. 2H) indicated that tRNACysGCA-A37 was modified only ∼57% in the tit1+ cells (Fig. 2G) {the ACL/body ratio was calculated as follows: [1 − (0.88/2.05)] × 100}. Accounting for incomplete modification suggests that i6A37 increased the specific activity of tRNACysGCA-A37 for its cognate codon almost 4-fold relative to that of unmodified tRNACysGCA. Quantification using U5 snRNA showed similar loading and relative expression levels of the ectopic tRNAs (Fig. 2I). Analysis of endogenous tRNATrp, tRNATyr, and tRNASerAGA on the same blot showed full modification (Fig. 2J to O), suggesting that the hypomodification in these strains was limited to the ectopic tRNACysGCA-A37.
In summary, the data for tit1+ cells indicate that tRNACysGCA-i6A37 competes better than tRNACysGCA-G37 for the cognate codon. The data for tit1-Δ cells argue that unmodified tRNACysGCA-A37 and -G37 compete equally against unmodified tRNATyrGUA. tRNACysGCA-G37 decreased lacZ-Y503C activity by 15% and 40% in the tit1+ and tit1-Δ strains, respectively, which is reflective of the differential competition by modified and unmodified tRNATyrGUA in the two strains. A conclusion is that the absence of i6A37 due to tit1 deletion disrupts the homeostasis of codon misreading in S. pombe.
Deletion of tit1+ causes sensitivity to rapamycin.
Increases in mistranslation can lead to positive or negative effects on growth (reviewed in references 12 and 42). Unlike growth of S. cerevisiae and human cells in vitro, normal growth of S. pombe is insensitive to rapamycin, a small-molecule drug that inhibits TOR kinases that control growth and proliferation in response to nitrogen, amino acids, and other nutrients (43, 44). Rapamycin does impose adverse effects on S. pombe, but they are not quite severe enough to slow growth, because its inhibition of the tor2+ component of TORC1 is only partial (45). Defects in nuclear tRNA metabolism in sla1-Δ mutants lacking the pre-tRNA chaperone known as La protein lead to a nutritional stress response manifested by leucine uptake deficiency, increased expression of amino acid-metabolizing (AAM) genes, and sensitivities to NH3 and rapamycin, indicative of TOR system perturbation (46).
While yYH1 (tit1+) and yNB5 (tit1-Δ) grew similarly on EMM, yNB5 growth was inhibited by rapamycin (Fig. 3A). Slow growth in rapamycin was rescued by ectopic tit1+ (yNB5 plus tit1+) and S. cerevisiae MOD5 but not by the previously characterized tit1-T12A catalytic mutant (18) (Fig. 3A).
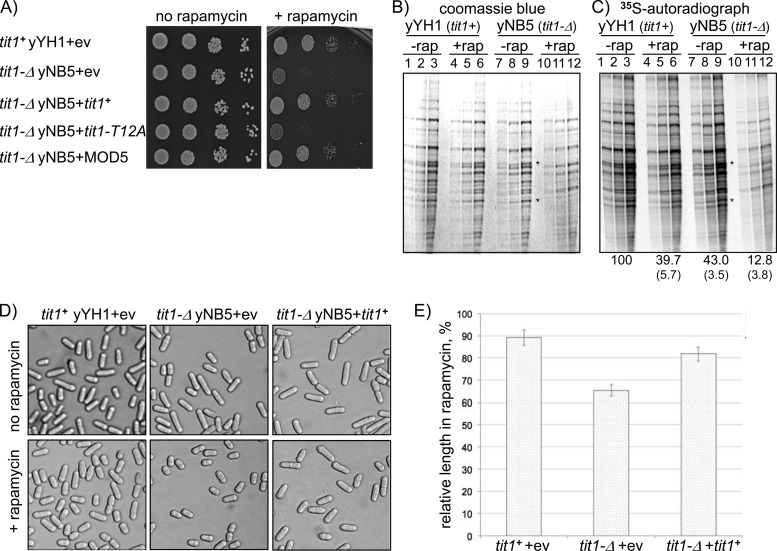
Fig 3.
Deletion of tit1+ causes growth and morphology phenotypes in rapamycin. (A) Comparative serial dilutions of tit1+ and tit1-Δ cells in EMM lacking uracil (left panel; no rapamycin) or EMM lacking uracil but containing 50 μg/ml rapamycin (right panel; + rapamycin), after transformation with empty vector (+ev) or the indicated ectopic plasmid-borne genes: S. pombe tit1+, the tit1-T12A catalytic mutant, and S. cerevisiae MOD5 (see the text). (B) Coomassie blue-stained SDS gel of extracts after labeling with [35S]methionine in the presence or absence of rapamycin (rap) as indicated above the lanes (each extract was loaded in triplicate, at 1×, 2×, and 4× concentrations). (C) The same gel as in panel B, but after drying and exposure to a phosphorimager to visualize 35S-labeled proteins. The asterisks indicate bands specifically absent for rapamycin-treated tit1-Δ cells (see the text). Quantitation results from 3 independent experiments are reported under the lanes. Lanes from gels after Coomassie blue staining were quantified with VisionWorks software, which confirmed that the signals were in the linear range. 35S was quantified with a FLA3000 phosphorimager. The 35S/blue ratios relative to that of tit1+ cells without rapamycin (100%) are reported. (D) Microscopic analysis of tit1+ and tit1-Δ cells containing empty vector (+ev) or ectopic tit1+, as indicated, in the absence (upper panels) or presence (lower panels) of rapamycin. (E) Quantitative analysis of cell length reduction by rapamycin for the three samples in panel D. Lengths of 50 cells from each of the samples represented by the six panels were measured. Cell length in the presence of rapamycin was expressed as % length in the absence of rapamycin. Error bars reflect standard deviations.
We examined incorporation of [35S]methionine into newly synthesized proteins. [35S]methionine was added to cells in log-phase growth in the presence or absence of rapamycin. Thirty minutes after addition of [35S]methionine, extracts were prepared, loaded in triplicate at 1×, 2×, and 4× concentrations, fractionated in SDS gels, stained with Coomassie blue, photographed (Fig. 3B), dried, and subjected to autoradiography (Fig. 3C). No significant differences in 35S incorporation were evident in tit1+ cells in the presence or absence of rapamycin or in tit1-Δ cells in its absence (Fig. 3B and C, lanes 1 to 9). tit1-Δ cells incorporated less 35S in the presence of rapamycin than did the other cells (Fig. 3B and C, compare lanes 7 and 8 to lanes 10 and 11), consistent with their decreased growth rate. The reproducible qualitative differences described below were more significant observations.
Inspection of the gels revealed a differential pattern in the rapamycin-treated tit1-Δ cell extract relative to the other three samples. Two major bands were specifically decreased in the rapamycin-treated tit1-Δ cells relative to the untreated tit1-Δ cells and the tit1+ cells, as apparent in the Coomassie gel and the autoradiograph (Fig. 3B and C, asterisks). This differential pattern is evidence of more than just a general reduction of protein synthesis and suggests a response to rapamycin that is specific to tit1-Δ cells. The identities of the relevant proteins are currently unknown and are beyond the scope of the present study.
We also observed a morphological response to rapamycin by tit1-Δ cells. While tit1-Δ and tit1+ cells appeared to be similar in the absence of rapamycin, tit1-Δ cells were relatively shortened after exposure to rapamycin (Fig. 3D). S. pombe cell shortening is a characteristic of the wee1 mutant phenotype and also occurs under certain stress conditions and in TOR mutants (47–49). The Wee1 kinase integrates signals that serve to coordinate cell size and cell cycle progression such that cells that have grown sufficiently in mass, reflective of adequate ribosome production, may undergo cell division and proliferation (50, 51). We measured the lengths of 50 cells in the presence of rapamycin and 50 cells in its absence for each of the strains/conditions represented by the panels in Fig. 3D and plotted their lengths relative to those of the no-rapamycin controls (Fig. 3E). While rapamycin reduced the length of tit1+ (yYH1) cells by 10%, it caused a more significant reduction in tit1-Δ cell length that was largely reversed by ectopic tit1+ (Fig. 3E).
tit1-Δ cells exhibit slow growth in glycerol.
Slow growth in media containing glycerol as the carbon source reflects mitochondrial deficiency (52). Mod5 is targeted to mitochondria and modifies mt-tRNAs (19, 53–55). It was not known if Tit1 is targeted to mitochondria, although multiple computational programs predicted that it would be (see Discussion). Because mod5 mutants do not exhibit slow growth in glycerol (56; A. Hopper, OSU, personal communication), it is significant that yNB5 does (Fig. 4A). This defect was rescued by ectopic tit1+ but not by the tit1-T12A catalytic mutant, and slightly less so by MOD5 (Fig. 4A).
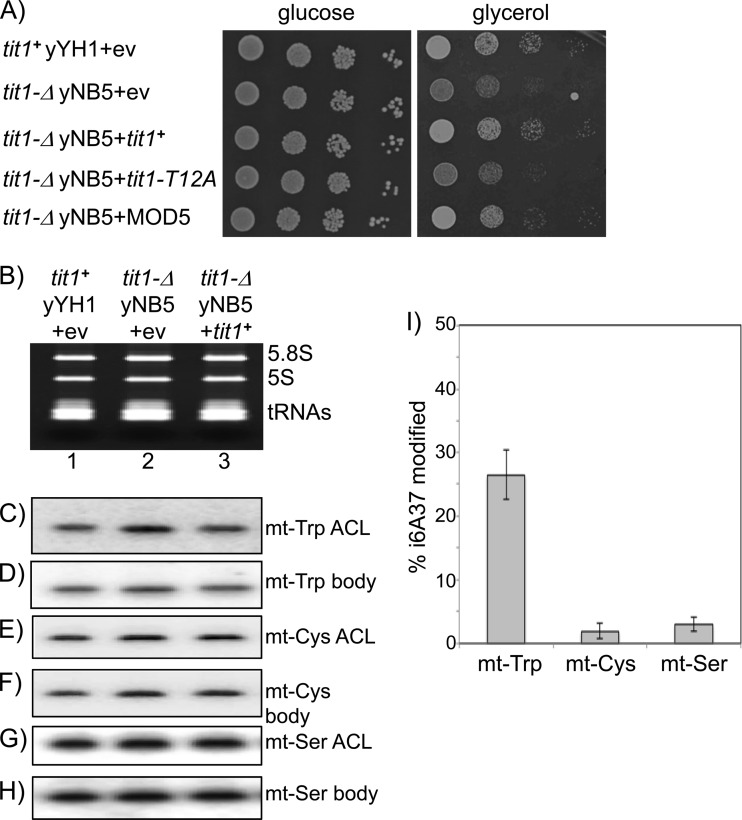
Fig 4.
Deletion of tit1+ causes i6A37 hypomodification of mt-tRNATrp and growth deficiency in glycerol. (A) Comparative serial dilutions of tit1+ and tit1-Δ cells in EMM lacking uracil (left panel; glucose) or EMM lacking uracil but containing glycerol instead of glucose (right panel; glycerol), after transformation with empty vector (+ev) or the indicated ectopic plasmid-borne genes: S. pombe tit1+, the tit1-T12A catalytic mutant, and S. cerevisiae MOD5. (B) Ethidium bromide-stained gel used to make a blot that was probed as shown in panels C to H. PHA6 Northern blot assays were used to compare the hybridization signals of ACL versus body probes for mitochondrial tRNAs mt-tRNATrp, mt-tRNACys, and mt-tRNASer, as indicated to the right of the panels. (I) Quantitative analysis of i6A37 modification of the 3 mt-tRNAs examined in panels B to H. Error bars reflect standard deviations for 5 experiments. Calculations for each mt-tRNA were performed as follows: % i6A37 modification = [1 − (ACLtit1+/BPtit1+)/(ACLtit1-Δ/BPtit1-Δ)] × 100. ACL, anticodon loop probe; BP, body probe.
MiaA and Tit1 modify tRNAs with the sequence AAA at ACL positions 36 to 38 (18, 21, 57). Examination of all S. pombe mitochondrion-encoded tRNA sequences for this feature identified mt-tRNATrp as a potential Tit1 substrate. Analysis of in vivo modification of mt-tRNATrp by the PHA6 assay indeed suggested significant modification (Fig. 4B, C, and D). Probing of the same blot for mt-tRNACys and mt-tRNASer, neither of which was predicted to be a Tit1 substrate, revealed no significant difference between the ACL and body probes (Fig. 4E to H).
Quantitative data from five independent experiments provided statistically significant evidence that mt-tRNATrp is modified, albeit partially, in tit1+ cells (Fig. 4I). Quantitative ACL and body probe data for the three mt-tRNA species examined were applied to the following formula: % modification = [1 − (ACLtit1+/BPtit1+)/(ACLtit1-Δ/BPtit1-Δ)] × 100, where “ACL” indicates the ACL probe and “BP” indicates the body probe. Note that this formula includes internal normalization by using the data from the tit1-Δ samples. The results suggest that although mt-tRNATrp was only partially modified in tit1+ cells under the conditions examined, its hypomodification in tit1-Δ cells may have contributed to their glycerol-sensitive slow growth. When tit1+ and tit-Δ cells were tested in YES (rich) medium or glycerol, the level of mt-tRNATrp modification was not significantly different from that for cells grown in EMM (not shown).
Identification of S. pombe mRNAs enriched in i6A37-tRNA cognate codons.
Although codon use frequency is usually based on genomic averages, it can also be calculated for individual mRNAs relative to the genomic average (6). In S. pombe, three amino acids are delivered to the translating ribosome by i6A37-tRNAs: a subset of Ser tRNAs plus Tyr and Trp tRNAs (18). We took three computational approaches to identify mRNAs with a biased composition of i6A37-cognate codons whose translation might be affected by tit1 deletion (6). We first examined the 5,006 S. pombe mRNAs for the most abundant Tit1 cognate codon, UCU (serine). tRNASerAGA-i6A37 is encoded by 7 genes, a number equal to that of all other serine tRNA genes combined, and its cognate UCU codon is the most used serine codon in S. pombe (41; http://gtrnadb.ucsc.edu/). We examined the mRNAs for gene-specific overuse of UCU. For example, rps1401 mRNA encodes the 40S ribosomal subunit protein S14, which is 140 amino acids long, and 4 of its 5 serine codons are UCU, representing an 80% use of UCU as the Ser codon, whereas the genomic average is 43%. This sorting identified ∼200 mRNAs with overuse of UCU and enrichment Z scores of 1.7 to 5.8, most of which are abundant mRNAs for ribosomal proteins, translation factors, and enzymes involved in mitochondrion-based energy metabolism. We note that although mRNA abundance was not a criterion for sorting, it had been noted previously that abundant codons, including UCU, are found in abundant mRNAs in S. cerevisiae (58). Table 2 lists the top 30 mRNAs, and the full list of mRNAs is available in Dataset S1 in the supplemental material. Among the top 75 mRNAs are multiple mRNAs for mitochondrial proteins.
Table 2.
Top 30 mRNAs enriched in UCU codons
| Locus | Gene name | Product | UCU Z score |
|---|---|---|---|
| SPBC19G7.03c | rps3002 | 40S ribosomal protein S30 | 5.830283744 |
| SPBC211.05 | sab10 | Splicing factor 3B | 5.830283744 |
| SPAC3F10.18c | rpl4102 | 60S ribosomal protein L41 | 5.830283744 |
| SPAC3G6.13c | rpl4101 | 60S ribosomal protein L41 | 5.830283744 |
| SPBC29A3.07c | sab14 | U2 snRNP-associated Sf3b14 homolog | 5.830283744 |
| SPAC11D3.01c | SPAC11D3.01c | Conserved fungal protein | 5.830283744 |
| SPCC1259.05c | cox9 | Cytochrome c oxidase subunit VIIa | 5.830283744 |
| SPBC23G7.05 | sui1 | Translation initiation factor eIF1 | 5.830283744 |
| SPBC25H2.07 | tif11 | Translation initiation factor eIF1A | 5.830283744 |
| SPAC1834.04 | hht1 | Histone H3 h3.1 | 4.590631652 |
| SPAC664.04c | rps1602 | 40S ribosomal protein S16 | 4.590631652 |
| SPAC13G6.07c | rps601 | 40S ribosomal protein S6 | 4.590631652 |
| SPCC576.11 | rpl15 | 60S ribosomal protein L15 | 4.495273799 |
| SPAC25G10.06 | rps2801 | 40S ribosomal protein S28 | 4.384022970 |
| SPAC959.08 | rpl2102 | 60S ribosomal protein L21 | 4.384022970 |
| SPBC21B10.10 | rps402 | 40S ribosomal protein S4 | 4.252544718 |
| SPAP27G11.13c | nop10 | snoRNP pseudouridylase H/ACA | 4.094770815 |
| SPAC3H5.05c | rps1401 | 40S ribosomal protein S14 | 4.094770815 |
| SPAC1071.10c | pma1 | P-type proton ATPase, P3-type Pma1 | 3.943856647 |
| SPAC4A8.15c | cdc3 | Profilin | 3.901936045 |
| SPBP8B7.06 | rpp201 | 60S acidic ribosomal protein P2A | 3.660892583 |
| SPAC1705.02 | SPAC1705.02 | Human 4F5S homolog | 3.660892583 |
| SPAC22A12.04c | rps2201 | 40S ribosomal protein S15a | 3.660892583 |
| SPAC5D6.01 | rps2202 | 40S ribosomal protein S15a | 3.660892583 |
| SPBC8D2.03c | hhf2 | Histone H4 h4.2 | 3.660892583 |
| SPBC1105.12 | hhf3 | Histone H4 h4.3 | 3.660892583 |
| SPAC1834.03c | hhf1 | Histone H4 h4.1 | 3.660892583 |
| SPBC336.10c | tif512 | Translation elongation factor eIF5A | 3.660892583 |
| SPCP1E11.09c | rpp103 | 60S acidic ribosomal protein Rpp1-3 | 3.660892583 |
| SPAC644.15 | rpp101 | 60S acidic ribosomal protein Rpp1-1 | 3.660892583 |
The output of this codon analysis was further analyzed using an S. pombe GO mapper. This revealed enrichments of the ribosomal subunit and of translation processes, with P values from 5.9 × 10−37 to 4 × 10−20, as well as metabolic processes, with P values from 7.6 × 10−8 to 5.4 × 10−7 (Table 3).
Table 3.
GO analysis of top 200 mRNAs enriched in UCU codons
| GO no. | GO term | P value |
|---|---|---|
| 44391 | Ribosomal subunit | 5.92E−37 |
| 5840 | Ribosome | 3.67E−36 |
| 2181 | Cytoplasmic translation | 1.55E−27 |
| 6412 | Translation | 4.07E−20 |
| 44237 | Cellular metabolic process | 7.60E−08 |
| 8152 | Metabolic process | 4.04E−07 |
| 19538 | Protein metabolic process | 4.47E−07 |
| 44267 | Cellular protein metabolic process | 5.49E−07 |
| 10467 | Gene expression | 7.23E−07 |
| 42254 | Ribosome biogenesis | 1.77E−06 |
| 9987 | Cellular process | 3.22E−05 |
| 22613 | RNP complex biogenesis | 3.79E−05 |
| 44238 | Primary metabolic process | 2.11E−04 |
| 44249 | Cellular biosynthetic process | 4.52E−04 |
| 1901576 | Organic substance biosynthetic process | 4.61E−04 |
| 71704 | Organic substance metabolic process | 6.93E−04 |
The second approach was to identify mRNAs enriched in i6A37-cognate codons relative to synonymous codons decoded by tRNAs that do not contain i6A37. This would be only the four UCN Ser codons, because the other two Ser codons (AGY) are decoded by tRNASerGCU, which is not modified by Tit1. All Tyr and Trp codons are decoded by i6A37-tRNAs, and synonymity is not an issue for these and therefore was not considered in this analysis. This identified ∼200 mRNAs, with enrichment Z scores of 1.5 to 2.5, including mRNAs for ribosomal proteins, translation factors, and enzymes involved in energy metabolism (see Dataset S1 in the supplemental material) that largely overlapped the mRNAs identified by the first approach.
While the number of mRNAs identified by these approaches that consider bias with regard to synonymous codon use represent only ∼5% of S. pombe genes, many of them, e.g., those encoding ribosomal proteins and translation factors, are expressed at high levels (see Dataset S1 in the supplemental material), suggesting the potential for significant impact.
The third approach was to identify mRNAs enriched in all i6A37-cognate codons relative to their overall genome averages for the four UCN Ser codons plus the two Tyr and one Trp codon, but without considering bias with regard to synonymous codon use. This identified 79 mRNAs, with enrichment Z scores of 2.08 to 7.78 (see Dataset S1 in the supplemental material), headed by the Wsc1 gene and followed by other genes related to the cell wall, including those encoding glycoproteins and enzymes involved in the major yeast cell wall component, 1,3-β-d-glucan. When these mRNAs were subjected to GO mapper analysis, the cell components identified were “cell surface” and “cell wall,” with P values of 3.18 × 10−12 and 2.25 × 10−10, respectively (see Dataset S1). Wsc1 is a cell wall-associated sensor of the cell wall integrity (CWI) pathway which communicates with TOR signaling (59).
Finally, we examined the top-ranked (highest Z scores) 160 mRNAs in the three sets of computationally derived mRNAs for their abundances, using the mRNA copy number values of Marguerat et al. (60). This revealed that mRNAs with i6A37-sensitive codons that are overused relative to the synonymous i6A37-insensitive codons (i.e., the four i6A37-Ser UCN codons versus the other two Ser [AGY] codons) are highly skewed toward high abundance relative to all other mRNAs (see Dataset S1 in the supplemental material). The mRNAs identified by the third computational approach, i.e., those enriched in all i6A37 codons relative to their genomic averages but without considering bias with regard to synonymous codon use, more closely reflect the abundance distribution of most cellular mRNAs (see Dataset S1).
Evidence of reduced translation efficiency in tit1-Δ cells.
Polypeptide step time during translation elongation, codon context sensitivity, and translational efficiency were reduced in Salmonella and E. coli strains lacking i6A37 and derivatives due to mutation of MiaA (28, 61).
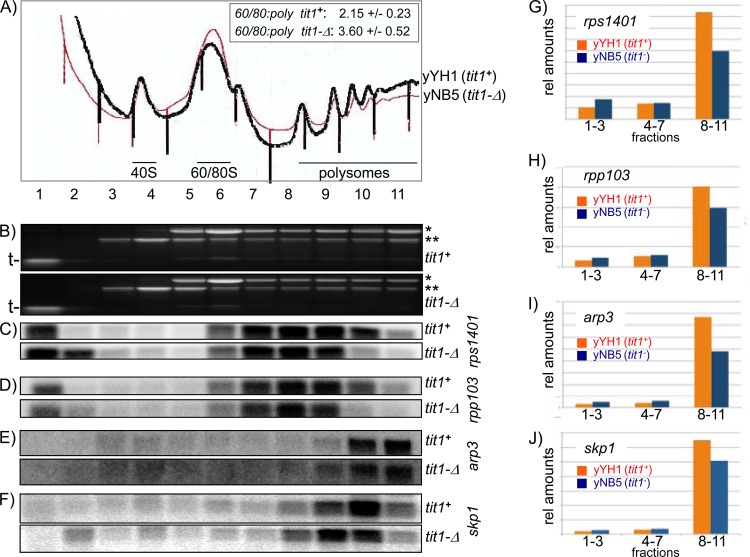
Analysis of polysome profiles has been used as an indicator of the efficiency of translation in yeast (62, 63). Lysates from yYH1 (tit1+) and yNB5 (tit1-Δ) cells grown in parallel were subjected to sucrose gradient sedimentation for polysome analysis (Fig. 5A). Although the 60S and 80S peaks are merged in the gradients, accounting for their increased combined peak height, by superimposing the similarly sized 40S peaks, we observed a difference in polysome levels for tit1+ and tit1-Δ cells, suggesting that tit1-Δ cells were compromised in translation (Fig. 5A). We quantified the ratios of 60S plus to 80S monosomes to polysomes for triplicate polysome profile experiments. As reported in the inset in Fig. 5A, the tit1-Δ cells had a significantly higher ratio than the tit1+ cells, consistent with compromised translation in the tit1-Δ cells.
Fig 5.
Deletion of tit1 leads to decreased translation efficiency. (A) Sucrose gradient sedimentation polysome profiles of extracts from wild-type (black tracing; yYH1 tit1+) and tit1-Δ (red tracing; yNB5) cells. Results of triplicate polysome profile quantitations are shown in the inset; the numbers report the means of three ratios of 60S plus 80S monosomes to polysomes. (B) Ethidium bromide-stained agarose gels of polysome fractions from yYH1 and yNB5 used for Northern blotting. Single and double asterisks to the right indicate 26S and 18S rRNAs, respectively, and “t-” to the left indicates the positions of the tRNAs. (C to F) Northern blots of RNAs isolated from the fractions in panel A, probed for rps1401, rpp103, arp3, and skp1 mRNAs, respectively, as indicated to the right. (G to J) Quantification of the mRNAs in the blots shown in panels C to F. After quantification of individual bands, the relative amounts in the pooled fractions indicated on the x axis were plotted.
Polysome profiles can also be used to compare translation efficiencies of specific mRNAs. An mRNA that is efficiently translated is engaged by more ribosomes and resides on larger polyribosomes to a greater extent than when it is less efficiently translated. We isolated RNAs from the polysome profile fractions shown in Fig. 5A (Fig. 5B). Northern blotting for rps1401 (encoding 40S ribosomal protein S14) and rpp103 (encoding 60S ribosomal protein L12P; also known as 60S acidic ribosomal protein P1) mRNAs revealed two notable differences in tit1+ and tit1-Δ cells. First, there were more rps1401 and rpp103 mRNAs in the pre-40S fractions 2 and 3 of tit1-Δ cells than in those of tit1+ cells, which is reflective of less efficient ribosome engagement. For both of these mRNAs, less than 9% of the total mRNA in all fractions was in the prepolysome fractions in tit1+ cells, and this was increased to 27 to 35% in tit1-Δ cells (Fig. 5G and H). Second, rps1401 (420 nucleotides [nt]; Z score = 4.1) and rpp103 (330 nt; Z score = 3.7) mRNAs were found on larger polyribosomes in tit1+ cells than in tit1-Δ cells (Fig. 5C and D). When these blots were probed for arp3 mRNA (1,284 nt; Z score = 0.2), there was a less obvious difference in the profiles, likely because the arp3 mRNA is longer, although quantitation clearly revealed a pattern of less efficient translation in tit1-Δ cells than in tit1+ cells, similar to the patterns for rps1401 and rpp103 (Fig. 5E and I). The pattern obtained on the same blots for skp1 mRNA (486 nt; Z score = −1.3) reflected a more efficient overall translation than that observed for rps1401 and rpp103, although the difference between tit1-Δ and tit1+ cells was smaller than those for the other mRNAs (Fig. 5F and J). These data are consistent with a complex pattern of altered translation in tit1-Δ cells, in part due to direct effects of decoding caused by lack of i6A37 and in part due to indirect effects related to a translational (and/or metabolic) stress response, as suggested by rapamycin sensitivity that is likely reflective of altered TOR signaling to the translation regulatory apparatus (see Discussion).
DISCUSSION
Here we report an analysis of tRNA-i6A37 function in fission yeast. While the most direct result of tit1+ deletion is i6A37 hypomodification of Tit1 tRNA substrates (18), we demonstrated defects in codon-specific decoding by using a lacZ reporter. The presence of i6A37 on tRNACysGCA increased its activity for the cognate codon. i6A37 increased the ability of tRNACysGCA to protect its cognate codon against misreading by tRNATyrGUA. Surprisingly, i6A37 also increased tRNATyrGUA misreading of a near-cognate Cys codon. Thus, a unifying activity appears to be that i6A37 increases affinity of the tRNA for the ribosome decoding center, at least when the tRNA is occupied by a cognate or near-cognate codon. In terms of fidelity, it appears that i6A37 increases fidelity at cognate codons and decreases fidelity at noncognate codons.
tit1 mutant cells appear to sense and react to an altered state of translation, manifested by two phenotypes: sensitivity to rapamycin, which inhibits the TOR signaling pathway, and slow growth in glycerol. The TOR pathway senses and reacts to the metabolic and growth states of cells. As alluded to above, a relationship between altered translation, metabolic activity, and TOR signaling has become apparent in S. pombe (46). TOR signaling affects the phosphorylation of S. pombe ribosomal protein S6 (Rps6) (64). In addition, Psk1, which is necessary for Rps6 phosphorylation, is regulated by TORC1 in S. pombe, in nutrient-dependent and rapamycin-sensitive manners (65). In summary, the results of this report support others that indicate that disruptions in tRNA homeostasis or function, nutrient metabolism, or rapamycin itself induce compensatory signaling to the translational machinery in S. pombe (46, 65). Both the rapamycin and glycerol phenotypes are rescued by ectopic tit1+ but not tit1-T12A, which is catalytically deficient for tRNA i6A transferase activity, providing evidence that they are due to a lack of tRNA modification. By computationally identifying the mRNAs most enriched in i6A37-cognate codons as those encoding ribosomal subunits and metabolic enzymes, the results help to explain the S. pombe phenotypes due to deletion of tit1+.
Sucrose gradient sedimentation showed increased levels of 60S plus 80S monosomes relative to polysomes in tit1-Δ cells, consistent with the idea that translation was globally compromised (Fig. 5A). This is consistent with the fact that the family of ribosomal subunit mRNAs identified as overenriched in i6A37-cognate codons are each present at very high abundance and therefore collectively engage a significant fraction of the translation machinery. We provided evidence, in the form of mRNA distributions on polysome profiles, that translation of the highly abundant cellular mRNAs rps1401 and rpp103 (encoding ribosomal subunits), which are enriched with i6A37-cognate codons, is compromised in tit1-Δ cells.
The skp1 and arp3 mRNAs, which have less i6A37-cognate codon enrichment and lower abundances, also showed evidence of less translational efficiency in tit1-Δ cells, although to a slightly lesser degree than that for rps1401 and rpp103. These analyses suggest that direct effects of i6A37 deficiency on cognate codon-enriched mRNAs are probably not the sole mechanism to account for the apparent translational compromise in tit1-Δ cells. We emphasize as an alternative that the effects of i6A37 deficiency may be due to both direct effects on translation efficiency and fidelity and indirect effects mediated by stress response signaling, in part through the TOR pathway, the sum of which is responsible for the observed phenotypes (66).
It is noteworthy that a 3-fold increase in tRNACysGCA-G37 levels resulted in only an ∼15% decrease in the tRNATyrGUA-i6A37-mediated misreading in the tit1+ cells (Fig. 2A). This disproportionately low level of decrease may reflect the fact that overexpression may have exhausted an activity critical to tRNACysGCA function. We also note that the 3-fold-overexpressed tRNACysGCA-A37 was hypomodified with i6A37, while endogenous substrates were fully modified. Nonetheless, the 3-fold increase in i6A37-modified tRNACysGCA-A37 led to 3-fold inhibition of tRNATyrGUA-i6A37-mediated misreading. It should also be considered that the tRNACysGCA-A37 was artificially altered at position 37 and that this might offset an otherwise unique feature of the tRNA that is important for its function (67). Future studies can address this by using the natural S. cerevisiae tRNACysGCA-A37 and lacZ-Y503 in S. cerevisiae MOD5 and mod5-Δ cells, as well as in S. pombe tit1+ and tit1-Δ cells.
A role for Tit1 in mitochondrial function.
Because slow growth on glycerol is common to mutants of mitochondrial function, it should be expected that this phenotype might be due, at least in part, to i6A37 hypomodification of mt-tRNATrp. However, hypomodification of cytoplasmic tRNAs involved in the translation of nucleus-encoded proteins that function in mitochondria might also contribute.
We used established computational programs to examine Tit1 and MOD5 sequences for predicted potential for mitochondrial targeting. According to the programs Predotar (68) and TargetP 1.1 (69), the fractions of total Tit1 and MOD5 proteins predicted for mitochondrial localization ranged from 18 to 36% for Tit1 and 23 to 47% for MOD5. Mito-prot-II (V1.101) (70) predicted the probability of export to mitochondria as 0.76 for Tit1 and 0.77 for MOD5. Thus, predictive programs and our results showing tit1+-dependent i6A37 modification of mitochondrion-encoded mt-tRNATrp provide evidence that Tit1 is targeted to mitochondria.
Species-specific subsets of tRNAs that carry i6A37 also extend to mitochondria.
Previous studies have shown that the subsets of tRNAs that carry i6A37 vary significantly, even among yeasts (18). Cytosolic tRNACysGCA carries i6A37 in S. cerevisiae but not in S. pombe, whereas cytosolic tRNATrpCCA carries i6A37 in S. pombe but not in S. cerevisiae (18), and the distribution in human cells differs from those in both yeasts (Lamichhane and Maraia, unpublished data). We can now extend these disparities to mitochondrial tRNAs. mt-tRNACys contains i6A37 in S. cerevisiae (19) but not in S. pombe, while mt-tRNATrp contains i6A37 in S. pombe but not in S. cerevisiae, and mt-tRNASer contains i6A37 in human cells but not in either yeast species (T. Lamichhane, unpublished data).
In addition to species-specific disparities in i6A37-tRNAs, there are global differences in overall tRNA gene copy number and the fractional composition of isoacceptors (16). tRNA gene copy number can vary much more than most other features of highly related genomes of related species (16). Striking reformations of the relative numbers of isoacceptor tRNA genes within synonymous codon groups have occurred in highly related genomes (16). These variables likely affect the degree to which different tRNAs compete for cognate versus near-cognate codons and, therefore, misreading rates. This suggests that although the same genetic code is shared by a vast number of species, its translation occurs in somewhat of a species-specific manner, governed at least in part by the composition and modification characteristics of the tRNAome. Similarly, codon biases in mRNAs that encode otherwise homologous proteins may be subjected to species-specific translation due to differential codon use and tRNAome composition. These genome variables would differentially sensitize subsets of mRNAs to Tit1 homologs in different species. Considerations of both the substrate tRNA isoacceptors and their cognate codon-sensitive mRNAs may help us to understand the variable pleiotropic effects of Tit1 deficiency in different species, including S. cerevisiae and S. pombe, and extending to the C. elegans life span gene GRO-1 and the human tumor suppressor, TRIT1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tom Dever for discussions and Marty Blum for media.
The Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, supports work in the Maraia lab. Work in the Begley lab is supported by the National Institutes of Health (grant 1R01ES017010).
Footnotes
Published ahead of print 28 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00278-13.
REFERENCES
- 1. Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev. 24: 1832– 1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bjork GR. 1995. Biosynthesis and function of modified nucleosides, p 165–206 In Söll D, RajBhandary UL. (ed), tRNA: structure, biosynthesis, and function. ASM Press, Washington, DC [Google Scholar]
- 3. Yokoyama S, Nishimura S. 1995. Modified nucleosides and codon recognition, p 207–223 In Söll D, RajBhandary UL. (ed), tRNA: structure, biosynthesis, and function. ASM Press, Washington, DC [Google Scholar]
- 4. Agris PF. 2008. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 9: 629– 635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Agris PF, Vendeix FA, Graham WD. 2007. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366: 1– 13 [DOI] [PubMed] [Google Scholar]
- 6. Begley U, Dyavaiah M, Patil A, Rooney JP, Direnzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. 2007. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 28: 860– 870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bauer F, Matsuyama A, Candiracci J, Dieu M, Scheliga J, Wolf DA, Yoshida M, Hermand D. 2012. Translational control of cell division by elongator. Cell Rep. 1: 424– 433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yarus M. 1982. Translational efficiency of transfer RNA's: uses of an extended anticodon. Science 218: 646– 652 [DOI] [PubMed] [Google Scholar]
- 9. Miyauchi K, Kimura S, Suzuki T. 2013. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 9: 105– 111 [DOI] [PubMed] [Google Scholar]
- 10. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341– 1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei F, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A, Matsui H, Atta M, Michiue H, Fontecave M, Yamagata K, Suzuki T, KT 2011. Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 121: 3598– 3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reynolds NM, Lazazzera BA, Ibba M. 2010. Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 8: 849– 856 [DOI] [PubMed] [Google Scholar]
- 13. Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ. 2010. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 16: 1797– 1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roy H, Ibba M. 2006. Molecular biology: sticky end in protein synthesis. Nature 443: 41– 42 [DOI] [PubMed] [Google Scholar]
- 15. Kramer EB, Farabaugh PJ. 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13: 87– 96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iben JR, Maraia RJ. 2012. Yeast tRNAomics: tRNA gene copy number variation and codon use provide bioinformatics evidence of a new wobble pair in a eukaryote. RNA 18: 1358– 1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwapisz M, Smagowicz WJ, Oficjalska D, Hatin I, Rousset JP, Zoladek T, Boguta M. 2002. Up-regulation of tRNA biosynthesis affects translational readthrough in maf1-delta mutant of Saccharomyces cerevisiae. Curr. Genet. 42: 147– 152 [DOI] [PubMed] [Google Scholar]
- 18. Lamichhane TN, Blewett NH, Maraia RJ. 2011. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA 17: 1846– 1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK. 1987. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 177– 184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lemieux J, Lakowski B, Webb A, Meng Y, Ubach A, Bussiere F, Barnes T, Hekimi S. 2001. Regulation of physiological rates in Caenorhabditis elegans by a tRNA-modifying enzyme in the mitochondria. Genetics 159: 147– 157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soderberg T, Poulter CD. 2000. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: essential elements for recognition of tRNA substrates within the anticodon stem-loop. Biochemistry 39: 6546– 6553 [DOI] [PubMed] [Google Scholar]
- 22. Spinola M, Galvan A, Pignatiello C, Conti B, Pastorino U, Nicander B, Paroni R, Dragani TA. 2005. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene 24: 5502– 5509 [DOI] [PubMed] [Google Scholar]
- 23. Spinola M, Colombo F, Falvella FS, Dragani TA. 2007. N6-isopentenyladenosine: a potential therapeutic agent for a variety of epithelial cancers. Int. J. Cancer 120: 2744– 2748 [DOI] [PubMed] [Google Scholar]
- 24. Persson BC, Esberg B, Olafsson O, Bjork GR. 1994. Synthesis and function of isopentenyl adenosine derivatives in tRNA. Biochimie 76: 1152– 1160 [DOI] [PubMed] [Google Scholar]
- 25. Laten H, Gorman J, Bock RM. 1978. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 5: 4329– 4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gefter ML, Russell RL. 1969. Role modifications in tyrosine transfer RNA: a modified base affecting ribosome binding. J. Mol. Biol. 39: 145– 157 [DOI] [PubMed] [Google Scholar]
- 27. Diaz I, Pedersen S, Kurland CG. 1987. Effects of miaA on translation and growth rates. Mol. Gen. Genet. 208: 373– 376 [DOI] [PubMed] [Google Scholar]
- 28. Ericson JU, Bjork GR. 1986. Pleiotropic effects induced by modification deficiency next to the anticodon of tRNA from Salmonella typhimurium LT2. J. Bacteriol. 166: 1013– 1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Esberg B, Bjork GR. 1995. The methylthio group (ms2) of N6-(4-hydroxyisopentenyl)-2-methylthioadenosine (ms2io6A) present next to the anticodon contributes to the decoding efficiency of the tRNA. J. Bacteriol. 177: 1967– 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janner F, Vogeli G, Fluri R. 1980. The antisuppressor strain sin1 of Schizosaccharomyces pombe lacks the modification isopentenyladenosine in transfer RNA. J. Mol. Biol. 139: 207– 219 [DOI] [PubMed] [Google Scholar]
- 31. Kohli J, Munz P, Soll D. 1989. Informational suppression, transfer RNA, and intergenic conversion, p 75–96 In Nasim A, Young P, Johnson BF. (ed), Molecular biology of the fission yeast. Academic Press, Inc., San Diego, CA [Google Scholar]
- 32. Jenner LB, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17: 555– 560 [DOI] [PubMed] [Google Scholar]
- 33. Nielsen DA, Chou J, MacKrell AJ, Casadaban MJ, Steiner DF. 1983. Expression of a preproinsulin-beta-galactosidase gene fusion in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 80: 5198– 5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang R, Gaidamakov SA, Xie J, Lee J, Martino L, Kozlov G, Crawford AK, Russo AN, Conte MR, Gehring K, Maraia RJ. 2011. LARP4 binds poly(A), interacts with poly(A)-binding protein MLLE domain via a variant PAM2w motif and can promote mRNA stability. Mol. Cell. Biol. 31: 542– 556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, AmiGO Hub, Web Presence Working Group 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288– 289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Diaz I, Ehrenberg M. 1991. ms2i6A deficiency enhances proofreading in translation. J. Mol. Biol. 222: 1161– 1171 [DOI] [PubMed] [Google Scholar]
- 37. Ring M, Bader DE, Huber RE. 1988. Site-directed mutagenesis of beta-galactosidase (E. coli) reveals that tyr-503 is essential for activity. Biochem. Biophys. Res. Commun. 152: 1050– 1055 [DOI] [PubMed] [Google Scholar]
- 38. Ring M, Huber RE. 1990. Multiple replacements establish the importance of tyrosine-503 in beta-galactosidase (Escherichia coli). Arch. Biochem. Biophys. 283: 342– 350 [DOI] [PubMed] [Google Scholar]
- 39. Penner RM, Roth NJ, Rob B, Lay H, Huber RE. 1999. Tyr-503 of beta-galactosidase (Escherichia coli) plays an important role in degalactosylation. Biochem. Cell Biol. 77: 229– 236 [DOI] [PubMed] [Google Scholar]
- 40. Hagervall TG, Pomerantz SC, McCloskey JA. 1998. Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol. 284: 33– 42 [DOI] [PubMed] [Google Scholar]
- 41. Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37: D93– D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johansson M, Zhang J, Ehrenberg M. 2012. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc. Natl. Acad. Sci. U. S. A. 109: 131– 136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weisman R, Choder M, Koltin Y. 1997. Rapamycin specifically interferes with the developmental response of fission yeast to starvation. J. Bacteriol. 179: 6325– 6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weisman R, Roitburg I, Nahari T, Kupiec M. 2005. Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. Genetics 169: 539– 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takahara T, Maeda T. 2012. TORC1 of fission yeast is rapamycin-sensitive. Genes Cells 17: 698– 708 [DOI] [PubMed] [Google Scholar]
- 46. Cherkasova V, Bahler J, Bacikova D, Pridham K, Maraia RJ. 2012. Altered nuclear tRNA metabolism in La-deleted Schizosaccharomyces pombe is accompanied by a nutritional stress response involving Atf1p and Pcr1p that is suppressible by Xpo-t/Los1p. Mol. Biol. Cell 23: 480– 491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nurse P. 2004. Wee beasties. Nature 432: 557. [DOI] [PubMed] [Google Scholar]
- 48. Nurse P, Thuriaux P. 1980. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics 96:627– 637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yanagida M, Ikai N, Shimanuki M, Sajiki K. 2011. Nutrient limitations alter cell division control and chromosome segregation through growth-related kinases and phosphatases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3508– 3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kellogg DR. 2003. Wee1-dependent mechanisms required for coordination of cell growth and cell division. J. Cell Sci. 116: 4883– 4890 [DOI] [PubMed] [Google Scholar]
- 51. Saracino F, Bassler J, Muzzini D, Hurt E, Agostoni Carbone ML. 2004. The yeast kinase Swe1 is required for proper entry into cell cycle after arrest due to ribosome biogenesis and protein synthesis defects. Cell Cycle 3: 648– 654 [PubMed] [Google Scholar]
- 52. Vassarotti A, Boutry M, Colson AM, Goffeau A. 1984. Independent loci for the structural genes of the yeast mitochondrial alpha and beta ATPase subunits. J. Biol. Chem. 259: 2845– 2849 [PubMed] [Google Scholar]
- 53. Gillman EC, Slusher LB, Martin NC, Hopper AK. 1991. MOD5 translation initiation sites determine N6-isopentenyladenosine modification of mitochondrial and cytoplasmic tRNA. Mol. Cell. Biol. 11: 2382– 2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Murawski M, Szczesniak B, Zoladek T, Hopper AK, Martin NC, Boguta M. 1994. maf1 mutation alters the subcellular localization of the Mod5 protein in yeast. Acta Biochim. Pol. 41: 441– 448 [PubMed] [Google Scholar]
- 55. Tolerico LH, Benko AL, Aris JP, Stanford DR, Martin NC, Hopper AK. 1999. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics 151: 57– 75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zoladek T, Vaduva G, Hunter LA, Boguta M, Go BD, Martin NC, Hopper AK. 1995. Mutations altering the mitochondrial-cytoplasmic distribution of Mod5p implicate the actin cytoskeleton and mRNA 3′ ends and/or protein synthesis in mitochondrial delivery. Mol. Cell. Biol. 15: 6884– 6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Motorin Y, Bec G, Tewari R, Grosjean H. 1997. Transfer RNA recognition by the Escherichia coli delta2-isopentenyl-pyrophosphate:tRNA delta2-isopentenyl transferase: dependence on the anticodon arm structure. RNA 3: 721– 733 [PMC free article] [PubMed] [Google Scholar]
- 58. Bennetzen JL, Hall BD. 1982. Codon selection in yeast. J. Biol. Chem. 257: 3026– 3031 [PubMed] [Google Scholar]
- 59. Fuchs BB, Mylonakis E. 2009. Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 8: 1616– 1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. 2012. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151: 671– 683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bouadloun F, Srichaiyo T, Isaksson LA, Bjork GR. 1986. Influence of modification next to the anticodon in tRNA on codon context sensitivity of translational suppression and accuracy. J. Bacteriol. 166: 1022– 1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hartwell LH, McLaughlin CS. 1969. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 62: 468– 474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. 1991. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 3203– 3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nakashima A, Sato T, Tamanoi F. 2010. Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J. Cell Sci. 123: 777– 786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nakashima A, Otsubo Y, Yamashita A, Sato T, Yamamoto M, Tamanoi F. 2012. Psk1, an AGC kinase family member in fission yeast, is directly phosphorylated and controlled by TORC1 and functions as S6 kinase. J. Cell Sci. 125: 5840– 5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. 2012. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3: 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC. 2005. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 12:788– 793 [DOI] [PubMed] [Google Scholar]
- 68. Small I, Peeters N, Legeai F, Lurin C. 2004. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581– 1590 [DOI] [PubMed] [Google Scholar]
- 69. Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953– 971 [DOI] [PubMed] [Google Scholar]
- 70. Claros MG. 1995. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput. Appl. Biosci. 11: 441– 447 [DOI] [PubMed] [Google Scholar]
- 71. Huang Y, Intine RV, Mozlin A, Hasson S, Maraia RJ. 2005. Mutations in the RNA polymerase III subunit Rpc11p that decrease RNA 3′ cleavage activity increase 3′-terminal oligo(U) length and La-dependent tRNA processing. Mol. Cell. Biol. 25: 621– 636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Intine RVA, Sakulich AL, Koduru SB, Huang Y, Pierstorrf E, Goodier JL, Phan L, Maraia RJ. 2000. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol. Cell 6: 339– 348 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.