Abstract
We previously showed that microRNA 181 (miR-181) can inhibit PRRSV replication by directly targeting its genomic RNA. Here, we report that miR-181 can downregulate the PRRSV receptor CD163 in blood monocytes and porcine alveolar macrophages (PAMs) through targeting the 3′ untranslated region (UTR) of CD163 mRNA. Downregulation of CD163 leads to the inhibition of PRRSV entry into PAMs and subsequently suppresses PRRSV infection. Our findings indicate that delivery of miR-181 can be used as antiviral therapy against PRRSV infection.
TEXT
Porcine reproductive and respiratory syndrome (PRRS) is one of the most important diseases in swine industry worldwide. PRRS virus (PRRSV) is an ∼15.4-kb positive-sense RNA virus belonging to the family Arteriviridae (1). Most recently, there have been devastating outbreaks in China of atypical PRRS, which is caused by a highly pathogenic PRRSV (HP-PRRSV) genotype (2). Currently, there are no effective ways to protect pigs from PRRSV infection (3).
Studies have revealed that microRNAs (miRNAs) participate in host antiviral responses, either by targeting cellular factors essential for virus replications (4, 5) or by directly targeting virus genomes (6–8). We previously showed that in blood monocytes, which are refractory to PRRSV infection, the microRNA 181 (miR-181) expression level is 9 times higher than that in porcine alveolar macrophages (PAMs) (9). Given that blood monocytes do not express the PRRSV receptor CD163 or express it at very low levels, we propose that miR-181 might downregulate CD163 and subsequently inhibit PRRSV infection.
To investigate whether miR-181 can target CD163, we first performed computational prediction analysis with RegRNA (10) to identify the potential target sites of miR-181 in CD163 mRNA. Three miR-181 seed binding sites were found, one in the 3′ untranslated region (UTR) (3544 to 3565) and two in the coding region (623 to 644 and 842 to 863).
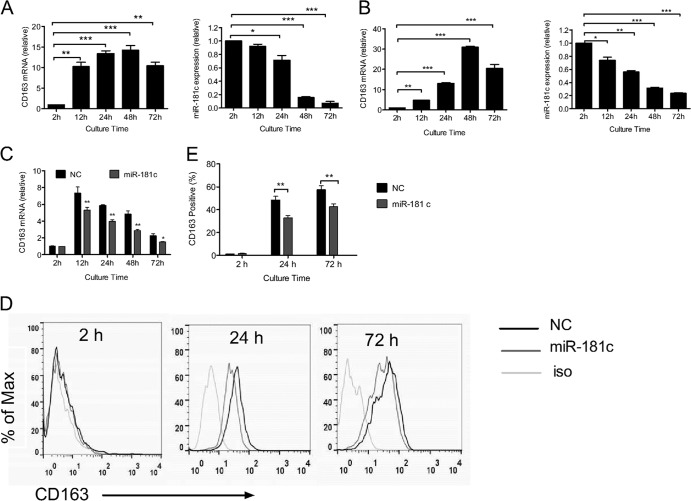
PRRSV infection in cultured monocytes is coordinated well with the increased CD163 (11). Thus, we examined the relationship of CD163 and miR-181c expression in cultured monocytes in the presence of L929 cell culture supernatant (12–14). Our results showed that CD163 mRNA was significantly upregulated (about 14 times higher at 48 h than at 2 h) (Fig. 1A, left). Meanwhile, the miR-181c expression level decreased gradually and was more than 80% lower in abundance at 48 h than at 2 h (Fig. 1A, right). We also analyzed CD163 and miR-181c expression levels in cultured monocytes without L929 cell culture supernatants. As shown in Fig. 1B, increased CD163 expression was also coordinated with the reduced miR-181c expression. These results suggest that there might be an inverse correlation between the expressions of miR-181c and CD163 during monocyte-macrophage differentiation. To further confirm this correlation, we transfected miR-181c mimics or negative-control (NC) mimics into 2-h-cultured monocytes and then detected CD163 mRNA expression using quantitative real-time PCR at different times. As shown in Fig. 1C, aberrant higher expression of miR-181c mimics significantly impaired the upregulation of CD163 (72% at 12 h, 67% at 24 h, 59% at 48, and 65% at 72 h relative to NC). We also analyzed CD163 protein expression on the cell surface using flow cytometry (Fig. 1D and E), and there were about 32% and 26% CD163+ cell reductions in miRNA181c-transfected cells at 24 h and 72 h compared to NC, respectively. These results suggested that miR-181c downregulated CD163 expression. We got similar results with other miR-181 members (data not shown), which are also lower in PAMs (9), and then focused on miR-181c in the following study.
Fig 1.
miR-181c downregulates CD163 expression in cultured blood monocytes (BMo). (A) During BMo differentiation in vitro upon the presence of L929 cell culture supernatant, CD163 mRNA was significantly upregulated (left) and the miR-181c expression level decreased gradually with increasing culture time (right). (B) Without the presence of L929 cell culture supernatant, CD163 mRNA (left) was also significantly upregulated and the miR-181c expression level (right) decreased gradually with increasing culture time. (C) Transfection of miR-181c mimics into 2-h-cultured BMo suppressed CD163 upregulation. miR-181c or negative-control (NC sequence, 5′-UUCUCCGAACGUGUCACGUTT-3′; not involved in regulation of CD163) mimics were transfected into BMo at a concentration of 60 nM using HiPerfect transfection reagents (Qiagen). CD163 mRNA and miR-181c expressions were normalized to the endogenous control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by quantitative real-time PCR at different culture times (12 h, 24 h, 48 h, and 72 h). (D) Blood monocytes were cultured and transfected with miR-181c or NC mimics as described in panel C, and CD163 expression at 2 h, 24 h, and 72 h was assessed by flow cytometry on cultured monocytes. Cells were incubated with mouse anti-pig CD163 monoclonal antibody (MAb) (AbDSerotec) or isotype control and then stained with Alexa Fluor 488-conjugated rabbit anti-mouse IgG (H+L) (Molecular Probes). (E) The percentage of CD163-positive cultured monocytes at 2 h, 24 h, and 72 h posttransfection was analyzed by flow cytometry. Experiments were performed with at least three independent replicates. Statistical analysis was performed using GraphPad Prism software, and differences in data were evaluated by Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
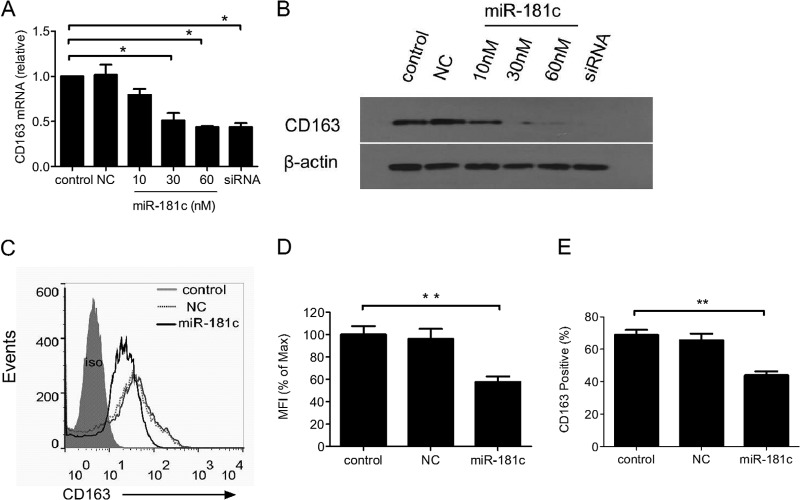
PAM is the natural host cell for PRRSV infection and has high expression of CD163 as well as extremely low expression of miR-181c (9, 11, 12, 15). To investigate whether overexpression of miR-181c reduces CD163 expression in PAMs, we transfected miR-181c mimics, NC mimics, or small interfering CD163 (siCD163) as a positive control into PAMs and analyzed CD163 expression using quantitative real-time PCR. As expected, CD163 mRNA was significantly downregulated by siCD163. Additional expression of miR-181c resulted in less CD163 mRNA dose dependently, and CD163 mRNA was reduced to about 80% and 40% of that in mock control cells when miR-181c mimics were at 10 nM and 60 nM, respectively (Fig. 2A), while NC mimics had no effect on CD163 expression. Next, we determined whether downregulation of CD163 mRNA leads to less CD163 protein. Immunoblot analysis showed that CD163 was reduced in a dose-dependent manner (Fig. 2B). Even though miR-181c impaired CD163 mRNA expression only modestly, the decrease of CD163 protein expression was more convincing. This suggested that miR-181c might reduce CD163 expression through both mRNA degradation and translation inhibition (16). The reduction of CD163 protein was also confirmed by flow cytometry (Fig. 2C). The mean fluorescence intensity (MFI) of stained CD163 in miR-181c-transfected PAMs was reduced to about 57% of that in control cells (Fig. 2D), and the percentage of CD163-positive cells was down to ∼43% from 68% in the mock or NC control group (Fig. 2E). Collectively, these results demonstrated that increasing expression of miR-181c in PAMs downregulated CD163 expression.
Fig 2.
Overexpression of miR-181c reduces CD163 mRNA and protein levels in PAMs. PAMs were transfected with miR-181c mimics at different concentrations, NC as a negative control, or siCD163 (a mix of equal amounts of the small interfering RNAs [siRNAs] 5′-GCCAUUAAUGCCACUGGUUTT-3′, 5′-GCUCAAGAUACACACAAAUTT-3′, and 5′-GCUGGAGUGAUUUGCUUAATT-3′) as a positive control using HiPerfect transfection reagents (Qiagen), and CD163 mRNA expression was determined by quantitative real-time PCR 72 h after transfection (A) and CD163 protein expression was analyzed using a Western blot in PAMs (B). (C) CD163 expression on PAMs at 72 h after transfection with NC or miR-181c mimics was assessed by flow cytometry (cells were stained as described in Fig. 1). (D) CD163 expression presented as MFI. (E) The percentage of CD163-positive PAMs at 72 h posttransfection was analyzed by flow cytometry. Data represent means and standard deviations (SD) of three independent experiments. Control, mock control cells; NC, negative microRNA control. Statistical analysis was performed using GraphPad Prism software, and differences in data were evaluated by Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
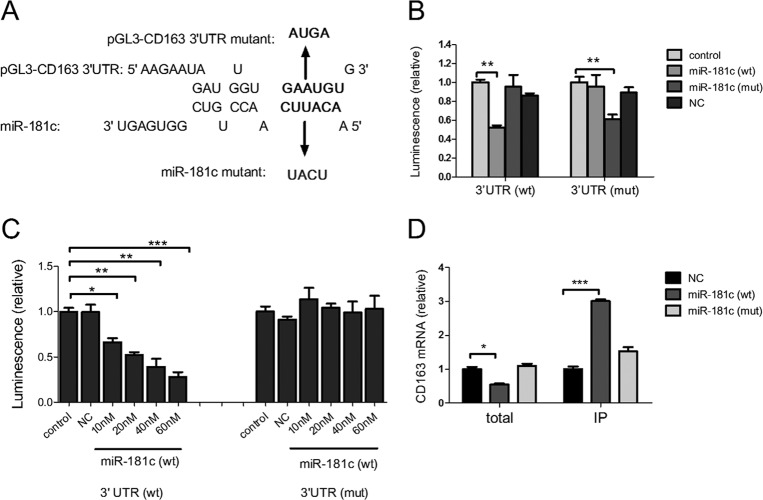
To investigate whether the diminished CD163 is due to direct targeting by miR-181c, we constructed firefly luciferase reporter pGL3-CD163 containing the wild-type sequence of the CD163 3′ UTR and also mutated four nucleotides in the predicted seed binding site and yielded pGL3-CD163 (mutant [mut]). In addition, we synthesized miR-181c (mut) with mutations in the seed sequence complementary to the mutation in the CD163 3′ UTR (Fig. 3A). Then a luciferase reporter assay was performed. We observed that miR-181c robustly downregulated the activity of firefly luciferase fused to the CD163 wild-type 3′ UTR whereas miR-181c (mut) did not (Fig. 3B). In the reciprocal experiment, we transfected HeLa cells with the mutant vector pGL3-CD163 (mut). As expected, transfection of miR-181c (mut) resulted in significantly less luciferase activity while miR-181c had no effect on it (Fig. 3B). Next, we examined the dose-dependent effect of miR-181c on luciferase reporters containing the wild-type or mutational CD163 3′ UTR. As shown in Fig. 3C, the luciferase activity of pGL3-CD163 was inversely proportional to the expression of miR-181c. The firefly luciferase activity of pGL3-CD163 declined to about 66%, 52%, 39%, and 28% of that in NC when miR-181c mimics were transfected at 10 nM, 20 nM, 40 nM, and 60 nM, respectively, while the luciferase activity of pGL3-CD163 (mut) was not affected by miR-181c (Fig. 3C). To obtain further evidence for the targeting of CD163 by miR-181c, we examined the physical interaction between CD163 mRNA and miR-181c using an RNA-induced silencing complex (RISC) immunoprecipitation assay (17, 18). In total cell lysates, the CD163 mRNA level in miR-181c-transfected cells was lower than that in NC- or miR-181c (mut)-transfected cells (Fig. 3D). In contrast, there was about 3-fold enrichment for CD163 mRNA in Argonaute (Ago)-bound mRNAs when miR-181c was overexpressed. As expected, no enrichment for CD163 mRNA was observed when miR-181c (mut) was transfected. Together, these results demonstrated that miR-181c can downregulate CD163 expression through directly targeting its mRNA.
Fig 3.
miR-181 targets the 3′ UTR of CD163 and physically binds to CD163 mRNA in the RISC. (A) Predicted binding site for miR-181c in the 3′ UTR of CD163 mRNA. Bold indicates the predicted seed region interaction site. Underlining indicates nucleotides replaced with the sequence indicated by the arrow in mutant reporter constructs or miR-181c mutants. (B) Dual-luciferase assay of HeLa cells transfected with miRNA-181c (wt), miRNA-181 (mut), or NC at 20 nM and luciferase reporter vector pGL3-CD163 (wt) or pGL3-CD163 (mut) as well as renilla vector; results are presented relative to the luminescence of renilla luciferase. (C) Dual-luciferase assay as described in panel B except for the use of 10 nM, 20 nM, 40 nM, or 60 nM miR-181c (wt) and 60 nM NC mimics. (D) Quantitative real-time PCR analysis of CD163 mRNA among RNAs extracted from RISC immunoprecipitates or total samples of the cells, normalized to GAPDH. Control, mock control cells; NC, negative microRNA control. Data are representative of three independent experiments. Statistical analysis was performed using GraphPad Prism software, and differences in data were evaluated by Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Given that CD163 functions as a cellular receptor for PRRSV, we next investigated whether downregulation of CD163 in PAMs led to the inhibition of PRRSV infection. To rule out the influence of miR-181c on the PRRSV genome, we constructed PRRSV-JXwn06 (mut), which had mutations in the target sequence of miR-181c in its genome (Fig. 4A), and then evaluated the effect of miR-181c on PRRSV infection in PAMs. We inoculated PAMs with PRRSV-JXwn06 (wt) or PRRSV-JXwn06 (mut) at a multiplicity of infection (MOI) of 5 at 72 h posttransfection with miR-181c mimics and then quantified intracellular PRRSV RNAs using quantitative real-time PCR at 3 h postinoculation at 37°C (it takes ∼3 h for PRRSV to enter PAMs). As shown in Fig. 4B, PRRSV RNAs in PAMs transfected with miR-181c were about 70% less than that in mock- or NC-transfected PAMs. However, no significant difference in RNA quantity existed between PRRSV-JXwn06 (wt)- and PRRSV-JXwn06 (mut)-infected cells. These results indicated that miR-181c remarkably blocked both PRRSV-JXwn06 (wt) and PRRSV-JXwn06 (mut) entry into PAMs. To further verify the inhibition of miR-181 on PRRSV infection and replication, we transfected PAMs with miR-181c or NC mimics and, 72 h later, inoculated the cells with PRRSV-JXwn06 (wild type [wt]) or PRRSV-JXwn06 (mut) at an MOI of 0.01. We then evaluated intracellular PRRSV RNAs at 3, 12, 24, and 36 h postinfection (Fig. 4C). Consistently with Fig. 4B, at 3 h postinfection, comparable PRRSV RNAs were detected in cells transfected with miR-181c for both PRRSV-JXwn06 (wt) and PRRSV-JXwn06 (mut). However, 12 h after infection, while PRRSV-JXwn06 (wt) and JXwn06 (mut) replicated comparably in control cells, PRRSV-JXwn06 (wt) replicated much more slowly than PRRSV-JXwn06 (mut) in miR-181c-transfected cells, and the amount of JXwn06 (wt) RNAs was about 8-fold lower than that of PRRSV-JXwn06 (mut). More importantly, 24 h later, PRRSV-JXwn06 (mut) replicated equally in miR-181c-transfected and control cells while PRRSV-JXwn06 (wt) was still inhibited robustly by miR-181c, and JXwn06 (wt) RNA was 100 times less than that in control cells. These data suggest that miR-181c can prevent PRRSV-JXwn06 (mut) from entering the cells but cannot impair its replication. The findings were also confirmed with a type 1 PRRSV strain which does not have miR-181 targeting sites in its genome (data not shown). Finally, we investigated the quantity of CD163 on PAMs in the presence or absence of miR-181c during PRRSV infection. PAMs were transfected with 60 nM miR-181c and then infected with PRRSV at an MOI of 0.01 at 12 h posttransfection. Cells were harvested at 12 h, 36 h, and 72 h posttransfection (relative to 0 h, 24 h, and 60 h after PRRSV infection, respectively) for CD163 analysis using a Western blot. As shown in Fig. 4D, CD163 expression on PAMs was downregulated by miR-181c during infection with PRRSV while PRRSV itself had no effect on it. Collectively, these results indicated that in addition to directly targeting the PRRSV genome, miR-181 inhibited PRRSV infection by downregulating its receptor, CD163.
Fig 4.
miR-181 inhibits PRRSV entry into PAMs. (A) Schematic construct of PRRSV-JXwn06 (mut) virus, which has mutations as the arrow indicates in the target sequence of miR-181c. (B) PAMs were transfected with NC or miR-181c (60 nM) 72 h before the inoculation of virus, and intracellular PRRSV RNAs in PAMs were evaluated by quantitative real-time PCR at 3 h after inoculation of PRRSV-JXwn06 (wt) or PRRSV-JXwn06 (mut) at 37°C. (C) Viral growth curves in PAMs transfected with NC or miR-181c mimics (60 nM) for 72 h and then infected with PRRSV JXwn06 or PRRSV-JXwn06 (mut) at an MOI of 0.01. Intracellular PRRSV RNA was analyzed by quantitative real-time PCR at 3 h, 12 h, 24 h, and 36 h after virus inoculation. (D) Expression levels of CD163 on PAM cells in the presence or absence of miR-181c during infection with PRRSV. PAMs were transfected with 60 nM miR-181c and then infected with PRRSV at an MOI of 0.01 at 12 h posttransfection. Cells were harvested at 12 h, 36 h, and 72 h posttransfection for CD163 analysis using a Western blot. Control, mock control cells; NC, negative microRNA control. Data represent means and SD of three independent experiments. Statistical analysis was performed using GraphPad Prism software, and differences in data were evaluated by Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
CD163, a macrophage-specific protein in the scavenger receptor cysteine-rich (SRCR) superfamily, functions as a cellular receptor for PRRSV, and the relationship between CD163 expression and PRRSV infection has been well established. Transfection of CD163 cDNA into several PRRSV nonpermissive cell lines renders them permissive for PRRSV infection and replication (19, 20). Pretreatment of PAMs or monocyte-derived macrophages (MDM) with tetradecanoyl phorbol acetate (TPA) or lipopolysaccharide (LPS) results in decreased CD163 expression and reduction of PRRSV infection (21), and anti-porcine CD163 antibody is shown to inhibit PRRSV infection of PAMs (22). Our results are consistent with these reports, showing that downregulation of CD163 by additional expression of miR-181 impairs PRRSV infection.
Viral tropism is a complicated issue influenced by many factors (23). Expression of CD163 is low in undifferentiated monocytes and gradually increases following cell activation and maturation (11, 14). Our data provide evidence that there is an inverse correlation between the expressions of miR-181 and CD163. PRRSV has a restricted tropism mainly for macrophages and dendritic cells but not for blood monocytes in vivo. Our findings here suggest that miR-181 affects PRRSV tropism by regulating CD163 expression. However, how miR-181 is regulated remains an important issue to be elucidated in the future. Nevertheless, our data here showed that miR-181 represses PRRSV infection through targeting its cell receptor, CD163.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (30770101) and the Faculty Starting Grant and State Key Laboratory of Agrobiotechnology (grants 2010SKLAB06-1 and 2012SKLAB01-6), China Agricultural University, Beijing, China.
Footnotes
Published ahead of print 5 June 2013
REFERENCES
- 1. Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. 10.1371/journal.pone.0000526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang YW, Meng XJ. 2010. Novel strategies and approaches to develop the next generation of vaccines against porcine reproductive and respiratory syndrome virus (PRRSV). Virus Res. 154:141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chiang K, Sung TL, Rice AP. 2012. Regulation of cyclin T1 and HIV-1 replication by microRNAs in resting CD4+ T lymphocytes. J. Virol. 86:3244–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sung TL, Rice AP. 2009. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog. 5:e1000263. 10.1371/journal.ppat.1000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560 [DOI] [PubMed] [Google Scholar]
- 7. Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, Das SC, Pattnaik AK, Beutler B, Han J. 2007. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27:123–134 [DOI] [PubMed] [Google Scholar]
- 8. Song L, Liu H, Gao S, Jiang W, Huang W. 2010. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 84:8849–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo XK, Zhang Q, Gao L, Li N, Chen XX, Feng WH. 2013. Increasing expression of microRNA 181 inhibits porcine reproductive and respiratory syndrome virus replication and has implications for controlling virus infection. J. Virol. 87:1159–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang HY, Chien CH, Jen KH, Huang HD. 2006. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res. 34:W429–W434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang L, Zhang H, Suo X, Zheng S, Feng WH. 2011. Increase of CD163 but not sialoadhesin on cultured peripheral blood monocytes is coordinated with enhanced susceptibility to porcine reproductive and respiratory syndrome virus infection. Vet. Immunol. Immunopathol. 141:209–220 [DOI] [PubMed] [Google Scholar]
- 12. Duan X, Nauwynck HJ, Pensaert MB. 1997. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch. Virol. 142:2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Englen MD, Valdez YE, Lehnert NM, Lehnert BE. 1995. Granulocyte/macrophage colony-stimulating factor is expressed and secreted in cultures of murine L929 cells. J. Immunol. Methods 184:281–283 [DOI] [PubMed] [Google Scholar]
- 14. Sanchez C, Domenech N, Vazquez J, Alonso F, Ezquerra A, Dominguez J. 1999. The porcine 2A10 antigen is homologous to human CD163 and related to macrophage differentiation. J. Immunol. 162:5230–5237 [PubMed] [Google Scholar]
- 15. Duan X, Nauwynck HJ, Pensaert MB. 1997. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV). Vet. Microbiol. 56:9–19 [DOI] [PubMed] [Google Scholar]
- 16. Fabian MR, Sonenberg N, Filipowicz W. 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79:351–379 [DOI] [PubMed] [Google Scholar]
- 17. Nonne N, Ameyar-Zazoua M, Souidi M, Harel-Bellan A. 2010. Tandem affinity purification of miRNA target mRNAs (TAP-Tar). Nucleic Acids Res. 38:e20. 10.1093/nar/gkp1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang WX, Wilfred BR, Hu Y, Stromberg AJ, Nelson PT. 2010. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA 16:394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, Welch SK. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 81:7371–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Welch SK, Calvert JG. 2010. A brief review of CD163 and its role in PRRSV infection. Virus Res. 154:98–103 [DOI] [PubMed] [Google Scholar]
- 21. Patton JB, Rowland RR, Yoo D, Chang KO. 2009. Modulation of CD163 receptor expression and replication of porcine reproductive and respiratory syndrome virus in porcine macrophages. Virus Res. 140:161–171 [DOI] [PubMed] [Google Scholar]
- 22. Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. 2008. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 89:2943–2953 [DOI] [PubMed] [Google Scholar]
- 23. Umbach JL, Cullen BR. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]