Abstract
The Myc oncoprotein is considered a master regulator of gene transcription by virtue of its ability to modulate the expression of a large percentage of all genes. However, mechanisms that direct Myc's recruitment to DNA and target gene selection to elicit specific cellular functions have not been well elucidated. Here, we report that the Pin1 prolyl isomerase enhances recruitment of serine 62-phosphorylated Myc and its coactivators to select promoters during gene activation, followed by promoting Myc's release associated with its degradation. This facilitates Myc's activation of genes involved in cell growth and metabolism, resulting in enhanced proproliferative activity, even while controlling Myc levels. In cancer cells with impaired Myc degradation, Pin1 still enhances Myc DNA binding, although it no longer facilitates Myc degradation. Thus, we find that Pin1 and Myc are cooverexpressed in cancer, and this drives a gene expression pattern that we show is enriched in poor-outcome breast cancer subtypes. This study provides new insight into mechanisms regulating Myc DNA binding and oncogenic activity, it reveals a novel role for Pin1 in the regulation of transcription factors, and it elucidates a mechanism that can contribute to oncogenic cooperation between Pin1 and Myc.
INTRODUCTION
The c-Myc (Myc) transcription factor is a potent regulator of most cellular processes, including cell cycle, cell growth, metabolism, apoptosis, and differentiation, through its orchestrated regulation of a vast number of target genes (1, 2). Myc heterodimerizes with its partner protein, Max, to bind to E box elements in promoters, where it can recruit coactivators, including histone acetyltransferases (HATs), and mediate transcriptional activation. At certain gene promoters, Myc/Max heterodimers can interact with and inhibit the Miz1 transcription factor at INR (initiator) elements, resulting in transcriptional repression (3). Myc represses other target genes through less defined mechanisms. In addition, Max heterodimerizes with Mad and Mnt family members, leading to the recruitment of histone deacetylases (HDACs) and transcriptional repression at E boxes. Together, the Myc/Max/Mad-Mnt network regulates a large percentage of the genome (4). Myc is highly expressed in most human tumors, tipping the balance toward Myc/Max over Mad-Mnt/Max heterodimers. However, whether Myc DNA binding and target gene regulation are largely controlled by Myc levels or whether other directed mechanisms exist for its recruitment to DNA requires further investigation.
The expression level of Myc is normally tightly controlled at multiple levels, including protein stability (5–8). Following cell growth stimulation, Myc levels peak, and this is partially due to sequential and interdependent phosphorylation at two conserved sites, threonine 58 (T58) and serine 62 (S62), that affect Myc stability in response to cell growth regulation (9–12). Myc stability increases upon phosphorylation at S62 (pS62) by extracellular signal-regulated kinase (ERK) and/or cyclin-dependent kinases (CDKs) following receptor tyrosine kinase activation (13, 14). Subsequent T58 phosphorylation (pT58) by glycogen synthase kinase 3β (GSK3β) then downregulates Myc (15, 16). This involves the peptidyl prolyl isomerase Pin1, which can isomerize proline 63 to facilitate removal of the stabilizing phosphate at S62 by the trans-specific phosphatase PP2A-B56α (17, 18). pT58-Myc is targeted by the E3 ubiquitin ligase SCFFbw7 and degraded by the proteasome (19–21). This process is coordinated by the scaffold protein Axin1, which can be detected at target gene promoters, suggesting degradation can occur at the promoter (9). Importantly, Myc phosphorylation at T58 and S62 can be deregulated in cancer cells, leading to enhanced pS62 and a constitutive increase in Myc stability (9, 22, 23).
While Pin1 can contribute to Myc turnover in normal cells, both Pin1 and Myc are often overexpressed in human cancer, and oncogenic activities have been reported for both proteins (24). As the only known phosphorylation-directed peptidyl prolyl isomerase, Pin1 plays a key role in the regulation of protein function by alterations in substrate conformation after phosphorylation. The result of Pin1-mediated isomerization varies depending on the substrate (25). For example, Pin1 has been reported to increase the stability of some targets, while it promotes the degradation of others. It has also been shown to affect the subcellular localization and activity of some transcription factors (26–28). Here, we report a novel role for Pin1 in facilitating Myc recruitment to target gene promoters, as well as Myc's subsequent release and degradation at the promoters. This dynamic DNA binding enhances Myc's association with transcriptional coactivators and trans activation of proproliferative Myc target genes to potentiate Myc's oncogenic activity. In cancer cells, where Myc turnover is impaired, Pin1 is cooverexpressed, and genes activated by Myc and Pin1 expression are enriched in poor-outcome breast cancer subtypes, as well as higher-stage and -grade breast tumors. Together, these studies reveal an elegant mechanism regulating Myc's transcriptional activity, provide critical new insight into the oncogenic role of Pin1, and suggest that Pin1 might be a viable cancer therapeutic target in tumors overexpressing Myc.
MATERIALS AND METHODS
Plasmids and shRNA/siRNA.
pDEST40-MycWT and pDEST40-MycS62A, cytomegalovirus (CMV) Myc, and the scrambled short hairpin RNA (shRNA) plasmids were generated as previously described (17, 18, 29). The 4x-Ebox-Luc construct and pGL2 were provided by Peter Hurlin (Shriners Hospital, Portland, OR). Plasmids encoding Xenopus wild-type Pin 1 (Pin1WT), WW mutant Pin1 (Pin1WW), and C109A mutant Pin1 (Pin1C109A or Pin1CM) were provided by Anthony Means (Duke University Medical Center, Durham, NC). Xenopus Pin1 (WT, WW mutant, and C109A mutant) cDNAs were moved from these plasmids into pDEST40 using Gateway technology (Invitrogen, Carlsbad, CA) as described previously (18). pLenti4/TO/V5-Dest-Myc was also generated using the Gateway technology. The small interfering RNA (siRNA) for Myc was a gift from Joe Gray (Oregon Health and Science University, Portland, OR [OHSU]).
Generation of Pin1 shRNAs.
For the generation of Pin1 shRNA-1, oligonucleotides encoding the sense ( 5′-AATTCCGGGAGAGGAGGACTTTGATTCAAGAGATCAAAGTCCTCCTCTCCCGC-3′ ) and antisense ( 5′-TCGAGCGGGAGAGGAGGACTTTGATCTCTTGAATCAAAGTCCTCCTCTCCCGG-3′ ) sequences were annealed and ligated into the pENTR-H1/TO (Invitrogen) expression vector as described by the manufacturer. Pin1 shRNA-2 in pSUPER was generated with the pSUPER RNA interference (RNAi) system (Oligoengine, Seattle, WA) using the following oligonucleotides: 5′-GATCCCCGCCATTTGAAGACGCCTCGTTCAAGAGACGAGGCGTCTTCAAATGGCTTTTTGGAAA-3′ (sense) and 5′-AGCTTTTCCAAAAAGCCATTTGAAGACGCCTCGTCTCTTGAACGAGGCGTCTTCAAATGGCGG-3′ (antisense).
Cell culture, transfection, and adenoviral infection.
MDA-MB-453, MDA-MB-436, human embryonic kidney HEK293, and 293M cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% standard fetal bovine serum (FBS), 2.5 mM l-glutamine, and 1× penicillin-streptomycin. MCF10A, MCF7, MDA-MB-436, MDA-MB-453, and HEK293 cells were purchased from the American Type Culture Collection (ATCC). 293M cells are a 293 derivative established in the laboratory for their high expression of pS62-Myc. The LY2 cell line was a gift from Lawrence Berkeley National Laboratory. MCF10A cells were grown in 45% DMEM, 45% Ham's F-12, 5% defined FBS, 2.5 mM l-glutamine, 20 ng/ml epidermal growth factor (EGF), 10 μg/ml insulin, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxin, and 1× penicillin-streptomycin. MCF7 cells were grown in 45% DMEM, 45% Ham's F-12, 10% FBS, 2.5 mM l-glutamine, and 1× penicillin-streptomycin. LY2 cells were grown in modified Iscove's modified Eagle's medium (IMEM) with 5% FBS, 2.5 mM l-glutamine, and 1× penicillin-streptomycin.
Generation of stable MCF10A-Myc cells.
Stable MCF10A-TR-MycWT (MCF10A-Myc) cells were generated by infecting a 100-mm dish of MCF10A cells with lentivirus encoding the Tet repressor, pLenti6/TR (Invitrogen), at a multiplicity of infection (MOI) of ∼10 in 5 ml MCF10A medium supplemented with 5% defined FBS and 6 μg/ml Polybrene (specialty medium) for 12 h. The medium was changed to medium supplemented with 5% defined FBS for 24 h. The cells were then split at a 1:10 dilution and maintained in medium supplemented with 5% defined FBS and 5 μg/ml blasticidin (Invitrogen) for 10 days until distinct colonies formed. The resulting colonies were then infected with lentivirus expressing V5-Myc (pLenti4/TO/V5-Dest-Myc) at an MOI of ∼10, as described above for the Tet repressor infection. Cells were selected in medium with 5% defined FBS, 5 μg/ml blasticidin, and 200 μg/ml zeocin (Invitrogen) for 10 days until distinct colonies formed. Six colonies were picked, expanded, and screened for the ability to express V5-Myc only when treated with 1 μg/ml doxycycline (Dox). The best clone was then used for further experiments and continuously maintained in medium with 5% defined FBS, 5 μg/ml blasticidin, and 200 μg/ml zeocin. Stable MCF10A-TR-MycT58A and stable MCF10A-TR-MycS62A cells were similarly constructed, except pLenti4/TO/V5-Dest-MycT58A or pLenti4/TO/V5-Dest-MycS62A was used.
Transfection and adenoviral infection.
For transfection of HEK293 cells, cells were plated to achieve 70 to 80% confluence 24 h postsplit. Transfections were performed using HEK-Fectin (Bio-Rad, Hercules, CA) according to the manufacturer's specifications. For luciferase assays, transfection mixtures included 200 ng of CMV β-galactosidase (β-Gal) to determine transfection efficiency. For Pin1 inhibition studies, cells were treated with 0.5 μM PiB (diethyl-1,3,6,8-tetrahydro-1,3,6,8-tetraoxobenzo[lmn]) dissolved in dimethyl sulfoxide (DMSO) or DMSO carrier alone for 18 h (pretreatment for colony-forming assays) or 48 h (quantitative chromatin immunoprecipitation [qChIP] and quantitative reverse transcription-PCR [qRT-PCR] experiments). The Pin1 adenovirus (AdPin1) was generated as previously described (18). For adenoviral infection of MCF10A and MCF10A-Myc cells, approximately 5 × 106 cells were trypsinized and resuspended in 500 μl serum-free DMEM. Green fluorescent protein adenovirus (AdGFP) or AdPin1 (MOI = 10) was added and mixed. The tubes were incubated at 37°C, 5% CO2 for 1 h with mixing every 10 min. Infected cells were then plated in 10-cm culture dishes containing 9.5 ml medium with serum and incubated for an additional 18 h.
Western blotting and antibodies.
Cell lysates were run on SDS-PAGE gels and transferred to Immobilon-FL membranes (Millipore). The membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE). Primary antibodies were diluted in 1:1 Odyssey blocking buffer-PBS with 0.05% Tween 20. Primary antibodies were detected with secondary antibodies labeled with the near-infrared fluorescent dyes IRDye800 (Rockland, Philadelphia, PA) and Alexa Fluor 680 (Molecular Probes, Eugene, OR). Secondary antibodies were diluted 1:10,000 in 1:1 Odyssey blocking buffer-PBS with 0.05% Tween 20. Blots were scanned with an Odyssey infrared imager (LI-COR Biosciences) to visualize proteins.
Mouse monoclonal V5 antibody was from Invitrogen. Myc antibody Y69 for Western blotting and qChIP was from Abcam (Cambridge, MA). Myc antibody N262 for qChIP was from Santa Cruz (Santa Cruz, CA). Pin1 mouse monoclonal antibody for Western blots was purchased from Genway Biotech, Inc. (San Diego, CA). Antibodies for qChIP were Pin1 from Santa Cruz, GSK3β from Cell Signaling Technology (Danvers, MA), Axin1 (C76H11) from Cell Signaling Technology, PP2Ac from Millipore (Temecula, CA), Fbw7 from Abcam, 19S (S6a) from Enzo Life Sciences, anti-acetyl-histone H4 from Upstate (Lake Placid, NY), p300 from Santa Cruz, hSNF5 from Abcam, GCN5 from Santa Cruz, Cdk9 from Santa Cruz, and total and pS2-RNA polymerase II from Covance (Princeton, NJ). β-Actin antibody was from Sigma-Aldrich, Inc. (St. Louis, MO). pS62-Myc antibody (33A12E10) used in Western blots and qChIP was from Abcam. For immunofluorescence, the following antibodies were used: pS62-Myc (generated in our laboratory) (30), rabbit polyclonal to Pin1 (Santa Cruz), Ki-67 (Novacastra, Newcastle upon Tyne, United Kingdom), and Alexa Fluor 594–donkey anti-rabbit IgG (Molecular Probes).
Luciferase assays.
Cell pellets were resuspended in 10× volumes of 1× cell lysis buffer (Promega, Madison, WI) with protease and phosphatase inhibitors. Cellular lysates were sonicated for 10 pulses at an output of 1 and a 10% duty cycle and incubated on ice for 20 min. The lysates were cleared by centrifugation at 14,000 rpm for 10 min at 4°C. Luciferase activity was measured using the Promega luciferase assay kit and a Berthold (Bundoora, Australia) luminometer. Luciferase activity was adjusted for β-Gal. Fold changes (FC) in luciferase activities were measured relative to empty vector or control transfections.
qRT-PCR.
RNA was isolated from cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. cDNA was generated using the High Capacity cDNA reverse transcription kit (ABI, Foster City, CA). qRT-PCR analysis was performed using the indicated primers (see Table S1 in the supplemental material) and SYBR green reagent (Invitrogen) on a Step-One real-time PCR machine (Applied Biosystems, Invitrogen) using the manufacturer's cycling conditions.
qChIP.
Cells were cross-linked with formaldehyde to a final concentration of 1% in medium and incubated at room temperature for 10 min. Cells were collected in 1× phosphate-buffered saline (PBS)–1 mM EDTA and pelleted by gentle centrifugation. The cells were resuspended in 700 μl ChIP lysis buffer (0.1% SDS, 0.5% Triton X-100, 20 mM Tris-HCl [pH 8.1], and 150 mM NaCl). The cell lysates were sonicated 6 times (output = 3.5; 30% duty cycle; 10 pulses) and then cleared by centrifugation at 14,000 rpm for 15 min at 4°C. Cell lysates were precleared with 50 μl of a 50% slurry of protein A beads for 1 h with rotation at 4°C. The lysates were again cleared by centrifugation at 14,000 rpm for 5 min at 4°C. Immunoprecipitations (IPs) were performed with 2 μg of each antibody overnight at 4°C. Two micrograms of normal rabbit IgG or normal mouse IgG (Santa Cruz Biotechnology) was used as a negative control. The immunoprecipitates were washed six times with ChIP lysis buffer and twice with 1× Tris-EDTA (TE). Samples were rotated for 15 min at 4°C for each wash step. The immunoprecipitates were eluted from the beads with elution buffer (0.1 M NaHCO3 and 1% SDS) by rotation for 15 min at room temperature. The elution products were transferred to new tubes, 5 M NaCl was added to a final concentration of 0.2 M, and samples were incubated at 65°C overnight. DNA was purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and used for quantitative PCR (qPCR) analysis (as described above) with specified primers (see Table S2 in the supplemental material). For quantitative ChIP experiments, primers to the promoter regions of Myc target genes, as well as internal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers, were used to amplify DNA. The internal GAPDH primers were used as a negative control. qPCR was used to measure signals in 1% of the input material, as well as each IP. The percentage of input was then calculated for each IP (control IgG and specific) as the IP signal above the input signal using the following formula: 100 × 2(input Ct − IP Ct), where CT is the threshold cycle. The relative level of bound DNA was then graphed as the percent input of the specific IP relative to the percent input of the mock IgG control using GraphPad Prism. For sequential ChIP, the first immunoprecipitation was carried out as described above. Protein-DNA complexes were eluted from the beads by incubation in 75 μl TE and 10 mM dithiothreitol (DTT) for 30 min at 37°C. The supernatant was then diluted 20 times in ChIP lysis buffer and utilized for the second round of IP. After washing, protein-DNA complexes were eluted again in TE and DTT as described above. Reactions were then de-cross-linked overnight as described above. For quantification of immunoprecipitated material, qPCR was used to measure signals in 1% of the input material, as well as each immunoprecipitation. The percentage of input was then calculated for each IP (control IgG and specific) as the IP signal above the input signal using the following formula: 100 × 2(input Ct − IP Ct). One percent of the input was boiled for 30 min in SDS sample buffer prior to electrophoresis to detect the indicated proteins by Western blotting.
Colony-forming assays.
MCF10A-Myc cells were infected with AdPin1 or AdGFP (MOI = 10) and induced with 1 μg/ml doxycycline for 18 h prior to plating. For drug treatments, cells were pretreated with DMSO or 1 μM PiB and induced with 1 μg/ml doxycycline for 18 h. As a control, each experiment included cells without doxycycline induction. 293M cells were also pretreated with DMSO or 1 μM PiB for 18 h. All soft-agar assays were carried out in triplicate in 6-well plates, with 24,000 cells per well. The base agar layer contained 0.8% Noble agar in 1.5× growth medium. Cells were harvested, and 24,000 cells were mixed with 0.6% Noble agar in 1.5× growth medium and plated on top of the base agar layer. The plates were fed twice a week with 2× medium containing 2 μg/ml doxycycline and/or 1.0 μM PiB. After 4 weeks, the plates were stained with 0.5 ml 0.005% crystal violet for 2 h, and the total number of colonies was determined.
RNA-seq.
For high-throughput RNA sequencing (RNA-seq), total RNA was isolated using TRIzol (Invitrogen) and purified using an RNAeasy minikit (Qiagen) according to the manufacturer's instructions. Poly(A) RNA was purified using oligotex-dT30 latex beads (Qiagen). First-strand cDNA synthesis was performed using Superscript III (Invitrogen) and Oligo(dT)20 (Invitrogen) according to the manufacturer's instructions. Second-strand synthesis was carried out according to standard methods, followed by double-stranded cDNA fragmentation with a 65-W pulse on a Misonix sonicator (fragment size, 100 to 700 bp) and polishing with the DNA terminator repair kit (Lucigen). A single A base was added with Klenow fragment (3′→5′ exo−) prior to ligation of genomic DNA adaptors (Illumina Solex Genomic 1G) at 22°C. Amplification of the library using 10 cycles of limited PCR with Phusion HF DNA polymerase (NEB) and genomic PCA primer (Illumina; Solexa Genomic primers 1.1 and 2.2) was conducted. Double-stranded cDNA libraries were sequenced on a Solexa G1 genome analyzer, and image analysis and base calling were conducted with the standard Illumina Analysis Pipeline 1.0 (Firecrest-Bustard). The χ2 statistic was applied to the counts of uniquely mapped reads within exons of RefSeq genes. Fold change calculations were based on the same tag counts after mean scaling of samples based on total tag counts in exons. A false discovery rate (FDR)-adjusted P value (q) of <0.01 and a fold change of 2.0 were used as thresholds for selection of significant genes. Significant genes that are “Myc target genes” were identified by cross-referencing a Myc target gene set composed of all genes listed on the Myc target gene database (http://www.myc-cancer-gene.org/site/mycTargetDB.asp), in addition to two ChIP-seq data sets (31, 32).
Kolmogorov-Smirnov statistical test.
Significantly regulated Ref-Seq genes were selected based on a q value of <0.01. This was cross-referenced with a Myc target gene data set that included all of the genes listed in the Myc target gene database (http://www.myc-cancer-gene.org) plus two other ChIP-based data sets (31, 32). Fold changes were calculated based on mean scaled tag counts within exons. The Kolmogorov-Smirnov (KS) statistical test was applied to evaluate enrichment of the significant gene list in two breast cancer data sets (see below): Gene Expression Omnibus (GEO) GSE2109 and The Cancer Genome Atlas (TCGA) Breast Invasive Carcinoma. A bootstrap version of the univariate Kolmogorov-Smirnov test (ks.boot from the R package:Matching) was used to measure the enrichment of the Myc/Pin1 versus Myc up genes in the genes at the top of the tumor gene list when the list is sorted by P value from most significant up to most significant down, with the up genes in the poorer-outcome group at the beginning of the list. This test provides correct coverage even when the distributions being compared are not entirely continuous. Ties are allowed with this test, unlike the traditional Kolmogorov-Smirnov test (33, 34; http://sekhon.berkeley.edu/). Enrichment at the other end of the list was also tested and reported as a “reverse” P value. A KS graph was generated to visualize the “D” score used in the KS test.
Public human breast cancer data sets.
This work was approved by the OHSU Institutional Review Board. Two public data sets, ID GSE2109 from GEO (http://www.ncbi.nlm.nih.gov/geo/) and a set of breast cancers from TCGA (http://cancergenome.nih.gov/), were used. The GEO set contains 352 breast samples with clinical annotation run on the Affymetrix Human Genome U133 Plus 2.0 Array. We processed the original Celestia raw data (CEL) files for these with the robust multiarray average (RMA) algorithm and identified the following groups based on the clinical annotation: 106 luminal A-like (Her2− with estrogen receptor positive [ER+] and/or progesterone receptor positive [PR+]), 47 triple negative (TN), 58 Her2+, 270 ductal, 39 lobular, 7 stage 4, 38 stage 1, 149 grade 3, and 32 grade 1. We obtained 430 breast samples from the TCGA run on the Agilent platform. They were molecularly subtyped based on PAM50 genes (35), resulting in 187 luminal A, 47 Her2+, 109 luminal B, and 79 basal. A total of 14,862 unique gene symbols common to these two data sets and our RNA-seq data set were identified. Direct comparison of expression values between the three different platforms was not practical, so a rank-based test (KS test) was applied to compare the enrichment of the significantly upregulated genes from our RNA-seq data set in the ranked list of genes from the other two data sets, where the poorer-outcome groups were compared to the better-outcome groups. For each comparison, all 14,862 unique genes were ranked based on q value, with the most significantly upregulated genes in the poor-outcome group at the start of the list and the most significantly downregulated genes at the end of the list.
Coimmunoprecipitation.
Cells were resuspended in 10× cell pellet volumes of co-IP buffer (20 mM Tris, pH 7.5, 12.5% glycerol, 0.5% NP-40, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, and 1 mM DTT) plus protease and phosphatase inhibitors. Cellular lysates were sonicated for 10 pulses (output = 1; 10 to 15% duty cycle), incubated on ice for 20 min, and cleared by centrifugation at 14,000 rpm for 10 min at 4°C. The cleared lysates were adjusted for transfection efficiency as measured by β-Gal activity and incubated with a 1:750 dilution of V5 antibody conjugated to protein A-Sepharose beads (Repligen, Waltham, MA). The immunoprecipitates were washed 3 times with 10× volumes of co-IP buffer and analyzed by Western blotting.
Xenografts.
For 293M xenografts, 1 × 106 cells in 25% Matrigel-75% growth medium were injected into the 4th mammary glands of 6- to 8-week-old nonobese diabetic (NOD)/SCID γ-chain-null mice. PiB treatments (0.6 mg/kg of body weight) were initiated on half the mice the day after xenografting into the xenograft site and continued until sacrifice (when tumors in the control group reached 2.0 cm or became ulcerated). Treatments were given daily for the first 2 weeks, followed by 3 times per week thereafter. Six mice/12 tumors were monitored for each group. All animal procedures were in accordance with the research guidelines for the use of laboratory animals adopted by the OHSU IACUC.
Immunofluorescence and TUNEL assays.
Serial paraffin sections from patients with matched normal and tumor formalin-fixed human breast cancer tissues (obtained from the archives of the OHSU Department of Pathology, IRB 2086) were incubated with the pS62-Myc (1:25)- or Pin1 (1:50)-specific antibodies overnight at 4°C. Sections were then incubated with Alexa Fluor 594–donkey anti-rabbit IgG (1:1,000) and mounted using antifade containing DAPI (4′,6-diamidino-2-phenylindole). Images were taken with a Hamamatsu digital camera mounted on a fluorescence microscope at the same magnification and exposure for each antibody. The immunofluorescence (IF) density was analyzed in photographs taken with a 40× lens using the Measure Density tool in the OpenLab 5.5 software according to the manufacturer's protocol. Staining intensity was measured in 10 random individual cells in each of 5 to 10 random fields of view, averaged for each sample, and graphed (±standard error). Formalin-fixed/paraffin-embedded SKBR3 xenograft tumor tissues were stained for Ki-67 (1:3,000) as described above. For quantification of Ki-67 staining, the average intensity of Ki-67 staining was normalized to the average DAPI signal from 10 random individual cells in each of 5 to 10 random fields of view. A terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed using the ApopTag Plus peroxidase in situ apoptosis detection kit (Chemicon, Billerica, MA) according to the manufacturer's instructions. TUNEL-positive cells in 6 random fields of view for each tumor sample were counted and expressed relative to the total number of cells in that field. Three untreated and three treated tumor samples were averaged and graphed using GraphPad Prism.
Generation of Blg-Cre; NeuNT; Myc mice.
RFS-MycWT mice were crossed with NeuNT and Blg-Cre mice (gifts from Owen Sansom, Beatson Institute for Cancer Research, Glasgow, United Kingdom) to produce mice that expressed both Myc and activated Neu in response to Cre-mediated recombination in the mammary gland. Once tumors became palpable, the mice were randomized into control or PiB-treated groups, and tumor growth was followed by daily caliper measurements. The PiB treatment was 0.6 mg/kg 3 times per week intraperitoneally (i.p.).
Data and statistics.
Three or more independent replicate experiments were performed in all cases. Average fold changes and standard errors were graphed using GraphPad Prism. P values were calculated using a standard Student's t test analysis (two-tailed distribution and two-sample unequal variance) to determine statistical significance as indicated in the graphs. Correlation coefficients were calculated using Microsoft Excel. P values for relevant comparisons are given. If no P value is shown, the comparison is not relevant or not significant.
RESULTS
Pin1 positively regulates Myc transcriptional activity and promoter binding.
Given reports of Pin1's oncogenic activity and the coupling of transcriptional activation with transcription factor turnover (36–38), we tested whether Pin1 could stimulate Myc-driven transcription. Using an E-box-driven luciferase (4xEbox-Luc) or control reporter plasmid (pGL2) transfected into HEK293 cells, we found that when Myc and Pin1 were coexpressed, luciferase activity significantly increased over that with Myc alone (Fig. 1A). This was not the result of increased Myc protein levels, as Myc levels were lower with coexpression of Pin1 (Fig. 1A, blots), consistent with Pin1 facilitating Myc turnover (18). In contrast, Pin1 knockdown with two different Pin1 shRNAs suppressed E-box-driven luciferase activity (Fig. 1B), despite an increase in endogenous Myc levels (Fig. 1B, blots). We previously observed an increase in the solubility of Myc upon nuclear extraction in Pin1-null (Pin1−/−) mouse embryonic fibroblasts (MEFs) (see Fig. S1A in the supplemental material), suggesting that Pin1 may regulate Myc chromatin association, which could help explain its effects on Myc transcriptional activity. We therefore used qChIP to examine Myc's association with an endogenous target gene in Pin1 wild-type (Pin1+/+) and Pin1−/− primary MEFs. We observed increased Myc target gene promoter binding in Pin1+/+ MEFs or Pin1−/− MEFs upon Pin1 reintroduction relative to Pin1−/− MEFs, despite a general reduction in Myc expression in the Pin1-expressing MEFs (Fig. 1C). We also observed that the induction of Myc target gene promoter binding in response to serum stimulation was reduced in the Pin1−/− versus Pin1+/+ MEFs (see Fig. S1B in the supplemental material). Thus, Pin1 increases Myc-driven transcription and enhances Myc DNA binding even with reduced Myc protein levels.
Fig 1.
Pin1 positively regulates Myc transcriptional activity and target gene binding. (A) HEK293 cells were transfected with the indicated expression plasmids, CMV β-Gal, and control luciferase reporter (pGL2) or 4xEbox-Luc. The transfected cells were grown in low serum for 48 h. Luciferase activity was normalized to β-Gal, and the β-Gal-normalized cell lysates were Western blotted. Myc/β-actin levels are shown. (B) Same as panel A but with the indicated shRNA plasmids. 4xEbox-Luc/pGL2 was graphed relative to the control shRNA. (C) Primary Pin1−/− and Pin1+/+ MEFs were infected with the indicated adenoviruses, and qChIP for endogenous Myc at the nucleolin promoter was performed as described in Materials and Methods. One percent of the input was used for Western blotting. Myc/β-actin levels are indicated. The fold enrichment of bound DNA was graphed as the percentage of input of the specific Myc IP relative to the percentage of input of the IgG control IP. (D) 293 cells were transfected with the indicated expression plasmids or shRNAs, and Myc binding to the indicated promoters was measured by qChIP as in panel C. ChIP inputs were Western blotted as in panel C. (E) RNA from 293 cells treated as in panel D was subjected to qRT-PCR, and expression of the indicated Myc target genes normalized to GAPDH is shown relative to the vector-only or scrambled shRNA (scr shRNA) transfection. (F) 293 cells were transfected with the indicated shRNAs and/or expression plasmids. qRT-PCR of nucleolin and E2F2 normalized to GAPDH and Western blotting for Pin1, Myc, and β-actin are shown. P values are shown for relevant significant comparisons. An asterisk indicates a P value of less than 0.001. The error bars indicate standard errors.
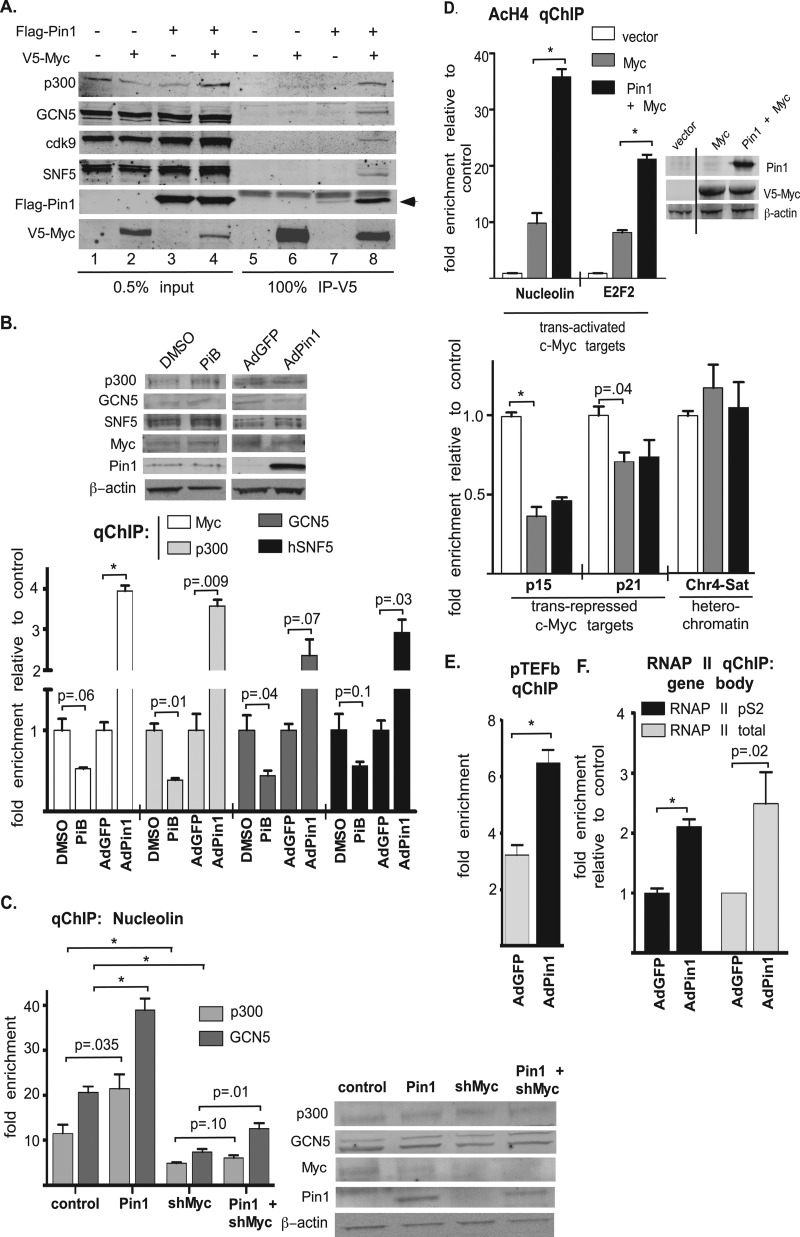
To further investigate the effects of Pin1 on Myc DNA binding at target gene promoters, we examined four trans-activated Myc targets involving three different polymerases (Pol) (Pol II [nucleolin and E2F2], Pol III [5S rRNA], and Pol I [rDNA]) and two trans-repressed Pol II transcribed targets (p15 and p21) in HEK293 cells after Pin1 overexpression or knockdown. Despite a decrease in Myc protein levels after Pin1 overexpression (Fig. 1D, blots, Pin1 versus vector), more Myc was bound to the four trans-activated promoters (Fig. 1D). Conversely, knockdown of Pin1 decreased Myc binding to the same promoters (Fig. 1D), although Myc levels were increased (Fig. 1D, blots, Pin1 shRNA versus scrambled [scr] shRNA). Manipulation of Pin1 levels had no effect on Myc binding to p15 and p21 promoters (Fig. 1D). Moreover, expression of these endogenous target genes correlated with Pin1-mediated changes in Myc DNA binding, where Pin1 overexpression increased expression of all four trans-activated targets and had no effect on the expression of p21 and p15 (Fig. 1E). The opposite results were seen after knockdown of Pin1, where Pin1 depletion decreased expression of the trans-activated targets without affecting p21 or p15 (Fig. 1E). To test the Myc dependency of the Pin1 effects on endogenous Myc target gene expression, we overexpressed Pin1 with or without Myc knockdown. While Pin1 overexpression increased the expression of nucleolin and E2F2, knockdown of Myc suppressed expression of these genes regardless of Pin1 overexpression (Fig. 1F). We performed similar experiments in human mammary epithelial MCF10A cells, where, similar to the HEK293 results, we observed that Pin1 overexpression increased the association of endogenous Myc with the four trans-activated promoters but had no effect on binding to the trans-repressed p21 promoter (see Fig. S1C in the supplemental material), while Pin1 inhibition with PiB, a small-molecule inhibitor of Pin1 (39), had the opposite effect, decreasing Myc binding to the trans-activated promoters without affecting binding to p21 (see Fig. S1C in the supplemental material). In addition, changes in Myc promoter binding correlated with changes in the expression of the endogenous Myc target genes in MCF10A cells (see Fig. S1D in the supplemental material). We also observed that Pin1 enhances Myc promoter binding at E-box sites (see Fig. S1E in the supplemental material), which could explain the lack of an effect of Pin1 on Myc binding to p15 and p21 promoters, since these are trans-repressed through other promoter elements (40, 41). Thus, Pin1 positively regulates Myc promoter binding at all trans-activated target genes tested, and this correlates with changes in their expression.
Pin1 regulation of Myc activity requires functional phospho-binding and isomerase domains in Pin1 and serine 62 phosphorylation of Myc.
To determine which domains of Pin1 are important for its ability to regulate Myc promoter binding and transcriptional activity, we expressed a Pin1WW domain mutant, which contains a point mutation in the WW domain (W33A) that inhibits its interaction with phosphorylated substrates (42), and a catalytically inactive Pin1 mutant, Pin1CM, which contains a point mutation in the isomerase domain (C109A) (43), in HEK293 cells and performed luciferase assays with the 4xEbox-Luc reporter to measure affects on Myc-driven transcriptional activity. We found that the Pin1WW and Pin1CM mutants were unable to stimulate the transcriptional activity of Myc like Pin1WT (Fig. 2A). In addition, neither mutant was able to regulate Myc DNA binding (Fig. 2B), demonstrating that Pin1's phospho-substrate binding domain and isomerase activity are required for its stimulation of Myc's transcriptional activity and DNA binding activity.
Fig 2.
Pin1 regulation of Myc activity requires functional phospho-binding and isomerase domains in Pin1 and serine 62 phosphorylation of Myc. (A) Luciferase assays were performed after expression of the indicated plasmids in 293 cells, as in Fig. 1A. (B) 293 cells were transfected with empty vector or the indicated Pin1 expression plasmids (V5-Pin1WT, V5-Pin1CM, or V5-Pin1WW). qChIP for endogenous Myc at the indicated promoters was performed. (C) Luciferase assay and Western blotting were carried out as described for Fig. 1A. The black line in the Western blot indicates that the samples were run on the same gel, but extra lanes were removed. (D) MCF10A-MycWT, -MycT58A, or -MycS62A cells were infected with AdGFP or AdPin1, and Myc expression was induced with doxycycline for 18 h. qChIP for ectopic MycWT, MycT58A, or MycS62A was performed at the E2F2 gene. (E) MCF10A-MycWT, -MycT58A, or -MycS62A cells were infected with AdGFP or AdPin1, and Myc expression was induced with doxycycline for 18 h. E2F2 expression levels were measured by qRT-PCR. Statistical significance is indicated as in Fig. 1. The error bars indicate standard errors.
Since Pin1 is reported to be a phosphorylation-directed prolyl isomerase and the WW phospho-binding domain is required for its regulation of Myc, we tested the role of Myc phosphorylation at T58 and S62 in Myc's response to Pin1. We performed luciferase assays with the Myc T58A and S62A phosphorylation mutants and found that Pin1 had reduced effects on the activity of MycT58A to drive E-box-mediated transcription and no effect on the activity of MycS62A (Fig. 2C). We also examined the ability of Pin1 to regulate MycS62A or MycT58A binding to chromatin using stable Dox-inducible MCF10A-MycWT, MCF10A-MycT58A, and MCF10A-MycS62A clones. Pin1 again enhanced MycWT binding to the E2F2 gene promoter, with reduced Myc protein expression (Fig. 2D, graph and blot, lane 2). However, Pin1 had a reduced effect on the binding of MycT58A and no effect on the binding of MycS62A, while also not affecting the expression of either mutant, consistent with our previous results showing that Pin1 does not regulate the degradation of these mutants (18). Furthermore, while Pin1 enhanced E2F2 mRNA expression driven by MycWT in these cells, it only moderately enhanced activation of E2F2 mRNA expression mediated by MycT58A and it had no effect on MycS62A activation of E2F2 (Fig. 2E). Thus, Pin1 activation of Myc involves S62 phosphorylation of Myc, Pin1's phospho-substrate binding domain, and its catalytic activity, suggesting that Pin1 interacts with and isomerizes pS62-Myc to enhance its DNA binding and transcriptional activities.
Pin1 associates with pS62-Myc at Myc-responsive promoters.
To test whether Pin1's regulation of Myc occurs at Myc target gene promoters, we assayed for the presence of Pin1 at Myc-regulated promoters. We detected V5-Pin1WT by qChIP at each of the Myc trans-activated target promoters, but not p15, using the same primers as for Myc qChIP, which span the Myc binding site(s) (Fig. 3A). In contrast, V5-Pin1WW was unable to associate with these promoters (Fig. 3A). We also detected endogenous Pin1 at the same Myc target promoters, and this binding could be reduced with Pin1 knockdown (see Fig. S2A in the supplemental material). In addition, the association of Pin1 with Myc target genes was enhanced by increased Myc expression (Fig. 3B). To verify that Pin1 is recruited to Myc-responsive promoters via Myc, we measured endogenous Pin1 binding to the E2F2 and nucleolin promoters after Myc knockdown in 293 cells. As shown in Fig. 3C, knockdown of Myc reduced Pin1 binding to these promoters. To test whether Myc and Pin1 can cooccupy Myc-responsive promoters, we performed sequential qChIP experiments. Ectopic Myc and Pin1 associated with the nucleolin and E2F2 promoters (Fig. 3D). Following release of the bound material from the Myc immunoprecipitation, we were able to sequentially qChIP Pin1 at these promoters (Fig. 3D). Similar results were found with endogenous Myc and Pin1, except the p21 promoter was also tested, and Myc, but not Pin1, was detected here (see Fig. S2B in the supplemental material). Furthermore, we were able to qChIP endogenous pS62-Myc at the nucleolin, E2F2, and p21 promoters, and sequentially, qChIP pS62-Myc and endogenous Pin1 at the nucleolin and E2F2 promoters but not at the p21 promoter (Fig. 3E). Thus, Pin1 binding to Myc trans-activated target promoters is Myc dependent, and Pin1 and pS62-Myc can cooccupy these promoters.
Fig 3.
Pin1 associates with Myc target gene promoters. (A) 293 cells were transfected with empty vector or the indicated Pin1 expression plasmids (V5-Pin1WT or V5-Pin1WW). qChIP for ectopic V5-Pin1 at the indicated promoters was performed, and 1% of the input was analyzed by Western blotting. (B) 293 cells were transfected with the indicated expression vectors, and qChIP for V5-Pin1 was performed at the indicated Myc target promoters. (C) 293 cells were transfected with the indicated shRNAs, endogenous Pin1 binding to the nucleolin and E2F2 promoters was measured by qChIP, and 1% of the input was analyzed by Western blotting. (D) 293 cells were used for individual qChIP or sequential qChIP with the indicated antibodies. (E) 293 cells were used for individual qChIP or sequential qChIP with the indicated antibodies. Statistical significance is indicated as in Fig. 1. The error bars indicate standard errors.
Pin1 increases the rate of recruitment of pS62-Myc to target gene promoters.
We next investigated whether the Pin1-mediated increase in Myc association with its target genes reflected a change in the rate of recruitment to the promoter. To examine the recruitment rate, we used the Dox-inducible MCF10A-MycWT cells to measure how quickly V5-Myc is recruited to chromatin by qChIP. Ectopic Myc was just detectable by Western blotting after 30 min of Dox treatment, and these levels continued to rise through the course of the experiment (Fig. 4A, blots). At each time point, the level of V5-Myc binding to the E2F2 promoter was measured by qChIP. In the control AdGFP-infected cells, V5-Myc slowly accumulated at the promoter over time after Dox induction (Fig. 4A, gray bars). In contrast, with Pin1 overexpression, V5-Myc was robustly detected at the E2F2 promoter within 15 min, with binding continuing to rise to high levels by 30 min before significantly decreasing at 45 min (Fig. 4A, black bars). A second wave of binding was observed at 60 to 120 min. The periodicity of this binding pattern was not reflected in the level of protein expression (Fig. 4A, middle right blot). We did not observe any Myc binding using our negative-control primers (internal GAPDH primers) (see Fig. S3A in the supplemental material). We observed corresponding changes in E2F2 gene expression (Fig. 4B). We also observed this cyclic binding with finer time intervals on the nucleolin promoter (see Fig. S3B in the supplemental material). Consistent with Pin1's lack of effect on Myc DNA binding at the p21 promoter, Pin1 expression had no effect on the recruitment rate of V5-Myc to the p21 promoter (Fig. 4C). Furthermore, consistent with the requirement for S62 phosphorylation for Pin1's regulation of Myc DNA binding, Pin1 had no effect on the rate of recruitment of V5-MycS62A in MCF10A-MycS62A cells to the E2F2 promoter (Fig. 4D), and the recruitment rate of MycS62A was delayed compared to MycWT.
Fig 4.
Pin1 increases the rate of recruitment of pS62-Myc to target gene promoters. (A) MCF10A-MycWT cells were infected with AdGFP or AdPin1 for 18 h and induced with 1 μg/ml Dox for 0 to 240 min. qChIP for V5-Myc at the E2F2 promoter was performed at the indicated time points after Dox induction of Myc, and 1% of the input was analyzed by Western blotting. (B) MCF10A-MycWT cells were treated as in panel A, and qRT-PCR for E2F2 was performed. (C) qChIP for Myc binding to the p21 promoter was performed in samples from panel A. (D) MCF10A-MycS62A cells were infected with AdGFP or AdPin1 for 18 h and induced with Dox for 0 to 120 min. qChIP for V5-MycS62A at the E2F2 promoter was performed at the indicated time points after Dox induction of MycS62A, and 1% of the input was analyzed by Western blotting. (E) MCF10A-MycWT cells were starved in 0.1% serum for 48 h and then stimulated with 2× serum and growth factors for 2 h in the presence or absence of 1 μM PiB or 10 μM U0126, and Myc expression was induced with Dox for 0 to 30 min as indicated. qChIP for V5-Myc at the nucleolin promoter was performed, and 1% of the input was analyzed by Western blotting. Statistical significance is indicated as in Fig. 1. The error bars indicate standard errors.
Since S62 phosphorylation of Myc occurs in response to serum stimulation, we tested whether Pin1 affects the rate of recruitment of Myc to promoters after serum stimulation. We found that inhibition of Pin1 with PiB prevented the early recruitment of Dox-induced Myc to the nucleolin promoter 2 h after serum stimulation. Treatment with U0126 (1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio] butadiene) to inhibit MEK and ERK signaling, known regulators of S62 phosphorylation, also prevented this rapid Myc recruitment in the stimulated cells (Fig. 4E). Furthermore, recruitment of MycS62A, which was slower and reduced in serum-stimulated cells compared to that of MycWT, was unaffected by either Pin1 inhibition or MEK inhibition (see Fig. S3C in the supplemental material). Thus, the rapid recruitment of Myc to target gene promoters following cell stimulation appears to involve both S62 phosphorylation and Pin1.
Pin1 enhances coactivator recruitment, Myc-mediated histone H4 acetylation, recruitment of pTEFb, and transcriptional elongation by RNA polymerase II.
Myc regulates the expression of its target genes through its recruitment of chromatin-remodeling proteins, including SNF5 (44) and the HATs GCN5 (45) and p300 (46). In addition, Myc recruits pTEFb, which phosphoryòlates the carboxyl-terminal domain (CTD) of RNA polymerase II (RNAP II) to allow transcriptional elongation (47). We examined whether Pin1 affects Myc's association with these transcriptional coactivators and found that, despite decreasing Myc protein levels with overexpression of Pin1 (Fig. 5A, compare input lanes 2 and 4), Myc association with p300, GCN5, cdk9 (a pTEFb subunit), and SNF5 increased (Fig. 5A, compare IP lanes 6 and 8). Consistent with this increased association, Pin1 expression enhanced the binding of coactivators to Myc-responsive promoters, as qChIP for p300, GCN5, and SNF5 in MCF10A cells showed increased binding of these proteins to Myc sites in target gene promoters with overexpression of Pin1 and reduced binding with Pin1 inhibition (Fig. 5B). We also observed that p300 recruitment to the E2F2 promoter after Dox induction of Myc followed the same cyclic kinetics as Myc binding in response to Pin1 overexpression (see Fig. S4 in the supplemental material), suggesting that p300 cycles at the promoter with Myc. As these coactivators can be recruited to chromatin by other transcription factors, we examined whether the increased recruitment of p300 and GCN5 to Myc-responsive promoters by Pin1 is Myc dependent. As shown in Fig. 5C, knockdown of Myc suppressed the enhanced binding of these factors with Pin1 overexpression.
Fig 5.
Pin1 enhances coactivator recruitment, Myc-mediated histone H4 acetylation, recruitment of pTEFb, and transcriptional elongation by RNA polymerase II. (A) 293 cells were transfected as indicated. Forty-eight hours posttransfection, V5-Myc was immunoprecipitated, and 0.5% of the input and coprecipitated proteins was detected by Western blotting. (B) MCF10A cells were treated with DMSO or PiB or infected with AdGFP or AdPin1 for 18 h. qChIP was performed using antibodies to the indicated endogenous proteins and primers specific for the Myc sites in the E2F2 promoter, and 1% of the input was analyzed by Western blotting. (C) 293 cells were transfected as indicated, qChIP for p300 and GCN5 at the nucleolin promoter was performed, and 1% of the input was analyzed by Western blotting. (D) 293 cells were transfected as indicated, and 1% of the input was analyzed by Western blotting. qChIP was performed using an AcH4 antibody and primers to the indicated genes. The black line in the Western blot indicates that extra lanes were removed. Primers to the satellite repeats of chromosome 4 (Chr4-Sat) were used as a heterochromatin control. (E) MCF10A-MycWT cells were infected with AdGFP or AdPin1, and Myc expression was induced with Dox for 4 h. pTEFb (cdk9) binding to the nucleolin promoter was measured with qChIP. (F) MCF10A-MycWT cells were infected with AdGFP or AdPin1 and Dox induced for 30 min. RNAP II (total and pS2) binding to the nucleolin gene body was measured with qChIP using downstream primers. Statistical significance is indicated as in Fig. 1. The error bars indicate standard errors.
To test the functional consequences of the enhanced HAT recruitment to Myc target gene promoters, we performed qChIP using an acetylated histone H4 antibody and primers specific for the Myc binding sites in target gene promoters after overexpression of Myc alone or Myc plus Pin1. At the tested trans-activated target promoters, expression of Myc increased H4 acetylation, a marker of active chromatin (48), and this increase was significantly potentiated by coexpression of Pin1 (Fig. 5D). In contrast, overexpression of Myc decreased acetylation at the trans-repressed p15 and p21 promoters, but in this case, Pin1 coexpression had no additional effect (Fig. 5D). In addition, Pin1 overexpression with Myc increased the binding of pTEFb (cdk9) to the nucleolin promoter (Fig. 5E), and this was associated with increased serine 2-phosphorylated (pS2) RNAP II in the body of the nucleolin gene 30 min after Myc induction (Fig. 5F). This is indicative of increased transcription elongation in response to Myc expression, together with Pin1 overexpression. Thus, in addition to increasing the rate of Myc recruitment to tested trans-activated promoters, Pin1 increases the level of Myc-dependent coactivator recruitment. This can affect an open chromatin state at these promoters and enhance RNAP II CTD phosphorylation to promote transcriptional elongation for increased target gene expression.
The disassociation of Myc from target gene promoters is facilitated by Pin1 and involves proteasomal degradation.
In our experiments examining the rate of recruitment of Myc to target gene promoters with Pin1 overexpression, we observed a cyclic pattern of Myc DNA binding (Fig. 4A). To further study this dynamic, we analyzed the rate at which Myc promoter binding is lost in the absence of new protein synthesis. We treated MCF10A-Myc cells with cycloheximide after Dox induction and measured V5-Myc binding to the nucleolin promoter by qChIP over time with or without Pin1 overexpression. Western blotting showed that Myc levels decreased over the course of the experiment, and this was mildly enhanced by Pin1 overexpression (Fig. 6A, blot). Likewise, the decrease in Myc promoter binding measured by qChIP was faster with Pin1 overexpression (Fig. 6A). Similar results were observed with endogenous Myc at the E2F2 promoter after cycloheximide treatment, where increased Myc turnover with Pin1 overexpression correlated with increased loss of Myc promoter binding, as well as with loss of target gene expression (see Fig. S5A to C in the supplemental material). Thus, the increase in Myc target gene promoter occupancy observed with Pin1 overexpression appears to result from an increased recruitment rate and not an increase in the retention time, since Pin1 accelerates Myc promoter dissociation.
Fig 6.
The disassociation of Myc from target gene promoters is facilitated by Pin1 and involves proteasomal degradation. (A) MCF10A-MycWT cells were infected with AdGFP or AdPin1 for 18 h, Dox induced for 2 h, and cycloheximide (CHX) treated (100 μg/ml) for between 0 and 120 min. qChIP for V5-Myc at the nucleolin promoter at the indicated time points after CHX treatment is graphed, and 1% of the input was analyzed by Western blotting. (B) MCF10A cells infected with AdGFP or AdPin1 were CHX treated for between 0 and 90 min with or without MG132 treatment. Myc qChIP at nucleolin was performed and graphed as the percentage remaining bound. (C) MCF10A-MycT58A cells were infected with AdGFP or AdPin1 for 18 h, Dox induced for MycT58A expression for 2 h, and CHX treated for between 0 and 120 min. qChIP for V5-Myc at the nucleolin promoter and Western blotting were performed. (D) Western blotting (left) and qChIP for the indicated proteins at the nucleolin promoter (right) were performed in MCF10A, MDA-MB-436, or 293M cells. Statistical significance is indicated as in Fig. 1. The error bars indicate standard errors.
In our previous work describing the role of Axin1 in promoting the assembly of a degradation complex for Myc containing GSK3β, PP2A, and Pin1, we observed that Axin1 could be detected at a Myc target gene promoter in nontransformed cells, suggesting that Myc may be turned over at its target gene promoters (9). Consistent with this idea, MG132 treatment lowered the rate of Myc promoter dissociation in MCF10A cells (Fig. 6B), indicating that proteasome activity helps to clear Myc from promoters. Furthermore, the MycT58A mutant, which has increased stability, showed slower dissociation from target gene promoters, and Pin1 had no effect on this rate, consistent with its inability to affect the turnover of the mutant (Fig. 6C) (18). Thus, timely dissociation of Myc from promoters involves Pin1, T58 phosphorylation, and proteasomal degradation.
We tested for the presence of proteins involved in Myc degradation at Myc target genes by qChIP. Analysis in MCF10A cells demonstrated that Myc, GSK3β, Pin1, PP2A, Axin1, Fbw7, and the 19S proteasome (19S) could all be detected at the Myc binding site in the nucleolin promoter (Fig. 6D, graph). In contrast, while Myc and Pin1 promoter association increased in two cancer cell lines, MDA-MB-436 and 293M (a 293 cell line derivative established in the laboratory with high pS62-Myc levels), association of proteins solely involved in Myc degradation (GSK3β, PP2A, Axin1, Fbw7, and 19S) was reduced (Fig. 6D, graph), and this was not due to changes in the expression levels of these proteins (Fig. 6D, blots). This result is consistent with our previous finding that Axin1 mRNA splicing is frequently altered in cancer cells and formation of the Axin1-mediated degradation complex for Myc is compromised (9, 23). Thus, in cancer cells, where Myc has increased stability (9, 23), recruitment of Pin1 and Myc is enhanced, but formation of the Axin1-mediated degradation complex at the promoter is reduced. This suggests that Pin1's activation function on Myc may be uncoupled from its degradation function in cancer cells.
Pin1 and Myc are cooverexpressed in cancer cells, where Pin1 enhances dynamic Myc DNA binding and target gene expression without affecting Myc turnover.
We have observed high pS62-Myc levels in many types of cancer cells, and this is often correlated with defects in the pS62/pT58 Myc degradation pathway downstream of Pin1 (9, 22, 23, 49). This could allow simultaneous high expression of pS62-Myc and Pin1 in cancer cells. As shown in Fig. 7A, we observed higher levels of both Pin1 and pS62-Myc, with lower pT58-Myc, in several human breast cancer cell lines and 293M cells than in the nontransformed MCF10A cells. Furthermore, we measured pS62-Myc, pT58-Myc, and Pin1 levels by immunofluorescence in 6 patient-matched pairs of normal and breast tumor tissue. We observed cooverexpression of pS62-Myc and Pin1 in breast tumor cells relative to matched normal ductal cells, with a high correlation coefficient (Fig. 7B; see Fig. S6A in the supplemental material). In contrast, pT58-Myc was reduced in tumors relative to matched normal cells (see Fig. S6B in the supplemental material). Thus, Pin1 levels correlate with pS62-Myc levels in tested human breast tumors.
Fig 7.
Pin1 and Myc are cooverexpressed in cancer cells, where Pin1 enhances dynamic Myc DNA binding and target gene expression without affecting its turnover. (A) The cell lines were subjected to Western blotting as indicated. Relative pS62- and pT58-Myc were measured as the intensity of the phospho-Myc/total Myc relative to MCF10A cells. Relative Pin1 levels were measured as Pin1/GAPDH levels. (B) Human breast cancer patient samples with matched normal samples were analyzed by IF for the expression of Pin1 and pS62-Myc. Representative images with areas of ductal carcinoma in situ (DCIS) and invasive adenocarcinoma are shown. (C) 293M cells were infected with AdGFP or AdPin1 and CHX treated (100 μg/ml) for 0 to 90 min, and Myc qChIP at the nucleolin promoter was performed at the indicated time points after CHX treatment. One percent of the input was analyzed by Western blotting. (D) 293M cells were treated with DMSO or PiB for 24 h and CHX for 0 to 90 min. qChIP for Myc at the nucleolin promoter was performed. (E) 293M cells were treated with DMSO or PiB for 24 h and CHX for 0 to 90 min. qChIP for pTEFb (cdk9) at the nucleolin promoter was performed. (F) 293M cells were treated with PiB for 24 h, and RNAP II (total and pS2) binding to the nucleolin gene body was measured with qChIP using downstream primers. (G) 293M cells were treated with DMSO or PiB and CHX treated for between 0 and 90 min. qRT-PCR for nucleolin was performed and normalized to GAPDH. Statistical significance is indicated as in Fig. 1. The error bars indicate standard errors.
We next examined the dissociation rate of Myc from promoters in cancer cells coexpressing high pS62-Myc and Pin1 levels. Cycloheximide treatment of 293M cells with and without ectopic Pin1 expression revealed that Myc turnover is slow in these cells (half-life greater than 90 min) and that expression of additional Pin1 does not affect Myc turnover (Fig. 7C, blots), consistent with reduced formation of the Axin1-mediated destruction complex for Myc in these cells (Fig. 6D). Interestingly, Myc protein still rapidly dissociated from the promoter by 30 min after cycloheximide treatment, but instead of continuing to diminish, as observed in MCF10A cells (Fig. 6A; see Fig. S5B in the supplemental material), a new peak of Myc binding was detected (Fig. 7C, graph). The kinetics of Myc DNA binding was not changed with additional ectopic Pin1, likely because Pin1 levels were already sufficiently high (Fig. 7C). In contrast, Pin1 inhibition, while also not affecting Myc turnover in these cells, reduced the starting level of Myc promoter binding and prevented the new peak of Myc binding (Fig. 7D). pTEFb showed similar kinetics of binding and inhibition by PiB treatment (Fig. 7E), and this was associated with decreased elongating Pol II (Fig. 7F). Moreover, target gene expression also followed kinetics similar to those of Myc and pTEFb promoter binding, and PiB again inhibited this (Fig. 7G). We also observed that PiB treatment reduced the overall levels of Myc binding and the cyclic nature of this binding in the breast cancer cell lines MDA-MB-453 (see Fig. S6C in the supplemental material) and MDA-MB-436 (see Fig. S6D in the supplemental material). Thus, in cancer cells with high pS62-Myc and Pin1 coexpression, Pin1 enhances the dynamics of Myc DNA binding, resulting in increased transcriptional activity of target genes without affecting Myc turnover.
Coexpression of Pin1 and Myc increases expression of cell growth and metabolism genes that are enriched in poor-outcome breast cancer.
To assess global changes in Myc target gene expression mediated by Pin1, we performed RNA-seq analysis, comparing MCF10A cells with Myc overexpression (Myc) to Myc plus Pin1 overexpression (Myc plus Pin1). We found that the majority of reported Myc target genes affected by Pin1 coexpression are upregulated (134 significantly upregulated and only 9 significantly downregulated Myc target genes; FC cutoff, 2.0; q value, less than 0.01) (Fig. 8A). This is in contrast to overexpression of Myc alone, which resulted in 11 upregulated and 40 downregulated Myc target genes using the same statistical cutoffs. We also found that the targets upregulated by coexpression of Myc and Pin1 are largely composed of genes involved in ribosome biogenesis/translation and metabolism based on gene ontology (GO) (Fig. 8A). We examined the expression of several genes identified in our screen that are involved in metabolic and ribosome biogenesis pathways by qRT-PCR upon coexpression of Myc and Pin1 and found them to be upregulated (see Fig. S7A in the supplemental material). We also used GoMiner analysis software (50) to score biological processes/gene pathway enrichment with Pin1 and Myc cooverexpression. This analysis also found that the most significantly enriched processes are translation/ribosome biogenesis and metabolism (Fig. 8B).
Fig 8.
Myc and Pin1 coexpression induces cell growth genes that are enriched in poor-outcome breast cancer. (A) MCF10A-Myc cells were infected with AdGFP or AdPin1, and Myc expression was induced with Dox for 2 h. RNA was harvested for RNA-seq, and functional classes of Myc target genes significantly altered (fold change cutoff, 2.0; FDR-adjusted q value cutoff, 0.01) with Pin1 overexpression are shown. (B) Functional pathway enrichment scores for Myc target genes upregulated by Pin1 overexpression using GoMiner software (NCI, Bethesda, MD). (C) Genes identified in the RNA-seq analysis as significantly upregulated by Myc plus Pin1 versus Myc alone (FC cutoff, 1.5; q value cutoff, 0.01) were used for KS statistical tests using 352 human breast cancer samples subtyped by immunohistochemical markers (GEO GSE2109). A representative KS plot is shown in which genes distinguishing TN tumors from ER+/Her2− (luminal A-like) tumors are ordered on the x axis and enrichment of genes upregulated by Pin1 plus Myc in the ordered genes are given a D score on the y axis. (D) KS tests were performed as in panel C. The P value for enrichment of the Myc-plus-Pin1 up genes in the first subtype relative to the second subtype and a “reverse” P value for enrichment in the second subtype relative to the first are shown. Statistical significance is indicated as in Fig. 1.
Since both Myc and Pin1 were cooverexpressed in the breast tumors we examined, we asked whether the Myc-plus-Pin1 gene signature is enriched in any specific subtypes of human breast cancer. We used a KS statistical test (similar to what is used in gene set enrichment analysis [GSEA] [51]) to determine the enrichment of our gene set in a publically available microarray data set from 352 human breast cancer samples subtyped by immunohistochemical markers (GEO GSE2109). A representative KS plot is shown in Fig. 8C, in which differentially regulated genes between TN tumors and ER+/Her2− (luminal A-like) tumors are ordered on the x axis, and genes in this list that are upregulated by Pin1 and Myc cooverexpression from our RNA-seq data are given a positive D score, shown on the y axis. A positive peak toward the left of the graph indicates enrichment of our gene set in the TN-upregulated genes. A P value was calculated for this level of enrichment using Monte Carlo simulations. A reverse P value for the opposite comparison (ER+/Her2− versus TN) was also calculated. Subtype comparisons between poor and good outcomes are summarized in Fig. 8D. We found that the Myc-plus-Pin1 gene signature was significantly enriched in the TN and Her2-positive human breast cancers versus the luminal A-like breast cancers, as well as in high-grade and -stage tumors (Fig. 8D; see Fig. S7B in the supplemental material). Similar results were obtained using a TCGA data set containing 430 human breast tumors molecularly subtyped by PAM50 (35). The Myc-plus-Pin1 signature was again enriched in the poorer-outcome Her2 and basal subtypes relative to the luminal A subtype (Fig. 8D; see Fig. S7B in the supplemental material). Thus, Pin1 and Myc cooverexpression activates genes that are enriched in poor-outcome breast cancer subtypes.
Pin1 enhances Myc's oncogenic potential.
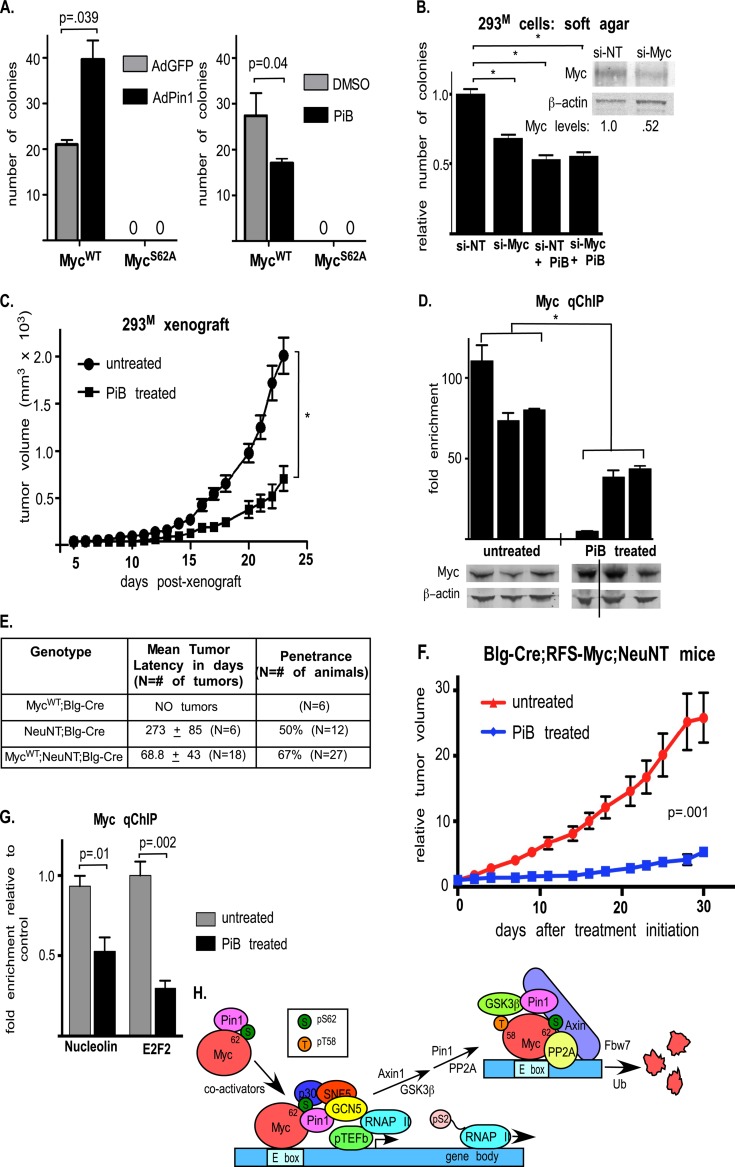
To investigate the effects of Pin1 on Myc-induced cell transformation, we performed soft-agar colony-forming assays using the MCF10A-Myc cells with or without Pin1 overexpression or inhibition. MCF10A cells are unable to form colonies in soft agar (52), but overexpression of MycWT induced a low level of colony formation (Fig. 9A). Pin1 overexpression increased the number of colonies induced by MycWT by about 50%, while treatment with the PiB inhibitor reduced colony formation (Fig. 9A). In addition, colony size was generally larger with Pin1 overexpression and smaller with Pin1 inhibition (see Fig. S8A in the supplemental material). In contrast, MCF10A-MycS62A cells were unable to form colonies in soft agar, and overexpression or inhibition of Pin1 in these cells had no effect (Fig. 9A). Thus, the Pin1 effect on transformation in MCF10A cells is Myc dependent and requires S62 phosphorylation.
Fig 9.
Pin1 regulates Myc oncogenic potential. (A) MCF10A-MycWT and MCF10A-MycS62A cells were infected with AdPin1 or AdGFP or pretreated with DMSO or 0.5 μg/ml PiB, as indicated, and induced with Dox for Myc expression for 18 h prior to plating in soft agar. The cells were grown in soft agar with continuous Dox and/or PiB treatment for 4 weeks, and the total number of colonies was determined. (B) 293M cells were transfected with a nontargeting (si-NT) or Myc-specific (si-Myc) siRNA for 48 h, treated with DMSO or PiB as indicated, and grown in soft agar as in panel A. (C) 293M-xenografted mice were treated by intratumoral injection of 0.6 mg/kg PiB. Tumor size was measured with calipers. Six mice (n = 12 tumors) were analyzed per group. (D) PiB-treated and untreated tumors from panel C were analyzed by Western blotting and by Myc qChIP at the nucleolin promoter. (E) Mice were generated to express MycWT knocked into the ROSA26 locus (RFS-MycWT) with activated Her2 (NeuNT) at its endogenous locus in the mammary gland in response to Cre recombinase (Blg-Cre). Tumor latency and penetrance are indicated. (F) Once mammary tumors were detected in the MycWT; NeuNT; Blg-Cre mice (Fig. 7H), they were randomized to the control untreated or PiB-treated study arm. The mice were treated with intraperitoneal injection of 0.6 mg/kg PiB three times per week (n = 6), and tumor growth was monitored by caliper measurements and compared to that of untreated animals (n = 12). (G) PiB-treated and untreated tumors from panel F were analyzed by Myc qChIP at the nucleolin and E2F2 promoters. (H) Model for the role of Pin1 in regulating Myc promoter binding and transcriptional activity. Statistical significance is indicated as in Fig. 1. The error bars indicate standard errors.
We also tested the effect of Pin1 inhibition on the tumorigenic growth of 293M cells in xenografts. We first examined the dependency of these cells on Myc for soft-agar growth. As shown in Fig. 9B, partial Myc knockdown reduced the growth of 293M cells in soft agar. PiB treatment also reduced growth (Fig. 9B). However, PiB had no additional growth-suppressing function when combined with Myc knockdown (Fig. 9B), suggesting that anchorage-independent growth of these cells is Myc dependent and that PiB exerts its action through Myc in this system. We also xenografted 293M cells into the 4th mammary gland of NOD/SCID γ-chain-null mice. As shown in Fig. 9C and in Fig. S8B and C in the supplemental material, PiB treatment substantially reduced tumor growth and final tumor size. In addition, treated tumors showed decreased proliferation (see Fig. S8D in the supplemental material) and increased apoptosis (see Fig. S8E in the supplemental material). Importantly, tumor cells from the treated animals had less Myc associated with the nucleolin promoter, as assessed by qChIP (Fig. 9D, graph), although they generally had more Myc protein (Fig. 9D, blots). Thus, Pin1 inhibition reduces tumorigenic growth of cells associated with suppressed Myc DNA binding.
Finally, we tested the effects of PiB treatment on tumor growth using a mouse model of Myc-induced spontaneous breast cancer that we have developed (23) (see the supplemental material). Mice were generated to express MycWT from the ROSA26 locus (RFS-MycWT), along with activated Her2 (NeuNT), in the mouse mammary gland in response to Cre recombinase (Blg-Cre) (53–55). Coexpression of MycWT along with NeuNT in this model accelerates mammary tumor formation (mean tumor latency, 69 days) compared to mice expressing NeuNT alone (273 days) or knocked-in MycWT alone, which do not develop tumors (Fig. 9E). Once mammary tumors were detected in the MycWT/NeuNT mice they were randomized to the control untreated or PiB-treated study arms. As shown in Fig. 9F, treatment with PiB significantly slowed tumor growth in this model. In addition, PiB-treated tumors showed decreased proliferation (see Fig. S8F in the supplemental material) and increased apoptosis (see Fig. S8G in the supplemental material) relative to control tumors. Importantly, tumor cells from the treated animals had less Myc associated with the nucleolin and E2F2 promoters, as assessed by qChIP (Fig. 9G). Thus, these data show a strong role for Pin1 in regulating the oncogenic potential of Myc both in vitro and in vivo and suggest that Pin1 is a potential therapeutic target for human cancer with high Pin1 and Myc levels.
DISCUSSION
A working model for the role of Pin1 in the coordinated activation and degradation of Myc.
Our data indicate that the Pin1 prolyl isomerase enhances the recruitment of S62-phosphorylated Myc to target gene promoters in response to cell growth stimulation and subsequently facilitates its release, associated with T58 phosphorylation and protein turnover at or near the promoter (Fig. 9H). This dynamic regulation of Myc DNA binding is associated with enhanced recruitment of several coactivators, resulting in changes in Myc-mediated chromatin acetylation patterns, as well as increased levels of elongating RNAP II. Thus, dynamic Myc DNA binding appears to contribute to optimal Myc transcriptional activity. Our results indicate that the Pin1 effect is primarily at trans-activated Myc target genes and, in particular, target genes involved in cell growth and enriched in poor-outcome breast cancer. This model is not incompatible with the recent report by Lin et al. in which Myc appears to amplify the expression of active genes in a given cell type (56). The spectrum of Pin1-responsive Myc target genes is consistent with ribosome biogenesis and metabolism genes known to be upregulated during cell proliferation and tumorigenesis (57–59).
Our data support a model where Pin1 regulates Myc at two points: (i) target gene promoter binding and cofactor recruitment, leading to transcriptional activation, and (ii) subsequent release from the promoter associated with degradation (Fig. 9H). In cancer cells with defects in the pS62/pT58 Myc degradation pathway downstream of Pin1, Pin1 no longer appears to facilitate Myc degradation, but it can still mediate Myc activation. Interestingly, we still observe a rapid dissociation of Myc from target gene promoters in cancer cells. However, unlike nontransformed cells, a new peak of Myc binding follows the rapid dissociation in the absence of new protein synthesis. This new Myc recruitment is Pin1 dependent and likely comes from remaining pools of pS62-Myc present in cancer cells. The dynamic interaction of Myc with target gene promoters appears to be important for maximal gene activation. We speculate that this cyclic pattern of Myc DNA binding plays some important role in continued firing of gene transcription, possibly through the release of paused polymerases (60, 61). At this point, we do not know the mechanism behind the rapid release of Myc following inhibition of protein synthesis in cancer cells with more stable Myc. We also do not know whether our experiments are measuring changes in the DNA binding of a single Myc molecule or whether Myc is moving very rapidly throughout the nucleus (62) and we are measuring more steady-state changes in the association of a pool of Myc with a given promoter.
Pin1 as a transcriptional coactivator for Myc.
Studies suggest that Myc regulates many, if not all, actively transcribed mammalian genes (56, 63). However, our findings and other published reports suggest that Myc activity is not simply controlled by Myc protein levels but that it is regulated by posttranslational modifications and recruitment of cooperating transcriptional coactivators (44, 45, 63–65). Our work suggests that Pin1 functions as a unique Myc coactivator, controlling the dynamics of Myc DNA binding, as well as facilitating Myc's recruitment of p300, GCN5, hSNF5, and pTEFb to promoters. In agreement with our data, Sanchez-Arevalo Lobo et al. have recently shown that Pin1 augments Myc's interaction with p300 to increase transcriptional activity (66). At this point, it is unclear whether Pin1's facilitation of Myc's association with transcriptional coactivators controls its dynamic interaction with DNA through their chromatin-remodeling activity or if Pin1-mediated isomerization of Myc facilitates its DNA interaction and this then helps stabilize its interaction with the other coactivators.
Another interesting question remaining to be addressed is how N-terminal proline isomerization events could affect Myc DNA binding mediated by the C-terminal basic helix-loop-helix–leucine zipper (B-HLH-LZ) domain. The amino terminus of Myc is thought to be mostly unstructured (67, 68); however, posttranslational modifications and proline isomerization can alter conformation states that can influence the activity of adjacent regions. Indeed, many Myc interactors make contact with multiple domains in both the N terminus and C terminus of Myc, including many coactivators. Specifically regarding our results, TRRAP (part of the GCN5 complex) interacts with MBII, as well as N-terminal amino acids adjacent to and partly overlapping MBI; pTEFb interacts with MBI; and p300 interacts with a large domain in the N terminus, including MBI and MBII, as well as the B-HLH-LZ domain in the C terminus (69).
It is also interesting to speculate that these results could apply to other transcription factors that are regulated by Pin1. Specifically, Pin1 is reported to influence the activity of p53, NF-κB, β-catenin, and hypoxia-inducible factor 1α through changes in their protein stability, nuclear localization, and level of chromatin binding, respectively (24, 25, 70–74). Pin1 was also shown to increase recruitment of the coactivator p300 to STAT3 (75) and p73 (71). In addition, it has been shown that Pin1 functions as a coactivator of the steroid receptor by affecting SRC-3 coactivator complex formation and turnover (28). However, none of these studies explored the dynamics of the transcription factor-DNA interactions. Direct analysis of the on-and-off rates of steroid receptor transcription factors has been performed (76), indicating a periodicity similar to what we observe here for Myc cycling at target gene promoters. Moreover, studies with the estrogen receptor have shown that inhibition of its release suppresses its transcriptional activity, suggesting that degradation facilitates activity by allowing loading of new transcription factor (76, 77). However, how steroid receptor cycling at promoters is controlled has not been reported. Given the similarities between our observations and some of these reports, our data could provide important contributions to these other systems.
Cooperation between Pin1 and Myc during tumorigenesis.
Several studies have indicated that ubiquitination of Myc is important for transcriptional activity (37, 78, 79), and the idea that the ubiquitin-proteasome system (UPS) promotes elimination of Myc transcriptional complexes is not new (63). Our work supports a model in which the UPS regulates the transcriptional activity of Myc. This is an attractive model for the regulation of Myc activity in normal cells, ensuring the tight control of Myc activity through coupled transcriptional activation and proteasomal degradation. However, this model does not address Myc regulation in tumor cells, where it is more stable. Our results suggest that Pin1 could still regulate dynamic Myc DNA binding in the presence of stable pools of pS62-Myc, and this could allow enhanced elongating polymerase and target gene expression.
Similar to Myc, Pin1 expression is elevated in many cancers, including breast, prostate, colorectal, brain, cervical, lung, ovarian, and liver cancers and melanoma (24, 80–83). Given that Pin1 can facilitate PP2A-mediated dephosphorylation of S62 and Myc degradation in nontransformed cells (18), it must be the case that, in these cancer cells and patient tumor samples, Myc dephosphorylation and degradation are blocked downstream of Pin1's first activation step. Indeed, we find high expression of pS62-Myc in multiple cancer types (9, 22, 23), and aside from Myc and Pin1, members of the Axin1-Myc destruction complex are reduced at target gene promoters in tested cancer cells. We have investigated defects in the pS62/pT58 Myc degradation pathway in breast cancer cell lines and patient samples and found reduced interaction of Myc with Axin1, GSK3β, and PP2Ac (9, 23). Moreover, in matched breast tumor and adjacent normal tissue, we have observed both a reduction in AXIN1 mRNA expression and a switch to a naturally occurring splice variant that is compromised in its ability to nucleate a destruction complex for Myc (23). Thus, in cancer cells, where S62 dephosphorylation and Myc degradation are inhibited, both pS62-Myc and Pin1 can be coexpressed at high levels, potentiating tumorigenesis.
Furthermore, given that the Myc-plus-Pin1 gene signature is enriched in breast cancer subtypes with traditionally poorer outcomes, these data suggest that Pin1 might be a good therapeutic target in these subtypes of breast cancer that overexpress Myc. Targeting Pin1 as a cancer therapeutic shows promise, as inhibition of Pin1 using juglone or dominant-negative mutants in cancer cells induces apoptosis and suppresses transformation (82, 84–86). In addition, cells from Pin1 knockout mice are impaired for cell cycle reentry, and Pin1 knockout combined with transgenic expression of v-Ha-Ras in the mammary gland reduces tumor formation (87). We show here that a specific Pin1 inhibitor, PiB (39), potently inhibits cell transformation and tumorigenesis in Myc-dependent models in vitro and in vivo. Furthermore, even though Pin1 has many targets in cells, the fact that Pin1-null mice are viable strengthens the therapeutic potential of Pin1 inhibitors as anticancer agents with low toxicity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karyn Taylor for her help with the preparation and ordering of reagents and Brittany Allen-Peterson for her helpful comments in preparing the manuscript. We also thank Devorah Goldman, Hilary Clark, and William H. Fleming for providing the NOD/SCID γ-chain-null mice and M. S. Dai for providing the acetylated histone H4 antibody. We thank Megan Troxell and Chris Corless for providing the matched normal and tumor breast cancer patient samples. We thank Paul Spellman for his suggestions regarding bioinformatic analysis of breast tumor samples and Anneleen Daemen for providing us the TCGA data set with associated subtyping. We also thank Jukka Westermarck for thoughtful discussions.
A.S.F. was supported by award number T32CA106195, “Training in the Molecular Basis of Skin/Mucosa Pathobiology,” to OHSU from the National Cancer Institute; a Collins Medical Trust Fund Award; and a Knight Cancer Institute Career Development Award. This work was also supported by R01/grant-num> CA100855 and CA129040, LLS 82325, DOD BC061306, and Komen BCTR0706821 to R.C.S.
Footnotes
Published ahead of print 28 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01455-12.
REFERENCES
- 1. Meyer N, Penn LZ. 2008. Reflecting on 25 years with MYC. Nat. Rev. Cancer 8: 976– 990 [DOI] [PubMed] [Google Scholar]
- 2. Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. 2004. Analysis of genomic targets reveals complex functions of MYC. Nat. Rev. Cancer 4: 562– 568 [DOI] [PubMed] [Google Scholar]
- 3. Kleine-Kohlbrecher D, Adhikary S, Eilers M. 2006. Mechanisms of transcriptional repression by Myc. Curr. Top. Microbiol. Immunol. 302: 51– 62 [DOI] [PubMed] [Google Scholar]
- 4. Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17: 1115– 1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flinn EM, Busch CM, Wright AP. 1998. myc boxes, which are conserved in myc family proteins, are signals for protein degradation via the proteasome. Mol. Cell. Biol. 18: 5961– 5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones TR, Cole MD. 1987. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol. Cell. Biol. 7: 4513– 4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelly K, Cochran BH, Stiles CD, Leder P. 1983. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell 35: 603– 610 [DOI] [PubMed] [Google Scholar]
- 8. Sears R, Leone G, DeGregori J, Nevins JR. 1999. Ras enhances Myc protein stability. Mol. Cell 3: 169– 179 [DOI] [PubMed] [Google Scholar]
- 9. Arnold HK, Zhang X, Daniel CJ, Tibbitts D, Escamilla-Powers J, Farrell A, Tokarz S, Morgan C, Sears RC. 2009. The Axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J. 28: 500– 512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lutterbach B, Hann SR. 1994. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol. Cell. Biol. 14: 5510– 5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pulverer BJ, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett JR. 1994. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene 9: 59– 70 [PubMed] [Google Scholar]
- 12. Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14: 2501– 2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bachireddy P, Bendapudi PK, Felsher DW. 2005. Getting at MYC through RAS. Clin. Cancer Res. 11: 4278– 4281 [DOI] [PubMed] [Google Scholar]
- 14. Sears RC. 2004. The life cycle of C-myc: from synthesis to degradation. Cell Cycle 3: 1133– 1137 [PubMed] [Google Scholar]
- 15. Bechard M, Dalton S. 2009. Subcellular localization of glycogen synthase kinase 3beta controls embryonic stem cell self-renewal. Mol. Cell. Biol. 29: 2092– 2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gregory MA, Qi Y, Hann SR. 2003. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J. Biol. Chem. 278: 51606– 51612 [DOI] [PubMed] [Google Scholar]
- 17. Arnold HK, Sears RC. 2006. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol. Cell. Biol. 26: 2832– 2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, Counter CM, Nevins JR, Means AR, Sears R. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6: 308– 318 [DOI] [PubMed] [Google Scholar]
- 19. Amati B. 2004. Myc degradation: dancing with ubiquitin ligases. Proc. Natl. Acad. Sci. U. S. A. 101: 8843– 8844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U. S. A. 101: 9085– 9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23: 2116– 2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malempati S, Tibbitts D, Cunningham M, Akkari Y, Olson S, Fan G, Sears RC. 2006. Aberrant stabilization of c-Myc protein in some lymphoblastic leukemias. Leukemia 20: 1572– 1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X, Farrell AS, Daniel CJ, Arnold H, Scanlan C, Laraway BJ, Janghorban M, Lum L, Chen D, Troxell M, Sears R. 2012. Mechanistic insight into Myc stabilization in breast cancer involving aberrant Axin1 expression. Proc. Natl. Acad. Sci. U. S. A. 109: 2790– 2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ryo A, Liou YC, Lu KP, Wulf G. 2003. Prolyl isomerase Pin1: a catalyst for oncogenesis and a potential therapeutic target in cancer. J. Cell Sci. 116: 773– 783 [DOI] [PubMed] [Google Scholar]
- 25. Yeh ES, Means AR. 2007. PIN1, the cell cycle and cancer. Nat. Rev. Cancer 7: 381– 388 [DOI] [PubMed] [Google Scholar]
- 26. Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, Kaplan F, Capobianco A, Pece S, Di Fiore PP, Del Sal G. 2009. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat. Cell Biol. 11: 133– 142 [DOI] [PubMed] [Google Scholar]
- 27. Takahashi K, Akiyama H, Shimazaki K, Uchida C, Akiyama-Okunuki H, Tomita M, Fukumoto M, Uchida T. 2007. Ablation of a peptidyl prolyl isomerase Pin1 from p53-null mice accelerated thymic hyperplasia by increasing the level of the intracellular form of Notch1. Oncogene 26: 3835– 3845 [DOI] [PubMed] [Google Scholar]
- 28. Yi P, Wu RC, Sandquist J, Wong J, Tsai SY, Tsai MJ, Means AR, O'Malley BW. 2005. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol. Cell. Biol. 25: 9687– 9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sears R, Ohtani K, Nevins JR. 1997. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol. Cell. Biol. 17: 5227– 5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Escamilla-Powers JR, Sears RC. 2007. A conserved pathway that controls c-Myc protein stability through opposing phosphorylation events occurs in yeast. J. Biol. Chem. 282: 5432– 5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106– 1117 [DOI] [PubMed] [Google Scholar]
- 32. Kim J, Lee JH, Iyer VR. 2008. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One 3: e1798. 10.1371/journal.pone.0001798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abadie A. 2002. Bootstrap tests for distributional treatment effects in instrumental variable models. J. Am. Stat. Assoc. 97:284– 292 [Google Scholar]
- 34. Sekhon J. 2011. Multivariate and propensity score matching software with automated balance optimization. J. Stat. Software. 42: 1– 52 [Google Scholar]
- 35. Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. 2009. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27: 1160– 1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Molinari E, Gilman M, Natesan S. 1999. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 18: 6439– 6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salghetti SE, Muratani M, Wijnen H, Futcher B, Tansey WP. 2000. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. U. S. A. 97: 3118– 3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomas D, Tyers M. 2000. Transcriptional regulation: kamikaze activators. Curr. Biol. 10: R341– R343 [DOI] [PubMed] [Google Scholar]
- 39. Uchida T, Takamiya M, Takahashi M, Miyashita H, Ikeda H, Terada T, Matsuo Y, Shirouzu M, Yokoyama S, Fujimori F, Hunter T. 2003. Pin1 and Par14 peptidyl prolyl isomerase inhibitors block cell proliferation. Chem. Biol. 10: 15– 24 [DOI] [PubMed] [Google Scholar]
- 40. Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, Tyner AL. 2001. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. U. S. A. 98: 4510– 4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Moroy T, Bartek J, Massague J, Hanel F, Eilers M. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 3: 392– 399 [DOI] [PubMed] [Google Scholar]
- 42. Liu T, Huang Y, Likhotvorik RI, Keshvara L, Hoyt DG. 2008. Protein Never in Mitosis Gene A Interacting-1 (PIN1) regulates degradation of inducible nitric oxide synthase in endothelial cells. Am. J. Physiol. Cell Physiol. 295: C819– C827 [DOI] [PubMed] [Google Scholar]
- 43. Winkler KE, Swenson KI, Kornbluth S, Means AR. 2000. Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science 287: 1644– 1647 [DOI] [PubMed] [Google Scholar]
- 44. Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. 1999. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 22: 102– 105 [DOI] [PubMed] [Google Scholar]
- 45. Kenneth NS, Ramsbottom BA, Gomez-Roman N, Marshall L, Cole PA, White RJ. 2007. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc. Natl. Acad. Sci. U. S. A. 104: 14917– 14922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vervoorts J, Luscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, Austen M, Luscher B. 2003. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 4: 484– 490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gargano B, Amente S, Majello B, Lania L. 2007. P-TEFb is a crucial co-factor for Myc transactivation. Cell Cycle 6: 2031– 2037 [DOI] [PubMed] [Google Scholar]
- 48. Hebbes TR, Thorne AW, Crane-Robinson C. 1988. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7: 1395– 1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, Sears R, Clurman BE, Look AT. 2007. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 204: 1813– 1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN. 2003. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 4: R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545– 15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miller FR. 2000. Xenograft models of premalignant breast disease. J. Mammary Gland Biol. Neoplasia 5: 379– 391 [DOI] [PubMed] [Google Scholar]
- 53. Andrechek ER, Hardy WR, Siegel PM, Rudnicki MA, Cardiff RD, Muller WJ. 2000. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 97: 3444– 3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Selbert S, Bentley DJ, Melton DW, Rannie D, Lourenco P, Watson CJ, Clarke AR. 1998. Efficient BLG-Cre mediated gene deletion in the mammary gland. Transgenic Res. 7: 387– 396 [DOI] [PubMed] [Google Scholar]
- 55. Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, Sears RC. 2011. Phosphorylation regulates c-Myc's oncogenic activity in the mammary gland. Cancer Res. 71: 925– 936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56– 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. 2008. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456: 971– 975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dang CV, Le A, Gao P. 2009. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15: 6479– 6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van Riggelen J, Yetil A, Felsher DW. 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 10: 301– 309 [DOI] [PubMed] [Google Scholar]
- 60. Giraud M, Yoshida H, Abramson J, Rahl PB, Young RA, Mathis D, Benoist C. 2012. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 109: 535– 540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141: 432– 445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arabi A, Rustum C, Hallberg E, Wright AP. 2003. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J. Cell Sci. 116: 1707– 1717 [DOI] [PubMed] [Google Scholar]
- 63. Luscher B, Vervoorts J. 2012. Regulation of gene transcription by the oncoprotein MYC. Gene 494: 145– 160 [DOI] [PubMed] [Google Scholar]
- 64. Agrawal P, Yu K, Salomon AR, Sedivy JM. 2010. Proteomic profiling of Myc-associated proteins. Cell Cycle 9: 4908– 4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4: 575– 580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sanchez-Arevalo Lobo VJ, Doni M, Verrecchia A, Sanulli S, Faga G, Piontini A, Bianchi M, Conacci-Sorrell M, Mazzarol G, Peg V, Losa JH, Ronchi P, Ponzoni M, Eisenman RN, Doglioni C, Amati B. 14 January 2013. Dual regulation of Myc by Abl. Oncogene. 10.1038/onc.2012.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dyson HJ, Wright PE. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6: 197– 208 [DOI] [PubMed] [Google Scholar]
- 68. Fladvad M, Zhou K, Moshref A, Pursglove S, Safsten P, Sunnerhagen M. 2005. N and C-terminal sub-regions in the c-Myc transactivation region and their joint role in creating versatility in folding and binding. J. Mol. Biol. 346: 175– 189 [DOI] [PubMed] [Google Scholar]
- 69. Cowling VH, Cole MD. 2006. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 16: 242– 252 [DOI] [PubMed] [Google Scholar]
- 70. Kim MR, Choi HS, Heo TH, Hwang SW, Kang KW. 2008. Induction of vascular endothelial growth factor by peptidyl-prolyl isomerase Pin1 in breast cancer cells. Biochem. Biophys. Res. Commun. 369: 547– 553 [DOI] [PubMed] [Google Scholar]
- 71. Mantovani F, Piazza S, Gostissa M, Strano S, Zacchi P, Mantovani R, Blandino G, Del Sal G. 2004. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol. Cell 14: 625– 636 [DOI] [PubMed] [Google Scholar]
- 72. Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X, Crook T, Del Sal G. 2007. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat. Struct. Mol. Biol. 14: 912– 920 [DOI] [PubMed] [Google Scholar]
- 73. Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP. 2003. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12: 1413– 1426 [DOI] [PubMed] [Google Scholar]
- 74. Takahashi K, Uchida C, Shin RW, Shimazaki K, Uchida T. 2008. Prolyl isomerase, Pin1: new findings of post-translational modifications and physiological substrates in cancer, asthma and Alzheimer's disease. Cell. Mol. Life Sci. 65: 359– 375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lufei C, Koh TH, Uchida T, Cao X. 2007. Pin1 is required for the Ser727 phosphorylation-dependent Stat3 activity. Oncogene 26: 7656– 7664 [DOI] [PubMed] [Google Scholar]
- 76. Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11: 695– 707 [DOI] [PubMed] [Google Scholar]
- 77. Lonard DM, Nawaz Z, Smith CL, O'Malley BW. 2000. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol. Cell 5: 939– 948 [DOI] [PubMed] [Google Scholar]
- 78. Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293: 1651– 1653 [DOI] [PubMed] [Google Scholar]
- 79. Zhang Q, Spears E, Boone DN, Li Z, Gregory MA, Hann SR. 2013. Domain-specific c-Myc ubiquitylation controls c-Myc transcriptional and apoptotic activity. Proc. Natl. Acad. Sci. U. S. A. 110: 978– 983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J, Wheeler TM, Lu KP, Bao L. 2003. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 63: 6244– 6251 [PubMed] [Google Scholar]
- 81. Lu KP. 2003. Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer Cell 4: 175– 180 [DOI] [PubMed] [Google Scholar]
- 82. Ryo A, Liou YC, Wulf G, Nakamura M, Lee SW, Lu KP. 2002. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol. Cell. Biol. 22: 5281– 5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, Lu KP. 2001. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 20: 3459– 3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lu KP, Hanes SD, Hunter T. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380: 544– 547 [DOI] [PubMed] [Google Scholar]
- 85. Rippmann JF, Hobbie S, Daiber C, Guilliard B, Bauer M, Birk J, Nar H, Garin-Chesa P, Rettig WJ, Schnapp A. 2000. Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ. 11: 409– 416 [PubMed] [Google Scholar]
- 86. Wulf G, Ryo A, Liou YC, Lu KP. 2003. The prolyl isomerase Pin1 in breast development and cancer. Breast Cancer Res. 5: 76– 82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. 2004. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 23: 3397– 3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.