Abstract
In this work, we characterized a new, 160-kb, blaOXA-48-harboring IncL/M-type plasmid isolated from a Klebsiella pneumoniae strain from France. Moreover, we report the transfer of a 60-kb OXA-48-encoding plasmid from Klebsiella pneumoniae to other Enterobacteriaceae in two patients.
TEXT
The emergence of carbapenemase-producing Enterobacteriaceae has become a major public health concern (1). Carbapenemases identified in Enterobacteriaceae are frequently class A β-lactamases, particularly KPC enzymes, and class B metallo-β-lactamases. However, the emergence of Enterobacteriaceae strains producing class D carbapenemases such as OXA-48 has increasingly been reported (2). This enzyme, initially detected in a carbapenem-resistant Klebsiella pneumoniae strain in Turkey (3), has been identified extensively in that country as well as in other surrounding Mediterranean countries (2, 4). Since then, it has also been identified in Senegal, Kuwait, the Sultanate of Oman, and India, as well as in several European countries (2, 5–10). Although OXA-48 has most often been found in K. pneumoniae, it has also been identified in Escherichia coli, Enterobacter spp., Klebsiella oxytoca, Citrobacter freundii, Providencia rettgeri, and Serratia marcescens (2, 7, 8, 10, 11). In these strains, blaOXA-48 is usually associated with a Tn1999-type transposon (12) and has been observed in self-conjugative 60- to 70-kb plasmids exhibiting similar restriction profiles (5, 6, 11, 12). It is considered that the current spread of blaOXA-48 is linked to the diffusion of a unique self-conjugative IncL/M plasmid, designated pOXA-48a, that was recently sequenced by Poirel et al. (13). However, the horizontal spread of blaOXA-48-harboring plasmids in a single patient has not yet been reported.
In this work, we report the transfer of a 60-kb OXA-48-encoding plasmid from K. pneumoniae to other Enterobacteriaceae in two patients. Moreover, we characterized a new, 160-kb, blaOXA-48-carrying conjugative plasmid from a K. pneumoniae isolate.
In 2010, we isolated six Enterobacteriaceae organisms resistant to ertapenem (MICs, 1.5 to 3 μg/ml) (Table 1) from samples from two patients hospitalized at the University Hospital Center of Nancy, France. There was no direct epidemiological link between the two patients, who were both colonized during an outbreak which affected our hospital center between 2010 and 2012 (10). For patient 1 (a 61-year-old man with type 2 diabetes), ertapenem-resistant K. pneumoniae KP1 was isolated from an intra-abdominal abscess and feces in October 2010. Two additional ertapenem-resistant strains, designated K. pneumoniae KP1a and E. coli EC1, were recovered 2 months later, from rectal and perineal wound swabs, respectively. For patient 2 (a 64-year-old man with type 2 diabetes who was admitted for acute cholangitis), the ertapenem-resistant K. pneumoniae strain KP2 was isolated from blood cultures, abdominal samples, and rectal swabs. Two months later, rectal swabs evidenced fecal carriage of two additional ertapenem-resistant Enterobacteriaceae: Enterobacter cloacae ECL2 and K. oxytoca KO2. MICs of β-lactams were determined by Etest (bioMérieux, Marcy l'Etoile, France) and interpreted according to CLSI guidelines (14). All strains displayed MIC values within the susceptible range for meropenem and doripenem (Table 1). A decreased susceptibility to imipenem was observed for EC1 and KO2. The six strains were resistant to penicillins but were susceptible to cefotaxime and ceftazidime, except for KP1, KP2, and ECL2. Extended-spectrum β-lactamase (ESBL) phenotypic detection was based on the use of combined double-disk synergy tests on Mueller-Hinton agar with and without cloxacillin, an AmpC inhibitor (15). The synergy test evidenced the production of an ESBL in KP1 and KP2, while ECL2 exhibited a phenotypic profile indicative of a high level of cephalosporinase expression. The modified Hodge test was positive for all strains.
Table 1.
Phenotypic and genotypic characteristics of OXA-48-producing isolates obtained from patients 1 and 2 and of transformants, transconjugants, and corresponding recipient strains used in this study
| Strain | Presence of blaOXA-48-carrying plasmid |
MIC (μg/ml) |
Modified Hodge test result | ||||
|---|---|---|---|---|---|---|---|
| 60 kb | 160 kb | Imipenem | Ertapenem | Meropenem | Doripenem | ||
| Clinical isolates | |||||||
| K. pneumoniae KPa | + | − | 0.75 | 3 | 0.38 | 0.5 | + |
| K. pneumoniae KP1a | − | + | 0.5 | 2 | 0.38 | 0.38 | + |
| E. coli EC1 | + | − | 2 | 3 | 0.38 | 0.75 | + |
| E. coli EC1a | − | − | 0.19 | 0.012 | 0.012 | 0.016 | − |
| K. pneumoniae KP2a | + | − | 0.75 | 4 | 0.75 | 0.75 | + |
| E. cloacae ECL2 | + | − | 6 | 3 | 0.38 | 0.25 | + |
| K. oxytoca KO2 | + | − | 0.75 | 1.5 | 0.75 | 0.25 | + |
| Other strains | |||||||
| E. coli DH10B | − | − | 0.19 | 0.012 | 0.016 | 0.023 | − |
| E. coli DH10B-T1 | + | − | 0.75 | 4 | 0.38 | 0.38 | + |
| E. coli DH10B-T2 | − | + | 0.75 | 2 | 0.38 | 0.25 | + |
| E. cloacae LBN600 | − | − | 0.25 | 0.25 | 0.023 | 0.032 | − |
| E. cloacae LBN600-T1 | − | + | 0.25 | 3 | 0.25 | 0.25 | + |
| K. oxytoca LBN548 | − | − | 0.19 | 0.023 | 0.016 | 0.023 | − |
| K. oxytoca LBN548-T1 | − | + | 0.38 | 4 | 0.38 | 0.38 | + |
Isolate harboring a blaCTX-M-1-group-carrying plasmid.
Molecular detection of β-lactamase-encoding genes was performed by using the Check-Points MDR CT102 array system (Check-Points, Wageningen, The Netherlands) (16). This DNA microarray hybridization suggested the presence of blaOXA-48 in all strains, as well as its association with blaCTX-M-1-group in strains KP1 and KP2. The results were confirmed by specific PCR experiments and sequencing, using primers oxa-48F (5′-AAGGAATGGCAAGAAAACAAAA-3′) and oxa-48R (5′-CCATAATCGAAAGCATGTAGCA-3′) for blaOXA-48 and primers ctx-mF (5′-ATCTGTTAAATCAGCGAGTTGAGAT-3′) and ctx-mR (5′-GTATTGCCTTTCATCCATGTCAC-3′) for blaCTX-M-1-group.
The strains were compared by pulsed-field gel electrophoresis (PFGE) with XbaI-digested genomic DNAs and a CHEF-DR III instrument (Bio-Rad, Marnes-la-Coquette, France). According to the criteria of Tenover et al. (17), KP1 and KP2 were clonally related, while KP1a displayed a different pulsotype. EC1 (from patient 1), which was resistant to ertapenem and intermediate to imipenem, exhibited the same PFGE profile as an E. coli isolate (EC1a) which was obtained from the perineal wound of patient 1 and was susceptible to all carbapenems tested.
The plasmid contents of strains were visualized after DNA linearization by the S1 nuclease followed by PFGE migration as described previously (18). S1-PFGE digests were transferred to a nylon membrane and hybridized with PCR-based probes labeled by use of a PCR DIG probe synthesis kit (Roche Diagnostics, Meylan, France). KP1 and KP2 contained a 50-kb plasmid carrying blaCTX-M-1-group. The hybridization specific to blaOXA-48 revealed the presence of a 60-kb plasmid carrying blaOXA-48 (pKPoxa-48N1) in both strains KP1 and EC1 of patient 1 and all strains of patient 2 (KP2, ECL2, and KO2) (Table 1). Although OXA-48 production has been observed mainly in K. pneumoniae, recent reports have shown that this gene may be encountered in other enterobacterial species. It has been hypothesized that this dissemination is probably related to the fact that blaOXA-48 is located on a transferable plasmid. The sequence of clinical events observed in this work and the microbiological findings strongly support the hypothesis that this interspecies spread occurred as the result of the mobilization of a 60-kb plasmid carrying blaOXA-48 in both patients.
However, Southern hybridization with strain KP1a indicated that blaOXA-48 was present, surprisingly, on a large plasmid of 160 kb (pKPoxa-48N2). This new OXA-48-encoding plasmid transferred into a cefoxitin-resistant E. cloacae strain (LBN600) and a sulfamethoxazole-trimethoprim-resistant K. oxytoca strain (LBN548) by liquid conjugation in Luria-Bertani (LB) broth at a donor/recipient ratio of 1:5 for 3 h at 37°C. The resulting transconjugants exhibited a significant increase of ertapenem MIC values (Table 1), and the presence of pKPoxa-48N2 was confirmed by S1-PFGE and blaOXA-48 hybridization. This result shows that the spread of blaOXA-48 in Enterobacteriaceae can be mediated by an additional plasmid.
The two transformants DH10B-T1 and DH10B-T2, containing pKPoxa-48N1 (60 kb) and pKPoxa-48N2 (160 kb), respectively, were obtained on LB agar plates containing 4 μg/ml ertapenem by the electroporation of KP1 and KP1a plasmid contents into E. coli DH10B by use of a MicroPulser electroporator (Bio-Rad). The complete nucleotide sequences of these plasmids were determined using a Roche Life Sciences 454 FLX genome sequencer platform (GATC Biotech, Konstanz, Germany). Sequencing generated 17,000 sequences (mean sequence size, 400 bp) for pKPoxa-48N1 and 19,000 sequences (mean sequence size, 390 bp) for pKPoxa-48N2. Sequences were assembled into one contig for pKPoxa-48N1 and into seven contigs for pKPoxa-48N2. The remaining gaps were closed by sequencing the PCR products obtained with primers designed from the extremities of each contig.
The complete sequencing of pKPoxa-48N1 showed that this circular plasmid of 62,583 bp (mean GC content of 51.1%) was an IncL/M plasmid which exhibited features very similar to those of pCTX-M360 (accession number NC_011641) and pOXA-48a from K. pneumoniae (accession number JN626286) at both the sequence and gene organization levels. The blaOXA-48 gene was the only antibiotic resistance gene present in pKPoxa-48N1. The overall sequence similarities between the common parts of this plasmid and pCTX-M360 or pOXA-48a were higher than 98% or 99%, respectively. The significant differences between pKPoxa-48N1 and pCTX-M360 were the absence of two transposable elements, i.e., a Tn2-type transposon containing blaTEM-1 and ISEcp1, associated with the blaCTX-M-3 gene (19), and the replacement of ISEcp1-blaCTX-M-3 by an IS1999-based composite transposon harboring blaOXA-48. In contrast to pOXA-48a, pKPoxa-48N1 contained an IS1R element inserted into the IS1999 element located upstream of blaOXA-48, as observed in the composite transposon Tn1999.2 (11).
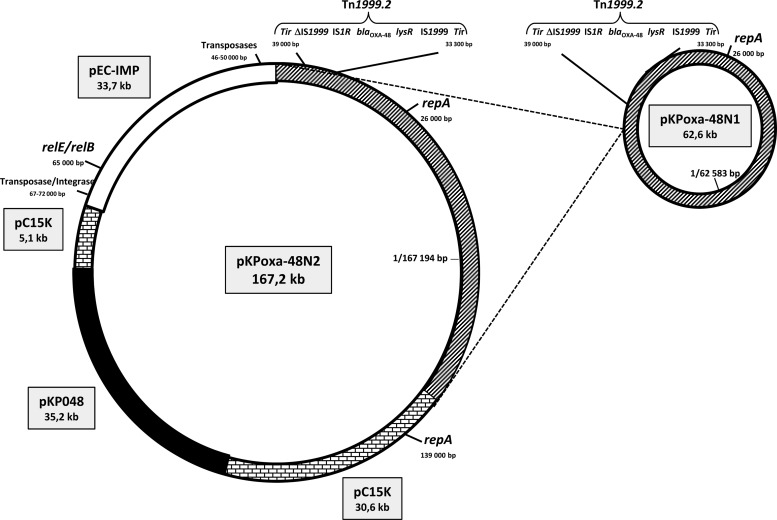
Total sequencing of pKPoxa-48N2 revealed an IncL/M plasmid of 167,194 bp with a mean GC content of 51%. The region containing blaOXA-48 (around 62.6 kb) was identical to that in pKPoxa-48N1 (Fig. 1). No other antibiotic resistance gene was found within pKPoxa-48N2. BLAST analysis showed that this plasmid consisted of three other regions, of 33.7, 35.7, and 35.2 kb, exhibiting high similarities (>98%) with respective regions of similar size in the plasmid pEC-IMP (318 kb; accession number NC_012555), found in E. cloacae, and two plasmids, pc15-k (96 kb; accession number NC_015154) and pKP048 (151 kb; accession number NC_014312), found in K. pneumoniae. The 62.2-kb region containing blaOXA-48 and the 33.7-kb pEC-IMP-like region were inserted into the 35.7-kb pc15-k-like region. pKPoxa-48N2 therefore resulted merely from multiple rearrangements involving different plasmids originating from different enterobacterial species. As for pKPoxa-48N1, blaOXA-48 was the only resistance gene observed in pKPoxa-48N2. Thus, all other resistance genes usually harbored by pEC-IMP, pc15-k, and pKP048 were lost during the events that led to the selection of pKPoxa-48N2. Interestingly, we observed that in contrast to pKPoxa-48N1, this plasmid harbored genes previously reported to encode the toxin-antitoxin systems RelE-StbE and RelB-DinJ (20). These genes, which were found to be located within the 33.7-kb pEC-IMP-like region, may therefore be responsible for a higher stability of pKPoxa-48N2 in bacteria.
Fig 1.
Schematic representation of the modular organization of pKPoxa-48N2 according to regions which are homologous to fragments from different plasmids: pKPoxa-48N1 (harboring blaOXA-48), pEC-IMP (harboring relE and relB), pC15K, and pKP048. Molecular sizes of the homologous fragments are indicated in kb.
In conclusion, we observed for the first time that a 60-kb blaOXA-48-harboring plasmid can be mobilized in different strains of Enterobacteriaceae species in vivo. We also showed that the mobilization of this gene among Enterobacteriaceae is not restricted to a single plasmid and may also rely on a 160-kb plasmid.
Nucleotide sequence accession numbers.
Complete nucleotide sequences of pKPoxa-48N1 and pKPoxa-48N2 were deposited in the GenBank database under accession numbers KC757416 and KC757417, respectively.
ACKNOWLEDGMENT
We are grateful to Isabelle Scholtus for her technical help.
Footnotes
Published ahead of print 3 June 2013
REFERENCES
- 1. Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18:263–272 [DOI] [PubMed] [Google Scholar]
- 2. Poirel L, Potron A, Nordmann PP. 2012. OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67:1597–1606 [DOI] [PubMed] [Google Scholar]
- 3. Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hammerum AM, Larsen AR, Hansen F, Justesen US, Friis-Møller AE, Lemming LE, Fuursted K, Littauer P, Schønning K, Gahrn-Hansen B, Ellermann-Eriksen S, Kristensen B. 2012. Patients transferred from Libya to Denmark carried OXA-48-producing Klebsiella pneumoniae, NDM-1-producing Acinetobacter baumannii and meticillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents 40:191–192 [DOI] [PubMed] [Google Scholar]
- 5. Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. 2011. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob. Agents Chemother. 55:2420–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Huang TD, Nordmann P. 2008. Plasmid-encoded carbapenem-hydrolyzing β-lactamase OXA-48 in an imipenem-susceptible Klebsiella pneumoniae strain from Belgium. Antimicrob. Agents Chemother. 52:3463–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dimou V, Dhanji H, Pike R, Livermore DM, Woodford N. 2012. Characterization of Enterobacteriaceae producing OXA-48-like carbapenemases in the UK. J. Antimicrob. Chemother. 67:1660–1665 [DOI] [PubMed] [Google Scholar]
- 8. Dortet L, Poirel L, Al Yaqoubi F, Nordmann P. 2012. NDM-1, OXA-48 and OXA-181 carbapenemase-producing Enterobacteriaceae in Sultanate of Oman. Clin. Microbiol. Infect. 18:EI44–EI48 [DOI] [PubMed] [Google Scholar]
- 9. Poirel L, Carbonnelle E, Bernabeu S, Gutmann L, Rotimi V, Nordmann P. 2012. Importation of OXA-48-producing Klebsiella pneumoniae from Kuwait. J. Antimicrob. Chemother. 67:2051–2052 [DOI] [PubMed] [Google Scholar]
- 10. Vaux S, Carbonne A, Thiolet JM, Jarlier V, Coignard B, RAISIN and Expert Laboratories Groups 2011. Emergence of carbapenemase-producing Enterobacteriaceae in France, 2004 to 2011. Euro Surveill. 16:19880 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19880 [DOI] [PubMed] [Google Scholar]
- 11. Carrër A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob. Agents Chemother. 54:1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carrër A, Poirel L, Eraksoy H, Cagatay AA, Badur S, Nordmann P. 2008. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 52:2950–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56:559–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing. CLSI M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 15. European Committee on Antimicrobial Susceptibility Testing 2012. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Consultation/EUCAST_guidelines_detection_of_resistance_mechanisms_121222
- 16. Naas T, Cuzon G, Bogaerts P, Glupczynski Y, Nordmann P. 2011. Evaluation of a DNA microarray (Check-MDR CT102) for rapid detection of TEM, SHV, and CTX-M extended-spectrum β-lactamases and of KPC, OXA-48, VIM, IMP, and NDM-1 carbapenemases. J. Clin. Microbiol. 49:1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 19. Zhu WH, Luo L, Wang JY, Zhuang XH, Zhong L, Liao K, Zeng Y, Lu YJ. 2009. Complete nucleotide sequence of pCTX-M360, an intermediate plasmid between pEL60 and pCTX-M3, from a multidrug-resistant Klebsiella pneumoniae strain isolated in China. Antimicrob. Agents Chemother. 53:5291–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen YT, Liao TL, Liu YM, Lauderdale TL, Yan JJ, Tsai SF. 2009. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob. Agents Chemother. 53:1235–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]