Abstract
There is an urgent need to develop new agents against mycobacterial infections, such as tuberculosis and other respiratory tract or skin affections. In this study, we have tested two human antimicrobial RNases against mycobacteria. RNase 3, also called the eosinophil cationic protein, and RNase 7 are two small cationic proteins secreted by innate cells during host defense. Both proteins are induced upon infection displaying a wide range of antipathogen activities. In particular, they are released by leukocytes and epithelial cells, contributing to tissue protection. Here, the two RNases have been proven effective against Mycobacterium vaccae at a low micromolar level. High bactericidal activity correlated with their bacterial membrane depolarization and permeabilization activities. Further analysis on both protein-derived peptides identified for RNase 3 an N-terminus fragment that is even more active than the parental protein. Also, a potent bacterial agglutinating activity was unique to RNase 3 and its derived peptide. The particular biophysical properties of the RNase 3 active peptide are envisaged as a suitable reference for the development of novel antimycobacterial drugs. The results support the contribution of secreted RNases to the host immune response against mycobacteria.
INTRODUCTION
Tuberculosis is still a global threat and one of the main infectious diseases, causing about 2 million deaths per year (1). Recently, the risk has been increased by the emergence of multidrug-resistant strains in hospitals and the growing population affected by AIDS (1–3). Tuberculosis is indeed an ancient plague, and there is even fossil evidence of hominid infection. Although only 10% of infected individuals develop the disease, about one-third of the world's population is estimated to be latently infected (4, 5).
Most of the species of Mycobacterium genera are environmental and nonpathogenic, whereas some, such as M. tuberculosis, are the cause of severe pulmonary diseases (6, 7). Moreover, there are also skin infections, such as leprosy, or other cutaneous infections caused by M. haemophilum, M. chelonae, or M. kansasii among others that threaten immunocompromised patients (8). Pathogenic mycobacteria invade and dwell inside human host targets, such as macrophages, successfully replicating inside the cells (9, 10). The final outcome of the host-pathogen first encounter is dependent on the host immune response, and a variety of antimicrobial proteins and peptides (AMPs) secreted by innate cells contribute to fight the intruder. Expression of antimycobacterial peptides is induced during the host response by a variety of innate cells, from blood to epithelial cells (4, 11). In particular, eosinophil and neutrophil granules are engulfed by infected macrophages (12–15). After the secreted AMPs and potential proteolytic products could target the macrophage intracellular dwelling pathogens (9, 12). Human-derived AMPs showing high targeted cytotoxicity but low immunogenicity are therefore promising antimycobacterial therapeutic agents (16). However, research on innate immunity during mycobacterial infection is still scarce, and only a few examples of characterized AMPs are available (11, 17). In particular, upon mycobacterial infection, high levels of cathelicidin, defensin, and hepcidin have been reported in macrophages and correlated to microbe growth inhibition (11, 18, 19). The upregulation in tuberculosis patients has been observed for α-defensins in eosinophils, and β-defensin secretion is triggered at airway epithelial cells (11, 15, 20). Both active cathelicidins and defensins can be released from precursors by in vivo proteolysis at the infection site (21–23).
Mycobacteria are also characterized by their unusual lipid-rich cell wall, composed of a variety of unique glycoconjugates and intercalating complex lipids, offering a highly impermeable barrier for common antibiotics. It is noteworthy that the mycolic acid outer layer provides a wax-like architecture to the cell wall that can hinder the uptake of many antimycobacterial drugs (24). Specific features of the antimicrobial peptides and proteins (AMPs), such as low molecular weight, high cationicity, amphipathic structure, and selective affinity to the prokaryotic negatively charged cell envelope, together with their immunomodulatory effects and diverse modes of action (25), make them an interesting source of novel antimycobacterial agents (11, 26).
In our laboratory, we are working on the mechanism of action of two human RNases that are secreted by key effector innate cells, which are known to contribute to the host response to mycobacterial infection (12, 15, 27, 28), and therefore sought to test their potential antimycobacterial activities. RNase 3 and RNase 7 (Fig. 1) are two representative members of the vertebrate-secreted RNase superfamily with a well-characterized cytotoxic action against a variety of pathogens (29–33). RNase 3, also called the eosinophil cationic protein (ECP), is a small highly cationic protein secreted by eosinophil secondary granules with potent antibacterial and antiparasitic activities (34, 35). RNase 3 protein expression has also been reported in stimulated neutrophils (36). We previously studied the RNase 3 antimicrobial mechanism of action against a wide range of Gram-positive and Gram-negative strains (37, 38) and designed peptide-derived pharmacophores (39, 40). Since eosinophils and neutrophils are potent host defense effector cells activated by mycobacterial infection (31, 41, 42), and RNase 3 was found to contribute to mycobacterial growth inhibition (15), we committed ourselves to characterize the protein activity. When eosinophilia was first linked to tuberculosis (43), eosinophils were regarded as mere offenders, exacerbating pulmonary inflammation. Notwithstanding, later bibliography evidenced their protective role contributing to bacterial clearance at the infection focus (28, 44). Eosinophils, together with neutrophils, are recruited in lung granulomas (15, 45), releasing their granule content into macrophages, where they can target intracellular pathogens (4, 13). Leukocyte granule proteins are therefore suitable weapons for eradicating macrophage-resident bacteria.
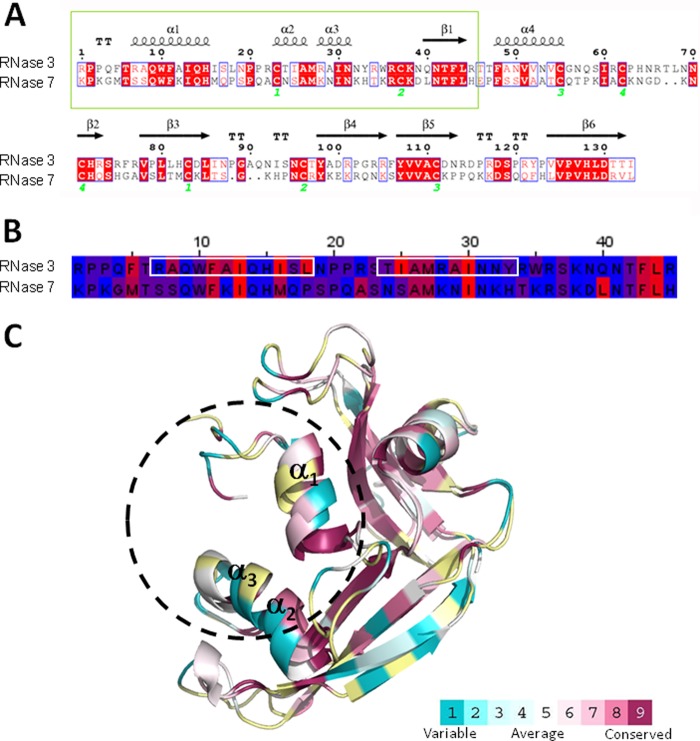
Fig 1.
(A) Comparison of the blast alignment of RNase 3 and RNase 7 primary sequences. The secondary structure of RNase 3 is depicted (4A2O PDB [76]). Strictly conserved residues are boxed in red, and conserved residues, as calculated by a similarity score, are boxed in white. The first 45 residues corresponding to RNase 3 and RNase 7 peptides are in a green box. Cysteine pairings for disulfide bridges are numbered below. The figure was created using the ESPript software (77). (B) Sequence alignment of RN3(1-45) and RN7(1-45) peptides. Residues are colored according to their hydrophobicity using the sequence alignment editor Jalview (78), and the aggregation prone regions predicted by both Aggregscan (79) and WALTZ (80) are boxed in white. (C) Graphic representation of RNase 3 and RNase 7 three-dimensional structures. The coordinates were obtained from 4A2O PDB (76) and 2HKY PDB (81), respectively. The surface representation was colored using the CONSURF web server (http://consurf.tau.ac.il/), featuring the relationships among the evolutionary conservation of amino acid positions inside the RNase A family. Residues were colored by their conservation score using the color-coding bar at the bottom image. Residues were colored in yellow when not enough information was available.
Complementarily, we have analyzed RNase 7 as an antimicrobial protein secreted by a variety of epithelial tissues (32, 33, 46–49). In particular, RNase 7 is abundantly secreted by keratinocytes and mainly contributes to the skin barrier protection (49, 50). Indeed, keratinocyte secreted proteins are mostly involved in the skin defense against infective microorganisms, such as M. leprae (51).
Finally, as a first approach to understand the underlying mechanism of action of both RNases, synthetic derived peptides have been characterized. Scarce experimental work has been applied thus far to enhance the antimycobacterial properties of natural compounds and very few examples of de novo designed peptides are currently available, such as cathelicidin or magainin analogs (19, 52). Here, we have analyzed an N terminus RNase 3-derived peptide as a suitable template toward further structure-based drug design applied therapy to mycobacterial diseases.
MATERIALS AND METHODS
Materials.
Escherichia coli BL21(DE3) cells and the pET11 expression vector were obtained from Novagen, (Madison, WI). A Live/Dead bacterial viability kit was purchased from Molecular Probes (Eugene, OR). The BacTiter-Glo assay kit was from Promega (Madison, WI). Sytox Green and DiSC3 (5) (3,3-dipropylthiacarbocyanine) were purchased from Invitrogen (Carlsbad, CA). Microplates 96-well type were purchased from Greiner (Wemmel, Belgium). The M. vaccae strain used, ATCC 15483 (CECT-3019T) (53, 54), was purchased at the Colección Española de Cultivo, Universidad de Valencia. Fmoc (9-fluorenylmethoxy carbonyl)-protected amino acids and 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) were obtained from Iris Biotech (Marktredwitz, Germany). Fmoc-Rink-amide (MBHA) resin was obtained from Novabiochem (Laüfelfingen, Switzerland). High-pressure liquid chromatography (HPLC)-grade acetonitrile (ACN) and peptide synthesis-grade N,N-dimethylformamide (DMF), N,N-diisopropylethylamine (DIEA), and trifluoroacetic acid (TFA) were obtained from Carlo Erba-SDS (Peypin, France). The cecropin A-melittin (CA-M) hybrid peptide, CA(1-8)-M(1-18) (KWKLFKKIGIGAVLKVLTTGLPALIS-NH2), was used as a control antimicrobial peptide.
Protein expression and purification.
Recombinant RNase 3 was expressed from a human synthetic gene (55). The cDNA from RNase 7 was a gift from Helene Rosenberg (National Institute of Allergy and Infectious Disease, National Institutes of Health, Bethesda, MD). Genes were cloned in pET11c. Protein expression in the E. coli BL21DE3 strain, folding of the protein from inclusion bodies, and purification were carried out as previously described (55).
Peptide synthesis and purification.
Peptides were designed based on the 1-45 N-terminus sequences of RNase 3, peptide RN3(1-45), and RNase 7, peptide RN7(1-45) (Fig. 1). Cys residues were substituted by Ser to avoid potential intra- and intermolecular disulfide bridges. A Ser residue was chosen as the best isosteric substitute for Cys. Peptides were synthesized as previously described (40). Briefly, solid-phase peptide synthesis was done by Fmoc-based chemistry on MBHA resin (0.1 mmol) in a model 433 synthesizer running FastMoc protocols. Couplings used an 8-fold molar excess each of Fmoc-amino acid and HBTU and a 16-fold molar excess of DIEA. Side chains of trifunctional residues were protected with tert-butyl (Ser, Thr, Tyr), tert-butyolxycarbonyl (Lys, Trp), 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl (Arg), and trityl (Asn, Gln, His) groups. After chain assembly, full deprotection and cleavage were carried out with TFA-water-triisopropylsilane (95:2.5:2.5 [vol/vol/vol], 90 min, at room temperature). Peptides were isolated by precipitation with cold diethyl ether, separated by centrifugation, dissolved in 0.1 M acetic acid, and lyophilized. Analytical reversed-phase HPLC was performed on a Luna C18 column. Linear gradients of solvent B (0.036% TFA in ACN) into solvent A (0.045% TFA in H2O) were used for elution at a flow rate of 1 ml/min and with UV detection at 220 nm. Preparative HPLC runs were performed on a Luna C18 column, using linear gradients of solvent (0.1% in ACN) into solvent A (0.1% TFA in H2O), as required, with a flow rate of 25 ml/min. Matrix-assisted laser desorption ionization–time of flight mass spectra were recorded in the reflector or linear mode in a Voyager DE-STR workstation using R-hydroxycinnamic acid matrix. Fractions of adequate (>90%) HPLC homogeneity and with the expected mass were pooled, lyophilized, and used in subsequent experiments. Peptide secondary structure and biophysical properties were predicted using the PSIPRED server (56).
MIC.
Antimicrobial activity was calculated as the 100% MIC (MIC100), defined as the lowest peptide concentration that completely inhibits microbial growth. The MIC of each protein and peptide [RNase 3, RNase 7, RN3(1-45), and RN7(1-45)] was determined from two independent experiments performed in triplicate for each concentration. A dilution of M. vaccae stock culture was plated onto agar petri dishes. A smooth colony was selected, and bacteria were incubated at 37°C in Corynebacterium broth (CB) medium and diluted to give approximately 5 × 105 CFU/ml. Bacterial suspension was incubated with proteins or peptides serially diluted from 50 to 0.1 μM at 37°C for 4 h in phosphate-buffered saline (PBS). Samples were plated onto petri dishes and incubated at 37°C for 48 h, and colonies were counted.
Alternatively, MIC100 of each protein and peptide was determined using the microdilution broth method according to National Committee for Clinical Laboratory Standards guidelines (57). Briefly, bacteria were incubated at 37°C in CB and diluted to give approximately 5 × 105 CFU/ml. MICs were determined in 96-well microplates. The bacterial suspensions were incubated with proteins or peptides at various concentrations (0.1 to 50 μM) at 37°C in CB. Bacterial growth was recorded by measuring the optical density at 550 nm (OD550) after incubation at 37°C for 48 h.
Bacterial viability assays.
Bacterial viability was assayed using the BacTiter-Glo microbial cell viability kit as described previously (38). Briefly, proteins or peptides were dissolved in PBS, serially diluted from 50 to 0.1 μM, and tested against M. vaccae (OD600 of ∼0.2) for 4 h of incubation time. An aliquot of 50 μl of culture was mixed with 50 μl of BacTiter-Glo reagent in a microtiter plate according to the manufacturer's instructions and incubated at 25°C for 15 min. Luminescence was read on a Victor3 plate reader (Perkin-Elmer, Waltham, MA) with a 1-s integration time. Fifty percent inhibitory concentrations (IC50) were calculated by fitting the data to a dose-response curve.
The kinetics of bacterial survival were determined using the Live/Dead bacterial viability kit in accordance with the manufacturer's instructions, as described previously (58). A Live/Dead bacterial viability kit is composed by the nucleic acid dyes Syto9, which can cross intact cell membranes, and propidium iodide (PI), which can only bind DNA and displace Syto9 when the cytoplasmic membrane is permeabilized. M. vaccae was grown at 37°C to an OD600 of 0.2, centrifuged at 5,000 × g for 5 min, and stained in a 0.85% NaCl solution containing the probes. The fluorescence intensity was continuously measured after protein or peptide addition (10 μM). To calculate bacterial viability, the signal in the range of 510 to 540 nm was integrated to obtain the Syto9 signal (live bacteria), and that in the range of 620 to 650 nm was integrated to obtain the PI signal (dead bacteria). The percentage of live bacteria was calculated at the final incubation time.
Bacterial cytoplasmic membrane depolarization assay.
Membrane depolarization was performed using the sensitive membrane potential DiSC3 (5) fluorescent probe as described previously (58). After interaction with intact cytoplasmic membrane, the fluorescent probe DiSC3 (5) was quenched. After incubation with the antimicrobial protein or peptide, the membrane potential was lost, and the probe was released to the medium ensuing in an increase of fluorescence that can be quantified and monitored as a function of time. Bacterial cultures were grown at 37°C to an OD600 of 0.2, centrifuged at 5,000 × g for 7 min, washed with 5 mM HEPES-KOH, 20 mM glucose (pH 7.2), and resuspended in 5 mM HEPES-KOH–20 mM glucose–100 mM KCl (pH 7.2) to an OD600 of 0.05. DiSC3 (5) was added to a final concentration of 0.4 μM, and changes in the fluorescence were continuously recorded after the addition of protein (10 μM) in a Victor3 plate reader. The time required to achieve maximum membrane depolarization was estimated from nonlinear regression analysis.
Bacterial cytoplasmic membrane permeation.
Bacterial cytoplasmic membrane permeation was monitored by using a Sytox Green uptake assay.
Sytox Green is a cationic cyanine dye (∼900 Da) that is not membrane permeable. When a cell's plasma membrane integrity is compromised, influx of the dye, and subsequent binding to DNA causes a large increase in fluorescence. For Sytox Green assays, M. vaccae bacterial cells were grown to mid-exponential growth phase (OD600 of 0.6) and then centrifuged, washed, and resuspended in PBS. Cell suspensions in PBS (OD600 of 0.2) were incubated with 1 μM Sytox Green for 15 min in the dark prior to the influx assay. At 2 to 4 min after initiating data collection, 10 μM concentrations of proteins or peptides were added to the cell suspension, and the increase in Sytox Green fluorescence was measured (excitation wavelength at 485 nm and emission at 520 nm) for 40 min in a Cary Eclipse spectrofluorimeter. Bacterial cell lysis with 10% Triton X-100 gives the maximum fluorescence reference value.
Minimal agglutination activity (MAC).
Bacterial cells were grown at 37°C to an OD600 of 0.2, centrifuged at 5,000 × g for 2 min, and resuspended either in PBS or CB media. An aliquot of 100 μl of the bacterial suspension was treated with increasing protein or peptide concentrations (from 0.01 to 50 μM) and incubated at 25°C for 1 h. The aggregation behavior was observed by visual inspection, and the agglutinating activity is expressed as the minimum agglutinating concentration of the sample tested, as previously described (38).
TEM.
Transmission electron microscopy (TEM) samples were prepared as previously described (59). M. vaccae was grown to an OD600 of 0.2 and incubated at 37°C with 10 μM proteins or peptides in PBS for 4 h. After treatment, bacterial pellets were prefixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer at pH 7.4 for 2 h at 4°C and postfixed in 1% osmium tetroxide buffered in 0.1 M cacodylate at pH 7.4 for 2 h at 4°C. The samples were dehydrated in acetone (50, 70, 90, 95, and 100%). The cells were immersed in Epon resin, and ultrathin sections were examined in a JEOL JEM 2011 instrument (JEOL, Ltd., Tokyo, Japan).
SEM.
Scanning electron microscopy (SEM) samples were prepared as previously described (59). Bacterial culture of M. vaccae were grown at 37°C to mid-exponential phase (OD600 of ∼0.2) and incubated with proteins or peptides (10 μM) in PBS at 37°C. Sample aliquots (500 μl) were taken after up to 4 h of incubation and prepared for SEM analysis. The micrographs were viewed at a 15-kV accelerating voltage on a Hitachi S-570 scanning electron microscope, and a secondary electron image of the cells for topography contrast was collected at several magnifications.
RESULTS AND DISCUSSION
A better understanding on the mechanism of action of AMPs effective against mycobacterial infection is a promising approach to developing alternative drugs, such as antituberculosis agents. Current treatments against tuberculosis are expensive, mostly long and cumbersome, and even occasionally ineffective (1). Unfortunately, only a few attempts have been made to apply peptide-based drugs in mycobacterial disease therapies (19). In the present study, we considered two human antimicrobial RNases, secreted by innate cells, such as eosinophils, neutrophils, and keratinocytes, which mostly contribute to fight mycobacterial infections.
Human host defense RNases against mycobacteria.
The bactericidal activity of RNase 3 and RNase 7 has been extensively documented against a wide range of Gram-negative and Gram-positive bacteria (30, 33, 38, 60). Here, both the eosinophil-secreted RNase 3 and the skin-derived RNase 7 were envisaged as good candidates to contribute to the host defense against mycobacterial infections. In order to assess their potential antimycobacterial activity, we evaluated the protein effect on bacterial viability. Mycobacterium vaccae was chosen as a rapidly growing nonvirulent and suitable working species model (61). Although the species infects cattle (62) and is generally considered nonpathogenic to humans, a few cases of cutaneous and pulmonary infection in farm workers have been reported (63).
Interestingly, experimental data indicated that RNase 3 and RNase 7 were indeed able to totally inhibit mycobacterial growth in a low micromolar range, showing MIC100s from 10 to 20 μM (Table 1). The same results were reproduced when these RNases tested in both PBS and CB broth, either plated in petri dishes or incubated in microtiter plates (results not shown). Thereafter, the microbial cell viability was assayed using the BacTiter-Glo luminiscent approach. Mycobacterial cells metabolically active, and thus viable, were measured by ATP quantification using a coupled luciferin/oxyluciferin in the presence of luciferase, where luminescence is proportional to ATP and hence to the number of viable cells in the culture. A comparison of IC50s for RNase 3 and RNase 7 yielded comparable results (Table 1). Therefore, the assays confirmed the antimycobacterial activity of the two tested human RNases; both are able to totally inhibit bacterial viability in a low micromolar range as indicated by the MIC and IC50 values. This is the first characterization of the antimycobacterial activity of human secreted RNases. The results reinforce the findings of previous preliminary studies on the role of eosinophils during mycobacterial infection and in particular on the contribution of eosinophil secretion proteins (15).
Table 1.
Antimicrobial and agglutinating activities of RNase 3, RNase 7, and their corresponding N-terminal derived peptides on M. vaccae
| Protein or peptide | Mean concn (μM) ± SEMa |
||
|---|---|---|---|
| MIC100 | IC50 | MAC | |
| RNase 3 | 20.0 ± 1.0 | 11.6 ± 0.2 | 1.0 ± 0.1 |
| RNase 7 | 20.0 ± 0.5 | 9.3 ± 1.2 | >50 |
| RN3(1-45) | 10.0 ± 0.5 | 4.2 ± 0.2 | 1.0 ± 0.1 |
| RN7(1-45) | 20.0 ± 0.8 | 9.5 ± 0.3 | >50 |
| CA-Mb | 20.0 ± 1.0 | 10.3 ± 0.3 | >50 |
The 100% MIC (MIC100), the 50% bacterial viability (IC50), and the minimal agglutinating activity (MAC) were calculated as described in Materials and Methods. MIC100 values were calculated by CFU counting on plated Petri dishes. All values are averaged from three replicates of two independent experiments.
The cecropin A-melittin hybrid peptide (CA-M) was used as a control.
Active N-terminal derived peptides.
We next sought to identify the proteins' functional domains. Both RNases, sharing a low sequence identity (∼40%), adopt the same three-dimensional overall fold, wherein nonconserved residues are mostly surface exposed (Fig. 1C). Previous works have outlined that the main determinants for the human RNases antimicrobial action are clustered at the N terminus region, and derived peptides were designed as potential lead pharmacophores (39, 40, 64, 65). Accordingly, synthetic peptides corresponding to the first 45 residues of both RNases, encompassing the first α1-α3 helices (Fig. 1), were tested against M. vaccae. Experimental data indicated that the peptides RN3(1-45) and RN7(1-45) retained most of the full protein antimicrobial properties. Interestingly, the RNase 3-derived peptide was even more effective than the parental protein, showing a very promising behavior.
Although the RN7(1-45) peptide emulated the MIC value of the whole protein, RN3(1-45) achieved MIC values at half-peptide concentrations, leading to total mycobacterial lethality at 10 μM. A cell viability assay corroborated that RN7(1-45) displayed the same effectiveness as the parental RNase 7 and that RN3(1-45) produced 50% of the mortality at a lower concentration (<5 μM). The peptide was even more active than the tested CA-M control peptide, a potent antimicrobial peptide with pore-forming ability that is effective against a wide range of bacterial strains (66, 67).
The RN3(1-45) peptide was previously shown to display a high antimicrobial activity on a wide range of Gram-negative and Gram-positive strains (40). To better interpret the particularly high bactericidal propensity of the RN3(1-45) peptide, its biophysical properties were analyzed in relation to its counterparts (Fig. 1). The peptide was observed to be mostly unstructured on aqueous solution and adopt a defined α-helix secondary structure on a lipid environment, as deduced from previous circular dichroism (CD) analysis (40) and nuclear magnetic resonance (NMR) studies (68). NMR spectroscopy identified a first α-helix matching protein α1 and a second α-helix covering the protein α2-α3 region (Fig. 1A) and expanding to the C terminus (68). Prediction of RNase 7 peptide secondary structure also suggested equivalent matching helical structures. Moreover, the CD spectrum of the RN7(1-45) peptide corroborated that its structuration is promoted by a lipid environment. A high affinity of both peptides for anionic phospholipids and a lipid bilayer disruption activity was registered when working on lipid vesicles as model membranes (40). Side-by-side comparison of the mechanism of action of both peptides on liposomes also supported a distinct behavior. In particular, a high lipid vesicle agglutination activity for the RN3(1-45) peptide, not shared by the RN7(1-45) peptide, was observed (38; M. Torrent, D. Pulido, J. Valle, M. V. Nogués, D. Andreu, and E. Boix, unpublished data). A hydrophobic patch, identified as an aggregation prone region, unique to the RNase 3 N terminus (Fig. 1B), could also facilitate its action at the lipid-rich mycobacterial wall level. Comparison of the two peptides physicochemical properties highlights the RNase 3 peptide amphipathic and cationic character, showing a higher pI (pI 12.61 versus pI 10.94) and positive net charge (+8 versus +7). The RN3(1-45) peptide amphipathic character is mostly enhanced by a pronounced alternating profile of cationic and hydrophobic residues (Fig. 1B). Moreover, when scanning both peptide sequences using the AMPA antimicrobial server (69), a wider propensity stretch was identified in RNase 3. A closer look at the respective amino acid composition reveals the presence of an unfavored anionic residue at the RNase 7 N terminus (Asp39), which would disrupt the antimicrobial region. Moreover, the RNase 3 N terminus includes a hyperexposed Trp residue (Trp35), which was proven to directly contribute to the protein membrane destabilization (70). On the other hand, we cannot disregard that the higher efficiency of the RNase 3 N terminus peptide in relation to the parental protein is indicative of a physiological role wherein the eosinophil granule protein once engulfed by macrophages can undergo proteolysis (9).
Bacterial viability assays.
The promising preliminary results encouraged us to further investigate the protein and peptide mechanisms of action at the bacterial cell level. Based on our previous characterization work on the action of RNase peptides on Gram-negative and Gram-positive bacteria (40, 64), we analyzed here the peptide cytotoxic mechanism on mycobacterial cells. We first compared the ability of proteins and N-terminus peptides to depolarize the mycobacterial cell membrane. Maximum depolarization values working at the IC50 were calculated. Comparative analysis showed a poor depolarization effect for both RNases (Table 2). The corresponding RNase 7 peptide, RN7(1-45), also depolarized as poorly as its parental protein, with only a 6.5% of the maximum reference value, suggesting a nontraditional pore-forming mechanism of action. The RN3(1-45) peptide was able to depolarize at values >60% (Table 2), significantly increasing its permeabilizing ability on mycobacterial cells compared to the parental protein. The RN3(1-45) effectiveness was even higher than the antimicrobial control peptide CA-M, with a high membrane depolarization activity against a wide range of Gram-positive and Gram-negative (67, 71). We suggest that the particular biophysical properties of the RNase 3 peptide can better overcome the complexity of the mycobacterial wall barrier, reaching more easily the cytoplasmic membrane.
Table 2.
Percent viability, membrane permeabilization, and membrane depolarization activities of RNase 3, RNase 7, and their N-terminal derived peptides on M. vaccae
| Protein or peptide | Mean % ± SEMa |
||
|---|---|---|---|
| Bacterial viability | Membrane permeabilization | Membrane depolarization | |
| RNase 3 | 55.1 ± 0.6 | 35.9 ± 0.1 | 8.8 ± 0.1 |
| RNase 7 | 48.7 ± 1.8 | 28.8 ± 0.1 | 4.1 ± 0.2 |
| RN3(1-45) | 6.8 ± 0.8 | 50.0 ± 1.2 | 63.2 ± 1.8 |
| RN7(1-45) | 55.1 ± 1.5 | 19.3 ± 0.1 | 6.5 ± 0.2 |
| CA-Mb | 44.8 ± 1.6 | 27.3 ± 0.1 | 44.7 ± 1.3 |
Bacterial viability was determined using a Live/Dead kit. Membrane permeabilization was determined by using the Sytox Green assay, and membrane depolarization activity was determined using the DiSC3(5) probe, as described in Materials and Methods. All values are averaged from three replicates of two independent experiments. For membrane permeabilization and membrane depolarization, the calculated percentages refer to the maximum values corresponding to the positive control (10% Triton X-100).
The cecropin A-melittin hybrid peptide (CA-M) was used as a control.
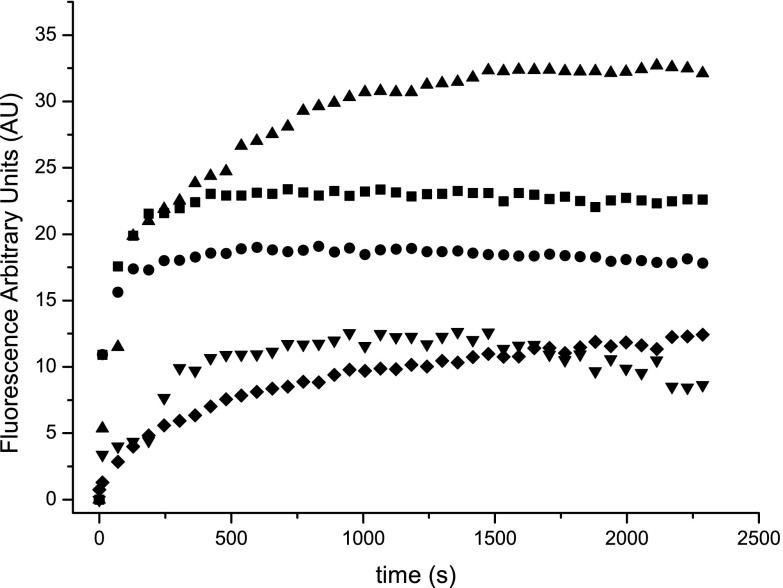
Further insight into the membrane-permeabilizing effect of both proteins and their derived peptides was performed using the Sytox Green assay. Sytox Green uptake/fluorescence was monitored as a function of time after adding 10 μM concentrations of protein and peptides (Fig. 2). The total permeabilizing effect was calculated after 40 min of incubation (Table 2) showing that the RN3(1-45) peptide presented the best permeabilizing effect with a 50% value, whereas both RNases achieved a lower permeabilizing value (ca. 30%) and the RNase 7 peptide only permeabilized the 20% of the total cell population. On the other hand, the membrane permeabilization time course profile indicated a similar timing for all of the tested samples, confirming that the protein interaction with cell membrane and subsequent permeabilizing effect is a rapid event, taking <5 min to produce half of the maximum membrane depolarization value.
Fig 2.
Membrane permeabilization was determined by SYTOX Green uptake after incubation of M. vaccae culture cells with 10 μM proteins and peptides. Results for RNase 3 (■), RNase 7(●), RN3(1-45) (▲), RN7(1-45) (▼), or CA-M (◆) are depicted as a function of time. A maximum fluorescence reference value of 64 ± 0.4 was achieved for 10% Triton X-100.
Then, in order to analyze the kinetics of the tested peptides on the mycobacterial population, we used a Live/Dead bacterial viability kit. The live bacterial population was estimated from the Syto 9 fluorescence dye, which can cross intact cell membranes, whereas dead bacteria, with damaged membranes, were stained with the PI fluorescent marker. By the integration of Syto 9 and PI fluorescence, we determined the viability percentage as a function of the incubation time upon addition of 10 μM proteins and peptides, monitoring the bacterial killing process. The viability percentage was calculated at the final incubation time. Similar reduction percentages of the mycobacterial population viability were registered for both RNases, the RN7(1-45) and CA-M peptides (Table 2). Again, the RN3(1-45) peptide was more effective and was able to almost abolish the mycobacterial population within the registered time, with only 6% final survival; the results are consistent with the aforementioned MIC and IC50 values (Table 1).
Bacterial agglutination assays.
Another key antimicrobial property thoroughly studied in our laboratory is the capacity of human RNase 3 to induce bacterial cell agglutination (37, 58, 59). The RNase 3 agglutinating activity, not shared with RNase 7, is specific toward Gram-negative bacteria and is dependent on the protein primary structure (37, 38). A sequence stretch responsible for the protein self-aggregation was spotted at the RNase 3 N terminus (72), and the N-terminal derived peptides partially retained the bacterial agglutinating ability (38, 40). In order to assess the agglutinating activity on M. vaccae cultures, we determined the minimal agglutination concentration (MAC), defined as the minimal concentration able to induce agglutination. Only RNase 3 and the corresponding RN3(1-45) peptide were able to induce mycobacterial cells to agglutinate at a 1 μM concentration in both PBS and CB broth media (Table 1). No agglutination was observed for RNase 7 and its derived peptide RN7(1-45) or for the CA-M reference peptide, even at the maximum concentration range tested. Complementary work on the peptides behavior on model membranes also revealed a specific vesicle agglutinating ability for the RNase 3 peptide that is not shared by the RNase 7 counterpart (Torrent et al., unpublished). Comparison of the peptide hydrophobicity and aggregation-prone profiles within the RNase A family context corroborated that the active segment is unique to the RNase 3 N terminus, explaining its cell agglutination properties (Fig. 1B). Moreover, the enhanced membrane destabilization activity of the RN3(1-45) peptide (Table 2) may partly rely on its aggregation propensity, where a local peptide self-aggregation at the mycobacterial surface could promote the membrane damage. We can also hypothesize that the induction of bacterial cell agglutination by eosinophil granule protein self-aggregation may trigger in vivo the autophagy path, contributing to mycobacterial clearance inside macrophages (73, 74).
Ultrastructural analysis of damage at the mycobacterial cell envelope.
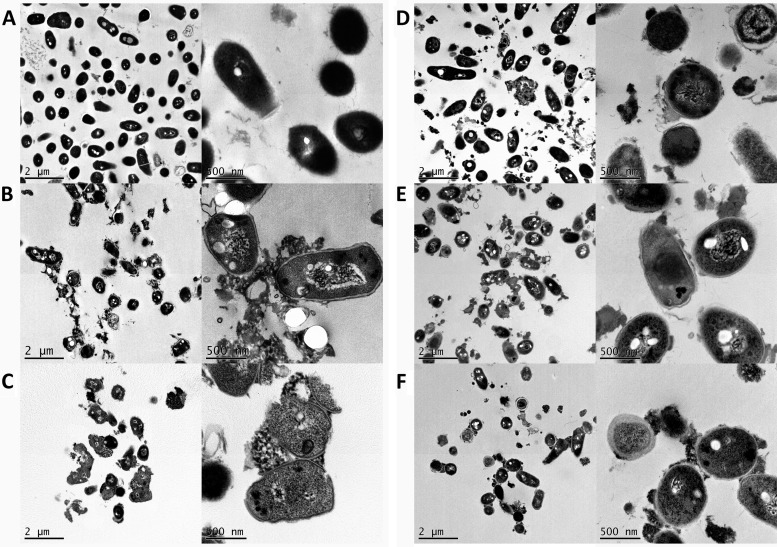
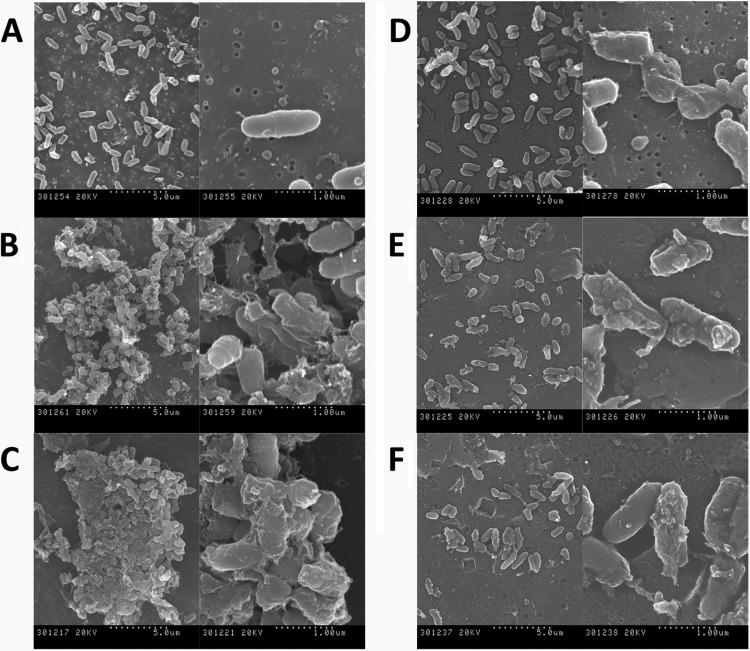
Finally, to better characterize the protein and peptide action at the mycobacterial cell envelope, electron microscopy techniques were applied. Treated cells were visualized by TEM and SEM. M. vaccae cells were examined by TEM after 4 h of incubation with 10 μM concentrations of both RNases and the RN3(1-45), RN7(1-45), and CA-M peptides (Fig. 3). All proteins and peptides under the assayed conditions produced a complete disruption of the cell integrity, bacterial swelling, intracellular material spillage, bacterial cell wall layer detachment, and alteration of cell morphology. Finally, we applied SEM to visualize the cell surface and the cell population behavior (Fig. 4). The methodology also proved useful for assessing the agglutination activity by evaluating simultaneously the size and density of the bacterial aggregates. Upon RNase 3 incubation, large, dense bacterial aggregates were observed, where cells were badly damaged, showing frequent blebs and partial loss of their baton-shape morphology. Cultures treated with the RN3(1-45) peptide also presented tight and dense aggregates, with visible loss of membrane integrity and cell morphology. On their side, RNase 7 and its derived peptide, RN7(1-45), displayed similar damage on mycobacterial cultures but without visible agglutination. Likewise, the CA-M antimicrobial peptide showed no mycobacterial agglutination, but severe cell damage, with blebbing and partial loss of cell content.
Fig 3.
Transmission electron micrographs of M. vaccae incubated for 4 h in the presence of 10 μM concentrations of proteins and peptides. (A) Control cells; (B) RNase 3; (C) RN3(1-45); (D) CA-M; (E) RNase 7; (F) RN7(1-45). The magnification scale is indicated at the bottom of each micrograph.
Fig 4.
Scanning electron micrographs of M. vaccae incubated for 4 h in the presence of 10 μM concentrations of proteins and peptides. (A) Control cells; (B) RNase 3; (C) RN3(1-45); (D) CA-M; (E) RNase 7; (F) RN7(1-45). The magnification scale is indicated at the bottom of each micrograph.
The high antimycobacterial and cell agglutinating activity of the RN3(1-45) peptide opens a new research field to explore its particular mechanism of action at the mycobacterial wall at the molecular level. In addition, considering our previous observation of amyloid-like aggregates at the bacterial surface (37) and the location of an amyloid-prone region at its N terminus (72), we are also planning to inspect in a mycobacterial infection model whether the eosinophil granule protein can undergo in vivo an ordered self-assembly process, as recently nicely reported for another human antimicrobial peptide contributing to innate immunity (75).
Conclusions.
Little is known about the mechanism of action of antimicrobial peptides against Mycobacterium species. In the present study we assessed the antimycobacterial activity of two human RNases that are secreted by innate cells during respiratory tract and skin infections. Eosinophil RNase 3 and skin RNase 7, together with their synthetic N terminus peptides, were assayed against M. vaccae to characterize their underlying mechanisms of action. The results represent the first characterization of the cytotoxicity of two RNase A family members toward mycobacteria. In particular, the RN3(1-45) peptide, showing both a high antimicrobial activity and agglutinating properties, offers new perspectives to develop antimycobacterial agents. Further work on other mycobacterial species with a more clinical approach is envisaged. We hypothesize that both innate cell secretion proteins may target in vivo the mycobacteria dwelling inside macrophages or other host cell types.
ACKNOWLEDGMENTS
TEM and SEM were performed at the Servei de Microscopia of the Universitat Autònoma de Barcelona (UAB). Spectrofluorescence assays were performed at the Laboratori d'Anàlisi i Fotodocumentació, UAB.
This study was supported by the Ministerio de Educación y Cultura (grant BFU2009-09371) and Ministerio de Economía y Competitividad (BFU2012-38965) and cofinanced by FEDER funds and by the Generalitat de Catalunya (2009 SGR 795). D.P. is a recipient of an FPU predoctoral fellowship (Ministerio de Educación y Cultura).
Footnotes
Published ahead of print 28 May 2013
REFERENCES
- 1. Dye C, Glaziou P, Floyd K, Raviglione M. 2012. Prospects for tuberculosis elimination. Annu. Rev. Public Health 14:3–16 [DOI] [PubMed] [Google Scholar]
- 2. Martinson NA, Hoffmann CJ, Chaisson RE. 2011. Epidemiology of tuberculosis and HIV: recent advances in understanding and responses. Proc. Am. Thorac. Soc. 8:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chiang CY, Yew WW. 2009. Multidrug-resistant and extensively drug-resistant tuberculosis. Int. J. Tuberc. Lung Dis. 13:304–311 [PubMed] [Google Scholar]
- 4. Saiga H, Shimada Y, Takeda K. 2011. Innate immune effectors in mycobacterial infection. Clin. Dev. Immunol. 2011:347594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Janssen S, Jayachandran R, Khathi L, Zinsstag J, Grobusch MP, Pieters J. 2012. Exploring prospects of novel drugs for tuberculosis. Drug Design Dev. Ther. 6:217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Groote MA, Huitt G. 2006. Infections due to rapidly growing mycobacteria. Clin. Infect. Dis. 42:1756–1763 [DOI] [PubMed] [Google Scholar]
- 7. Brown-Elliott BA, Nash KA, Wallace RJ. 2012. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin. Microbiol. Rev. 25:545–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elston D. 2009. Nontuberculous mycobacterial skin infections recognition and management. Am. J. Clin. Dermatol. 10:281–285 [DOI] [PubMed] [Google Scholar]
- 9. Liu PT, Modlin RL. 2008. Human macrophage host defense against Mycobacterium tuberculosis. Curr. Opin. Immunol. 20:371–376 [DOI] [PubMed] [Google Scholar]
- 10. Pinheiro RO, Salles JD, Sarno EN, Sampaio EP. 2011. Mycobacterium leprae-host cell interactions and genetic determinants in leprosy: an overview. Future Microbiol. 6:217–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shin D-M, Jo E-K. 2011. Antimicrobial peptides in innate immunity against mycobacteria. Immune Netw. 11:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tan BH, Meinken C, Bastian M, Bruns H, Legaspi A, Ochoa MT, Krutzik SR, Bloom BR, Ganz T, Modlin RL, Stenger S. 2006. Macrophages acquire neutrophil granules for antimicrobial activity against intracellular pathogens. J. Immunol. 177:1864–1871 [DOI] [PubMed] [Google Scholar]
- 13. Soehnlein O. 2009. Direct and alternative antimicrobial mechanisms of neutrophil-derived granule proteins. J. Mol. Med. 87:1157–1164 [DOI] [PubMed] [Google Scholar]
- 14. Lasco TM, Turner OC, Cassone L, Sugawara I, Yamada H, McMurray DN, Orme IM. 2004. Rapid accumulation of eosinophils in lung lesions in guinea pigs infected with Mycobacterium tuberculosis. Infect. Immun. 72:1147–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Driss V, Legrand F, Hermann E, Loiseau S, Guerardel Y, Kremer L, Adam E, Woerly G, Dombrowicz D, Capron M. 2009. TLR2-dependent eosinophil interactions with mycobacteria: role of alpha-defensins. Blood 113:3235–3244 [DOI] [PubMed] [Google Scholar]
- 16. Mendez-Samperio P. 2008. Role of antimicrobial peptides in host defense against mycobacterial infections. Peptides 29:1836–1841 [DOI] [PubMed] [Google Scholar]
- 17. Mendez-Samperio P. 2010. The human cathelicidin hCAP18/LL-37: a multifunctional peptide involved in mycobacterial infections. Peptides 31:1791–1798 [DOI] [PubMed] [Google Scholar]
- 18. Sow FB, Florence WC, Satoskar AR, Schlesinger LS, Zwilling BS, Lafuse WP. 2007. Expression and localization of hepcidin in macrophages: a role in host defense against tuberculosis. J. Leukoc. Biol. 82:934–945 [DOI] [PubMed] [Google Scholar]
- 19. Sonawane A, Santos JC, Mishra BB, Jena P, Progida C, Sorensen OE, Gallo R, Appelberg R, Griffiths G. 2011. Cathelicidin is involved in the intracellular killing of mycobacteria in macrophages. Cell. Microbiol. 13:1601–1617 [DOI] [PubMed] [Google Scholar]
- 20. Mendez-Samperio P, Miranda E, Trejo A. 2006. Mycobacterium bovis bacillus Calmette-Guerin (BCG) stimulates human beta-defensin-2 gene transcription in human epithelial cells. Cell. Immunol. 239:61–66 [DOI] [PubMed] [Google Scholar]
- 21. Lehrer RI, Ganz T. 2002. Cathelicidins: a family of endogenous antimicrobial peptides. Curr. Opin. Hematol. 9:18–22 [DOI] [PubMed] [Google Scholar]
- 22. Zanetti M. 2004. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 75:39–48 [DOI] [PubMed] [Google Scholar]
- 23. Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. 1999. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286:113–117 [DOI] [PubMed] [Google Scholar]
- 24. Mishra AK, Driessen NN, Appelmelk BJ, Besra GS. 2011. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol. Rev. 35:1126–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fjell CD, Hiss JA, Hancock REW, Schneider G. 2012. Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 11:37–51 [DOI] [PubMed] [Google Scholar]
- 26. Kapoor R, Eimerman PR, Hardy JW, Cirillo JD, Contag CH, Barron AE. 2011. Efficacy of antimicrobial peptoids against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55:3058–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martineau AR, Newton SM, Wilkinson KA, Kampmann B, Hall BM, Nawroly N, Packe GE, Davidson RN, Griffiths CJ, Wilkinson RJ. 2007. Neutrophil-mediated innate immune resistance to mycobacteria. J. Clinical Invest. 117:1988–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akuthota P, Xenakis JJ, Weller PF. 2011. Eosinophils: offenders or general bystanders in allergic airway disease and pulmonary immunity? J. Innate Immun. 3:113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boix E, Torrent M, Sánchez D, Nogués MV. 2008. The antipathogen activities of eosinophil cationic protein. Current Pharm. Biotech. 9:141–152 [DOI] [PubMed] [Google Scholar]
- 30. Boix E, Salazar VA, Torrent M, Pulido D, Nogués MV, Moussaoui M. 2012. Structural determinants of the eosinophil cationic protein antimicrobial activity. Biol. Chem. 393:801–815 [DOI] [PubMed] [Google Scholar]
- 31. Malik A, Batra JK. 2012. Antimicrobial activity of human eosinophil granule proteins: involvement in host defence against pathogens. Crit. Rev. Microbiol. 38:168–181 [DOI] [PubMed] [Google Scholar]
- 32. Spencer JD, Schwaderer AL, DiRosario JD, McHugh KM, McGillivary G, Justice SS, Carpenter AR, Baker PB, Harder J, Hains DS. 2011. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. 80:175–181 [DOI] [PubMed] [Google Scholar]
- 33. Koten B, Simanski M, Glaser R, Podschun R, Schroder JM, Harder J. 2009. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS One 4:e6424. 10.1371/journal.pone.0006424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Venge P, Bystrom J, Carlson M, Hakansson L, Karawacjzyk M, Peterson C, Seveus L, Trulson A. 1999. Eosinophil cationic protein (ECP): molecular and biological properties and the use of ECP as a marker of eosinophil activation in disease. Clin. Exp. Allergy 29:1172–1186 [DOI] [PubMed] [Google Scholar]
- 35. Bystrom J, Amin K, Bishop-Bailey D. 2011. Analysing the eosinophil cationic protein: a clue to the function of the eosinophil granulocyte. Respir. Res. 12:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monteseirin J, Vega A, Chacon P, Camacho MJ, El Bekay R, Asturias JA, Martinez A, Guardia P, Perez-Cano R, Conde J. 2007. Neutrophils as a novel source of eosinophil cationic protein in IgE-mediated processes. J. Immunol. 179:2634–2641 [DOI] [PubMed] [Google Scholar]
- 37. Torrent M, Pulido D, Nogués MV, Boix E. 2012. Exploring new biological functions of amyloids: bacteria cell agglutination mediated by host protein aggregation. PLoS Pathog. 8:e1003005. 10.1371/journal.ppat.1003005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pulido D, Moussaoui M, Andreu D, Nogues MV, Torrent M, Boix E. 2012. Antimicrobial action and cell agglutination by the eosinophil cationic protein are modulated by the cell wall lipopolysaccharide structure. Antimicrob. Agents Chemother. 56:2378–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Torrent M, de la Torre BG, Nogues VM, Andreu D, Boix E. 2009. Bactericidal and membrane disruption activities of the eosinophil cationic protein are largely retained in an N-terminal fragment. Biochem. J. 421:425–434 [DOI] [PubMed] [Google Scholar]
- 40. Torrent M, Pulido D, De la Torre BG, Garcia-Mayoral MF, Nogues MV, Bruix M, Andreu D, Boix E. 2011. Refining the eosinophil cationic protein antibacterial pharmacophore by rational structure minimization. J. Med. Chem. 54:7. [DOI] [PubMed] [Google Scholar]
- 41. Borelli V, Vita F, Shankar S, Soranzo MR, Banfi E, Scialino G, Brochetta C, Zabucchi G. 2003. Human eosinophil peroxidase induces surface alteration, killing, and lysis of Mycobacterium tuberculosis. Infect. Immun. 71:605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Driss V, Hermann E, Legrand F, Loiseau S, Delbeke M, Kremer L, Guerardel Y, Dombrowicz D, Capron M. 2012. CR3-dependent negative regulation of human eosinophils by Mycobacterium bovis BCG lipoarabinomannan. Immunol. Lett. 143:202–207 [DOI] [PubMed] [Google Scholar]
- 43. Vijayan VK, Reetha AM, Jawahar MS, Sankaran K, Prabhakar R. 1992. Pulmonary eosinophilia in pulmonary tuberculosis. Chest 101:1708–1709 [DOI] [PubMed] [Google Scholar]
- 44. Kita H. 2011. Eosinophils: multifaceted biological properties and roles in health and disease. Immunol. Rev. 242:161–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Castro AG, Esaguy N, Macedo PM, Aguas AP, Silva MT. 1991. Live but not heat-killed mycobacteria cause rapid chemotaxis of large numbers of eosinophils in vivo and are ingested by the attracted granulocytes. Infect. Immun. 59:3009–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harder J, Schroder JM. 2002. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 277:46779–46784 [DOI] [PubMed] [Google Scholar]
- 47. Zhang J, Dyer KD, Rosenberg HF. 2003. Human RNase 7: a new cationic ribonuclease of the RNase A superfamily. Nucleic Acids Res. 31:602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zanger P, Holzer J, Schleucher R, Steffen H, Schittek B, Gabrysch S. 2009. Constitutive expression of the antimicrobial peptide RNase 7 is associated with Staphylococcus aureus infection of the skin. J. Infect. Dis. 200:1907–1915 [DOI] [PubMed] [Google Scholar]
- 49. Simanski M, Köten B, Schröder JM, Gläser R, Harder J. 2012. Antimicrobial RNases in cutaneous defense. J. Innate Immun. 4:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bernard JJ, Gallo RL. 2011. Protecting the boundary: the sentinel role of host defense peptides in the skin. Cell. Mol. Life Sci. 68:2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Casanova JL, Abel L. 2002. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 20:581–620 [DOI] [PubMed] [Google Scholar]
- 52. Jiang ZQ, Higgins MP, Whitehurst J, Kisich KO, Voskuil MI, Hodges RS. 2011. Anti-tuberculosis activity of alpha-helical antimicrobial peptides: de novo designed L- and D-enantiomers versus L-and D-LL37. Protein Peptide Lett. 18:241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boenickse R, Juhasz E. 1964. Description of the new species Mycobacterium vaccae n. sp. Zentralbl. Bakteriol. Orig. 192:133–135 [PubMed] [Google Scholar]
- 54. Skerman VBD, McGowan V, Sneath PHA. 1980. Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30:225–420 [PubMed] [Google Scholar]
- 55. Boix E, Nikolovski Z, Moiseyev GP, Rosenberg HF, Cuchillo CM, Nogues MV. 1999. Kinetic and product distribution analysis of human eosinophil cationic protein indicates a subsite arrangement that favors exonuclease-type activity. J. Biol. Chem. 274:15605–15614 [DOI] [PubMed] [Google Scholar]
- 56. Buchan DWA, Ward SM, Lobley AE, Nugent TCO, Bryson K, Jones DT. 2010. Protein annotation and modeling servers at University College London. Nucleic Acids Res. 38:W563–W568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. National Committee for Clinical Laboratory Standards 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 58. Torrent M, Badia M, Moussaoui M, Sanchez D, Nogues MV, Boix E. 2010. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 277:1713–1725 [DOI] [PubMed] [Google Scholar]
- 59. Torrent M, Navarro S, Moussaoui M, Nogues MV, Boix E. 2008. Eosinophil cationic protein high-affinity binding to bacteria-wall lipopolysaccharides and peptidoglycans. Biochemistry 47:3544–3555 [DOI] [PubMed] [Google Scholar]
- 60. Singh A, Batra JK. 2011. Role of unique basic residues in cytotoxic, antibacterial and antiparasitic activities of human eosinophil cationic protein. Biol. Chem. 392:337–346 [DOI] [PubMed] [Google Scholar]
- 61. Ho YS, Adroub SA, Abadi M, Al Alwan B, Alkhateeb R, Gao G, Ragab A, Ali S, van Soolingen D, Bitter W, Pain A, Abdallah AM. 2012. Complete genome sequence of Mycobacterium vaccae type strain ATCC 25954. J. Bacteriol. 194:6339–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koehne G, Maddux R, Britt J. 1981. Rapidly growing mycobacteria associated with bovine mastitis. Am. J. Vet. Res. 42:1238–1239 [PubMed] [Google Scholar]
- 63. Hachem R, Raad I, Rolston KVI, Whimbey E, Katz R, Tarrand J, Libshitz H. 1996. Cutaneous and pulmonary infections caused by Mycobacterium vaccae. Clin. Infect. Dis. 23:173–175 [DOI] [PubMed] [Google Scholar]
- 64. Sanchez D, Moussaoui M, Carreras E, Torrent M, Nogues V, Boix E. 2011. Mapping the eosinophil cationic protein antimicrobial activity by chemical and enzymatic cleavage. Biochimie 93:331–338 [DOI] [PubMed] [Google Scholar]
- 65. Wang H, Schwaderer AL, Kline J, Spencer JD, Kline D, Hains DS. 2013. Contribution of structural domains to ribonuclease 7's activity against uropathogenic bacteria. Antimicrob. Agents Chemother. 57:766–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Andreu D, Ubach J, Boman A, Wahlin B, Wade D, Merrifield RB, Boman HG. 1992. Shortened cecropin-A melittin hybrids: significant size reduction retains potent antibiotic activity. FEBS Lett. 296:190–194 [DOI] [PubMed] [Google Scholar]
- 67. Milani A, Benedusi M, Aquila M, Rispoli G. 2009. Pore forming properties of cecropin-melittin hybrid peptide in a natural membrane. Molecules 14:5179–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. García-Mayoral MF, Moussaoui M, de la Torre BG, Andreu D, Boix E, Nogués MV, Rico M, Laurents DV, Bruix M. 2010. NMR structural determinants of eosinophil cationic protein binding to membrane and heparin mimetics. Biophys. J. 98:2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Torrent M, Di Tommaso P, Pulido D, Nogués M, Notredame C, Boix E, Andreu D. 2012. AMPA: an automated web server for prediction of protein antimicrobial regions. Bioinformatics 28:130–131 [DOI] [PubMed] [Google Scholar]
- 70. Torrent M, Cuyas E, Carreras E, Navarro S, Lopez O, de la Maza A, Nogues MV, Reshetnyak YK, Boix E. 2007. Topography studies on the membrane interaction mechanism of the eosinophil cationic protein. Biochemistry 46:720–733 [DOI] [PubMed] [Google Scholar]
- 71. Saugar JM, Rodriguez-Hernandez MJ, de la Torre BG, Pachon-Ibanez ME, Fernandez-Reyes M, Andreu D, Pachon J, Rivas L. 2006. Activity of cecropin A-melittin hybrid peptides against colistin-resistant clinical strains of Acinetobacter baumannii: molecular basis for the differential mechanisms of action. Antimicrob. Agents Chemother. 50:1251–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Torrent M, Odorizzi F, Nogues MV, Boix E. 2010. Eosinophil cationic protein aggregation: identification of an N-terminus amyloid prone region. Biomacromolecules 11:1983–1990 [DOI] [PubMed] [Google Scholar]
- 73. Cemma M, Brumell JH. 2012. Interactions of pathogenic bacteria with autophagy systems. Curr. Biol. 22:R540–R545 [DOI] [PubMed] [Google Scholar]
- 74. Mostowy S, Cossart P. 2012. Bacterial autophagy: restriction or promotion of bacterial replication? Trends Cell Biol. 22:283–291 [DOI] [PubMed] [Google Scholar]
- 75. Chu HT, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, Winter MG, Winter SE, Wehkamp J, Shen B, Salzman NH, Underwood MA, Tsolis RM, Young GM, Lu WY, Lehrer RI, Baumler AJ, Bevins CL. 2012. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337:477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Boix E, Pulido D, Moussaoui M, Nogues MV, Russi S. 2012. The sulfate-binding site structure of the human eosinophil cationic protein as revealed by a new crystal form. J. Struct. Biol. 179:1–9 [DOI] [PubMed] [Google Scholar]
- 77. Gouet P, Courcelle E, Stuart DI, Metoz F. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305–308 [DOI] [PubMed] [Google Scholar]
- 78. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Conchillo-Sole O, de Groot NS, Aviles FX, Vendrell J, Daura X, Ventura S. 2007. AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Maurer-Stroh S, Debulpaep M, Kuemmerer N, de la Paz ML, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, Schymkowitz JWH, Rousseau F. 2010. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods 7:237–U109 [DOI] [PubMed] [Google Scholar]
- 81. Huang YC, Lin YM, Chang TW, Wu SJ, Lee YS, Chang MD, Chen C, Wu SH, Liao YD. 2007. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. J. Biol. Chem. 282:4626–4633 [DOI] [PubMed] [Google Scholar]