Abstract
The antimicrobial mechanism of a lactococcal bacteriocin, lacticin Q, can be described by the toroidal pore model without any receptor. However, lacticin Q showed different degrees of activity (selective antimicrobial activity) against Gram-positive bacteria even among related species. The ability of lacticin Q to induce pore formation in liposomes composed of lipids from different indicator strains indicated that its selective antimicrobial activity could not be attributed only to membrane lipid composition. We investigated the accumulation of deleterious hydroxyl radicals after exposure to lacticin Q as a contributing factor to cell death in the indicator strains. When lacticin Q of the same concentration as the MIC or minimum bactericidal concentration was added to the indicator cultures, high levels of hydroxyl radical accumulation were detected. Treatment with hydroxyl radical scavengers, thiourea and 2,2′-bipyridyl, decreased the levels of hydroxyl radical accumulation and recovered cell viability. These results suggest that, with or without pore formation, the final antimicrobial mechanism of lacticin Q is the accumulation of hydroxyl radicals, which varies by strain, resulting in the selective antimicrobial activity of lacticin Q.
INTRODUCTION
Bacteria produce ribosomally synthesized antimicrobial peptides or proteins referred to as bacteriocins, which generally inhibit species closely related to the producer bacterium (1). Some bacteriocins display antimicrobial activity against even phylogenetically distant Gram-positive and Gram-negative bacteria (2, 3). Especially, there is a great interest in the bacteriocins produced by lactic acid bacteria (LAB) because they are generally regarded as safe (4). Many LAB bacteriocins show antimicrobial activity at nanomolar concentration against a wide range of food-borne Gram-positive bacterial strains such as Listeria monocytogenes, Staphylococcus aureus, and Bacillus cereus (1, 4, 5). Thus far, a large number of bacteriocins have been identified as promising biopreservatives.
Some bacteriocins such as nisin, pediocin PA-1, and lacticin 3147 are characterized by an antimicrobial mechanism that uses a docking molecule as a receptor. Nisin and lacticin 3147 use lipid II, a precursor of peptidoglycan, as the docking molecule to form pores on bacterial cytomembranes (6, 7, 8). Nisin has also been found to inhibit peptidoglycan synthesis upon interaction with lipid II (9). A recent study has demonstrated that nisin also interacts with lipid III and lipid IV, cell wall teichoic acid (WTA) precursors, to block the WTA biosynthesis (10). Pediocin PA-1 exerts antimicrobial activity in a similar manner by using a docking molecule, MptD, a membrane protein, to form pores on bacterial cytomembranes (11, 12).
Recently, a novel LAB bacteriocin, termed lacticin Q, has been identified; it is produced by Lactococcus lactis QU 5 isolated from corn (13). Lacticin Q possesses strong antimicrobial activity at nanomolar concentrations against a broad spectrum of bacteria. Lacticin Q directly induces inner molecule efflux from large unilamellar vesicles (LUVs) by pore formation without any specific receptor (14). The action mechanism of lacticin Q is proposed as the huge toroidal pore (HTP) model. This model is described as follows. First, lacticin Q molecules bind to negatively charged membrane leaflets. Then, they form pores on the membrane, causing leakage of cell contents with lipid flip-flop. Finally, lacticin Q molecules migrate from the outer to the inner membrane leaflets (15). Lacticin Q does not exert antibacterial activity against Gram-negative bacteria, because it cannot induce permeability of the outer membrane of Gram-negative bacteria (16). In contrast, lacticin Q shows inhibitory activity against almost all the Gram-positive bacteria. However, the intensity of the activity varied by species and, in some cases, even by strain. Although this selective antimicrobial activity phenomenon is also observed for most bacteriocins, the underlying mechanism(s) remains unclear.
Reactive oxygen species (ROS) such as superoxide anions, hydrogen peroxide, and hydroxyl radicals are natural by-products of oxidative energy metabolism and are a permanent threat to living organisms (17). The hydroxyl radical is a highly reactive and short-lived ROS. There is no enzyme that can detoxify the hydroxyl radical, making it extremely toxic and lethal. Recently, considerable interest has been focused on the accumulation of ROS, which plays an important role in bactericidal antibiotic-induced bacterial cell death (18, 19). Although bactericidal antibiotics against Escherichia coli and Staphylococcus aureus have different primary targets in these cells, studies have suggested that hydroxyl radical accumulation caused by bactericidal antibiotics mediates cell death.
In this study, we analyzed the factors that influence the selective antimicrobial activity of lacticin Q against Gram-positive bacteria. We focused on the membrane lipid composition and examined the different abilities of indicator strains to resist pore formation. The results indicated that an antimicrobial mechanism(s) other than pore formation could influence the selective antimicrobial activity. We examined whether accumulation of hydroxyl radicals as a lethal factor was induced by lacticin Q and whether this accumulation was essential for cell death.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and materials.
L. lactis QU 5, used as the lacticin Q-producing strain, was grown at 30°C in de Man, Rogosa, and Sharpe (MRS) broth (Oxoid, Basingstoke, United Kingdom) for 18 h (20). Lacticin Q was purified from L. lactis QU 5 culture supernatant, as described earlier (13). The strains used as indicator strains were incubated at 30°C or 37°C in MRS broth, except for Streptococcus mutans JCM 5705T, which was grown at 37°C in tryptic soy broth (BD, Detroit, MI, USA) supplemented with 0.6% (wt/wt) yeast extract (Nacalai Tesque, Kyoto, Japan). Cultures were maintained in 15% (vol/vol) glycerol at −80°C. The L. lactis IL1403 resistant strain (R) was obtained from the L. lactis IL1403 wild-type strain (W) with the medium containing lacticin Q of the same concentrations as the MICs and by increasing the lacticin Q concentrations to 1.25- and 1.5-fold MICs according to a previously described method (21).
Lacticin Q activity assay.
The MICs and minimum bactericidal concentrations (MBCs) of lacticin Q against the indicator strains were determined by the turbidimetric assay method (22) with some modifications. Overnight-incubated indicator strains were reinoculated in liquid medium under their optimal conditions and incubated until an optical density at 600 nm (OD600) of 1.0 was reached. They were adjusted to a final OD600 of 0.05 in a 96-well microplate, and lacticin Q was added at arbitrary serial concentrations. Then, the respective medium was added to a volume of 100 μl. The indicator strains were cultured until the controls (without lacticin Q) reached an OD600 of 0.2 to 0.3 under optimal conditions for each indicator strain. Then, all the wells were diluted with 100 μl of the respective media, and the OD600 was monitored for each strain with a microplate reader (Immuno Mini NJ-2300; Inter Medical, Tokyo, Japan). The concentrations of lacticin Q that decrease the OD600 to less than 0.025 were defined as the MICs of lacticin Q. Similarly, the indicator strains were incubated under optimal conditions for 24 h, and then the concentrations of lacticin Q that caused at least a 99% decrease in viable cells (CFU/ml) were defined as the MBCs of lacticin Q. The experiments for MIC and MBC changes by hydroxyl radical scavengers, hydroxyl radical quencher (thiourea, 0.15 M) (Tokyo Chemical Industry Co., Tokyo, Japan), and iron chelator (2,2′-bipyridyl, 0.5 mM) (Tokyo Chemical Industry Co.) were performed similarly as described above. Triplicate assays were performed, and each result was confirmed to be identical.
Lipid extraction.
Lipid extraction of indicator strains was performed according to the procedure of Bligh and Dyer (23) with some modifications. Indicator strains were grown under optimal conditions until mid-log phase. Cells were harvested by centrifugation (8,000 × g for 20 min). Then, the frozen cell pellets in methanol-chloroform (2:1, vol/vol) were broken with a Sonifier 250D (Branson Sonic Power Co., Danbury, CT, USA). After 30 min, Milli-Q water–chloroform (1:1, vol/vol) was added to give a final ratio of 2:2:1 (methanol–chloroform–Milli-Q water). Then, the mixture was centrifuged (1,800 × g for 10 min), and the chloroform phase was collected. A second extraction was performed by washing the remaining composition with chloroform. The chloroform of the combined chloroform phase was evaporated in a rotary evaporator and completely removed by vacuum drying overnight. Resultant lipids were weighed and redissolved in chloroform to determine concentrations. Lipids were flushed with a stream of nitrogen and stored at −20°C.
Preparation of LUVs and calcein leakage.
LUVs were prepared according to a previously described method (15). The chloroform in the lipids was removed by an evaporator and vacuum dried. The dried lipids were resuspended to a final concentration of 0.5 mg/ml in buffer A (10 mM Tris-HCl, 75 mM NaCl, 1 mM EDTA, pH 7.4). For the calcein leakage experiments, a final concentration of 70 mM calcein (Dojindo, Kumamoto, Japan) was added to buffer A at the same time as hydrated lipid film, and finally, untrapped calcein was removed by gel filtration. Calcein leakage from LUVs was examined at an excitation wavelength (EX) of 490 nm and an emission wavelength (EM) of 520 nm on an F-7000 spectrofluorometer (Hitachi High-Technologies, Tokyo, Japan). Fluorescence by the addition of buffer A was determined as 0% calcein efflux and that by the addition of 0.1% (vol/vol) Triton X-100 (Wako Pure Chemical Industries, Osaka, Japan) was determined as 100% calcein efflux.
Disruption of membrane potential.
Disruption of membrane potential by lacticin Q was analyzed according to the method of Breukink et al. (24). Disruption of membrane potential was monitored by the intensity of the fluorescence of the dye DiSC3 (5) (3,3′-dipropylthiadicarbocyanine iodide) (Invitrogen, Carlsbad, CA, USA) using a spectrofluorometer, and EX and EM were set as 622 and 675 nm, respectively. The mid-log-phase cells of indicator strains were collected by centrifugation (8,000 × g for 10 min), washed with buffer D (250 mM sucrose, 5 mM MgSO4, 10 mM potassium phosphate, pH 7.0), and then resuspended in buffer D at a final OD600 of 0.05. For each measurement, 100 μl of 20 μM DiSC3 (5) was added to 2 ml of buffer D to reach a final concentration of 1 μM. After stabilization of the baseline, 100 μl of lacticin Q of a decided concentration was added. When the fluorescence intensity became stable, the fluorescence intensity was considered the maximum disruption of the membrane potential. Fluorescence by the addition of buffer D was determined as 0% disruption of membrane potential, and that by the addition of 0.1% (vol/vol) Triton X-100 was determined as 100% disruption of membrane potential.
Hydroxyl radical accumulation assay.
Hydroxyl radical levels were measured using 5 μM HPF {2-[6-(4′-hydroxy)phenoxy-3H-xanthen-3-on-9-yl] benzoic acid} (Sekisui Medical Co., Tokyo, Japan), which is a fluorescent dye oxidized by hydroxyl radicals with high specificity (25). Hydroxyl radical levels were monitored using a spectrofluorometer, and EX and EM were set as 490 and 515 nm, respectively. Indicator strains were incubated until an OD600 of 0.3 was reached, and then antimicrobials were added. Hydroxyl radical levels were measured every hour for 3 h. Indicator strains incubated with water instead of antimicrobials were used as a negative control. Ampicillin was reported as a hydroxyl radical inducer (19). The hydroxyl radical levels induced by the respective MICs of ampicillin sodium salt (Nacalai Tesque, Kyoto, Japan) (L. lactis ATCC 19435T, 0.55 μM; L. lactis IL1403 W, 16.9 μM; L. lactis IL1403 R, 2.1 μM; Pediococcus pentosaceus JCM 5890T, 67.5 μM; P. pentosaceus JCM 5885, 135 μM; S. mutans JCM 5705T, 1.05 μM) were used as positive controls. The viability rescue experiments were performed similarly as described above. For investigation of the effect of hydroxyl radical scavengers, the indicator strains were incubated with lacticin Q and hydroxyl radical scavengers, hydroxyl radical quencher (thiourea, 0.15 M), and iron chelator (2,2′-bipyridyl, 0.5 mM).
RESULTS
Lacticin Q activity assay.
The MICs and MBCs of lacticin Q against various Gram-positive indicator strains are listed in Table 1. The indicator strains showed marked differences in the MICs and MBCs of lacticin Q. L. lactis IL1403 R showed a MIC and an MBC 100 times higher than those of its parent strain, L. lactis IL1403 W.
Table 1.
Antimicrobial spectra of lacticin Q by the turbidimetric assay method
| Indicator straina | MIC (nM) | MBC (nM) |
|---|---|---|
| Lactococcus lactis ATCC 19435T | 5 | 50 |
| Lactococcus lactis IL1403 W | 100 | 500 |
| Lactococcus lactis IL1403 Rb | 10,000 | 50,000 |
| Pediococcus pentosaceus JCM 5890T | 750 | 1,500 |
| Pediococcus pentosaceus JCM 5885 | 1,000 | 5,000 |
| Streptococcus mutans JCM 5705T | 10,000 | 50,000 |
ATCC, American Type Culture Collection, Rockville, MD; JCM, Japan Collection of Microorganisms, Wako, Japan.
L. lactis IL1403 R was obtained by adapting L. lactis IL1403 W gradually to lacticin Q.
Influence of lipid composition on pore formation.
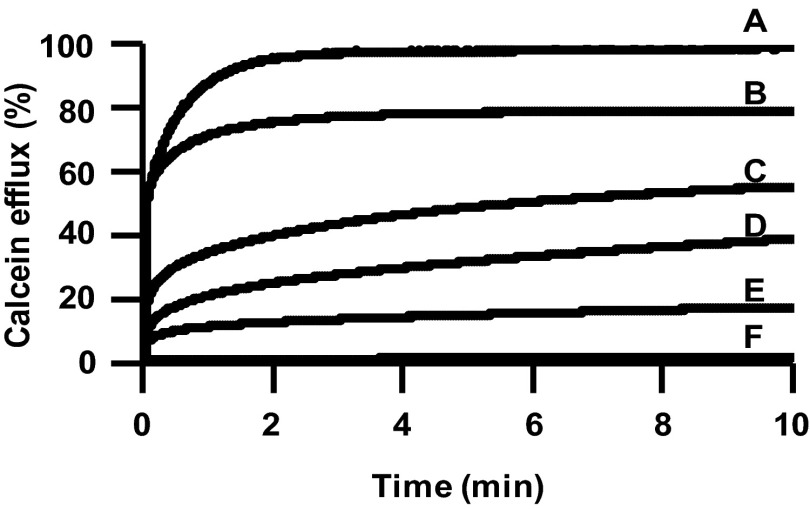
Lacticin Q acts on the cell membrane without any receptors (15). Hence, membrane lipid composition could be a factor that influences selective antibacterial activity. Using the extracted total lipids from the indicator strains shown in Table 1, we constructed the respective LUVs. The same size and concentration of calcein-trapped LUVs were subjected to 500 nM lacticin Q, and the calcein efflux was observed, as shown in Fig. 1. P. pentosaceus JCM 5885 (Fig. 1, line A) and L. lactis IL1403 R (Fig. 1, line B) showed almost 100% and around 80% calcein efflux, respectively. Therefore, these two strains were considered to have membranes more susceptible to pore formation by lacticin Q than the other strains. However, both of these strains showed high MICs and MBCs, indicating that they were not highly sensitive to lacticin Q (Table 1). In contrast, some sensitive strains had rigid membranes unsusceptible to pore formation, such as L. lactis IL1403 W (Fig. 1, line F) and L. lactis ATCC 19435T (Fig. 1, line E). Surprisingly, changes in the membrane lipid composition of L. lactis IL1403 R were observed, allowing pore formation easily by lacticin Q. These results strongly suggest that besides lipid composition, other important factors influence cell viability and selective antimicrobial activity.
Fig 1.
Calcein efflux from the liposomes composed of lipids extracted from the six indicator strains. The lines show calcein efflux from different liposomes with the addition of 500 nM lacticin Q. As controls, 0% calcein efflux and 100% calcein efflux were obtained by the addition of buffer and 0.1% (vol/vol) Triton X-100, respectively. The final lipid concentration was 0.5 mg/ml. Indicator strains: A, P. pentosaceus JCM 5885; B, L. lactis IL1403 R; C, S. mutans JCM 5705T; D, P. pentosaceus JCM 5890T; E, L. lactis ATCC 19435T; F, L. lactis IL1403 W.
Disruption of membrane potentials of indicator cells.
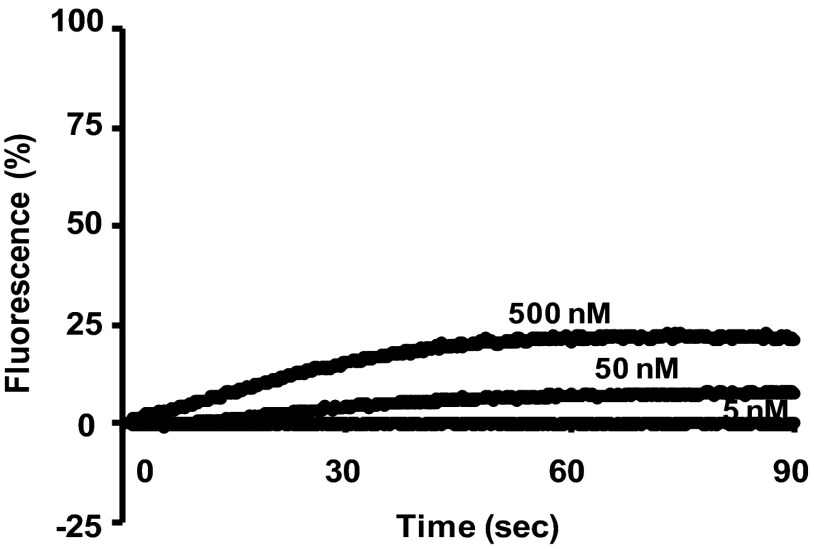
To estimate the influence of pore formation on selective antimicrobial activity in vivo, we monitored disruption of the membrane potential by lacticin Q in the six indicator strains. The maximum levels of disruption of the membrane potential (percent fluorescence) of the six indicator strains are shown in Table 2. When 500 nM lacticin Q was added, most of the indicator strains showed similar disruptions of membrane potential of about 10 to 20% fluorescence; however, they showed quite different MICs and MBCs (Table 1). This finding indicated that the strains were capable of adapting themselves to pore formation induced by lacticin Q. Therefore, the selective antimicrobial activity was also dependent on the pore formation characteristics of the indicator strains. In some cases, such as nisin, antimicrobial activity is not a single process (9). There may be antibacterial mechanisms of lacticin Q other than pore formation, as observed in the most sensitive indicator strain, L. lactis ATCC 19435T (Fig. 2). Lacticin Q at a MIC of 5 nM caused no pore formation (i.e., no disruption of membrane potential) in L. lactis ATCC 19435T. This result suggested that against L. lactis ATCC 19435T, there were some other mechanisms more effective than pore formation.
Table 2.
Maximum of disruption of membrane potential in six indicator strains induced by 500 nM lacticin Qa
| Indicator strain | Fluorescence (%) |
|---|---|
| Lactococcus lactis ATCC 19435T | 21.36 ± 0.4 |
| Lactococcus lactis IL1403 W | 9.52 ± 0.1 |
| Lactococcus lactis IL1403 R | 10.07 ± 0.3 |
| Pediococcus pentosaceus JCM 5890T | 14.25 ± 0.1 |
| Pediococcus pentosaceus JCM 5885 | 13.30 ± 0.2 |
| Streptococcus mutans JCM 5705T | 21.46 ± 0.2 |
L. lactis IL1403 R was obtained by adapting L. lactis IL1403 W gradually to lacticin Q. As control, 0% fluorescence and 100% fluorescence were obtained by the addition of buffer and 0.1% (vol/vol) Triton X-100 respectively. Values of fluorescence (%) are means for averages of triplicate experiments ± standard deviation.
Fig 2.
Disruption of the membrane potential of L. lactis ATCC 19435T by treatment with 5, 50, and 500 nM lacticin Q. As controls, 0% fluorescence and 100% fluorescence were obtained by the addition of buffer and 0.1% (vol/vol) Triton X-100, respectively.
Accumulation of hydroxyl radicals formed by lacticin Q.
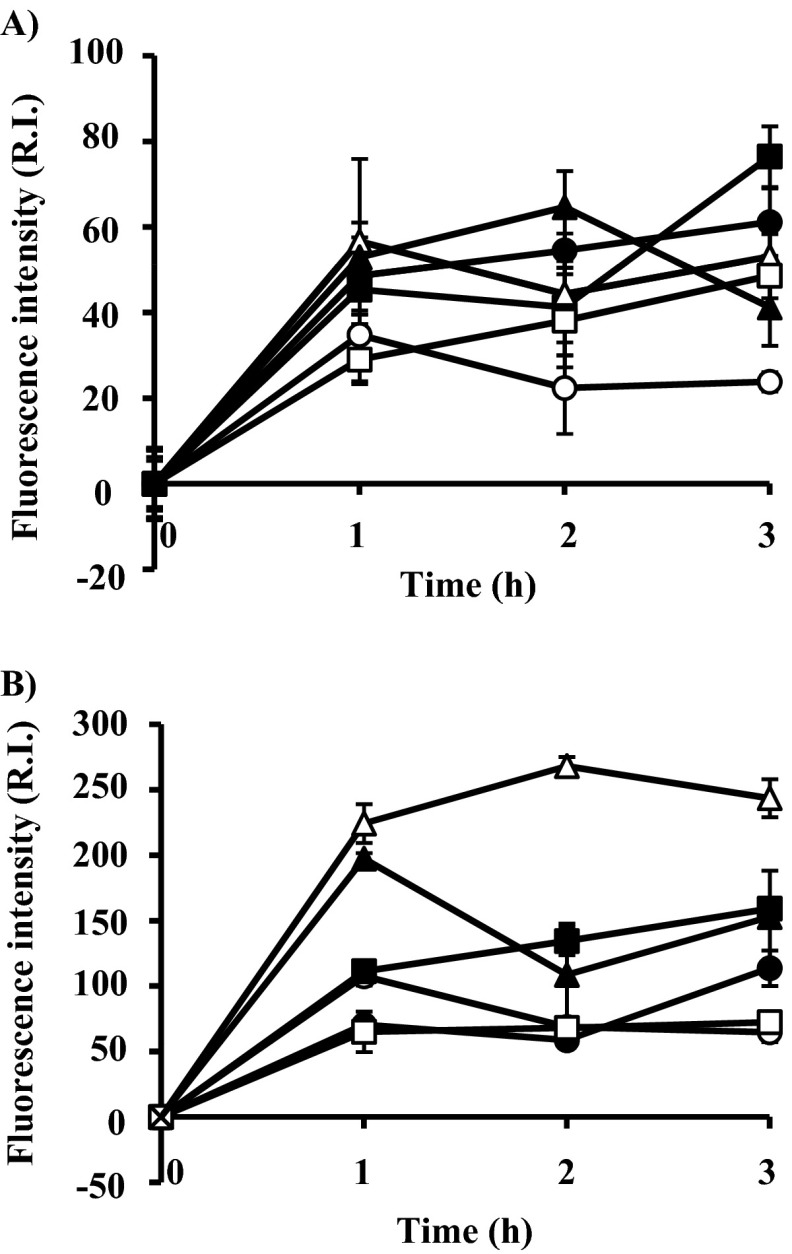
Previous studies showed that bactericidal antibiotics could promote formation of a ROS (such as a hydroxyl radical) that contributes to cell death (18, 19). In this study, lacticin Q also showed bactericidal activity (MBCs; Table 1). We investigated whether hydroxyl radical formation could contribute to cell death induced by lacticin Q. In the six indicator strains, accumulation of hydroxyl radicals induced by the respective MICs or MBCs of lacticin Q was confirmed, as shown in Fig. 3. When MICs of lacticin Q were added, relatively lower hydroxyl radical production levels were detected, as shown in Fig. 3A. On the other hand, when MBCs of lacticin Q were added, higher hydroxyl radical production levels were observed, as shown in Fig. 3B. These results suggest that both MICs and MBCs of lacticin Q induced accumulation of hydroxyl radicals and that hydroxyl radical accumulation is an important factor in lacticin Q-induced cell death.
Fig 3.
Hydroxyl radical production induced by the respective MICs (A) and MBCs (B) of lacticin Q. Fluorescence intensity (R.I., relative intensity) was measured with a spectrofluorometer for 3 h after the addition of lacticin Q. Open circles, L. lactis ATCC 19435T; filled circles, L. lactis IL1403 W; open triangles, L. lactis IL1403 R; filled triangles, P. pentosaceus JCM 5890T; open squares, P. pentosaceus JCM 5885; filled squares, S. mutans JCM 5705T. Indicator strains incubated without lacticin Q addition were used as a negative control. Error bars indicate standard deviations for replicate samples of triplicate experiments.
Effect of hydroxyl radical scavengers on lacticin Q activity.
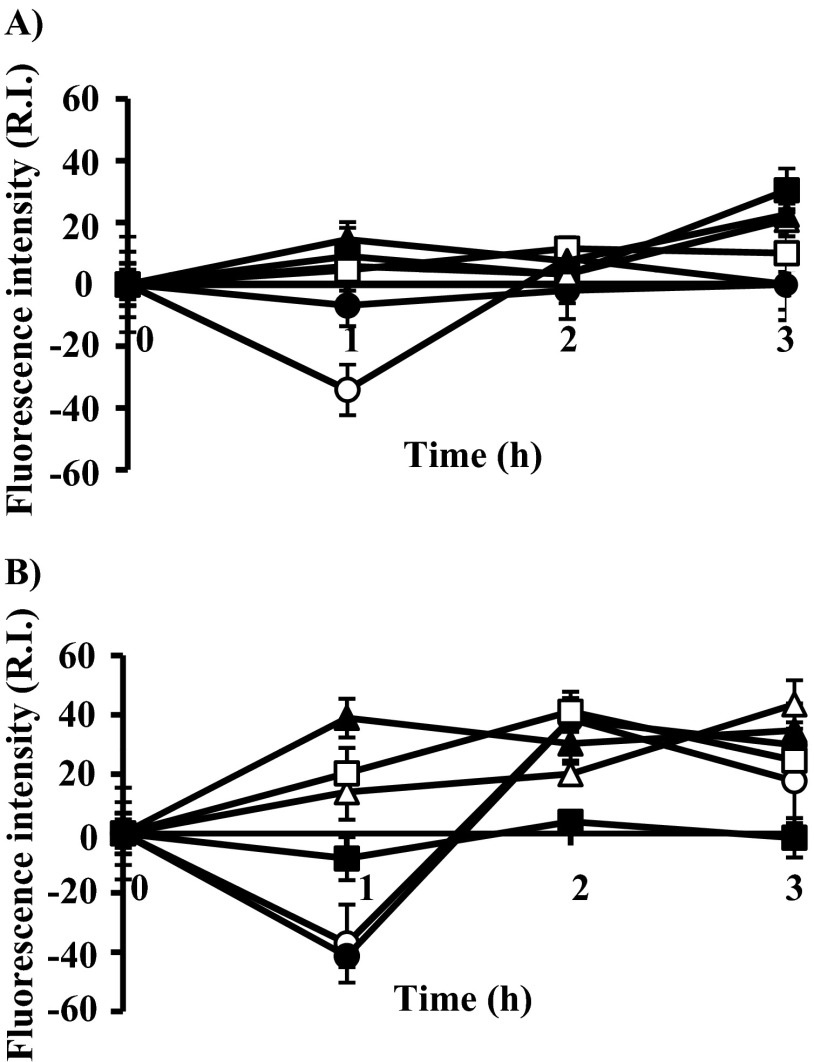
To confirm whether accumulation of hydroxyl radicals is induced by lacticin Q, we evaluated the effects of hydroxyl radical scavengers on hydroxyl radical production in the six indicator strains, and the results are shown in Fig. 4. In all these six indicator strains, when hydroxyl radical scavengers were added, the accumulation levels of hydroxyl radicals were lower than those observed without the scavenger treatment. MICs of lacticin Q induced very low or no hydroxyl radical production (Fig. 4A), and MBCs of lacticin Q induced hydroxyl radical production lower than a relative intensity (RI) of 50 (Fig. 4B). To confirm the effect of hydroxyl radical scavengers on lacticin Q activity, we measured the MICs and MBCs of these six indicator strains treated with hydroxyl radical scavengers (Table 3). When hydroxyl radical scavengers were added, the MICs increased almost 2-fold. A similar phenomenon was observed for MBCs. All the six indicator strains were rescued to a certain extent by treatment with hydroxyl radical scavengers. These results indicated that the accumulation of hydroxyl radicals is an important factor in lacticin Q-induced cell death.
Fig 4.
Hydroxyl radical production induced by the respective MICs (A) and MBCs (B) of lacticin Q with the addition of hydroxyl radical scavengers. Fluorescence intensity (R.I., relative intensity) was measured with a spectrofluorometer for 3 h after the addition of lacticin Q and the scavengers. Open circles, L. lactis ATCC 19435T; filled circles, L. lactis IL1403 W; open triangles, L. lactis IL1403 R; filled triangles, P. pentosaceus JCM 5890T; open squares, P. pentosaceus JCM 5885; filled squares, S. mutans JCM 5705T. Indicator strains incubated without lacticin Q addition were used as a negative control. Error bars indicate standard deviations for replicate samples of triplicate experiments.
Table 3.
MIC and MBC changes by addition of hydroxyl radical scavengers in six indicator strains
| Indicator strain | TBa/water (fold increase) |
|
|---|---|---|
| MIC | MBC | |
| Lactococcus lactis ATCC 19435T | 4 | 2 |
| Lactococcus lactis IL1403 W | 2 | 1.5 |
| Lactococcus lactis IL1403 R | 2 | 1.2 |
| Pediococcus pentosaceus JCM 5890T | 1.3 | 1.3 |
| Pediococcus pentosaceus JCM 5885 | 3 | 1.5 |
| Streptococcus mutans JCM 5705T | 5 | 1.1 |
TB, thiourea and 2,2′-bipyridyl (hydroxyl radical scavengers). Fold increase is that over original MICs and MBCs.
DISCUSSION
In this study, we investigated the in vitro and in vivo factors that influence the selective antimicrobial activity of lacticin Q and elucidated a new antimicrobial mechanism of lacticin Q. To the best of our knowledge, this is the first report on the accumulation of hydroxyl radicals induced by LAB bacteriocins.
Nisin-resistant Listeria strains have been well researched. Nisin-resistant Listeria strains contain more straight-long-chain and fewer branched-short-chain fatty acids and thereby have enhanced membrane rigidity. The resistant strains also have reduced amounts of the anionic lipid phosphatidylglycerol and contain significantly more zwitterionic phosphatidylethanolamine, as well as altered fatty acid components in their membrane (26). Cationic lipids prevent cationic bacteriocins from binding to the membrane, thus inhibiting their killing activity. Mutants resistant to class IIa bacteriocins were also found to have a modified lipid composition that enhanced membrane rigidity (27, 28). Only Gram-positive bacteria show sensitivity to lacticin Q. Other types of cells are insensitive to lacticin Q because of their different membrane lipid composition (16). In vitro, we tested calcein efflux from LUVs constructed from total lipids extracted from indicator cells (Fig. 1). We expected the LUVs with more rigid membranes to counteract the effects of lacticin Q, and thus, the indicator strains should show lower antimicrobial activity (higher MICs or MBCs). Some indicator strains such as L. lactis IL1403 W and L. lactis ATCC 19435T were shown to have rigid membranes that were unsusceptible to pore formation by lacticin Q. However, they showed lower MICs or MBCs than did the other indicator strains. Conversely, resistant indicator strains such as L. lactis IL1403 R constructed flexible membranes that are susceptible to pore formation. These results were highly unexpected and suggested that membrane lipid composition is not the only factor in selective antimicrobial activity of lacticin Q.
This hypothesis was supported also by the results of our in vivo analysis, where the disruption of membrane potential was measured. As shown in Table 2, all of the indicator strains showed similar disruptions of membrane potential, indicating similar degrees of pore formation. The survival rates of the indicator strains were evaluated by a CFU count, and the insensitive strains showed higher survival rates even when they showed a similar degree of pore formation (data not shown). We hypothesized that the insensitive indicator strains possessed high durability or resilience against pore formation and that there are other mechanisms underlying antimicrobial activity, especially in sensitive indicator strains. This hypothesis was supported by the results of L. lactis ATCC 19435T, a sensitive indicator strain. L. lactis ATCC 19435T had a more rigid membrane (Fig. 1, line E) and formed no pores with the addition of MICs of lacticin Q (Fig. 2).
Recently, a new viewpoint has emerged that the overproduction or accumulation of hydroxyl radicals plays a role in bacterial cell death induced by bactericidal antibiotics (18, 19). In the present study, we detected accumulation of hydroxyl radicals mediated by lacticin Q at both MICs and MBCs (Fig. 3). A previous report showed that the bactericidal antibiotic ampicillin induced cell death by mediating accumulation of hydroxyl radicals (19). To compare the accumulation levels of hydroxyl radicals, we used ampicillin as a positive control (data not shown). Accumulations of hydroxyl radicals induced by lacticin Q were similar to or higher than those induced by ampicillin. These results suggest that the accumulation of hydroxyl radicals is an antimicrobial mechanism of lacticin Q. We observed an interesting phenomenon that when 5 nM lacticin Q was added, the accumulation occurred in strain L. lactis ATCC 19435T (Fig. 3A, open circles) as observed in other indicator strains, although there was no pore formation (Fig. 2). This mechanism seemed to be more effective than pore formation against the strain L. lactis ATCC 19435T.
ROS are very important products as terminal electron acceptors of oxygen during the cellular processes (metabolic pathway) and occur widely in nature (17). Oxygen participates in the oxidoreduction step of NADH to NAD+. Hydrogen peroxide is formed through the action of NADH oxidase, pyruvate oxidase, and α-glycerophosphate oxidase (29). The hydroxyl radicals are formed under oxidative stress by using released iron together with hydrogen peroxide, and this step is the so-called Fenton reaction (H2O2 + Fe2+→·OH + OH− + Fe3+) (30). Therefore, the iron chelator 2,2′-bipyridyl and the hydroxyl radical quencher thiourea were used to test the rescue from lacticin Q treatment. As shown in Fig. 4 and Table 3, an obvious rescue was observed in all six indicator strains. In addition, the generation of hydrogen peroxide by lacticin Q was also evaluated in these six indicator strains. Lacticin Q caused accumulation of hydrogen peroxide in all the indicator strains (data not shown). These results suggested that lacticin Q also showed activity involving accumulation of hydroxyl radicals by the Fenton reaction.
We also evaluated the levels of hydroxyl radical production induced by the addition of lacticin Q at lower concentration (lower than MICs). Low hydroxyl radical production was detected only at the beginning of the first hour, and then after 2 h, the hydroxyl radical level decreased to the same level as that of the negative control (data not shown). Some cellular proteins such as OxyR and NADH peroxidase were reported to detect and scavenge hydrogen peroxide in bacterial cells (17, 29). Therefore, we supposed that such scavenging proteins could decrease hydroxyl radicals in the indicator strains. In fact, some indicator strains were rescued from low accumulation of hydroxyl radicals and cell death was prevented. This result suggested that the hydroxyl radical accumulation induced by lacticin Q depends on the potential of different indicator strains to scavenge the deleterious hydroxyl radicals. In other words, the indicator strains will survive until the lacticin Q concentration, which differs by strain, becomes high enough to cause hydroxyl radical accumulation. It also suggested that accumulation of hydroxyl radicals was the final antimicrobial mechanism of lacticin Q. Therefore, the antimicrobial activity of lacticin Q differs by strain, resulting in selective activity.
This is the first study to report that the antimicrobial mechanism of LAB bacteriocins involves the accumulation of hydroxyl radicals. The selective activity of lacticin Q against Gram-positive bacteria depends not only on the pore formation but also on the indicator strains' capacity to scavenge hydroxyl radicals. The accumulation of hydroxyl radicals is the final antimicrobial mechanism of lacticin Q. This finding would have significant implications for the practical use of lacticin Q as an industrial food preservative and as an antimicrobial.
ACKNOWLEDGMENTS
This work was partially supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (JSPS) and by a research project for utilizing advanced technologies in agriculture, forestry, and fisheries by the Ministry of Agriculture, Forestry and Fisheries of Japan.
Footnotes
Published ahead of print 3 June 2013
REFERENCES
- 1. Klaenhammer TR. 1988. Bacteriocins of lactic acid bacteria. Biochimie 70:337–349 [DOI] [PubMed] [Google Scholar]
- 2. Messaoudi S, Kergourlay G, Rossero A, Ferchichi M, Prévost H, Drider D, Manai M, Dousset X. 2011. Identification of lactobacilli residing in chicken ceca with antagonism against Campylobacter. Int. Microbiol. 14:103–110 [DOI] [PubMed] [Google Scholar]
- 3. Messaoudi S, Kergourlay G, Dalgalarrondo M, Choiset Y, Ferchichi M, Prévost H, Pilet MF, Chobert JM, Manai M, Dousset X. 2012. Purification and characterization of a new bacteriocin active against Campylobacter produced by Lactobacillus salivarius SMXD51. Food Microbiol. 32:129–134 [DOI] [PubMed] [Google Scholar]
- 4. Cleveland J, Montville TJ, Nes IF, Chikindas ML. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1–20 [DOI] [PubMed] [Google Scholar]
- 5. Gálvez A, Abriouel H, López RL, Ben Omar N. 2007. Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 120:51–70 [DOI] [PubMed] [Google Scholar]
- 6. Brötz H, Josten M, Wiedemann I, Schneider U, Götz F, Bierbaum G, Sahl HG. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317–327 [DOI] [PubMed] [Google Scholar]
- 7. Gross E, Morell JL. 1971. The structure of nisin. J. Am. Chem. Soc. 93:4634–4635 [DOI] [PubMed] [Google Scholar]
- 8. Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772–1779 [DOI] [PubMed] [Google Scholar]
- 9. Hasper HE, Kramer NE, Smith JL, Hillman JD, Zachariah C, Kuipers OP, de Kruijjf B, Breukink E. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636–1637 [DOI] [PubMed] [Google Scholar]
- 10. Müller A, Ulm H, Reder-Christ K, Sahl HG, Schneider T. 2012. Interaction of type A lantibiotics with undecaprenol-bound cell envelope precursors. Microb. Drug Resist. 18:261–270 [DOI] [PubMed] [Google Scholar]
- 11. Diep DB, Skaugen M, Salehlan Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gravesen A, Ramnath M, Rechinger KB, Andersen N, Jänsch L, Héchard Y, Hastings JW, Knøchel S. 2002. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 148:2361–2369 [DOI] [PubMed] [Google Scholar]
- 13. Fujita K, Ichimasa S, Zendo T, Koga S, Yoneyama F, Nakayama J, Sonomoto K. 2007. Structural analysis and characterization of lacticin Q, a novel bacteriocin belonging to a new family of unmodified bacteriocins of gram-positive bacteria. Appl. Environ. Microbiol. 73:2871–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoneyama F, Imura Y, Ichimasa S, Fujita K, Zendo T, Nakayama J, Matsuzaki K, Sonomoto K. 2009. Lacticin Q, a lactococcal bacteriocin, causes high-level membrane permeability in the absence of specific receptors. Appl. Environ. Microbiol. 75:538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoneyama F, Imura Y, Ohno K, Zendo T, Nakayama J, Matsuzaki K, Sonomoto K. 2009. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob. Agents Chemother. 53:3211–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoneyama F, Ohno K, Imura Y, Li M, Zendo T, Nakayama J, Matsuzaki K, Sonomoto K. 2011. Lacticin Q-mediated selective toxicity depending on physicochemical features of membrane components. Antimicrob. Agents Chemother. 55:2446–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X, Zhao X. 2009. Contribution of oxidative damage to antimicrobial lethality. Antimicrob. Agents Chemother. 53:1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 20. de Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23:130–135 [Google Scholar]
- 21. Kramer NE, Smid EJ, Kok J, de Kruijff B, Kuipers OP, Breukink E. 2004. Resistance of Gram-positive bacteria to nisin is not determined by lipid II levels. FEMS Microbiol. Lett. 239:157–161 [DOI] [PubMed] [Google Scholar]
- 22. Yoneyama F, Fukao M, Zendo T, Nakayama J, Sonomoto K. 2008. Biosynthetic characterization and biochemical features of the third natural nisin variant, nisin Q, produced by Lactococcus lactis 61-14. J. Appl. Microbiol. 105:1982–1990 [DOI] [PubMed] [Google Scholar]
- 23. Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 24. Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl HG, de Kruijff B. 1999. Use of the cell wall precursor lipid II by a pore-formation peptide antibiotic. Science 286:2361–2364 [DOI] [PubMed] [Google Scholar]
- 25. Setsukinai K, Urano Y, Kakinuma K, Majima HJ, Nagano T. 2003. Development of novel florescence probes that can reliably detect reactive oxygen species and distinguish specific species. J. Biol. Chem. 278:3170–3175 [DOI] [PubMed] [Google Scholar]
- 26. Crandall AD, Montville TJ. 1998. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl. Environ. Microbiol. 64:231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naghmouchi K, Drider D, Kheadr E, Lacroix C, Prevost H, Fliss I. 2006. Multiple characterizations of Listeria monocytogenes sensitive and insensitive variants to divergicin M35, a new pediocin-like bacteriocin. J. Appl. Microbiol. 100:29–39 [DOI] [PubMed] [Google Scholar]
- 28. Vadyvaloo V, Hastings JW, van der Merwe MJ, Rautenbach M. 2002. Membranes of class IIa bacteriocin-resistant Listeria monocytogenes cells contain increased levels of desaturated and short-acyl-chain phosphatidylglycerols. Appl. Environ. Microbiol. 68:5223–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyoshi A, Rochat T, Gratadoux JJ, Le Loir Y, Oliveira SC, Langella P, Azevedo V. 2003. Oxidative stress in Lactococcus lactis. Genet. Mol. Res. 2:348–359 [PubMed] [Google Scholar]
- 30. Jang S, Imlay JA. 2007. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282:929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]