Abstract
The multiresistance gene cfr was identified for the first time in streptococci, namely, in porcine Streptococcus suis isolate S10. The cfr gene was detected on the ∼100-kb plasmid pStrcfr, where it was bracketed by two copies of the novel insertion sequence ISEnfa5, located in the same orientation. The detection of a cfr- and ISEnfa5-containing amplicon by inverse PCR suggests that ISEnfa5 may play a role in the dissemination of cfr.
TEXT
The cfr gene, encoding a 23S rRNA methyltransferase, confers resistance to five chemically unrelated antimicrobial classes, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (1), as well as decreased susceptibility to the 16-membered macrolides spiramycin and josamycin (2). Since its first identification in a Staphylococcus sciuri isolate of bovine origin (3), this multiresistance gene has been detected in isolates of the genera Staphylococcus, Bacillus, Enterococcus, Macrococcus, Jeotgalicoccus, Proteus, and Escherichia (4). Although florfenicol and lincosamides are used for the control of Streptococcus suis infections, the gene cfr has not yet been reported in streptococci. In a routine surveillance study of antimicrobial resistance of bacteria from farm animals in 2012, a hemolytic, florfenicol-resistant (MIC ≥ 16 μg/ml) streptococcal isolate, S10, was obtained from the nasal swab of an apparently healthy pig at a conventional farm in the Beijing, China, area. Gram staining, colony morphology, and ATB Rapid ID 32 Strep analysis (bioMérieux, Craponne, France) identified isolate S10 as Streptococcus suis. Moreover, 16S rRNA gene sequences, obtained using both universal prokaryotic primers (3) and S. suis-specific primers (5), showed 100% identity to the 16S rRNA gene sequence of S. suis strain NYFC (GenBank accession no. FJ660465.1).
S. suis is a global pathogen that causes a wide variety of diseases in swine, such as meningitis, pneumonia, endocarditis, septicemia, and arthritis (6). Of the 35 official serotypes described to date for S. suis, S. suis serotype 2 (SS2) is the most virulent and frequently isolated from diseased animals (7). A PCR assay (5) to detect virulence genes in SS2 isolates, including capsular polysaccharide (cps2J), muramidase-released protein (mrp), extracellular factor (ef), and suilysin (sly), yielded negative results for isolate S10. Rapid serotype-specific PCR screening to detect 15 serotypes of S. suis using previously described primers (8, 9) revealed that S. suis S10 does not belong to any of these serotypes.
S. suis S10 was investigated for the presence of the genes cfr, fexA, and fexB, all of which confer florfenicol resistance in Gram-positive bacteria, using previously described primers (3). Both cfr and fexA were detected in S. suis S10, and the nucleotide sequence of cfr in isolate S10 showed 100% identity to that of the cfr gene on plasmid pSCFS1 of S. sciuri (GenBank accession no. NC_005076). To the best of our knowledge, this is the first report of either the cfr or the fexA gene in a Streptococcus isolate. Isolate S10 exhibited high MIC values for florfenicol (64 μg/ml), clindamycin (128 μg/ml), tiamulin (64 μg/ml), tetracycline (64 μg/ml), and erythromycin (128 μg/ml) but showed low MIC values for ciprofloxacin (1 μg/ml), ampicillin (0.25 μg/ml), and ceftiofur (0.25 μg/ml). In addition, isolate S10 exhibited a linezolid MIC of 4 μg/ml, which corresponds to linezolid MIC values previously observed among cfr-carrying enterococcal and staphylococcal isolates of porcine origin (3, 10). S1 nuclease pulsed-field gel electrophoresis (PFGE) revealed at least three plasmid bands ranging from 50 to 140 kb in size in isolate S10, while Southern blot analysis (11) indicated that both cfr and fexA were located on an ∼100-kb plasmid, designated pStrcfr (Fig. 1). However, pStrcfr could not be transferred into recipient strain Enterococcus faecalis JH2-2 or Staphylococcus aureus RN4220 either by conjugation or by transformation (3), suggesting that this cfr-carrying plasmid may not be conjugative and/or able to replicate in bacterial species other than streptococci.
Fig 1.
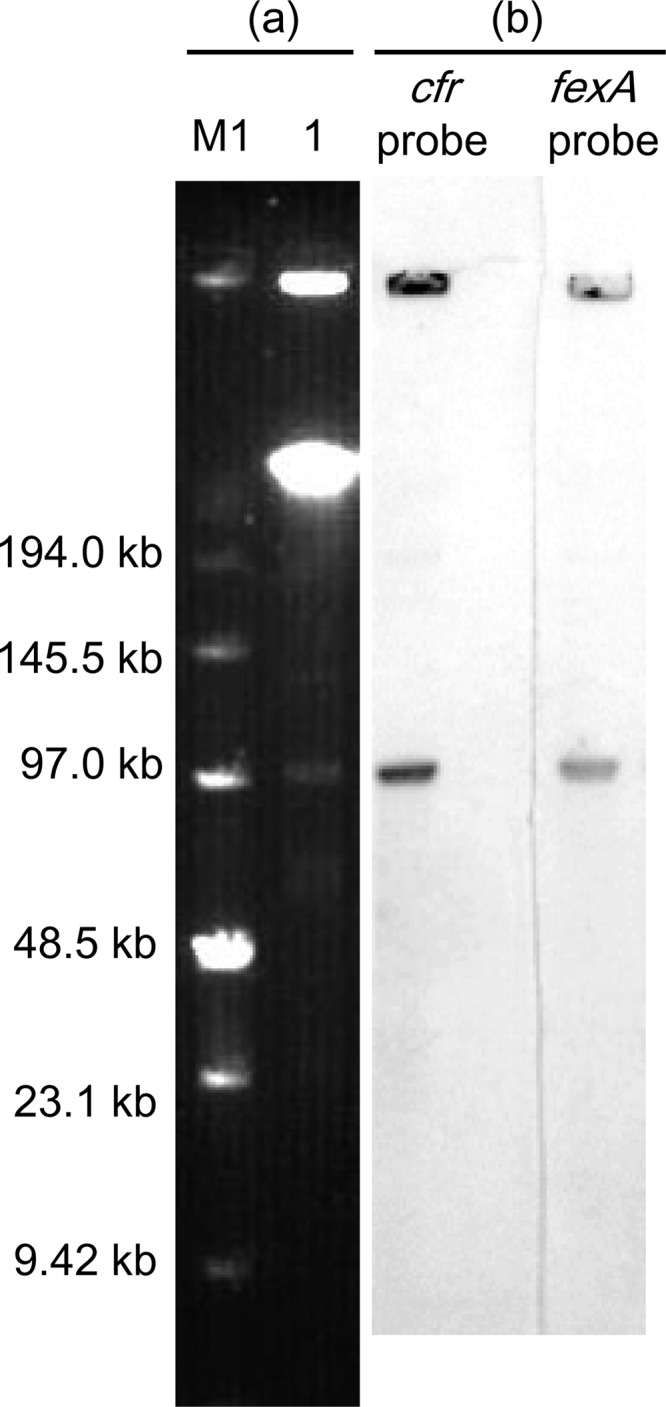
Identification of cfr and fexA genes in the plasmids of S. suis S10. Plasmids from S10 were characterized by S1 nuclease PFGE (a) or Southern blot hybridization of S10 with cfr or fexA probes (b). Lane M1, low-range pulsed-field gel marker (New England BioLabs, Beverly, MA); lane 1, S. suis S10.
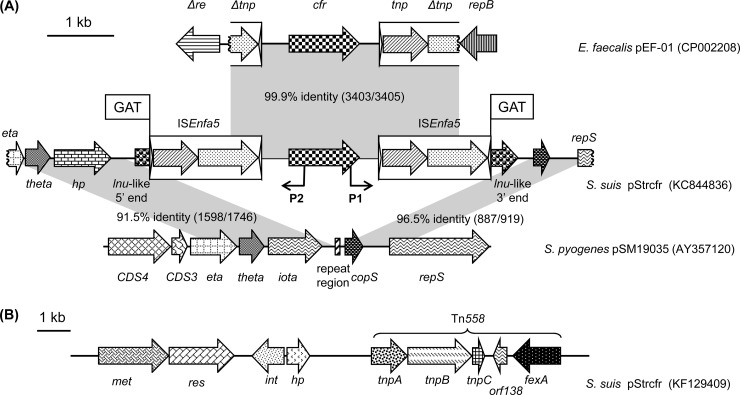
To determine the flanking regions of cfr in pStrcfr, the genetic environment of the cfr gene was sequenced by a modified random primer walking strategy (12), starting from each end of the cfr gene. A segment of 8,762 bp was determined (Fig. 2A). Within this fragment, the cfr gene was bracketed by two identical insertion sequence (IS)-like structures that were in the same orientation. Interestingly, the region containing the two IS-like elements and the cfr gene in pStrcfr showed 99.9% identity to the corresponding cfr-carrying segment of pEF-01 in E. faecalis (GenBank accession no. NC_014508) (11). This high level of similarity between the streptococcal and enterococcal cfr-carrying segments suggested the spread of the cfr gene between these two genera. The IS-like element, which was 1,686 bp in length and contained two open reading frames, was submitted to the IS database (http://www-is.biotoul.fr/is.html) and received the designation ISEnfa5. ISEnfa5 is related to ISSth1 (GenBank accession no. FR875178) from Streptococcus thermophilus JIM 8232 and belongs to the IS3 family. ISEnfa5 is slightly larger than ISSth1 (1684 bp). The first open reading frame of ISEnfa5 coded for a protein of 224 amino acids (aa), which had 42.1% (96/228) identity and 62.3% (142/228) similarity to transposase Orf1 of ISSth1. The second open reading frame overlapped the first one by 4 bp and coded for a 299-aa protein, which exhibited 48.8% (143/292) identity and 68.8% (201/292) similarity to transposase Orf2 of ISSth1. Each ISEnfa5 element had imperfect inverted repeats (IR) of 39 bp (right IR) and 40 bp (left IR) at its ends. In addition, direct target site duplications (5′-GAT-3′) were detected immediately upstream of the left ISEnfa5 and downstream of the right ISEnfa5. Analysis of the up- and downstream flanking regions by primer walking suggested that the integration of the ISEnfa5-cfr-ISEnfa5 segment occurred within an lnu-like gene. Further up- and downstream of the ISEnfa5-cfr-ISEnfa5 segment in pStrcfr, a 1,746-bp region and a 919-bp region exhibited 91.5% and 96.5% nucleotide sequence identity, respectively, to the corresponding regions of plasmid pSM19035 (GenBank accession no. AY257120) from Streptococcus pyogenes (Fig. 2A). The 852-bp hp gene in pStrcfr showed 85.7% (730/852) nucleotide sequence identity to the 807-bp iota gene from pSM19035.
Fig 2.
(A) Genetic environment of the cfr gene in plasmid pStrcfr from strain S10 and structural comparison with plasmids pEF-01 from E. faecalis EF-01 and pSM19035 from S. pyogenes; (B) genetic environment of the fexA gene in pStrcfr. The arrows indicate the positions and directions of transcription of the genes. Different genes are indicated by different shadings. Regions of >90% homology are marked by gray shading. Black arrows labeled P1 and P2 indicate the primers used for inverse PCR.
To determine the possible existence of a free circular form containing ISEnfa5-flanked sequences, an inverse PCR assay was performed using previously designed primers P1 and P2 that were located within the cfr gene (11) and plasmid DNA purified from S. suis S10 as the template. A PCR product of the expected size of 3,405 bp was obtained, and sequence analysis revealed that the amplicon contained the cfr gene sequences, the sequences between cfr and two ISEnfa5 elements, and one intact ISEnfa5 element (data not shown). This observation suggested that recombination between the two ISEnfa5 copies may occur, resulting in looping out of the cfr-carrying segment including one ISEnfa5 element. Taking into account the knowledge that ISSth1-related insertion sequences, including ISEnfa5, produce 3-bp duplications at their target site, the acquisition of the cfr-carrying segment in S. suis might have occurred in two steps. In step 1, an ISEnfa5 element integrated into the lnu-like gene of plasmid pStrcfr, thereby producing the 3-bp target site duplication at its integration site. In step 2, a DNA segment containing the cfr gene and another copy of ISEnfa5 underwent recombination with the already present ISEnfa5. This resulted in an integrated cfr gene flanked by two ISEnfa5 elements located in the same orientation in pStrcfr. However, the structure ISEnfa5-cfr-ISEnfa5 also resembles a composite transposon, and the possibility of a direct mobilization of this putative composite transposon into the coding sequence of the lnu-like gene via a classical transposition mechanism cannot be excluded.
Considering the coexistence of fexA and cfr genes on plasmid pStrcfr, a 15,519-bp segment carrying the fexA gene was obtained by primer walking (Fig. 2B). An intact Tn558 transposon including the fexA gene in this segment exhibited 99.9% (5667/5673) nucleotide sequence identity to the corresponding region of cfr-carrying plasmid pSS-01 (GenBank accession no. JQ041372) in Staphylococcus cohnii. Immediately upstream of Tn558, a gene (hp) coding for a hypothetical protein of 230 aa and an integrase gene (int) were detected. The 319-aa integrase protein showed 89.0% (284/319) identity and 91.8% (293/319) similarity to an integrase from Enterococcus faecium (GenBank accession no. ELA55919). A putative restriction-modification (RM) system was found further downstream of the int gene. The 670-aa protein encoded by the res gene exhibited 39.1% (263/673) identity and 59.3% (399/673) similarity to the Alwl restriction endonuclease from Streptococcus pneumoniae NP070 (GenBank accession no. EHD56118). The 700-aa protein encoded by the met gene revealed 99.4% (696/700) identity and 100% (700/700) similarity to a DNA methyltransferase from Enterococcus faecalis (GenBank accession no. ADD81212). As the presence of an RM gene complex in a plasmid increases the stability of plasmid maintenance in bacterial cells (13), the RM system detected in this study may play a role in the stabilization of plasmid pStrcfr in streptococci.
In conclusion, we report the first identification of a plasmid-borne cfr gene in streptococci of porcine origin and present evidence for the involvement of ISEnfa5 in the mobility of this multiresistance gene. This observation extends the current knowledge of bacterial hosts capable of acquiring cfr, as well as of the role of IS elements in its dissemination. Attention should be paid to the potential risks of transfer of the cfr gene from S. suis S10 to other porcine streptococci. Therefore, routine surveillance for the presence of cfr in streptococci of porcine origin is warranted.
Nucleotide sequence accession numbers.
The 8,762-bp segment of the cfr gene bracketed by two identical IS-like structures and the 15,519-bp segment carrying the fexA gene were submitted to GenBank and can be found under accession numbers KC844836 and KF129409, respectively.
ACKNOWLEDGMENTS
The work was funded by grants from the National Basic Research Program of China (no. 2013CB127200) and the National Natural Science Foundation of China (no. 31001087) and also by grant number 01KI1014D (MedVet-Staph) of the German Federal Ministry of Education and Research (BMBF) provided through the German Aerospace Center (DLR).
Footnotes
Published ahead of print 3 June 2013
REFERENCES
- 1. Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith LK, Mankin AS. 2008. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52:1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Y, Wang Y, Schwarz S, Li Y, Shen Z, Zhang Q, Wu C, Shen J. 2013. Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob. Agents Chemother. 57:42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shen J, Wang Y, Schwarz S. 29 March 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. [Epub ahead of print.] 10.1093/jac/dkt092 [DOI] [PubMed] [Google Scholar]
- 5. Zhu Y, Tan Z, Zhu L, Pan H, Zhang X, Xu L, Qian H, Gu L, Ye X, Dong C, Bao C, Zhu R, Zhu F, Wang H. 2008. A Streptococcus suis serotype 2 caused streptococcal toxic shock syndrome (STSS) in a patient. J. Nanjing Med. Univ. 22:313–316 [Google Scholar]
- 6. King SJ, Heath PJ, Luque I, Tarradas C, Dowson CG, Whatmore AM. 2001. Distribution and genetic diversity of suilysin in Streptococcus suis isolated from different diseases of pigs and characterization of the genetic basis of suilysin absence. Infect. Immun. 69:7572–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Higgins R, Gottschalk M. 1999. Streptococcal diseases, p 563–570 In Straw BE, D'Allaire S, Mengeling WL, Taylor DJ. (ed), Diseases of swine. Iowa State University Press, Ames, IA [Google Scholar]
- 8. Wang K, Sun X, Lu C. 2012. Development of rapid serotype-specific PCR assays for eight serotypes of Streptococcus suis. J. Clin. Microbiol. 50:3329–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kerdsin A, Dejsirilert S, Akeda Y, Sekizaki T, Hamada S, Gottschalk M, Oishi K. 2012. Fifteen Streptococcus suis serotypes identified by multiplex PCR. J. Med. Microbiol. 61:1669–1672 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Zhang W, Wang J, Wu C, Shen Z, Fu X, Yan Y, Zhang Q, Schwarz S, Shen J. 2012. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 56:1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Wang Y, Wu C, Shen Z, Schwarz S, Du XD, Dai L, Zhang W, Zhang Q, Shen J. 2012. First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob. Agents Chemother. 56:1650–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang K, McClure JA, Elsayed S, Conly JM. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naito T, Kusano K, Kobayashi I. 1995. Selfish behavior of restriction-modification systems. Science 267:897–899 [DOI] [PubMed] [Google Scholar]