Abstract
The cfr gene encodes the Cfr methyltransferase that primarily methylates C-8 in A2503 of 23S rRNA in the peptidyl transferase region of bacterial ribosomes. The methylation provides resistance to six classes of antibiotics of clinical and veterinary importance. The rlmN gene encodes the RlmN methyltransferase that methylates C-2 in A2503 in 23S rRNA and A37 in tRNA, but RlmN does not significantly influence antibiotic resistance. The enzymes are homologous and use the same mechanism involving radical S-adenosyl methionine to methylate RNA via an intermediate involving a methylated cysteine in the enzyme and a transient cross-linking to the RNA, but they differ in which carbon atom in the adenine they methylate. Comparative sequence analysis identifies differentially conserved residues that indicate functional sequence divergence between the two classes of Cfr- and RlmN-like sequences. The differentiation between the two classes is supported by previous and new experimental evidence from antibiotic resistance, primer extensions, and mass spectrometry. Finally, evolutionary aspects of the distribution of Cfr- and RlmN-like enzymes are discussed.
INTRODUCTION
Cfr was first reported in 2000 (1), and RlmN was first reported in 2008 (2). Cfr confers resistance to antibiotics binding to the peptidyl transferase center on the ribosome, defining a PhLOPSa phenotype reflecting resistance to phenicol, lincosamide, oxazolidinone, pleuromutilin, and streptogramin A antibiotic classes (3), and Cfr also provides resistance to some large macrolide antibiotics (4). The cfr gene is thus a health threat when spreading in pathogenic bacteria because many clinically important antibiotics become useless for treatment. The cfr gene with only minor sequence differences has now been found worldwide in various bacteria isolated from humans and animals (5, 6, and as summarized in 7). It is also evident that the cfr gene can be horizontally transferred to its hosts, as it is always found either on plasmids or together with insertion sequences. Competition experiments involving wild-type and inactivated Cfr indicate only a small fitness cost upon expression of Cfr and no cost related to the C-8 methylation itself (8). Recently, we have cloned three cfr-like genes from the order Bacillales and confirmed that they indeed confer resistance like the original Cfr methyltransferase (7). This indicates that there is a natural reservoir of cfr-like genes.
The primary product of the Cfr-mediated methylation is 8-methyladenosine (m8A), a new natural RNA modification (9) that has so far not been seen at sites other than A2503 in 23S rRNA. In addition, Cfr provides 2,8-dimethyladenosine, although at a lower efficiency. It was also established by site-directed mutagenesis that Cfr is a radical S-adenosylmethionine (SAM) enzyme (9), as already suggested by previous sequence comparison (10).
The 2-methyladenosine (m2A) modification at Escherichia coli 23S RNA was discovered in 1995 (11), and the responsible “housekeeping” RlmN methyltransferase was subsequently shown by phylogenetic comparisons to be similar to Cfr (2). The phenotypic effects of RlmN are uncertain, but small effects on antibiotic binding as well as fitness have been presented (2, 12, 13). It has also been suggested that the modification fine-tunes interactions between ribosomes and nascent peptides involved in stalling (14). Recently, it was shown that RlmN is a dual-specificity enzyme that also methylates A37 in tRNA (15), and it was proposed that the loss of A2503 23S RNA modification causes reduced proofreading in protein synthesis.
Both RlmN and Cfr are radical SAM enzymes, a superfamily that catalyzes a diverse set of reactions that involve cleavage of unreactive C-H bonds by a 5-deoxyadenosyl radical generated by reductive cleavage of SAM (16, 17). A new mechanism involving protein methylation and transitory cross-linking has recently been proposed to explain the mechanism of methylation by Cfr and RlmN (18–20). Also, an X-ray structure of RlmN with an Fe-S cluster, both with and without a SAM ligand, has been published (21). Cfr can be modeled on this structure, showing important differences between the enzymes (21). It has been suggested that the cfr gene evolved from the rlmN gene via gene duplication (22), but the lineage in which the duplication occurred is unknown.
In this study, we use bioinformatics to identify RlmN and Cfr homologs and identify strongly conserved sequence differences between these classes of enzymes. Our phylogenetic analysis shows that Cfr-like proteins form a distinct, well-supported group within the RlmN family. The theoretical differentiation of these enzymes' function is supported by previously obtained functional evidence together with new findings from gene cloning, followed by determination of antibiotic resistance as well as modification analysis by primer extension and mass spectrometry. Our sequence searching and phylogenetic classification also reveal other distinct groups within the RlmN family, including eukaryotic and bacterial clusters of unknown function.
MATERIALS AND METHODS
Data set assembly.
PSI-BLAST was carried out against the NCBI RefSeq protein database using Staphylococcus sciuri Cfr as the query. Three iterations were run with an E value cutoff of 0.01. The resulting 5,101 sequences were aligned using MAFFT v6.864b (23).
FastTree (24) was used to construct a phylogenetic tree of the alignment. This showed a group corresponding to Cfr- and RlmN-like sequences, along with other, more distantly related homologs: pyruvate formate lyase activating enzyme and nitrogenase iron-molybdenum cofactor biosynthesis protein, molybdenum cofactor biosynthesis protein A, and coenzyme pyrroloquinoline quinine (PQQ) synthesis protein. The more distant relatives in the tree and extremely truncated partial sequences were removed to create a data set of Cfr plus RlmN family sequences which was realigned.
Consensus sequences were generated with the Python script Consensus Finder (25). The selection was based on the following principles: only two of the plasmid-borne Cfrs are included, as these are almost identical and would be overrepresented in the data set. Those genes, for which some functional evidence or other knowledge of the protein is present, are included. In addition, the RlmN sequences are selected to sample broadly across the tree.
Phylogenetic analyses.
Sites were selected for phylogenetic analysis using SeqFIRE (26) with lenient settings (the BLOSUM62 substitution group, a similarity threshold of 50%, the maximum size of a nonconserved block set to 25, and a minimum block size of one amino acid). The resulting Cfr and RlmN (Cfr+RlmN) data set dimensions were 214 sites from 1,978 sequences. A maximum likelihood tree was constructed with RAxML-HPC2 (27) on the CIPRES portal (28) using the WAG+PROTCAT model and 100 bootstrap replicates. A cut-down phylogeny of the Cfr+RlmN family was constructed with RaxML using the same parameters. This data set was based on the sequences selected for consensus sequence analysis plus additional Cfr-like sequences identified in the NCBI nr database by BLASTP searching with S. sciuri Cfr and an E value cutoff of 100 (data set dimensions were 75 taxa and 319 positions after SeqFIRE streamlining).
Strains used for transformation, expression, MIC analysis, and methylation analysis.
The E. coli TOP10 strain (Invitrogen) was used for transformation of ligated plasmids. The hyperpermeable E. coli AS19 strain (29) was used for MIC analysis because it is much more sensitive to antibiotics than are other E. coli strains. The RlmN-lacking strain E. coli JW2501-1 (30) was used for methylation analysis to facilitate identification of Cfr methylation by avoiding interference from the RlmN methylation at the same position.
Construction of plasmids encoding cfr- and rlmN-like genes.
Plasmids were constructed similarly to the pCfrhis plasmid (9, 22) except that no histidine tag was added. The plasmids bearing inducible Cfr-like genes or RlmN-like genes were constructed by PCR amplification of the genes from genomic DNA or synthetic genes, followed by cloning into plasmid pLJ102 (31) for expression of the proteins. The Brevibacillus brevis gene GI 226313314 (nlbb) was amplified for cloning by a two-step PCR amplification. The first PCR on genomic DNA used the primers 5′-CCACCCATACCATCTGCTAC-3′ and 5′-AAATGCCACTCCTTTGCC-3′, and then a second PCR added NdeI or HindIII sites for cloning with the primers 5′-GGATGTGGAGATCATATGCCGTTAACGACATTTAC-3′ and 5′-CGATTTCCAAAGCTTCACCCCACGGTTTC-3′. The PCR fragment was then cloned as previously described (7) to construct pBbRlmN (see Table 2). The construction of pRlmN with RlmN from E. coli was done similarly. The synthetic genes were similarly cloned directly from the plasmid provided by GenScript (Piscataway, NJ). pClCs contains a coding sequence for ClCs identical to the coding sequence from Clostridium sporogenes (GI 187776707), and pClPa contains the coding sequence for ClPa from Paenibacillus sp. Y412MC10 (GI 261407206), but both have another coding usage (see the supplemental material). Plasmids were retransformed into E. coli strains AS19 and JW2501-1. All three plasmid constructs were sequenced at the inserted gene to verify the identity of the cloned genes.
Table 2.
MICs of E. coli AS19 strains in the presence or absence of plasmids expressing cfr, rlmN, or cfr- or rlmN-like genesa
| Plasmid | Gene expressed | MIC (μg/ml) |
||||
|---|---|---|---|---|---|---|
| Florfenicol | Clindamycin | Linezolid | Tiamulin | Quinupristin-dalfopristin | ||
| None | 1 | 16–32 | 8 | 0.5 | 2–4 | |
| pLJ102 | 1 | 16–32 | 8 | 0.5 | 4 | |
| pCfrhis | cfr | 16–32 | >128 | 128 | 32–64 | 64 |
| pClPa | clpa | 4 | 128 | 16–32 | 2 | 4–8 |
| pClCs | clcs | 1 | 32 | 4–8 | 0.25–0.5 | 2–4 |
| pBbRlmN | nlbb | 1 | 16–32 | 8 | 0.25 | 2 |
| pRlmN | rlmN | 1 | 16–32 | 8 | 0.5 | 2 |
The tabulated MICs are given in units of micrograms/ml and are the average of at least three independent experiments. An interval is given when no clear distinction between the values was obtained. Only >2-fold differences are considered significant.
Verification of expression of plasmid-carried genes by SDS gel analysis.
E. coli AS19 cells harboring the plasmids with the cfr-like genes were grown at 37°C to an optical density at 450 nm (OD450) of 0.2 to 0.3, followed by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (to 1 mM) for induction of the genes. Cells were harvested after 3 to 3.5 h of growth and stored at −80°C. Samples were dissolved in 1× SDS/dithiothreitol (DTT) loading buffer, boiled for 5 min, and loaded on standard SDS gels along with standard markers. Gels were run at 180 V and then stained with Coomassie brilliant blue G.
Antibiotic susceptibility testing of strains.
Drug susceptibility testing was done essentially as described previously (7) using a microtiter plate format and measuring optical density values at 450 nm with a Victor 3 spectrophotometer (PerkinElmer). Overnight cultures in LB were diluted to an OD450 value of 0.01, followed by mixing of 100 μl diluted culture with 100 μl of antibiotic solution in a series with 2-fold concentration steps. Expression of cfr and the cfr-like genes was induced at the dilution step by adding 1 mM IPTG to all samples. The plasmids bear a gene for a repressor (LacIq) of the IPTG-inducible promoter, which keeps the gene silent without IPTG addition. The tested concentration ranges were as follows: for florfenicol, 0.5 to 32 μg/ml; for clindamycin and linezolid, 2 to 128 μg/ml; for tiamulin, 0.25 to 128 μg/ml; and for quinupristin-dalfopristin (Synercid), 1 to 64 μg/ml. The MIC was defined as the drug concentration at which the growth of the cultures was absent after 24 h of incubation at 37°C.
Primer extension analysis to verify modification at A2503 23S RNA.
After induction of Cfr and the Cfr-like proteins in E. coli JW2501-1 strains, the bacteria were grown for about 3 h to allow new rRNA to be transcribed, modified, and incorporated into ribosomes. Then RNA was extracted with a GeneJET RNA purification kit (Fermentas). Methylation at A2503 was monitored by primer extension analysis with AMV reverse transcriptase (Finnzymes) and a Cy5-labeled deoxyoligonucleotide primer (5′-GAACAGCCATACCCTTG-3′), complementary to nucleotides 2,540 to 2,556 of E. coli 23S rRNA. The cDNA extension products were separated on 6% polyacrylamide sequencing gels. The positions of the stops were visualized by a fluorescence scan and identified by referencing to dideoxynucleotide sequencing reactions on 23S rRNA that were electrophoresed in parallel.
Mass spectrometric analysis of RNA.
23S rRNA subfragments of around 50 nucleotides were isolated by hybridizing the rRNA with an oligodeoxynucleotide complementary to the region around A2503, followed by digestion with mung bean nuclease as described in reference 32. The oligodeoxynucleotides used had the sequence GCC CCA GGA TGC GAC GAG CCG ACA TCG AGG TGC CAA ACC TCC CCG CC for Thermus thermophilus, the sequence GCC CCA GGA TGC GAT GAG CCG ACA TCG AGG TGC CAA ACC TCC CCG TCG for Bacillus subtilis, and the sequence GCC CCA GGA TGT GAT GAG CCG ACA TCG AGG TGC CAA ACA CCG CCG TCG for E. coli.
After subfragment purification, the RNA was digested with RNase T1 for matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectrometric (MS) analysis as previously reported (33). Briefly, 1 to 2 pmol rRNA subfragment was RNase T1 digested to completion and analyzed directly using 3-hydroxypicolinic acid as matrix. Mass spectra were recorded in positive ion mode with a reflectron time of flight mass analyzer on a PerSeptive Voyager-DE STR instrument (Applied Biosystems).
MALDI tandem mass spectrometry of the E. coli/pBbRlmN A2503UG2505 23S rRNA RNase T1 digestion product was performed on a MicroMass MALDI Q-TOF Ultima instrument (Waters, Manchester, United Kingdom) in positive ion mode as previously described (34).
The E. coli/pClPa, T. thermophilus, and B. subtilis rRNA subfragments were reduced to nucleosides by enzymatic digestion and separated by liquid chromatography on a graphitized carbon column using an Agilent LC/Chip Cube system. Detection was done online with an Agilent XCT Ultra 6340 ion trap mass spectrometer, with which tandem MS analysis was done to the MS5 level. Experimental details were identical to the ones previously described (9).
RESULTS AND DISCUSSION
A natural chromosomal host for Cfr has not yet been identified, as the cfr gene is hitherto found on plasmids together with transferable sequences or in a few cases on chromosomes but flanked by transferable sequences. Until recently, all cfr genes found were very similar, with only one or two amino acid changes. It is now clear that more-divergent cfr-like genes are present on the chromosome of some bacteria in the Bacillales order (7). This study also pointed to the difficulty in predicting without any experimental evidence whether a gene codes for the worrisome Cfr methyltransferase conferring antibiotic resistance or the harmless RlmN methyltransferase.
Alignment and phylogenetic analysis of Cfr- and RlmN-like sequences.
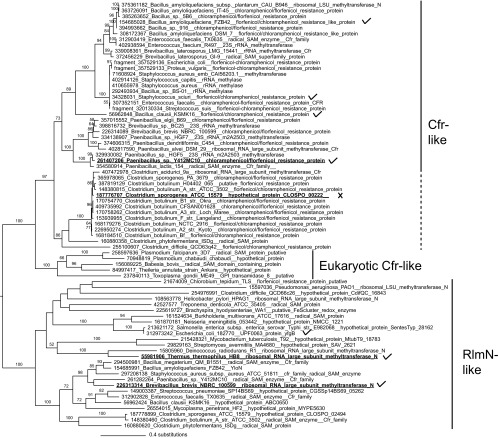
Sequences similar to Cfr were retrieved from the NCBI RefSeq protein database using PSI-BLAST with Staphylococcus sciuri Cfr as the query. The resulting data set contained Cfr, the close relative RlmN, and more-distant relatives, such as the pyruvate formate lyase activating enzyme, nitrogenase iron-molybdenum cofactor biosynthesis protein, molybdenum cofactor biosynthesis protein A, and coenzyme PQQ synthesis protein. The data set was reduced to a family of Cfr-like and RlmN-like proteins, which were aligned and subjected to maximum likelihood phylogenetic analysis. A phylogenetic tree of 1,978 sequences is presented in Fig. S1 in the supplemental material. This tree places the Cfrs from clinical and veterinary samples in a strongly supported (85% maximum likelihood bootstrap support) group within the RlmN family, together with 22 sequences from Bacillales and Clostridia and a single Enterococcus strain. There is, however, no statistical support for the Cfr-like group's specific placement within the RlmN family. Therefore, it is not possible to infer the source of Cfr, beyond the fact that it evolved from an RlmN, or an RlmN-like protein.
To investigate the differences between Cfr and RlmN on the amino acid level, we selected two sets of bacterial genes from the alignment for deducing consensus sequences. Criteria for selection (see Materials and Methods) were employed to maximize the likelihood of sampling only “true” Cfr and RlmN sequences and to sample broadly across the RlmN family phylogeny. Alignments showing Cfr-like and RlmN-like sequences and their consensus sequences are presented in Fig. S2 in the supplemental material (the whole reduced data set) and Fig. 1 (a further reduced alignment of representative sequences). The alignment clearly shows that the proteins are alignable across their full lengths, with some specific differences. Fifty-seven amino acids are strongly conserved for both classes (indicated by gray shading in Fig. 1). Thirteen amino acids are specific and conserved for each class, and these are boxed in Fig. 1. These differences are likely to be associated with important functional differences between the Cfr and RlmN families. The reduced alignment of the RlmN family was used to create a cut-down version of the RlmN plus Cfr family tree, presented in Fig. 2, that also includes newly added Cfr-like sequences from the NCBI nr database. The tree shows a clear distinction between Cfr-like genes and RlmN-like genes and an early divergence of eukaryotic Cfr-like sequences. Also to be noted is the divergence of clostridial Cfr-like proteins that will be discussed below.
Fig 1.
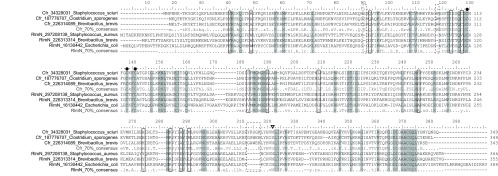
Cfr and RlmN consensus sequences. Cfr and RlmN consensus sequences together with examples of sequences used for determining the consensus—the full list is presented in Fig. S2 in the supplemental material. The black-line boxes show the 13 positions that are selectively conserved in the designated Cfr and RlmN alignment, meaning that >70% have a specific amino acid in all Cfrs and another specific amino acid in RlmNs. Gray shading marks the 57 positions that are totally conserved according to the same criteria. The dashed boxes show the sites of insertions that might play a role in the Cfr/RlmN distinction. The black-line triangle marks the position of an N-to-D change in Clostridia at an otherwise conserved position. Dots above the sequence indicate the CxxxCxxC motif cysteines involved in binding of the Fe-S cluster.
Fig 2.
Maximum likelihood phylogenetic tree of a subset of Cfr- and RlmN-like sequences. Maximum likelihood bootstrap percentage (MLBP) support is indicated on branches. Only branches with >50% MLBP support are labeled. The scale bar below the tree shows the evolutionary distance, expressed as the number of substitutions per site. Numbers in taxon names are NCBI GI numbers. Underlined genes are investigated in this study. A check mark indicates a function/modification as predicted, and X indicates a function that was tested and not verified.
We also looked at eukaryotic Cfr-like and RlmN-like sequences from the 1,978-sequence alignment to determine their relationship to Cfr and RlmN. A sister group to the Cfr-like group in the phylogenetic tree contains a small group of genes from alveolate eukaryotes, including Plasmodium parasites. This relationship of alveolate Cfr-like and Cfr clades has full support in the phylogenies (100% bootstrap percentage) (see Fig. S1 and S2 in the supplemental material). Alignment of these eukaryotic sequences with the Cfr and RlmN bacterial consensus sequences (see Fig. S2 in the supplemental material) shows that four of the 13 strongly differentially conserved sites are identical to the Cfr consensus (positions 45, 154, 216, and 282 in Fig. S2 in the supplemental material) and three (positions 223, 320, and 327 in Fig. S2 in the supplemental material) are identical to RlmN consensus while the remaining six (positions 53, 99, 158, 244, 310, and 324 in Fig. S2 in the supplemental material) are different from both Cfr and RlmN or not well conserved. In general, there is a good correlation between conservation in Cfr and RlmN and that in the eukaryotic lookalikes, especially around the C-terminal G.DIdAACGQL sequence. Although the sequence conservation suggests that these proteins are RNA methyltransferases with a function similar to that of Cfr and RlmN, their specific function and molecular target are not predictable from this analysis.
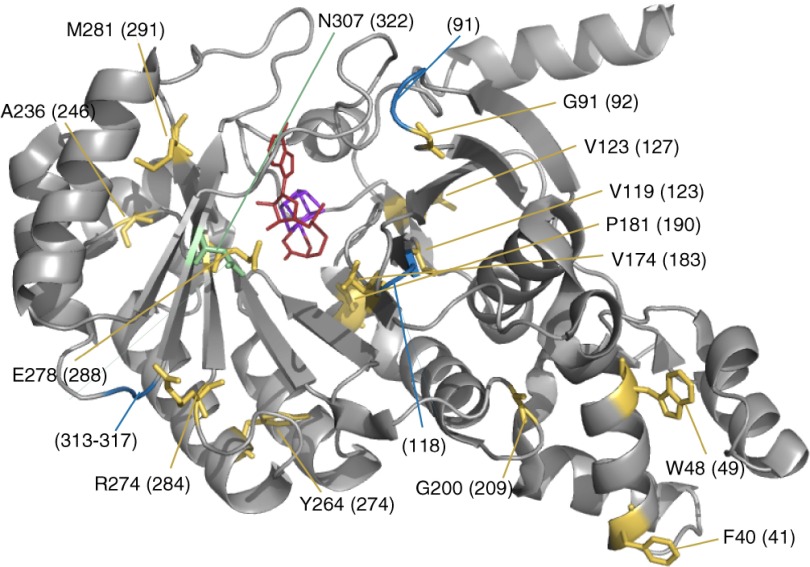
The X-ray model structure of RlmN with an Fe-S cluster and a SAM ligand (21) presented in Fig. 3 makes it possible to relate the site-specific differential conservation of Cfr/RlmN to structural features. It shows the positions of the 13 Cfr/RlmN-specific amino acids from Fig. 1 in the RlmN structure model from Boal et al. (21). The active center, if defined as the Fe-S/SAM site, is positioned at the end of a funnel-like structure, and eight of the Cfr/RlmN-specific amino acid sites (G91, V119, V123, V174, P181, R274, E278, and M281) are located at or very close to this binding pocket (Fig. 3). Three of these (V174, R274, and E278) are particularly noteworthy, as they line the funnel with side chains oriented to interact with incoming molecules and may contribute to m2A/m8A specificity. Three sites (F40, W48, and G200) are positioned far away from—but on the same site as—the “entrance” to the active site and on the outside of the enzymes. These sites might thus be implicated in target binding/target positioning, although further structural and mutational analyses are required to confirm this. There are three well-conserved sequence insertions in Cfr relative to RlmN (dotted line boxes in Fig. 1), and the structural location of these insertions is shown in blue in Fig. 3. Two of these insertions (alignment positions 18 and 313 to 317) (Fig. 1) are at the rim of the entrance to the binding pocket and may allow an alternative positioning of the entering target adenine approving for methylation at the C-8 position. Another potentially important position is the differentially conserved V123 in RlmN (alignment position 127), which is oriented away from the binding pocket and is replaced by cysteine in Cfr (Fig. 1). The residue shown in green in the structure is specifically changed in Clostridia at an otherwise conserved position and will be discussed below.
Fig 3.
Model structure of RlmN with marking of Cfr/RlmN-specific amino acids. X-ray model structure of RlmN (21) with an Fe-S cluster in purple and a SAM ligand in red. The 13 Cfr/RlmN-specific amino acids from Fig. 1 are marked in yellow and numbered according to the RlmN sequence, with alignment numbers according to Fig. 1 in parentheses. The blue markings are the positions of three insertions in Cfrs relative to RlmNs (marked with dashed boxes in Fig. 1). The green residue is the site of an N-to-D substitution in Clostridia at an otherwise conserved position.
Verification of the dispersal of Cfr methyltransferase genes among bacteria.
A reliable classification of the Cfr and RlmN classes of enzymes is desirable to identify cfr resistance genes in various organisms and to get a better understanding of the Cfr and RlmN functions and evolution. The predicted Cfr sequences are presented in the tree in Fig. 2. The clades with Cfr and Cfr-like proteins with evidence as resistance determinants are marked Cfr-like, and those with direct investigation of the gene function are marked with check marks. The Cfr-like proteins from Bacillus amyloliquefaciens, Bacillus clausii, and Brevibacillus brevis provide PhLOPSa resistance and are thus true Cfr-like proteins (7). The rest of the Cfr-like group comprises genes from Enterococcus, Paenibacillus, and Clostridium that diverge before these previously experimentally confirmed Cfr proteins (Fig. 2). We hypothesized that the well-supported (97% bootstrap support) group of Cfr-like enzymes contains all true Cfrs.
To functionally test our hypothesis based on phylogenetic classification of Cfr sequences, we cloned genes from Paenibacillus and Clostridium (see details in Table 1), expressed them in E. coli, and investigated resistance. The first experiments to isolate the genes starting from genomic DNA from the hosts failed, and therefore, synthetic genes coding for the same proteins were used and cloned in a plasmid behind an inducible promoter as previously done with the S. sciuri cfr gene (9, 22). The plasmids named pClPa (with a cfr-like Paenibacillus gene) and pClCs (with a cfr-like Clostridium gene) (Table 1) were transformed into E. coli AS19 (29), which shows an increased sensitivity to most antibiotics.
Table 1.
Origin of cfr-like and rlmN-like genes and modification activities at A2503 in 23S RNA
| Gene label | Protein name | Host organisma | Modification(s) | Reference(s) |
|---|---|---|---|---|
| cfr | Cfr | Staphylococcus (Ec) | m8A2503 and m2A2503 | 1, 9 |
| clba | ClBa | Bacillus amyloliquefaciens (Ec) | m8A2503b | 7 |
| clbc | ClBc | Bacillus clausii (Ec) | m8A2503b | 7 |
| clbb | ClBb | Brevibacillus brevis NBRC 100599 (Ec) | m8A2503b | 7 |
| clcs | ClCs | Clostridium sporogenes ATCC 15579c (Ec) | None | This study |
| clpa | ClPa | Paenibacillus sp. Y412MC10c (Ec) | m8A2503 | This study |
| rlmN/yfgB | RlmN | E. coli | m2A2503 | 2, 11, 37 |
| Staphylococcus aureus NWMN_1128 | mA2503d | 2 | ||
| Thermus thermophilus HB8 | m2A2503 | 36, this study | ||
| Bacillus subtilis subsp. subtilis 168 | m2A2503 | This study | ||
| Deinococcus radiodurans | m2A2503 | 35 | ||
| nlbb | NlBb | Brevibacillus brevis NBRC 100599 (Ec) | m2A2503e | This study |
Ec is added in parentheses if the modification was investigated in E. coli.
Based on phenotype and primer extension, no information on additional m2A2503 modification.
The gene sequences have been modified to suit codon usage in E. coli for optimal expression.
Methylation assignment based on the primer extension stop.
C-2 methylation is assumed as it is an adenine methylation but not m8A.
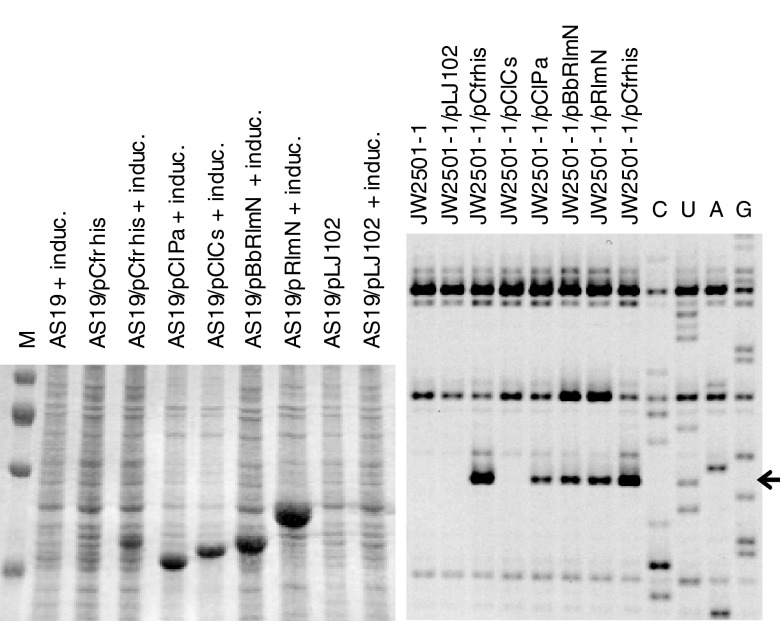
In the expression system used, protein production is strongly dependent on addition of IPTG, as previously shown by primer extension analysis for Cfr (22). Also, there is no significant growth effect from IPTG induction as well as only a very minor effect of the presence of the plasmid (data not shown). Expression was investigated by SDS gel analysis, and as shown in Fig. 4A, strong protein bands appear with expression of ClPa and ClCs—the Cfr-like proteins from Paenibacillus and Clostridium (Table 1). The ClPa protein appears at a lower mass than expected, but its identity was verified by peptide mass fingerprinting (data not shown). To establish if the Cfr-like proteins ClPa and ClCs confer a resistance pattern similar to that of the Cfr methyltransferase, MICs were determined with the five antibiotics florfenicol, clindamycin, linezolid, tiamulin, and quinupristin-dalfopristin as done previously (7). These represent the five antibiotic classes in the PhLOPSa phenotype conferred by the Cfr methyltransferase (3). The MICs are summarized in Table 2 together with controls of strains without plasmid, with the parent pLJ102 plasmid, or with pCfrhis expressing His-tagged Cfr from Staphylococcus (9). ClPa confers some resistance to the PhLOPSa antibiotics, verifying that it has a Cfr-type function, although it apparently is less effective than the Staphylococcus plasmid-coded Cfr (Table 2). The Clostridia ClCs does not mediate MIC changes. The RNA methylation at A2503 in 23S rRNA was also checked by primer extension as described in our previous study (7) to prove the relationship between modification and phenotype. This was done by transforming the plasmids into JW2501-1, an E. coli RlmN-lacking strain (30) (as the inherent m2A methylation mediated by RlmN causes a minor primer extension stop at A2503 that interferes with detection of the m8A methylation from Cfr-like enzymes). The analysis is presented in Fig. 4B and shows a clear stop at A2503 of 23S RNA from ClPa containing E. coli JW2501-1 in line with the resistance observed in the MIC experiment. The stronger stop in the Cfr-encoding strains is consistent with the higher resistance observed in these strains (Table 2). Finally, the exact identity of the methylation by ClPa was checked by mass spectrometric analysis. The chromatographic retention time and the MS4 behavior of the methylated A-nucleoside were identical to an 8-methyladenosine standard (data not shown), in accordance with our previous report on the identification of m8A2503 (9), thus confirming that the clpa gene from Paenibacillus encodes an m8A2503 methyltransferase. An independent MALDI-TOF mass spectrometric analysis revealed a substoichiometric methylation of A2503—less than 50%, as judged by MS signal intensities (data not shown)—in accordance with the low antibiotic resistance observed (Table 2) and the relatively weak extension stop (Fig. 4B). Hence, the Paenibacillus clpa gene encodes an m8A2503 methyltransferase, but the enzyme does not exhibit high efficiency when functioning in the ectopic E. coli system.
Fig 4.
Gels with protein expression profiles and RNA primer extension stops. (A) Analysis of whole-cell extracts by SDS-PAGE showing expression of Cfr, ClCs, ClPa, RlmN, and BbRlmN. The extracts are from E. coli AS19 harboring the plasmids listed on top of the gel. Lanes marked with “+ induc.” contain samples with IPTG-induced expression. M indicates size markers at 100, 70, 55, and 35 kDa (from the top). (B) Primer extension analysis of reverse transcriptase stops on 23S rRNA from E. coli JW2501-1 strains harboring the plasmids expressing Cfr, ClCs, ClPa, RlmN, and BbRlmN. The region shown is limited to the nucleotides flanking A2503, which is methylated by Cfr and RlmN. Lanes 1 to 8 show primer extension reactions on total RNA from cells harboring the indicated plasmids and induced by IPTG. Lanes 9 to 12, marked C, U, A, and G, refer to dideoxynucleotide sequencing reactions. The arrow points to the A2503 stop (one position below the sequencing position, as the extension stops before the modified nucleotide) mediated by methylation from Cfr, ClPa, RlmN, and BrRlmN.
There is no stop in the ClCs-containing strain, consistent with no observed MIC changes. Thus, the Clostridia ClCs protein does not methylate E. coli 23S RNA. Indeed, the tree in Fig. 2 shows that the clostridial sequences branch off early in the Cfr-like clade, suggesting that the Cfr-type methylation function may have evolved after the divergence of the clostridial sequences. However, we cannot conclude that ClCs does not have a Cfr-like function altogether, as this protein may be able to methylate Clostridium 23S RNA but not E. coli 23S RNA due to sequence differences in RNA or r-proteins or in the ribosome assembly process. All the Clostridium Cfr-like sequences in Fig. S2 in the supplemental material show a sequence change from N to D at position 289/307 (Cfr/RlmN numbering) of an otherwise conserved amino acid (alignment position 322) that is mapped at the entrance to the active site as shown in green in Fig. 3. A future study will be directed to investigate the significance of this particular change for methylation of A2503 in 23S RNA in E. coli.
Verification of the dispersal of RlmN methyltransferase genes among bacteria.
RlmN was identified as the enzyme responsible for the m2A2503 23S RNA modification in E. coli in 2008 (2). A similar protein from the clinical Staphylococcus aureus strain Newman exhibits 36% sequence identity to E. coli protein RlmN, and a primer extension stop at A2503 of 23S rRNA from this strain (E. coli numbering used for all organisms) is absent when the corresponding gene is inactivated (2), suggesting that it is also an RlmN. More recently, Deinococcus radiodurans has also been shown to have the m2A2503 modification (35). T. thermophilus was reported to have methylation at A2503 in 23S RNA (36), knockout of the rlmN-like gene abolishes A2503 methylation (A. Rasmussen, H. Park, and F. Kirpekar, unpublished data), and Bacillus species are also methylated at A2503 (B. T. Porse, and F. Kirpekar, unpublished data). To verify that these methylations are in fact m2A, we purified approximately 50-nucleotide-long 23S rRNA subfragments around position A2503 from T. thermophilus and B. subtilis. These subfragments were digested to nucleosides and analyzed by liquid chromatography/ion trap mass spectrometry as previously described (9). The chromatographic retention time and the MS5 behavior of the methylated A nucleosides were identical to m2A2503 obtained from E. coli and distinctly different from an 8-methyladenosine standard (data not shown), a finding which strongly suggests that the RlmN-like genes in T. thermophilus and B. subtilis encode m2A2503 methyltransferases.
Our phylogenetic analysis shows that most bacteria have an rlmN-like gene and that all those that have a cfr-like gene also have an rlmN-like gene. To verify the diverse function, we investigated RlmN function in Cfr-containing Brevibacillus brevis. We recently investigated the B. brevis cfr-like gene clBb (GI 226314089) and observed antibiotic resistance as well as a stop at position 2503 with a primer extension on 23S RNA indicating methylation (7). Here, we cloned the Brevibacillus brevis rlmN-like gene nlbb (Fig. 1 and Table 1) in a similar way and investigated its function. As a control, we also cloned the E. coli rlmN gene in the same way. NlBb, coded by nlbb, was investigated as described above for ClPa and ClCs. As expected, the protein NlBb was expressed (Fig. 4A) and had no effect on PhLOPSa MICs (Table 2) but provided a primer extension stop at A2503 23S RNA similar to the one provided by E. coli RlmN expressed the same way (Fig. 4B). Finally, it was verified by MALDI tandem mass spectrometry that the A2503UG2505 fragment of 23S RNA from the RlmN-lacking E. coli JW2501-1 strain expressing BbRlmN contains an additional methyl group on A2503 (data not shown).
The origin and the dissemination of Cfr- and RlmN-like proteins.
Even though the plasmid-borne Cfr has been found worldwide and in different bacteria, we have yet no indications of the direct origin of the Cfr that has been found in clinical and veterinary samples. According to our previous study (7) and the present phylogenetic analysis and the ClPa data, there are natural Cfr-like proteins in bacilliates, but they are apparently not widespread beyond that. The function of the clostridial Cfr-like proteins remains an unresolved question. It is worth taking into account that the databases represent only a tiny part of the diversity of bacterial life, and this information is biased from the interest of academia and industry. It is likely that more Cfr and Cfr-like sequences will be revealed with future genome sequencing.
The rlmN-like genes seem to be widespread in bacteria, with additional divergent paralogous groups of unknown function. At present, Cfr is the only paralogous subgroup within the RlmN-like family tree with known function. The phylogenetic analysis also revealed two particularly taxonomically limited groups (highlighted by yellow boxes in Fig. S1 in the supplemental material) that are strongly supported. One group comprises Aquificae, one deferribacterium, and one deltaproteobacterium, while the other contains members of beta- and gammaproteobacteria. The long branches separating these groups from the main RlmN backbone are suggestive of functional divergence, similar to what has occurred in the evolution of Cfr. In addition to the Cfr-like eukaryotic clade, there are other eukaryotic subgroups throughout the RlmN-like tree, most widespread in plants and algae. Genes for RlmN-like proteins are also found in two species of archaea: Nitrosopumilus maritimus and “Candidatus Nitrosoarchaeum limnia.” The multiple paralogous groups of RlmN-like microbial sequences suggest that horizontal gene transfer has played a significant role in the distribution of these proteins. Gene duplication and differential lineage sorting in the RlmN family tree are also likely to have contributed. With the new knowledge about the dual specificity of RlmN mediating m2A2503 on 23S rRNA and m2A37 on tRNA (15), one might speculate that the main substrate is tRNA for some RlmN-like enzymes. It is also possible that Cfr has one or more additional molecular targets. Another interesting question is which part of the enzymes accounts for the specificity. We need more studies to obtain knowledge about the specificity of the various enzymes to clarify their exact function, target, and dissemination.
Concluding remarks.
It is not obvious where the cfr gene circulation on plasmids and transposons came from or how it evolved. Our analyses suggest that cfr-like genes are limited to a small subset of bacterial species. However, their presence in known pathogens, their mobility, and their action against multiple antibiotics even in heterologous systems make them a matter of concern for antibiotic resistance. The similar rlmN genes, which do not confer significant resistance, are abundant, but it remains to be established if they all have the same targets or whether there is diversity toward 23S rRNA, tRNA, and even other RNAs. The Cfr- and RlmN-specific conserved sites provide a very good indication of whether a gene is Cfr-like or RlmN-like. The classification can then be verified by phylogenetic analysis, as we have carried out in this study.
Supplementary Material
ACKNOWLEDGMENTS
The Danish Council for Independent Research—Natural Sciences and the Lundbeck Foundation are gratefully acknowledged for financial support. G.C.A. is supported by the European Social Fund through the Estonian Science Foundation programs Mobilitas (MJD99) and ETF (I9020). T.T. is supported by the European Regional Development Fund through the Center of Excellence in Chemical Biology.
We also thank Peter Højrup for mass spectrometry analysis of ClCs, Wanida Skibsted for construction of pRlmN, and Jesper Foged Havelund for help with the mass spectrometry analysis of methylation by ClPa.
Footnotes
Published ahead of print 10 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00448-13.
REFERENCES
- 1. Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Toh SM, Xiong L, Bae T, Mankin AS. 2008. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA 14:98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith LK, Mankin AS. 2008. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52:1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y, Wang Y, Schwarz S, Shen Z, Zhou N, Lin J, Wu J, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J. Antimicrob. Chemother. 67:1824–1827 [DOI] [PubMed] [Google Scholar]
- 6. Cai JC, Hu YY, Zhang R, Zhou HW, Chen GX. 2012. Linezolid-resistant clinical isolates of meticillin-resistant coagulase-negative staphylococci and Enterococcus faecium from China. J. Med. Microbiol. 61:1568–1573 [DOI] [PubMed] [Google Scholar]
- 7. Hansen LH, Planellas MH, Long KS, Vester B. 2012. The order Bacillales hosts functional homologs of the worrisome cfr antibiotic resistance gene. Antimicrob. Agents Chemother. 56:3563–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. LaMarre JM, Locke JB, Shaw KJ, Mankin AS. 2011. Low fitness cost of the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 55:3714–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giessing AM, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F. 2009. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15:327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57:1064–1073 [DOI] [PubMed] [Google Scholar]
- 11. Kowalak JA, Bruenger E, McCloskey JA. 1995. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23S ribosomal RNA. J. Biol. Chem. 270:17758–17764 [DOI] [PubMed] [Google Scholar]
- 12. Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee HW, Hong JL, Hartland EL, Stinear TP, Howden BP. 2010. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 6:e1000944. 10.1371/journal.ppat.1000944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. LaMarre JM, Howden BP, Mankin AS. 2011. Inactivation of the indigenous methyltransferase RlmN in Staphylococcus aureus increases linezolid resistance. Antimicrob. Agents Chemother. 55:2989–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. 2010. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 29:3108–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benitez-Paez A, Villarroya M, Armengod ME. 2012. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA 18:1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang SC, Frey PA. 2007. S-Adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem. Sci. 32:101–110 [DOI] [PubMed] [Google Scholar]
- 18. Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. 2011. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science 332:604–607 [DOI] [PubMed] [Google Scholar]
- 19. Grove TL, Radle MI, Krebs C, Booker SJ. 2011. Cfr and RlmN contain a single [4Fe-4S] cluster, which directs two distinct reactivities for S-adenosylmethionine: methyl transfer by SN2 displacement and radical generation. J. Am. Chem. Soc. 133:19586–19589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan F, Fujimori DG. 2011. RNA methylation by radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc. Natl. Acad. Sci. U. S. A. 108:3930–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. 2011. Structural basis for methyl transfer by a radical SAM enzyme. Science 332:1089–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaminska KH, Purta E, Hansen LH, Bujnicki JM, Vester B, Long KS. 2010. Insights into the structure, function and evolution of the radical-SAM 23S rRNA methyltransferase Cfr that confers antibiotic resistance in bacteria. Nucleic Acids Res. 38:1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26:1899–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Atkinson GC, Baldauf SL. 2011. Evolution of elongation factor G and the origins of mitochondrial and chloroplast forms. Mol. Biol. Evol. 28:1281–1292 [DOI] [PubMed] [Google Scholar]
- 26. Ajawatanawong P, Atkinson GC, Watson-Haigh NS, Mackenzie B, Baldauf SL. 2012. SeqFIRE: a web application for automated extraction of indel regions and conserved blocks from protein multiple sequence alignments. Nucleic Acids Res. 40:W340–W347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 28. Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees, p 1–8 Proc. Gateway Comput. Environ. Workshop (GCE), New Orleans, LA, 14 November 2010). [Google Scholar]
- 29. Sekiguchi M, Iida S. 1967. Mutants of Escherichia coli permeable to actinomycin. Proc. Natl. Acad. Sci. U. S. A. 58:2315–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. 2006. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell 23:173–182 [DOI] [PubMed] [Google Scholar]
- 32. Andersen TE, Porse BT, Kirpekar F. 2004. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA 10:907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Douthwaite S, Kirpekar F. 2007. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 425:3–20 [DOI] [PubMed] [Google Scholar]
- 34. Mengel-Jorgensen J, Kirpekar F. 2002. Detection of pseudouridine and other modifications in tRNA by cyanoethylation and MALDI mass spectrometry. Nucleic Acids Res. 30:e135. 10.1093/nar/gnf135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Havelund JF, Giessing AM, Hansen T, Rasmussen A, Scott LG, Kirpekar F. 2011. Identification of 5-hydroxycytidine at position 2501 concludes characterization of modified nucleotides in E. coli 23S rRNA. J. Mol. Biol. 411:529–536 [DOI] [PubMed] [Google Scholar]
- 36. Mengel-Jorgensen J, Jensen SS, Rasmussen A, Poehlsgaard J, Iversen JJ, Kirpekar F. 2006. Modifications in Thermus thermophilus 23S ribosomal RNA are centered in regions of RNA-RNA contact. J. Biol. Chem. 281:22108–22117 [DOI] [PubMed] [Google Scholar]
- 37. Rozenski J, Crain PF, McCloskey JA. 1999. The RNA Modification Database: 1999 update. Nucleic Acids Res. 27:196–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.