Abstract
Eukaryotic RNA viruses are known to utilize host factors; however, the identity of these factors and their role in the virus life cycle remain largely undefined. Here, we report a method to identify proteins bound to the viral RNA during amplification in cell culture: thiouracil cross-linking mass spectrometry (TUX-MS). TUX-MS relies on incorporation of a zero-distance cross-linker into the viral RNA during infection. Proteins bound to viral RNA are cross-linked prior to cell lysis, purified, and identified using mass spectrometry. Using the TUX-MS method, an unbiased screen for poliovirus (PV) host factors was conducted. All host and viral proteins that are known to interact with the poliovirus RNA were identified. In addition, TUX-MS identified an additional 66 host proteins that have not been previously described in poliovirus amplification. From these candidates, eight were selected and validated. Furthermore, we demonstrate that small interfering RNA (siRNA)-mediated knockdown of two of these uncharacterized host factors results in either a decrease in copy number of positive-stranded RNA or a decrease in PV translation. These data demonstrate that TUX-MS is a robust, unbiased method to identify previously unknown host cell factors that influence virus growth. This method is broadly applicable to a range of RNA viruses, such as flaviviruses, alphaviruses, picornaviruses, bunyaviruses, and coronaviruses.
INTRODUCTION
Positive-sense RNA viruses encompass one-third of the viral genera and include numerous pathogens, such as severe acute respiratory syndrome (SARS) coronavirus, hepatitis C virus (HCV), West Nile virus, Chikungunya virus, and western, eastern, and Venezuelan equine encephalitis viruses. In view of the significant impact of RNA viruses on human health, considerable effort has been made toward the development of suitable therapeutics. Vaccines or combination antivirals are available for several RNA viruses, such as poliovirus (PV), influenza virus, HCV, and human immunodeficiency virus (HIV) (1–5). Nonetheless, there are many RNA viruses for which there are no effective treatments or vaccines, such as dengue virus, West Nile virus, alphaviruses, and the SARS virus, to name a few. Identification of host proteins required for viral amplification would help identify possible targets for antivirals and increase our understanding of the biology of viral infection. While much is known about the roles of viral proteins in viral amplification, viruses also require cellular host factors (6–10). When viruses enter a cell, they subvert, modify, or inhibit host cell processes in order to replicate. Identification of host factors required for viral amplification has been laborious; however, some studies have demonstrated that host proteins can and do play a direct role in viral amplification. For example, injection of polioviral and rhinoviral RNA into frog oocytes does not result in the production of infectious viral particles unless proteins are also expressed from human mRNAs in the oocytes (8, 9). Similarly, rabbit reticulocyte lysate (RRL) translates poliovirus RNA inefficiently unless human proteins are added (11–13).
Drug discovery has traditionally targeted viral proteins to develop antivirals. However, RNA viruses rapidly develop drug-resistant variants in response to monotherapy. This is due to the error-prone nature of the enzyme that synthesizes the viral genome, the RNA-dependent RNA polymerase. Misincorporation of nucleotides during viral replication results in rapid evolution of the viral genome, while selective pressure by the drug results in an outgrowth of drug-resistant variants. Therefore, new antivirals or combination therapies are required. An alternative approach for developing effective antivirals is to target host proteins that are required for viral amplification. Host factors may represent a higher barrier for viruses to evolve resistant variants against. The major obstacle to this approach is our limited knowledge of which host factors are required for viral amplification. Therefore, there is a great need to identify host factors required for RNA viral replication and to better understand their role in viral amplification. In particular, host factors that are not essential to the host can be exploited for development of antivirals.
Among the positive-sense single-stranded RNA viruses that constitute important human pathogens are enteroviruses from the Picornaviridae family, including poliovirus (PV), enterovirus 71 (EV71), and coxsackievirus B3 (CVB3). These viruses produce acute infections that can cause debilitating sequelae, such as paralysis and myocarditis (14, 15). There are effective vaccines available for PV. However, following eradication of PV and cessation of vaccination, an antiviral will be necessary to address sporadic infections or elimination of PV from persistent excretors (i.e., people who continue to shed PV throughout their lives, thus maintaining a worldwide reservoir of PV) (16). Vaccines do not exist for the majority of enteroviruses, and antivirals would be a first line of defense in preventing disease progression to more severe forms.
Poliovirus has a positive-sense single-stranded RNA (ssRNA) genome encapsidated by viral proteins. The viral genome encodes a large polyprotein that is proteolytically cleaved by viral proteases. Viral replication occurs in the cytoplasm by synthesis of a minus-strand RNA that is used as a template for synthesis of the plus-strand RNA genomes that are packaged. Virions are released following cell lysis (17). Using PV as a model, we have developed a cell-based method to identify host proteins that bind to viral RNA during viral infection: thiouracil cross-linking mass spectrometry (TUX-MS). Briefly, 4-thiouridine (4sU), a zero-distance cross-linker, is incorporated into viral RNA, and any proteins bound to the viral RNA are cross-linked to it. Cross-linked RNA-protein complexes are isolated under denaturing conditions, and the proteins are identified by mass spectrometry. We have demonstrated the utility of this method by identifying 15 previously known host proteins plus an additional 66 putative host proteins that bind to the PV RNA during infection in HeLa cells. Validation of a subset of these putative host factors in secondary assays demonstrated that the majority of them affect viral amplification. Furthermore, we show that knockdown of two host proteins, NONO (non-POU domain-containing octamer-binding protein) or CNBP (cellular nucleic acid-binding protein), identified using TUX-MS results in either a decrease in the number of copies of PV positive-stranded RNA or a reduction in PV translation. Therefore, TUX-MS is a robust method to identify host factors for RNA viruses.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
Transduction of HeLa cells with uracil phosphoribosyltransferase (UPRT) gene-containing lentiviruses generated HeLaUPRT cells. Lentivirus was generated by cotransfection of HEK293T cells with a LNCX plasmid containing myc-tagged UPRT and plasmids expressing tat and vesicular stomatitis virus G protein (VSVG) (a generous gift from Edward Mocarski). HeLaUPRT and 911 (human embryonic retinoblast) cells were grown at 37°C and 5% CO2 in complete Dulbecco's modified minimum essential medium (DMEM) with 10% fetal bovine serum (FBS) and penicillin-streptomycin). The HeLaUPRT cells were supplemented with 1 mg/ml G418 to select for the UPRT gene. The Mahoney PV, mengovirus, and adenovirus type 5 were propagated on HeLaUPRT cells, and titers were determined on either HeLaUPRT (PV) or 911 (adenovirus) cells. Viral infections were performed in CPBS (phosphate-buffered saline [PBS], 0.1 mg/ml MgCl2 and CaCl2) for 1 h. Where indicated, 1 mM 4-thiouracil (4TU) (Sigma-Aldrich) or 10 μg/ml actinomycin D (Act D) was added. Then the cells were washed with PBS, and infections were carried out in complete medium for the indicated times. To generate the poliovirus internal ribosome entry site (IRES) dicistronic reporter (pRFPV), the PV IRES (nucleotides [nt] 53 to 739 of the poliovirus type I Mahoney genome) was amplified using the forward and reverse PV_pRF specific primers (Table 1). The PCR product was digested with EcoRI and PciI and ligated to the EcoRI and NcoI sites in the pRF dual-luciferase plasmid, which has been described elsewhere (18). The dicistronic reporter was expressed under the control of the simian virus 40 (SV40) promoter. As an additional control for cap-dependent translation, a reporter plasmid containing the β-galactosidase gene was cotransfected with the IRES construct (19).
Table 1.
The gene product and primer designations, accession numbers, and primer sequences used in this study
| Gene product or primer designation | Accession no. | Primer sequence (5′→3′) |
|
|---|---|---|---|
| Forward | Reverse | ||
| ACTB | NM_001101.3 | GCACTCTTCCAGCCTTCC | TGTCCACGTCACACTTCATG |
| GAPDH | NM_002046.3 | ACATCGCTCAGACACCATG | TGTAGTTGAGGTCAATGAAGGG |
| HNRNPL | NM_001533.2 | TCAGTGAATCCCGGAACAATCGGT | TCCCAGCTCATCGCAGATCTCAAA |
| RSL1D1 | NM_015659.2 | AGAGAAGTGGGAGAGCGTGAAACT | CAGCAATTGGGATGAAGCCACCAA |
| XRCC6 | NM_001469.3 | GTCTTCTGTCCAAGTTGGTCGCTT | GGCGATGAAGAAGCAGAGGAAGAA |
| DDX17 | NM_006386.4 | CAACAGGATAGGGACATCAGA | CACTGCATTCTTTGGCTAAGG |
| DDX5 | NM_004396.3 | AGCGTGACTGGGTTCTAAATG | AGGAGTTAGGGTAGTCATAATTGATG |
| NONO | NM_001145408.1 | AAGAAGAAATGATGCGGCGACAGC | TGGGAGGTGCTATGGGCATAAACA |
| CNBP | NM_001127192.1 | GCCATCAACTGCAGCAAGACAAGT | TTGCACGGGAATGCACAATTGAGG |
| PVpos | V01148.1 | ATGTTCCTGTCGGTGCTGTG | CACTGTCCTGCTCTGGTTGG |
| PVneg | V01148.1 | GCGGGAACACAAAGGCATTC | ACTCCTGACAACAACCAGACATC |
| PV_pRF | V01148.1 | TACGAATTCACTCCGGTATTGCGGTACCCTTGTACG | ATACATGTTGATACAATTGTCTGATTGAAATAACTG |
Northern blotting.
For Northern blotting, 5 μg of total RNA isolated from UV cross-linked cells using TRIzol (Invitrogen) or 5 μl of the unconcentrated poly(A) fraction was separated on a denaturing formaldehyde agarose gel and transferred to a Zeta-probe membrane (Bio-Rad) as described previously (20).
Northwestern blotting.
A total of 5 × 106 HeLaUPRT cells were either infected with poliovirus (multiplicity of infection [MOI] of 10) or not (mock infected) at 37°C and 5% CO2 in CPBS (phosphate-buffered saline with 0.1 mg/ml MgCl2 and CaCl2) with or without 10 μg/ml actinomycin D (Sigma) for 1 h. The medium was replaced with DMEM with 10% FBS, with or without 10 μg/ml actinomycin D and with or without 1 mM 4-thiouracil (Sigma-Aldrich). After 5 h, total RNA was isolated using TRIzol reagent (Invitrogen). Five micrograms of total RNA was biotinylated with pyridyldithiol-biotin (biotin-HPDP) (Pierce) in TE (10 mM Tris-HCl [pH 7.4], 1 mM EDTA) for 1.5 h at 25°C in the dark. Then RNA was precipitated with isopropanol, centrifuged at 20,000 × g for 20 min, resuspended in RNA loading buffer (1× MOPS [morpholinepropanesulfonic acid] buffer [pH 7.0] [see below], with 50% formamide, 18% formaldehyde, and 0.4 mg/ml ethidium bromide) and separated by electrophoresis on a denaturing 6% formaldehyde–0.8% agarose gel in MOPS (40 mM MOPS, 10 mM NaOAc, 1 mM EDTA [pH 7.0]). The gel was transferred to Hybond N+ membrane (GE Healthcare) and cross-linked to the membrane using a UV Stratalinker 2400 (Stratagene). Then the blot was blocked (blocking solution: 125 mM NaCl, 17 mM Na2HPO4, 7.3 mM NaH2PO4, 1% sodium dodecyl sulfate [SDS]), probed with streptavidin-horseradish peroxidase (HRP) (Pierce) (1:10,000 dilution in blocking solution), and washed 2 times with wash I (12.5 mM NaCl, 1.7 mM Na2HPO4, 0.73 mM NaH2PO4, 0.1% SDS) and 2 times with wash II (10 mM Tris-HCl, 10 mM NaCl, 2.1 mM MgCl2). HRP was detected by enhanced chemiluminescence (ECL) Western blotting substrate (Pierce) and autoradiography.
TUX-MS.
A total of 1 × 108 to 2 × 108 HeLaUPRT cells were either PV infected or not in the presence of Act D as described above. After 1 h, the medium was replaced with DMEM, 10% FBS, 10 μg/ml Act D, and 1 mM 4-thiouracil. Five hours postinfection (5 hpi) the cells were washed with PBS, and just enough PBS was left on the cells to cover them. The cells were exposed to 365 nm UV for 20 min, followed by lysis (100 mM Tris-HCl [pH 7.5], 500 mM lithium chloride [LiCl], 10 mM EDTA [pH 8], 0.1% lithium dodecyl sulfate [LDS], 1% NP-40, 1% deoxycholic acid [DOC], 5 mM dithiothreitol [DTT], 1× cOmplete protease inhibitor cocktail [Roche]). Lysates were incubated with Dynabead oligo(dT)25 beads (Invitrogen) twice. The eluates were concentrated to 10 μl using Amicon Ultra-0.5 3-kDa columns (Millipore) and treated with 10 ng bovine RNase A (Fisher Scientific) per sample. Cross-linking of the [35S]methionine-labeled viral proteins was performed exactly as described, except for the following changes. At 2.5 hpi, the cells were incubated in starvation medium (DMEM minus l-methionine and l-cysteine [Mediatech] plus 10% dialyzed FBS, 10 μg/ml Act D, and 1 mM 4-thiouracil) for 30 min. Then at 3 hpi, the cells were incubated in starvation medium plus 0.2 mCi/ml [35S]methionine/cysteine (PerkinElmer). At 5 hpi, cells were cross-linked and RNA-protein complexes were isolated using Dynabead oligo(dT)25 beads (Invitrogen) (as described above). The proteins were analyzed by SDS-PAGE and autoradiography.
LC-MS/MS and data analysis.
Proteins were digested in solution with trypsin (Promega), separated by reverse-phase nano-liquid chromatography (nano-LC) using a linear 3-h liquid chromatography gradient (Ultimate 3000 RSLC; Dionex), and detected online by tandem mass spectrometry (MS/MS) performed on an electrospray ionization linear trap quadrupole (ESI-LTQ) Orbitrap XL mass spectrometer (Thermofisher Scientific), as described previously (21). Peptide precursors were detected at a resolution of 60,000 and selected for collision-induced dissociation (CID) fragmentation in the LTQ by a data-dependent Top10 method. Thermo RAW data files were processed, and MS2 spectra were searched by Proteome Discoverer/SEQUEST (v. 1.2) against the human subset of UniProt-Swiss-Prot, appended with common contaminants and custom PV protein sequences. Probabilistic calculation of false-positive rates was performed by Scaffold/X! Tandem (v.3.0; Proteome Software) using the Peptide Prophet algorithm (22). Protein identifications were accepted at ≥99.0% protein probability, as assigned by the Protein Prophet algorithm (23), with at least 2 unique peptides having >90% peptide probability in one biological replicate and at minimum one unique peptide having >90% peptide probability in the other biological replicate. These criteria resulted in an estimated global false discovery rate (FDR) of <1% at the protein and peptide level. Spectral counting analyses were performed for proteins with ≥4 assigned spectra, with a threshold of ≥2-fold increase in spectral counts classified as a significant enrichment in PV-infected samples. Gene ontology (GO) classification was performed using either ProteinCenter (version 3.7), the BiNGO Cytoscape plugin (24, 25) using external ontology and gene association annotations (downloaded from www.geneontology.org in August 2011), or by ClueGO (26). For gene ontology enrichment analysis using ProteinCenter, the reference gene set (35,613 entries) was from the Swiss-Prot database. Functional network analysis was performed and visualized by STRING (www.string-db.org) and Cytoscape, respectively, using default settings, except text mining evidence was disabled and a combined STRING confidence score of >0.5 was required to retain functional associations.
siRNA transfections.
For PCBP2, hnRNP L, RSL1D1, XRCC6, DDX17, DDX5, NONO, and CNBP (protein designations defined below), HeLaUPRT cells were transfected with either the DS negative scrambled control (IDT), PCBP2 (E2 siRNA) (27) hnRNP L (Ambion Silencer Select s6741; Applied Biosystems), RSL1D1 (s25187), XRCC6 (s5457), DDX17 (p72/p82 siRNA) (28), DDX5 (s4008), NONO (s9614), NONO_s2 (s9613), or CNBP (s230174) siRNAs using the Neon transfection system (Invitrogen). Two days after transfection, the cells were infected with PV or adenovirus 5 (MOI of 0.1). Alternatively, Silencer Select siRNA against hnRNP U (s6745) or control siRNA was transfected twice at 0 and 3 days and at 5 days. HeLaUPRT cells were infected with PV, mengovirus, or adenovirus 5 at an MOI of 0.1. At either 6 (poliovirus), 5.5 (mengovirus), or 30 (adenovirus 5) hpi, virus was harvested and titers were determined using HeLaUPRT cells. Knockdown efficiency was determined at the time of infection by either Western analysis or quantitative reverse transcription (qRT)-PCR using iQ SYBR green Supermix (Bio-Rad) and gene-specific primers (Table 1). Total protein was obtained by cell lysis in E1 lysis buffer (50 mM HEPES [pH 7.0], 250 mM NaCl, 0.1% NP-40). Protein was separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membrane (Millipore), and probed with either anti-PCBP2 (29), mouse anti-β-actin (sc-47778; Santa Cruz Biotechnology), mouse anti-hnRNP U 3G6 (30), or chicken anti-β-actin (ab14001; Ambion). Secondary antibodies were either HRP conjugated (sc-2005; Santa Cruz Biotechnology) or IRDye 680CW conjugated (LI-COR), and detection was performed by ECL substrate (Pierce) and autoradiography or LI-COR Odyssey, respectively. Cell viability was measured 48 h after siRNA-mediated knockdown of individual host factors. Briefly, cells were transfected via electroporation with 20 nM indicated siRNA and plated either at 5 × 104 cells per well in a 24-well plate for RNA isolation or at 8.5 × 103 cells per well in a 96-well plate for cell viability. 48 hpi, qRT-PCR of total RNA isolated using TRIzol (Invitrogen) was performed to measure the efficiency of the knockdown, and cell viability was measured using the Vybrant MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay kit (Invitrogen) according to the manufacturer's protocol. Cell viability for each knockdown was reported relative to negative-control siRNA (set to 100%; n = 3).
Real-time qPCR.
Total RNA was extracted from cells using TRIzol (Ambion). One microgram of RNA was used to generate cDNA with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega) as described by the manufacturer using a PV-specific primer, either PVpos reverse or PVneg reverse, for the plus or minus strand of poliovirus, respectively (Table 1). qPCR was performed using iQ SYBR green Supermix (Bio-Rad) and complementary strand-specific primers (PVpos forward and reverse or PVneg forward and reverse) (Table 1). Copy number was calculated using a standard curve generated from known quantities of in vitro-transcribed plus- or minus-strand poliovirus RNA.
Immunoprecipitation.
At 5 hpi, cells were harvested and resuspended in 1 ml of FA lysis buffer (50 mM HEPES KOH [pH 7.5], 140 mM NaCl, 0.1% [wt/vol], sodium deoxycholate, 1% Triton X-100, 1 mM EDTA, 1× cOmplete protease inhibitors, EDTA free [Roche Applied Science], 1 μl/ml RNasin [Promega]) on ice for 15 min. Lysates were clarified by centrifugation (15000 × g, 10 min, 4°C). RNA coimmunoprecipitations (co-IP) were carried out essentially as described previously (31). Briefly, lysates were precleared with 75 μl of a 50% protein A-Sepharose bead (Sigma-Aldrich) in TE (10 mM Tris-Cl [pH 8.0], 1 mM EDTA) for 1 h at 4°C. The beads were removed via centrifugation (4,000 × g, 1.5 min, 4°C). Ten percent of the precleared lysate was set aside for the input. Aliquots of the remaining precleared lysate were incubated overnight at 4°C rotating with 1 μg polyclonal antibody to NONO (ab70335; AbCam), 1 μg monoclonal c-Myc antibody (SC40; Santa Cruz Biotechnology), or no antibody. Protein A-Sepharose beads (25 μl of a 50% slurry in TE) were rotated for 90 min at 4°C. Beads were pelleted by centrifugation (4,000 × g, 1.5 min, 4°C) and washed with FA lysis buffer, FA500 lysis buffer (FA lysis buffer with 500 mM NaCl), with LiCl buffer (10 mM Tris-Cl [pH 8.0], 250 mM LiCl, 0.5% [wt/vol] sodium deoxycholate, 0.5% NP-40, 1 mM EDTA, 1× cOmplete protease inhibitors, 1 μl/ml RNasin), and with TE plus 1 μl/ml RNasin. Then coimmunoprecipitates were eluted with 100 μl of RIP elution buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 1% [wt/vol] SDS, 1 μl/ml RNasin). NaCl was brought up to 300 mM and 20 μg of proteinase K (Sigma-Aldrich) for input, supernatant, and coimmunoprecipitate and incubated at 42°C for 1 h and then at 65°C for 15 min. The coimmunoprecipitates were pelleted (4,000 × g, 1.5 min, 4°C), and RNA was extracted using TRIzol (Invitrogen). cDNA was generated from equivalent amounts (1/10 of starting material) input, supernatant, and coimmunoprecipitate using MMLV reverse transcriptase (Promega) and random hexamers. cDNA (2.5 μl) was amplified by PCR with PV-specific primers (32) (PVpos forward and reverse; Table 1) and visualized by ethidium bromide on an agarose gel.
Translation assays.
HeLaUPRT cells were transfected with scrambled control, NONO, CNPB, or PCBP2 siRNAs as described above and plated at 5 × 104 cells/well in a 24-well plate. At 48 h post-siRNA transfection, the cells were cotransfected with both the pRFPV plasmid and the β-galactosidase reporter plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols with 0.4 μg of each plasmid per well. Twenty-four hours later (72 h post-siRNA transfection), the cells were washed once with PBS and lysed in 100 μl of Tropix lysis solution (100 mM potassium phosphate [pH 7.8], 0.2% Triton X-100 [Applied Biosystems]). Renilla and firefly luciferase activities were measured using 4 μl of lysate and the dual-luciferase kit (Promega) according to the manufacturer's protocols. β-Galactosidase activity was measured using the Galacto-Light Plus kit (Applied Biosystems) and the manufacturer's protocols. All activities were measured using an FB 12 luminometer (Berthold), all assays were measured in duplicate, and experiments were performed in triplicate. IRES activity was normalized to β-galactosidase activity and expressed as a percentage of that of the scrambled control. Knockdown efficiency was determined as described previously.
Nucleotide sequence accession numbers.
Sequences were submitted to the Swiss-Prot database under the following accession numbers: RSL1D1 (ribosomal L1 domain-containing protein 1), O76021; CNBP (cellular nucleic acid-binding protein), P62633; XRCC6 (X-ray repair cross-complementing protein 6), P12956; hnRNP U (heterogeneous nuclear ribonucleoprotein U), Q00839; DDX5 (probable ATP-dependent RNA helicase), P17844; NONO (non-POU domain-containing octamer-binding protein), Q15233; DDX17 (probable ATP-dependent RNA helicase DDX17), Q92841; hnRNP L (heterogeneous nuclear ribonucleoprotein L), P14866; PCBP2 [poly(rC)-binding protein 2], Q15366; PV (poliovirus), P03300; and mengovirus, P12296.
RESULTS
Viral RNA can be exclusively labeled in infected cells.
Cells that stably express uracil phosphoribosyltransferase (UPRT) from Toxoplasma gondii will incorporate 4-thiouridine (4sU) into all newly synthesized RNA (Fig. 1A, lane 1) when 4-thiouracil (4TU) is present in the medium (33). Briefly, UPRT converts 4TU into UMP, which is then converted by cellular kinases to thiouridine triphosphate (4sUTP). The cellular and viral polymerases can use 4sUTP as a substrate during RNA synthesis, resulting in incorporation of 4sU into newly synthesized RNAs. This analog only differs from uracil by the exchange of the 4-keto oxygen atom for a sulfur atom.
Fig 1.
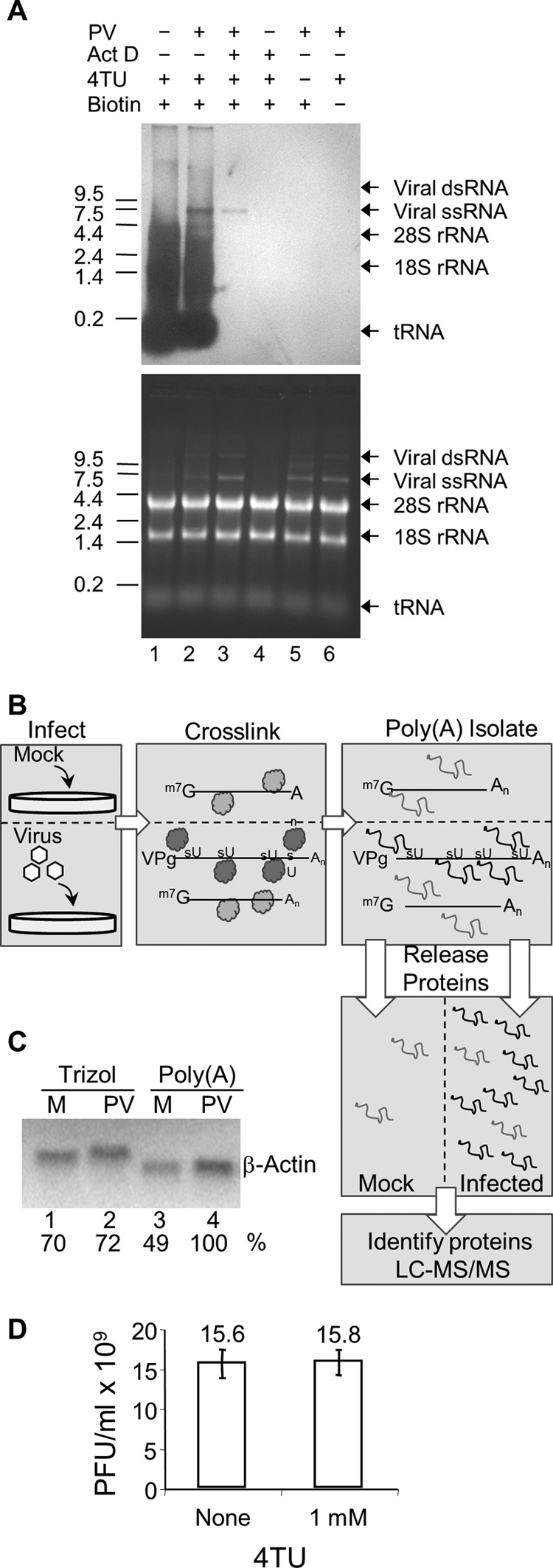
The TUX-MS method. (A) 4-Thiouracil can be exclusively incorporated into poliovirus RNA in vivo. HeLaUPRT cells were incubated with actinomycin D (Act D), poliovirus, or 4TU (30 min after addition of Act D or PV) as indicated. Total RNA was isolated and cross-linked to a thio-reactive biotin (lanes 1 to 5), separated on a denaturing formaldehyde-agarose gel, transferred to a membrane, and probed with streptavidin-HRP to detect thio-containing RNAs (top). Total RNA was visualized by ethidium bromide staining of the gel (bottom) prior to transfer to the membrane. (B) Diagram of the TUX-MS method. HeLaUPRT cells are mock infected or PV infected in the presence of 4TU and Act D. 4sU is exclusively incorporated into PV viral RNA. UV cross-linking of bound proteins to the thio-containing viral RNA (represented as dark gray balls or lines) is shown. Proteins that are bound to the non-thio-containing mRNA are not cross-linked (represented as light gray balls or lines). Poly(A) RNA is isolated using oligo(dT)25 magnetic beads under denaturing conditions. The RNA is degraded with RNase A releasing the proteins from the complex. The proteins are trypsinized, and the peptides are identified by LC-MS/MS. (C) Prior to RNase A digestion, mock-infected (lanes M) and PV-infected samples are normalized to one another based on levels of cellular transcripts. RNA from mock- and PV-infected cells was isolated either by TRIzol or by the TUX-MS method, and either equal micrograms (total mRNA) or equal volumes [poly(A) selected] of RNA were subjected to β-actin Northern analysis. Band intensities were quantified on a PhosphorImager, and relative mRNA levels are indicated. Similar results were obtained with α-tubulin (data not shown). (D) Cells were infected with PV in the presence or absence of 4TU. At 8 hpi, virus was harvested and the titer was determined by plaque assay.
When HeLaUPRT cells were incubated with 4TU, all newly synthesized RNA incorporated 4sU, as seen by the heterogeneous distribution of mRNA detected by Northwestern analysis (Fig. 1A, top; lane 1). If cells were infected with PV for 5 h, an additional viral RNA band appears at 7.5 kb, corresponding to newly synthesized viral RNA (lane 2). Addition of actinomycin D (Act D), which inhibits cellular transcription, but not the viral polymerase results in labeling of only the viral RNA (lane 3) (34). Therefore, 4sU can be exclusively incorporated into PV RNA. If 4TU was omitted from the medium or the RNA was not labeled with biotin, then no RNA was detected on the Northwestern blot (Fig. 1A, top, lanes 5 and 6), despite the presence of equal amounts of RNA in all lanes (Fig. 1A, bottom). This demonstrates that this labeling method is specific. PV RNA is visible as a single-stranded form and a double-stranded replication intermediate (Fig. 1A, bottom, lanes 2, 3, 5, and 6) (35). However, only the single-stranded RNA appears on the Northwestern blot from the infected cells (Fig. 1A, lanes 2 and 3) but was not present in the mock-infected cells (Fig. 1A, lane 4), since the reactive sulfur participates in the hydrogen bonds required for base pairing in the double-stranded form, making it inaccessible for biotin labeling. Importantly, incorporation of 4TU into the viral RNA had no effect on viral amplification (Fig. 1D).
Isolation and identification of proteins that interact with viral RNA using TUX-MS.
Incorporation of 4sU into RNA functions as a zero-distance cross-linker upon exposure to long-wave UV light. Cross-linking ensures that only proteins that are bound to the viral RNA under physiological conditions in the cell will be isolated. This reduces potential background from proteins that nonspecifically bind to viral RNA after the loss of spatial organization during cell lysis. In addition, protein-protein cross-linking is very inefficient at long UV wavelengths in the absence of photoreactive cross-linkers (36).
To identify proteins that bind directly to the viral RNA during infection, cells were either mock infected or PV infected in the presence of Act D and 4TU (Fig. 1B). Then 5 h postinfection (hpi), cells were irradiated to cross-link proteins bound to the 4sU-containing RNA prior to cell lysis. Since PV RNA is polyadenylated, cross-linked RNA-protein complexes were isolated from lysates using oligo(dT)25 magnetic beads under denaturing conditions. This purification resulted in the isolation of both viral RNA and cellular mRNAs. However, since Act D was present, only the viral RNA incorporated 4sU (Fig. 1A, lane 3) and thus cross-linked to proteins. The mock-infected sample served to identify nonspecific coisolated proteins. We found that the PV-infected samples prepared from oligo(dT)25-based capture consistently yielded more cellular mRNAs than uninfected cells. However, RNA extracted by TRIzol consistently yielded equivalent cellular mRNA levels, suggesting that recovery of RNA using the lysis conditions that are compatible with oligo(dT)25-based isolation was biased (i.e., more efficient) toward PV-infected cells (Fig. 1C). The most likely explanation is that this difference is due to the significant cytopathic effect (CPE) by 5 hpi in PV-infected cells, which leads to a more efficient lysis of the PV-infected cells under these conditions. Therefore, equal amounts of mock-infected and infected samples were used based on the levels of cellular mRNA determined by Northern analysis (Fig. 1C, lanes 3 and 4). This normalization allowed comparison of relative protein abundance in mock- versus PV-infected samples using label-free quantitative mass spectrometry. Proteins were released from the purified RNA-protein complexes by digestion with RNase A, digested in solution with trypsin, and identified using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Two PV-infected samples were prepared independently for TUX-MS analysis and were found to be highly reproducible (see Data Set S1 in the supplemental material).
Identification of viral proteins that interact with the poliovirus RNA.
The PV-infected samples yielded an average of 75 unique peptides (129 assigned spectra) for the PV type 1 polyprotein, representing 46% sequence coverage (Fig. 2A). The PV RNA encodes a single polyprotein that is proteolytically cleaved into mature proteins. However, several precursor (uncleaved) proteins (such as 3CD) have functions in the viral life cycle independent of the fully processed proteins (3Cpro and 3D). Mature and precursor proteins can be distinguished by mass spectrometry if peptides are identified that have a tryptic cleavage on one end and viral protease cleavage on the other or the peptides clearly bridge a viral cleavage site, respectively. Therefore, the presence of mature viral proteins was experimentally determined by [35S]methionine labeling prior to cross-linking to the viral RNA. Since PV infection shuts down translation of host mRNAs, the only proteins that are [35S]methionine labeled are viral proteins (Fig. 2B, compare lanes 1 to 3 with 6 to 8). The viral proteins that cross-linked to PV RNA are VP0, VP3, VP1, 2C, 3CD, 3Cpro, and 3Dpol (Fig. 2B). 3B (VPg), the genome-linked protein, was most likely run off the bottom of the gel due to its small size (<3 kDa). However, a single peptide was detected by LC-MS/MS that corresponded to a region within 3B (data not shown). Nearly all of these proteins or their precursors have been shown to bind to PV RNA in vitro (29, 37–42). Importantly, TUX-MS identified peptides that cover all of the viral proteins expected to interact with the viral RNA and confirmed their association with the PV RNA in cell culture.
Fig 2.
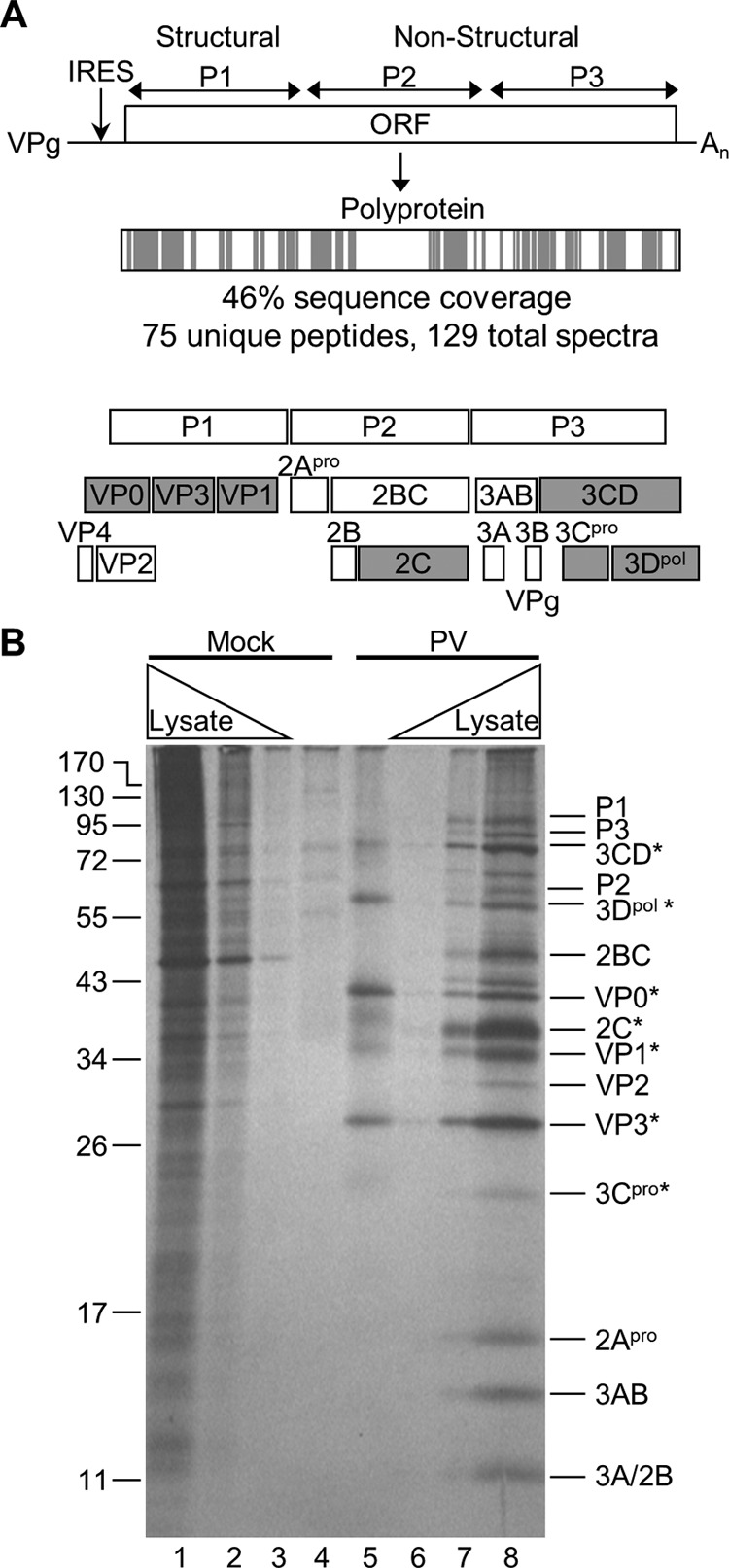
PV proteins identified by TUX-MS. (A) The PV genome encodes a polyprotein that is cleaved by viral proteases into functional precursors and mature proteins. The PV polyprotein is shaded to represent the location of the identified peptides (gray) that were detected by TUX-MS in samples from PV-infected cells. The cleavage of the precursor and mature PV proteins is shown below. The PV proteins that are shaded gray were experimentally identified in panel B. (B) Detection of PV proteins that cross-link to the viral RNA in cells. Cells were mock infected (lanes 1 to 4) or PV infected (lanes 5 to 8) in the presence of Act D and 4TU. Proteins were pulse-labeled with [35S]methionine for 2 h prior to cross-linking at 5 hpi, and cells were lysed. Mock-infected (lane 4) and PV-infected (lane 5) lysates were purified using oligo(dT)25 magnetic beads, RNase A treated, separated by SDS-PAGE, and visualized by autoradiography (lanes 4 and 5). Increasing amounts of whole-cell [35S]methionine-labeled lysates were obtained from mock-infected (lanes 1 to 3) and PV-infected (lanes 6 to 8) cells. PV proteins detected in the infected whole-cell lysate are indicated (right); asterisks indicate the PV proteins that cross-linked to the viral RNA (lane 5).
Identification of cellular proteins that interact with the PV RNA.
Aside from viral proteins, several cellular proteins are known to act as viral RNA-binding host factors for PV and related viruses in the Picornaviridae family (43–45). All of the 15 known host proteins were identified using TUX-MS (Table 2). These served as positive controls, which validated the TUX-MS method and demonstrated that it is robust. In addition, proteins that are involved in protein synthesis, such as initiation factors (eukaryotic initiation factors eIF4G, eIF4A, eIF3, and eIF4H), elongation factors (eIF5A and eEF2), and ribosomal proteins (S10, S2, S3, S14, S26, S5, S9, L22, L18, L3, L4, L6, L7, and L8) were also detected (see Data Set S1 in the supplemental material), as would be expected for viral RNAs that were being actively translated.
Table 2.
Known picornaviral host factors identified by TUX-MS
| Protein name | Designation | Accession no.a | Fold increase (no. of spectra)b | Cellular localizationc | Function(s) |
Reference | |
|---|---|---|---|---|---|---|---|
| Cellulard | Virale | ||||||
| Nucleolin | NCL | P19338 | 50 (50) | N/C | RNA binding | Translation | 79 |
| Lupus La protein | SSB | P05455 | 11 (11) | N | RNA binding | Translation | 13 |
| Interleukin enhancer-binding factor 2 (NF45) | ILF2 | Q12905 | 3.9 (16) | N/C | Transcriptional regulation | Translation, inhibition | 80 |
| ATP-dependent RNA helicase A | DHX9 | Q08211 | 3.3 (56) | N/C | RNA helicase | Replication | 81 |
| Splicing factor, arginine/serine-rich 3 (SRP20) | SFRS3 | P84103 | 3.3 (20) | N | RNA binding | Translation | 82 |
| Interleukin enhancer-binding factor 3 (DRBP76) | ILF3 | Q12906 | 2.1 (65) | N/C | Transcriptional regulation | Translation, inhibition | 80 |
| Heterogeneous nuclear ribonucleoprotein K | HNRNPK | P61978 | 2.1 (54) | N/C | RNA binding | Replication | 56 |
| Poly(rC)-binding protein 2 | PCBP2 | Q15366 | 1.9 (21) | N/C | RNA binding | Translation, replication | 83 |
| Poly(rC)-binding protein 1 | PCBP1 | Q15365 | 1.7 (22) | N/C | RNA binding | Replication | 60 |
| Heterogeneous nuclear ribonucleoproteins C1/C2 | HNRNPC | P07910 | 1.6 (28) | N | RNA binding | Replication | 84 |
| Cold shock domain-containing protein E1 (UNR) | CSDE1 | O75534 | 1.2 (32) | C | RNA binding | Translation | 85 |
| Polypyrimidine tract-binding protein 1 | PTBP1 | P26599 | 1.2 (25) | N | RNA binding | Translation | 86 |
| Far upstream element-binding protein 2 | KHSRP | Q92945 | 1.1 (28) | N/C | RNA binding | Translation | 87 |
| Polyadenylate-binding protein 1 | PABPC1 | P11940 | 0.8 (55) | N/C | Poly(A) binding | Translation | 88 |
| Proliferation-associated protein 2G4 (ITAF45) | PA2G4 | Q9UQ80 | N/A (3) | N/C | RNA binding | Translation | 89 |
UniProt-SwissProt accession number.
Average fold increase in total spectra for PV-infected sample versus mock-infected control.
Cellular localization is indicated as nuclear (N) or cytoplasmic (C). N/C, proteins known to shuttle between the nucleus and the cytoplasm.
General cellular functions of the protein.
Stage of the virus life cycle in which the protein is active.
The primary goal of using TUX-MS was to identify host factors not previously described to be involved in PV amplification. Therefore, in an unbiased fashion, proteins identified by TUX-MS were evaluated by spectral count enrichment as a measure of relative protein abundance in PV- versus mock-infected samples. In total, 82 cellular proteins (see Data Set S1 in the supplemental material) passed strict inclusion criteria (see Materials and Methods). Then, we applied several bioinformatics strategies to evaluate the ability of TUX-MS to enrich for RNA-interacting host factors and also to uncover potential novel protein functions or pathways involved in PV amplification. The respective gene ontology (GO) terms and protein family (Pfam) domains represented by these 82 host proteins were statistically compared to GO terms represented by the background human genome (see Materials and Methods). We found a significant overrepresentation of nucleic acid-binding (79/82) and RNA processing (50/82) gene ontology terms and the RRM1 (RNA recognition motif 1) domain (39/80) (Fig. 3C), suggesting that these proteins have RNA binding functions, as would be expected for proteins identified by the TUX-MS method. However, using TUX-MS we identified less than 2.6% of the cellular proteins with RNA binding capability, demonstrating the selectivity for this method. Furthermore, the isolated proteins have a diverse range of abundances, ranging from less than 1 ppm to over 9,000 ppm; therefore, isolation was not based on protein abundance. Sixteen of the 82 PV-enriched proteins are translation factors, ribosomal proteins, or are known to be involved in PV amplification. The remaining 66 host proteins (Table 3) have not previously been implicated in enterovirus amplification, but 63 are known or predicted to contain conserved nucleic acid binding domains. Notably, 23 proteins were exclusively detected in the samples isolated from the PV-infected cells (Table 3; INF), while 43 proteins were enriched in the RNP complexes isolated from PV-infected cells at least 2-fold over mock-infected cells using spectral counting analysis. Half of the known host factors, such as PCBP2, did not meet these strict inclusion criteria. This implies that there may be additional potential host factors in the complete data set (see Data Set S1) that did not pass the spectral counting-based enrichment threshold. Nevertheless, we opted for a more stringent filtering to focus on those host factors that are most predominantly associated with viral RNA.
Fig 3.
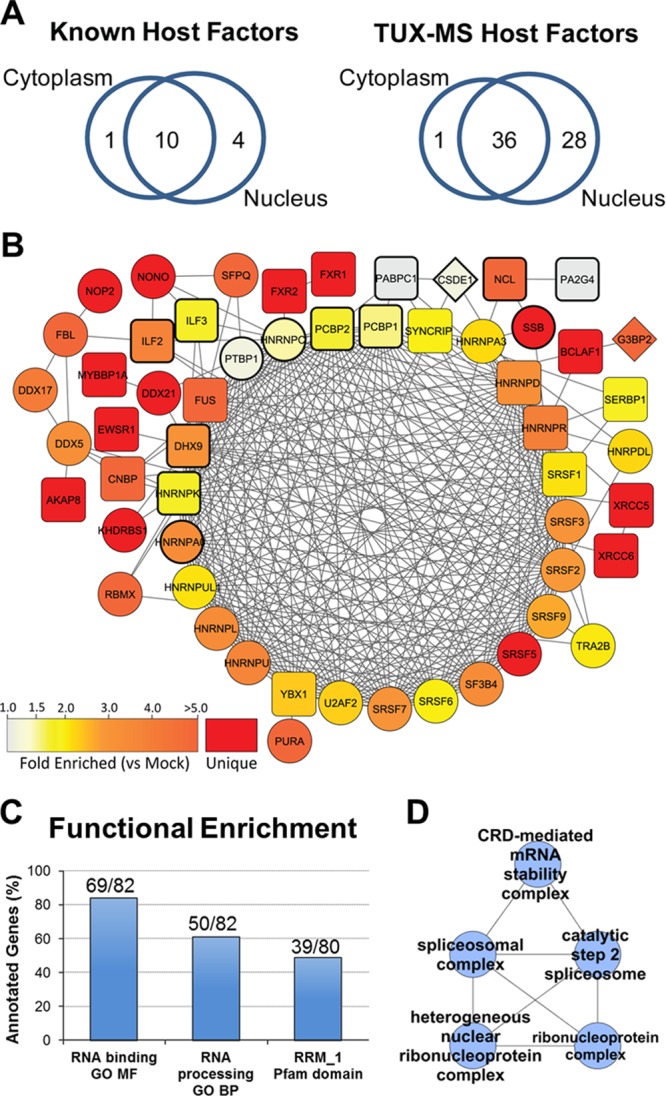
Functional analysis of known and putative host factors reveals shared cellular functions in nucleocytoplasmic shuttling, spliceosome assembly, and pre-mRNA editing. (A) Venn diagrams were constructed based on the subcellular localization(s) of known (left) and TUX-MS (right) host factors classified by gene ontology annotation. (B) STRING functional association network representing 54/81 host proteins (see Tables 1 and 2). Nodes are labeled with the proteins' primary gene names. Bold node outlines indicate proteins previously identified as PV host factors (Table 1). Node colors represent a range of average spectral count fold enrichment (1.0- to ≥5-fold) in PV-infected versus mock-infected samples (n = 2). Red nodes correspond to host factors detected only in PV-infected samples. Node shape corresponds to its cellular localization: circles, nucleus; diamonds, cytoplasm; squares, both. Functional associations were retained with a combined score of >0.5. Associations represented by multiple lines of evidence were collapsed to a single edge. (C) Proteins identified as unique or enriched in PV-infected samples (n = 82) were uploaded to ProteinCenter (version 3.7). Functional overrepresentation was determined versus the Swiss-Prot reference gene set (35,613 entries). The most statistically significant terms for molecular function (GO MF), biological processes (GO BP), and Pfam domains are indicated as percentages of annotated genes. RRM_1, RNA recognition motif, RNP-1. FDR-corrected P values were 1.8e−60, 5.7e−39, and 2.7e−47, respectively. (D) Proteins from panel C were analyzed by ClueGO functional clustering. The network highlights functional clusters of ribonucleoprotein, spliceosomal, and CRD-mediated mRNA stability complexes.
Table 3.
The 66 TUX-MS-identified host factors that have not been previously implicated in the enteroviral life cyclea
| Protein name | Designation | Accession no.b | Mol mass (kDa) | No. of unique peptidesc | Total no. of spectrad | Fold incorporatede | Gene ontologyf | Cellular localizationg | Relative abundanceh | Interaction |
Reference(s) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virusi | IRESj | |||||||||||
| Non-POU domain-containing octamer-binding protein | NONO | Q15233 | 54.2 | 14 | 21 | INF | RRM | N | ++++ | HIV-1, HDV | c-Myc | 90–92 |
| Nucleolar RNA helicase 2 | DDX21 | Q9NR30 | 87.3 | 15 | 17 | INF | RBP | N | +++ | BDV | 93 | |
| Ribosomal L1 domain-containing protein 1 | RSL1D1 | O76021 | 55.0 | 13 | 15 | INF | RBP | N/C | +++ | |||
| Ribonucleases P/MRP protein subunit POP1 | POP1 | Q99575 | 114.7 | 12 | 14 | INF | RBP | N/C | ++ | |||
| Myb-binding protein 1A | MYBBP1A | Q9BQG0 | 148.9 | 12 | 12 | INF | DBP | N/C | +++ | |||
| SAFB-like transcription modulator | SLTM | Q9NWH9 | 117.2 | 8 | 11 | INF | RRM | N | ++ | |||
| KH domain-containing, RNA-binding, signal transduction-associated protein 1 | KHDRBS1 | Q07666 | 48.2 | 10 | 11 | INF | RBP | N | ++++ | HIV-1 | 94 | |
| TATA-binding protein-associated factor 2N | TAF15 | Q92804 | 61.8 | 4 | 10 | INF | RRM | N/C | +++ | |||
| Cellular nucleic acid-binding protein (ZNF9) | CNBP | P62633 | 19.5 | 7 | 9 | INF | RBP | N/C | ++++ | JCV | ODC | 95–97 |
| Putative rRNA methyltransferase NOP2 | NOP2 | P46087 | 89.3 | 8 | 9 | INF | RBP | N | ++ | |||
| Fragile X mental retardation syndrome-related protein 1 | FXR1 | P51114 | 69.7 | 7 | 7 | INF | RBP | N/C | +++ | |||
| Fragile X mental retardation syndrome-related protein 2 | FXR2 | P51116 | 74.2 | 4 | 7 | INF | RBP | N/C | ++ | |||
| X-ray repair cross-complementing protein 5 | XRCC5 | P13010 | 82.7 | 6 | 7 | INF | DBP | N/C | ++++ | |||
| X-ray repair cross-complementing protein 6 | XRCC6 | P12956 | 69.8 | 5 | 6 | INF | DBP | N/C | ++++ | PDGF2, VEGF | 98 | |
| Heterogeneous nuclear ribonucleoprotein L-like | HNRPLL | Q8WVV9 | 60.1 | 4 | 5 | INF | RRM | N | ++ | |||
| RNA-binding protein 28 | RBM28 | Q9NW13 | 85.7 | 5 | 5 | INF | RRM | N/C | ++ | |||
| Bcl-2-associated transcription factor 1 | BCLAF1 | Q9NYF8 | 106.1 | 5 | 5 | INF | DBP | N/C | +++ | |||
| Histone H2B type 1-D | HIST1H2BD | P58876 | 13.9 | 2 | 5 | INF | DBP | N | +++++ | |||
| RNA-binding protein EWS | EWSR1 | Q01844 | 68.5 | 4 | 5 | INF | RRM | N/C | +++ | HCV | HCV | 99 |
| Splicing factor, arginine/serine-rich 5 | SFRS5 | Q13243 | 31.3 | 3 | 4 | INF | RRM | N | +++ | |||
| Importin subunit alpha-2/karyopherin alpha 2 | KPNA2 | P52292 | 57.9 | 4 | 4 | INF | RBP | N/C | ++++ | |||
| A-kinase anchor protein 8 | AKAP8 | O43823 | 76.1 | 4 | 4 | INF | DBP | N/C | ++ | |||
| Nucleolysin TIA-1 isoform p40 | TIA1 | P31483 | 43.0 | 3 | 10 | INF | RRM | N/C | ++ | |||
| RNA-binding protein FUS | FUS | P35637 | 53.4 | 12 | 26 | 25.5 | RRM | N/C | +++ | |||
| Polyubiquitin-B | UBB | P0CG47 | 25.8 | 7 | 25 | 25 | N/C | ++++ | ||||
| Splicing factor, proline- and glutamine-rich (PSF) | SFPQ | P23246 | 76.1 | 15 | 21 | 21.0 | RRM | N | ++++ | HDV | c-Myc | 92, 100 |
| Zinc finger protein 326 | ZNF326 | Q5BKZ1 | 65.7 | 10 | 12 | 11.5 | DBP | N | +++ | |||
| Zinc finger RNA-binding protein | ZFR | Q96KR1 | 117.0 | 10 | 11 | 11.0 | RBP | N/C | ++ | |||
| Zinc finger protein 638 | ZNF638 | Q14966 | 220.6 | 15 | 18 | 8.8 | RRM | N/C | + | |||
| Heterogeneous nuclear ribonucleoprotein U-like protein 2 | HNRNPUL2 | Q1KMD3 | 85.1 | 15 | 21 | 7.0 | RBP | N | +++ | |||
| Heterogeneous nuclear ribonucleoprotein G | RBMX | P38159 | 42.3 | 18 | 35 | 5.8 | RRM | N | +++ | |||
| Ras GTPase-activating protein-binding protein 2 | G3BP2 | Q9UN86 | 54.1 | 5 | 6 | 5.5 | RRM | C | +++ | |||
| rRNA 2′-O-methyltransferase fibrillarin | FBL | P22087 | 33.8 | 5 | 6 | 5.5 | RBP | N | +++ | |||
| Transcriptional activator protein Pur-alpha | PURA | Q00577 | 34.9 | 5 | 5 | 5.0 | DBP | N/C | +++ | |||
| RNA-binding protein 4 | RBM4 | Q9BWF3 | 40.3 | 8 | 9 | 4.3 | RRM | N/C | +++ | |||
| Probable ATP-dependent RNA helicase DDX17 | DDX17 | Q92841 | 72.4 | 23 | 50 | 4.1 | RBP | N | +++ | |||
| Heterogeneous nuclear ribonucleoprotein R | HNRNPR | O43390 | 70.9 | 22 | 65 | 4.0 | RRM | N/C | ++++ | |||
| Zinc finger CCCH-type antiviral protein 1 | ZC3HAV1 | Q7Z2W4 | 101.4 | 11 | 12 | 4.0 | RBP | N/C | +++ | RNA viruses | 65 | |
| Heterogeneous nuclear ribonucleoprotein U (SAF-A) | HNRNPU | Q00839 | 90.5 | 55 | 138 | 3.8 | RBP | N/C | ++++ | |||
| Heterogeneous nuclear ribonucleoprotein L | HNRNPL | P14866 | 64.1 | 25 | 102 | 3.5 | RRM | N/C | ++++ | HDV, HSV, HCV | HCV, Cat-1 | 67–69, 91, 101 |
| RNA-binding protein 39 | RBM39 | Q14498 | 59.4 | 4 | 4 | 3.5 | RRM | N/C | +++ | |||
| Splicing factor 3B subunit 4 | SF3B4 | Q15427 | 44.4 | 3 | 4 | 3.5 | RRM | N | ++ | |||
| Heterogeneous nuclear ribonucleoprotein A0 | HNRNPA0 | Q13151 | 30.8 | 9 | 11 | 3.5 | RRM | N | ++++ | |||
| Protein FAM98A | FAM98A | Q8NCA5 | 55.4 | 3 | 4 | 3.5 | +++ | |||||
| Peptidyl-prolyl cis-trans-isomerase A | PPIA | P62937 | 18.0 | 3 | 4 | 3.5 | N/C | +++++ | ||||
| Probable ATP-dependent RNA helicase DDX5 | DDX5 | P17844 | 69.1 | 28 | 51 | 3.4 | RBP | N | +++ | HCV, IAV | 66, 102 | |
| Heterogeneous nuclear ribonucleoprotein D0 (AUF1) | HNRNPD | Q14103 | 38.4 | 17 | 31 | 3.4 | RRM | N/C | ++++ | |||
| Ubiquitin-associated protein 2-like | UBAP2L | Q14157 | 114.5 | 9 | 14 | 3.4 | DBP | N | +++ | |||
| La-related protein 1 | LARP1 | Q6PKG0 | 123.5 | 9 | 10 | 3.3 | RBP | N/C | ++ | |||
| Serine/arginine-rich splicing factor 10 | SFRS13A | O75494 | 31.3 | 5 | 7 | 3.3 | RRM | N/C | ++ | |||
| Splicing factor, arginine/serine-rich 7 | SFRS7 | Q16629 | 27.3 | 8 | 13 | 3.3 | RRM | N | ++++ | HSV-1 | 103 | |
| RNA-binding protein 47 | RBM47 | A0AV96 | 64.1 | 5 | 6 | 3.0 | RRM | N | + | |||
| Serine/arginine-rich splicing factor 2 | SFRS2 | Q01130 | 25.5 | 3 | 9 | 3.0 | RRM | N | +++ | |||
| RNA-binding protein Raly | RALY | Q9UKM9 | 32.5 | 5 | 6 | 3.0 | RRM | N | +++ | |||
| Splicing factor, arginine/serine-rich 9 | SFRS9 | Q13242 | 25.5 | 6 | 9 | 2.8 | RRM | N | +++ | |||
| Nuclease-sensitive element-binding protein 1 | YBX1 | P67809 | 35.9 | 17 | 51 | 2.6 | RBP | N/C | ++++ | DEN | c-Myc | 92, 104 |
| Splicing factor U2AF 65-kDa subunit | U2AF2 | P26368 | 53.5 | 6 | 8 | 2.5 | RRM | N | ++++ | |||
| Nucleolysin TIAR | TIAL1 | Q01085 | 41.6 | 13 | 18 | 2.5 | RRM | N/C | + | |||
| Heterogeneous nuclear ribonucleoprotein D-like | HNRPDL | O14979 | 46.4 | 13 | 29 | 2.4 | RRM | N/C | ++++ | NRF | 105, 106 | |
| Heterogeneous nuclear ribonucleoprotein A3 | HNRNPA3 | P51991 | 39.6 | 25 | 43 | 2.4 | RRM | N | ++++ | |||
| Serine/arginine-rich splicing factor 1 (ASF-1, SF2-P33) | SFRS1 | Q07955 | 27.7 | 11 | 19 | 2.3 | RRM | N/C | ++++ | HDV | 91 | |
| Heterogeneous nuclear ribonucleoprotein U-like protein 1 | HNRNPUL1 | Q9BUJ2 | 96.0 | 15 | 23 | 2.3 | RBP | N | +++ | Adenovirus | 107 | |
| Transformer-2 protein homolog beta | TRA2B | P62995 | 33.7 | 6 | 9 | 2.3 | RRM | N | +++ | |||
| Serine/arginine-rich splicing factor 6 | SFRS6 | Q13247 | 39.6 | 8 | 11 | 2.2 | RRM | N | +++ | |||
| Heterogeneous nuclear ribonucleoprotein Q | SYNCRIP | O60506 | 69.6 | 30 | 64 | 2.2 | RRM | N/C | ++++ | HCV | HCV, BiP | 108–110 |
| Plasminogen activator inhibitor 1 RNA-binding protein | SERBP1 | Q8NC51 | 45.0 | 17 | 22 | 2.2 | RBP | N/C | ++++ | |||
The mass spectroscopy was performed on two poliovirus-infected samples, and the data presented represent an average of results between those data sets. The data sets were highly reproducible (see Data Set S1 in the supplemental material).
UniProt-SwissProt accession number.
Average number of unique peptides identified by MS.
Average total number of spectra that were assigned to that protein from the MS analysis.
Average fold increase in total spectra for PV-infected versus mock-infected control. For proteins only detected in the PV-infected samples, fold change is indicated as “infinite” (INF).
Most of the identified host factors are RNA binding proteins (RBP) or have RNA recognition motifs (RRMs), which are indicated in the gene ontology column. Some are DNA binding proteins (DBP). Blank table cells indicate they are not known to bind either RNA or DNA to date.
Cellular localization is indicated as nuclear (N) or cytoplasmic (C). N/C, proteins known to shuttle between the nucleus and the cytoplasm.
Protein abundances according to the Protein Abundance Across Organisms Database (pax-db.org). Abundances are expressed relative to β-actin (5,120 ppm). +++++ represents the range of 9,999 to 1,000 ppm. Stepwise 10-fold decreases down to 0.99 to 0.1 ppm (+) are indicated.
Viruses whose life cycle has been shown to be impacted in some manner by the host factor. HCV, hepatitis C virus; HDV, hepatitis D virus; BDV, Borna disease virus; JCV, JC virus; HSV, herpes simplex virus; IAV, influenza A virus; DEN, dengue virus.
Host factors that have been implicated as an IRES-trans-acting factor (ITAF) for other IRESs are indicated. ODC, ornithine decarboxylase; PDGF2, platelet-derived growth factor 2; VEGF, vascular endothelial growth factor; NRF, nuclear respiratory factor.
The vast majority of the known host factors (Table 2) are either hnRNPs (heterogeneous ribonucleoproteins), nuclear proteins, or proteins that shuttle between the cytoplasm and the nucleus (43, 44) (Fig. 3A, left). Interestingly, a similar trend was observed for host factors identified by TUX-MS. Over half of the proteins identified using TUX-MS (36/65) can localize to either the nucleus or cytoplasm, while the remainder are predominantly localized to the nucleus (Fig. 3A, right). (One protein did not have a known localization.) Although PV replicates in the cytoplasm, PV infection of cells results in the cytoplasmic accumulation of a variety of shuttling and nonshuttling nuclear proteins, such as nucleolin (46), La (13, 47), Sam68 (48), hnRNP A1, hnRNP K, hnRNP C (49), SRp20 (50), and PTB (51). Therefore, the fact that most of the proteins identified by TUX-MS are nuclear is not surprising and demonstrates a common feature with the known host factors (Fig. 3A, left). Given this commonality, known and putative host factors (Tables 2 and 3) were further examined for shared functional relationships. Using the STRING database of global protein interaction networks, both experimental evidence and computational prediction evidence were utilized to assemble functional associations, including direct protein-protein interaction data, curated pathways, and homology-based predictions (52). As illustrated in Fig. 3B, STRING analysis of 81 host factors (66 putative and 15 known from Tables 3 and 2, respectively) resulted in a single functional network composed of 54 host factors. Notably, 14 out of 15 known host factors (nodes outlined in bold) were in the network, which also contained the majority of putative host factors identified by TUX-MS. Within this network, we observed an interconnected core set of 25 proteins, the majority of which were host factors annotated to ribonucleoprotein and spliceosome complexes (Fig. 3B and D). Additionally, functional GO clustering analysis of host factors found an enrichment in the coding region instability determinant (CRD)-mediated mRNA stability complex (Fig. 3D). Specifically, we identified four out of the five characterized members of this complex, DHX9, YBX1, SYNCRIP, and hnRNP U, which is a cytoplasmic RNP that has been shown to control c-Myc stability (53). Overall, bioinformatic evaluation of the proteins identified by TUX-MS provided multiple lines of computational evidence, including protein domain and function analysis, which support their potential role as cellular host factors involved in PV amplification.
siRNA knockdown of host factors identified by TUX-MS affects poliovirus amplification in cells.
To determine what effect, if any, the identified host factors had on PV amplification in cell culture, the host factors were knocked down using siRNAs (Fig. 4A), and the effects on viral amplification were determined. Specifically, following siRNA knockdown of a single host factor, cells were infected with PV at a low multiplicity of infection (MOI) of 0.1, and the titer of the virus was determined after a single round of amplification at 6 hpi. PCBP2 [poly(rC) binding protein 2] was used as a positive control in our knockdown assay since it has been shown to be involved in both translation and replication of PV (54, 55), although siRNA knockdowns have not been previously demonstrated for PCBP2. Knockdown of PCBP2 (Fig. 4A, left) led to a decrease in viral amplification by 71% (Fig. 4B). To determine how reliable the TUX-MS method is for identification of host factors required for PV infection, eight host factors were chosen for experimental validation from Table 3. Host factors were chosen throughout the list, with three identified exclusively in the PV sample and five demonstrating an enrichment in the PV sample. For these proteins, we observed efficient knockdown, without an impact on cell viability (Fig. 4A, bottom). Knockdown of heterogeneous nuclear ribonucleoprotein L (hnRNP L), ribosomal L1 domain-containing protein 1 (RSL1D1), X-ray repair cross-complementing protein 6 (XRCC6), probable ATP-dependent RNA helicase DDX5, non-POU domain-containing octamer-binding protein (NONO), or cellular nucleic acid binding protein (CNBP) resulted in a decrease in PV titers (Fig. 4B, white bars). The greatest effects on PV amplification (64% and 53% decreases) were observed when CNBP or NONO was knocked down, respectively (Fig. 4B). Multiple cycle amplification assays that measured either the number of plaques or the size of the plaques when CNBP was knocked down yielded similar results (data not shown). DDX17 was the only protein examined that resulted in an increase in viral titers, suggesting that this protein inhibits PV amplification. The heterogeneous nuclear ribonucleoprotein U (hnRNP U) was the only host factor tested that did not have a significant effect on PV amplification, suggesting that the virus either does not require it, or there is another host factor with a redundant function (such as the hnRNPU-like proteins in Table 3). Nevertheless, it was confirmed by Western analysis that hnRNP U specifically cross-links to RNA in PV-infected cells (Fig. 4C). This specificity demonstrates that it indeed interacts with viral RNA and is not merely an abundant nonspecific binding protein.
Fig 4.
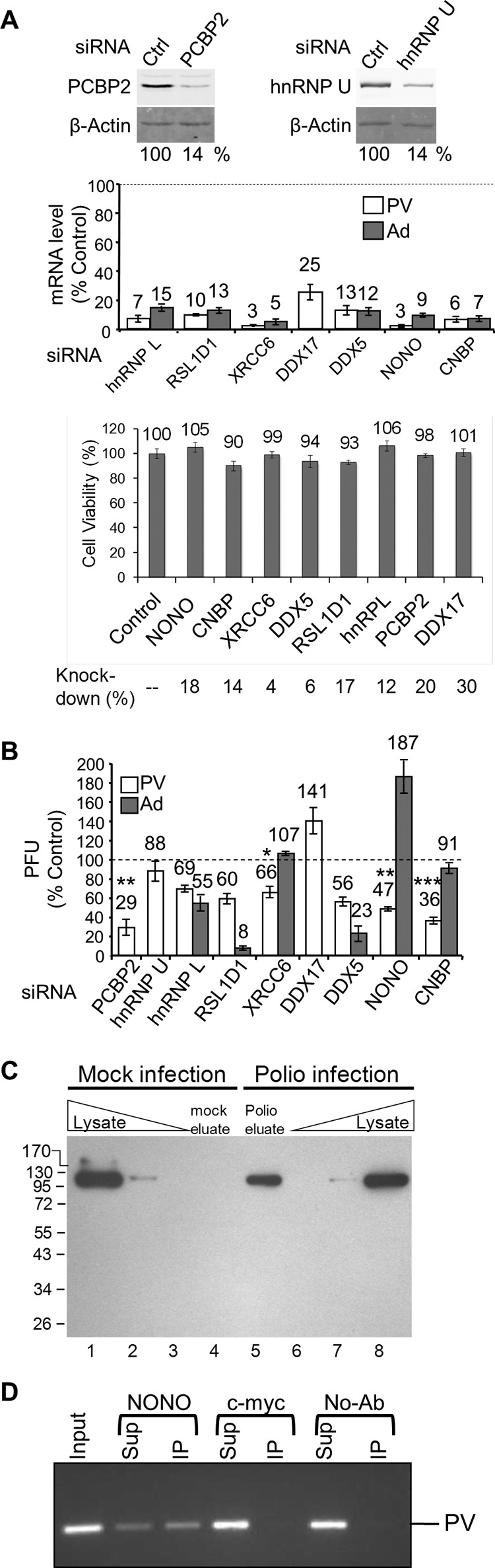
Depletion of host proteins found to interact with PV RNA has an impact on PV amplification. HeLa cells were transfected with either control or specific siRNAs as indicated. (A) Quantitative Western analysis of PCBP2 and hnRNP U knockdown. (Top) A β-actin Western blot is shown as a loading control. (Middle) qRT-PCR of mRNA levels following knockdowns normalized to both β-actin and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA levels. (Bottom) Cell viability was measured using an MTT assay 48 h following knockdown by the indicated siRNAs; the amount of mRNA remaining following knockdown as determined by qRT-PCR is indicated. (B) Knockdown cells were infected at an MOI of 0.1 with PV (white bars) or adenovirus 5 (dark gray bars), and the virus titer after a single round of amplification (6 or 30 hpi, respectively) was determined by plaque assay. (C) Cells were mock infected (lanes 1 to 4) or PV infected (lanes 5 to 8) in the presence of Act D and 4TU. Cross-linking with long-wavelength UV light was performed at 5 hpi prior to cell lysis. RNA-protein complexes were isolated using oligo(dT25) magnetic beads, and then RNA was degraded with RNase A and the proteins were separated by SDS-PAGE (lanes 4 and 5) along with total protein whole-cell lysates from mock-infected (lanes 1 to 3) and poliovirus-infected (lanes 6 to 8) cells. hnRNP U was detected by Western analysis. (D) PV RNA immunoaffinity purifies with NONO. PV-infected HeLaUPRT cells were harvested at 5 hpi and subject to immunoprecipitation with NONO, c-myc, or no antibody (No-Ab). RNA was isolated from input, supernatant (Sup), and immunoprecipitation (IP) and detected by reverse transcription and PCR. Shown is a representative result (n = 3).
Investigation of adenovirus amplification indicates specificity of host factors for poliovirus infection.
To test whether knockdown of the host factors impaired the overall cellular fitness for viral amplification, the effects of siRNA knockdown on adenovirus amplification were determined. PV uses an internal ribosomal entry site (IRES) to translate a long polyprotein that is proteolytically cleaved into precursor and mature proteins. Since many of the known PV host factors are IRES trans-acting factors, adenovirus was chosen because it is a DNA virus that is not known to contain an IRES. Effects on adenovirus amplification were determined in cells depleted for the host factors that demonstrated a decrease in poliovirus titers. Similar to PV, adenovirus titers were decreased for hnRNP L, RSL1D1, and DDX5 (Fig. 4B, dark gray bars), suggesting that either these proteins are essential and their depletion decreases the overall fitness of the cell, or they are required for amplification of PV and adenovirus. Interestingly, NONO depletion results in an 87% increase in adenovirus titers, while CNBP or XRCC6 depletion does not significantly affect the ability of the cells to amplify adenovirus (Fig. 4B, dark gray bars). This suggests that NONO may function to inhibit adenovirus amplification.
NONO directly binds to PV RNA to promote PV amplification.
Identification of NONO by the TUX-MS method, which uses a zero-distance cross-linker, suggests that NONO is directly bound to the viral RNA. To confirm this, we performed immunoaffinity purification of NONO from PV-infected cells and used reverse transcription (RT)-PCR to detect coisolation of PV RNA. We found that PV RNA was specifically coisolated with anti-NONO antibody, but not with anti-c-myc antibody or no antibody (Fig. 4D). Taken together, these data suggest that NONO directly interacts with PV RNA to enhance PV amplification.
Host factors accelerate viral amplification.
It is worth noting that virus harvested from cells depleted of a host factor did not alter the plaque morphology or size following a single cycle of replication (data not shown). Our studies show that knockdown of factors identified by TUX-MS triggers a 2- to 3-fold decrease in viral titers at 6 hpi, in agreement with previously reported decreases in viral amplification for other host factors (32, 56). In particular, the effects of knocking down CNBP and NONO on viral titers at 6 hpi are consistent with effects on viral amplification observed for La, PTB, hnRNP K, and hnRNP C1/C2 (32, 56–58). As 6 hpi represents a late stage of infection, we next asked if assessment of the impact on virus titers at an earlier time point of infection would reveal delays in virus production. For this earlier time point, we elected to focus on two proteins, the uncharacterized factor, NONO, and the positive control, PCBP2. Indeed, we observed a greater reduction in PV titers at the earlier time point of infection, as NONO and PCBP2 knockdowns triggered a 10- to 30-fold decrease in titers at 4 hpi. (Fig. 5A). We confirmed that at 4 hpi, two independent siRNAs that target NONO resulted in a decrease in PV amplification (Fig. 5A). Consistent with these observations, if the titer of virus is determined on cells following NONO or CNBP knockdowns, then there is a corresponding decrease in plaque number and size (data not shown). Altogether, these results demonstrate that in the absence of the host factors there is a delay in viral production.
Fig 5.
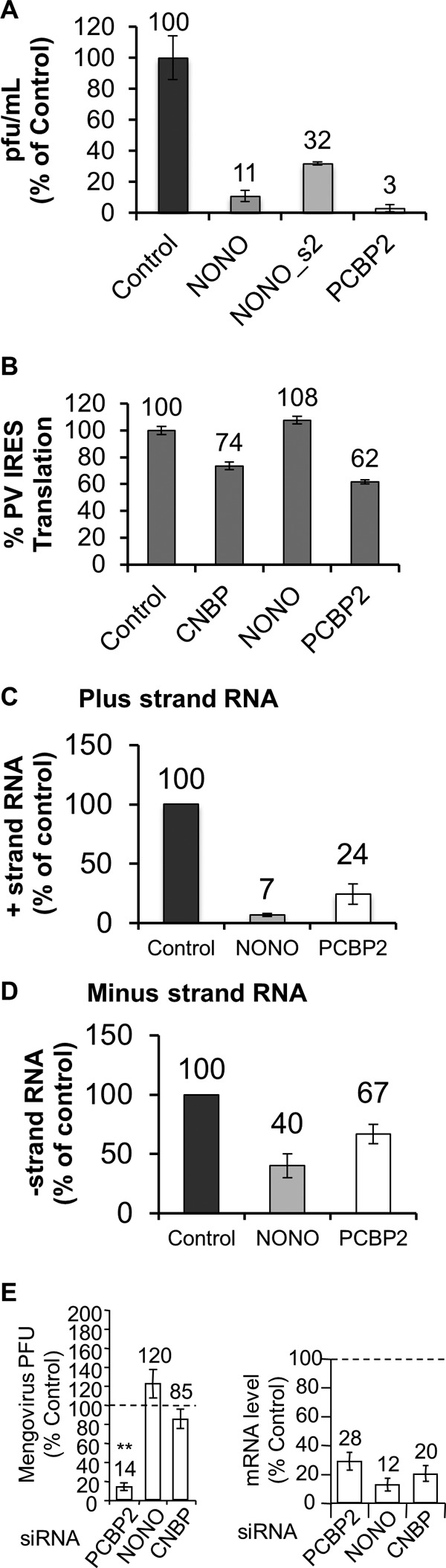
Knockdown of NONO or CNBP affects PV replication or translation but not mengovirus amplification. (A). HeLaUPRT cells were transfected with scrambled control, NONO, or PCBP2 siRNAs. Forty-eight hours posttransfection of the siRNA, the cells were infected with PV at an MOI of 0.1. Virus was harvested at 4 hpi, and the titer was determined by plaque assay on HeLaUPRT cells. (B) HeLaUPRT cells were transfected with scrambled control, NONO, CNBP, or PCBP2 siRNAs, and 48 h posttransfection, the cells were transfected with a dicistronic reporter plasmid containing the PV IRES in the intercistronic region and a control plasmid expressing a cap-dependent β-galactosidase. Twenty-four hours later, luciferase and β-galactosidase activities were determined. The amount of mRNA remaining following knockdown was determined by qRT-PCR (CNBP, 9.9%; NONO, 30%; and PCBP2, 23%). IRES-driven translation was normalized to β-galactosidase activity and expressed as a percentage of the control siRNA. (C and D) HeLaUPRT cells were transfected with scrambled control, NONO, or PCBP2 siRNA. Forty-eight hours posttransfection, the cells were infected with PV at an MOI of 0.1. qRT- PCR was used to determine the number of plus-strand (C) and minus-strand (D) RNAs at 4 hpi. Copy numbers were determined using an in vitro-transcribed template of a known amount (plus strand, control, 7.7 × 109, NONO, 5.4 × 108, and PCBP2, 1.4 × 109 copies/ng RNA; minus strand, control, 3.0 × 104, NONO, 1.3 × 104, and PCBP2, 2 × 104 copies/ng RNA). (E) Knockdown cells were infected at an MOI of 0.1 with mengovirus (MV), and the virus titer after a single round of amplification (5.5 hpi) was determined by plaque assay (left). mRNA levels after qRT-PCR of mRNA following knockdown were normalized to β-actin (right). The same results were obtained by normalization to GAPDH mRNA levels. qRT-PCR and titer results are percentages relative to the control siRNA (represented by the horizontal dotted lines). Errors bars are standard errors for n ≥ 3. The P value for viral amplification compared to the control siRNA is indicated. **, P < 0.005.
Knockdowns of NONO or CNBP distinctly affect PV translation and PV RNA replication.
In order to determine if knockdown of NONO or CNBP had an effect on PV translation, a dicistronic reporter assay was used to measure PV IRES activity. Knockdown of NONO had no effect on PV IRES activity (Fig. 5B). In contrast, knockdown of PCBP2 and CNBP reduced PV IRES activity by 38% and 26%, respectively. This finding is consistent with previous reports that demonstrated that PCBP2 is required for efficient PV IRES activity (54, 58–60). Since NONO had no effect on PV translation, we next tested whether knockdown of NONO impacted the synthesis of PV plus- or minus-strand RNA. NONO knockdown triggered a 10-fold decrease in positive-stranded RNA (Fig. 5C) and a 2-fold decrease in minus-strand RNA (Fig. 5D). These results demonstrate a more significant effect on viral replication for NONO than the previously known host factor, PCBP2. Taken together, these results suggest that NONO, while not affecting viral translation, plays a role in enhancing PV RNA replication.
Knockdown of NONO does not have an effect on mengovirus amplification.
To determine whether knockdown of host factors that were shown to have an effect on poliovirus can also affect a distantly related picornavirus, we performed a single-round amplification experiment using mengovirus. Mengovirus belongs to the genus Cardiovirus and contains a type II IRES, similar to encephalomyocarditis virus (EMCV), which means that the ribosome is recruited to the start codon. This is in contrast to the type I IRES of polioviruses, which involves the ribosome scanning to a downstream start codon (61). PCBP2 knockdown has a significant effect on mengovirus amplification (Fig. 5E). While it is known that PCBP2 is not required for mengovirus translation (59), it may still play a role in mengovirus replication, since it is known to be involved in PV replication. To our knowledge, this is the first demonstration that PCBP2 is required for mengovirus amplification. In contrast, knockdown of NONO or CNBP did not significantly affect amplification of mengovirus (Fig. 5E). This suggests that these host factors may be specific to poliovirus or within the Enterovirus genus. Taken together, these data support a significant role for CNBP and NONO in PV amplification.
DISCUSSION
Using the TUX-MS method, we identified 66 previously unknown host factors that bind to PV RNA following infection, in addition to confirming all of the previously reported host factors. Further validation of eight of the novel host factors revealed that all but one played roles as enhancers or inhibitors of PV amplification. For the one exception, hnRNP U, our validation results nevertheless demonstrated that, while not having an effect on PV amplification, hnRNP U was specifically associated with PV RNA during infection. Knockdown of NONO or CNBP resulted in a decrease in PV amplification at 6 hpi equivalent to those of other known host factors, such as PCBP2 (Fig. 4), La, PTB, and hnRNP C1/C2 (32, 57, 58). Importantly, this effect was accentuated at an earlier time point of infection for NONO, suggesting that NONO accelerates virus production. Further analysis of CNBP and NONO revealed that they are required for efficient translation and plus-strand RNA synthesis, respectively. Immunoprecipitation of NONO revealed that it specifically bound to the PV RNA. Taken together, these results demonstrate that TUX-MS is a highly effective method for host factor identification with a low false-positive rate.
Most of the host factors that we validated were enhancers of PV amplification, with two exceptions—an inhibitor (DDX17) and a specific RNA binding protein that did not affect viral amplification (hnRNP U). DDX17 is a binding partner and cofactor for zinc finger CCCH-type antiviral protein 1 (ZAP), which was also detected by TUX-MS (Table 3) (62). ZAP is an antiviral protein that targets the RNA of retroviruses, filoviruses, and alphaviruses for degradation (63–65) and potentially poliovirus (Table 3). The majority of the assayed host factors appear to be enhancers of PV amplification, such as the DEAD box RNA helicase DDX5. DDX5 has been shown to participate in RNA replication during HCV infection (66). hnRNP L, XRCC6, CNBP, and RSLID1 have either been shown to enhance other IRESs or be involved in translation (67–70). NONO, which we found had a role in viral replication, has two RNA recognition motifs and a coiled-coiled protein interaction domain (71, 72). It is known to be involved in a number of nuclear functions and forms monomers and heteromers to bind to double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), and RNA (73, 74). Interestingly, NONO is known to form a complex with SFPQ (splicing factor, proline- and glutamine-rich) and Matrin-3 (MATR3) (73, 75), which were also identified by TUX-MS (see Data Set S1 in the supplemental material).
Although poliovirus is a cytoplasmic replicating virus, most of its known host factors are either nuclear or cycle between the nucleus and the cytoplasm. The relocalization of known host factors from the nucleus to the cytoplasm during poliovirus infection has been observed for a number of host factors (13, 46–51, 76). PV infection results in degradation of several nucleoporins, which disrupts nuclear-cytoplasmic transport pathways (49, 77). This is associated with a number of predominately nuclear host proteins being relocalized to the cytoplasm either due to retention in the cytoplasm, due to binding to the PV RNA to proteins, or because import is impaired (50, 78). Therefore, the fact that the majority of the proteins identified by TUX-MS are either nuclear or cycle between the nucleus and the cytoplasm is entirely consistent with the known host factors and relocalization of proteins from the nucleus to the cytoplasm during viral infection.
Since all eight of the host proteins we tested were validated, this suggests that TUX-MS is a robust assay to identify host proteins that bind the viral RNA during infection in cell culture. There are a number of reasons that likely contribute to the low false-positive rate and the efficient discovery of all known and 66 novel host factors. First, the cross-linking was performed prior to cell lysis, and therefore cross-links were established prior to the disruption of cellular compartmentalization. Second, the RNA-protein complexes were isolated under denaturing conditions, reducing the possibility that nonspecific RNA binding proteins would remain associated with the RNA during isolation. Third, the use of a mock control allowed for the elimination of potential false positives that may either bind to host RNA or the resin under denaturing conditions. There is a possibility that some factors are present by nonspecific association with viral RNA, while not binding to host RNA in the mock samples. However, the successful validation of selected uncharacterized factors suggests that TUX-MS has a low false-positive rate and is a valuable methodology with the potential to provide us the biological picture of PV in infected cells. Fourth, the RNA levels were normalized prior to RNase digestion, ensuring that mock-infected and infected samples were comparable with respect to the RNA-protein complexes. Taken together, this suggests that TUX-MS can be used to identify additional host factors with a low false-positive discovery rate even for a well-studied virus, such as poliovirus.
Although we identified all of the known host factors, several did not meet our stringent criteria using spectral counting analysis (e.g., PCBP2); these criteria were selected to identify new potential candidates with relatively high confidence. Spectral counting analysis was utilized for relative quantification for TUX-MS as it is computationally facile, well suited to detect large differences in relative abundance, and can be readily integrated into most proteomics workflows. Yet, one disadvantage is the lack of sensitivity for proteins that generate low spectral counts, either due to low abundance or poor ability to be detected by MS. As a consequence, we expect a subset of excluded proteins to be false negatives, and so cannot eliminate the possibility that additional host factors are present in the complete data set (see Data Set S1 in the supplemental material). On the other hand, it is also likely that numerous excluded proteins were correctly classified as nonspecific since (i) many proteins were also identified in isolations from mock-infected cells and (ii) they include highly abundant and/or known immunoisolation contaminants, such as actin. It is likely that integration of complementary quantitative approaches, such as isotope labeling, as well as targeted studies will be required to determine the specificity of the excluded proteins and further expand the known host factors required for PV amplification.
Taken together, our results suggest that TUX-MS is an effective, unbiased method to identify host factors that are functionally associated with the viral RNA. This method is broadly applicable to other RNA viral families that do not require host cell transcription or a DNA-dependent RNA polymerase during viral infection, such as picornaviruses, flaviviruses, coronaviruses, alphaviruses, and bunyaviruses. While the method we presented here used Act D to inhibit host transcription to allow exclusive incorporation of 4sU into the viral RNA, we think that the TUX-MS method could also be modified to be performed without inhibition of host transcription and therefore be expanded to be used on viruses that require host transcription (e.g., influenza virus and HIV-1). This modification would entail labeling of all RNA in the cell with 4sU followed by isolation of the viral RNA-protein complexes using a virus-specific oligonucleotide(s) rather than oligo(dT). Furthermore, the use of mass spectrometry in TUX-MS also provides the ability to incorporate isotope-coded tags that enable sample multiplexing and could be used to perform global, quantitative profiling of RNA interactions with viral and host proteins throughout the viral life cycle. Thus, extension of TUX-MS to additional viruses could reveal virus-specific RNA-host factor interactions, providing new host candidates to design selective or broadly applicable pharmacological interventions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Prevelige, Louise Chow, and R. Curtis Hendrickson for critical reading of the manuscript. We also thank others who generously supplied reagents: LNCX UPRT-myc, tat, and VSVG plasmids (Edward Mocarski), anti-hnRNP U 3G6 antibody (Gideon Dreyfuss), anti-PCBP2 (Raul Andino), and mengovirus (Ann Palmenberg).
This work was supported by Public Health Service grants R01GM084547 (S.R.T), DP1DA026192 (I.M.C), R21AI102187 (S.R.T. and I.M.C.), and 5T32AI007150-30 (E.M.L) from the National Institutes of Health and the Human Frontier Science Program RGY0079/2009-C (I.M.C.).
Footnotes
Published ahead of print 5 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00950-13.
REFERENCES
- 1. Dore GJ, Matthews GV, Rockstroh J. 2011. Future of hepatitis C therapy: development of direct-acting antivirals. Curr. Opin. HIV AIDS 6:508–513 [DOI] [PubMed] [Google Scholar]
- 2. Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N. Engl. J. Med. 338:853–860 [DOI] [PubMed] [Google Scholar]
- 3. Mocroft A, Ledergerber B, Katlama C, Kirk O, Reiss P, d'Arminio Monforte A, Knysz B, Dietrich M, Phillips AN, Lundgren JD. 2003. Decline in the AIDS and death rates in the EuroSIDA study: an observational study. Lancet 362:22–29 [DOI] [PubMed] [Google Scholar]
- 4. Osterholm MT, Kelley NS, Sommer A, Belongia EA. 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis. 12:36–44 [DOI] [PubMed] [Google Scholar]
- 5. Chumakov K, Ehrenfeld E, Wimmer E, Agol VI. 2007. Vaccination against polio should not be stopped. Nat. Rev. 5:952–958 [DOI] [PubMed] [Google Scholar]
- 6. Cristea IM, Carroll JW, Rout MP, Rice CM, Chait BT, MacDonald MR. 2006. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 281:30269–30278 [DOI] [PubMed] [Google Scholar]
- 7. Cristea IM, Rozjabek H, Molloy KR, Karki S, White LL, Rice CM, Rout MP, Chait BT, MacDonald MR. 2010. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J. Virol. 84:6720–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gamarnik AV, Andino R. 1996. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 15:5988–5998 [PMC free article] [PubMed] [Google Scholar]
- 9. Gamarnik AV, Boddeker N, Andino R. 2000. Translation and replication of human rhinovirus type 14 and mengovirus in Xenopus oocytes. J. Virol. 74:11983–11987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frolova E, Gorchakov R, Garmashova N, Atasheva S, Vergara LA, Frolov I. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 80:4122–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dorner AJ, Semler BL, Jackson RJ, Hanecak R, Duprey E, Wimmer E. 1984. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 50:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325 [DOI] [PubMed] [Google Scholar]
- 13. Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esfandiarei M, McManus BM. 2008. Molecular biology and pathogenesis of viral myocarditis. Annu. Rev. Pathol. 3:127–155 [DOI] [PubMed] [Google Scholar]
- 15. Ho M. 2000. Enterovirus 71: the virus, its infections and outbreaks. J. Microbiol. Immunol. Infect. 33:205–216 [PubMed] [Google Scholar]
- 16. Halsey NA, Pinto J, Espinosa-Rosales F, Faure-Fontenla MA, da Silva E, Khan AJ, Webster AD, Minor P, Dunn G, Asturias E, Hussain H, Pallansch MA, Kew OM, Winkelstein J, Sutter R. 2004. Search for poliovirus carriers among people with primary immune deficiency diseases in the United States, Mexico, Brazil, and the United Kingdom. Bull. World Health Organ. 82:3–8 [PMC free article] [PubMed] [Google Scholar]
- 17. Racaniello V. 2007. Picornaviridae: the viruses and their replication, p 795–838 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 18. Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE. 2000. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 19:899–905 [DOI] [PubMed] [Google Scholar]
- 19. Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. 1998. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16:423–428 [DOI] [PubMed] [Google Scholar]
- 20. Thompson SR, Sarnow P. 2003. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology 315:259–266 [DOI] [PubMed] [Google Scholar]
- 21. Kramer T, Greco TM, Enquist LW, Cristea IM. 2011. Proteomic characterization of pseudorabies virus extracellular virions. J. Virol. 85:6427–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 23. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658 [DOI] [PubMed] [Google Scholar]
- 24. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449 [DOI] [PubMed] [Google Scholar]
- 26. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Woolaway K, Asai K, Emili A, Cochrane A. 2007. hnRNP E1 and E2 have distinct roles in modulating HIV-1 gene expression. Retrovirology 4:28. 10.1186/1742-4690-4-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jalal C, Uhlmann-Schiffler H, Stahl H. 2007. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 35:3590–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gamarnik AV, Andino R. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882–892 [PMC free article] [PubMed] [Google Scholar]
- 30. Dreyfuss G, Choi YD, Adam SA. 1984. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol. Cell. Biol. 4:1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Selth LA, Gilbert C, Svejstrup JQ. 2009. RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harbor Protoc. 10.1101/pdb.prot5234 [DOI] [PubMed] [Google Scholar]
- 32. Ertel KJ, Brunner JE, Semler BL. 2010. Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J. Virol. 84:4229–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. 2005. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 23:232–237 [DOI] [PubMed] [Google Scholar]
- 34. Reich E, Franklin RM, Shatkin AJ, Tatum EL. 1962. Action of actinomycin D on animal cells and viruses. Proc. Natl. Acad. Sci. U. S. A. 48:1238–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hewlett MJ, Rozenblatt S, Ambros V, Baltimore D. 1977. Separation and quantitation of intracellular forms of poliovirus RNA by agarose gel electrophoresis. Biochemistry 16:2763–2767 [DOI] [PubMed] [Google Scholar]
- 36. Suchanek M, Radzikowska A, Thiele C. 2005. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods 2:261–267 [DOI] [PubMed] [Google Scholar]
- 37. Pfister T, Egger D, Bienz K. 1995. Poliovirus subviral particles associated with progeny RNA in the replication complex. J. Gen. Virol. 76:63–71 [DOI] [PubMed] [Google Scholar]
- 38. Banerjee R, Echeverri A, Dasgupta A. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71:9570–9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiang W, Harris KS, Alexander L, Wimmer E. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paul AV, van Boom JH, Filippov D, Wimmer E. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280–284 [DOI] [PubMed] [Google Scholar]
- 41. Andino R, Rieckhof GE, Baltimore D. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369–380 [DOI] [PubMed] [Google Scholar]
- 42. Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124–1134 [PMC free article] [PubMed] [Google Scholar]
- 43. Lin JY, Chen TC, Weng KF, Chang SC, Chen LL, Shih SR. 2009. Viral and host proteins involved in picornavirus life cycle. J. Biomed. Sci. 16:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pacheco A., Martinez-Salas E. 2010. Insights into the biology of IRES elements through riboproteomic approaches. J. Biomed. Biotechnol 2010:458927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. King HA, Cobbold LC, Willis AE. 2010. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 38:1581–1586 [DOI] [PubMed] [Google Scholar]
- 46. Waggoner S, Sarnow P. 1998. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 72:6699–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shiroki K, Isoyama T, Kuge S, Ishii T, Ohmi S, Hata S, Suzuki K, Takasaki Y, Nomoto A. 1999. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol. 73:2193–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McBride AE, Schlegel A, Kirkegaard K. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. U. S. A. 93:2296–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gustin KE, Sarnow P. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fitzgerald KD, Semler BL. 2011. Re-localization of cellular protein SRp20 during poliovirus infection: bridging a viral IRES to the host cell translation apparatus. PLoS Pathog. 7:e1002127. 10.1371/journal.ppat.1002127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Back SH, Kim YK, Kim WJ, Cho S, Oh HR, Kim JE, Jang SK. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3Cpro. J. Virol. 76:2529–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39:D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weidensdorfer D, Stohr N, Baude A, Lederer M, Kohn M, Schierhorn A, Buchmeier S, Wahle E, Huttelmaier S. 2009. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15:104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Blyn LB, Towner JS, Semler BL, Ehrenfeld E. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gamarnik AV, Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin JY, Li ML, Huang PN, Chien KY, Horng JT, Shih SR. 2008. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J. Gen. Virol. 89:2540–2549 [DOI] [PubMed] [Google Scholar]
- 57. Costa-Mattioli M, Svitkin Y, Sonenberg N. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 24:6861–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Florez PM, Sessions OM, Wagner EJ, Gromeier M, Garcia-Blanco MA. 2005. The polypyrimidine tract binding protein is required for efficient picornavirus gene expression and propagation. J. Virol. 79:6172–6179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Walter BL, Nguyen JH, Ehrenfeld E, Semler BL. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5:1570–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Walter BL, Parsley TB, Ehrenfeld E, Semler BL. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thompson SR. 2012. Tricks an IRES uses to enslave ribosomes. Trends Microbiol. 20:558–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen G, Guo X, Lv F, Xu Y, Gao G. 2008. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc. Natl. Acad. Sci. U. S. A. 105:4352–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Karki S, Li MM, Schoggins JW, Tian S, Rice CM, MacDonald MR. 2012. Multiple interferon stimulated genes synergize with the zinc finger antiviral protein to mediate anti-alphavirus activity. PLoS One 7:e37398. 10.1371/journal.pone.0037398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 77:11555–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu Y, Gao G. 2008. ZAP-mediated mRNA degradation. RNA Biol. 5:65–67 [DOI] [PubMed] [Google Scholar]
- 66. Goh PY, Tan YJ, Lim SP, Tan YH, Lim SG, Fuller-Pace F, Hong W. 2004. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J. Virol. 78:5288–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang C, Caprara MG, Venema RC, Komar AA, Snider MD, Hatzoglou M. 2009. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol. Cell. Biol. 29:2899–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hwang B, Lim JH, Hahm B, Jang SK, Lee SW. 2009. hnRNP L is required for the translation mediated by HCV IRES. Biochem. Biophys. Res. Commun. 378:584–588 [DOI] [PubMed] [Google Scholar]
- 69. Hahm B, Kim YK, Kim JH, Kim TY, Jang SK. 1998. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 72:8782–8788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rojas M, Farr GW, Fernandez CF, Lauden L, McCormack JC, Wolin SL. 2012. Yeast Gis2 and its human ortholog CNBP are novel components of stress-induced RNP granules. PLoS One 7:e52824. 10.1371/journal.pone.0052824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Passon DM, Lee M, Rackham O, Stanley WA, Sadowska A, Filipovska A, Fox AH, Bond CS. 2012. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. U. S. A. 109:4846–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dong B, Horowitz DS, Kobayashi R, Krainer AR. 1993. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 21:4085–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shav-Tal Y, Zipori D. 2002. PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett. 531:109–114 [DOI] [PubMed] [Google Scholar]
- 74. Mathur M, Tucker PW, Samuels HH. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21:2298–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang Z, Carmichael GG. 2001. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106:465–475 [DOI] [PubMed] [Google Scholar]
- 76. Brunner JE, Ertel KJ, Rozovics JM, Semler BL. 2010. Delayed kinetics of poliovirus RNA synthesis in a human cell line with reduced levels of hnRNP C proteins. Virology 400:240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Park N, Skern T, Gustin KE. 2010. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J. Biol. Chem. 285:28796–28805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Belov GA, Lidsky PV, Mikitas OV, Egger D, Lukyanov KA, Bienz K, Agol VI. 2004. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 78:10166–10177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Izumi RE, Valdez B, Banerjee R, Srivastava M, Dasgupta A. 2001. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res. 76:17–29 [DOI] [PubMed] [Google Scholar]
- 80. Merrill MK, Gromeier M. 2006. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J. Virol. 80:6936–6942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lawrence P, Rieder E. 2009. Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus. J. Virol. 83:11356–11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bedard KM, Daijogo S, Semler BL. 2007. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 26:459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 93:11115–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Brunner JE, Nguyen JH, Roehl HH, Ho TV, Swiderek KM, Semler BL. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 79:3254–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hunt SL, Hsuan JJ, Totty N, Jackson RJ. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 13:437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Borman A, Howell MT, Patton JG, Jackson RJ. 1993. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J. Gen. Virol. 74:1775–1788 [DOI] [PubMed] [Google Scholar]
- 87. Lin JY, Li ML, Shih SR. 2009. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 37:47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bradrick SS, Dobrikova EY, Kaiser C, Shveygert M, Gromeier M. 2007. Poly(A)-binding protein is differentially required for translation mediated by viral internal ribosome entry sites. RNA 13:1582–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028–2045 [PMC free article] [PubMed] [Google Scholar]
- 90. Zolotukhin AS, Michalowski D, Bear J, Smulevitch SV, Traish AM, Peng R, Patton J, Shatsky IN, Felber BK. 2003. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell. Biol. 23:6618–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sikora D, Greco-Stewart VS, Miron P, Pelchat M. 2009. The hepatitis delta virus RNA genome interacts with eEF1A1, p54(nrb), hnRNP-L, GAPDH and ASF/SF2. Virology 390:71–78 [DOI] [PubMed] [Google Scholar]
- 92. Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE. 2008. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 28:40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Watanabe Y, Ohtaki N, Hayashi Y, Ikuta K, Tomonaga K. 2009. Autogenous translational regulation of the Borna disease virus negative control factor X from polycistronic mRNA using host RNA helicases. PLoS Pathog. 5:e1000654. 10.1371/journal.ppat.1000654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. McLaren M, Asai K, Cochrane A. 2004. A novel function for Sam68: enhancement of HIV-1 RNA 3′ end processing. RNA 10:1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sammons MA, Antons AK, Bendjennat M, Udd B, Krahe R, Link AJ. 2010. ZNF9 activation of IRES-mediated translation of the human ODC mRNA is decreased in myotonic dystrophy type 2. PLoS One 5:e9301. 10.1371/journal.pone.0009301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liu M, Kumar KU, Pater MM, Pater A. 1998. Identification and characterization of a JC virus pentanucleotide repeat element binding protein: cellular nucleic acid binding protein. Virus Res. 58:73–82 [DOI] [PubMed] [Google Scholar]
- 97. Gerbasi VR, Link AJ. 2007. The myotonic dystrophy type 2 protein ZNF9 is part of an ITAF complex that promotes cap-independent translation. Mol. Cell. Proteomics 6:1049–1058 [DOI] [PubMed] [Google Scholar]
- 98. Silvera D, Koloteva-Levine N, Burma S, Elroy-Stein O. 2006. Effect of Ku proteins on IRES-mediated translation. Biol. Cell 98:353–361 [DOI] [PubMed] [Google Scholar]
- 99. Lu H, Li W, Noble WS, Payan D, Anderson DC. 2004. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J. Proteome Res. 3:949–957 [DOI] [PubMed] [Google Scholar]
- 100. Greco-Stewart VS, Thibault CS, Pelchat M. 2006. Binding of the polypyrimidine tract-binding protein-associated splicing factor (PSF) to the hepatitis delta virus RNA. Virology 356:35–44 [DOI] [PubMed] [Google Scholar]
- 101. Guang S, Felthauser AM, Mertz JE. 2005. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol. Cell. Biol. 25:6303–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jorba N, Juarez S, Torreira E, Gastaminza P, Zamarreno N, Albar JP, Ortin J. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8:2077–2088 [DOI] [PubMed] [Google Scholar]
- 103. Escudero-Paunetto L, Li L, Hernandez FP, Sandri-Goldin RM. 2010. SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology 401:155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Paranjape SM, Harris E. 2007. Y box-binding protein-1 binds to the dengue virus 3′ untranslated region and mediates anti-viral effects. J. Biol. Chem. 282:30497–30508 [DOI] [PubMed] [Google Scholar]
- 105. Reboll MR, Oumard A, Gazdag AC, Renger I, Ritter B, Schwarzer M, Hauser H, Wood M, Yamada M, Resch K, Nourbakhsh M. 2007. NRF IRES activity is mediated by RNA binding protein JKTBP1 and a 14-nt RNA element. RNA 13:1328–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Omnus DJ, Mehrtens S, Ritter B, Resch K, Yamada M, Frank R, Nourbakhsh M, Reboll MR. 2011. JKTBP1 is involved in stabilization and IRES-dependent translation of NRF mRNAs by binding to 5′ and 3′ untranslated regions. J. Mol. Biol. 407:492–504 [DOI] [PubMed] [Google Scholar]
- 107. Blackford AN, Bruton RK, Dirlik O, Stewart GS, Taylor AM, Dobner T, Grand RJ, Turnell AS. 2008. A role for E1B-AP5 in ATR signaling pathways during adenovirus infection. J. Virol. 82:7640–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Liu HM, Aizaki H, Choi KS, Machida K, Ou JJ, Lai MM. 2009. SYNCRIP (synaptotagmin-binding, cytoplasmic RNA-interacting protein) is a host factor involved in hepatitis C virus RNA replication. Virology 386:249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kim KS, Chapman NM, Tracy S. 2008. Replication of coxsackievirus B3 in primary cell cultures generates novel viral genome deletions. J. Virol. 82:2033–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cho S, Park SM, Kim TD, Kim JH, Kim KT, Jang SK. 2007. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol. Cell. Biol. 27:368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


