Abstract
For most parainfluenza viruses, a virus type-specific interaction between the hemagglutinin-neuraminidase (HN) and fusion (F) proteins is a prerequisite for mediating virus-cell fusion and cell-cell fusion. The molecular basis of this functional interaction is still obscure partly because it is unknown which region of the F protein is responsible for the physical interaction with the HN protein. Our previous cell-cell fusion assay using the chimeric F proteins of parainfluenza virus 5 (PIV5) and simian virus 41 (SV41) indicated that replacement of two domains in the head region of the PIV5 F protein with the SV41 F counterparts bestowed on the PIV5 F protein the ability to induce cell-cell fusion on coexpression with the SV41 HN protein while retaining its ability to induce fusion with the PIV5 HN protein. In the study presented here, we furthered the chimeric analysis of the F proteins of PIV5 and SV41, finding that the PIV5 F protein could be converted to an SV41 HN-specific chimeric F protein by replacing five domains in the head region with the SV41 F counterparts. The five SV41 F-protein-derived domains of this chimera were then divided into 16 segments; 9 out of 16 proved to be not involved in determining its specificity for the SV41 HN protein. Finally, mutational analyses of a chimeric F protein, which harbored seven SV41 F-protein-derived segments, revealed that replacement of at most 21 amino acids of the PIV5 F protein with the SV41 F-protein counterparts was enough to convert its HN protein specificity.
INTRODUCTION
The parainfluenza viruses (PIVs), which belong to the genus Rubulavirus, Avulavirus, or Respirovirus in the family Paramyxoviridae, have two kinds of glycoprotein spikes on the envelope: hemagglutinin-neuraminidase (HN) protein tetramers and fusion (F) protein trimers (1). The attachment protein HN is responsible for binding to the sialoconjugate receptors on the cell surface and for enzymatic destruction of the receptors, while the F protein mediates membrane fusion, such as cell-cell fusion or virus-cell fusion. Cleavage of the F precursor (F0) by cellular proteases into disulfide-linked subunits F1 and F2 is a prerequisite for its fusion activity, similar to the other class I viral fusion proteins (1).
Unlike most of the other class I fusion proteins, however, the F protein does not have an apparent receptor-binding function. Moreover, most F proteins require a fusion-promoting function of the attachment protein HN in a virus type-specific manner (2, 3). Although it is not well-known how the HN protein promotes the F-protein-mediated membrane fusion, it is believed that the fusion is induced through a series of conformational changes of the F protein that are initiated by its specific interaction with the homologous HN protein (4–6). The fusion-promoting function of the HN protein appears to depend on it being bound to its cellular receptors (7–10), and the HN protein seems to promote the F-protein-mediated fusion in manner that is dependent on the balance between its inherent F-triggering efficiency and receptor-attachment regulatory functions (binding and destruction), as suggested by Porotto et al. (11). Furthermore, the presumptive signal-transducing activity of the HN protein may be required for the induction of cell-cell fusion (12).
On the one hand, the stalk region of the HN protein, which is a tetrameric coiled coil bundle structure (13, 14), is inferred to contain the site that determines the F-protein specificity in promoting fusion (15–17), while the head region carries both the receptor-binding and -destroying activities (18, 19). The involvement of the HN stalk region of an avulavirus, Newcastle disease virus, in the physical interaction with the F protein has been certified by coimmunoprecipitation analysis (20, 21). On the other hand, our previous chimeric analyses of the F proteins of two closely related rubulaviruses, human parainfluenza virus 2 (HPIV2) and simian virus 41 (SV41), suggested that a 144-amino-acid region (designated the middle region) in the ectodomain of the HPIV2 F protein contains the site(s) that determines its specificity for the HPIV2 HN protein in the induction of cell-cell fusion (22). Recently, we found that replacement of two domains in the head region of the PIV5 F protein with the SV41 F counterparts bestowed on the PIV5 F protein the ability to induce cell-cell fusion on coexpression with the SV41 HN protein without significantly affecting its ability to induce fusion with the PIV5 HN protein (23). Similarly, mutations of four amino acids in the head region of the F protein of canine distemper virus (CDV) strain Onderstepoort impair the ability to induce cell-cell fusion with the coexpressed measles virus (MV) hemagglutinin (H) protein without significantly affecting the ability to induce fusion with the CDV H protein (24). CDV and MV are closely related members of the genus Morbillivirus in the family Paramyxoviridae (1).
It is noteworthy that replacement of the stalk region of a parainfluenza virus HN protein with that of another HN protein results in conversion of the F protein specificity (17, 23), being consistent with the involvement of the stalk region in the specific interaction with the homologous F protein. However, such a region of the F protein has not been identified. In the study presented here, we furthered the chimeric analysis of the F proteins of PIV5 and SV41 and found that replacement of 21 amino acids on the PIV5 F trimer surface with the SV41 F counterparts resulted in full conversion of HN protein specificity.
MATERIALS AND METHODS
Cells and recombinant plasmids.
Monolayers of HeLa and BHK cells were maintained in Eagle's minimum essential medium (MEM) supplemented with 5% fetal calf serum (25, 26). The recombinant SRα plasmid encoding the HN or F protein of SV41 was described previously (17). The recombinant SRα plasmid encoding the HN protein of PIV5 strain W3A or that encoding the F protein of PIV5 strain WR was described elsewhere (27, 28). The WR F protein was used because it induced fusion only when coexpressed with the HN protein, whereas the W3A F protein was not used because it induced fusion even in the absence of the HN protein (27). The W3A HN protein was used instead of the WR HN protein because the former exhibited higher fusion-promoting activity than the latter when coexpressed with the PIV5 (WR) F protein (our unpublished data).
Generation of chimeric F proteins.
To generate a chimeric F protein of SV41 and PIV5, a desired domain of the SV41 F-encoding plasmid was amplified by PCR, while both of the flanking regions of the corresponding domain of the PIV5 F-encoding plasmid were amplified while simultaneously introducing the restriction enzyme site for SpeI or PstI at the distal end, as described recently (23). These three fragments were then combined stepwise by fusion PCR and inserted into the PIV5 F-protein-encoding plasmid by utilizing restriction enzyme sites SpeI and PstI in the PIV5 F-encoding cDNA (Fig. 1A).
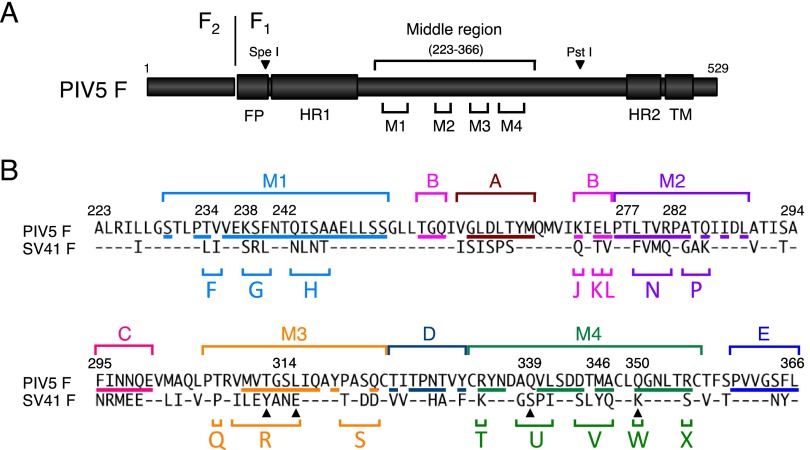
Fig 1.
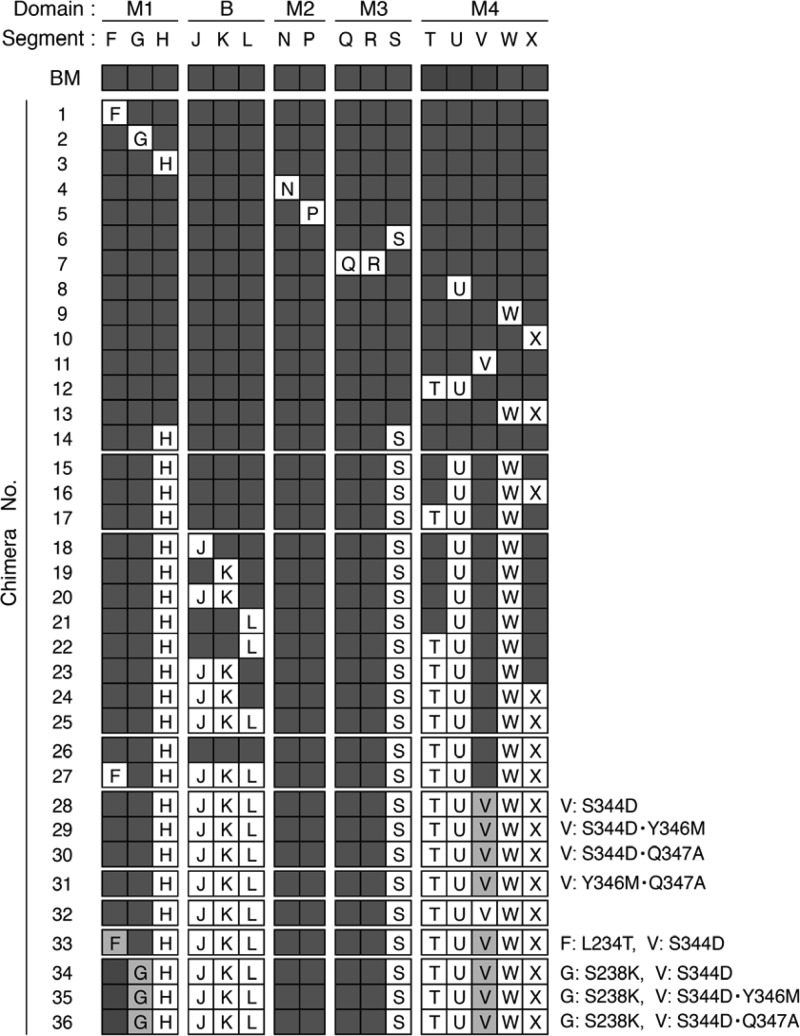
Division of five domains in the middle region of the PIV5 (WR) F protein into 16 segments. (A) Schematic diagram of the PIV5 F protein. The positions of four major domains (M1, M2, M3, and M4) in the middle region are indicated. The names of the restriction enzymes (SpeI and PstI), whose sites are present in the PIV5 F-encoding cDNA, indicate the corresponding positions in the PIV5 F polypeptides. The positions of heptad repeat regions 1 and 2 (HR1 and HR2) are based on the secondary structure of the HPIV3 F protein in its postfusion form (39). FP, fusion peptide; TM, transmembrane domain. (B) Amino acid sequence alignment of the middle regions of the F proteins of PIV5 and SV41. The positions of the four major domains and five minor domains (A, B, C, D, and E) are indicated above the amino acid sequences. Segments in each domain are indicated as capital letters below the amino acid sequences. Amino acid residues of the PIV5 F protein, which are exposed on the trimer surface, as described elsewhere (23), are underlined. Dashes in the SV41 F sequence indicate amino acids identical to those of the PIV5 F protein. The four filled triangles below the amino acid sequences of segments R, U, and W indicate the positions of the PIV5 F residues whose MV F counterparts are considered responsible for the H-F interaction (31).
Quantification of cell-cell fusion.
Subconfluent HeLa or BHK cell monolayers in six-well culture plates were transfected with 2 μg/well of F-encoding plasmid and 1 μg/well of HN-encoding plasmid by using Lipofectamine LTX reagent (Invitrogen) and an X-tremeGENE 9 DNA transfection kit (Roche), respectively. After 24 h (HeLa cells) or 12 h (BHK cells) of incubation at 37°C, the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), washed three times with PBS, and stained with Giemsa's solution. A photomicrograph which involved approximately 2.5 × 104 cells was taken, and the areas (number of pixels) occupied by the fused cells (or syncytia) were measured with the aid of graphics software, NIH ImageJ, version 1.45s. Then, the extent of cell fusion was estimated as the percentage of the syncytial areas to the total area of the photograph. Ten randomly taken photographs were measured for each sample, and the average fusion index (in percent) and standard deviation were determined.
Western blotting.
Subconfluent HeLa or BHK cell monolayers were transfected with 2 μg/well of F-encoding plasmid as described above. After 24 h (HeLa cells) or 12 h (BHK cells) of incubation at 37°C, the cells were lysed on ice with 500 μl/well of lysis buffer (50 mM HEPES [pH 7.3], 10 mM lauryl maltoside, 1 mM phenylmethylsulfonyl fluoride, 100 mM NaCl), as reported previously (29). An aliquot (15 μl) of each cell lysate was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions, and the separated proteins were electroblotted to a nitrocellulose membrane (Whatman). The membrane was then successively treated with monoclonal antibody 1D1 specific for a region (residues 448 to 452) immediately upstream of the heptad repeat 2 region of the PIV5 F1 subunit (29), biotinylated horse immunoglobulin specific for mouse IgG (Vector Laboratories), and streptavidin-biotin-peroxidase complex (Vector Laboratories). The F-protein bands were then visualized by enhanced chemiluminescence (ECL) using a Western blotting luminol reagent (Santa Cruz Biotechnology), followed by exposure to X-ray film (Konica, Tokyo, Japan).
Cell surface biotinylation.
Subconfluent HeLa or BHK cell monolayers in six-well culture plates were transfected with 2 μg/well of F-encoding plasmid as described above. After 24 h (HeLa cells) or 12 h (BHK cells) of incubation at 37°C, the cells were treated with 0.3 mg/ml of EZ-link sulfo-NHS-LC-biotin (Thermo Scientific) in PBS supplemented with 0.1 mM CaCl2 and 1 mM MgCl2 at 23°C for 30 min and lysed on ice with 500 μl/well of lysis buffer. The biotinylated proteins in the cell lysates (150 μl) were then immunoprecipitated with rabbit antiserum specific for the PIV5 F2 subunit (28) and subjected to SDS-PAGE under nonreducing conditions, because we could not identify the F-protein bands under reducing conditions due to comigrating cellular bands precipitated by the anti-F2 peptide rabbit serum. The biotinylated and immunoprecipitated F proteins were then electroblotted to a nitrocellulose membrane and detected by ECL after treating with streptavidin-biotin-peroxidase complex. The intensity of the F-protein band was quantified with the aid of graphics software, NIH ImageJ, version 1.45s, and the relative surface expression level was estimated.
RESULTS
Replacement of five domains in the middle region of the PIV5 F protein with those of the SV41 F protein converts its HN protein specificity.
Previously, we performed chimeric analysis of the F proteins of PIV5 and SV41 in order to determine the site(s) that determines the HN protein specificity of the F protein. In brief, the middle region of the PIV5 F protein was divided into four trimer surface-exposed major domains and five minor domains (23) (Fig. 1A and B). These nine domains were then replaced with the SV41 F counterparts individually or in various combinations, and the resultant chimeric F proteins were expressed in HeLa cells together with the HN protein of PIV5 or SV41. The results indicated that replacement of the domains M1 and M2, which harbored 16 noncontiguous amino acids that were not conserved between the F proteins, enabled the PIV5 F protein to functionally interact with the SV41 HN protein, inducing cell-cell fusion in HeLa cells (23). In addition to the two domains identified, M1 and M2, the SV41 F-derived domains M4 and B seemed to be involved in the functional interaction with the SV41 HN protein (23). However, any chimeric PIV5 F proteins that could induce fusion with the SV41 HN protein also induced fusion with the PIV5 HN protein (23). We thus intended to create a chimeric PIV5 F protein which would show clear specificity for the SV41 HN protein by replacing its four domains (M1, M2, M4, and B), which had been regarded to be important for determining the HN protein specificity, as described above, with the corresponding domains of the SV41 F protein. To our disappointment, the resultant chimera, B + M(1 + 2 + 4) did not induce cell-cell fusion in HeLa cells either with the SV41 HN protein or with the PIV5 HN protein (see Fig. 3D), presumably because the cleaved form (F1 + 2) of this chimera was poorly expressed on the cell surface (see Fig. 3B). After our several attempts, however, it turned out that replacement of the domain M3, in addition to the four domains mentioned above, resulted in the chimera B + M(1 + 2 + 3 + 4), which was able to induce weak fusion (approximate fusion index, 1%) only when coexpressed with the SV41 HN protein (see Fig. 3D), though its cleaved form was weakly expressed on the cell surface, like that of B + M(1 + 2 + 4) was (see Fig. 3B and Fig. 5A). Interestingly, however, when we employed BHK cells for the cell-cell fusion assay, B + M(1 + 2 + 3 + 4) induced remarkable fusion (fusion index, 20.6% ± 1.9%) only when coexpressed with the SV41 HN protein (see Fig. 3E and Fig. 5B). In view of the findings that the extent of fusion induced by coexpression of the SV41 HN and F proteins in BHK cells was 19.6% ± 0.8% (data not shown), while that induced by coexpression of the PIV5 HN and F proteins was 12.6% ± 1.4% (see Fig. 5B), we have concluded that B + M(1 + 2 + 3 + 4) is an SV41 HN-specific protein similar to the SV41 F protein. This chimera harbored 41 SV41 F-derived amino acids, 32 of which were regarded as being exposed on the F trimer surface (Fig. 1B and Fig. 2). It was noteworthy, on the one hand, that the above-mentioned B + M(1 + 2 + 4) could not also induce fusion in BHK cells, whereas its cleaved form was expressed on the cell surface more abundantly than that of B + M(1 + 2 + 3 + 4) was (Fig. 3C and E).
Fig 3.
Replacement of five domains in the middle region of the PIV5 F protein with those of the SV41 F protein converts HN protein specificity. (A) Schematic diagram of chimeric F proteins. Dark boxes represent amino acid regions derived from the PIV5 F protein, while bright boxes represent the domains derived from the SV41 F protein. (B and C) Detection of chimeric F proteins in plasmid-transfected cells. Subconfluent HeLa (B) or BHK (C) cell monolayers in six-well culture plates were transfected with the recombinant plasmid encoding each F protein and lysed at 24 h (B) or 12 h (C) posttransfection. To detect the F proteins in the plasmid-transfected cells, the total cell lysates were subjected to SDS-PAGE under reducing conditions, followed by Western blotting with an anti-F1 monoclonal antibody (top). To detect the F proteins expressed on the cell surface, the plasmid-transfected cells were biotinylated, the cell lysates were subjected to immunoprecipitation with anti-F2 rabbit serum, and the precipitates were analyzed by SDS-PAGE under nonreducing conditions; the biotinylated F-protein band detected by immunoprecipitation represents the cleaved from (F1 + 2), as reported previously (29) (bottom). The relative surface expression (RSE) level of the F proteins is presented below each lane. (D and E) Fusion activity of the chimeric F proteins. For analyzing the HN protein specificity of the F proteins in a cell-cell fusion assay, subconfluent HeLa (D) or BHK (E) cell monolayers in six-well culture plates were transfected with 2 μg/well of the recombinant SRα plasmid encoding each F protein together with 1 μg/well of the recombinant SRα plasmid encoding the SV41 HN protein or the PIV5 HN protein or with the SRα plasmid (no HN). After 24 h (D) or 12 h (E), the cells were fixed with 4% paraformaldehyde and the average fusion index was determined as described in Materials and Methods; error bars indicate standard deviations.
Fig 5.
Segment S is not involved in determining HN protein specificity. (A) Detection of chimeric F proteins in plasmid-transfected BHK cells. Western blotting (top) and immunoprecipitation (bottom) were carried out as described in the legend for Fig. 3C. The number above each lane corresponds to the number of the chimera indicated in Fig. 4. 5F, PIV5 F protein; BM, B + M(1 + 2 + 3 + 4); V, vector. (B) Fusion activity of chimeric F proteins. The average fusion index was determined as described in the legend for Fig. 3E. The numbers on the abscissa axis correspond to the chimera numbers presented in panel A.
Fig 2.
Positions of SV41 F-derived segments or amino acids in chimeric PIV5 F proteins. (Top) Side views of the F trimers; (bottom) bottom views of the F trimers. The SV41 F-derived segments and amino acids, which are exposed on the trimer surface, are indicated in the colors representing the domains they belong to (Fig. 1B); for simplicity, those in only one monomer are shown. The amino acids shown in black indicate those that intervene among the SV41 F-derived amino acids in chimera no. 36. The figure was prepared with the aid of the Deep View Swiss-Pdb Viewer program on the basis of the crystal structure of the PIV5 (W3A) F-cleavage-site mutant FR3 (Protein Data Bank accession number 2B9B) (40).
These results have indicated that the PIV5 F protein can be converted to an SV41 HN-specific chimeric F protein by replacement of its five domains (M1, M2, M3, M4, and B) with those of the SV41 F protein.
On the other hand, B + M2 and B + M(1 + 2) showed clear specificity for the PIV5 HN protein in HeLa cells (Fig. 3D). Interestingly, they induced prominent fusion in BHK cells irrespective of the presence of the HN protein (Fig. 3E). Nonetheless, their specificity for the PIV5 HN protein also seemed to be apparent in BHK cells, because their fusion indices with the PIV5 HN protein were significantly higher than those with the SV41 HN protein, which were similar to the fusion indices without the HN protein (Fig. 3E). Consistent with this observation, we have recently reported that the HN-independent fusion activity of the chimera B + M(1 + 2) was further promoted by the SV41 HN protein but not by the Newcastle disease virus HN protein (23). Accordingly, when a certain chimeric F protein exhibited HN-independent fusion activity, we regarded the fusion index to be the background and its HN protein specificity could be estimated only when it induced fusion with either of the HN proteins at a level significantly higher than the background.
Segment S is not involved in determining HN protein specificity.
In order to identify the SV41 F-derived amino acids that would be responsible for converting the PIV5 F protein to an SV41 HN-specific protein, the 41 amino acids in the five domains (M1, M2, M3, M4, and B), which were not shared by the F proteins of PIV5 and SV41, as described above, were grouped into 16 segments (Fig. 1B). Then, the SV41 F-derived segments of B + M(1 + 2 + 3 + 4) were then replaced with the PIV5 F counterparts individually or in combination, and the HN protein specificity of the resulting chimeric F proteins was investigated by using BHK cells; their chimeric structures are shown as schematics in Fig. 4.
Fig 4.
Structures of chimeric PIV5 F proteins. The chimeric structure of each chimera is shown as a set of 16 squares which correspond to the 16 segments shown in Fig. 1B. Dark gray squares, SV41 F-derived segment; white squares, PIV5 F-derived segment; light gray squares, SV41 F-derived segment F, G, or V with a mutation(s), details of which are shown on the right side of each chimera. The name of each segment is indicated only in the white and gray squares. BM, B + M(1 + 2 + 3 + 4).
To begin with, we focused on the eight segments in the domains M1, M2, and M3. As shown in Fig. 5B, SV41 F-derived segment S of B + M(1 + 2 + 3 + 4) could be replaced with the PIV5 F counterpart without affecting the prominent specificity for the SV41 HN protein, as represented by chimera no. 6, indicating that segment S is not involved in determining HN protein specificity. In contrast, chimera no. 2 exhibited low but apparent fusion activity (approximately 12%) with either of the HN proteins, indicating that SV41 F-derived segment G plays a critical role in determining HN protein specificity. Although chimera no. 1 induced moderate fusion even in the absence of the HN proteins, it induced fusion with the SV41 HN protein but not with the PIV5 HN protein at a level significantly higher than this background. This result suggested that segment F was not involved in determining HN protein specificity; we verified this assumption later on. On the other hand, chimeras no. 3, no. 4, and no. 5, whose cleaved forms were weakly expressed on the cell surface, did not show fusion activity with either of the HN proteins; thus, the role of segments H, N, and P could not be evaluated. The role of segments Q and R in determining HN protein specificity also remained unclear, since the corresponding chimera, no. 7, did not induce fusion with either of the HN proteins, even though the amount of its cleaved form on the cell surface was larger than that of chimera no. 6.
Segments H, T, U, W, and X are not involved in determining HN protein specificity.
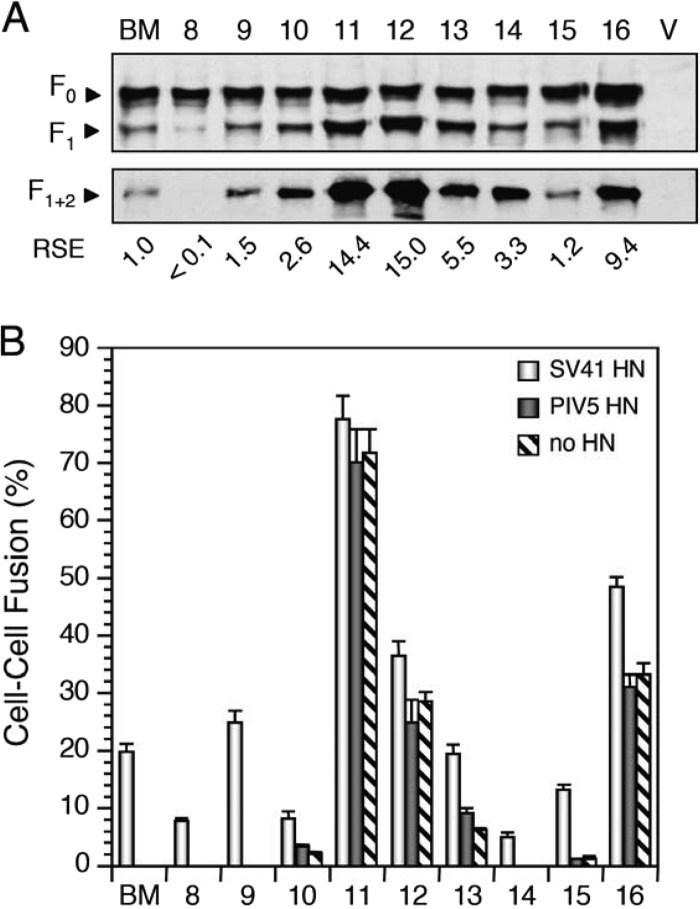
We then focused on the five segments in the domain M4 and found that SV41 F-derived segments U and W of B + M(1 + 2 + 3 + 4) could be replaced with the PIV5 HN counterparts without affecting the specificity for the SV41 HN protein, as represented by chimeras no. 8 and no. 9, respectively (Fig. 6B). Furthermore, replacement of segment X did not seem to affect the HN protein specificity of chimeras B + M(1 + 2 + 3 + 4) and no. 9, because the resultant chimeras, no. 10 and no. 13, respectively, induced fusion with the SV41 HN protein at levels significantly higher than their backgrounds but not with the PIV5 HN protein. These results suggested that segments U, W, and X are not involved in determining HN protein specificity. In contrast, replacement of segment V of B + M(1 + 2 + 3 + 4) resulted in chimera no. 11, which exhibited very high fusion activity (approximately 75%) independently of the coexpression with the HN protein, and thus, the role of V in determining HN protein specificity remained to be evaluated; we address this issue later on. Interestingly, replacement of segment T of chimera no. 8 resulted in chimera no. 12, which induced moderate HN-independent fusion (Fig. 6B). However, chimera no. 12 induced fusion with the SV41 HN protein at a level significantly higher than this background but not with the PIV5 HN protein. This result suggests that segment T is not involved in determining HN protein specificity.
Fig 6.
Segments H, T, U, W, and X are not involved in determining HN protein specificity. (A) Detection of chimeric F proteins in plasmid-transfected BHK cells. Western blotting (top) and immunoprecipitation (bottom) were carried out as described in the legend for Fig. 3C. The number above each lane corresponds to the number of the chimera indicated in Fig. 4. BM, B + M(1 + 2 + 3 + 4); V, vector. (B) Fusion activity of chimeric F proteins. The average fusion index was determined as described in the legend for Fig. 3E. The numbers on the abscissa axis correspond to the chimera numbers presented in panel A.
As we described above, chimeras no. 6, no. 8, and no. 9 induced fusion only when coexpressed with the SV41 HN protein. Importantly, chimeras no. 6 and no. 9 induced fusion specifically with the SV41 HN protein even more prominently than the parent B + M(1 + 2 + 3 + 4) did. We then reassessed the role of segment H in domain M1 in the context of chimera no. 6, revealing that replacement of segment H with the PIV5 F counterpart reduced the fusion activity but did not affect the specificity for the SV41 HN protein, as represented by chimera no. 14 (Fig. 6B). This result thus suggested that segment H is not is not involved in determining HN protein specificity.
Taken together, the findings obtained so far have suggested that six segments (H, S, T, U, W, and X) are not involved in determining HN protein specificity. We then replaced these six segments of B + M(1 + 2 + 3 + 4) with those of the PIV5 F protein. As expected, the resultant chimera, no. 26, exhibited apparent specificity for the SV41 HN protein but it also showed moderate HN-independent fusion activity (Fig. 7B). Similarly, replacement of five segments (H, S, U, W, and X) of the six resulted in SV41 HN-specific chimera no. 16, which also showed HN-independent fusion activity (Fig. 6B). Intriguingly, when four segments (H, S, U, and W) of the six were replaced, the resultant chimera, no. 15, showed minimal HN-independent fusion and induced fusion specifically with the SV41 HN protein (Fig. 6B). Finally, replacement of segment T of chimera no. 15 resulted in chimera no. 17, which induced fusion specifically with the SV41 HN protein at a reduced level but showed no HN-independent fusion activity (Fig. 7B). These results were consistent with the six segments not being involved in determining HN protein specificity.
Fig 7.
Domain B is not involved in determining HN protein specificity. (A) Detection of chimeric F proteins in plasmid-transfected BHK cells. Western blotting (top) and immunoprecipitation (bottom) were carried out as described in the legend for Fig. 3C. The number above each lane corresponds to the number of the chimera indicated in Fig. 4. BM, B + M(1 + 2 + 3 + 4); V, vector. (B) Fusion activity of chimeric F proteins. The average fusion index was determined as described in the legend for Fig. 3E. The numbers on the abscissa axis correspond to the chimera numbers presented in panel A.
Domain B is not involved in determining HN protein specificity.
We then investigated the three segments J, K, and L in domain B in the context of SV41 HN-specific chimera no. 15, which showed negligible HN-independent fusion activity, as described above. As shown in Fig. 7B, segment J and/or segment K of chimera no. 15 could be replaced with the PIV5 F counterparts without affecting HN protein specificity, as represented by chimeras no. 18, no. 19, and no. 20, which induced fusion with the SV41 HN protein at levels significantly higher than their backgrounds but not with the PIV5 HN protein. In contrast, replacement of segment L of chimera no. 15 resulted in chimera no. 21, which induced fusion specifically with the SV41 HN protein but at a reduced level (Fig. 7B). Similar to chimera no. 15, replacement of segments J and K of chimera no. 17 resulted in chimera no. 23, which showed high HN-independent fusion activity but retained specificity for the SV41 HN protein, while replacement of segment L of chimera no. 17 resulted in chimera no. 22, which induced fusion specifically with the SV41 HN protein but at a reduced level (Fig. 7B). These results have suggested, unexpectedly, that the three segments in domain B are not involved in determining HN protein specificity.
Interestingly, we found after several attempts that replacement of segment X of chimera no. 23 reduced the HN-independent fusion activity without significantly affecting its fusion activity with the SV41 HN protein, as represented by chimera no. 24 (Fig. 7B). More interestingly, replacement of segment L of chimera no. 24 resulted in chimera no. 25, which induced fusion specifically with the SV41 HN protein to a similar extent but showed remarkably reduced HN-independent fusion activity (Fig. 7B and Fig. 8B). Notably, when domain B of chimera no. 25 was replaced with the SV41 F counterpart, the resultant chimera, no. 26, showed decreased fusion activity with the SV41 HN protein and increased HN-independent fusion activity (Fig. 7B).
Fig 8.
Segment V and residue 238 in segment G are not involved in determining HN protein specificity. (A) Detection of chimeric F proteins in plasmid-transfected BHK cells. Western blotting (top) and immunoprecipitation (bottom) were carried out as described in the legend for Fig. 3C. The number above each lane corresponds to the number of the chimera indicated in Fig. 4. V, vector. (B) Fusion activity of chimeric F proteins. The average fusion index was determined as described in the legend for Fig. 3E. The numbers on the abscissa axis correspond to the chimera numbers presented in panel A.
Segment V and residue 238 in segment G are not involved in determining HN protein specificity.
As described above, chimera no. 25 exhibited prominent specificity for the SV41 HN protein. This chimera contained 19 trimer surface-exposed amino acids derived from the SV41 F protein, as calculated from Fig. 1B; the uppermost amino acid among the 19 was residue 234 in segment F (Fig. 2). We then intended to see whether replacement of SV41 F-derived segment F, in which only L234 is exposed on the trimer surface (Fig. 1B), with the PIV5 F counterpart, T234, would affect HN protein specificity. As shown in Fig. 7B, the resultant chimera, no. 27, induced moderate fusion either with the SV41 HN protein or with the PIV5 HN protein, indicating that the presence of SV41 F-derived segment F (most likely, L234) is critical for chimera no. 25 to exhibit clear specificity for the SV41 HN protein, whereas it is not critical in the context of B + M(1 + 2 + 3 + 4), as represented by chimera no. 1 (Fig. 5B). On the other hand, the lowermost amino acid among the 19 was residue S344 in segment V of chimera no. 25 (Fig. 2). Interestingly, replacement of SV41 F-derived S344 of chimera no. 25 with the PIV5 F counterpart, D344, resulted in chimera no. 28, which induced fusion with the SV41 HN protein more prominently than chimera no. 25 did (Fig. 8B). Furthermore, replacement of SV41 F-derived Y346 or Q347 in segment V of chimera no. 28 resulted in chimeras no. 29 and no. 30, respectively, which induced fusion specifically with the SV41 HN protein similarly to chimeras no. 28 and no. 25, respectively (Fig. 8B). On the other hand, replacement of both Y346 and Q347 of chimera no. 25 resulted in chimera no. 31, which induced fusion specifically with the SV41 HN protein but at a reduced level (Fig. 8B). Finally, although replacement of all the amino acids in segment V resulted in chimera no. 32, which showed increased HN-independent fusion activity, the SV41 HN protein could promote chimera no. 32-mediated fusion to a level significantly higher than this background, whereas the PIV5 HN protein could not. Taken together, these results suggest that segment V is not involved in determining HN protein specificity.
We then reassessed the role of SV41 F-derived L234 in segment F in the context of chimera no. 28, since this chimera exhibited clearer specificity for the SV41 HN protein than chimera no. 25 did, as described above. As shown in Fig. 8B, when SV41 F-derived L234 of chimera no. 28 was replaced with the PIV5 F counterpart, T234, the resultant chimera, no. 33, showed reduced specificity for the SV41 HN protein, indicating that the presence of SV41 F-derived L234 is important for chimera no. 28 in order to exhibit clear specificity for the SV41 HN protein, the same as it is for chimera no. 25. It should be pointed out, in this context, that SV41 F-derived L234 is in contact with SV41 F-derived S238 in segment G, as shown in Fig. 2 (no. 25, top). We therefore replaced SV41 F-derived S238 of chimera no. 28 with the PIV5 F counterpart, K238. Notably, the resultant chimera, no. 34, did not show HN-independent fusion activity, whereas it retained clear specificity for the SV41 HN protein (Fig. 8B).
Finally, we reassessed the roles of SV41 F-derived Y346 and Q347 in segment V in the context of chimera no. 34. Unfortunately, replacement of SV41 F-derived Y346 with the PIV5 F counterpart, M346, resulted in a fusion-deficient chimera, no. 35 (Fig. 8B), whose cleaved form was weakly expressed on the cell surface (Fig. 8A). Most notably, however, SV41 F-derived Q347 of chimera no. 34 could be replaced with the PIV5 F counterpart, A347, without affecting its clear specificity for the SV41 HN protein, as represented by chimera no. 36 (Fig. 8B). As shown in Fig. 1B and Fig. 2, chimera no. 36 harbored 21 SV41 F-derived amino acids, of which 16 were exposed on the trimer surface.
Taken together, the SV41 F-derived 21 amino acids of B + M(1 + 2 + 3 + 4), a chimeric PIV5 F protein that harbored 41 SV41 F-derived amino acids, could be replaced with the PIV5 F counterpart without affecting its clear specificity for the SV41 HN protein. In other words, replacement of the corresponding 21 amino acids of the PIV5 F protein with those of the SV41 F protein resulted in an F protein that exhibited clear specificity for the SV41 HN protein.
DISCUSSION
In the present study, we have shown that replacement of 21 amino acids of the PIV5 F protein with the SV41 F counterparts is enough to convert the PIV5 F protein to a protein specific for the SV41 HN protein, as represented by chimera no. 36. Importantly, 16 out of the 21 amino acids are exposed on the trimer surface (Fig. 1B and 2) and thus could be regarded as the candidates that directly mediate the physical interaction of the F protein with the HN protein. Interestingly, these 16 amino acids form into a noncontiguous and nearly perpendicular line on the lateral side of the head region of chimera no. 36 (Fig. 2), which is approximately 6 nm in length, as deduced from the total length (12 nm) of the PIV5 F trimer (30). The uppermost residue of this line is residue 234 in segment F, while the lowermost residue is residue 346 in segment V or residue 314 in segment R. Uppermost residue 234 has been proved to be critical for determining HN protein specificity, as represented by chimera no. 33. In contrast, lowermost residue 346 is not involved in determining HN protein specificity, as represented by chimeras no. 29, no. 31, and no. 32; evaluation of this residue as a determinant of the HN protein specificity would be our future task. Therefore, the candidate amino acids of the F protein that would mediate physical HN-F interaction are 15 in number. It should be pointed out, on the other hand, that residue 282 in domain M2 is located between segments N and P of chimera no. 36 (Fig. 2). This residue is shared by the F proteins of SV41 and PIV5 (Fig. 1B) and thus does not seem to be responsible for determining HN protein specificity. However, since it protrudes from the midst of the above-described perpendicular line in the head region of chimera no. 36 (Fig. 2), we cannot rule out the possibility that this conserved residue (proline) somehow takes part in the physical HN-F interaction. Similarly, three conserved residues, which belong to domain M1 (residues 241 and 242) or domain M2 (residue 277), are located among segments F (residue 234), G (residues 239 and 240), and N of chimera no. 36 (Fig. 2); they may also take part in the physical HN-F interaction.
On the basis of the interface propensity and the conservation of physical chemical properties among F proteins, it has been proved that six residues at the base of the MV F head region receive the signal from the MV H protein (31), suggesting that these residues are involved in the H-F interaction. It is worth noting, in this context, that two (Q322 and E325) of the identified six residues correspond to T312 and L315 in PIV5 F segment R, respectively (Fig. 1B and Fig. 2), which are the constituents of the 15 candidates that would mediate the HN-F interaction, as described above. However, another two (Y349 and R360) of the six residues correspond to Q339 in segment U and Q350 in segment W, respectively (Fig. 1B), which proved not to be important for the HN-F interaction. The remaining two (Q383 and L394) correspond to PIV5 F residues D373 and L384, respectively, which are not included in the middle region and thus may not be involved in the HN-F interaction. Notably, in this context, three CDV F residues that are considered to be involved in the H-F interaction are located in F2 or in the fusion peptide (24). Taken together, these findings suggest that the sites of the rubulavirus F protein interacting with the HN protein are only partially identical to the sites of the morbillivirus F protein interacting with the H protein, which would reflect a difference in the mechanisms of F activation between the two genera (4).
Our previous chimeric analysis revealed that a chimeric PIV5 F protein which harbored four SV41 F-derived domains (M1, M2, M3, and M4), or, in other words, 38 SV41 F-derived amino acids (Fig. 1B), induced fusion either with the PIV5 HN protein or with the SV41 HN protein (23). We found in the present study, however, that replacement of only 21 amino acids in these four domains is enough to convert the PIV5 F protein to an SV41 HN-specific protein. Thus, whether a given chimeric PIV5 F protein interacts with the SV41 HN protein or with the PIV5 HN protein does not simply reflect the number of SV41 F-derived amino acids, suggesting that the tertiary and/or quaternary structure of the putative HN-interacting site of the chimeric PIV5 F protein seems to be extremely important for determining its HN protein specificity. In other words, the role of a given SV41 F-derived domain or amino acid in the interaction with the SV41 HN protein may vary in a manner that is dependent on the structure of the respective chimeric PIV5 F protein, since it is very likely that the conformation and/or relative position of the SV41 F-derived domains or amino acids in the context of a chimeric F protein may differ from that in another chimeric F protein or from that in the SV41 F protein. Such a difference would be enough to discriminate between an SV41 HN-specific structure and a cross-reactive structure.
Although it is appreciated that the HN-F interaction is virus type specific, the HPIV2 HN protein is able to substitute for the HN proteins of SV41 and HPIV4A (17), while the HN protein of mumps virus can substitute for the HN proteins of HPIV2 and PIV5 (22, 27). Such cross-reactivity of some HN proteins cannot simply be explained by their proximity to the substitutable HN proteins in terms of primary structure, since the converse heterotypic combinations of the HN and F proteins result in no fusion (17, 22). We thus speculate that similarity in the tertiary and/or quaternary structure between the stalk regions of the HN proteins is also crucial for their compatibility; a subtle difference might result in the one-way cross-reactivity, as we have suggested elsewhere (32). It is important to point out, in this context, that a given HN (or F) protein has the potential to substitute for another HN (or F) protein when the overall amino acid sequence identity between the proteins is 39% (or 36%) or greater (22, 32). It is thus important to note that the SV41 F protein cannot substitute for the PIV5 F protein and vice versa, despite the fact that their amino acid sequence identity is 49.9% (23, 33), which, fortunately, enabled us to perform the chimeric analyses of their F proteins.
A number of chimeric F proteins created in the present study displayed HN-independent fusion activity; above all, chimera no. 11 exhibited hyperfusogenic activity in the absence of the HN protein (Fig. 6B), indicating that SV41 F-derived segment V somehow contributes to this phenotype. Consistently, replacement of segment V of chimera no. 25 with the SV41 F counterpart resulted in chimera no. 32, which showed increased HN-independent fusion activity (Fig. 8B). Similarly, SV41 F-derived segments F, J, K, and X also seemed to contribute to HN-independent fusion activity in the context of some chimeras, for example, no. 1, no. 18, no. 19, and no. 16, respectively. With regard to this issue, it is known that introduction of several mutations into the PIV5 F protein bestows HN-independent fusion activity on the F protein (27, 34, 35). These mutations are considered to destabilize the F protein so that it can undergo the conformational changes that lead to fusion in the absence of the HN protein (36). Therefore, if the physical interaction between the HN stalk region and the putative HN-interacting region of the F protein results in destabilization of the F protein, then mutation or chimeric recombination of the HN-interacting region or its franking region might result in destabilization of the F protein in some cases, as we have recently postulated (23).
Intriguingly, the fusion activities of most chimeric F proteins in BHK cells at 12 h posttransfection are much higher than those in HeLa cells at 24 h posttransfection, as represented by the chimeras shown in Fig. 3D and E. Notably, B + M(1 + 2) and B + M(2 + 4) induced hyperfusogenic activity in BHK cells independently of the coexpressed HN protein. This finding indicates, importantly, that the difference in the fusion activity of a given chimera between the two cell lines cannot simply be explained by the possible difference in the surface expression level of the chimera and/or in the amount of cellular receptors for the HN protein. Rather, it seems likely that these cell lines differ in the lipid composition of the plasma membrane, in the integrity of the cytoskeleton, such as that of cortical actin, or in the amount of a putative receptor(s) for the F protein, any one of which would account for the different susceptibilities of the two cell lines to the intrinsic fusion activity of the F proteins. Importantly, in this context, it has been reported that cellular transmembrane molecules, such as interferon-inducible transmembrane protein 1 (IFITM1) or CD98 heavy chain, regulate the cell-cell fusion induced by viral glycoproteins (37, 38). Possible differences in the functions of these cellular fusion regulatory proteins between the two cell lines may account for the higher fusion activity of the chimeras in BHK cells. Intriguingly, on the other hand, the difference in the surface expression levels between the chimeras in BHK cells seems much less than that in HeLa cells (Fig. 3B and C), suggesting that there may be differences in the transport machinery and/or in the quality control mechanism between the two cell lines.
The cleaved form (F1 + 2) of some chimeras, including B + M(1 + 2 + 3 + 4), is poorly expressed on the BHK cell surface compared to the expression of the PIV5 F protein, but this phenotype does not seem to correlate either with the chimeric structure, with the HN protein specificity, or with HN independence. Interestingly, on the one hand, the data from Western blotting of the total cell lysate indicate that the amounts of the cleaved form (F1) of these chimeras are also considerably smaller than those of the chimeras whose F1 + 2 forms are efficiently expressed on the cell surface. In contrast, the amounts of the uncleaved form (F0) of most chimeras are similar regardless of their surface expression efficiency. These findings thus indicate that the above-mentioned paucity in the amount of the cell surface-expressed F1 + 2 cannot simply be explained by low cleavage efficiency. Provided that there is no difference in translation efficiency, it is possible that those chimeras whose F1 + 2 forms are poorly expressed on the cell surface are degraded after cleavage. In all likelihood, their F1 + 2 forms may be structurally labile, which would result in their transport to the late endosome/lysosome for degradation either directly from the trans-Golgi network or from the cell surface. Although it is possible that these chimeras are prone to undergo misfolding in the endoplasmic reticulum and thus are retrotranslocated to the cytoplasm for degradation in the proteasome, such degradation would result in small amounts not only of the cleaved form but also of the uncleaved form.
ACKNOWLEDGMENT
This work was supported by a Grant-in-Aid for Scientific Research (grant 23590538) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Lamb RA, Parks GD. 2007. Paramyxoviridae: the viruses and their replication, p 1449–1496 Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Heminway BR, Yu Y, Galinski MS. 1994. Paramyxovirus mediated cell fusion requires co-expression of both the fusion and hemagglutinin-neuraminidase glycoproteins. Virus Res. 31:1–16 [DOI] [PubMed] [Google Scholar]
- 3. Hu X, Ray R, Compans RW. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iorio RM, Melanson VR, Mahon PJ. 2009. Glycoprotein interactions in paramyxovirus fusion. Future Virol. 4:335–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lamb RA, Jardetzky TS. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morrison TG. 2003. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta 1614:73–84 [DOI] [PubMed] [Google Scholar]
- 7. McGinnes LW, Morrison TG. 2006. Inhibition of receptor binding stabilizes Newcastle disease virus HN and F protein-containing complexes. J. Virol. 80:2894–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moscona A, Peluso RW. 1991. Fusion properties of cells persistently infected with human parainfluenza virus type 3: participation of hemagglutinin-neuraminidase in membrane fusion. J. Virol. 65:2773–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moscona A, Peluso RW. 1992. Fusion properties of cells infected with human parainfluenza virus type 3: receptor requirements for viral spread and virus-mediated membrane fusion. J. Virol. 66:6280–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Russell CJ, Jardetzky TS, Lamb RA. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Porotto M, Murrell M, Greengard O, Doctor L, Moscona A. 2005. Influence of the human parainfluenza virus 3 attachment protein's neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 79:2383–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsurudome M, Nishio M, Ito M, Tanahashi S, Kawano M, Komada H, Ito Y. 2008. Effects of hemagglutinin-neuraminidase protein mutations on cell-cell fusion mediated by human parainfluenza type 2 virus. J. Virol. 82:8283–8295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bose S, Welch BD, Kors CA, Yuan P, Jardetzky T, Lamb RA. 2011. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J. Virol. 85:12855–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2011. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. U. S. A. 108:14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deng R, Mirza AM, Mahon PJ, Iorio RM. 1997. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch. Virol. 13(Suppl):115–130 [DOI] [PubMed] [Google Scholar]
- 16. Tanabayashi K, Compans RW. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsurudome M, Kawano M, Yuasa T, Tabata N, Nishio M, Komada H, Ito Y. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213:190–203 [DOI] [PubMed] [Google Scholar]
- 18. Mirza AM, Sheehan JP, Hardy LW, Glickman RL, Iorio RM. 1993. Structure and function of a membrane anchor-less form of the hemagglutinin neuraminidase glycoprotein of Newcastle disease virus. J. Biol. Chem. 268:21425–21432 [PubMed] [Google Scholar]
- 19. Thompson SD, Portner A. 1987. Localization of functional sites on the hemagglutinin-neuraminidase glycoprotein of Sendai virus by sequence analysis of antigenic and temperature-sensitive mutants. Virology 160:1–8 [DOI] [PubMed] [Google Scholar]
- 20. Melanson VR, Iorio RM. 2004. Amino acid substitutions in the F-specific domain in the stalk of the Newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J. Virol. 78:13053–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Melanson VR, Iorio RM. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 80:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsurudome M, Ito M, Nishio M, Kawano M, Okamoto K, Kusagawa S, Komada H, Ito Y. 1998. Identification of regions on the fusion protein of human parainfluenza virus type 2 which are required for haemagglutinin-neuraminidase proteins to promote cell fusion. J. Gen. Virol. 79:279–289 [DOI] [PubMed] [Google Scholar]
- 23. Tsurudome M, Ito M, Nishio M, Nakahashi M, Kawano M, Komada H, Nosaka T, Ito Y. 2011. Identification of domains on the fusion (F) protein trimer that influence the hemagglutinin-neuraminidase specificity of the F protein in mediating cell-cell fusion. J. Virol. 85:3253–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee JK, Prussia A, Paal T, White LK, Snyder JP, Plemper RK. 2008. Functional interaction between paramyxovirus fusion and attachment proteins. J. Biol. Chem. 283:16561–16572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ito M, Nishio M, Kawano M, Komada H, Ito Y, Tsurudome M. 2009. Effects of multiple amino acids of the parainfluenza virus 5 fusion protein on its haemagglutinin-neuraminidase-independent fusion activity. J. Gen. Virol. 90:405–413 [DOI] [PubMed] [Google Scholar]
- 26. Tsurudome M, Ito M, Nishio M, Kawano M, Komada H, Ito Y. 2001. Hemagglutinin-neuraminidase-independent fusion activity of simian virus 5 fusion (F) protein: difference in conformation between fusogenic and nonfusogenic F proteins on the cell surface. J. Virol. 75:8999–9009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ito M, Nishio M, Kawano M, Kusagawa S, Komada H, Ito Y, Tsurudome M. 1997. Role of a single amino acid at the amino terminus of the simian virus 5 F2 subunit in syncytium formation. J. Virol. 71:9855–9858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ito M, Nishio M, Komada H, Ito Y, Tsurudome M. 2000. An amino acid in the heptad repeat 1 domain is important for the haemagglutinin-neuraminidase-independent fusing activity of simian virus 5 fusion protein. J. Gen. Virol. 81:719–727 [DOI] [PubMed] [Google Scholar]
- 29. Tsurudome M, Ito M, Nishio M, Kawano M, Komada H, Ito Y. 2006. A mutant fusion (F) protein of simian virus 5 induces hemagglutinin-neuraminidase-independent syncytium formation despite the internalization of the F protein. Virology 347:11–27 [DOI] [PubMed] [Google Scholar]
- 30. Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. 2006. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 103:17903–17908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Apte-Sengupta S, Negi S, Leonard VH, Oezguen N, Navaratnarajah CK, Braun W, Cattaneo R. 2012. Base of the measles virus fusion trimer head receives the signal that triggers membrane fusion. J. Biol. Chem. 287:33026–33035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsurudome M. 2011. Virus entry: parainfluenza viruses, p 35–61 Ming L. (ed), Negative strand RNA virus. World Scientific, Hackensack, NJ [Google Scholar]
- 33. Tsurudome M, Bando H, Kawano M, Matsumura H, Komada H, Nishio M, Ito Y. 1991. Transcripts of simian virus 41 (SV41) matrix gene are exclusively dicistronic with the fusion gene which is also transcribed as a monocistron. Virology 184:93–100 [DOI] [PubMed] [Google Scholar]
- 34. Russell CJ, Jardetzky TS, Lamb RA. 2004. Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J. Virol. 78:13727–13742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seth S, Vincent A, Compans RW. 2003. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin-neuraminidase protein requirement for membrane fusion. J. Virol. 77:167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paterson RG, Russell CJ, Lamb RA. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17–30 [DOI] [PubMed] [Google Scholar]
- 37. Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC, Gratton E, Cohen FS, Liu SL. 2013. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9:e1003124. 10.1371/journal.ppat.1003124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsurudome M, Ito Y. 2000. Function of fusion regulatory proteins (FRPs) in immune cells and virus-infected cells. Crit. Rev. Immunol. 20:167–196 [PubMed] [Google Scholar]
- 39. Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 102:9288–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]