Abstract
Epstein-Barr virus (EBV) infection leads to lifelong viral persistence through its latency in B cells. EBV-specific T cells control reactivations and prevent the development of EBV-associated malignancies in most healthy carriers, but infection can sometimes cause chronic disease and malignant transformation. Epstein-Barr nuclear antigen 1 (EBNA-1) is the only viral protein consistently expressed during all forms of latency and in all EBV-associated malignancies and is a promising target for a therapeutic vaccine. Here, we studied the EBNA-1-specific immune response using the EBV-homologous rhesus lymphocryptovirus (rhLCV) infection in rhesus macaques. We assessed the frequency, phenotype, and cytokine production profiles of rhLCV EBNA-1 (rhEBNA-1)-specific T cells in 15 rhesus macaques and compared them to the lytic antigen of rhLCV BZLF-1 (rhBZLF-1). We were able to detect rhEBNA-1-specific CD4+ and/or CD8+ T cells in 14 of the 15 animals screened. In comparison, all 15 animals had detectable rhBZLF-1 responses. Most peptide-specific CD4+ T cells exhibited a resting phenotype of central memory (TCM), while peptide-specific CD8+ T cells showed a more activated phenotype, belonging mainly to the effector cell subset. By comparing our results to the human EBV immune response, we demonstrate that the rhLCV model is a valid system for studying chronic EBV infection and for the preclinical development of therapeutic vaccines.
INTRODUCTION
Epstein-Barr virus (EBV), also called human herpesvirus 4 (HHV4), is a double-stranded DNA virus of the gammaherpesvirus family and the Lymphocryptovirus (LCV) genus. More than 95% of the human population is infected with EBV by adulthood (1). While primary infection in children is commonly asymptomatic, it may cause infectious mononucleosis if the initial infection is delayed until adolescence or young adulthood. EBV is transmitted orally and infects both B lymphocytes and squamous pharyngeal epithelial cells. Its 172-kb genome encodes two distinct programs of gene expression that are broadly characterized as either lytic or latent. The viral gene products associated with latent infection have growth-transforming activities that immortalize B cells and maintain stable, nonproductive EBV infection. While the majority of individuals develop strong cellular immune responses against both lytic and latent antigens that control viral replication, the virus is never fully eliminated. Instead, by downregulating most protein expression, EBV establishes lifelong, persistent infections in a small number of memory B cells. EBV can assume different stages of latency (I to III), and the Epstein-Barr nuclear antigen-1 (EBNA-1) is the only viral protein consistently detected in even the most dormant stage (2). EBV periodically reactivates, but primary infections as well as reactivations are controlled by the adaptive immune system (3).
Although EBV infections and reactivation events are in general benign, EBV contributes to the development of about 1% of all human cancers (1, 4). The predominant type of EBV-associated cancer varies depending on geographic regions. In central Africa, EBV is tightly linked to endemic forms of Burkitt's lymphoma, while in southern China and some populations of Eskimos and Greenlanders, it is linked to nasopharyngeal carcinomas (5). In other populations EBV has been associated with B-cell malignancies such as Hodgkin's lymphoma and lymphoproliferative diseases, especially in immunocompromised individuals (6, 7). EBV infections have been implicated in several autoimmune diseases such as multiple sclerosis, systemic lupus erythematosus, rheumatoid arthritis, and others (8). Autoimmune reactions are assumed to be triggered by antigenic mimicry (9), and antiviral drugs have been shown to cause remission of autoimmune disorders in some patients (10).
Vaccines to block primary EBV infection or eliminate cells carrying persisting EBV are currently unavailable. Preventative vaccines should be based on antigens that elicit neutralizing antibodies such as membrane antigen gp350 (11, 12), while therapeutic vaccines need to target antigens produced during latency, such as EBNA-1. EBNA-1 is a particularly important target as it is the only antigen expressed during all stages of viral infection, including every form of latency, lytic infection, and all EBV-associated cancers (13). EBNA-1 is a virus-encoded DNA binding protein that functions to maintain the viral episome during latency and is essential for viral DNA replication during latency (14). This phosphoprotein is separated into C- and N-terminal domains, which are linked by an internal glycine-alanine-rich repeat domain. The repeat domain stabilizes the protein and inhibits EBNA-1's degradation by cellular proteasomes. As a result, both the generation of peptide sequences for association with major histocompatibility complex (MHC) class I molecules and the induction of EBNA-1-specific CD8+ T cells are impaired (15). Nevertheless, most EBV-infected humans develop CD8+ T cells in response to EBNA-1 (14). These EBNA-1-specific cytotoxic T lymphocytes (CTLs) have been shown to prevent the outgrowth of infected B cells in vitro and to secrete gamma interferon (IFN-γ) in response to stimulation, thus suggesting that they play a role in controlling infections in vivo (16). Additionally, EBNA-1 is an immunodominant target of EBV-specific CD4+ T cells that are capable of inhibiting virus-induced B-cell proliferation in vitro (17, 18). In support of the importance of EBNA-1-specific T-cell responses, loss of EBNA-1-specific CD4+ or CD8+ T cells has been correlated with numerous diseases, including non-Hodgkin's lymphoma, nasopharyngeal carcinoma, and Burkitt's lymphoma (19–23).
The animal model based on rhesus lymphocryptovirus (rhLCV) is a particularly powerful tool for studying various aspects of EBV due to its unusually high degree of biological, genetic, and pathogenic similarity to EBV and the likeness of the immune responses between humans and nonhuman primates (24, 25). Infection of nonhuman primates strongly resembles EBV infection of humans (26–28). Most animals are infected during infancy through the oral route, and a large majority of the target population is naturally infected by adulthood (29). The virus, which can be detected in blood or upon activation in saliva, persists for life as a latent infection in B cells and immortalizes B cells in tissue culture, and, like EBV, it has been associated with virus-positive B cell lymphomas during immunosuppression (30). Furthermore, rhLCV has a high degree of sequence homology to EBV, molecular organization is well conserved between the two viruses, and they have an identical repertoire of latent and lytic genes (29, 31). Thus, the biology, molecular virology, and immunology of rhLCV infection in rhesus macaques are very similar to EBV infection in humans, making this model appropriate for studying EBV immunology as well as for preclinical testing of EBV therapeutics.
Using the rhLCV model, we have characterized the frequency and functionality of the latent rhEBNA-1-specific immune response in healthy rhLCV-infected rhesus macaques. We compare this to the response directed against the highly immunogenic, immediate-early lytic rhBZLF-1 protein, which has been studied extensively in humans and to some degree in rhesus macaques. Although some studies have been conducted to assess immune responses to rhLCV (27, 32), the scope of such studies has been more limited. Here, we characterized peptide-specific T-cell responses in blood from a cohort of 15 captivity-kept rhesus macaques using multicolor flow cytometry, which allowed us to define dominant CD4+ and CD8+ T-cell subsets and cytokine production profiles. Our results show that most rhLCV-seropositive animals carry both rhEBNA-1- and rhBZLF-1-specific T cells. rhEBNA-1-specific CD4+ T cells were highly functional, with both central memory and effector memory T cells (TCM, expressing high levels of CD28, CD95, and CCR7 [CD28hi CD95hi CCR7hi], and TEM, expressing high levels of CD28 and CD95 and low levels of CCR7 [CD28hi CD95hi CCR7lo], respectively) and a wide range of cytokine responses, while the CD8+ T-cell response was composed primarily of more activated gamma interferon (IFN-γ)-producing effector (EFF; CD28lo CD95hi) cells. In contrast, rhBZLF-1-specific CD4+ T cells maintained a resting phenotype of central memory T cells and differed in cytokine production compared to those specific to rhEBNA-1, primarily producing single-cytokine responses. As with rhEBNA-1, the CD8+ T-cell response against rhBZLF-1 was more activated than the CD4+ T-cell response but contained a large proportion of memory cells as well. The similarities between our findings and the human immune response against these two very important proteins serve to further validate the rhLCV model as a useful and highly relevant animal model for studying various aspects of EBV. Furthermore, our study provides a baseline for future studies of therapeutic interventions in the rhLCV model.
MATERIALS AND METHODS
Nonhuman primates (NHPs).
Adult, healthy, and simian immunodeficiency virus (SIV)-uninfected Indian-origin Macaca mulatta animals were housed at the Yerkes National Primate Research Center (YNPRC; Atlanta, GA). Most animals with the exception of PH1019 and PWw were born at YNPRC. Animals were either housed at a field station or at the main station. All animals were female adults ranging from 7 to 20 years of age at the start of the study. Animals were tested for Mamu-A*01, -A*02, -A*08, -B*01, -B*04, -B*08, and -B*17. All animals tested positive for rhLCV infection by serologic testing for serum antibodies against the rhLCV small viral capsid antigen (33). Additional samples were obtained from five rhLCV-seronegative rhesus macaques in the extended specific-pathogen-free colony housed at the New England National Primate Research Center, Harvard Medical School, Southborough, MA. All procedures involving handling of animals were performed according to approved protocols and upon review by the Institutional Animal Care and Use Committees.
Isolation and preservation of lymphocytes.
Peripheral blood mononuclear cells (PBMCs) were isolated from blood as described previously (34). They were tested immediately after isolation or frozen in 90% fetal bovine serum (FBS) and 10% dimethyl sulfoxide (DMSO) (Sigma, St. Louis, MO) at −80°C until testing.
ICS.
The function of rhEBNA-1- and rhBZLF-1-specific T cells was assessed by intracellular cytokine staining (ICS) after stimulation with peptide pools (35). rhEBNA-1 was used at a final concentration of 2 μg of each peptide per ml, and rhBZLF-1 was used at a final concentration of 1 μg of each peptide per ml. Frozen cells were thawed and immediately washed with Hank's balanced salt solution (HBSS) supplemented with 2 units/ml DNase I, resuspended with RPMI medium, and stimulated for 6 h with anti-CD28 (clone CD28.2), anti-CD49d (clone 9F10), and brefeldin A. Cells were stained with violet-fluorescent reactive dye Pacific blue (Invitrogen, Carlsbad, CA), anti-CD14-Pacific blue (clone M5E2), anti-CD16-Pacific blue (clone 3G8), anti-CD8-allophycocyanin (APC)-H7 (clone SK1), anti-CD4-Alexa 700 (clone OKT4), anti-CD95-phycoerythrin (PE)-Cy5 (clone DX2), and anti-CD28-Texas red (clone CD28.2) (Beckman Coulter, Fullerton, CA) for 30 min at 4°C. Additionally, cells were stained with anti-CCR7-PE (clone 150503). After fixation and permeabilization with Cytofix/Cytoperm (BD Biosciences, San Jose, CA) for 30 min at 4°C, cells were stained with anti-IFN-γ–APC (clone B27), anti-interleukin-2–fluorescein isothiocyanate (anti-IL-2–FITC; clone MQ1-17H12), anti-tumor necrosis factor alpha (TNF-α)–PE–Cy7 (clone MAb11; R&D Systems), and anti-CD3-peridinin chlorophyll protein (PerCp)-Cy5.5 (clone SP34-2) for 30 min at 4°C. Cells were washed twice, fixed with BD Stabilizing Fixative (BD Biosciences, San Jose, CA), and then analyzed by fluorescence-activated cell sorting (FACS) using an LSRII instrument (BD Biosciences, San Jose, CA) and DiVa software. Flow cytometric acquisition and analysis of samples were performed on at least 500,000 events. Postacquisition analyses were performed with FlowJo software (TreeStar, Ashland, OR). Data shown on graphs represent values of peptide-stimulated cells from which background values have been subtracted and are the average value of two time points collected 2 months apart. We define a positive response as any value greater than the mean plus 2 standard deviations of total CD4+ (183 CD4+ cells per 106 CD3+ T cells) or CD8+ (216 CD8+ cells per 106 CD3+ T cells) cytokine responses from five seronegative subjects (data not shown). Polyfunctionality pie chart graphs were generated using SPICE software (NIH, Bethesda, MD). Single-color controls used CompBeads Anti-Mouse Igk (BD Biosciences, San Jose, CA) for compensation. Unless otherwise noted, antibodies were purchased from BD (BD Biosciences, San Jose, CA).
Synthetic peptides.
The rhEBNA-1 peptide pool consists of 85 15-mer peptides overlapping by 10 amino acids except for the GA repeat domain, which overlaps by 5 amino acids (Genemed Synthesis, Inc., San Antonio, TX; NeoBioSci, Cambridge, MA). To account for its large size, rhEBNA-1 was divided into two pools, one with peptides 1 to 42 and the other with peptides 43 to 85; responses were combined after normalization and subtraction of background values. The rhBZLF-1 peptide pool consists of 60 15-mer peptides with an 11-amino-acid overlap and was provided by A. Kaur. Peptides were reconstituted in DMSO.
Statistical analysis.
rhEBNA-1- and rhBZLF-1-specific immune responses were compared between CD4+ and CD8+ T cells. IFN-γ, IL-2, and TNF-α cytokine production as well as effector, central memory (TCM), and effector memory (TEM) subset analyses within both CD4+ and CD8+ T-cell populations was also compared. All comparisons were performed using a two-tailed Wilcoxon's signed-rank test unless otherwise noted. A Bonferroni adjustment was applied to all multiple comparisons in order to control for a type I error rate. A P value of <0.05 was considered statistically significant. GraphPad Prism, version 6 (La Jolla, CA), and SPICE, version 5.22 (NIH, Bethesda, MD), were used for calculations and illustrations.
RESULTS
Frequency and magnitude of rhEBNA-1- and rhBZLF-1-specific T-cell responses.
We used intracellular cytokine staining (ICS) to measure T-cell responses to rhEBNA-1. For comparison, we also measured T-cell responses to the highly immunogenic immediate-early antigen rhBZLF-1, which regulates the switch from latent infection to lytic replication and is not expressed during latency (36, 37). Peripheral blood mononuclear cells (PBMCs) were isolated and tested for CD4+ and CD8+ T cells producing IFN-γ, IL-2, and/or TNF-α in response to a short in vitro stimulation with overlapping rhEBNA-1 or rhBZLF-1 peptide pools. Background activity was measured by culturing cells in medium containing DMSO for the same length of time. Upon staining, cells were analyzed by flow cytometry. Postacquisition analyses were conducted by FlowJo software. Figure 1A shows the gating strategy. Briefly, single cells were gated from PBMCs (Fig. 1A, panel a) followed by lymphocytes (b), live CD3+ T cells (c), and CD4+ versus CD8+ T cells (d) and differentiation within each T-cell population (e). Figure 1B and C show responses to the rhEBNA-1 peptide pool exemplified by one rhLCV-seropositive (Fig. 1B) and one rhLCV-seronegative (Fig. 1C) animal. Results were normalized to numbers of responding cells per 106 live CD3+ cells. To calculate the sum of the peptide-specific responses, we subtracted normalized background activity and then summed the seven possible different combinations of functions. We define a positive response as anything higher than the mean background plus 2 standard deviations of total CD4+ (183 CD4+ cells per 106 CD3+ T cells) or CD8+ (216 CD8+ cells per 106 CD3+ T cells) cytokine responses from the five seronegative animals from the New England Primate Research Center (data not shown). Peptide-specific responses were measured at two time points spaced 2 months apart, and analyses were based on the average response of each subject. rhEBNA-1- and rhBZLF-1-specific total cytokine responses for each time point can be seen in Tables 1 and 2, respectively.
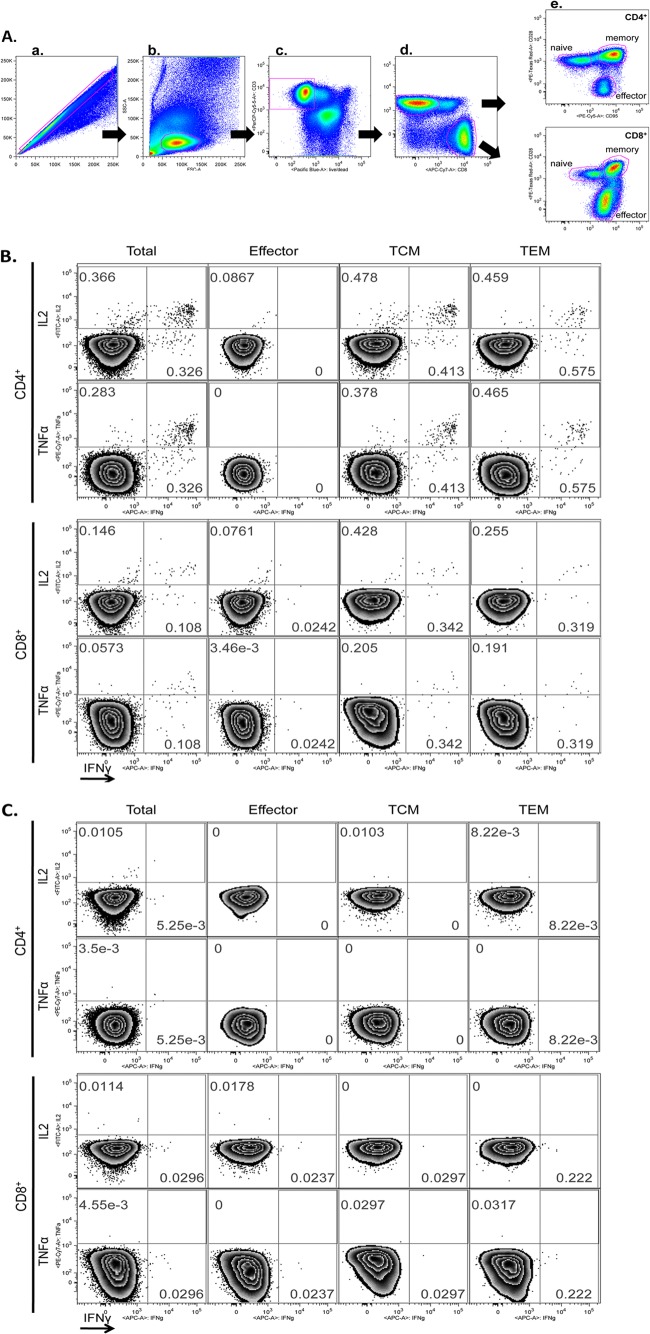
Fig 1.
Gating strategy for multiparameter flow cytometry and representative examples. (A) The following gating strategy was used to identify rhEBNA-1-specific T cells. Cells were initially gated on the basis of their forward scatter (FSC) area versus FSC height profile to remove doublets (a), followed by a lymphocyte gate based on the FSC area versus side scatter (SSC) area profile (b). Dead cells were then excluded by gating CD3 versus a live-cell stain (Pacific blue). The amine reactive live/dead stain was in the same channel as CD14+, CD16+, and/or CD20+ cells so that non-T cell populations were also gated out at this step (c). CD4+ and CD8+ T-cell differentiation (d) was based on CD28 and CD95 expression (effector, CD28lo CD95hi; memory, CD28hi CD95hi) (e). Memory cells were further differentiated into central memory and effector memory with CCR7 (central memory, CD28hi CD95hi CCR7hi; effector memory, CD28hi CD95hi CCR7lo). A gate was then set for each stain identifying intracellular-accumulated IFN-γ, IL-2, or TNF-α. EBNA-1-specific CD4+ and CD8+ T-cell multisubset analysis from peripheral blood of one seropositive (B) and one seronegative (C) animal was performed. The analyses were performed with samples immediately frozen after blood collection and PBMC isolation. Phorbol myristate acetate and ionomycin were used as positive controls, and medium with DMSO was used as a negative control (data not shown). Numbers represent the frequency of cytokine-producing cells in the corresponding quadrant. Samples were analyzed by FACS using LSRII and DiVa software. Postacquisition analyses were performed with FlowJo.
Table 1.
Total cytokine-secreting rhEBNA-1- specific CD4+ and CD8+ T cells for each animal at both collection points
| Subject | No. of T cells by cell populationa |
|||
|---|---|---|---|---|
| CD4 |
CD8 |
|||
| Bleed 1 | Bleed 2 | Bleed 1 | Bleed 2 | |
| RNw9 | 461 | 498 | 284 | 235 |
| RQt5 | 562 | 294 | 393 | 74 |
| RYc3 | 63 | 1,334 | 238 | 410 |
| RRi9 | 146 | 538 | 342 | 21 |
| RLy9 | 103 | 394 | 44 | 62 |
| RTp4 | 739 | 159 | 725 | 34 |
| RCj7 | 351 | 744 | 764 | 623 |
| RTn5 | 268 | 208 | 0 | 60 |
| RZi7 | 67 | 91 | 89 | 14 |
| RLz5 | 1,380 | 760 | 450 | 465 |
| PH1019 | 71 | 122 | 451 | 149 |
| PWw | 158 | 150 | 545 | 149 |
| RVw6 | 1,666 | 2,713 | 267 | 603 |
| RCv5 | 469 | 714 | 130 | 200 |
| RYa6 | 378 | 368 | 457 | 111 |
Responses at both collection points (bleed 1 and bleed 2) are displayed as counts per 106 live CD3+ cells after subtraction of background values.
Table 2.
Total cytokine-secreting rhBZLF-1- specific CD4+ and CD8+ T cells for each animal at both collection points
| Subject | No. of T cells by cell populationa |
|||
|---|---|---|---|---|
| CD4 |
CD8 |
|||
| Bleed 1 | Bleed 2 | Bleed 1 | Bleed 2 | |
| RNw9 | 260 | 191 | 2,395 | 1,973 |
| RQt5 | 323 | 427 | 324 | 246 |
| RYc3 | 32 | 1,893 | 434 | 556 |
| RRi9 | 698 | 579 | 1,331 | 614 |
| RLy9 | 1,566 | 845 | 0 | 364 |
| RTp4 | 152 | 255 | 937 | 610 |
| RCj7 | 342 | 254 | 464 | 361 |
| RTn5 | 255 | 327 | 352 | 779 |
| RZi7 | 137 | 40 | 336 | 294 |
| RLz5 | 438 | 178 | 465 | 163 |
| PH1019 | 144 | 405 | 1,756 | 1,190 |
| PWw | 211 | 730 | 498 | 425 |
| RVw6 | 133 | 605 | 411 | 768 |
| RCv5 | 1,337 | 957 | 190 | 535 |
| RYa6 | 1,007 | 851 | 583 | 234 |
Responses at both collection points (bleed 1 and bleed 2) are displayed as counts per 106 live CD3+ cells after subtraction of background values.
We were able to detect rhEBNA-1-specific CD4+ and/or CD8+ T cells in 14 of the 15 rhesus macaques (93%) from the Yerkes National Primate Research Center (Fig. 2A). Twelve of the 15 animals (80%) had rhEBNA-1-specific CD4+ T-cell responses above the detection limit, with a magnitude that ranged between 238 and 2,190 responding cells per 106 CD3+ T cells. Slightly fewer animals had detectable rhEBNA-1-specific CD8+ T-cell responses (10 of the 15, or 67%) with a lower magnitude that ranged from 233 to 693 cells per 106 CD3+ T cells. The mean rhEBNA-1-specific CD4+ T-cell response of 638 cells/106 CD3+ T cells was significantly higher than the mean CD8+ T-cell response of 387 cells/106 CD3+ T cells (P = 0.0391). One subject, RZi7, had neither CD4+ nor CD8+ rhEBNA-1-specific T cells above the detection limit. Eight of the remaining 14 animals had both CD4+ and CD8+ rhEBNA-1-specific T cells; in 7 of these animals the CD4+ response was higher. We were unable to detect a peptide-specific CD8+ T-cell response in four animals, and two animals did not have a detectable CD4+ T-cell response.
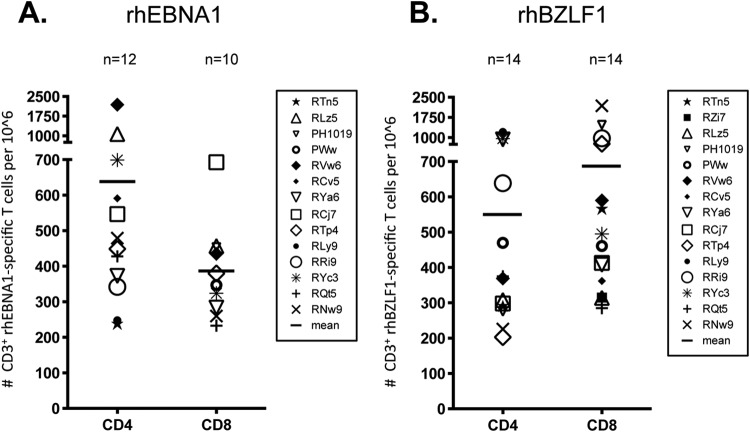
Fig 2.
Numbers of rhEBNA-1- and rhBZLF-1-specific T cells in PBMCs. We assessed the frequency and magnitude of rhEBNA-1- and rhBZLF-1-specific T-cell responses in 15 rhesus macaques by stimulating PBMCs with an overlapping rhEBNA-1 or rhBZLF-1 peptide pool, phorbol myristate acetate/ionomycin (positive control), or medium with DMSO (negative control) in the presence of brefeldin A and then performing ICS. All values are represented as peptide-specific T-cell counts per million live CD3+ T cells. To calculate the sum of the peptide-specific responses, we subtracted normalized background activity and then summed the seven possible different combinations of functions. Values below the mean plus 2 standard deviations of total cytokine CD4+ or CD8+ T cells from the average of five seronegative samples (CD4+, 183 cells per 106 CD3+ T cells; CD8+, 216 cells per 106 CD3+ T cells [data not shown]) were considered below the detection limit and excluded from the figures. The average sum of total cytokine-secreting rhEBNA-1-specific (A) or rhBZLF-1-specific (B) CD4+ and CD8+ T cells after subtraction of background values is shown. Data for each animal reflect the mean of two time points spaced 2 months apart; values for each time point can be seen in Tables 1 and 2. Bars indicate the mean value of all positive animals within each group. Standard deviations are as follows: rhEBNA-1 CD4, 538 cells; rhEBNA-1 CD8, 134 cells; rhBZLF-1 CD4, 358 cells; rhBZLF-1 CD8, 538 cells. There was a significant difference between total rhEBNA-1 CD4+ and CD8+ T-cell cytokine responses (P = 0.0391). P values were calculated using a two-sided Wilcoxon's matched-pairs signed-rank test, with a P value of <0.05 considered significant. All multiple comparisons were Bonferroni adjusted to control for a type I error rate.
In contrast to results for rhEBNA-1, we detected rhBZLF-1-specific CD4+ and/or CD8+ T cells in all 15 of the screened animals (Fig. 2B). Fourteen of these macaques (93%) had rhBZLF-1-specific CD4+ T-cell responses above the detection limit, and the magnitude ranged from 226 to 1,206 rhBZLF-1-specific cells per 106 CD3+ T cells. RZi7 was the only animal with no detectable CD4+ T-cell response to rhBZLF-1. We were also able to detect rhBZLF-1-specific CD8+ T-cell responses in 14 animals (93%), and the magnitude of the response was larger than the rhBZLF-1 CD4+ T-cell response, ranging from 285 to 2,184 rhBZLF-1-specific CD8+ T cells per 106 CD3+ T cells. The one subject with undetectable rhBZLF-1 CD8+ T-cell responses had undetectable rhEBNA-1 CD8+ T cells as well. While the difference between the mean CD4+ (550 cells/106 CD3+ T cells) and CD8+ (687 cells/106 CD3+ T cells) T-cell responses was not significant, there seemed to be a trend toward higher CD8+ responses, as noted in 7 of the 13 animals with both CD4+ and CD8+ peptide-specific T cells. Of the remaining six animals, four had larger CD4+ than CD8+ T-cell responses, and two had similar levels of both CD4+ and CD8+ rhBZLF-1-specific T cells (a difference of fewer than 10 cells per 106 CD3+ T cells).
Numbers of rhEBNA-1- and rhBZLF-1-specific CD4+ and CD8+ T cells varied between individual animals; some rhesus macaques had over 500 rhEBNA-1- or rhBZLF-1-specific circulating CD4+ or CD8+ T cells per 106 CD3+ T cells, while the majority of animals had lower numbers, averaging 200 to 400 cells. Animals with high rhEBNA-1 responses did not necessarily have high rhBZLF-1 responses and vice versa. In contrast, animals with high responses against one peptide pool tended to have lower responses against the other.
Peptide-specific CD4+ and CD8+ T-cell responses differ in phenotypes and types of cytokine production.
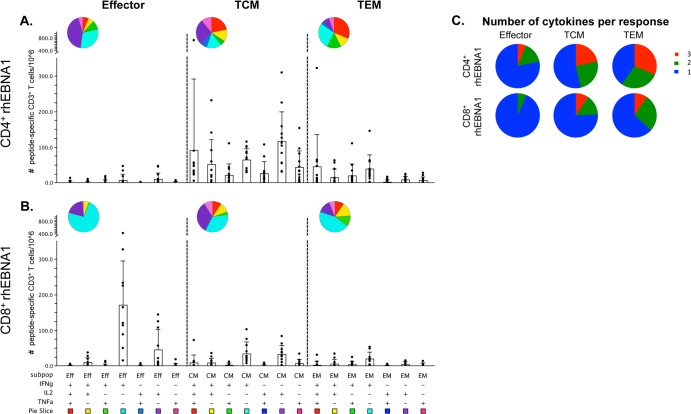
We next assessed rhEBNA-1- and rhBZLF-1-specific subset phenotypes (effector, central memory [TCM], and effector memory [TEM]) and their corresponding functionalities based on the production of IFN-γ, IL-2, and/or TNF-α. The average CD4+ T-cell response to rhEBNA-1 was composed largely of TCM cells (mean, 426 cells per 106 CD3+ T cells); TEM cells were detectable at about a one-third lower frequency (mean, 138 cells per 106 CD3+ T cells), and effector CD4+ T cells were virtually absent (mean, 28 cells per 106 CD3+ T cells) (Fig. 3A). Differences between each subset were significant (for TCM versus EFF cells, P = 0.0015; for TCM versus TEM cells, P = 0.003; for EFF versus TEM cells, P = 0.0015).
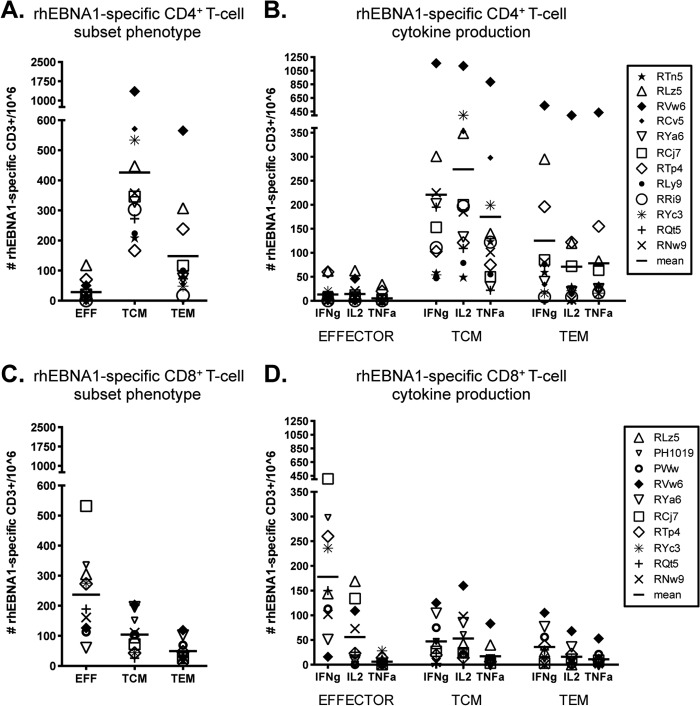
Fig 3.
Subset analysis of rhEBNA-1-specific T-cell responses. The average total counts of rhEBNA-1-specific cytokine-producing CD4+ (A) and CD8+ (C) T cells within the effector (CD28lo CD95hi), central memory (TCM; CD28hi CD95hi CCR7hi), and effector memory (TEM; CD28hi CD95hi CCR7lo) cell populations of all positive animals are shown. There was a significant difference between each CD4+ T-cell subset (TCM versus EFF, P = 0.0015; TCM versus TEM, P = 0.003; and EFF versus TEM, P = 0.0015). The CD8+ TEM subset was significantly smaller than both CD8+ effector and CD8+ TCM populations (TEM versus EFF cells, P = 0.0177; TEM versus TCM cells, P = 0.0294). The numbers of rhEBNA-1-specific CD4+ (B) and CD8+ (D) effector, TCM, and TEM cells producing IFN-γ, IL-2, and TNF-α were determined. Bars indicate the averages. Within the CD4+ response there were significant differences between TCM IL-2 production versus TNF-α production (P = 0.0102), TEM IFN-γ production versus IL-2 production (P = 0.006), and TEM IFN-γ production versus TNF-α production (P = 0.0117). Within the CD8+ response there were significant differences between effector IFN-γ production versus TNF-α production (P = 0.006), TCM TNF-α production versus IFN-γ production (P = 0.0411), TCM TNF-α production versus IL-2 production (P = 0.0117), and TEM IFN-γ production versus TNF-α production (P = 0.0294). All P values were calculated using a two-sided Wilcoxon's matched-pairs signed-rank test, with a P value of <0.05 considered significant. All multiple comparisons were Bonferroni adjusted to control for a type I error rate.
Both TCM and TEM populations produced all three cytokines (Fig. 3B). There were subtle differences in the profiles of each. On average, central memory cells produced IL-2 with the highest frequency (IL-2 versus TNF-α, P = 0.0102), followed by IFN-γ and then TNF-α. In contrast, TEM cells most often produced IFN-γ (41%; IFN-γ versus IL-2, P = 0.006; IFN-γ versus TNF-α, P = 0.0117), followed by IL-2 and TNF-α, which were about the same.
The rhEBNA-1 CD8+ T-cell response differed from the CD4+ T-cell response in terms of both phenotype and function. In contrast to the large CD4+ TCM response, 60% of rhEBNA-1-specific CD8+ T cells had an effector phenotype with a mean of 237 cells per 106 CD3+ T cells (Fig. 3C). T cells with a TCM phenotype were detected at about one-half the frequency of the effector response, with a mean of 104 cells per 106 CD3+ T cells. The TEM response was significantly smaller, contributing only 12.5% to the total CD8+ response, with a mean of 49 cells per 106 CD3+ T cells (TEM versus EFF cells, P = 0.0177; TEM versus TCM cells, P = 0.0294). rhEBNA-1-specific CD8+ effector T cells mainly produced IFN-γ (74%; IFN-γ versus TNF-α, P = 0.006) (Fig. 3D). Cytokine production within the TCM and TEM subsets was low, but the profile of the average response was similar to the rhEBNA-1 CD4+ T-cell response; TNF-α was produced by the fewest TCM cells (TNF-α versus IFN-γ, P = 0.0411; TNF-α versus IL-2, P = 0.0117), and IFN-γ was produced by the most TEM cells (IFN-γ versus TNF-α, P = 0.0294).
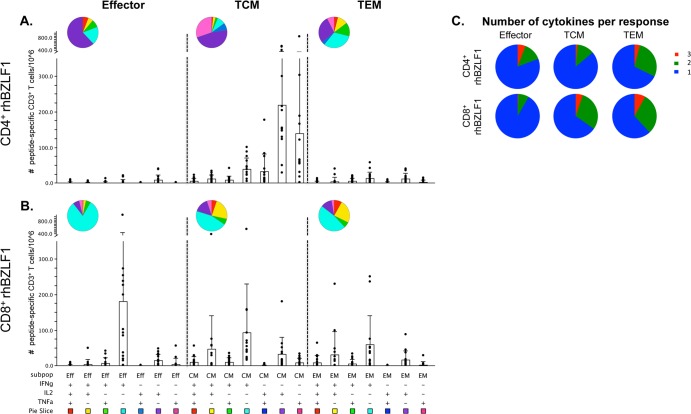
The CD4+ T-cell response to rhBZLF-1 was nearly exclusively composed of TCM cells (mean, 467 cells per 106 CD3+ T cells) (Fig. 4A). The TCM response contributed to 88% of the total rhBZLF-1 CD4+ T-cell response, while the effector memory and effector subsets represented only 9% and 3% of the total response, respectively. Differences between each subset were significant (TCM versus EFF cells, P = 0.0003; TCM versus TEM cells, P = 0.0003; and EFF versus TEM cells, P = 0.0018). The dominant functions of the rhBZLF-1 CD4+ TCM response differed from that of rhEBNA-1. While all three cytokines were still detected, IFN-γ production was significantly lower than both IL-2 (51%) and TNF-α production (37%), comprising only 12% of the total response (IFN-γ versus IL-2, P = 0.0018) (Fig. 4B).
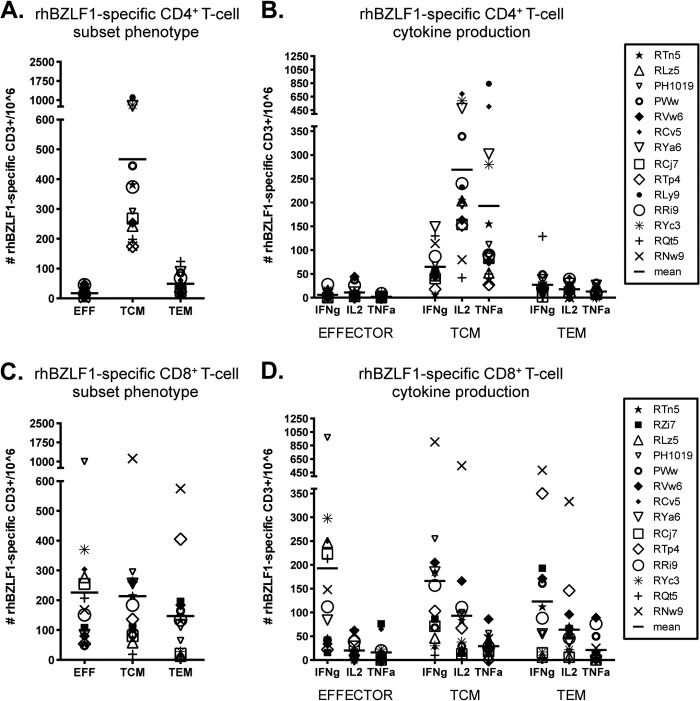
Fig 4.
Subset analysis of rhBZLF-1-specific T-cell responses. The average total counts of rhBZLF-1-specific cytokine-producing CD4+ (A) and CD8+ (C) T cells within the effector (CD28lo CD95hi), central memory (TCM; CD28hi CD95hi CCR7hi), and effector memory (TEM; CD28hi CD95hi CCR7lo) cell populations of all positive animals were determined. There were significant differences between each CD4+ T-cell subset (TCM versus EFF cells, P = 0.0003; TCM versus TEM cells, P = 0.0003; and EFF versus TEM cells, P = 0.0018). No significant differences were detected between the CD8+ subsets. The numbers of rhBZLF-1-specific CD4+ (B) and CD8+ (D) effector, TCM, and TEM cells producing IFN-γ, IL-2, and TNF-α were determined. Bars indicate the averages. Within the CD4+ TCM response, IFN-γ production was significantly lower than IL-2 production (P = 0.0018). Within the CD8+ response IFN-γ was highest in all three subsets (effector IFN-γ production versus IL-2 and TNF-α production, P = 0.0054 and 0.0036, respectively; TCM IFN-γ production versus IL-2 and TNF-α production, P = 0.0006 and 0.0018, respectively; TEM IFN-γ production versus IL-2 and TNF-α production, P = 0.003 and 0.0006, respectively). All P values were calculated using a two-sided Wilcoxon's matched-pairs signed-rank test, with a P value of <0.05 considered significant. All multiple comparisons were Bonferroni adjusted to control for a type I error rate.
The rhBZLF-1 CD8+ T-cell response was larger than the rhEBNA-1 CD8+ T-cell response. It was composed of more equal proportions of effector (mean, 226 cells per 106 CD3+ T cells), TCM (mean, 214 cells per 106 CD3+ T cells), and TEM (mean, 147 cells per 106 CD3+ T cells) subsets (Fig. 4C). Although there were no significant differences between the average magnitudes of each response, the TEM subset contributed only 25% of the total rhBZLF-1 CD8+ T-cell response, while the effector and TCM subsets were each 38.5% and 36.5%, respectively. In terms of cytokine production, all three subsets produced IFN-γ most frequently (in EFF cells [84%], IFN-γ versus IL-2 and TNF-α, P = 0.0054 and 0.0036, respectively; in TCM cells [58%], IFN-γ versus IL-2 and TNF-α, P = 0.0006 and 0.0018, respectively; in TEM cells [59%], IFN-γ versus IL-2 and TNF-α, P = 0.003 and 0.0006, respectively) (Fig. 4D). IL-2 was produced with a frequency of about 30% within each memory subset, while only 10% of memory responses were positive for TNF-α.
Polyfunctional analyses reveal distinct cytokine combinations within each subpopulation.
Because increased polyfunctionality has been associated with increased protection and therefore better control of viral replication in some chronic viral infections (38, 39), we further assessed peptide-specific T-cell functionality using Boolean gating to analyze the seven possible combinations of IFN-γ, IL-2, and TNF-α production. Comparing the cytokine profiles of the most common rhLCV-specific T-cell subsets, rhEBNA-1-specific TCM CD4+ T cells were highly polyfunctional and exhibited an array of different cytokine production profiles. While we detected substantial responses within every combination, CD4+ TCM cells most often produced either all three cytokines or IL-2 alone (Fig. 5A). The rhEBNA-1-specific CD4+ TEM responses were quite similar, producing either all three cytokines or IFN-γ alone most often, but the frequency of IL-2 and TNF-α production (either alone or together) was substantially reduced in the CD4+ TEM responses. In contrast, the most common rhEBNA-1-specific CD8+ T-cell subset of effector CD8+ T cells was strongly dominated by cells producing IFN-γ only (Fig. 5B). There was also a small population of IL-2-producing effector cells. Based on the degree of double- and triple-cytokine-producing cells, the percentage of polyfunctional rhEBNA-1-specific CD4+ TCM responses was significantly larger than the percentage of polyfunctional CD8+ TCM responses (P = 0.0021) (Fig. 5C). A similar trend was observed between CD4+ and CD8+ TEM responses (P = 0.0519). Furthermore, CD4+ TCM and TEM responses were significantly more polyfunctional than CD4+ effector responses (EFF versus TCM cells, P = 0.0366; EFF versus TEM cells, P = 0.0279). We observed a similar trend within the rhEBNA-1-specific CD8+ T-cell response as well (EFF versus TCM cells, P = 0.0177).
Fig 5.
Multifunction analysis of rhEBNA-1-specific T-cell responses. Fifteen rhesus macaques were assessed for polyfunctional rhEBNA-1-specific T-cell responses. Graphs show responses as counts per 106 live CD3+ cells after subtraction of background values for all positive animals. (A and B) All possible combinations of IFN-γ, IL-2, and TNF-α production by rhEBNA-1-specific CD4+ and CD8+ effector, TCM, and TEM cells. Black circles represent the average number of T cells comprising each cytokine combination in a single animal. Bars represent the mean value ± the standard deviation of each group. Each combination of cytokine responses is represented by plus (+) and minus (−) signs below the x axis, which is also associated with a pie slice color. The same colors are reflected in the pie charts which display the average ratio of the various cytokine combinations within each subset. (C) Pie charts display the proportion of triple (red), double (green), and single (blue) cytokine-producing CD4+ (top) and CD8+ (bottom) peptide-specific responses based on average production of IFN-γ, IL-2, and/or TNF-α. Percentages of polyfunctional CD4+ TCM and TEM cells were significantly larger than CD4+ effector responses (TCM versus EFF cells, P = 0.0366; TEM versus EFF cells, P = 0.0279). The percentage of polyfunctional CD8+ TCM cells was significantly larger than CD8+ effector responses (P = 0.0177). CD4+ TCM cells also exhibited a larger degree of polyfunctionality than CD8+ TCM cells (P = 0.0021). P values within groups or between CD4+ and CD8+ responses were calculated using a two-sided Wilcoxon's matched-pairs signed-rank test or two-sided Mann-Whitney test, respectively. A P value of <0.05 was considered significant. All multiple comparisons were Bonferroni adjusted to control for a type I error rate. Graphs were generated using SPICE software (exon.niaid.nih.gov/spice/).
In contrast to the high degree of polyfunctionality within the rhEBNA-1 CD4+ memory T-cell response, rhBZLF-1-specific CD4+ TCM cells were largely monofunctional, most often producing only IL-2 or TNF-α (Fig. 6A). rhBZLF-1-specific CD8+ effector T cells were strongly dominated by cells producing IFN-γ only (Fig. 6B). While effector CD8+ T-cell responses to rhEBNA-1 and rhBZLF-1 exhibited similar functions, we also detected rhBZLF-1-specific TCM and TEM CD8+ T cells that produced either IFN-γ only, IFN-γ in combination with IL-2, or IL-2 alone. While the rhBZLF-1 CD8+ TCM response was significantly more polyfunctional than the CD4+ response (P = 0.0081), the majority of both CD4+ and CD8+ T-cell responses were composed of single-cytokine-producing cells and therefore exhibited only a low degree of polyfunctionality (Fig. 6C).
Fig 6.
Multifunction analysis of rhBZLF-1-specific T-cell responses. Fifteen rhesus macaques were assessed for polyfunctional rhBZLF-1-specific T-cell responses. Graphs show responses as counts per 106 live CD3+ cells after subtraction of background values for all positive animals. (A and B) All possible combinations of IFN-γ, IL-2, and TNF-α production by rhBZLF-1-specific CD4+ and CD8+ effector, TCM, and TEM cells. Black circles represent the average number of T cells comprising each cytokine combination in a single animal. Bars represent the mean value ± the standard deviation of each group. Each combination of cytokine responses is represented by plus (+) and minus (−) signs below the x axis, which is also associated with a pie slice color. The same colors are reflected in the pie charts which display the average ratio of the various cytokine combinations within each subset. (C) Pie charts display the proportion of triple (red), double (green), and single (blue) cytokine-producing CD4+ (top) and CD8+ (bottom) peptide-specific responses based on average production of IFN-γ, IL-2, and/or TNF-α. CD8+ TCM cells exhibited a larger degree of polyfunctionality than CD4+ TCM cells (P = 0.0081). P values were calculated using a two-sided Mann-Whitney test, with a P value of <0.05 considered significant. All multiple comparisons were Bonferroni adjusted to control for a type I error rate. Graphs were generated using SPICE software (exon.niaid.nih.gov/spice/).
DISCUSSION
rhLCV and EBV share a high degree of sequence homology, similar molecular structures, and comparable pathogenicities (24). rhLCV could thus be the model of choice for preclinical studies aimed at preventing EBV or treating EBV-associated malignancies, but more detailed understanding of rhLCV-specific T-cell responses is needed to firmly establish the rhLCV model. Here, we characterized the ex vivo T-cell response to rhEBNA-1 in latently infected rhesus macaques and directly compared it to the T-cell response of the lytic antigen rhBZLF-1 within the same population of animals. We show that both rhEBNA-1- and rhBZLF-1-specific T cells can be detected directly ex vivo in blood of EBV-seropositive rhesus macaques kept in captivity. rhEBNA-1-specific CD4+ and CD8+ T cells are detected at 80% and 67%, respectively, in adult animals naturally infected with rhLCV. In comparison, rhBZLF-1-specific CD4+ and CD8+ T cells are both present in 93% of the animals. rhEBNA-1-specific CD4+ T cells are composed of both TCM and TEM subsets that produce IFN-γ, IL-2, and TNF-α, while rhEBNA-1-specific CD8+ T cells are primarily activated effector cells that produce IFN-γ. This may suggest that repeated in situ reactivation of rhLCV or the persistent presence of latently infected cells expressing rhEBNA-1 primarily maintains activation of CD8+ rather than CD4+ T cells. In comparison, rhBZLF-1-specific CD4+ T cells are primarily of the TCM phenotype and produce IL-2 and TNF-α, while the CD8+ T-cell response is composed of both effector and memory subsets that mainly produced IFN-γ, followed by IL-2. rhEBNA-1-specific CD4+ T cells are more polyfunctional than rhEBNA-1-specific CD8+ T cells and produce multiple cytokine combinations. In contrast, rhBZLF-1-specific CD4+ T cells produce mostly IL-2 or TNF-α alone. rhEBNA-1- and rhBZLF-1-specific CD8+ T cells have similar functional properties within the effector cell population although rhBZLF-1-specific CD8+ TCM cells exhibit a higher degree of polyfunctionality. Unlike responses in humans (40), responses to rhEBNA-1 in rhesus macaques were not influenced by the Mamu genotype although additional studies with larger cohorts and more extensive genotyping as well peptide mapping would be required to firmly establish the effects of MHC class I restricting elements on T-cell responses.
Both rhEBNA-1 and rhBZLF-1 T-cell responses have previously been studied in rhesus macaques. Using IFN-γ enzyme-linked immunosorbent spot (ELISpot) assays, Fogg et al. (32) measured antigen-specific responses to individual latent rhLCV proteins directly ex vivo by culturing PBMCs with recombinant vaccinia viruses. While responses were detected against all latent proteins tested, rhEBNA-1-specific CD8+ T cells were identified most frequently in nearly half of the animals (11 of 23) and with the highest mean responses of the latent proteins tested. The same group detected ex vivo rhBZLF-1-specific CD8+ T-cell responses at a frequency of 63% (36 of 57) (36). Orlova et al. (27) showed by IFN-γ ELISpot assays of T-cell lines from healthy seropositive rhesus macaques that rhBZLF-1-specific T cells were present in 53% (8 of 15) of the animals tested, 40% of which had specific CD8+ T cells. Using a more sensitive method that allows for simultaneous screening of several T-cell cytokines, we show that every rhLCV-seropositive rhesus macaque carries rhBZLF-1-specific T cells, while the majority of animals (14 of 15) have circulating rhEBNA-1-specific T cells. Such T cells could not be detected at significant frequencies in rhLCV-seronegative animals.
EBNA-1- and BZLF-1-specific T cells have been studied in more depth in the human system and with a variety of techniques and functional readouts in diverse cohorts. Early studies based on ELISpot assays for IFN-γ revealed EBNA-1-specific T-cell responses in less than 20% (2 of 13) of healthy human adults, while responses to another latent antigen, EBNA-3, were detected in nearly 80% of this cohort (41). In contrast, Fogg et al. (19) detected EBNA-1-specific T cells ex vivo in over 90% (12 of 13) of human adults, and ∼ 50% of those responses included EBNA-1-specific CD8+ T cells. Bickham et al. (42) studied the EBNA-1 CD4+ T-cell response and found detectable responses in only 37% (7 of 19) of the tested individuals directly ex vivo; upon in vitro expansion EBNA-1-specific CD4+ T cells became detectable in 95% of those individuals. Similarly, Munz et al. (17) detected EBNA-1-specific CD4+ T-cell responses in all human adults (10 of 10) upon initial in vitro expansion of specific cells.
BZLF-1 is considered an immunodominant target of the EBV-specific T-cell response (1, 43). Prevalence of T cells in response to this antigen within human populations is commonly studied using specific HLA backgrounds and immunodominant epitopes, and so their presence and characteristics in a truly diverse population are not well documented. Houssaint et al. (43) detected BZLF-1-specific CD8+ T cells in 60% of polyclonal T-cell lines from human subjects, but epitope-specific responses have been detected at a much higher rate when cells are tested with the appropriate HLA background. For example, Tan et al. and Woodbury et al. both reported BZLF-1-specific responses in all individuals of their HLA-specific cohorts (44, 45). Less is known about BZLF-1-specific CD4+ T cells, but Long et al. (46) used IFN-γ ELISpot assays to detect responses in 79% of EBV-seropositive individuals. The various techniques and corresponding results have been reviewed elsewhere (1, 37, 46), but, in general, BZLF-1 is known to induce a stronger CD8+ than CD4+ T-cell response, which is similar to what we observed in the current study.
Other studies have examined subset phenotypes and cytokine production of human EBV-specific T cells by flow cytometry. Heller et al. (47) reported the presence of proliferation-competent IFN-γ-producing EBNA-1-specific CD4+ T cells in 90% (18 of 20) of tested individuals; slightly more than half belonged to the TCM subset, and the remaining belonged to the TEM subset. Both the frequency and subset distribution are quite similar to our findings in rhesus macaques. Guerreiro et al. (48) described responses in eight EBV-seropositive individuals and found the majority of directly ex vivo-tested EBNA-1-specific CD4+ and CD8+ T cells and BZLF-1-specific CD4+ T cells belonged to the TCM subset and produced IL-2 and/or TNF-α, while BZLF-1-specific CD8+ T cells were more activated and produced mainly IFN-γ and TNF-α. While we describe similar subset distributions, we found that rhesus EBNA-1 CD4+ T cells were more polyfunctional and more commonly produced IFN-γ either alone or in combination with other cytokines. We also detected a more activated rhEBNA-1 CD8+ T-cell response based on the large effector population. It is possible that some of these differences are due to the fact that Guerreiro et al. analyzed the cytokine profiles of only two of the highest responders. Not only may the small sample size sway the response, but also the profile of high responders may naturally differ. Finally, Ning et al. (49) compared BZLF-1 polyfunctional responses to a combination of latent EBV antigens, adding MIP1-α and CD107a to the staining panel. Although they did not analyze subset distributions, they found polyfunctional responses within both human CD4+ and CD8+ T-cell populations similar to results of our studies in rhesus macaques.
The immunodominance of EBV lytic and latent proteins in healthy virus carriers has been reviewed elsewhere (1, 50), but it is most interesting that trends observed in our study parallel the responses observed in humans; EBNA-1-specific CD4+ T-cell responses are more dominant than CD8+ T-cell responses, and the opposite is observed for BZLF-1. Upon primary infection and subsequent reactivations, BZLF-1-specific CD8+ T cells are probably seeing more antigen and have a more direct role in controlling reactivations than the CD4+ response, which is mostly central memory. On the other hand, EBNA-1-specific CD8+ T cells may be less sensitive in detecting antigen due to the glycine-alanine repeat domain, and this may somehow trigger a larger and more functional CD4+ T-cell response.
All of the studies in humans as well as rhesus macaques demonstrate that a natural infection with EBV or rhLCV induces detectable CD4+ and CD8+ T-cell responses to both EBNA-1 and BZLF-1. Differences in subset distribution or functionality of EBNA-1-specific T-cell responses in different studies may in part reflect differences in the employed assays. We have chosen to assess antigen-specific responses with overlapping peptide pools and a short 6-h stimulation because we feel that this method provides the most accurate reflection of the response in vivo. Prolonged stimulation of T cells by their ex vivo expansion not only changes T-cell functions but also leads to selective proliferation of some T cells and concomitant loss of others. Furthermore, studies that use only IFN-γ as an output of response specificity are likely minimizing the actual frequencies of the responses. Variances may also reflect different study populations from diverse geographic regions, different age ranges, or different underlying diseases. While humans are more frequently exposed to infectious agents and more commonly encounter perturbations of their immune systems that may support EBV reactivation, such as infections (51), autoimmunity (52), or stress (53, 54), the nonhuman primates of our study were housed in a relatively controlled environment. Furthermore, it is important to note that most primates become naturally infected with rhLCV during the first few years of life. Exposure of humans in developed countries is more sporadic and can be delayed until adolescence or even adulthood although infections occur earlier in less developed countries such as Africa (55). Some studies suggest that responses against lytic and latent antigen in healthy seropositive individuals are influenced by the length of time since primary exposure (45, 56), and this is much more variable in the human response. Nonetheless, other data failed to demonstrate a correlation between age and magnitude of T-cell responses to antigens of EBV using cohorts from Africa (57).
The immunological responses to EBV are complex yet critical for understanding their role in human carcinogenesis and related diseases. Although primary infection is in general benign, the persistence of this oncogenic virus can lead to multiple cancer types, typically exacerbated by immunological dysfunction or immunosuppression associated with human immunodeficiency virus type 1 (HIV-1) infection or solid-organ transplants. Recent evidence also supports a role for EBV infection in several autoimmune disorders, suggesting that EBV has a complex interaction with the host immune system. The natural rhLCV infection of rhesus macaques provides an ideal animal model to study host immunological responses to EBV-like viruses and is also the system of choice for preclinical evaluation of EBV vaccines. Therefore, the importance of this study is twofold: first, by comparing rhesus macaque EBNA-1 and BZLF-1 T-cell responses to human EBNA-1 and BZLF-1 T-cell responses, we have further validated the rhLCV model as an appropriate and useful system for studying EBV. Second, an improved understanding of the rhEBNA-1 immune response will benefit the preclinical development of vaccines that could potentially prevent or control EBV-associated malignancies.
ACKNOWLEDGMENTS
This study was supported by grants from the NIH/NCI to P.M.L. (1RC2CA148325) and to the Yerkes National Primate Research Center (P51OD11132). R.M.L. was further supported by the University of Pennsylvania Training Grant in Vaccines and Immune Therapy (grant 5T32AI70099-4) awarded from the GTV graduate program of the University of Pennsylvania. Services at the New England Primate Research Center were supported by a base grant (P51OD011103).
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Landais E, Saulquin X, Houssaint E. 2005. The human T cell immune response to Epstein-Barr virus. Int. J. Dev. Biol. 49:285–292 [DOI] [PubMed] [Google Scholar]
- 2. Kerr BM, Lear AL, Rowe M, Croom-Carter D, Young LS, Rookes SM, Gallimore PH, Rickinson AB. 1992. Three transcriptionally distinct forms of Epstein-Barr virus latency in somatic cell hybrids: cell phenotype dependence of virus promoter usage. Virology 187:189–201 [DOI] [PubMed] [Google Scholar]
- 3. Schooley RT, Arbit DI, Henle W, Hirsch MS. 1984. T-lymphocyte subset interactions in the cell-mediated immune response to Epstein-Barr virus. Cell Immunol. 86:402–412 [DOI] [PubMed] [Google Scholar]
- 4. Parkin DM. 2006. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 118:3030–3044 [DOI] [PubMed] [Google Scholar]
- 5. Henle W, Henle G. 1974. The Epstein-Barr Virus (EBV) in Burkitt's lymphoma and nasopharyngeal carcinoma. Ann. Clin. Lab Sci. 4:109–114 [PubMed] [Google Scholar]
- 6. Mueller N. 1987. Epidemiologic studies assessing the role of the Epstein-Barr virus in Hodgkin's disease. Yale J. Biol. Med. 60:321–332 [PMC free article] [PubMed] [Google Scholar]
- 7. Quadrelli C, Barozzi P, Riva G, Vallerini D, Zanetti E, Potenza L, Forghieri F, Luppi M. 2011. β-HHVs and HHV-8 in lymphoproliferative disorders. Mediterr. J. Hematol. Infect. Dis. 3:e2011043. 10.4084/MJHID.2011.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Posnett DN. 2008. Herpesviruses and autoimmunity. Curr. Opin. Investig. Drugs 9:505–514 [PubMed] [Google Scholar]
- 9. Garzelli C, Pacciardi A, Carmignani M, Conaldi G, Basolo F, Falcone G. 1989. A human monoclonal autoantibody isolated from a patient with infectious mononucleosis reactive with both self antigens and Epstein-Barr virus nuclear antigen (EBNA). Immunol. Lett. 22:211–216 [DOI] [PubMed] [Google Scholar]
- 10. Dreyfus DH. 2011. Autoimmune disease: a role for new anti-viral therapies? Autoimmun. Rev. 11:88–97 [DOI] [PubMed] [Google Scholar]
- 11. Epstein MA, Randle BJ, Finerty S, Kirkwood JK. 1986. Not all potently neutralizing, vaccine-induced antibodies to Epstein-Barr virus ensure protection of susceptible experimental animals. Clin. Exp. Immunol. 63:485–490 [PMC free article] [PubMed] [Google Scholar]
- 12. Lockey TD, Zhan X, Surman S, Sample CE, Hurwitz JL. 2008. Epstein-Barr virus vaccine development: a lytic and latent protein cocktail. Front. Biosci. 13:5916–5927 [DOI] [PubMed] [Google Scholar]
- 13. Sivachandran N, Wang X, Frappier L. 2012. Functions of the Epstein-Barr virus EBNA1 protein in viral reactivation and lytic infection. J. Virol. 86:6146–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munz C. 2004. Epstein-Barr virus nuclear antigen 1: from immunologically invisible to a promising T cell target. J. Exp. Med. 199:1301–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. U. S. A. 94:12616–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee SP, Brooks JM, Al-Jarrah H, Thomas WA, Haigh TA, Taylor GS, Humme S, Schepers A, Hammerschmidt W, Yates JL, Rickinson AB, Blake NW. 2004. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J. Exp. Med. 199:1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Munz C, Bickham KL, Subklewe M, Tsang ML, Chahroudi A, Kurilla MG, Zhang D, O'Donnell M, Steinman RM. 2000. Human CD4+ T lymphocytes consistently respond to the latent Epstein-Barr virus nuclear antigen EBNA1. J. Exp. Med. 191:1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nikiforow S, Bottomly K, Miller G, Munz C. 2003. Cytolytic CD4+-T-cell clones reactive to EBNA1 inhibit Epstein-Barr virus-induced B-cell proliferation. J. Virol. 77:12088–12104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fogg MH, Wirth LJ, Posner M, Wang F. 2009. Decreased EBNA-1-specific CD8+ T cells in patients with Epstein-Barr virus-associated nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. U. S. A. 106:3318–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fu T, Voo KS, Wang RF. 2004. Critical role of EBNA1-specific CD4+ T cells in the control of mouse Burkitt lymphoma in vivo. J. Clin. Invest. 114:542–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heller KN, Arrey F, Steinherz P, Portlock C, Chadburn A, Kelly K, Munz C. 2008. Patients with Epstein Barr virus-positive lymphomas have decreased CD4+ T-cell responses to the viral nuclear antigen 1. Int. J. Cancer 123:2824–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moormann AM, Heller KN, Chelimo K, Embury P, Ploutz-Snyder R, Otieno JA, Oduor M, Munz C, Rochford R. 2009. Children with endemic Burkitt lymphoma are deficient in EBNA1-specific IFN-gamma T cell responses. Int. J. Cancer 124:1721–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Piriou E, van Dort K, Nanlohy NM, van Oers MH, Miedema F, van Baarle D. 2005. Loss of EBNA1-specific memory CD4+ and CD8+ T cells in HIV-infected patients progressing to AIDS-related non-Hodgkin lymphoma. Blood 106:3166–3174 [DOI] [PubMed] [Google Scholar]
- 24. Moghaddam A, Rosenzweig M, Lee-Parritz D, Annis B, Johnson RP, Wang F. 1997. An animal model for acute and persistent Epstein-Barr virus infection. Science 276:2030–2033 [DOI] [PubMed] [Google Scholar]
- 25. Rivailler P, Carville A, Kaur A, Rao P, Quink C, Kutok JL, Westmoreland S, Klumpp S, Simon M, Aster JC, Wang F. 2004. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood 104:1482–1489 [DOI] [PubMed] [Google Scholar]
- 26. Blake NW, Moghaddam A, Rao P, Kaur A, Glickman R, Cho YG, Marchini A, Haigh T, Johnson RP, Rickinson AB, Wang F. 1999. Inhibition of antigen presentation by the glycine/alanine repeat domain is not conserved in simian homologues of Epstein-Barr virus nuclear antigen 1. J. Virol. 73:7381–7389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Orlova N, Wang F, Fogg MH. 2011. Persistent infection drives the development of CD8+ T cells specific for late lytic infection antigens in lymphocryptovirus-infected macaques and Epstein-Barr virus-infected humans. J. Virol. 85:12821–12824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ruf IK, Moghaddam A, Wang F, Sample J. 1999. Mechanisms that regulate Epstein-Barr virus EBNA-1 gene transcription during restricted latency are conserved among lymphocryptoviruses of Old World primates. J. Virol. 73:1980–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carville A, Mansfield KG. 2008. Comparative pathobiology of macaque lymphocryptoviruses. Comp. Med. 58:57–67 [PMC free article] [PubMed] [Google Scholar]
- 30. Wang F, Rivailler P, Rao P, Cho Y. 2001. Simian homologues of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rivailler P, Cho YG, Wang F. 2002. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J. Virol. 76:12055–12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fogg MH, Kaur A, Cho YG, Wang F. 2005. The CD8+ T-cell response to an Epstein-Barr virus-related gammaherpesvirus infecting rhesus macaques provides evidence for immune evasion by the EBNA-1 homologue. J. Virol. 79:12681–12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rao P, Jiang H, Wang F. 2000. Cloning of the rhesus lymphocryptovirus viral capsid antigen and Epstein-Barr virus-encoded small RNA homologues and use in diagnosis of acute and persistent infections. J. Clin. Microbiol. 38:3219–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoy K, Tatsis N, Korioth-Schmitz B, Lasaro MO, Hensley SE, Lin SW, Li Y, Giles-Davis W, Cun A, Zhou D, Xiang Z, Letvin NL, Ertl HC. 2007. Effect of preexisting immunity to adenovirus human serotype 5 antigens on the immune responses of nonhuman primates to vaccine regimens based on human- or chimpanzee-derived adenovirus vectors. J. Virol. 81:6594–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lasaro MO, Haut LH, Zhou X, Xiang Z, Zhou D, Li Y, Giles-Davis W, Li H, Engram JC, Dimenna LJ, Bian A, Sazanovich M, Parzych EM, Kurupati R, Small JC, Wu TL, Leskowitz RM, Klatt NR, Brenchley JM, Garber DA, Lewis M, Ratcliffe SJ, Betts MR, Silvestri G, Ertl HC. 2011. Vaccine-induced T cells provide partial protection against high-dose rectal SIVmac239 challenge of rhesus macaques. Mol. Ther. 19:417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elliott SL, Pye SJ, Schmidt C, Cross SM, Silins SL, Misko IS. 1997. Dominant cytotoxic T lymphocyte response to the immediate-early trans-activator protein, BZLF1, in persistent type A or B Epstein-Barr virus infection. J. Infect. Dis. 176:1068–1072 [DOI] [PubMed] [Google Scholar]
- 37. Fogg MH, Garry D, Awad A, Wang F, Kaur A. 2006. The BZLF1 homolog of an Epstein-Barr-related gamma-herpesvirus is a frequent target of the CTL response in persistently infected rhesus macaques. J. Immunol. 176:3391–3401 [DOI] [PubMed] [Google Scholar]
- 38. Kannanganat S, Ibegbu C, Chennareddi L, Robinson HL, Amara RR. 2007. Multiple-cytokine-producing antiviral CD4 T cells are functionally superior to single-cytokine-producing cells. J. Virol. 81:8468–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Precopio ML, Betts MR, Parrino J, Price DA, Gostick E, Ambrozak DR, Asher TE, Douek DC, Harari A, Pantaleo G, Bailer R, Graham BS, Roederer M, Koup RA. 2007. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. J. Exp. Med. 204:1405–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stuber G, Dillner J, Modrow S, Wolf H, Szekely L, Klein G, Klein E. 1995. HLA-A0201 and HLA-B7 binding peptides in the EBV-encoded EBNA-1, EBNA-2 and BZLF-1 proteins detected in the MHC class I stabilization assay. Low proportion of binding motifs for several HLA class I alleles in EBNA-1. Int. Immunol. 7:653–663 [DOI] [PubMed] [Google Scholar]
- 41. Subklewe M, Chahroudi A, Bickham K, Larsson M, Kurilla MG, Bhardwaj N, Steinman RM. 1999. Presentation of Epstein-Barr virus latency antigens to CD8+, interferon-gamma-secreting, T lymphocytes. Eur. J. Immunol. 29:3995–4001 [DOI] [PubMed] [Google Scholar]
- 42. Bickham K, Munz C, Tsang ML, Larsson M, Fonteneau JF, Bhardwaj N, Steinman R. 2001. EBNA1-specific CD4+ T cells in healthy carriers of Epstein-Barr virus are primarily Th1 in function. J. Clin. Invest. 107:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Houssaint E, Saulquin X, Scotet E, Bonneville M. 2001. Immunodominant CD8 T cell response to Epstein-Barr virus. Biomed. Pharmacother. 55:373–380 [DOI] [PubMed] [Google Scholar]
- 44. Tan LC, Gudgeon N, Annels NE, Hansasuta P, O'Callaghan CA, Rowland-Jones S, McMichael AJ, Rickinson AB, Callan MF. 1999. A re-evaluation of the frequency of CD8+ T cells specific for EBV in healthy virus carriers. J. Immunol. 162:1827–1835 [PubMed] [Google Scholar]
- 45. Woodberry T, Suscovich TJ, Henry LM, Davis JK, Frahm N, Walker BD, Scadden DT, Wang F, Brander C. 2005. Differential targeting and shifts in the immunodominance of Epstein-Barr virus—specific CD8 and CD4 T cell responses during acute and persistent infection. J. Infect. Dis. 192:1513–1524 [DOI] [PubMed] [Google Scholar]
- 46. Long HM, Leese AM, Chagoury OL, Connerty SR, Quarcoopome J, Quinn LL, Shannon-Lowe C, Rickinson AB. 2011. Cytotoxic CD4+ T cell responses to EBV contrast with CD8 responses in breadth of lytic cycle antigen choice and in lytic cycle recognition. J. Immunol. 187:92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heller KN, Upshaw J, Seyoum B, Zebroski H, Munz C. 2007. Distinct memory CD4+ T-cell subsets mediate immune recognition of Epstein Barr virus nuclear antigen 1 in healthy virus carriers. Blood 109:1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guerreiro M, Na IK, Letsch A, Haase D, Bauer S, Meisel C, Roemhild A, Reinke P, Volk HD, Scheibenbogen C. 2010. Human peripheral blood and bone marrow Epstein-Barr virus-specific T-cell repertoire in latent infection reveals distinct memory T-cell subsets. Eur. J. Immunol. 40:1566–1576 [DOI] [PubMed] [Google Scholar]
- 49. Ning RJ, Xu XQ, Chan KH, Chiang AK. 2011. Long-term carriers generate Epstein-Barr virus (EBV)-specific CD4+ and CD8+ polyfunctional T-cell responses which show immunodominance hierarchies of EBV proteins. Immunology 134:161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hislop AD, Taylor GS, Sauce D, Rickinson AB. 2007. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu. Rev. Immunol. 25:587–617 [DOI] [PubMed] [Google Scholar]
- 51. Sugano N, Ikeda K, Oshikawa M, Idesawa M, Tanaka H, Sato S, Ito K. 2004. Relationship between Porphyromonas gingivalis, Epstein-Barr virus infection and reactivation in periodontitis. J. Oral Sci. 46:203–206 [DOI] [PubMed] [Google Scholar]
- 52. Angelini DF, Serafini B, Piras E, Severa M, Coccia EM, Rosicarelli B, Ruggieri S, Gasperini C, Buttari F, Centonze D, Mechelli R, Salvetti M, Borsellino G, Aloisi F, Battistini L. 2013. Increased CD8+ T cell response to Epstein-Barr virus lytic antigens in the active phase of multiple sclerosis. PLoS Pathog. 9:e1003220. 10.1371/journal.ppat.1003220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Glaser R, Pearl DK, Kiecolt-Glaser JK, Malarkey WB. 1994. Plasma cortisol levels and reactivation of latent Epstein-Barr virus in response to examination stress. Psychoneuroendocrinology 19:765–772 [DOI] [PubMed] [Google Scholar]
- 54. Stowe RP, Pierson DL, Feeback DL, Barrett AD. 2000. Stress-induced reactivation of Epstein-Barr virus in astronauts. Neuroimmunomodulation 8:51–58 [DOI] [PubMed] [Google Scholar]
- 55. Minhas V, Brayfield BP, Crabtree KL, Kankasa C, Mitchell CD, Wood C. 2010. Primary gamma-herpesviral infection in Zambian children. BMC Infect. Dis. 10:115. 10.1186/1471-2334-10-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Scherrenburg J, Piriou ER, Nanlohy NM, van Baarle D. 2008. Detailed analysis of Epstein-Barr virus-specific CD4+ and CD8+ T cell responses during infectious mononucleosis. Clin. Exp. Immunol. 153:231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chattopadhyay PK, Chelimo K, Embury PB, Mulama DH, Sumba PO, Gostick E, Ladell K, Brodie TM, Vulule J, Roederer M, Moormann AM, Price DA. 2013. Holoendemic malaria exposure is associated with altered Epstein-Barr virus-specific CD8+ T-cell differentiation. J. Virol. 87:1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]