Abstract
Several posttranscriptional modifications of bacterial rRNAs are important in determining antibiotic resistance or sensitivity. In all Gram-positive bacteria, dimethylation of nucleotide A2058, located in domain V of 23S rRNA, by the dimethyltransferase Erm(B) results in low susceptibility and resistance to telithromycin (TEL). However, this is insufficient to produce high-level resistance to TEL in Streptococcus pneumoniae. Inactivation of the methyltransferase RlmAII, which methylates the N-1 position of nucleotide G748, located in hairpin 35 of domain II of 23S rRNA, results in increased resistance to TEL in erm(B)-carrying S. pneumoniae. Sixteen TEL-resistant mutants (MICs, 16 to 32 μg/ml) were obtained from a clinically isolated S. pneumoniae strain showing low TEL susceptibility (MIC, 2 μg/ml), with mutation resulting in constitutive dimethylation of A2058 because of nucleotide differences in the regulatory region of erm(B) mRNA. Primer extension analysis showed that the degree of methylation at G748 in all TEL-resistant mutants was significantly reduced by a mutation in the gene encoding RlmAII to create a stop codon or change an amino acid residue. Furthermore, RNA footprinting with dimethyl sulfate and a molecular modeling study suggested that methylation of G748 may contribute to the stable interaction of TEL with domain II of 23S rRNA, even after dimethylation of A2058 by Erm(B). This novel finding shows that methylation of G748 by RlmAII renders S. pneumoniae TEL susceptible.
INTRODUCTION
In prokaryotic cells, rRNAs and ribosomal proteins undergo specific posttranscriptional modifications by a wide variety of enzymes during maturation (1). Although the functions of most of these modifications are currently unknown, many are believed to modulate rRNA maturation and to affect its stability. In Escherichia coli, the 23S rRNA contains 25 known modifications, of which 14 are methylations, 9 are pseudouridines, 1 is a methylated pseudouridine, and 1 is unknown (1, 2). Approximately one-third of modified residues of the 23S rRNA are clustered around the nascent peptide exit tunnel (NPET), which starts at the peptidyltransferase center (PTC) and spans the body of the large ribosomal subunit (3). Macrolide antibiotics bind in the NPET, close to the PTC, hindering the passage of the newly synthesized polypeptides through the NPET (3).
However, some modifications play important roles in determining antibiotic resistance or sensitivity (4). Among the proteins involved, the Erm family of proteins are known as methyltransferases that methylate the N-6 position of nucleotide A2058 in 23S rRNA domain V (5). Methylation of A2058, which forms an interaction site for the MLSB (macrolide, lincosamide, and streptogramin B) antibiotics, results in MLSB resistance (6). Erm methyltransferases have been classified into 2 types according to whether they add one or two methyl groups to this nucleotide position. Dimethylation confers the MLSB type II phenotype, with high resistance to all MLSB antibiotics, whereas monomethylation confers the MLSB type I phenotype, with high resistance to lincosamides and low to moderate resistance to macrolides and streptogramin B antibiotics (4, 6). Production of Erm dimethyltransferases in many pathogenic bacteria is the most common mechanism of macrolide resistance.
Telithromycin (TEL) is a semisynthetic derivative of the 14-member ring macrolide erythromycin (ERY), which was the first ketolide approved for clinical use. Its high efficacy has been demonstrated against Streptococcus pneumoniae isolates that cause community-acquired respiratory tract disease (7–9). TEL binds more strongly to ribosomes than ERY does, largely because of the alkyl-aryl substituent extending from positions 11 and 12 of the macrolactone ring (10, 11). The alkyl-aryl arm stacks on nucleotides A752 and U2609 to form a base pair that bridges domains II and V in 23S rRNA (12). Chemical probing and primer extension analyses have shown that nucleotide A752 is also involved in TEL binding in E. coli and Staphylococcus aureus (12). This may allow TEL to overcome common macrolide resistance mechanisms, including target modifications directed by Erm monomethyltransferases and mutations in the 23S rRNA and ribosomal proteins that interrupt macrolide binding (13, 14). However, the production of Erm dimethyltransferases causes TEL resistance in some Gram-positive bacteria, e.g., Streptococcus pyogenes (15).
S. pneumoniae resistance to macrolides has increased worldwide. PROTEKT year 5 data (2003 to 2004) show that the most common resistance mechanism globally has been the erm(B)-encoded Erm dimethyltransferase, which is present in 55% of macrolide-resistant isolates (16). Furthermore, the PneumoWorld study in Europe and the Asian Network for Surveillance of Resistant Pathogens (ANSRP) have shown that among erythromycin-resistant S. pneumoniae isolates, ∼80% carry the erm(B) gene (17, 18). Although high-level TEL resistance can easily be created from an erm(B)-carrying strain in a laboratory (19), many erm(B)-carrying strains of S. pneumoniae remain clinically sensitive to TEL (15). A few TEL-resistant strains of erm(B)-carrying S. pneumoniae have been isolated (20, 21). A large deletion upstream of the erm(B) gene resulted in constitutively high dimethylation at A2058, leading to TEL resistance (MICs, 8 to 16 μg/ml) (21). On the other hand, a clinical isolate of S. pneumoniae that is highly resistant to TEL (MIC, >256 μg/ml) carries an erm(B) gene encoding a truncated leader peptide and has mutations in the ribosomal protein L4 (20). Therefore, it remains unclear which determinant is essential for the high-level TEL resistance in erm(B)-carrying S. pneumoniae.
We have investigated the strong antibacterial activity of TEL that is effective even against erm(B)-carrying S. pneumoniae. TEL-resistant mutants were initially generated in vitro from a clinically isolated strain showing a low TEL susceptibility by introducing mutations into the regulatory region of erm(B) mRNA, and the whole-genome sequences of mutants were analyzed by next-generation sequencing. We thereby identified a mutation in the gene encoding RlmAII, which methylates position N-1 of nucleotide G748 in 23S rRNA domain II (22). Further analyses showed that methylation of G748 by RlmAII renders erm(B)-carrying S. pneumoniae TEL susceptible.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are shown in Tables 1 and 2, respectively. S. pneumoniae strain S1 revealing reduced TEL susceptibility (MIC, 2 μg/ml) was clinically isolated in Japan in 2010 and provided by Miroku Medical Laboratory Co., Ltd. (Nagano, Japan). Pneumococci were routinely cultured at 37°C and 5% CO2 in brain heart infusion plus 0.5% yeast extract (BHI-Y) broth and BHI-Y agar supplemented with 5% horse blood. E. coli was grown in L broth (1% Bacto tryptone, 0.5% Bacto yeast extract, 0.5% sodium chloride, pH 7.4) and L agar. When necessary, the medium was supplemented with kanamycin (25 to 500 μg/ml), tetracycline (10 μg/ml), erythromycin (25 μg/ml), spectinomycin (100 μg/ml), and ampicillin (25 μg/ml). For inducible conditions for erm(B), 25 μg/ml ERY was added during growth.
Table 1.
Bacterial strains used in this study
| Strain | Relevant characteristics | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | F− recA endA gyrA thi hsdR supE relA Δ(lacZYA-argF) deoR ϕ80dlacZΔM15 | Laboratory collection |
| DH5αZ1 | Same as DH5α, but tetR lacIq Specr | Laboratory collection |
| Streptococcus pneumoniae strains | ||
| TIGR4 | Prototroph | ATCC |
| S1 | TEL-resistant clinical isolate | Miroku Medical Laboratory Co., Ltd. |
| Sp32 to Sp41 | TEL-resistant mutants of S1 isolated from 8-μg/ml TEL-containing BHI-Y agar plates | This study |
| Sp42 to Sp47 | TEL-resistant mutants of S1 isolated from 16-μg/ml TEL-containing BHI-Y agar plates | This study |
| Sp182 | S1 erm(B)::aad(9) | This study |
| Sp274 | S1 tlrB::aad(9) | This study |
| Sp303 | TIGR4 tlrB::aad(9) | This study |
Table 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| pUC18 | Cloning vector | Laboratory collection |
| pT7Blue | Cloning vector | Novagen |
| pLZ12-Km2 | Shuttle vector | 55 |
| pTKY858 | pT7Blue with 618-bp erm(B) fragment | 31 |
| pTKY862 | pLZ12-Km2 with aad(9) Sp resistance cassette | 31 |
| pTKY1041 | pLZ12-Km2 with 1,401-bp erm(B) fragment from S1 | This study |
| pTKY1042 | pT7Blue with disrupted erm(B) fragment by insertion of aad(9) Sp resistance cassette | This study |
| pTKY1109 | pUC18 with 328-bp tlrB fragment | This study |
| pTKY1110 | pUC18 with disrupted rlmAII fragment by insertion of an aad(9) Sp resistance cassette | This study |
| pTKY1111 | pLZ12-Km2 with 1,065-bp tlrB fragment from S1 | This study |
Antimicrobial susceptibility testing.
Susceptibility to antibiotics was determined by the serial 2-fold dilution method, using Mueller-Hinton agar plates supplemented with 5% lysed horse blood. Susceptibility or resistance of pneumococci to TEL and ERY was assessed in accordance with the recommendations of the Clinical and Laboratory Standards Institute (23).
Transformation.
Synthetic competence-stimulating peptide (CSP) 1 or 2 and the method of Iannelli and Pozzi (24) were used to transform S. pneumoniae S1 or TIGR4, respectively, into transformation-competent states.
Construction of erm(B) and tlrB disruption mutants.
Disruption of erm(B) was performed as follows. The pTKY858 plasmid, carrying part of the erm(B) gene, was cleaved with StyI. The overhanging ends were blunted with T4 polymerase and ligated to a fragment containing the spectinomycin resistance gene (Sp), which was generated from pTKY862 after digestion with BamHI followed by blunting with T4 DNA polymerase. The resulting plasmid, pTKY1042, was used to replace Δerm(B)::Sp in S. pneumoniae strain S1.
Disruption of the tlrB gene, which encodes RlmAII, was performed as follows. A fragment including part of the tlrB gene was amplified from chromosomal DNA prepared from S. pneumoniae strain S1 by a PCR using the forward primer 5′-CTAGAATTCAAATCTCAAGCCCAAACTTC-3′ and the reverse primer 5′-CCAATGCATGCCCAGTTGGGTTCAC-3′. The PCR product was digested with EcoRI-SphI, and the fragment was cloned into pUC18. The resultant plasmid, pTKY1109, was cleaved with HincII and ligated to the Sp fragment. The resulting plasmid, pTKY1110, was used to replace ΔtlrB::Sp in S. pneumoniae. Double-crossover events in all constructed mutants were assessed by PCR and Southern hybridization.
Construction of plasmids.
To construct the pTKY1041 plasmid, carrying erm(B), the gene was amplified from the chromosome of S1 by using the primers erm(B)LP3-EcoRI-F (5′-CGGAATTCCTTGCTTCGTTG-3′) and erm(B)-R5-BamHI (5′-CGCGGATCCTTAGCTCCTTGGAA-3′). The generated fragment was cleaved with EcoRI at the 5′ end and BamHI at the 3′ end and then cloned into the pLZ12-Km2 vector. To construct the pTKY1111 plasmid, carrying tlrB, the tlrB gene was amplified from the chromosome of S1 by using the primers rlmAII-XhoI-F (5′-CTGTACTCGAGTACGGCAAGGCGACG-3′) and rlmAII-ApaI-R (5′-GGTTTGGGGCCCTGTTCTTATGCGTTTTG-3′). The generated fragment was cleaved with XhoI at the 5′ end and ApaI at the 3′ end and then cloned into the pLZ12-Km2 vector.
Next-generation sequencing and data analysis.
Relative mutations in the bacterial genome were estimated by sequencing the genomic DNA samples extracted from each strain. Total DNA was extracted by the method of Blue and Mitchell (25). For preparation of the library DNA, 500 ng of each total DNA was sheared to 800 bp by Covaris (M&S). The sheared DNA products were purified using a MiniElute PCR purification kit (Qiagen) and then were used as templates for pyrosequencing with a GS Junior platform (Roche). The data were treated by a data analysis pipeline. Both reads from S1 and Sp36 were assembled into contigs by the de novo assembler. The nucleotide sequence polymorphisms among these strains were listed by mapping sequence reads on the assembled contigs with a reference mapper program. We collected nucleotide substitutions meeting the following 2 conditions: (i) the substitution was supported by >5 sequencing reads, and (ii) >90% of the reads supported the substitution. To find substitutions affecting protein function, we predicted open reading frames (ORFs) on the assembled contigs by using the RAST Web server, version 3.0 (http://rast.nmpdr.org/), and annotated the relationship between substitutions and ORFs. Correspondence between substitutions and ORFs was assigned by a custom Perl script implemented with Bioperl 1.6 (26).
DMS modification of ribosomes in vivo.
Dimethyl sulfate (DMS) modification of ribosomes in vivo was based on a previously published protocol (27, 28). Bacterial cells of strain S1 were grown in BHI-Y broth to the exponential growth phase. TEL (25 μM) was added to one aliquot of each culture for 1 h, followed by collection of the cells by centrifugation. Each pellet was resuspended in 80 μl DMS buffer (50 mM sodium cacodylate, pH 7.0, 1 mM EDTA), with or without TEL. DMS solution was added (final concentration, ∼50 mM) to each culture, and the cultures were incubated for 45 min. Reactions were stopped by the addition of 20 μl ice-cold stop buffer (1.5 M sodium acetate, pH 7.0, 1 M β-mercaptoethanol). Cells were harvested at 4°C for extraction of total RNAs and primer extension as described below.
Primer extension to detect methylated G748 and dimethylated A2058 in 23S rRNA.
The degree of methylation of each RNA was assayed by a primer extension method initially described by Morgan and colleagues (29). Briefly, bacterial cultures were grown in BHI-Y broth to the exponential growth phase. Total RNAs were extracted from bacterial cultures by use of RNeasy minikits (Qiagen). Two micrograms of total RNA, a 400 nM concentration of an oligonucleotide primer containing 5′-linked fluorescein isothiocyanate (FITC), and 500 μM (each) dATP, dCTP, dTTP, and ddGTP for G748 or dGTP, dCTP, dTTP, and ddATP for A2058 were mixed, heated to 95°C for 1 min, and cooled to allow annealing at 53°C. A mixture containing SuperScript III reverse transcriptase (Invitrogen) was added, and the reaction was continued at 53°C for an additional 20 min. The primers were complementary to 23S rRNA bases 756 to 790 (5′-TTTGGAATTTCTCCGCTACCCACAAGTCATCCAAG-3′) for m1G748 and bases 2066 to 2101 (5′-CAGACACTCAATATCAAACTGCAGTAAAGCTCCATG-3′) for m2A2058. Reactions were terminated by the addition of Novex Tris-borate-EDTA (TBE)–urea sample buffer (Invitrogen) after incubation at 70°C for 15 min. Extension products were resolved in a 10% TBE-urea gel and visualized using a TyphoonFLA9000 photoimager (GE Healthcare).
Molecular modeling and molecular mechanics.
The three-dimensional (3-D) crystal structure of the 50S subunit of the 70S ribosome with TEL bound from E. coli (Protein Data Bank entry 3OAT) was used as a template for the initial structure. The methylated form of G748 was generated by the Leap module of the AMBER 11.0 program package. Each model was placed in a rectangular box filled with ∼775,000 TIP3P water molecules. The molecular mechanics calculations for methylated and unmethylated forms of G748 in 23S rRNA were carried out using the AMBER 11.0 program package, together with the AMBER parm10 force field and all_modrna08 force field. Partial charges for the TEL atoms were calculated using the quantum chemical calculation in GAUSSIAN 03 (Gaussian, Inc., Carnegie, PA) followed by reduced electrostatic potential fitting, implemented in the antechamber module of AMBER. The 2 systems were energetically minimized for 10,000 steps (5,000 steps of the steepest-descent method followed by 5,000 steps of the conjugate gradient method). A cutoff distance of 10 Å for nonbonded interactions was used. The π-CH interaction energy was estimated by the formula proposed by Yuki et al. (30).
RESULTS
Mutations in the regulatory region of erm(B) mRNA result in highly increased dimethylation of A2058 in 23S rRNA in an S. pneumoniae clinical isolate.
The S. pneumoniae isolate S1, which has reduced TEL susceptibility (MIC, 2 μg/ml), has mutations (A138G, T389C, C724T, and A1745T) in the 4 alleles encoding 23S rRNA and a mutation (S20N) in ribosomal protein L4, which have often been detected in many macrolide-resistant S. pneumoniae strains (31–33). Strain S1 also has an erm(B) gene in the Tn3872-like structure (Fig. 1A). Tn3872 is a Tn916-like conjugative transposon with an erm(B) gene, which is carried in a Tn917-like element (34). Tn917 carries an erm(B) gene (accession no. M11180) but does not mediate resistance to other antimicrobial agents, such as tetracycline (35). A 5′ leader sequence of 259 nucleotides that has the potential to encode a leader peptide [erm(B)L] of 27 amino acids is the regulatory region of erm(B) mRNA involved in mRNA stabilization and translational attenuation, through the formation of alternative mRNA structures (36). Since mutation in the regulatory region of erm(B) results in TEL resistance (21, 36), we sequenced the region carrying erm(B). Two nucleotide differences [C242G and T253G; nucleotide numbers in erm(B) mRNA (36)] were found (Fig. 1A and B). A 182-bp deletion in the region upstream of erm(B) [positions −87 to −269 relative to the ATG start codon of erm(B)L] that included the −35 region of the putative promoter was found (Fig. 1A).
Fig 1.
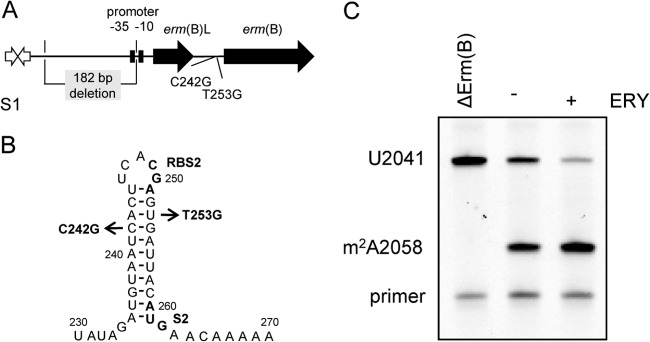
Effects of mutations in the erm(B) region on dimethylation at A2058 in 23S rRNA. (A) Diagram representing the erm(B) region in Tn3872 and the mutant erm(B) region of S1. (B) Secondary structure of translationally inactive form of the regulatory region of erm(B) mRNA and mutations detected in the parental strain S1. RBS2, ribosome-binding site; S2, start codon of the methyltransferase structural gene. (C) Levels of dimethylation at A2058 in 23S rRNAs extracted from strain S1 cultured with and without ERY. Dimethylated A2058 was detected by reverse transcriptase extension from bases 2066 to 2101. Dimethylation at A2058 aborts the extension.
To examine the contribution of the erm(B) region to the reduced TEL sensitivity of strain S1, we constructed an erm(B)-disrupted mutant by inserting the spectinomycin resistance gene aad(9) into erm(B) of strain S1 and determined the TEL MIC of the resultant mutant, Sp182. Disruption of erm(B) resulted in a significant decrease in the TEL MIC, to the level for strain ATCC 49619 (<0.03 μg/ml), which was used as a standard drug-susceptible strain. TEL sensitivity was complemented when the erm(B) gene was provided by plasmid pTKY1041. To define the essential nature of erm(B) for the low TEL susceptibility of strain S1, the laboratory strain TIGR4, which is susceptible to all antibiotics, was also transformed with S1 erm(B). Susceptibility testing of the transformants showed that TIGR4 derivatives with pTKY1041 had low TEL sensitivity (MIC, 1 μg/ml), suggesting that an erm(B) gene with nucleotide alterations (C247G and T253G) and a 182-bp deletion determines the low TEL susceptibility of strain S1.
The increased level of A2058 dimethylated by Erm in 23S rRNA is related to an increase of TEL resistance (37, 38). The methyltransferase encoded by erm(B) is constitutively or inducibly synthesized (6). It is necessary to constitutively synthesize Erm(B) for TEL resistance, because TEL does not induce the synthesis of methyltransferases (39). To show whether A2058 in the 23S rRNA of strain S1 is constitutively dimethylated by Erm(B), the levels of dimethylation at A2058 in strain S1 cultured with and without ERY, which acts as an inducer of erm(B) (36), were assayed by a primer extension assay. Dimethylation at A2058 arrests the progress of reverse transcription at position 2059, whereas transcription on unmethylated or monomethylated A2058 proceeds until it is terminated by incorporation of ddATP at U2041. A large percentage (71.0% ± 8.4%) of the A2058 in strain S1 was dimethylated even in the absence of ERY (Fig. 1C), suggesting that there was constitutive expression of erm(B). However, the erm(B) gene seems to continue to be inducible, because ERY exposure increased the level of dimethylation to 76.7% ± 9.7%. This slight increase in dimethylation by ERY exposure did not increase the TEL MIC (2 μg/ml). In strain TIGR4 harboring the pTKY1041 plasmid, the level of constitutive dimethylation at A2058 was also similar to that in strain S1 (data not shown). These results suggest that the highly increased dimethylation at A2058 in 23S rRNA due to mutations in the regulatory region of erm(B) reduces TEL sensitivity in S. pneumoniae strain S1.
Isolation of TEL-resistant mutants of clinical isolate S1.
TEL-resistant mutants were generated from strain S1. A total of 16 mutants were isolated from strain S1 (MIC, 2 μg/ml) by selection with 8 μg/ml (10 mutants; Sp32 to Sp41) and 16 μg/ml (6 mutants; Sp42 to Sp47) of TEL. All the mutants had increased MICs (16 to 32 μg/ml) (Table 3). ERY exposure did not increase the TEL MIC of any of the mutants. To examine whether the increased resistance to TEL was stable, the mutants were subcultured in drug-free BHI-Y broth at 37°C and 5% CO2 in air for 10 generations. The mutants were stable, such that subculturing them without the selective pressure of TEL did not generate derivatives with decreased TEL MICs. The primer extension assay showed that the level of dimethylated A2058 in each TEL-resistant mutant was similar to that in strain S1 under conditions both with and without ERY (Table 3), suggesting that a mutation unrelated to erm(B) function results in high-level TEL resistance.
Table 3.
Mutations in RlmAII proteins of TEL-resistant S. pneumoniae strains
| Mutation in RlmAII | Strain | MICa (μg/ml) |
Level (%) of dimethylation at A2058b |
||
|---|---|---|---|---|---|
| −ERY | +ERY | −ERY | +ERY | ||
| Wild type | S1 | 2 | 2 | 71.0 ± 8.4 | 76.7 ± 9.7 |
| Cys23Arg (67tGT69 → cGT) | Sp44 | 32 | 32 | 87.5 ± 5.6 | 81.9 ± 13.2 |
| Cys36Arg (106tGC108 → cGC) | Sp47 | 32 | 32 | 77.3 ± 2.2 | 66.7 ± 29.7 |
| Leu113Pro (337CtA339 → CcA) | Sp42, Sp43 | 32 | 32 | 90.4 ± 7.7 | 82.6 ± 16.4 |
| Ala135Thr (403gCT405 → aCT) | Sp39 | 32 | 32 | 89.8 ± 7.9 | 75.7 ± 17.2 |
| Leu164Pro (490CtG491 → CcG) | Sp33 | 16 | 16 | 69.2 ± 6.3 | 94.2 ± 9.4 |
| Leu165Pro (492CtT494 → CcT) | Sp37 | 32 | 32 | 79.8 ± 17.5 | 70.8 ± 7.2 |
| Arg200Cys (598cGT600 → tGT) | Sp36, Sp46 | 32 | 32 | 82.5 ± 9.0 | 80.8 ± 14.8 |
| Tyr212Cys (634TaT636 → TgT) | Sp32, Sp34 | 16 | 32 | 91.2 ± 5.7 | 81.7 ± 11.4 |
| Leu248Pro (739CtA741 → CcA) | Sp45 | 32 | 32 | 77.1 ± 7.8 | 80.27 ± 7.5 |
| Trp142stop codon (424TgG426 → TaG) | Sp38 | 16 | 16 | 75.8 ± 11.4 | 80.9 ± 10.0 |
| Frame shift by insertion of an A between nt 355 and 356 | Sp35, Sp40, Sp41 | 32 | 32 | 87.9 ± 7.9 | 82.6 ± 8.8 |
| ΔRlmAII | Sp274 | 16 | 16 | 73.7 ± 10.0 | 85.7 ± 11.5 |
Values for at least 3 independent experiments are given.
Quantification of the band corresponding to dimethylation at A2058 relative to the total band. Mean values and standard deviations for at least 3 independent experiments are given.
To determine mutations involved in increased TEL resistance, whole-genome sequencing of strains S1 and Sp36 was performed by next-generation sequencing technology, and the sequences of ORFs were analyzed for the two strains. As a result, 2 mutations were found in Sp36. One mutation was in a gene encoding a glycosyltransferase, leading to an Ala21Val alteration. The genomic sequences that have been reported show that the gene encoding the glycosyltransferase is not conserved among S. pneumoniae strains, suggesting that this mutation may be unrelated to the common mechanism of TEL resistance. The other was in the tlrB gene, encoding RlmAII, which is a methyltransferase; this mutation results in an Arg200Cys alteration. To detect alterations in RlmAII proteins from other TEL-resistant mutants, DNA fragments containing the tlrB genes from TEL-resistant mutants were generated by PCR amplification and sequenced. Each TEL-resistant mutant had one nucleotide difference in the tlrB gene (Table 3). The tlrB gene consists of 849 nucleotides (from ATG to TAA) which are translated to a polypeptide of 282 amino acid residues with a molecular mass of 35.1 kDa. In strain Sp38, a G752A difference changed Trp142 to a stop codon. In strains Sp35, Sp40, and Sp41, one adenine base insertion between nucleotides 355 and 356 generated a frameshift mutation creating amino acid changes in residues 121 to 125, with a stop codon at residue 126. In the other strains, each point mutation caused one amino acid change (Table 3). RlmAII methylates the N-1 position of nucleotide G748 in 23S rRNA domain II of S. pneumoniae (40). To reveal the level of N-1 methylation at G748 in TEL-resistant mutants, total RNAs were extracted from the parental strain S1, TEL-resistant mutants, and an RlmAII-deficient mutant, Sp274, which was constructed by inserting aad(9) into the tlrB gene of strain S1. The synthesis of a cDNA strand was initiated by annealing an oligonucleotide to positions 756 to 790 of the 23S rRNA. The reverse transcriptase stops synthesizing the DNA strand when it reaches the nucleotide preceding an m1G nucleotide, since the N-1 of guanosine required for proper Watson-Crick base pairing to a cytosine is blocked by a methyl group (29). In the absence of methylation at position 748, the reverse transcriptase stops synthesizing the DNA strand when it reaches the cytosine at nucleotide 744, because of the presence of ddGTP in the mixture. The top band in the strain S1 lane in Fig. 2 corresponds to m1G748, whereas the top band in the RlmAII-deficient mutant Sp274 lane corresponds to C744. For all TEL-resistant strains, bands corresponding to C744 appeared (Fig. 2). These results indicate that the function of RlmAII disappeared in TEL-resistant mutants.
Fig 2.
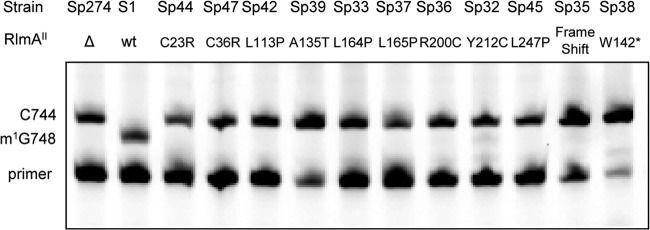
Levels of methylation at G748 in 23S rRNAs extracted from TEL-resistant S. pneumoniae strains. rRNAs were sequenced by reverse transcriptase extension from bases 756 to 790. Methylation at G748 N-1 aborts the extension.
To confirm that the inactivation of RlmAII confers TEL resistance, we determined the TEL MIC of the RlmAII-deficient mutant Sp274 derived from strain S1 (Table 3). Susceptibility testing showed that disruption of RlmAII resulted in resistance of S. pneumoniae to TEL (MIC, 16 μg/ml). TEL resistance due to RlmAII disruption was complemented (MIC, 1 μg/ml) in strain Sp274 when the pTKY1111 plasmid, carrying tlrB of strain S1, was provided, suggesting that RlmAII is involved in the TEL susceptibility of strain S1. On the other hand, the RlmAII disruption in the laboratory strain TIGR4 did not cause TEL resistance, i.e., both mutant strain Sp303 and parental strain TIGR4 showed TEL MICs of <0.03 μg/ml. From these results, we conclude that RlmAII functioning is involved in low TEL susceptibility of S. pneumoniae and that the loss of function in combination with the constitutive dimethylation of A2058 by Erm(B) confers high-level TEL resistance in S. pneumoniae.
The methyl group at the N-1 position of G748 contributes to stable interaction of TEL with the ribosome.
The alkyl-aryl side chain of TEL increases the affinity of the ketolide scaffold for the ribosome by a factor of several hundred (10). A structural study of TEL binding to the 70S ribosome from E. coli showed that this side chain closely approaches the loop of hairpin 35 in domain II of 23S rRNA and establishes a stacking interaction with the A752-U2609 base pair (12). However, the orientation of this side chain is dependent on the nucleotide sequence of 23S rRNA (12, 41). In S. pneumoniae 23S rRNA, the sequence of the A752-U2609 base pair is conserved. A deletion of A752 leads to resistance to TEL (42). This suggests that the A752-U2609 base pair contributes to the proper interaction of the alkyl-aryl side chain with the S. pneumoniae 23S rRNA. To examine the interaction of TEL with the A752-U2609 base pair after dimethylation of A2058 by Erm(B), we probed the S1 ribosome in solution by RNA footprinting for TEL protection of A752, because binding of the antibiotic to E. coli and S. aureus ribosomes protects A752 from chemical modification with DMS (12). Nucleotide A752 was protected from chemical modification in the S1 ribosome treated with TEL (Fig. 3), indicating that A752 is involved in the specific binding of TEL, even after dimethylation of A2058 by Erm(B).
Fig 3.
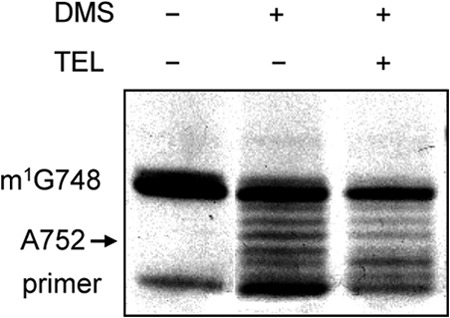
RNA footprinting for TEL protection of A752 in 23S rRNA containing dimethylated A2058. DMS was added to bacterial cultures of RlmAII-positive strain S1 after treatment with and without TEL. Reactions were quenched, and total RNAs were purified and used in primer extension assays to detect base modifications near the TEL interaction site in 23S rRNA. Unmodified RNA was used as a control (DMS −, TEL −). The gel shows the primer extension products from bases 756 to 790.
The effect of methylation at G748 was investigated theoretically by comparing two energetically optimized conformations of TEL-binding regions, with and without a methyl group at G748 (Fig. 4). In the energetically minimized structure of the unmethylated-G748 model, the distances from the center of the 5-member ring and that of the 6-member ring of A752 to the center of the aromatic ring of the alkyl-aryl group of TEL were both 3.9 Å (Fig. 4A). In the methylated-G748 model, these distances were estimated to be 3.9 Å and 3.7 Å, respectively (Fig. 4B). The methyl group of G748 could push the alkyl-aryl group of TEL toward the aromatic rings of A752. As a result, tighter π-π packing between the alkyl-aryl ring of TEL and the aromatic rings of A752 was formed in the methylated-G748 model than in the unmethylated-G748 model. In contrast, 2 hydrogen bonds, formed by one of the hydrogen atoms on the amino group at the N-6 position of A2058 bonding with the oxygen atom of a hydroxyl group and the nitrogen atom of the N(CH3)2 group on the cladinose sugar of TEL, were not affected by methylation of G748 (Fig. 4). This suggests that the affinity of TEL for the ribosome decreases when G748 is not methylated due to the inactivation of RlmAII. Furthermore, the methyl group of G748 pointing toward the alkyl-aryl group of TEL also formed a π-CH interaction with the aromatic ring of the alkyl-aryl group. The distance between the carbon atom of the methyl group and the center of the aromatic ring of the alkyl-aryl group was estimated to be 4.0 Å, corresponding to an interaction energy of −0.61 kcal/mol (Fig. 4B). This direct interaction between the methyl group of G748 and TEL may also contribute to stabilization of TEL binding to the ribosome.
Fig 4.
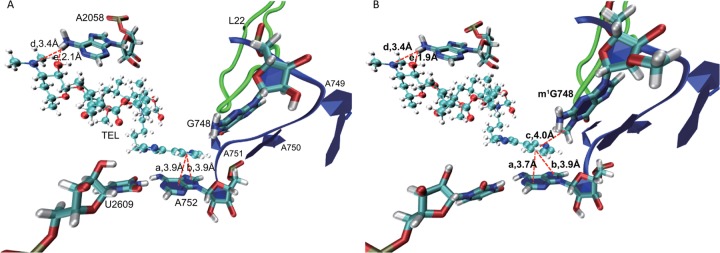
Energetically minimized structures of TEL binding to the 50S subunit of the ribosome containing unmethylated (A) and methylated (B) G748. Residues 748 to 752 in hairpin 35 are represented by a ribbon model (blue). Ribosomal protein L22 is shown by a tube model (green). TEL is represented by a ball-and-stick model. Dashed lines show the distances from the center of the aromatic ring of the alkyl-aryl chain of TEL to the center of each aromatic ring of A752 (a and b) and to the carbon atom of the methyl group at the N-1 position of G748 (c) and from a hydrogen atom of the amino group at the N-6 position of A2058 to the nitrogen atom (d) and to the oxygen atom of the hydroxyl oxygen group (e) on the cladinose sugar of TEL.
DISCUSSION
Inactivation of the methyltransferase RlmAII, which methylates the N-1 position of nucleotide G748, located in hairpin 35 of domain II of 23S rRNA, results in increased resistance to TEL when A2058 is constitutively dimethylated by Erm(B) in S. pneumoniae. Constitutive dimethylation of A2058 by Erm(B) causes low susceptibility to TEL in S. pneumoniae (Fig. 1). The central macrolactone ring of TEL establishes hydrophobic interactions with nucleotides 2057, 2058, and 2611 in domain V, which partially form the tunnel wall on the side of the PTC (3). Both the oxygen and nitrogen atoms of the cladinose sugar of TEL form hydrogen bonds with one of the hydrogen atoms of the amino group on A2058 (Fig. 4) (12). Each hydrogen atom of the amino group of A2058 may contribute to these interactions. In the case of monomethylation, one hydrogen atom maintains the interactions with TEL. Therefore, constitutive dimethylation is necessary for resistance and low susceptibility to TEL.
We found 2 nucleotide differences (C242G and T253G) in the regulatory region of the erm(B) gene of strain S1 (Fig. 1B). Two stem-loop structures are formed in the regulatory region of erm(B) mRNA and are altered due to ERY binding to the ribosome (36). A stem-loop downstream of these structures sequesters the translational initiation region of the methylase structural gene. Thus, it is necessary for the translational initiation of the methylase to collapse this stem-loop. Both nucleotide differences are located in the downstream stem-loop (Fig. 1B). A T253A nucleotide difference has been identified in laboratory-derived mutants of S. pneumoniae showing resistance to TEL (19) and in clinical isolates of TEL-resistant enterococci (36). Min et al. (36) showed that the T253A mutation increases the minimum free energy of the stem-loop structure from −13.2 to −8.5 kcal/mol, which results in destabilization of the structure. The minimum free energy of the structure with the C242G and T253G changes is increased further. Therefore, these differences result in stable expression of Erm(B), leading to an increase in the dimethylation level of A2058 and in low susceptibility to TEL. Furthermore, a 182-bp deletion was found in the upstream region of erm(B) compared with the sequence of Tn3872 (Fig. 1A). The −35 region of the erm(B) promoter was included in the deleted region (35). Transcription of the erm(B) gene of plasmid pIP1527 starts from 3 different sites following 3 overlapping promoters, which are recognized by both E. coli and Bacillus subtilis RNA polymerases (43). Two promoters are also located within the upstream region of erm(B) of strain S1. Thus, the erm(B) gene of strain S1 could be transcribed from these promoters.
The loop in hairpin 35 in domain II of 23S rRNA, which consists of 8 nucleotides (745GUUGAAAA752), is close to the NPET construction formed by ribosomal proteins L4 and L22 and portions of domain V of 23S rRNA (Fig. 4) (3). The side chains of TEL and a 16-member ring macrolide, tylosin, can also interact with hairpin 35 in addition to interacting with domain V. Thus, the interaction of both TEL and tylosin with the ribosome persists even after monomethylation of A2058 by Erm (11, 44). However, interaction of TEL with hairpin 35 differs from that of tylosin (45). The C14-linked mycinose of tylosin interacts with 2 nucleotides, G748 and A751, in hairpin 35 (46). Thus, methylation of G748 by RlmAII, which was first identified in Streptomyces fradiae, an organism that naturally produces tylosin (47), inhibits the binding of tylosin to the ribosome when A2058 is monomethylated by Erm (44). Similarly, in S. pneumoniae, methylation of G748 by RlmAII is assumed to inhibit the binding of tylosin, because disruption of RlmAII increases sensitivity of the S. pneumoniae strain to tylosin when nucleotide A2058 is methylated by Erm (our unpublished data). In the interaction with TEL, the methylation of G748 by RlmAII may increase the proper interaction of the alkyl-aryl chain with hairpin 35, even after dimethylation of A2058 (Fig. 3). The alkyl-aryl ring of TEL is locked in its proper place because of the interaction with the aromatic rings of A752 by π-π packing (Fig. 4) (12). In the S. pneumoniae ribosome, the π-CH interaction can be formed between the alkyl-aryl ring and the methyl group of G748 (Fig. 4B). Moreover, π-π interactions within A752 are more stable in the methylated-G748 ribosome model than in the unmethylated-G748 ribosome model (Fig. 4). Thus, constitutive dimethylation of A2058 by Erm alone is insufficient to confer TEL resistance in S. pneumoniae.
Hairpin 35 may be involved in the interaction of ERY (11, 48, 49). However, mutations and a deletion of A752 do not affect the ERY MIC in E. coli (45). In contrast, A752 deletion in S. pneumoniae 23S rRNA results in a significant increase in MICs of the other macrolides, in addition to TEL (42). Increased MICs of clindamycin and pristinamycin are moderate compared with those of the other macrolides (42). This result suggests that hairpin 35 in S. pneumoniae 23S rRNA is involved in the direct interaction of the central macrolactone ring.
Comparison of the amino acid sequences of RlmAI and RlmAII indicates that these enzyme classes are homologous; ∼29% of the residues are conserved across species (40, 50). In this study, several mutants generating an amino acid change in RlmAII were isolated, all of which reduced RlmAII activity (Fig. 2). Each amino acid function was considered according to the results of a structural study of RlmAI (50). Residues Cys23 and Cys36 are conserved among RlmA classes. They are located within a Zn-binding domain and coordinate with a single Zn ion (50). Residues Leu113, Ala135, Arg200, and Leu247 are located in α-helices α3, α4, α6, and α7. Residues Leu164 and Leu165 are located in the β8 β-strand. RlmAI works as a dimer, because it contains a wide, W-shaped cleft—a putative binding site for the rRNA substrate. Helices α3 and α4 and the β8 strand may be involved in the maintenance of this cleft. Helices α6 and α7 may be involved in the interaction within the dimer. Therefore, amino acid changes within these regions reduce RlmAII activity because of weaker binding to rRNA. Despite functional similarity, the target nucleotide of RlmAII is different from that of RlmAI (40). In hairpin 35 in E. coli 23S rRNA, other modifications are known in addition to G745 methylation by RlmAI (3). Nucleotides U746 and U747 are modified to pseudouridine by RluA and via C-5 methylation by RlmC, respectively (51, 52). In some bacterial species, hairpin 35 is unmodified (53). BLAST searches show that both RluA and RlmC homologs are identified in S. pneumoniae. Alignments of these proteins between S. pneumoniae and E. coli show few similarities. YefA, a homolog of RlmC in Bacillus subtilis, methylates U747 and U1939 (54). Therefore, hairpin 35 and other regions of S. pneumoniae 23S rRNA may be modified in a different pattern from those of other bacterial species, which might affect the binding of macrolides and ketolides in S. pneumoniae.
ACKNOWLEDGMENTS
We thank N. Kitagawa, Y. Kuroe, K. Sone, K. Nagamatsu, and A. Kimura for technical assistance. Theoretical calculations were partly performed using the Research Centre for Computational Science, Okazaki, Japan.
This work was supported by a grant (grant H24-Shinkou-Ippan-010) from the Ministry of Health, Labor and Welfare of Japan.
Footnotes
Published ahead of print 28 May 2013
REFERENCES
- 1. Kaczanowska M, Ryden-Aulin M. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71:477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chow CS, Lamichhane TN, Mahto SK. 2007. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2:610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kannan K, Mankin AS. 2011. Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Ann. N. Y. Acad. Sci. 1241:33–47 [DOI] [PubMed] [Google Scholar]
- 4. Poehlsgaard J, Douthwaite S. 2005. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 3:870–881 [DOI] [PubMed] [Google Scholar]
- 5. Skinner R, Cundliffe E, Schmidt FJ. 1983. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J. Biol. Chem. 258:12702–12706 [PubMed] [Google Scholar]
- 6. Weisblum B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fogarty PF, Dunbar CE. 2003. Resetting the immune system in immune thrombocytopenic purpura: immunoablative strategies. Clin. Adv. Hematol. Oncol. 1:365–371 [PubMed] [Google Scholar]
- 8. Van Bambeke F, Harms JM, Van Laethem Y, Tulkens PM. 2008. Ketolides: pharmacological profile and rational positioning in the treatment of respiratory tract infections. Expert Opin. Pharmacother. 9:267–283 [DOI] [PubMed] [Google Scholar]
- 9. Bozdogan B, Appelbaum PC, Kelly LM, Hoellman DB, Tambic-Andrasevic A, Drukalska L, Hryniewicz W, Hupkova H, Jacobs MR, Kolman J, Konkoly-Thege M, Miciuleviciene J, Pana M, Setchanova L, Trupl J, Urbaskova P. 2003. Activity of telithromycin and seven other agents against 1034 pediatric Streptococcus pneumoniae isolates from ten central and eastern European centers. Clin. Microbiol. Infect. 9:653–661 [DOI] [PubMed] [Google Scholar]
- 10. Hansen LH, Mauvais P, Douthwaite S. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31:623–631 [DOI] [PubMed] [Google Scholar]
- 11. Xiong L, Shah S, Mauvais P, Mankin AS. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633–639 [DOI] [PubMed] [Google Scholar]
- 12. Dunkle JA, Xiong L, Mankin AS, Cate JH. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U. S. A. 107:17152–17157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maglio D, Nicolau DP, Nightingale CH. 2003. Impact of pharmacodynamics on dosing of macrolides, azalides, and ketolides. Infect. Dis. Clin. North Am. 17:563–577 [DOI] [PubMed] [Google Scholar]
- 14. Farrell DJ, Felmingham D. 2004. Activities of telithromycin against 13,874 Streptococcus pneumoniae isolates collected between 1999 and 2003. Antimicrob. Agents Chemother. 48:1882–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jalava J, Kataja J, Seppala H, Huovinen P. 2001. In vitro activities of the novel ketolide telithromycin (HMR 3647) against erythromycin-resistant Streptococcus species. Antimicrob. Agents Chemother. 45:789–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farrell DJ, Couturier C, Hryniewicz W. 2008. Distribution and antibacterial susceptibility of macrolide resistance genotypes in Streptococcus pneumoniae: PROTEKT year 5 (2003–2004). Int. J. Antimicrob. Agents 31:245–249 [DOI] [PubMed] [Google Scholar]
- 17. Reinert RR, van der Linden M, Al-Lahham A. 2005. Molecular characterization of the first telithromycin-resistant Streptococcus pneumoniae isolate in Germany. Antimicrob. Agents Chemother. 49:3520–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim SH, Song JH, Chung DR, Thamlikitkul V, Yang Y, Wang H, Lu M, So TM, Hsueh PR, Yasin RM, Carlos CC, Pham HV, Lalitha MK, Shimono N, Perera J, Shibl AM, Baek JY, Kang CI, Ko KS, Peck KR. 2012. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob. Agents Chemother. 56:1418–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walsh F, Willcock J, Amyes S. 2003. High-level telithromycin resistance in laboratory-generated mutants of Streptococcus pneumoniae. J. Antimicrob. Chemother. 52:345–353 [DOI] [PubMed] [Google Scholar]
- 20. Wolter N, Smith AM, Low DE, Klugman KP. 2007. High-level telithromycin resistance in a clinical isolate of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51:1092–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wolter N, Smith AM, Farrell DJ, Northwood JB, Douthwaite S, Klugman KP. 2008. Telithromycin resistance in Streptococcus pneumoniae is conferred by a deletion in the leader sequence of erm(B) that increases rRNA methylation. Antimicrob. Agents Chemother. 52:435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Douthwaite S, Crain PF, Liu M, Poehlsgaard J. 2004. The tylosin-resistance methyltransferase RlmAII (TlrB) modifies the N-1 position of 23S rRNA nucleotide G748. J. Mol. Biol. 337:1073–1077 [DOI] [PubMed] [Google Scholar]
- 23. CLSI 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI M100-S17, vol 27 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 24. Iannelli F, Pozzi G. 2004. Method for introducing specific and unmarked mutations into the chromosome of Streptococcus pneumoniae. Mol. Biotechnol. 26:81–86 [DOI] [PubMed] [Google Scholar]
- 25. Blue CE, Mitchell TJ. 2003. Contribution of a response regulator to the virulence of Streptococcus pneumoniae is strain dependent. Infect. Immun. 71:4405–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehvaslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12:1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moore SD, Sauer RT. 2008. Revisiting the mechanism of macrolide-antibiotic resistance mediated by ribosomal protein L22. Proc. Natl. Acad. Sci. U. S. A. 105:18261–18266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moazed D, Robertson JM, Noller HF. 1988. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334:362–364 [DOI] [PubMed] [Google Scholar]
- 29. Sigmund CD, Ettayebi M, Borden A, Morgan EA. 1988. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 164:673–690 [DOI] [PubMed] [Google Scholar]
- 30. Yuki H, Tanaka Y, Hata M, Ishikawa H, Neya S, Hoshino T. 2007. Implementation of π-π interactions in molecular dynamics simulation. J. Comput. Chem. 28:1091–1099 [DOI] [PubMed] [Google Scholar]
- 31. Takaya A, Kitagawa N, Kuroe Y, Endo K, Okazaki M, Yokoyama E, Wada A, Yamamoto T. 2010. Mutational analysis of reduced telithromycin susceptibility of Streptococcus pneumoniae isolated clinically in Japan. FEMS Microbiol. Lett. 307:87–93 [DOI] [PubMed] [Google Scholar]
- 32. Tait-Kamradt A, Davies T, Cronan M, Jacobs MR, Appelbaum PC, Sutcliffe J. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cattoir V, Merabet L, Legrand P, Soussy CJ, Leclercq R. 2007. Emergence of a Streptococcus pneumoniae isolate resistant to streptogramins by mutation in ribosomal protein L22 during pristinamycin therapy of pneumococcal pneumonia. J. Antimicrob. Chemother. 59:1010–1012 [DOI] [PubMed] [Google Scholar]
- 34. McDougal LK, Tenover FC, Lee LN, Rasheed JK, Patterson JE, Jorgensen JH, LeBlanc DJ. 1998. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2312–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shaw JH, Clewell DB. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Min YH, Kwon AR, Yoon EJ, Shim MJ, Choi EC. 2008. Translational attenuation and mRNA stabilization as mechanisms of erm(B) induction by erythromycin. Antimicrob. Agents Chemother. 52:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu M, Douthwaite S. 2002. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob. Agents Chemother. 46:1629–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Douthwaite S, Jalava J, Jakobsen L. 2005. Ketolide resistance in Streptococcus pyogenes correlates with the degree of rRNA dimethylation by Erm. Mol. Microbiol. 58:613–622 [DOI] [PubMed] [Google Scholar]
- 39. Ackermann G, Rodloff AC. 2003. Drugs of the 21st century: telithromycin (HMR 3647)—the first ketolide. J. Antimicrob. Chemother. 51:497–511 [DOI] [PubMed] [Google Scholar]
- 40. Liu M, Douthwaite S. 2002. Methylation at nucleotide G745 or G748 in 23S rRNA distinguishes Gram-negative from Gram-positive bacteria. Mol. Microbiol. 44:195–204 [DOI] [PubMed] [Google Scholar]
- 41. Bulkley D, Innis CA, Blaha G, Steitz TA. 2010. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. U. S. A. 107:17158–17163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Canu A, Malbruny B, Coquemont M, Davies TA, Appelbaum PC, Leclercq R. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brisson-Noel A, Arthur M, Courvalin P. 1988. Evidence for natural gene transfer from gram-positive cocci to Escherichia coli. J. Bacteriol. 170:1739–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fish SA, Cundliffe E. 1996. Structure-activity studies of tylosin-related macrolides. J. Antibiot. (Tokyo) 49:1044–1048 [DOI] [PubMed] [Google Scholar]
- 45. Novotny GW, Jakobsen L, Andersen NM, Poehlsgaard J, Douthwaite S. 2004. Ketolide antimicrobial activity persists after disruption of interactions with domain II of 23S rRNA. Antimicrob. Agents Chemother. 48:3677–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117–128 [DOI] [PubMed] [Google Scholar]
- 47. Liu M, Kirpekar F, Van Wezel GP, Douthwaite S. 2000. The tylosin resistance gene tlrB of Streptomyces fradiae encodes a methyltransferase that targets G748 in 23S rRNA. Mol. Microbiol. 37:811–820 [DOI] [PubMed] [Google Scholar]
- 48. Garza-Ramos G, Xiong L, Zhong P, Mankin A. 2001. Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA. J. Bacteriol. 183:6898–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poulsen SM, Kofoed C, Vester B. 2000. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 304:471–481 [DOI] [PubMed] [Google Scholar]
- 50. Das K, Acton T, Chiang Y, Shih L, Arnold E, Montelione GT. 2004. Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site. Proc. Natl. Acad. Sci. U. S. A. 101:4041–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raychaudhuri S, Niu L, Conrad J, Lane BG, Ofengand J. 1999. Functional effect of deletion and mutation of the Escherichia coli ribosomal RNA and tRNA pseudouridine synthase RluA. J. Biol. Chem. 274:18880–18886 [DOI] [PubMed] [Google Scholar]
- 52. Madsen CT, Mengel-Jorgensen J, Kirpekar F, Douthwaite S. 2003. Identifying the methyltransferases for m5U747 and m5U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 31:4738–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mengel-Jorgensen J, Jensen SS, Rasmussen A, Poehlsgaard J, Iversen JJ, Kirpekar F. 2006. Modifications in Thermus thermophilus 23S ribosomal RNA are centered in regions of RNA-RNA contact. J. Biol. Chem. 281:22108–22117 [DOI] [PubMed] [Google Scholar]
- 54. Desmolaize B, Fabret C, Bregeon D, Rose S, Grosjean H, Douthwaite S. 2011. A single methyltransferase YefA (RlmCD) catalyses both m5U747 and m5U1939 modifications in Bacillus subtilis 23S rRNA. Nucleic Acids Res. 39:9368–9375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Okada N, Tatsuno I, Hanski E, Caparon M, Sasakawa C. 1998. Streptococcus pyogenes protein F promotes invasion of HeLa cells. Microbiology 144:3079–3086 [DOI] [PubMed] [Google Scholar]


