Abstract
Two multidrug-resistant Bacteroides fragilis clinical isolates contain and express a novel nim gene, nimJ, that is not recognized by the “universal” nim primers and can confer increased resistance to metronidazole when introduced into a susceptible strain on a multicopy plasmid. HMW615, an appendiceal isolate, contains at least two copies of nimJ on its genome, while HMW616, an isolate from a patient with sepsis, contains one genomic copy of nimJ. B. fragilis NimJ is phylogenetically closer to Prevotella baroniae NimI and Clostridium botulinum NimA than to the other known Bacteroides Nim proteins. The predicted protein structure of NimJ, based on fold recognition analysis, is consistent with the crystal structures derived for known Nim proteins, and specific amino acid residues important for substrate binding in the active site are conserved. This study demonstrates that the “universal” nim primers will not detect all nim genes with the ability to confer metronidazole resistance, but nimJ alone cannot account for the very high metronidazole MICs of these resistant clinical isolates.
INTRODUCTION
Bacteroides spp. are Gram-negative anaerobic bacteria that are usually present as a commensal in the human and animal gut microbiomes. A recent metagenomic analysis (1–3) reported that Bacteroides species account for 27.8% (mean) of the gut bacteria with a broad standard deviation (16.6%) and range (0.1 to 64.9%) (2). The metagenomic studies (3) also support the earlier reports that the species Bacteroides fragilis is a relatively small proportion of the gut Bacteroides (4–6), although identification to the species level, data collection, and analyses have become much more complex. When B. fragilis moves out of its niche in the gut, it becomes an opportunistic pathogen and can cause serious infection. B. fragilis has been implicated in almost every type of infection and is the anaerobe most frequently isolated from patients with intra-abdominal sepsis, necrotizing skin, perforated and gangrenous appendicitis, and soft tissue infections (1, 7).
B. fragilis is inherently resistant to a wide variety of drugs (1). B. fragilis strains that are resistant to previously effective agents are being isolated with increasing frequency and are challenging current therapeutic regimens. These strains are often associated with adverse outcomes, including increased morbidity and mortality. To date, metronidazole is among the few drugs still considered reliable for the treatment of B. fragilis infections and is the most commonly prescribed drug worldwide for this purpose (8). In the last several years, however, metronidazole-resistant strains have been isolated more frequently and have often been associated with adverse outcome, including death (9) or amputation (10). Metronidazole was first introduced against protozoal infections in the middle of the 20th century (11). As administered, it is an inactive prodrug, and activation requires the partial reduction of the nitro group (12, 13) in metronidazole to the toxic nitroso radical intermediate that then binds to DNA, causing single- and double-stranded DNA breakage (14). Most pathogens do not have this activation mechanism and are thus intrinsically resistant. In sensitive organisms, “active” metronidazole resistance is often attributed to the nitroimidazole resistance gene (nim). While the exact mechanism is not known, the generally accepted premise is that nim codes for a nitroimidazole reductase that reduces the nitro group of 4- or 5-nitroimidazole to an amino group to make the inactive compound 5-aminoimidazole, thus avoiding the formation of toxic nitroso radicals that are essential for antimicrobial activity (15). The nim homologs are found in both Gram-positive and -negative genera of aerobic and anaerobic bacteria and archaea, suggesting that the nim gene family is ancient and widespread. The nim genes can be found on the chromosome, or more significantly, on mobilizable plasmids that pose a significant threat to the continuing utility of 5-nitroimidazole drugs.
Molecular detection of nim genes in Bacteroides isolates was described in 1996 using specific primers that were assumed to detect conserved sequences in all of the nim gene types (16). Identification of the nim type was accomplished by restriction analysis of the amplicon produced by these primers with Hsp92II, resulting in unique banding patterns for nimA to -F (17). To date, eight nim genes (nimA to -H) have been described in B. fragilis (18, 19), and an additional nimI gene was described in Prevotella (a related anaerobic Gram-negative genus) (19). To our knowledge, most clinical studies determine if a strain is nim positive or nim negative based on the outcome of a strain-specific PCR using these primers.
However, it has become increasingly clear that the mere presence of the nim gene is not the sole determining factor for the metronidazole resistance observed in an isolate. Increasing numbers of nim-negative metronidazole-resistant strains have been found (18, 20). Furthermore, metronidazole-resistant strains can be easily induced from nim-negative B. fragilis (20). Thus, although nim-based resistance is generally considered the most important mechanism of resistance to metronidazole in Bacteroides species, it is already acknowledged that other mechanisms are involved, such as increased efflux gene transcription levels, alterations in the DNA repair system, metabolic changes, and lack of activation of the metronidazole molecule (21–24).
In previous studies, we investigated two nim-negative, metronidazole-resistant (and multidrug-resistant) clinical isolates of B. fragilis (10). HMW615 (aka WAL 272) was a clinical isolate from a pediatric patient with appendicitis and was originally obtained from the R. M. Alden Research Laboratory, Los Angeles, CA (23). Detailed examination of B. fragilis HMW615 indicated that no single mechanism of those evaluated could account for the extremely high levels of clinical resistance observed and that either this was caused by multiple simultaneous resistance mechanisms, or some as yet unknown factor was involved. A second isolate, B. fragilis HMW616 (aka W1), was originally isolated from a patient in the United Kingdom who eventually died of sepsis (25). This isolate was resistant to metronidazole, β-lactams, β-lactam–β-lactamase inhibitor combinations, carbapenems, macrolides, and tetracyclines. Although microbiological cure was apparently achieved with linezolid, the patient ultimately died. No nim genes (9) were detected using “universal” nim primers and restriction fragment length polymorphism (RFLP) profiling (18). B. fragilis HMW610 was isolated from an American soldier who was injured in a blast accident while deployed in Afghanistan; after being thrown into a sewage-infested river he developed leg abscesses with multidrug-resistant B. fragilis (10); the leg ultimately was amputated, but the patient survived. Two plasmids were isolated from B. fragilis HMW610: (i) an 8.3-kb plasmid shown to carry a nimE gene with confirmed similarity to the sequenced fragment of pBF388c (an 8.3-kb plasmid present in a metronidazole-resistant B. fragilis strain isolated in Kuwait) (26) and (ii) a 5.5-kb plasmid identical to pHAG1 (isolated from HMW616 [previously W1]) (9) and the class III plasmid pBFB35, which is widespread throughout Bacteroides strains (27). This report describes a new nim gene in B. fragilis, nimJ, that is not recognized by universal nim primers. nimJ can confer increased resistance to metronidazole in B. fragilis 638R (a metronidazole-susceptible, nim-negative laboratory isolate) when introduced on a multicopy plasmid. The data indicate, however, that the presence of the nimJ gene is not solely responsible for the high metronidazole MICs seen in the two clinical isolates and that there are other unknown factors contributing to increased metronidazole resistance in these isolates.
MATERIALS AND METHODS
Strains and culture conditions.
All strains used are listed in Table 1. All strains were grown at 37°C as described previously (28) using brain heart infusion medium supplemented with 15 μg/ml hemin (BHIS) for Bacteroides isolates (Anaerobe Systems, Morgan Hill, CA) and Luria-Bertani (LB) agar or broth (Sigma) for Escherichia coli. Bacteroides isolates were incubated anaerobically (5% carbon dioxide, 5% hydrogen, 90% nitrogen), and E. coli was incubated either aerobically or anaerobically. E. coli DH5α was used as the host to determine whether the pSPORT vector containing the nim homolog could confer metronidazole resistance in anaerobically grown E. coli. Ampicillin (50 μg/ml), erythromycin (10 μg/ml), rifampin (10 μg/ml), gentamicin (10 μg/ml), and kanamycin (40 μg/ml) were used for selection as indicated.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Use in this study | Source or reference |
|---|---|---|
| Strains | ||
| B. fragilis | ||
| ATCC 25285 | ATCC type strain | |
| 638R | Clinical isolate (51) | |
| HMW610 | Clinical isolate (10) | |
| HMW615 | Clinical isolate (23) | |
| HMW616 | Clinical isolate (9) | |
| 638R/pMCL140 | This study | |
| 638R/pMCL140:610nimE | Transcription and activity of nim gene in B. fragilis | This study |
| 638R/pMCL140:615nimJ | Transcription and activity of nim gene in B. fragilis | This study |
| 638R/pMCL140:616nimJ | Transcription and activity of nim gene in B. fragilis | This study |
| E. coli | ||
| DH5α | Invitrogen | |
| Stellar | Invitrogen | |
| Stellar/pMCL140:610nimE | Used to introduce pMCL140::nim into B. fragilis | This study |
| Stellar/pMCL140:615nimJ | Used to introduce pMCL140::nim into B. fragilis | This study |
| Stellar/pMCL140:616nimJ | Used to introduce pMCL140::nim into B. fragilis | This study |
| DH5α/pSportI | This study | |
| DH5α/pSportI:610nimE | This study | |
| DH5α/pSportI:615nimJ | This study | |
| DH5α/pSportI:616nimJ | This study | |
| HB101/pRK231 | E. coli helper with mobilizing plasmid | |
| Plasmids | ||
| pMCL140 | Plasmid for overexpression in B. fragilis | L. Comstock (35) |
| pMCL140:610nimE | This study | |
| pMCL140:615nimJ | This study | |
| pMCL140:616nimJ | This study | |
| pSportI | Plasmid for overexpression in E. coli |
Molecular methods.
DNA preparation, restriction digestions, gel electrophoresis, and analysis were done as previously described (28).
Genome sequencing.
B. fragilis strains HMW610, HMW615, and HMW616 were submitted to the Broad Institute and sequenced as part of the Human Microbiome Project, Bacteroides Group Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/). The Broad Institute sequencing project utilized the 454 whole-genome shotgun methodology and Newbler (454 Life Sciences) assembly. This sequencing project was supported by the National Institute of Allergy and Infectious Disease/National Institutes of Health-funded Genome Sequencing Center for Infectious Diseases at the Broad Institute. The clinical isolates HMW610, -615, and -616 have been given the Broad Institute designations HMPREF1203, HMPREF1204, and HMPREF1205, respectively. Fasta files of the genome sequences and associated annotations were downloaded from the Broad Institute.
Genomic analysis.
The RAST Annotation Server (29) was used for comparative genome analysis. This server assigns protein-encoding genes (PEGs) to subsystems, a subsystem being a set of functional roles that an annotator has decided should be thought of as related. For RAST analysis, all submitted sequences were downloaded from either the Broad Institute (for the clinical isolates) or NCBI (for the reference strains) and submitted to the RAST server. Genome comparisons were done using the sequence comparison feature of the SEED server (30). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 5 (31). Amino acid sequences were aligned using ClustalW, and the phylogenetic tree was generated by the maximum likelihood method with bootstrap replications set to 500.
Prediction of NimJ protein structure.
Predicted protein sequences were submitted to the Phyre Protein Fold recognition server (32), and the predicted models were viewed with Jmol (an open-source Java viewer for chemical structures in three dimensions [3D]). The predicted Protein Data Bank (PDB) structure generated by Phyre was submitted to the Dali server (http://ekhidna.biocenter.helsinki.fi/dali_server/start); this program compares a query structure supplied by the user against the database of known structures (PDB) carrying out automatic comparisons of protein structures determined by X-ray crystallography or nuclear magnetic resonance (NMR) and returns the list of structural neighbors (33). The predicted PDB structure for NimJ was also threaded directly on the known crystal structure of NimA from Deinococcus radiodurans (DrNimA) soaked with metronidazole, pyruvate, and acetate (1W3R.pdb) (34) using the Dali Lite server (http://ekhidna.biocenter.helsinki.fi/dali_lite/start) by the sum-of-pairs method. The similarity of compared intramolecular distances was measured by Dali Z scores with structures having significant similarities being assigned a Z score above 2 (resulting from similar folds).
Transcription levels of nimJ in HMW610, -615, and -616.
Transcription levels of nimJ were determined using quantitative reverse transcription-PCR (qRT-PCR). Total RNA for qRT-PCR was prepared from early to mid-log growth strains (optical density at 600 nm [OD600] of 0.6 to 0.8) using the RNeasy minikit with RNAprotect bacterial reagent (Qiagen, Chatsworth, CA). Contaminating DNA was removed with the RNA-free DNase kit (Qiagen) using the manufacturer's recommended protocol. qRT-PCR studies were done with the Power SYBR green RT-PCR mix and the StepOnePlus instrument (Applied Biosystems, Chatsworth, CA). The primers used for qRT-PCR analysis are listed in Table 2. The comparative threshold cycle (ΔΔCT) method was used to determine the relative transcription levels using 16S RNA as the endogenous control. The analysis program is part of the StepOne software.
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| Cloning primers for expression in B. fragilis | |
| 610nimE-pMCL140-F | GAACGTTGGATCCCCGGGTAATACTAAAGATCAGTAATATGTTCAGAGAAATGC |
| 610nimE-pMCL140-R | TCGAGCTCGGTACCCGGGTACTCATCTAAATCGTTTTTTCAAGAGGTTTTCTC |
| 615nimJ-pMCL140-F | GAACGTTGGATCCCCGGGAGTGTTGCGGAATTAAGGCTATGAATG |
| 615nimJ-pMCL140-R | TCGAGCTCGGTACCCGGGTTGTCATATACGTACCTCTATTACTTTTGATGC |
| 616nimJ-pMCL140-F | GAACGTTGGATCCCCGGGTAAGGAGTATTAGGATTGCTATGAGTGAATTTAG |
| 616nimJ-pMCL140-R | TCGAGCTCGGTACCCGGGTTGTCATATACGTACCTCTATTACTTTTGATGC |
| Cloning primers for expression in E. coli | |
| 610nimE-pSport-F | CGGTCCGGAATTCCCGGGTAATACTAAAGATCAGTAATATG |
| 610nimE-pSport-R | TGAGCTCGT CGACCCGGG TACTCATCT AAATCGTTTTTTC |
| 615nimJ-pSport-F | CGGTCCGGAATTCCCGGGAGTGTTGCGGAATTAAGGC |
| 615nimJ-pSport-R | TGAGCTCGTCGACCCGGGTTGTCATATACGTACCTCTAT |
| 616nimJ-pSport-F | CGGTCCGGAATTCCCGGGTAAGGAGTATTAGGATTGCTA |
| 616nimJ-pSport-R | TGAGCTCGTCGACCCGGGTTGTCATATACGTACCTCTAT |
| qRT-PCR primers | |
| 610nimE-qRT-F | TATCGTTTTGCGTTGTGGAA |
| 610nimE-qRT-R | CATCAGACAAACCTGGCTCA |
| 615-616nimJ-qRT-F | TGACAAGGCTTCGTTCTGTG |
| 615-616nimJ-qRT-R | GTCGAAACGAATCATCAGCA |
Cloning the nim gene in an expression vector and plasmid mobilization to B. fragilis 638R.
The expression vector pMCL140, noted for its robust transcription levels of inserted genes (35), was a kind gift from Laurie Comstock (Channing Laboratory, Harvard Medical School). SmaI (New England BioLabs, Ipswich, MA) was used to linearize pMCL140. nim genes were PCR amplified using nimJ- or nimE-specific cloning primers (Table 2) and Phusion DNA polymerase (New England BioLabs, Ipswich, MA) with template DNA from appropriate B. fragilis strains. Cloning was done using Phusion DNA polymerase (New England BioLabs) and the In-Fusion HD kit (Clontech, Mountain View, CA). After the In-Fusion HD reaction, the mix was used to transform E. coli Stellar cells, and the transformant mixture was plated on ampicillin-containing LB (50 μg/ml). Ampicillin-resistant colonies were selected, and the presence of the cloned nim gene was confirmed by PCR analysis and subsequent sequencing of the plasmid insert. The nim-containing vectors were then introduced to B. fragilis 638R by a triparental mating technique using E. coli DH5α/pMCL140::nimJ (HMW615 or HMW616, respectively) and E. coli DH5α/pMCL140::nimE (HMW610) as donors, B. fragilis 638R as the recipient host, and E. coli HB101/pRK231 as the mobilizer (36, 37). Transconjugants were selected on BHIS plates containing erythromycin, rifampin, and gentamicin (10 μg/ml of each). The nimJ gene was also cloned into pSport (Invitrogen, NY) using the In-Fusion HD cloning kit as described above. pSPORT-nimJ was mobilized into E. coli DH5α for the MIC evaluation.
Screening for nimJ in laboratory constructs and in other clinical isolates.
Colony PCR analysis was done using OneTaq DNA polymerase (New England Biolabs) following the manufacturer's recommendation. The primers used for detection or cloning of the particular nim gene were the qRT-PCR or cloning primers listed in Table 2, respectively. The parameters of the PCR for the presence or absence of nim were 30 s at 94°C, 30 s at 55°C, and 30 s at 68°C for 31 cycles with a final cycle of 11 min at 68°C. PCR-generated amplicons were sequenced by Laragen (Culver City, CA).
MICs.
MICs were determined using the Etest methodology (bioMérieux SA Marcy l'Etoile, France) which is comparable to the NCCLS agar dilution method for testing anaerobic bacteria (38, 39). The cells were adjusted to 1 McFarland standard (OD600 of 0.257) in phosphate saline buffer (pH 7.4). The cells were spread on Brucella blood agar plates (Anaerobe Systems, Morgan Hill, CA) using sterile cotton swabs. The Etest strip was placed in the center, and the plates were incubated anaerobically for 16 to 24 h.
RESULTS
Genome sequencing of multidrug-resistant clinical isolates.
Metronidazole-resistant B. fragilis strains HMW615 and -616 were found to be nim negative based on PCR using the previously published “universal” nim primers (16). In order to gain more information about the nature of the metronidazole resistance seen, these strains were submitted to the Broad Center for genome sequencing (Bacteroides Group Sequencing Project, Broad Institute of Harvard and MIT). At the same time, we submitted B. fragilis HMW610, also a virulent and metronidazole (and multidrug)-resistant clinical isolate that was previously designated nim positive using the universal nim primers (16); the strain carried the nimE gene on an 8.3-kb plasmid (10). Genome sequencing confirmed the absence of nimA to -H genes in HMW615 and HMW616. Since the Broad Center annotated a large part of the B. fragilis genome(s) as hypothetical proteins, we decided to reannotate using the RAST annotation server (http://rast.nmpdr.org), which assigns genes to subsystem-based protein families. While studying the RAST genome annotations, we noted that HMW615 peg9 was annotated as a gene coding for a pyridoxamine 5′-phosphate oxidase-related protein. (This is the family that includes nim genes.) The Annotation Overview tool then detected three apparent homologs of this gene (named nimJ) within the HMW615 genome (Table 3). Using BLAST analysis with this gene within the RAST program, we found a nearly identical gene in the HMW616 with 99% nucleotide (495/498) and 98% amino acid (163/165) identical residues. The previously described nimE gene detected in HMW610 (10) has 56% nucleotide and 57% amino acid similarity with nimJ. The presence of the nimJ sequences in B. fragilis HMW615 and HMW616 was further confirmed by PCR followed by sequencing.
Table 3.
nim genes in B. fragilis HMW610, -615, and -616
| Strain (nim homologs) | Supercontig no. | Position |
Broad Institute locus taga | RAST IDb | Recognized by universal primersc | |
|---|---|---|---|---|---|---|
| Start | Stop | |||||
| HMW615 | 1.1 | 238 | 735 | HMPREF1204_00002 | fig|1073387.3.peg0.9 | No |
| HMW615 | 1.2 | 1550932 | 1551429 | HMPREF1204_02912 | fig|1073387.3.peg.1957 | No |
| HMW615 | 1.5 | 215 | 712 | HMPREF1204_04081 | fig|1073387.3.peg.4096 | No |
| HMW616 | 1.2 | 1736505 | 1736008 | HMPREF1205_01450 | fig|6666666.20506.peg.2695 | No |
| HMW610 | 1.5 | 1033 | 458 | HMPREF1203_04663 | fig|6666666.20571.peg.4745 | Yes |
HMPREF1204_00002, HMPREF1204_02912, and HMPREF1204_04081 are annotated in the Broad annotation as Bacteroides fragilis HMW615 hypothetical protein (498 nucleotides [nt]). HMPREF1205_01450 is annotated in the Broad annotation as Bacteroides fragilis HMW616 hypothetical protein (498 nt). HMPREF1203_04663 is annotated in the Broad annotation as Bacteroides fragilis HMW610 hypothetical protein (513 nt). All of these genes are annotated by the RAST server as coding for pyridoxamine 5′-phosphate oxidase-related, FMN binding.
fig, fellowship for interpretation of genomes; peg, protein-encoding gene.
NIM-3 (5′-ATG TTC AGA GAA ATG CGG CGT AAG CG-3′) and NIM-5 (5′-GCT TCC TTG CCT GTC ATG TGC TC-3′).
Alignments of the predicted NimJ protein sequences to those of other Bacteroides Nim proteins and phylogenetic analysis of NimJ.
Phylogenetic analysis of the predicted NimJ protein sequence indicated that it is more closely related to Nim proteins from Prevotella baroniae and Clostridium botulinum than to other Nim proteins described in Bacteroides (40, 41). Based on these results, these sequences were included for the subsequent phylogenetic analysis and tree building (Fig. 1). HMW610 Nim has an identical amino acid sequence to the published sequence for B. fragilis NimE, whereas NimJ from HMW615 and HMW616 is closest to P. baroniae NimI and similar to C. botulinum NimA.
Fig 1.
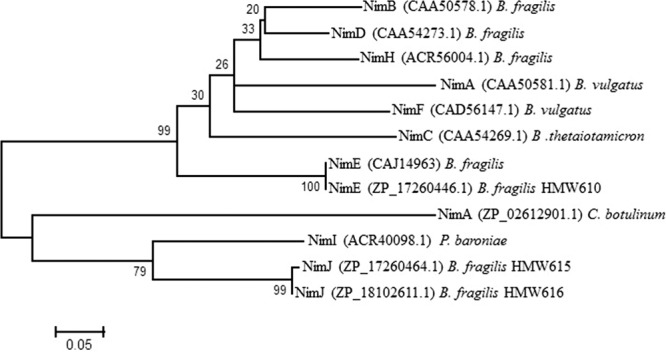
Phylogenetic analysis of B. fragilis NimJ. Phylogenetic analysis was performed with the MEGA 5 program using a MUSCLE alignment. The phylogenetic tree was reconstructed using the maximum likelihood method, and reliability for internal branching was assessed using the bootstrapping method (500 bootstrap replicates).
The predicted protein structure of NimJ is consistent with that of known Nim proteins.
The predicted protein sequence of NimJ was submitted to the Phyre Protein Fold recognition server (32), and the predicted models were viewed with Jmol (an open-source Java viewer for chemical structures in 3D). Molecules that belong to the PNPOx-like superfamily and catalyze flavin mononucleotide (FMN)-mediated redox reactions, including Nim reductases and other flavin-nucleotide-binding (FMN) proteins, exist as dimers, have a characteristic fold, and possess beta-barrel structural elements (42, 43). Recent crystal studies identified specific, conserved residues (Pro45, His59, and Tyr94) that are part of the active site of Nim proteins (42, 43). Phyre analysis of the predicted protein sequence for NimJ predicted a similar fold structure for NimJ, including the predicted beta barrel. The conserved residues that are found in the active site are also present in the predicted protein sequence of NimJ in the predicted active site.
The predicted PDB structure file generated by Phyre was submitted to the Dali server, which compared the query structure against the database of known structures (PDB) (33). Matches were found with multiple PDB structures of NIM-like proteins with the highest match (Z score, 23.3) to 2furA/2, a putative FMN-binding protein (TA1372) from Thermoplasma acidophilum. The predicted superimposition of the two structures is shown in Fig. 2A.
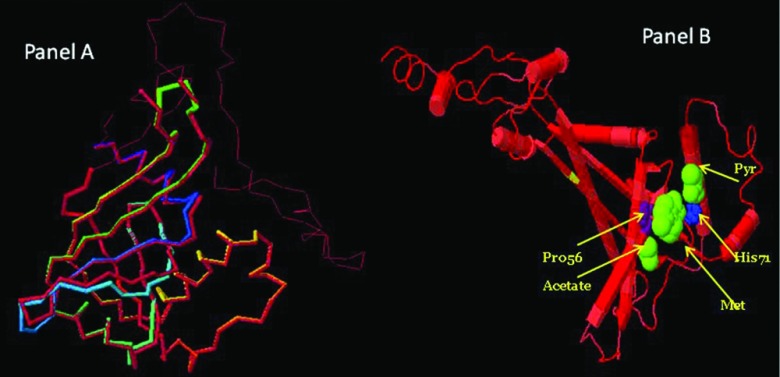
Fig 2.
(A) Dali Lite superimposed models of NimJ and closest PDB match. Phyre-predicted PDB structure of B. fragilis of NimJ superimposed on the closest match in the PDB database. The highest match was 2furA2 (Z score, 23.3). The NimJ backbone is rainbow colored from the N terminal to the C terminal. (B) Dali pairwise superimposed models: 1W3R.pdb (DR NimA soaked with metronidazole, pyruvate, and acetate) and PDB file of HMW615 NimJ (generated from Phyre server). The yellow band is tyrosine 111. The positions of metronidazole, pyruvate, and acetate molecules are indicated. DR NimA is in the cartoon mode, colored by sequence conservation (identical sequence is in red) with HMW615 NimJ.
The crystal structure of the Nim protein has been shown to be slightly altered when crystalized with its natural ligands, metronidazole, pyruvate, and acetate (43). Therefore, the Dali Lite server was also used to thread the predicted PDB file for NimJ onto the known crystal structure of DR NimA soaked with metronidazole, pyruvate, and acetate (1W3R.pdb) (34). The Dali Z score was 12.8. (structures that have significant similarities have a Z score above 2 and usually have similar folds). The predicted superimposition of the two structures is shown in Fig. 2B.
Prevalence of nimJ in clinical isolates.
B. fragilis clinical isolates from the United States (n = 15) and from Groote Schuur Hospital, South Africa (n = 23) (44), were examined using the nimJ qRT PCR primers, but the gene was not found in any of these clinical isolates. A search of the Broad database of unfinished Bacteroides genome sequences in the Human Microbiome Project with the nimJ sequence also did not find any other strains with the nimJ gene.
Overexpression of nimE and nimJ confers higher MICs of metronidazole for B. fragilis 638R.
The nim alelles (610nimE, 615nimJ, and 616nimJ) were cloned and expressed in B. fragilis 638R using pMCL140, and the overexpression was confirmed using qRT-PCR (for detection of transcription levels). In related studies, the levels of transcription of a gene insert in pMCL140 introduced into B. fragilis 638R were found to be ∼40-fold above the baseline (data not shown). In these studies, the expression of nim from the chromosomes of HMW610 (nimE), HMW615 (nimJ), and HMW616 (nimJ) was compared to those of the corresponding alleles carried on the pMCL140 vector introduced into B. fragilis 638R. Transcription levels were ∼2- to 4-fold higher in the clinical isolate than the corresponding levels from the strain with a plasmid carrying the gene (Table 4).
Table 4.
Difference in transcription levels of nim genes between clinical isolates and B. fragilis 638R carrying pMCL140::nim
| Sample | Target | RQ rangea | Avg fold change (RQ avg)b |
|---|---|---|---|
| 638R/pMCL140::610nimE | 610nimE | 0.7–1 | 3.41 |
| HMW610 | 610nimE | 2.37–3.46 | |
| 638R/pMCL140::615nimJ | 615-616nimJ | 0.22–1 | 4.26 |
| HMW615 | 615-616nimJ | 2.47–2.73 | |
| 638R/pMCL140::616nimJ | 615-616nimJ | 0.76–1 | 1.76 |
| HMW616 | 615-616nimJ | 1.27–1.83 |
The comparative CT (ΔΔCT) method was used to determine the relative quantitation (RQ) using 16S RNA as the endogenous control in the samples and in the reference sample. (B. fragilis 638R/pMCL:nim was chosen as the reference sample for each set.) Two biological and two technical replicates were used for each determination.
The RQ values of the two biologic repeats were averaged and then normalized to 1 to determine the fold difference between the clinical isolate and the corresponding B. fragilis 638 isolate containing its nimJ gene on a multicopy plasmid.
Determination of MICs.
All four biological replicates of each allele conferred ∼4- to 6-fold increases in the MICs of metronidazole when introduced into B. fragilis 638R compared to 4 biological replicates of B. fragilis 638R harboring the vector pMCL140 (Table 5). There was no difference in the metronidazole MIC for E. coli DH5α carrying pSport/nimJ compared to the MIC for E. coli DH5α carrying pSportI (data not shown).
Table 5.
MICS of metronidazole for B. fragilis clinical isolates and laboratory constructs
| Strain | MIC or MIC range (μg/ml) | Fold changea |
|---|---|---|
| ATCC 25285 | 0.19–0.25 | |
| 638R/pMCL140 | 0.03 | |
| 638R/pMCL140:610nimE | 0.17 | ∼4 |
| 638R/pMCL140:615nimJ | 0.19 | ∼4 |
| 638R/pMCL140:616nimJ | 0.14 | ∼4 |
| HMW610 | 8–16 | |
| HMW615 | 6–8 | |
| HMW616 | 8–24 |
Fold change between strain carrying empty pMCL140 plasmid and strain carrying plasmid with nim insert.
DISCUSSION
These studies were a continuation of the previous analyses of three virulent, multidrug-resistant and metronidazole-resistant clinical isolates of Bacteroides fragilis (HMW610, HMW615, and HMW616). Metronidazole is currently the most commonly prescribed antibiotic worldwide for infections involving Bacteroides. Since the time that nim genes were first described in Bacteroides, the presence of this gene (particularly with a related insertion sequence) has been considered the premier mechanism for metronidazole resistance. Moreover, the presence of nim genes on plasmids suggests the potential for horizontal gene transfer (45), especially in the gastrointestinal tract, which is a very favorable environment for gene transfer and the likely source for B. fragilis isolates. Although nim-based resistance is an important mechanism of resistance to metronidazole, it has already been acknowledged that other mechanisms must be involved. B. fragilis HMW615 and B. fragilis HMW616, both nim-negative isolates when tested with the “universal” primers, had previously been analyzed. They showed increased efflux gene transcription levels, and their high metronidazole MICs were somewhat lowered by addition of carbonyl cyanide m-chlorophenylhydrazone (CCCP; an inhibitor of the energy-driven resistance-nodulation-division [RND] family of Bacteroides multidrug efflux [bme] pumps) (9, 23). However, since metronidazole efflux systems alone did not cause the high levels of clinical resistance observed, we concluded that they were clearly not the only contributing factor and other, as yet unknown, factors were involved. The current genome sequence analysis of B. fragilis HMW615 and B. fragilis HMW616 strains was therefore undertaken to identify genomic differences that might account for the increased resistance to metronidazole. The clinical isolate B. fragilis HMW610 [nimE positive] was submitted at the same time for sequencing and subsequent characterization.
The three isolates came from different parts of the world and were all isolated as pathogenic strains; all strains were multidrug resistant and metronidazole resistant. We added the RAST annotation to that provided by Broad and found that many proteins annotated only as “hypothetical proteins” in Broad could be assigned to a specific family in RAST; this was the case with the multiple alleles of the nim-like gene (now called nimJ) observed in HMW615 as well as the close homolog in HMW616. The universal nim primers did not amplify the nimJ sequences. Interestingly, although the gene sequences of the “universal nim” primers and nimJ are different, the predicted amino acid sequence at the site of primer binding in NimJ is identical to that of the “classical” Nim proteins (FREMRRK). Differences in codon usage in nimJ for several of the amino acids, therefore, result in the amino acid sequences at these sites being identical.
The predicted protein sequence of NimJ is highly homologous to that of the classical Nim proteins, and the predicted fold structure analysis identified the Nim crystal structure as the best match. The FMN domain and conserved residues (Pro45, His59, and Tyr94) that are considered to be important for the reductase reaction are all conserved. This initial observation strongly suggested that nimJ could code for a 5-nitroimidazole reductase, as is suggested for the other well-studied Nim proteins, and we therefore proceeded with further study of the functional characteristics and phenotype conferred by this gene.
Previous studies have shown that the plasmid-carried nimA (pIP417 from B. vulgatus) and nimC (pIP417 from B. thetaiotaomicron) genes transferred metronidazole resistance to metronidazole-sensitive B. fragilis strain 638R (46). It was also shown that a 1- to 2-kb fragment (containing nimA or nimC, respectively) of the native plasmids cloned into pBI191 (46) and introduced into B. fragilis 638R transferred levels of metronidazole resistance comparable to that of the clinical isolate. In these cases, the cloned fragment included known Bacteroides insertion sequence (IS) elements located a few base pairs upstream of the nim gene; these IS elements were shown to promote expression of the nim genes (47, 48). Chromosomal fragments containing the nimB gene from plasmid-free nim-positive isolate B. fragilis 8 could be cloned into plasmids and transfer metronidazole resistance to B. fragilis 638R (49); the transferred resistance determinant was located on a 1.6-kb fragment. In the present study, the nimJ gene (cloned together with 20 flanking base pairs into the strong promoter expression vector pMCL140) was introduced to the metronidazole-sensitive recipient B. fragilis 638R and was able to increase metronidazole resistance ∼4- to 6-fold. No obvious IS elements upstream of the nim genes were detected. To our knowledge, this is the first time that the nim gene was cloned on an expression plasmid without accompanying IS elements and shown to confer increased MICs of metronidazole for the host strain. It is possible that inclusion of the sequence upstream of nimJ could result in a stronger transcription signal, and this is currently being evaluated.
Both Broad analysis and RAST analysis of HMW615 identified three nimJ homologs (HMPREF1204_00002 (supercontig 1.1), HMPREF1204_02912 (supercontig 1.2), and HMPREF1204_04081 (supercontig 1.5). It could not definitively be proved that all three homologs were unique genes on the HMW615 genomes because the HMW615 supercontigs have not been assembled into a finished genome, and there may be overlap between them. HMPREF1204_00002 and HMPREF1204_04081 could be distinguished from each other based on differences in a region 9 kb downstream of their respective nimJ genes. Supercontig 1.2 containing HMPREF1204_2912 ended before this region, so this strategy could not be used to distinguish between it and the others. Upstream sequence was not available for HMPREF1204_00002 and HMPREF1204_04081 to enable differences or similarities between the homologs to be identified.
The presence of adjacent insertion elements has been associated with nim genes in Bacteroides (45). A gene coding for an IS4 family transposase that shares 99 to 100% similarity with other IS4 family transposases was found upstream of HMPREF1204_2912 (in the opposite orientation). In pWAL610, there is also a transposase of the IS4/5 class adjacent to nimE, in the opposite orientation; however, this transposase has little similarity at the nucleotide level to the transposase upstream of HMPREF1204_02912. The closest homolog to nimJ, nimI of Prevotella baroniae, had no known insertion sequence elements detected upstream of nimI (19). Genes coding for NimI were found in all of the clinical isolates tested. It is noteworthy that nimI was found in all P. baroniae isolates tested, but not in 33 type strains belonging to other Prevotella species. The authors commented that NimI formed a new homogeneous group distant from the other Nim types involved in metronidazole resistance in anaerobic bacteria confirmed by the phylogenetic analysis reported in this work. Since the species P. baroniae has not yet been sequenced, no further conclusions can be reached about the genomic context of nimI in relation to possible similarities to nimJ.
In summary, we have identified a new nim gene, nimJ, that increases resistance to metronidazole when introduced into B. fragilis 638R. However, overexpression of the nimJ gene in B. fragilis 638R does not confer the same degree of resistance found on the original clinical isolate, suggesting that the isolates carry additional unknown resistance factors. The data are consistent with the published literature concluding that screening for nim is not advisable for diagnosis of or screening for metronidazole resistance clinical isolates (50). The work presented here has clearly demonstrated that at least one nim gene, nimJ, will not be recognized by the universal primers, and this may also be the case with other unknown nim alleles. In addition, it is clear that the mere presence and transcription of a nim gene are not solely responsible for the high MICs seen in the metronidazole-resistant clinical isolates. These studies clearly suggest that MIC determination is more reliable screening method for metronidazole resistance.
The discovery of the contribution of the nimJ to metronidazole resistance is significant in the context of clinical drug resistance surveillance studies and should be taken into account when predicting clinical outcomes. These studies confirm that metronidazole resistance in clinical isolates of B. fragilis is multifactorial. Hopefully, the current generation of genomic and transcriptomic data combined with functional analysis will lead to a better understanding of this problem and may suggest therapeutic directions.
ACKNOWLEDGMENTS
This research is based upon work supported in part by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development and in part by NIAID (NIH) grant no. 1R56AI083649-01A2. V.A. and R.M. are funded by the South African National Research Foundation (NRF), the South African Medical Research Council, and the University of Cape Town.
Footnotes
Published ahead of print 28 May 2013
REFERENCES
- 1. Wexler HM. 2007. Bacteroides: the Good, the Bad, and the Nitty-Gritty. Clin. Microbiol. Rev. 20:593–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arumugam M, Raes J, Pelletier E, Le PD, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le RK, Maguin E, Merieux A, Melo MR, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qin J, Li R, Raes J, Arumugam M, Burgdorf K, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende D, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen H, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich S, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moore W, Holdeman L. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 75:961–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Polk BF, Kasper DL. 1977. Bacteroides fragilis subspecies in clinical isolates. Ann. Intern. Med. 86:569–571 [DOI] [PubMed] [Google Scholar]
- 6. Salyers AA. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293–313 [DOI] [PubMed] [Google Scholar]
- 7. Brook I. 2010. The role of anaerobic bacteria in bacteremia. Anaerobe 16:183–189 [DOI] [PubMed] [Google Scholar]
- 8. Lofmark S, Edlund C, Nord CE. 2010. Metronidazole is still the drug of choice for treatment of anaerobic infections. Clin. Infect. Dis. 50(Suppl 1):S16–S23 [DOI] [PubMed] [Google Scholar]
- 9. Pumbwe L, Wareham DW, Aduse-Opoku J, Brazier JS, Wexler HM. 2007. Genetic analysis of mechanisms of multidrug resistance in a clinical isolate of Bacteroides fragilis. Clin. Microbiol. Infect. 13:183–189 [DOI] [PubMed] [Google Scholar]
- 10. Sherwood JE, Fraser S, Citron DM, Wexler HM, Blakely GW, Jobling K, Patrick S. 2011. Multi-drug resistant Bacteroides fragilis recovered from blood and severe leg wounds caused by an improvised explosive device (IED) in Afghanistan. Anaerobe 17:152–155 [DOI] [PubMed] [Google Scholar]
- 11. Watt L, Jennison RF. 1960. Clinical evaluation of metronidazole: a new systemic trichomonacide. Br. Med. J. 2:902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knight R, Skolimowski I, Edwards D. 1978. The interaction of reduced metronidazole with DNA. Biochem. Pharmacol. 27:2089–2093 [DOI] [PubMed] [Google Scholar]
- 13. Edwards D. 1993. Nitroimidazole drugs—action and resistance mechanisms. I. Mechanisms of action. J. Antimicrob. Chemother. 31:9–20 [DOI] [PubMed] [Google Scholar]
- 14. Zahoor A, Lafleur M, Knight R, Loman H, Edwards D. 1987. DNA damage induced by reduced nitroimidazole drugs. Biochem. Pharmacol. 36:3299–3304 [DOI] [PubMed] [Google Scholar]
- 15. Carlier J, Sellier N, Rager M, Reysset G. 1997. Metabolism of a 5-nitroimidazole in susceptible and resistant isogenic strains of Bacteroides fragilis. Antimicrob. Agents Chemother. 41:1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trinh S, Reysset G. 1996. Detection by PCR of the nim genes encoding 5-nitroimidazole resistance in Bacteroides spp. J. Clin. Microbiol. 34:2078–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lofmark S, Fang H, Hedberg M, Edlund C. 2005. Inducible metronidazole resistance and nim genes in clinical Bacteroides fragilis group isolates. Antimicrob. Agents Chemother. 49:1253–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gal M, Brazier JS. 2004. Metronidazole resistance in Bacteroides spp. carrying nim genes and the selection of slow-growing metronidazole-resistant mutants. J. Antimicrob. Chemother. 54:109–116 [DOI] [PubMed] [Google Scholar]
- 19. Alauzet C, Mory F, Teyssier C, Hallage H, Carlier JP, Grollier G, Lozniewski A. 2010. Metronidazole resistance in Prevotella spp. and description of a new nim gene in Prevotella baroniae. Antimicrob. Agents Chemother. 54:60–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schaumann R, Petzold S, Fille M, Rodloff AC. 2005. Inducible metronidazole resistance in nim-positive and nim-negative Bacteroides fragilis group strains after several passages metronidazole containing Columbia agar plates. Infection 33:368–372 [DOI] [PubMed] [Google Scholar]
- 21. Steffens L, Nicholson S, Paul L, Nord C, Patrick S, Abratt V. 2010. Bacteroides fragilis RecA protein overexpression causes resistance to metronidazole. Res. Microbiol. 161:346–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abratt VR, Zappe H, Woods DR. 1993. A reporter gene vector to investigate the regulation of glutamine synthetase in Bacteroides fragilis Bf1. J. Gen. Microbiol. 139:59–65 [DOI] [PubMed] [Google Scholar]
- 23. Pumbwe L, Chang A, Smith RL, Wexler HM. 2007. BmeRABC5 is a multidrug efflux system that can confer metronidazole resistance in Bacteroides fragilis. Microb. Drug Resist. 13:96–101 [DOI] [PubMed] [Google Scholar]
- 24. Diniz CG, Farias LM, Carvalho MA, Rocha ER, Smith CJ. 2004. Differential gene expression in a Bacteroides fragilis metronidazole-resistant mutant. J. Antimicrob. Chemother. 54:100–108 [DOI] [PubMed] [Google Scholar]
- 25. Wareham DW, Wilks M, Ahmed D, Brazier JS, Millar M. 2005. Anaerobic sepsis due to multidrug-resistant Bacteroides fragilis: microbiological cure and clinical response with linezolid therapy. Clin. Infect. Dis. 40:e67–e68 [DOI] [PubMed] [Google Scholar]
- 26. Soki J, Fodor E, Hecht DW, Edwards R, Rotimi VO, Kerekes I, Urban E, Nagy E. 2004. Molecular characterization of imipenem-resistant, cfiA-positive Bacteroides fragilis isolates from the USA, Hungary and Kuwait. J. Med. Microbiol. 53:413–419 [DOI] [PubMed] [Google Scholar]
- 27. Soki J, Wareham DW, Ratkai C, Aduse-Opoku J, Urban E, Nagy E. 2010. Prevalence, nucleotide sequence and expression studies of two proteins of a 5.6kb, class III, Bacteroides plasmid frequently found in clinical isolates from European countries. Plasmid 63:86–97 [DOI] [PubMed] [Google Scholar]
- 28. Pumbwe L, Ueda O, Yoshimura F, Chang A, Smith R, Wexler HM. 2006. Bacteroides fragilis BmeABC efflux systems additively confer intrinsic antimicrobial resistance. J. Antimicrob. Chemother. 58:37–46 [DOI] [PubMed] [Google Scholar]
- 29. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1,000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamura KF, Peterson DF, Peterson NF, Stecher GF, Nei MF, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 33. Holm L, Kaariainen S, Wilton C, Plewczynski D. 2006. Using Dali for structural comparison of proteins. Curr. Protoc. Bioinformatics Chapter 5:Unit 5.5. 10.1002/0471250953.bi0505s14 [DOI] [PubMed] [Google Scholar]
- 34. Holm L, Kaariainen S, Rosenstrom P, Schenkel A. 2008. Searching protein structure databases with DaliLite v. 3. Bioinformatics 24:2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chatzidaki-Livanis M, Weinacht KG, Comstock LE. 2010. Trans locus inhibitors limit concomitant polysaccharide synthesis in the human gut symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. U. S. A. 107:11976–11980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guiney DG, Jr, Hasegawa P, Davis CE. 1984. Homology between clindamycin resistance plasmids in Bacteroides. Plasmid 11:268–271 [DOI] [PubMed] [Google Scholar]
- 37. Veeranagouda Y, Husain F, Wexler HM. 2012. Transposon mutagenesis of the anaerobic commensal, Bacteroides fragilis, using the EZ::TN5 transposome. FEMS Microbiol. Lett. 333:94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. CLSI 2012. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, 8th ed CLSI document M11-A8. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 39. Rosenblatt JE, Gustafson DR. 1995. Evaluation of the Etest for susceptibility testing of anaerobic bacteria. Diagn. Microbiol. Infect. Dis. 22:279–284 [DOI] [PubMed] [Google Scholar]
- 40. Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. 10.1186/1471-2148-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leiros HK, Tedesco C, McSweeney SM. 2008. High-resolution structure of the antibiotic resistance protein NimA from Deinococcus radiodurans. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64:442–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leiros HK, Brandsdal BO, McSweeney SM. 2010. Biophysical characterization and mutational analysis of the antibiotic resistance protein NimA from Deinococcus radiodurans. Biochim. Biophys. Acta 1804:967–976 [DOI] [PubMed] [Google Scholar]
- 44. Galvao BP, Meggersee RL, Abratt VR. 2011. Antibiotic resistance and adhesion potential of Bacteroides fragilis clinical isolates from Cape Town, South Africa. Anaerobe 17:142–146 [DOI] [PubMed] [Google Scholar]
- 45. Soki J, Gal M, Brazier JS, Rotimi VO, Urban E, Nagy E, Duerden BI. 2006. Molecular investigation of genetic elements contributing to metronidazole resistance in Bacteroides strains. J. Antimicrob. Chemother. 57:212–220 [DOI] [PubMed] [Google Scholar]
- 46. Reysset G, Haggoud A, Su WJ, Sebald M. 1992. Genetic and molecular analysis of pIP417 and pIP419: Bacteroides plasmids encoding 5-nitroimidazole resistance. Plasmid 27:181–190 [DOI] [PubMed] [Google Scholar]
- 47. Trinh S, Haggoud A, Reysset G, Sebald M. 1995. Plasmids pIP419 and pIP421 from Bacteroides: 5-nitroimidazole resistance genes and their upstream insertion sequence elements. Microbiology 141:927–935 [DOI] [PubMed] [Google Scholar]
- 48. Haggoud A, Reysset G, Azeddoug H, Sebald M. 1994. Nucleotide sequence analysis of two 5-nitroimidazole resistance determinants from Bacteroides strains and of a new insertion sequence upstream of the two genes. Antimicrob. Agents Chemother. 38:1047–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haggoud A, Reysset G, Sebald M. 1992. Cloning of a Bacteroides fragilis chromosomal determinant coding for 5-nitroimidazole resistance. FEMS Microbiol. Lett. 74:1–5 [DOI] [PubMed] [Google Scholar]
- 50. Dubreuil L, Odou MF. 2010. Anaerobic bacteria and antibiotics: what kind of unexpected resistance could I find in my laboratory tomorrow? Anaerobe 16:555–559 [DOI] [PubMed] [Google Scholar]
- 51. Privitera GF, Dublanchet AF, Sebald M. 1979. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J. Infect. Dis. 139:97–101 [DOI] [PubMed] [Google Scholar]