Abstract
In recent decades, pathogenic fungi have become a serious threat to human health, leading to major efforts aimed at characterizing new agents for improved treatments. Promising in this context are antimicrobial peptides produced by animals and plants as part of innate immune systems. Here, we describe an antifungal defensin, NaD1, with activity against the major human pathogen Candida albicans, characterize the mechanism of killing, and identify protection strategies used by the fungus to survive defensin treatment. The mechanism involves interaction between NaD1 and the fungal cell surface followed by membrane permeabilization, entry into the cytoplasm, hyperproduction of reactive oxygen species, and killing induced by oxidative damage. By screening C. albicans mutant libraries, we identified that the high-osmolarity glycerol (HOG) pathway has a unique role in protection against NaD1, while several other stress-responsive pathways are dispensable. The involvement of the HOG pathway is consistent with induction of oxidative stress by NaD1. The HOG pathway has been reported to have a major role in protection of fungi against osmotic stress, but our data indicate that osmotic stress does not contribute significantly to the adverse effects of NaD1 on C. albicans. Our data, together with previous studies with human beta-defensins and salivary histatin 5, indicate that inhibition of the HOG pathway holds promise as a broad strategy for increasing the activity of antimicrobial peptides against C. albicans.
INTRODUCTION
The yeast Candida albicans represents the most common cause of fungal infection in humans. This organism is commensal but can become pathogenic in immunocompromised individuals, causing serious life-threatening disease from which mortality can reach up to 40% (1–3). Relatively few options exist for treatment of systemic fungal disease, and the emergence of resistance is limiting these options further. This has created a need for new antifungal drugs (4). In this context, antimicrobial peptides represent a promising avenue (5).
Antimicrobial peptides are abundant throughout nature. They are generally small, cationic peptides that are components of the innate immune system in many organisms. They are part of the first line of defense against pathogens, including bacteria, viruses, and fungi, and operate by a range of diverse mechanisms (6, 7). Many of these peptides act solely by disrupting target membranes (8). However, more complicated mechanisms also exist, involving uptake into the cytoplasm and action within the target cell (9). In the case of fungal pathogens, several antimicrobial peptides have been investigated for inhibitory activity, including human beta-defensins, histatin 5, LL-37, and plant defensins (10–12). The mechanisms by which these polypeptides act against C. albicans vary considerably. Salivary histatin 5 induces fungal cell death via the release of cellular ATP and activation of P2X receptors (13), in a process that involves an interaction with cell wall proteins (14) and a potassium channel (15). Human beta-defensins display some shared mechanisms with histatin 5, yet synergy between some beta-defensins and histatin 5 indicates that not all targets are shared (16). The human cathelicidin LL-37 acts via disruption of the plasma membrane. Treatment of C. albicans with LL-37 causes fragmentation of the plasma membrane into vesicle-like structures and release of intracellular molecules (17). For a review of the mechanisms of action of naturally occurring antifungal peptides, see the work of van der Weerden et al. (18).
Our interest is in plant defensins that target fungal pathogens. Plant defensins are numerous and represent an excellent alternative to conventional antifungal therapies. These peptides are small (∼5-kDa), basic proteins which are expressed in many plant tissues, including seeds, leaves, flowers, and fruit (19). They have a highly conserved structure consisting of an α-helix and three β-strands that are linked by three disulfide bonds (19, 20). This stable structure is strengthened by a fourth disulfide bond that connects the N and C termini, forming a pseudocyclic backbone (19). The three-dimensional structure of plant defensins shares similarities with those of human and insect defensins (5, 19, 20). Despite this conserved structure, defensins have a wide variety of biological functions, and even those with antifungal activity have different mechanisms of action (7). RsAFP2, a defensin from radish, induces cell wall stress and septin mislocalization (21) as well as metacaspase-independent cell death (22). HsAFP1, from Heuchera sanguinea, also induces cell death through a mechanism that is dependent on the presence of functional mitochondria (23). Lipid binding is a crucial step in the activity of some defensins. RsAFP2 from Raphanus sativus and DmAMP1 from Dahlia merckii interact with sphingolipids (24, 25), whereas PsD1 from Pisum sativum binds to phosphatidylcholine (26). PvD1, from Phaseolus vulgaris, induces reactive oxygen and nitrogen oxide production as well as permeabilization of the fungal membrane (27). New defensins are being identified regularly, and the diversity of biological phenomena associated with their activity continues to expand. Understanding the mechanism by which these molecules act to inhibit fungal growth is essential to their development as antifungal therapeutics.
We have reported that the plant defensin NaD1, which is produced in the flowers of the ornamental tobacco Nicotiana alata (28), has potent antifungal activity against a number of filamentous fungal pathogens that have a severe impact on important agricultural crops (29). In the current study, we have examined the effects of NaD1 on the growth and survival of a major human pathogen, the yeast C. albicans. We show that NaD1 is internalized into C. albicans cells and that it exhibits fungicidal activity at least in part through an oxidative mechanism involving reactive oxygen species (ROS) production. We further show that the high-osmolarity glycerol (HOG) stress response pathway functions in fungal protection against NaD1, suggesting that modulation of this pathway might represent a means to augment the efficacy of NaD1 activity against C. albicans.
MATERIALS AND METHODS
Protein sources.
NaD1 was extracted from Nicotiana alata flowers based on the method described in reference 29. DmAMP1 was expressed using the methylotrophic yeast Pichia pastoris. A DNA fragment encoding the mature DmAMP1 along with a XhoI restriction endonuclease site and a KEX2 cleavage site at the 5′ end and a NotI restriction endonuclease site and a stop codon at the 3′ end was purchased from Genscript for cloning into the pPIC9 vector. The STE13 protease site between the KEX2 cleavage site and the mature protein was removed to prevent the addition of Glu-Ala repeats at the N terminus of DmAMP1. An alanine was added to the N terminus to ensure efficient cleavage by KEX2. DNA was then inserted into the pPIC9 plasmid via the XhoI and NotI restriction sites by standard protocols. The pPIC9 plasmid containing the gene of interest was linearized using the SalI restriction enzyme. Electrocompetent P. pastoris GS115 cells (Invitrogen) were prepared as described by Chang et al. (30), and linearized DNA was transformed into these cells using standard protocols. DmAMP1 was then expressed in buffered minimal medium according to the manufacturer's instructions and purified via cation-exchange chromatography and reverse-phase high-performance liquid chromatography (RP-HPLC). The protein concentration was determined using the bicinchoninic acid (BCA) protein assay (Pierce).
Fungal strains.
The libraries of C. albicans homozygous, transposon insertion mutants in genes affecting cell wall synthesis and structure, kinases, and transcription factors were from the Mitchell laboratory and are described in references (31 to 33). The mutants were constructed in the C. albicans strain background BWP17 (ura3::imm434/ura3::imm434 iro1/iro1::imm434 his1::hisG/his1::hisG arg4/arg4). The mutant in the HOG1 kinase is a homozygous deletion mutant (31). The libraries were obtained from the Fungal Genetics Stock Center (FGSC). Cryptococcus and Aspergillus strains were obtained from Dee Carter (The University of Sydney, Australia) and were as follows: Cryptococcus neoformans var. grubii H99, Cryptococcus neoformans var. neoformans JEC20 and JEC21 (isogenic strains differing in mating type; JEC20 is MATa and JEC21 is MATα), Cryptococcus gattii WM276 (molecular type VGI) and R265 (molecular type VGIIa), Aspergillus niger 5181, and Aspergillus flavus 5310. All Saccharomyces cerevisiae experiments were performed in the haploid BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) background. The standard condition for growth of yeast strains was growth in yeast extract-peptone-dextrose (YPD) at 30°C with constant shaking (250 rpm). For C. albicans mutant strains, YPD was supplemented with 80 μg/ml uridine. For the growth of Aspergillus strains, spores were added to half-strength potato dextrose agar (PDA; Becton, Dickinson) and grown at room temperature for 14 days.
For construction of complemented C. albicans strains, the HOG1 and PBS2 open reading frames plus promoter and terminator regions were amplified from genomic DNA of wild-type C. albicans (BWP17 strain background) and were cloned into the pDDB78 plasmid and integrated at the HIS1 locus of the respective mutant strains. As the control, an empty pDDB78 vector was integrated into the mutant genomes, to restore His1 prototrophy. For the library screens, the wild-type strain was DAY286 (ARG4+ URA3+ his1−), which was matched for markers with the library mutants, whereas the complemented strains and HIS1+ mutants were tested against the DAY185 reference strain (ARG4+ URA3+ HIS1+). Primers for amplification of PBS2 were TTCACACAGGAAACAGCTATGACCATGATTACGCCAAGCTTAAAACAGTTTCTAAATAGTCTG and TCGACCATATGGGAGAGCTCCCAACGCGTTGGATGCATAGGTCGAATTAATTAGACGTGTT.
The HOG1 gene was amplified using the primers TTCACACAGGAAACAGCTATGACCATGATTACGCCAAGCTCTTAAAGATTCATCCAATGATGG and TCGACCATATGGGAGAGCTCCCAACGCGTTGGATGCATAGCAGAAGACAATCTTTTGAACTAT.
Preparation of S. cerevisiae [rho0] cells was based on the procedure outlined in reference 34. Briefly, BY4741 cells were cultured in 5 ml YPD with 20 μg/ml ethidium bromide at 30°C in the absence of light for 3 days. Cells were plated on YPD agar. Respiratory deficiency was confirmed by lack of growth on the nonfermentable carbon source glycerol.
Growth inhibition and survival assays.
For C. albicans growth inhibition assays, cells were grown in YPD overnight (30°C, 250 rpm). Cells were counted using a hemocytometer and diluted to 5,000 cells/ml in half-strength potato dextrose broth (PDB; Becton, Dickinson). Diluted cells (80 μl) were added to the wells of a microtiter plate along with 20 μl of protein solution to give a 0 to 10 μM final concentration. Growth of cells was monitored by measuring absorbance at 595 nm in a SpectraMAX M5e plate reader (Molecular Devices), using a 9-well scan. Measurements were taken at t = 0 and t = 24 h during incubation at 30°C. For testing activity of NaD1 in serum or in the presence of ascorbate, the C. albicans DAY185 strain was used, and activity was tested in the presence or absence of 5% fetal calf serum (FCS) or 50 mM sodium ascorbate. For testing of activity at 37°C, the method was performed as described above but with 24 h of incubation at 30°C or 37°C. S. cerevisiae assays were performed using a similar protocol with an optical density at 600 nm (OD600) of 0.01 (approximately 3 × 105 cells/ml).
For Cryptococcus inhibition assays, cells were grown overnight in YPD (30°C, 250 rpm). Cells were counted with a hemocytometer and diluted to 1.5 × 106 cells/ml in half-strength PDB. The rest of the method was performed as described above.
For Aspergillus growth inhibition assays, spores were collected from PDA plates by flooding them with 20 ml half-strength PDB. The medium was removed from the plate, and the spore suspension was spun at 14,000 rpm for 15 min and resuspended in approximately 1 ml of half-strength PDB. Spores were counted using a hemocytometer and diluted to 5 × 104 spores/ml with half-strength PDB. The remainder of the method was performed as described above.
For survival assays, C. albicans DAY185 overnight cultures were diluted to an OD600 of 0.2 in half-strength PDB (10 ml) and incubated for 5 h (30°C, 250 rpm). Cells were then diluted to an OD600 of 1.0 and treated with NaD1 (2, 3.5, 5, 10, 20, and 50 μM) for 15 min (30°C, 250 rpm). Survival was determined by counting CFU of 1/1,000 and 1/10,000 dilutions after overnight growth on YPD plates at 30°C. For sorbitol survival assays, cells were pretreated for an hour with 1 M sorbitol in half-strength PDB (30°C, 250 rpm).
Labeling of NaD1 with BODIPY fluorescent tag.
NaD1 was labeled with BODIPY-FL-EDA (Life Technologies) essentially as described in reference 35. Briefly, NaD1 (1 mM) and BODIPY-FL-EDA (10 mM) were dissolved in 0.1 M MES buffer (2-morpholinoethanesulfonic acid, pH 4.5; Amresco), together with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC; Sigma; 2 mM). The solution was incubated at room temperature for 2 h. Insoluble material was removed by centrifugation at 13,000 rpm for 10 min before excess BODIPY-FL-EDA and EDC were removed using a 3,000-molecular-weight-cutoff (MWCO) Vivaspin 500 column (Sartorius). The protein concentration was determined using the BCA protein assay (Pierce).
Confocal microscopy of BODIPY-labeled NaD1 and DHR 123.
C. albicans DAY185 overnight cultures were diluted to an OD600 of 0.1 in half-strength PDB, and cells were treated with 5 μM propidium iodide (PI; Sigma) for 10 min prior to visualization. For the BODIPY-NaD1 experiments, 10 μM labeled NaD1 was added. For the dihydrorhodamine 123 (DHR 123; Molecular Probes) experiments, the probe was added at a 25 μM concentration along with native NaD1. Cells were monitored using a Zeiss LSM510/ConfoCor confocal microscope with argon and diode-pumped solid-state (DPSS) lasers. BODIPY or DHR 123 was excited at 488 nm, and emission was detected at 505 to 530 nm. PI was excited at 561 nm, and fluorescence was monitored at 575 to 615 nm. Images were captured using Zen2009 (Zeiss) software and analyzed using FIJI. Brightness and contrast for Fig. 3A were adjusted using the auto-brightness/contrast function of FIJI.
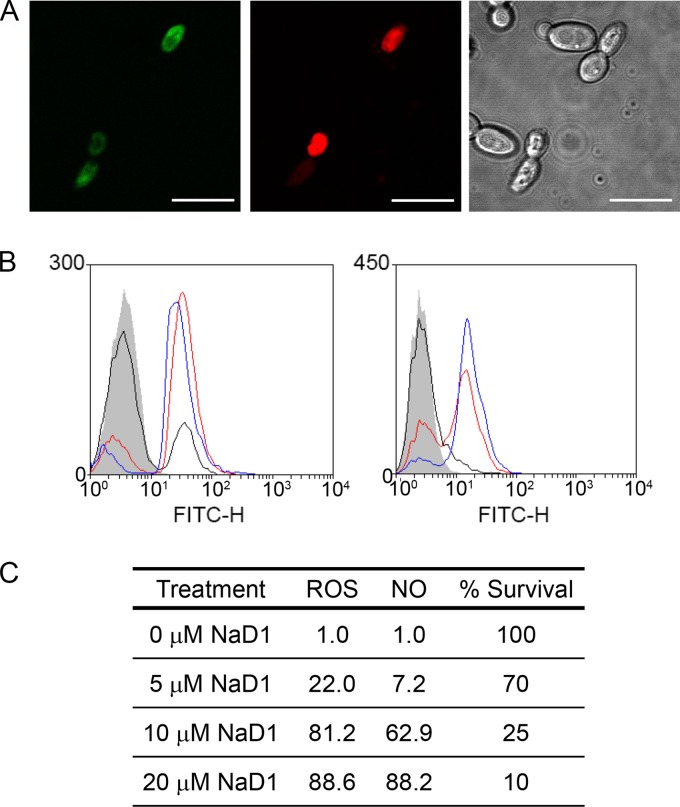
Fig 3.
Production of reactive oxygen species (ROS) and nitric oxide (NO) by C. albicans in response to treatment with NaD1. (A) Confocal microscopy showing ROS production (DHR 123, green fluorescence) and propidium iodide uptake (red fluorescence) in C. albicans cells after treatment with 10 μM NaD1 for 30 min. Scale bars, 10 μm. (B) Flow cytometry of cells showing ROS (DHR 123, left panel) and NO (DAF-FM, right panel) production in response to treatment with 5 μM (black lines), 10 μM (red lines), and 20 μM (blue lines) NaD1. A no-protein control is represented by gray shading. The vertical and horizontal axes represent cell counts and fluorescence intensity, respectively. Data are a representative example of three independent experiments, which gave equivalent results. FITC-H, fluorescein isothiocyanate. (C) The percentage of cells that produced ROS (DHR 123) and NO (DAF-FM) in response to treatment of C. albicans with 0, 5, 10, or 20 μM NaD1, calculated from flow cytometry. Data are a representative example of three independent experiments, which gave equivalent results. Cell survival assays were also performed at an OD600 of 0.1 to match the OD600 of cells used in flow cytometry analysis (n = 3) and are shown as percentages of the value for the no-protein control.
Screen of C. albicans mutant libraries.
C. albicans mutants were inoculated into YPD (100-μl cultures in microtiter plates) and grown overnight, at 30°C with constant shaking (250 rpm). Cells were then added to half-strength PDB (100 μl) to a concentration of about 5,000 cells/ml (1/20,000 dilution from overnight culture). NaD1 (in water) was added to a 2 μM or 5 μM final concentration. Plates were incubated for 24 h at 30°C, and growth was monitored by measuring OD595 using a SpectraMAX M5e plate reader (Molecular Devices). Growth of mutants was compared to that of no-protein controls. The screen was repeated three times, and positive hits were confirmed by using growth inhibition assays, as described above.
Detection of HOG1 phosphorylation in response to NaD1.
Overnight cultures of C. albicans DAY185 and hog1Δ/Δ cells were diluted to an OD600 of 0.2 in half-strength PDB and grown for 5 h at 30°C (250 rpm). Cells were then diluted to an OD600 of 1.0, and 10 ml of the cell suspension was treated with NaD1 (10 or 20 μM) or 1 M NaCl for 5, 10, or 15 min. Cells were pelleted by centrifugation (3,000 rpm, 5 min) and washed in 500 μl of cold Milli-Q water. The pellets were frozen with liquid nitrogen. Trichloroacetic acid (TCA; 20%, 100 μl) was added to the frozen pellet, and cells were disrupted by vortexing in the presence of glass beads. Glass beads were washed further with 500 μl 10% TCA. Precipitated proteins were pelleted by centrifugation at 14,000 rpm (5 min), and then the pellet was washed with 1 ml ice-cold acetone. Proteins were separated on Bio-Rad AnyKD mini-Tris-glycine gels and transferred to polyvinyl difluoride (PVDF) membranes. Membranes were probed with phospho-p38 mitogen-activated protein kinase (MAPK) (Thr180/Tyr182) (Cell Signaling) or HOG1 (y-215) (Santa Cruz Biotechnology) antibodies that had been diluted 1/200 in 5% skim milk in Tris-buffered saline–Tween (TBST) at 4°C, overnight. After washing, the membranes were incubated overnight at 4°C with the secondary antibody (enhanced chemiluminescence [ECL] anti-rabbit IgG linked to horseradish peroxidase; GE Healthcare; 1/20,000 dilution). Detection was with ECL detection reagent (GE Healthcare) with a Bio-Rad ChemiDoc MP imaging system. Hog1 levels were quantified using Bio-Rad Image Lab 3.0, and levels of phosphorylated Hog1 were calculated relative to total Hog1 levels.
ROS and NO production.
An overnight culture (10 ml) of C. albicans DAY185 was diluted to an OD600 of 0.2 in half-strength PDB and grown to an OD600 of 1.0 (30°C, 250 rpm) before cells were diluted again to an OD600 of 0.1. Cells (200 μl) were incubated for 1 h (30°C, shaking, in the dark) with DHR 123 (for assaying ROS production) or DAF-FM (for assaying nitric oxide [NO] production). Both compounds were from Molecular Probes and were used at a 25 μM concentration. DHR 123 is a nonfluorescent molecule which can diffuse into the cell and is oxidized to fluorescent rhodamine 123. DAF-FM diacetate is also nonfluorescent and cell permeant and becomes fluorescent after reaction with nitric oxide. The use of these probes to detect ROS and NO production in yeast has previously been described in reference 36. In order to gain sufficient cell numbers for flow cytometry analysis, 106 cells/ml were used. NaD1 was added 15 min prior to flow cytometry. Cells were analyzed using a BD FACSCanto II cytometer with excitation at 488 nm and emission detected using a 530/30 filter. Data were analyzed using Weasel v3.0 (Walter and Eliza Hall Institute).
Similarly, S. cerevisiae cultures were incubated overnight in YPD and diluted to an OD600 of 0.05 in half-strength PDB, with and without 25 μM DHR 123 or DAF-FM, and incubated for 45 min at 30°C with shaking and protection from light. Further preparation and flow cytometry analysis were performed as outlined for C. albicans above.
RESULTS
NaD1 displays antifungal activity against the yeasts Candida albicans, Saccharomyces cerevisiae, and Cryptococcus species.
To find conditions under which NaD1 displayed antifungal activity against yeasts, we tested several media (PDB, half-strength PDB, and YPD). Consistent with our previous report, NaD1 had a 50% inhibitory concentration (IC50) of >10 μM against C. albicans in YPD (29); however, it displayed strong antifungal activity against both C. albicans and S. cerevisiae in half-strength PDB medium at 30°C. In half-strength PDB, NaD1 inhibited the growth of C. albicans with an IC50 of 2.3 ± 0.6 (Fig. 1A); in full-strength PDB, the IC50 was 3.6 ± 0.2 μM. Similar growth inhibition by NaD1 was also observed for the model yeast S. cerevisiae (Fig. 1B). Moreover, NaD1 displayed activity against C. neoformans and C. gattii strains at low-micromolar concentrations (Table 1). To broaden our understanding of the antifungal properties of NaD1, we further tested its activity against Aspergillus niger and A. flavus. NaD1 inhibited the growth of A. niger with an IC50 of 2.06 μM but was significantly less effective against A. flavus, with an IC50 greater than 10 μM (Table 1). The inhibitory activity of NaD1 against A. niger is consistent with our previously reported finding that NaD1 has antifungal activity against Aspergillus nidulans (29).
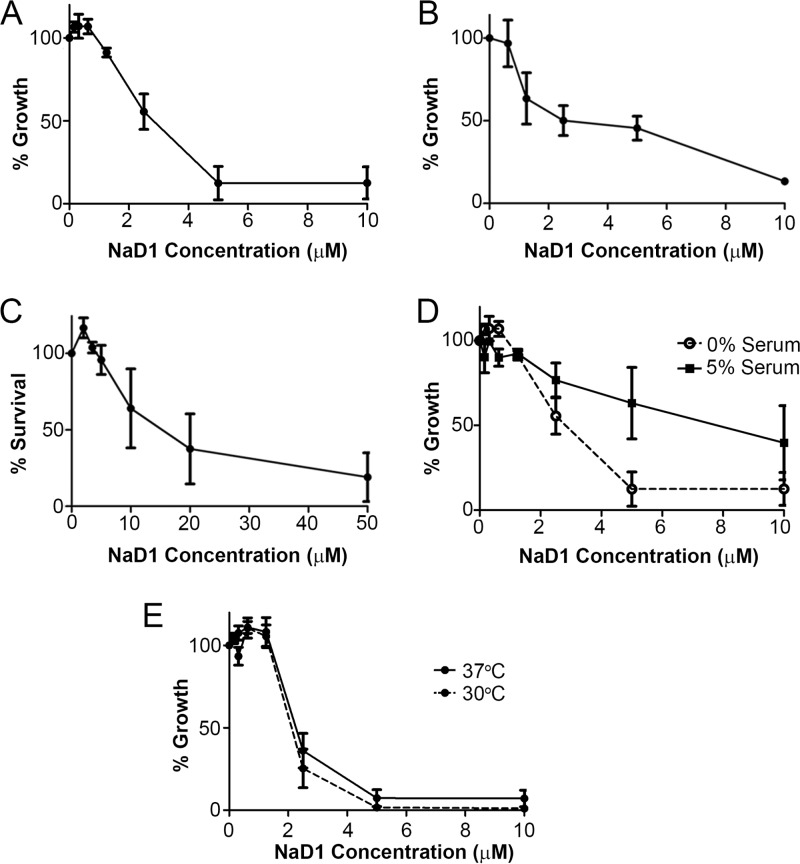
Fig 1.
Antifungal activity of NaD1. (A and B) Growth inhibition of C. albicans DAY185 (A) and S. cerevisiae BY4741 (B) at various concentrations of NaD1, relative to a no-protein control at 30°C. Error bars represent standard errors of the means (n = 3). (C) Cell survival of C. albicans DAY185 treated with various concentrations of NaD1 for 15 min, relative to a no-protein control. Error bars represent standard errors of the means (n = 3). (D) NaD1 activity on C. albicans in the presence of 5% fetal calf serum compared to a serum-free control (30°C). Error bars represent standard errors of the means (n = 3). (E) Growth inhibition of C. albicans DAY185 by NaD1 at 30°C and 37°C. NaD1 was added at various concentrations, relative to a no-protein control. Error bars represent standard errors of the means (n = 3).
Table 1.
Growth inhibition of Cryptococcus and Aspergillus strains by NaD1
| Strain (n = 3) | NaD1 IC50 (μM; mean ± SD) |
|---|---|
| C. neoformans var. grubii H99 | 2.04 ± 0.15 |
| C. neoformans var. neoformans JEC20 | 0.91 ± 0.81 |
| C. neoformans var. neoformans JEC21 | 2.38 ± 1.01 |
| C. gattii WM276 | 1.45 ± 0.59 |
| C. gattii R265 | 1.57 ± 0.57 |
| A. niger 5181 | 2.06 ± 0.76 |
| A. flavus 5310 | >10 |
Further characterization of NaD1 activity was performed using C. albicans. To test whether the effects of NaD1 were cytotoxic, we incubated C. albicans with NaD1 for 15 min at increasing concentrations and determined cell viability. NaD1-treated cells lost viability when plated on YPD, suggesting that NaD1 is fungicidal, rather than fungistatic (Fig. 1C). After 15 min of treatment with 15 μM NaD1, only 50% of the C. albicans DAY185 cells remained viable. The higher IC50 in cell survival assays is due to a higher initial cell density (3,000 times) used than the cell density in growth inhibition assays. We next tested two conditions which are relevant in the human host: the presence of serum and temperature of 37°C. NaD1 retained activity against C. albicans in the presence of 5% serum, although the IC50 was higher than that in the no-serum controls (the IC50 of 2.3 μM in the absence of serum increased to approximately 6 μM following addition of serum) (Fig. 1D). NaD1 retained activity at 37°C (Fig. 1E).
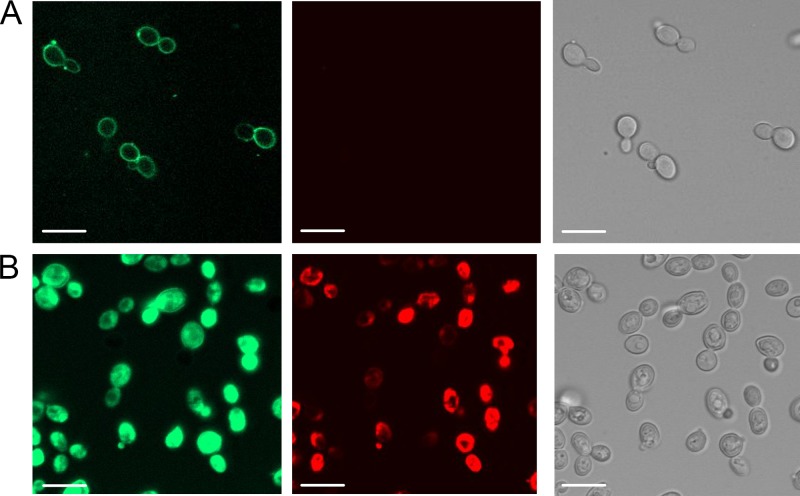
To study the interaction of NaD1 with C. albicans cells, we labeled the defensin with BODIPY and examined its location in treated cells using confocal microscopy. NaD1 accumulated on the surface of C. albicans cells after 5 min (Fig. 2A). At this stage the cells were viable, as assessed by a lack of propidium iodide (PI) fluorescence. After 20 min of treatment, BODIPY-labeled NaD1 had entered the cytoplasm, and at that stage PI had also entered the cells, indicating that the integrity of the plasma membrane had been compromised (Fig. 2B). Thus, NaD1 interacts with the fungal cell surface before it disrupts the plasma membrane and enters the cytoplasm, killing the cell.
Fig 2.
Uptake of NaD1 into C. albicans cells. Confocal microscopy of C. albicans DAY185 5 min (A) and 20 min (B) after addition of BODIPY-labeled NaD1 (10 μM, green; left) together with propidium iodide (red, middle). At 5 min, NaD1 was bound to the surface of the cell. After 20 min, both the NaD1 and propidium iodide had entered the cells. Right panels are light micrographs. Scale bars, 10 μm.
Treatment of C. albicans with NaD1 results in ROS and NO production and oxidative killing.
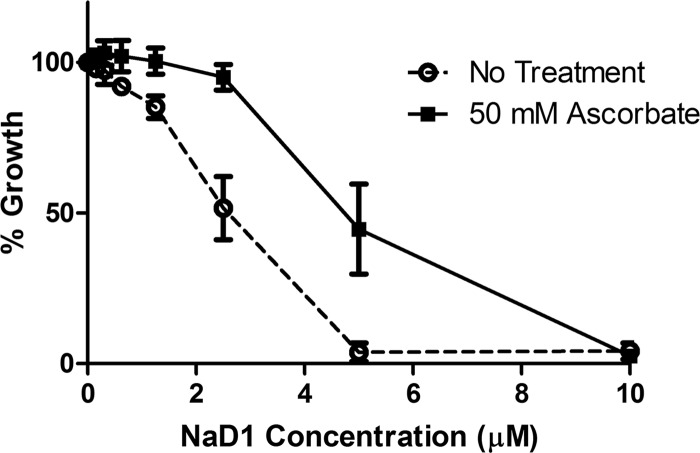
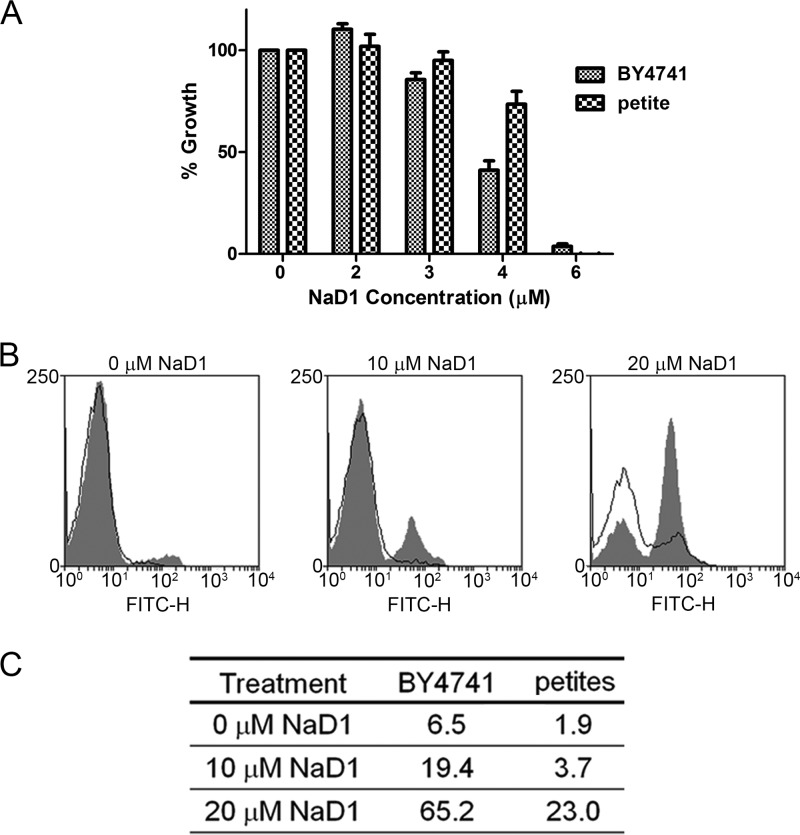
Many antimicrobial peptides have been reported to act via membrane disruption, but there are increasing reports of peptides that induce cell death by stimulating production of reactive oxygen species (ROS) (27, 29, 37, 38). Moreover, hydrogen peroxide-induced oxidative stress has been linked to production of nitric oxide (NO) during apoptosis-like cell death in yeast (36), and the plant defensin PvD1 has been shown to induce NO production by fungi (27). Therefore, we tested whether ROS and NO generation is part of the mechanism that NaD1 employs against C. albicans. First, production of ROS was monitored by confocal microscopy using the fluorescent dye DHR 123. ROS production was evident 20 min after NaD1 treatment, and this ROS production occurred only in cells with compromised plasma membranes (PI positive) (Fig. 3A). Production of ROS and NO was also monitored by flow cytometry using the fluorescent dyes DHR 123 and DAF-FM, respectively. Treatment of C. albicans (OD600 of 0.1) with NaD1 resulted in a dose-dependent increase in ROS and NO levels, ranging from 22% ROS-positive cells at 5 μM NaD1 to 81.2% and 88.6% at 10 μM and 20 μM, respectively. Few cells produced NO at 5 μM NaD1. However, after exposure to 10 and 20 μM NaD1, 62.9% and 88.2% of cells, respectively, had produced NO (Fig. 3B and C). Under these conditions, cell survival after 10 min of exposure to NaD1 was 70% at 5 μM, 25% at 10 μM, and 10% at 20 μM NaD1 (Fig. 3C). To test the link between ROS production and NaD1-induced cell death, we asked whether the antioxidant ascorbate could rescue C. albicans from the effect of NaD1. Ascorbate treatment helped protect C. albicans from the adverse effects of NaD1 treatment, increasing the IC50 by about 2-fold, from 2.3 to 5 μM (Fig. 4). The respiratory chain in the mitochondrial inner membrane is the main site of ROS production. To further probe these connections, we used the model yeast S. cerevisiae, which, unlike C. albicans, is petite positive and allows for inactivation of mitochondrial respiratory activity by deletion of the mitochondrial genome, thus creating the so-called [rho0] mutants. The [rho0] mutant of S. cerevisiae was more resistant to NaD1 than were the wild-type cells with proficient mitochondrial respiration (Fig. 5A), and there was less NaD1-induced ROS production in [rho0] mutant cells than in the wild type at a 4 μM concentration of NaD1 (Fig. 5B and C). These data further strengthen the link between NaD1 and oxidative damage. We note that at 6 μM NaD1, a concentration almost fully inhibitory for growth, the petite mutant was not more resistant than the wild type, indicating that NaD1 has effects additional to induction of ROS production.
Fig 4.
Effect of ascorbate on NaD1 activity against C. albicans. Wild-type C. albicans (DAY185) was treated with various concentration of NaD1 in the presence or absence of 50 mM ascorbate. Data are expressed relative to a no-NaD1 ascorbate control. Error bars represent standard errors of the means (n = 3).
Fig 5.
Action of NaD1 against S. cerevisiae [rho0] mutants. (A) Inhibition of S. cerevisiae BY4741 and petites ([rho0] mutants) by NaD1. At 4 μM NaD1, the growth of petite mutants was greater than the growth of the wild type. Error bars are standard errors of the means (n = 3). (B) Flow cytometry showing ROS (DHR 123) production by wild-type S. cerevisiae (BY4741, gray shading) and [rho0] mutants (black lines) in response to 10 and 20 μM NaD1. The vertical and horizontal axes represent cell counts and fluorescence intensity, respectively. Data are a representative example of three independent experiments, which gave equivalent results. (C) The percentage of ROS (DHR 123)-positive S. cerevisiae BY4741 and [rho0] cells after treatment with 0, 10, or 20 μM NaD1. Percentages were calculated using flow cytometry data. Data are a representative example of three independent experiments, which gave equivalent results.
The C. albicans Hog1 pathway is required for protection against NaD1.
To further understand how NaD1 acts on C. albicans cells, we screened several C. albicans mutant libraries for sensitivity or resistance to the defensin. The libraries consisted of mutants in kinase, transcription factors, and cell wall (synthesis and structure) genes, all of which were constructed by the Mitchell laboratory (31–33), and obtained from the Fungal Genetics Stock Center. Most of the mutants in the kinase and cell wall libraries and some mutants in the transcription factor library were represented by two independent disruption clones per gene. Only genes for which both clones showed altered sensitivity were scored as positive. Positive hits from the library screen were tested further in several repeat experiments to confirm the altered sensitivity to NaD1 and to obtain IC50s. The mutants that were reproducibly resistant or sensitive to NaD1 in the growth inhibition assays were confirmed as hits and are listed in Table 2. Mutants lacking Hog1 or its upstream kinase Pbs2, which both act in the HOG pathway, were most sensitive to NaD1 in the growth inhibition assays, with IC50s of 0.8 μM and 1.0 μM, respectively (the IC50 of the wild type was 2.3 μM). Importantly, complementation of the mutants with wild-type PBS2 or HOG1 genes led to restoration of the wild-type phenotype (Fig. 6). In addition to NaD1, the hog1 and pbs2 mutants were also more sensitive to another plant defensin, DmAMP1 (Table 2). The only reproducibly resistant strain identified was a mutant lacking the putative bZIP transcription factor encoded by orf19.6845 (IC50 of 4.2 μM [Table 2]). The orf19.6845 mutant was resistant to NaD1, but not to DmAMP1, suggesting that NaD1 and DmAMP1 act by different mechanisms (Table 2).
Table 2.
IC50s of C. albicans mutants
| C. albicans strain | Function | IC50 (μM)a (P) |
|
|---|---|---|---|
| NaD1 | DmAMP1 | ||
| Wild type (DAY286) | 2.3 ± 0.6 | 0.8 ± 0.5 | |
| pbs2−/− mutant | MAPKK, regulation of Hog1 | 1.0 ± 0.6 (0.001) | 0.1 (0.010) |
| hog1Δ/Δ mutant | MAPK, osmotic and oxidative stress response | 0.8 ± 0.3 (0.000) | 0.1 ± 0.02 (0.011) |
| orf19.6845−/− mutant | bZIP transcription factor | 4.2 ± 0.6 (0.000) | 0.5 ± 0.2 (0.210) |
| mkc1−/− mutant | MAPK, cell wall integrity and stress | 1.7 ± 0.05 (0.125) | 0.3 ± 0.02 (0.032) |
IC50s are means ± standard deviations from at least three independent experiments. P values were calculated using an independent sample t test (IBM SPSS Statistics v19).
Fig 6.
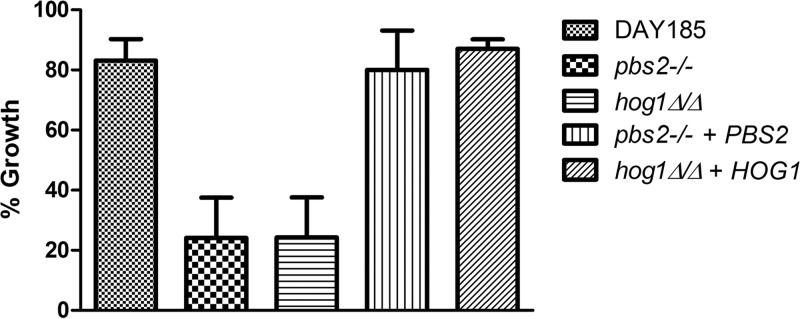
The function of Pbs2 and Hog1 in protection of C. albicans against NaD1. The C. albicans pbs2−/− and hog1Δ/Δ mutants were three times more sensitive to 1.25 μM NaD1 treatment than was wild-type DAY185, and the phenotypes were complemented by reintroduction of wild-type PBS2 and HOG1 genes into the respective mutants. Percent growth was calculated based on growth of no-protein controls. Error bars represent standard errors of the means (n = 3).
Our data indicated that NaD1 interacts with the surface of C. albicans cells prior to entering the cells and causing cell death (Fig. 2). Also, a recent study found that the plant defensin RsAFP2 causes cell wall stress (21). Collectively, this evidence prompted us to test more directly the involvement of the MKC1 gene in NaD1 tolerance. MKC1 encodes the downstream kinase of the protein kinase C (PKC)-dependent cell wall integrity (CWI) pathway and has also been shown to be phosphorylated in a Hog1-dependent manner in response to oxidative stress (39). The mkc1 mutant was not sensitive to NaD1. However, it was sensitive to DmAMP1, with an IC50 of approximately 0.3 μM (compared to 0.8 μM for the wild type [Table 2]), again suggesting that these two defensins act by different mechanisms.
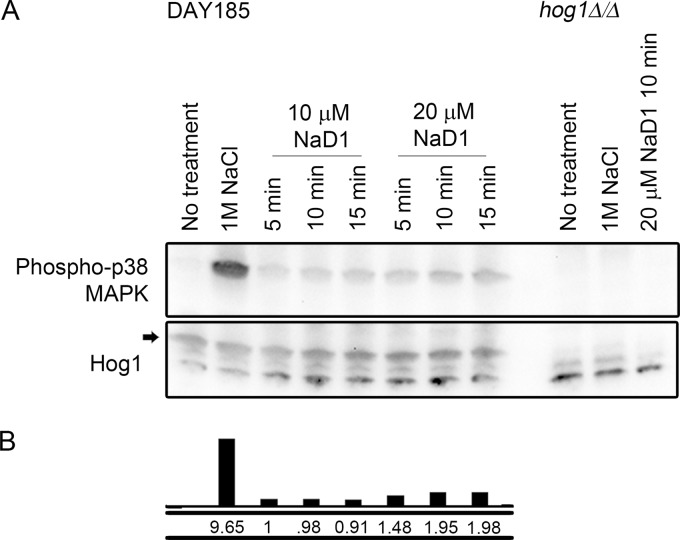
To further probe the involvement of the Hog1 pathway in the response of C. albicans to NaD1, phosphorylation of Hog1 was monitored after treatment of cells with NaD1 (Fig. 7). The HOG pathway is the main stress-responsive pathway that protects fungi from osmotic stress (40), and therefore, as a positive control, we also measured Hog1 phosphorylation after treatment with 1 M NaCl. Hog1 was phosphorylated as early as 5 min after treatment with 10 μM NaD1, although to a lesser extent than that in response to 1 M NaCl. Treatment with 20 μM NaD1 led to about 2-fold-more phosphorylation of Hog1 than that observed with 10 μM NaD1.
Fig 7.
Treatment of C. albicans cells with NaD1 leads to Hog1 phosphorylation. (A) Western blots showing levels of phosphorylated Hog1 (detected with antibodies against phospho-p38 MAPK) and total Hog1 levels. The band corresponding to total Hog1 is indicated with an arrow. Hog1 phosphorylation was induced after 10 min of treatment with 1 M NaCl and by treatment with 10 μM or 20 μM NaD1. No p-Hog1 was detected in the hog1Δ/Δ strain (negative control) or in the absence of treatment. (B) Density of p-Hog1 bands in panel A was calculated relative to total Hog1 levels and is shown compared to the 5-min, 10 μM NaD1 treatment, which was set as 1.
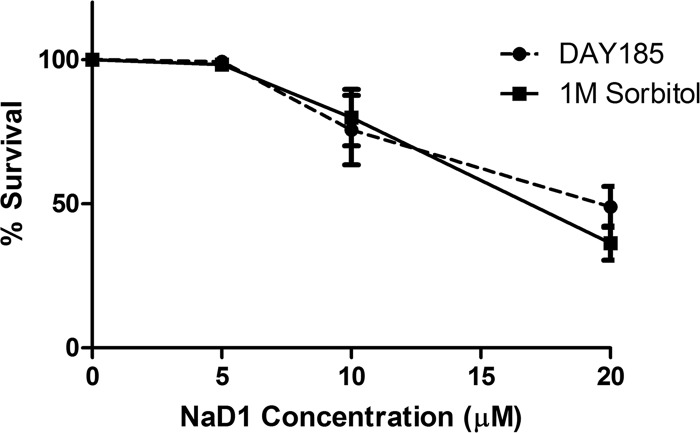
Our observations that NaD1 induces ROS production and oxidative killing (Fig. 3 and 4) are consistent with the known role for the HOG pathway in response to oxidative stress in C. albicans (41). However, the core role of the HOG pathway in all tested fungal species is the response to osmotic stress (40, 42). Therefore, to test if NaD1 activation of the HOG pathway was generated by osmotic stress, we asked if NaD1 toxicity could be rescued by pretreatment with sorbitol. This approach has been used previously to link the effects of the antimicrobial peptide histatin 5 to osmotic stress in C. albicans (43). As illustrated in Fig. 8, we found that pretreatment with sorbitol had no protective role against NaD1-dependent growth inhibition. These data suggest that osmotic stress is unlikely to contribute in a significant manner to the activity of NaD1 against C. albicans.
Fig 8.
Effect of sorbitol on NaD1 activity against C. albicans. Cell survival of C. albicans DAY185 pretreated with 1 M sorbitol followed by treatment with various concentrations of NaD1 for 15 min. Data are shown relative to a no-protein control. Error bars represent standard errors of the means (n = 3).
DISCUSSION
In this report, we show that the plant defensin NaD1 has fungicidal activity against the human pathogen C. albicans and is also active against other important human pathogens, namely, C. neoformans and C. gattii. The antifungal activity of NaD1 was comparable to that of other plant defensins and antimicrobial peptides. In this study, we found that both NaD1 and DmAMP1 inhibit the growth of C. albicans at low-micromolar concentrations. Previous studies with DmAMP1 show the IC50 to be approximately 2 μM against S. cerevisiae (44). This activity is on par with the activity of NaD1 against this yeast. The potency of the antifungal activity of NaD1 against C. albicans is also comparable to those of the peptides histatin 5, LL-37, and human beta-defensin 2. Although it is difficult to directly compare the exact IC50s because of differences in experimental conditions (most notably fungal cell densities and times of treatment), histatin 5, LL-37, beta-defensin 2, and NaD1 are all fungicidal against C. albicans at lower micromolar concentrations. For example, histatin 5 and LL-37 were shown to kill 50% of C. albicans cells at concentrations less than 10 μM, after treatment for 1 h (17). Human beta-defensin 2 is able to kill 50% of C. albicans cells with a 1 μM treatment for 3 h (45), and NaD1 is able to kill 50% of cells with a 15 μM treatment for only 15 min (this study).
Following binding to the cell surface of C. albicans, NaD1 permeabilizes the membrane and is internalized into fungal cells, causing killing by a mechanism that, at least in part, depends on oxidative damage. These results in C. albicans parallel our data with NaD1 activity against the filamentous pathogen Fusarium oxysporum. NaD1 is fungicidal against F. oxysporum, permeabilizes hyphae, enters into the cytoplasm of cells, and induces ROS (29). Collectively, these data indicate that NaD1 is active against both filamentous and yeast pathogens, with a conserved mechanism. In addition to NaD1, other plant defensins (RsAFP2, HsAFP1, and PvD1) also kill C. albicans by oxidative damage related to induction of ROS and NO production (23, 27, 38), as do other antimicrobial peptides (for example, histatin 5) (37), suggesting that this is a broad mechanism of action for antifungal compounds.
NaD1 does not disrupt artificial liposomes, indicating that it does not cause membrane permeabilization via a canonical mechanism that involves nonspecific insertion into the membrane (35). In F. oxysporum, perturbation of cell wall integrity by treatment with proteinase K or β-glucanase inhibited permeabilization of the cells and killing by NaD1 (35). Our data suggest that, as in F. oxysporum, interaction of NaD1 with the surface of C. albicans cells precedes membrane permeabilization and killing. That is, the defensin concentrates on the cell surface, before the plasma membrane is disrupted, as evidenced by the lack of propidium iodide fluorescence at this first stage of the interaction (Fig. 1E). In this study, we have screened a cell wall mutant library of C. albicans, which contains mutants in genes encoding several cell wall proteins, as well as genes with functions in cell wall biogenesis and remodeling, such as chitinases, mannosyltransferases, glucosyltransferases, and glycosidases (33). Given the link between NaD1 and cell walls, it was surprising to find no NaD1-resistant or NaD1-sensitive strains in the cell wall mutant collection. Our previous observations that proteinase K treatment protected F. oxysporum hyphae from the toxic effects of NaD1 led to the hypothesis that a cell wall protein was acting as a receptor for NaD1 (35). None of the cell wall protein mutants in the C. albicans collection were resistant to NaD1 treatment, but the library was not inclusive of all cell wall proteins. The complete list of the cell wall mutants in the library can be downloaded as an Excel file from the Fungal Genetics Stock Center website at http://www.fgsc.net. Our screen suggests that the cell wall proteins for which mutants were present in the library (33) are not receptors for NaD1 on C. albicans cells, but it is also possible that several cell wall proteins are involved and that an effect will not be observed by single gene disruptions. Further studies will be required to identify the receptor for NaD1 on fungal cell walls, including biochemical approaches. Another plant defensin, RsAFP2, which interacts with glucosylceramides present in fungal membranes as well as cell walls, was recently shown to cause cell wall stress, activate the PKC CWI pathway, and influence the organization of the septin ring (21). The kinase library from the work of Blankenship et al. (31) that we screened contains mutants in the Bck1 and Mkc1 kinases of the PKC CWI pathway, as well as kinases that are required for septin assembly (31). None of these mutants were sensitive to NaD1 in our experiments. As shown in Table 2, mkc1 mutants were hypersensitive to DmAMP1, a protein that is 50% identical to RsAFP2 and also requires interaction with sphingolipids for activity. In contrast, these mutants were not hypersensitive to NaD1, indicating that NaD1 acts on C. albicans cells by a mechanism distinct from that of RsAFP2 and DmAMP1.
The Hog1 MAPK pathway was the only stress-responsive pathway that we identified as being required for survival of C. albicans at lower concentrations of NaD1. Mutants in the genes encoding Hog1 and its upstream kinase Pbs2 were sensitive to NaD1, and we could show phosphorylation of Hog1 in response to NaD1 treatment. The Hog1 pathway is a central stress-responsive pathway in fungi (46). In C. albicans specifically, this pathway is activated by a range of stresses, including osmotic and oxidative stress (41, 42). Importantly for our study, the Hog1 pathway is also activated after exposure of C. albicans to the antimicrobial peptides histatin 5 and human beta-defensins (43, 47). Furthermore, the Hog1 pathway is also the only stress-responsive pathway implicated in the resistance of C. albicans to histatin 5 and human beta-defensins (43, 47). Interestingly, deletion of the PTC1 gene, which is a negative regulator of the Hog1 pathway through dephosphorylation of Hog1p kinase, causes hypersensitivity to the plant defensin HsAFP1 (23). Therefore, increased Hog1 activity is detrimental to the fungus during exposure to HsAFP1. This report is in direct contrast to what has previously been reported for histatin 5, human beta-defensins, and now NaD1.
The factors that activate the Hog1 pathway in response to NaD1 are not known. Our observations that NaD1 induces ROS and that the antioxidant ascorbate can in part rescue the susceptibility of C. albicans to NaD1 are consistent with induction of oxidative stress. The Mkc1 kinase is phosphorylated when C. albicans is treated with hydrogen peroxide, and this event requires the presence of Hog1; however, unlike Hog1, Mkc1 is not required for growth in response to hydrogen peroxide-induced oxidative stress (39). Similarly, only Hog1, and not Mkc1, is required for tolerance to treatment with NaD1. The transcription factor Cap1 is a key factor required for activation of oxidative stress response genes in C. albicans. Cap1 acts independently of Hog1 (41). The transcription factor library that we screened contains a cap1 mutant, but this mutant was not sensitive to NaD1, arguing that the Cap1 pathway is not involved in survival upon NaD1 treatment. Vylkova et al. (43) have reported that histatin 5 induces glycerol production in C. albicans as well as activation of the Hog1-dependent osmotic stress response. Their model suggests that histatin 5 killing involves initial osmotic stress, followed by secondary oxidative stress. Pretreatment of C. albicans with sorbitol reduced the toxicity of histatin 5, indicating that osmotic stress is involved. Oxidative stress induced by H2O2 did not affect histatin 5 activity. In contrast to results with histatin 5, NaD1 toxicity was not rescued by pretreatment with sorbitol (Fig. 8), but it was reduced by treatment with the antioxidant ascorbate at all NaD1 concentrations tested. Also, S. cerevisiae [rho0] mutants with decreased mitochondrial function and decreased ROS production were more resistant to NaD1 treatment. Interestingly, despite showing resistance to NaD1 treatment at 4 μM, [rho0] mutants are still inhibited fully at higher concentrations (Fig. 5). This could suggest that the action of NaD1 is biphasic and that the mechanism of action at higher concentrations involves additional adverse effects to which mitochondrial mutants are sensitive. Taken together, these data are consistent with oxidative stress, not osmotic stress, as the primary response of C. albicans cells to NaD1 treatment.
The observation that the human antimicrobial peptides histatin 5 and beta-defensins 2 and 3 activate the HOG pathway has led to the suggestion that modulation of this pathway might be a promising strategy for increasing the activity of antimicrobial peptides against pathogenic fungi (43, 47). Our report now adds an antifungal plant defensin to the list of antimicrobial peptides for which inactivation of the HOG pathway might prove beneficial. In addition to being active against C. albicans, NaD1 displays potent activity against a number of agriculturally important fungal pathogens (29). It will be important to explore whether the Hog1 pathway operates in response to NaD1 in these plant pathogens. Previous work with the plant defensins MsDef1, MtDef2, and RsAFP2 against the filamentous plant pathogen Fusarium graminearum showed that the Hog1 pathway was dispensable for protection and that the Ste11 MAPK pathway and the homolog of Mkc1 were involved in modulating susceptibility to these proteins (48). These observations show that the stress-responsive pathways involved in protecting fungal pathogens differ depending on the nature of the antifungal peptides and the pathogens. The HOG pathway is certainly worth considering in the context of developing future antifungal therapies based on antimicrobial peptides, and future studies with a range of antimicrobial peptides and fungal pathogens should provide a comprehensive understanding of the protective mechanisms used by pathogenic fungi to defend themselves from antimicrobial peptides and provide the knowledge foundation for designing combinatorial therapy approaches.
ACKNOWLEDGMENTS
We thank Dee Carter for fungal strains.
This work was supported by a Discovery Project from the Australian Research Council (ARC, DP120102694) and funds from Hexima Ltd. Work in the A.T. lab on C. albicans is funded by the Australian National Health and Medical Research Council (NHMRC) and the Monash University Researcher Accelerator (MRA) grant.
Footnotes
Published ahead of print 20 May 2013
REFERENCES
- 1. Wey SB, Mori M, Pfaller MA, Woolson RF, Wenzel RP. 1988. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch. Intern. Med. 148:2642–2645 [DOI] [PubMed] [Google Scholar]
- 2. Ortega M, Marco F, Soriano A, Almela M, Martínez JA, Pitart C, Mensa J. 2010. Candida spp. bloodstream infection: influence of antifungal treatment on outcome. J. Antimicrob. Chemother. 65:562–568 [DOI] [PubMed] [Google Scholar]
- 3. Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Butts A, Krysan DJ. 2012. Antifungal drug discovery: something old and something new. PLoS Pathog. 8:e1002870. 10.1371/journal.ppat.1002870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aerts AM, Francois IEJA, Cammue BPA, Thevissen K. 2008. The mode of antifungal action of plant, insect and human defensins. Cell. Mol. Life Sci. 65:2069–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pasupuleti M, Schmidtchen A, Malmsten M. 2012. Antimicrobial peptides: key components of the innate immune system. Crit. Rev. Biotechnol. 32:143–171 [DOI] [PubMed] [Google Scholar]
- 7. van der Weerden NL, Anderson MA. 2013. Plant defensins: common fold, multiple functions. Fungal Biol. Rev. 26:121–131 [Google Scholar]
- 8. Hancock REW, Lehrer R. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82–88 [DOI] [PubMed] [Google Scholar]
- 9. Nicolas P. 2009. Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J. 276:6483–6496 [DOI] [PubMed] [Google Scholar]
- 10. Tavares PM, Thevissen K, Cammue BPA, Francois IEJA, Barreto-Bergter E, Taborda CP, Marques AF, Rodrigues ML, Nimrichter L. 2008. In vitro activity of the antifungal plant defensin RsAFP2 against Candida isolates and its in vivo efficacy in prophylactic murine models of candidiasis. Antimicrob. Agents Chemother. 52:4522–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong JH, Ng TB, Legowska A, Rolka K, Hui M, Cho CH. 2011. Antifungal action of human cathelicidin fragment (LL13–37) on Candida albicans. Peptides 32:1996–2002 [DOI] [PubMed] [Google Scholar]
- 12. Ruissen ALA, Groenink J, Helmerhorst EJ, Walgreen-Weterings E, van't Hof W, Veerman ECI, Nieuw Amerongen AV. 2001. Effects of histatin 5 and derived peptides on Candida albicans. Biochem. J. 356:361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koshlukova SE, Lloyd TL, Araujo MWB, Edgerton M. 1999. Salivary histatin 5 induces non-lytic release of ATP from Candida albicans leading to cell death. J. Biol. Chem. 274:18872–18879 [DOI] [PubMed] [Google Scholar]
- 14. Li XS, Reddy MS, Baev D, Edgerton M. 2003. Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J. Biol. Chem. 278:28553–28561 [DOI] [PubMed] [Google Scholar]
- 15. Baev D, Rivetta A, Li XS, Vylkova S, Bashi E, Slayman CL, Edgerton M. 2003. Killing of Candida albicans by human salivary histatin 5 is modulated, but not determined, by the potassium channel TOK1. Infect. Immun. 71:3251–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vylkova S, Nayyar N, Li W, Edgerton M. 2007. Human b-defensins kill Candida albicans in an energy-dependent and salt-sensitive manner without causing membrane disruption. Antimicrob. Agents Chemother. 51:154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Den Hertog A, van Marle J, van Veen H, van't Hof W, Bolscher J, Veerman E, Nieuw Amerongen A. 2005. Candidacidal effects of two antimicrobial peptides: histatin 5 causes small membrane defects, but LL-37 causes massive disruption of the cell membrane. Biochem. J. 388:689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Weerden NL, Bleackley MR, Anderson MA. 5 February 2013. Properties and mechanisms of action of naturally occurring antifungal peptides. Cell. Mol. Life Sci. [Epub ahead of print.] 10.1007/s00018-013-1260-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lay FT, Anderson MA. 2005. Defensins—components of the innate immune system in plants. Curr. Protein Pept. Sci. 6:85–101 [DOI] [PubMed] [Google Scholar]
- 20. Thomma BPHJ, Cammue BPA, Thevissen K. 2002. Plant defensins. Planta 216:193–202 [DOI] [PubMed] [Google Scholar]
- 21. Thevissen K, de Mello Tavares P, Xu D, Blankenship J, Vandenbosch D, Idkowiak-Baldys J, Govaert G, Bink A, Rozental S, de Groot PW, Davis TR, Kumamoto CA, Vargas G, Nimrichter L, Coenye T, Mitchell A, Roemer T, Hannun YA, Cammue BP. 2012. The plant defensin RsAFP2 induces cell wall stress, septin mislocalization and accumulation of ceramides in Candida albicans. Mol. Microbiol. 84:166–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aerts AM, Carmona-Gutierrez D, Lefevre S, Govaert G, Francois IE, Madeo F, Santos R, Cammue BP, Thevissen K. 2009. The antifungal plant defensin RsAFP2 from radish induces apoptosis in a metacaspase independent way in Candida albicans. FEBS Lett. 583:2513–2516 [DOI] [PubMed] [Google Scholar]
- 23. Aerts AM, Bammens L, Govaert G, Carmona-Gutierrez D, Madeo F, Cammue BP, Thevissen K. 2011. The antifungal plant defensin HsAFP1 from Heuchera sanguinea induces apoptosis in Candida albicans. Front. Microbiol. 2:47. 10.3389/fmicb.2011.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thevissen K, François IEJA, Takemoto JY, Ferket KKA, Meert EMK, Cammue BPA. 2003. DmAMP1, an antifungal plant defensin from dahlia Dahlia merckii, interacts with sphingolipids from Saccharomyces cerevisiae. FEMS Microbiol. Lett. 226:169–173 [DOI] [PubMed] [Google Scholar]
- 25. Thevissen K, Warnecke DC, Francois IEJA, Leipelt M, Heinz E, Ott C, Zahringer U, Thomma BPHJ, Ferket KKA, Cammue BPA. 2004. Defensins from insects and plants interact with fungal glucosylceramides. J. Biol. Chem. 279:3900–3905 [DOI] [PubMed] [Google Scholar]
- 26. de Medeiros LN, Angeli R, Sarzedas CG, Barreto-Bergter E, Valente AP, Kurtenbach E, Almeida FCL. 2010. Backbone dynamics of the antifungal Psd1 pea defensin and its correlation with membrane interaction by NMR spectroscopy. Biochim. Biophys. Acta 1798:105–113 [DOI] [PubMed] [Google Scholar]
- 27. Mello E, Ribeiro SF, Carvalho A, Santos I, Cunha M, Santa-Catarina C, Gomes V. 2011. Antifungal activity of PvD1 defensin involves plasma membrane permeabilization, inhibition of medium acidification, and induction of ROS in fungi cells. Curr. Microbiol. 62:1209–1217 [DOI] [PubMed] [Google Scholar]
- 28. Lay FT, Brugliera F, Anderson MA. 2003. Isolation and properties of floral defensins from ornamental tobacco and petunia. J. Plant Physiol. 131:1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van der Weerden NL, Lay FT, Anderson MA. 2008. The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J. Biol. Chem. 283:14445–14452 [DOI] [PubMed] [Google Scholar]
- 30. Chang T, Schroder LA, Thomson JM, Klocman AS, Tomasini AJ, Stromhaug PE, Dunn WA., Jr 2005. PpATG9 encodes a novel membrane protein that traffics to vacuolar membranes, which sequester peroxisomes during pexophagy in Pichia pastoris. Mol. Biol. Cell 16:4941–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752. 10.1371/journal.ppat.1000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nobile CJ, Mitchell AP. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150–1155 [DOI] [PubMed] [Google Scholar]
- 33. Norice CT, Smith FJ, Solis N, Filler SG, Mitchell AP. 2007. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot. Cell 6:2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slonimski PP, Perrodin G, Croft JH. 1968. Ethidium bromide induced mutation of yeast mitochondria: complete transformation of cells into respiratory deficient non-chromosomal “petites.” Biochem. Biophys. Res. Commun. 30:232–239 [DOI] [PubMed] [Google Scholar]
- 35. van der Weerden NL, Hancock REW, Anderson MA. 2010. Permeabilization of fungal hyphae by the plant defensin NaD1 occurs through a cell wall-dependent process. J. Biol. Chem. 285:37513–37520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Almeida B, Buttner S, Ohlmeier S, Silva A, Mesquita A, Sampaio-Marques B, Osório NS, Kollau A, Mayer B, Leão C, Laranjinha J, Rodrigues F, Madeo F, Ludovico P. 2007. NO-mediated apoptosis in yeast. J. Cell Sci. 120:3279–3288 [DOI] [PubMed] [Google Scholar]
- 37. Helmerhorst EJ, Troxler RF, Oppenheim FG. 2001. The human salivary peptide histatin 5 exerts its antifungal activity through the formation of reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A. 98:14637–14642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aerts AM, Francois IE, Meert EM, Li QT, Cammue BP, Thevissen K. 2007. The antifungal activity of RsAFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. J. Mol. Microbiol. Biotechnol. 13:243–247 [DOI] [PubMed] [Google Scholar]
- 39. Navarro-García F, Eisman B, Fiuza SM, Nombela C, Pla J. 2005. The MAP kinase Mkc1p is activated under different stress conditions in Candida albicans. Microbiology 151:2737–2749 [DOI] [PubMed] [Google Scholar]
- 40. Smith DA, Morgan BA, Quinn J. 2010. Stress signalling to fungal stress-activated protein kinase pathways. FEMS Microbiol. Lett. 306:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alonso-Monge R, Navarro-García F, Román E, Negredo AI, Eisman B, Nombela C, Pla J. 2003. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2:351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. San José C, Monge RA, Pérez-Díaz R, Pla J, Nombela C. 1996. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 178:5850–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vylkova S, Jang WS, Li W, Nayyar N, Edgerton M. 2007. Histatin 5 initiates osmotic stress response in Candida albicans via activation of the Hog1 mitogen-activated protein kinase pathway. Eukaryot. Cell 6:1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thevissen K, Osborn RW, Acland DP, Broekaert WF. 2000. Specific binding sites for an antifungal plant defensin from dahlia (Dahlia merckii) on fungal cells are required for antifungal activity. Mol. Plant Microbe Interact. 13:54–61 [DOI] [PubMed] [Google Scholar]
- 45. Feng Z, Jiang B, Chandra J, Ghannoum M, Nelson S, Weinberg A. 2005. Human beta-defensins: differential activity against Candidal species and regulation by Candida albicans. J. Dent. Res. 84:445–450 [DOI] [PubMed] [Google Scholar]
- 46. Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJP, Quinn J. 2006. Role of Hog-1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17:1018–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Argimon S, Fanning S, Blankenship JR, Mitchell AP. 2011. Interaction between the Candida albicans high-osmolarity glycerol (HOG) pathway and the response to human b-defensins 2 and 3. Eukaryot. Cell 10:272–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramamoorthy V, Zhao X, Snyder AK, Xu JR, Shah DM. 2007. Two mitogen-activated protein kinase signalling cascades mediate basal resistance to antifungal plant defensins in Fusarium graminearum. Cell. Microbiol. 9:1491–1506 [DOI] [PubMed] [Google Scholar]