Abstract
Viral integrations are important in human biology, yet genome-wide integration profiles have not been determined for many viruses. Adeno-associated virus (AAV) infects most of the human population and is a prevalent gene therapy vector. AAV integrates into the human genome with preference for a single locus, termed AAVS1. However, the genome-wide integration of AAV has not been defined, and the principles underlying this recombination remain unclear. Using a novel high-throughput approach, integrant capture sequencing, nearly 12 million AAV junctions were recovered from a human cell line, providing five orders of magnitude more data than were previously available. Forty-five percent of integrations occurred near AAVS1, and several thousand novel integration hotspots were identified computationally. Most of these occurred in genes, with dozens of hotspots targeting known oncogenes. Viral replication protein binding sites (RBS) and transcriptional activity were major factors favoring integration. In a first for eukaryotic viruses, the data reveal a unique asymmetric integration profile with distinctive directional orientation of viral genomes. These studies provide a new understanding of AAV integration biology through the use of unbiased high-throughput data acquisition and bioinformatics.
INTRODUCTION
Genomic viral integrations are critically important in human biology, playing roles in normal physiology and evolution, viral diseases, cancer, and gene therapy (1). Adeno-associated virus serotype 2, a nonenveloped single-stranded DNA virus, has long been considered unique among known mammalian viruses due to its capacity to integrate site-preferentially (2). AAV has also been highly successful in nonintegrating gene therapy applications (3, 4). In addition to its success as a vector, the AAV integration machinery has been actively investigated for targeted integration strategies (5–7). AAV, therefore, presents an intriguing biological paradigm for both viral and vector integration into the human genome.
AAV integration has two exogenous requirements: trans-acting large viral replication proteins, Rep68 and Rep78 (8–10), and cis-acting DNA elements containing Rep binding sites, such as those present in the replication origin of the viral inverted terminal repeat (ITR) or the viral P5 promoter (11–13). Preferential integration occurs at a locus on human chromosome 19q13.4, in the first exon of protein phosphatase 1 regulatory subunit 12C (PPP1R12C), a site termed AAVS1 (14–17). Rep binding and endonuclease sites, sequence features characteristic of the AAV replication origin, are also present in the human genome, most notably as the defining sequence element of AAVS1 (8, 18, 19).
The large nonstructural Rep proteins are key mediators of virus biology, influencing viral gene expression, replication, and integration. Both isoforms contain an N-terminal DNA binding/endonuclease domain linked to an AAA+ SF3 helicase domain (20, 21). In replication origins, four tandem imperfect GAGC tetranucleotides provide the DNA binding domain for Rep 68/78 (22). The large Rep proteins undergo DNA facilitated oligomerization, where the linker between the DNA binding domain and helicase are critical for complex formation (23–25). Recent crystal structure and cryo-electron microscopy (cryo-EM) studies have revealed that AAV Rep68/78 can form double octameric or hexameric rings, with the rings facing opposite directions (21, 24, 26). DNA binding, endonuclease, helicase activity, and Rep oligomerization are required for both viral replication and integration (9, 27).
Our current understanding of the genomic sites targeted by AAV integration is based on a spectrum of low-throughput studies that have generated a small number of junction sequences, approximately 200 from the entire literature, using a variety of biased strategies. Studies originally demonstrated targeted integration through Southern blot analysis, fluorescence in situ hybridization (FISH), and AAVS1-specific PCR (2, 9, 10). Two studies have used low-throughput genomic approaches, involving enzyme digestion, ligation-mediated PCR, and cloning, to investigate AAV integration (28, 29). One study was unable to detect any integrants in AAVS1 (28). The other study found that AAVS1 integrations in exon 1 of PPP1R12C represented less than one percent of events, while integration in the general vicinity (within 100 kb) of AAVS1 accounted for less than 10 percent (29). Efforts to apply computational techniques to AAV integration have been limited by the small and biased data pools, which preclude thorough bioinformatics (29). Therefore, in spite of a large body of research on the topic, the true nature of AAV integration and its determinants remains to be established.
In this study, we present integrant capture sequencing (IC-Seq), a novel genome-wide high-throughput technique to elucidate viral integrations. We acquired 12 million AAV integration events and identified over 150,000 unique integration sites. AAVS1 was the primary integration target site accounting for 45% of events, which are distributed in a distinctive single-sided peak-and-tail configuration. Our data reveal an unprecedented two-stage directional integration of AAV genomes, which places new demands on the configuration of a Rep-dependent integration model. Nearly 2,500 hotspots of integration were computationally determined and found to be predominantly associated with genes. Hotspot distribution was primarily correlated with the presence of Rep DNA binding motifs and high levels of gene expression. These studies provide a new understanding of viral integration through the use of unbiased high-throughput data acquisition and bioinformatics.
MATERIALS AND METHODS
Cell culture and wtAAV infection.
HeLa cells (ATCC) were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle medium supplemented with 10% Cosmic calf serum (HyClone). Twenty-four hours prior to infection, cells were seeded in 10 wells of 24-well plates at 1 × 105 cells/well; therefore, upon infection, approximately 2 × 105 HeLa cells were present per well (2 × 106 cells per experiment). HeLa cells were infected with purified wtAAV generated by plasmid cotransfection (Applied Viromics) at 1 × 104 viral genomes/cell. After a 48-hour incubation, cells were harvested and plated in a 75-cm2 flask. Upon reaching confluence, these flasks were harvested and plated into two 150-cm2 flasks. Cells were grown for the remainder of the 3 weeks postinfection, with passaging as needed into two fresh 150-cm2 flasks.
Integrant capture sequencing. (i) DNA oligonucleotide sequences.
Sequences of the pLinker primer and asymmetric linker oligonucleotides were described previously (30, 31). The AAV primer sequences were as previously described (29); the external primer was modified with 5′-Bio-TEG.
(ii) Genomic DNA library generation.
Five aliquots of 2.5 million HeLa cells, containing ∼250 μg of genomic DNA in total, were harvested by trypsinization and washed with PBS. Aliquots were lysed in proteinase K buffer (100 mM Tris [pH 8], 0.2% SDS, 200 mM NaCl, 5 mM EDTA) with 200 μg/ml proteinase K. Genomic DNA was purified by phenol-chloroform extraction and ethanol precipitation. Sonication was conducted using a Bioruptor (Diagenode) to generate DNA smears of 500 to 1,200 bp, with an 850-bp core. DNA was polished using an End-It DNA repair kit (Epicenter), purified, and dA tailed utilizing the 3′-5′ exo-Klenow fragment (NEB). Then fragments were ligated to 200 pmol of annealed linkers.
(iii) Viral junction amplification.
All PCRs were conducted using Herculase II Fusion DNA polymerase (Agilent Technologies) according to manufacturer specifications. Pooled, linker-ligated DNA was divided into 800-ng aliquots and subjected to linear amplification (single-primer) PCR with biotinylated SP-1, as follows: 98°C for 3 min, 12 cycles of 98°C for 40 s, 65°C for 30 s, and 72°C for 45 s, and then 72°C for 1 min. Reaction mixtures were then spiked with pLinker and subjected to exponential PCR amplification, as follows: 98°C for 3 min; 35 cycles of 98°C for 40 s, 65°C for 30 s, and 72°C for 45 s; and 72°C for 5 min. Amplification products of 400 bp to 1.2 kb were isolated by agarose gel electrophoresis, and virus primer-specific products were enriched by magnetic streptavidin bead pull-down. Seminested PCR was performed with SP-2 and pLinker as follows: 98°C for 3 min; 35 cycles of 98°C for 40 s, 65°C for 30 s, and 72°C for 40 s; and 72°C for 5 min). Amplification products of 400 bp to 1.2 kb were isolated by agarose gel electrophoresis.
(iv) Paired-end library production and sequencing.
Linkers were digested with AscI and removed by agarose gel purification. Fragments were then polished, purified, and dA tailed as after the genomic DNA sonication. Fragments were then ligated to Illumina paired-end adapters and isolated by agarose gel electrophoresis. A final, 30-cycle library PCR was conducted utilizing Illumina primers PE1.0 and PE2.0 according to manufacturer specifications, and amplification products of 350 bp to 1 kb were isolated by agarose gel electrophoresis. The final libraries were submitted to 50 × 50 paired-end deep sequencing using an Illumina HiSeq 2000.
(v) Sanger sequencing of viral junction clones.
Subsequent to either the seminested junction or final library PCRs, a small aliquot of the pooled products for each sample were dA tailed and cloned using the TOPO TA kit (Invitrogen). Clones were grown and sequenced using M13 forward/reverse primers (Biotic Solutions). Sequences were considered for additional analysis if they met the inclusion criteria for high-throughput reads as described below.
Computational analysis. (i) Read validation and alignment.
Each end of paired-end reads was 3′ trimmed to 36 bp and validated using Bowtie to ensure that correct priming and processing had occurred. Viral ends required the 25-bp SP2 and the next 11 bp of viral sequence that is contiguous with the primer, allowing two mismatches. On the target side, presence of a perfect match to the remaining 7 bp of linker sequence was required. The 29-bp remainder of the target side was aligned with the human genome (hg18/NCBI Build 36.1) using Bowtie. Up to 2 mismatches were allowed, and unique alignments in the best alignment stratum were required. Identical target alignments, same strand and position, were combined into a single putative unique integration event, and any event supported by a single alignment was not considered in further analyses. Integration positions were given as the 5′ end of target alignment reads.
(ii) Determination of integration hotspots.
Integration hotspots were defined as a region of at least three integration events for which the frequency of events differed in a statistically significant fashion; P was <1 × 10−9, as determined by a negative binomial test, from a random distribution along the genome (30, 31). Hotspots with 100% repeat overlap, as defined by RepeatMasker, or present on the Y chromosome were removed from consideration as probable artifacts. Circos was used to generate circular whole-genome visualizations (32).
(iii) Hotspot correlations.
Hotspots with genomic features, expression, copy number, etc., were correlated utilizing BedTools, PyBedTools, and R (33).
(iv) Gene ontology.
The gene ontology network map was constructed using the BiNGO plugin of Cytoscape (34, 35). The ontology file utilized was GO_Molecular_Function, applying a hypergeometric test for significance with the Benjamini-Hochberg false discovery correction.
All data sets and lists are available upon request.
RESULTS
Integrant capture sequencing.
To determine the location, frequency, and structure of AAV integrations in human chromosomal DNA, we developed IC-Seq, an assay to capture, enrich, and sequence viral insertion events (Fig. 1). The HeLa cell line, a human cervical carcinoma line, was utilized in this study because it is the most published model system for AAV infection and integration, and an abundance of relevant bioinformatics data sets are available. Moreover, AAV is commonly associated with human reproductive tissue (36, 37). HeLa cells were infected with AAV and grown for 3 weeks with no selection, while maintaining high redundancy, to diminish background free viral DNA prior to DNA extraction (9, 38, 39). Viral-chromosomal junctions were recovered by seminested ligation-mediated PCR from randomly fragmented genomic DNA (850-bp average fragment size), a method modified from translocation capture sequencing (30, 31). AAV primers (Fig. 1A) were selected to bind in the viral P5 promoter located upstream of the inverted terminal repeat, amplifying the region containing the highest density of previously reported junctions (40). The linker-tag primer was derived from the translocation capture protocol (30).
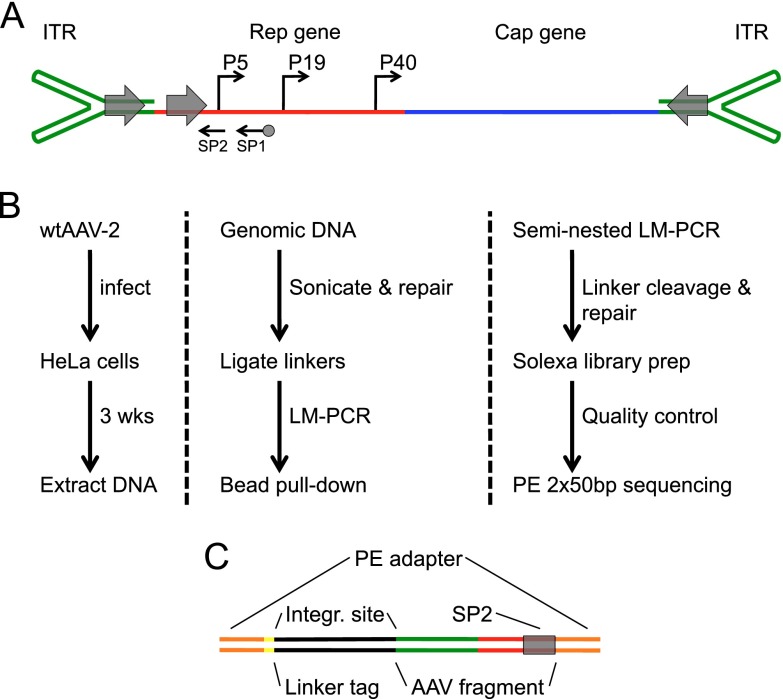
Fig 1.
AAV genome organization and integration capture sequencing schematic. (A) AAV genome features. The inverted terminal repeats (green) form the ends of the single-strand 4.7-kb viral genome. The AAV promoters (P5, P19, and P40) drive expression of two genes, Rep (red) and Cap (blue). Viral replication protein binding sites (gray arrows) are located in each ITR and in the P5 promoter. SP1 and SP2 (black arrows) are locations for sequencing primer 1 and 2 binding (SP1 is biotinylated). (B) IC-Seq outline. HeLa cells infected with wtAAV were grown for 3 weeks prior to DNA extraction. Genomic DNA was sonicated, blunted, A-tailed, and ligated to T-tailed asymmetric linkers. Integrations were amplified by seminested ligation-mediated PCR, incorporating bead pull-down target enrichment, followed by linker cleavage, Illumina linker ligation, and paired-end high-throughput sequencing. (C) Diagrammatic representation of elements present in final IC-Seq DNA library products submitted for paired-end sequencing.
Sonication generates unique linker ligation points for each integration event, allowing independent events to be studied without sequencing through viral-chromosomal junctions (Fig. 1C). As part of our quality control, small portions of the junction libraries were cloned and sequenced. Of 80 clones, 65% contained the appropriate linker tag and P5 sequence structure, and 36.5% of these contained viral-chromosomal junctions. The viral breakpoint occurred most frequently in the ITR hairpins, viral deletions were rare, and intervening unclassifiable sequences were not observed. Thus, IC-Seq efficiently captures wtAAV integration events with little background. After quality control, junction libraries were submitted for high-throughput paired-end Illumina sequencing. This generated a total of 702 million reads from two biological replicates (Fig. 2A). Samples were computationally validated for correct AAVp5 and linker tag sequences (as described in Materials and Methods) and were then aligned to the human genome.
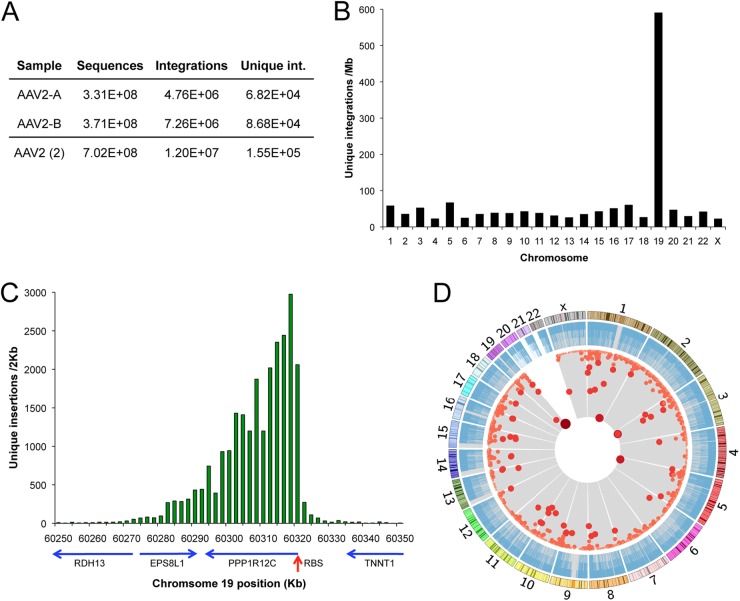
Fig 2.
Chromosomal distribution of integration events and hotspots. (A) Summary of IC-Seq sample A and B data. (B) Unique integration events per mappable megabase of human chromosomes. (C) Profile of unique integrations around AAVS1 in 2-kb intervals, with genes and gene orientation (blue arrows). RBS, Rep binding site of AAVS1. (D) Genome-wide view of all unique insertion events (blue bars) and mathematically determined integration hotspots (red dots). Darkness, size, and proximity to the center correspond to increasing insertions per hotspot. Chromosomal size and banding patterns are represented in the outer ring.
AAV insertions.
We mapped almost 12 million viral integrations to the human genome, which represented 154,976 unique nucleotide positions that possessed an average of 80 events per site (Fig. 2A). To minimize the effects of PCR amplification efficiency, unique nucleotide positions, rather than total reads, were used for further analysis. Unique AAV integrants were found on every chromosome (Fig. 2B) with 37,673 (24.3%) unique events in chromosome 19, an integration frequency per mappable Mb 10-fold higher than other chromosomes. On chromosome 19, 87.7% of events occurred within a 100-kb region proximal to the canonical AAVS1 (Fig. 2C). This region spans several genes and displays a distinctive single-sided peak-and-tail frequency distribution. As described below, this asymmetric profile was a characteristic feature of wtAAV integration loci in general.
Integration hotspots.
We next examined the human genome for loci of high-density AAV integration. Integration hotspots were defined as a region of at least three integration events for which the frequency of events differed from a random genomic distribution in a statistically significant fashion, with a P of <1 × 10−9, as determined by a negative binomial test (30, 31).
The two biological replicates were subjected to hotspot analysis independently, to determine the similarity between samples. Overlapping hotspots, present in both replicates, contained 81.6% of all hotspot-derived integration events, demonstrating the high level of experimental reproducibility. Due to this similarity, sequencing data from the replicates were combined and used to establish our highest-resolution hotspot map. This analysis revealed a total of 2,456 hotspots for wtAAV integration in the human genome (Fig. 2D). Each chromosome contained dozens to hundreds of hotspots, with the exception of chromosomes 20, 21, and 22. To determine the impact of HeLa cell aneuploidy on hotspot chromosomal distribution, a high-resolution locus copy number map was generated from single-nucleotide-polymorphism arrays and used to compare the copy number at loci bearing hotspots with the distribution expected by chance. HeLa aneuploidy did not appear to bias the genome-wide hotspot profile, as the average copy number at hotspots was 2.48, compared to 2.42 for the entire array. The largest hotspot was localized to AAVS1 (PPP1R12C), covering over 100 kb and representing 17.2% of all unique integrations, while the second largest, in PTH1R, contained only 2.0% (Table 1). Only two genomic loci, other than AAVS1, have been described in previous studies as AAV integration targets, 5p13.3 and 3p24.3 (29). These loci correspond to LOC729862 and FGD5, our third- and eighth-ranked hotspots (Table 1).
Table 1.
Top wtAAV-2 integration hotspotsa
| Rank | Chromosome | Geneb | Integrations |
Span (kb)c | |||
|---|---|---|---|---|---|---|---|
| Unique |
Total |
||||||
| No. | % | No. | % | ||||
| 1 | 19 | PPP1R12C | 25,068 | 17.23 | 5,180,608 | 45.02 | 102.9 |
| 2 | 3 | PTH1R | 2,843 | 1.95 | 2,053,921 | 17.85 | 54.9 |
| 3 | 5 | LOC729862 | 2,430 | 1.67 | 431,855 | 3.75 | 29.1 |
| 4 | 1 | RGL1 | 1,389 | 0.95 | 111,743 | 0.97 | 25.4 |
| 5 | 19 | ACSBG2 | 956 | 0.66 | 105,279 | 0.91 | 34.4 |
| 6 | 1 | NFIA | 802 | 0.55 | 71,147 | 0.62 | 31.4 |
| 7 | 14 | SYT16 | 563 | 0.39 | 50,711 | 0.44 | 32.4 |
| 8 | 3 | FGD5 | 537 | 0.37 | 47,576 | 0.41 | 33.8 |
| 9 | 4 | PCDH7 | 492 | 0.34 | 42,923 | 0.37 | 28.7 |
| 10 | 1 | CASZ1 | 478 | 0.33 | 27,624 | 0.24 | 19.4 |
| 11 | X | TBL1X | 384 | 0.26 | 52,679 | 0.46 | 8.6 |
| 12 | 1 | POGZ | 370 | 0.25 | 34,088 | 0.30 | 23.6 |
| 13 | 1 | WNT4 | 369 | 0.25 | 2,247 | 0.02 | 0.8 |
| 14 | 10 | MGMT | 293 | 0.20 | 2,305 | 0.02 | 0.5 |
| 15 | 1 | EMBP1 | 286 | 0.20 | 9,554 | 0.08 | 1.3 |
The 15 largest wtAAV-2 integration hotspots are shown.
Some hotspots cover multiple genes or are outside of genes; in these cases, the designation represents the nearest gene.
Hotspots within 10 kb of each other were considered part of the same event for this analysis.
Overall, we found good correlation between unique integrations and total integrations in hotspots (Table 1); however, the top three hotspots presented a notable exception. For these hotspots, we found that the extreme frequency of integration in the peak region led to an underestimate of their impact on the insertion profile of wtAAV. This was based on two observations: (i) every nucleotide position was targeted at these peaks, and therefore saturation was reached, and (ii) the number of observed events/site in peak domains (cluster number of 800) greatly exceeded (>10×) that for the average sequence (P < 1 × 10−5), indicating substantial oversaturation. Therefore, for the top three hotspots, total reads provide a more accurate representation of integration frequency than unique nucleotide positions. Analyzed in this manner, 5.2 million reads, or 45% of all integrant sequences, occur in AAVS1 (PPP1R12C) (Table 1). The second largest hotspot, in PTH1R, contributes 2.1 million sequences, almost 18%, while the third largest, near LOC729862, represents about 3.8% of the total integrant sequences. In our estimation, these data provide the most accurate measure to date of the top AAV hotspots and indicate that the largest three hotspots alone represent about 67% of all integrations.
Hotspots and Rep binding sites (RBS).
The AAV replication proteins Rep 68 and 78 bind DNA at tandem GAGC sequences, which are RBS (27). To determine whether genomic RBS drive hotspot localization, we investigated the integration profile around these sites. Using the chromosomal frequency of GAGC trimers for modeling, we first asked how well hotspots correlate with RBS chromosomal distribution (Fig. 3A). The analysis revealed that the number of RBS per chromosome explains roughly 80% of the variability in the chromosomal hotspot distribution.
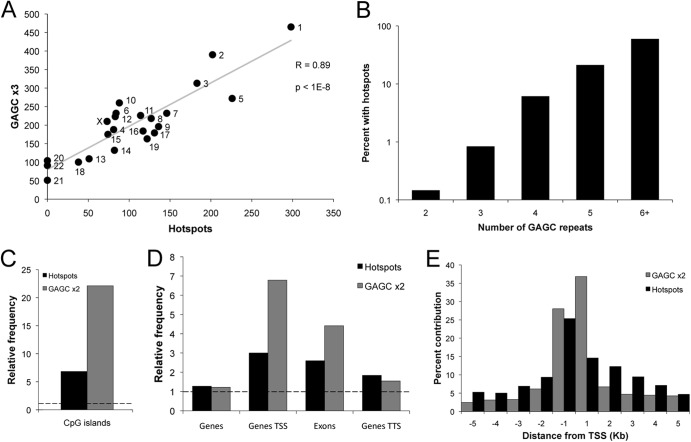
Fig 3.
Integration hotspots colocalize with Rep binding sites (GAGC repeats). Computational analysis of hotspots and various GAGC repeat elements, where n in GAGC ×n represents the number of GAGC tetranucleotide repeats (see Materials and Methods). (A) Integration hotspots per chromosome as a function of GAGC ×3 sequences, with simple linear regression in gray. P < 1 × 10−8 (t test). (B) Percent of genomic GAGC ×n sites that are within 50 bp of an integration hotspot. Sites that exceeded the GAGC count of each bin were subtracted. P < 0.001 for all categories (permutation test). (C) Relative frequency of hotspots and GAGC ×2 sequences intersecting CpG islands. Relative frequency is defined as the fold enrichment compared to a random distribution (see Materials and Methods). The dashed line indicates expected frequency based on a random model. P was <0.001 for both (permutation test). (D) Relative frequency of hotspots and GAGC ×2 sequences intersecting genes and specific gene regions. TSS, transcription start site; TTS, transcription termination site. The dashed line indicates expected frequency based on a random model. P was <0.001 for all categories (permutation test). (E) Composite density profile of integration hotspots and GAGC ×2 sequences proximal to transcription start sites.
We next asked if increasing GAGC copy number predicts the probability of generating a hotspot, requiring the hotspot to be within 50 bp of the RBS (Fig. 3B). For this analysis, computational data sets include only the exact RBS repeat number specified. For loci with two GAGC copies (GAGC ×2 loci), we found that a statistically significant 0.1% of sites were occupied by hotspots. Increasing numbers of GAGC repeats had a corresponding increase in occupancy, reaching 59.5% for GAGC ×6+ loci. The greatest change occurred from GAGC ×3 to GAGC ×4, with a 7.3-fold occupancy increase. Subsequent additions yield large, but diminishing, returns: GAGC ×5 and GAGC ×6+ result in only 3.5- and 2.8-fold enhancements, respectively. Thus, we conclude that AAV Rep binding sites are the primary determinant of AAV integration and that a dose-dependent response to GAGC sequences exists.
A second sequence element present in the ITRs, the terminal resolution site (TRS), is the specific site in the viral genome cleaved by the Rep endonuclease (41). We observed a 3.29-fold enrichment (P < 0.001) of hotspots around TRS sequences (GGCCAACT). However, we were unable to detect an enhancement in the probability of hotspot localization to RBS bearing canonical minimal TRS (CAAC/GTTG) compared to RBS alone. This lack of TRS correlation with RBS is consistent with in vitro experimentation that has found that constraints on this sequence exist but are minimal and difficult to define (22, 42, 43). Additionally, the spacing between the TRS and RBS as well as secondary structure may contribute to the complexity of determining a TRS influence (19, 44). Thus, the presence of a TRS may function in a modest capacity as an independent factor influencing hotspot localization.
Hotspots, genomic features, and transcription.
The human genome is relatively G/C poor, containing only ∼40% G/C, and CpG dinucleotides are further underrepresented (45, 46). Regions of high G/C content exist but are not randomly distributed in the genome (47). Consequently, Rep binding sites (GAGC ×n), which are 75% G/C and contain CpGs, are highly correlated with G/C-rich genomic features, especially CpG islands (Fig. 3C). Furthermore, hotspots and GAGC sequences are significantly overrepresented in active genes. Over 56% of hotspots overlap transcription units versus 44% expected by chance (P < 0.001). Transcription start sites (TSS), exons, and transcription termination sites (TTS) correlate with RBS and hotspots (Fig. 3D). AAV integration hotspots and GAGC repeats are highly represented at TSS, while decreasing markedly on either side (Fig. 3E). Thus, we conclude that G/C rich genomic features, which occur predominately near the beginning of genes, are likely to possess Rep binding sites and attract AAV integration.
Functional genomic markers that define transcriptional activity and accessible DNA were highly correlated with both hotspots and RBS (Fig. 4A). Most of these features, such as DNase-hypersensitive regions, H3K4me3, and H3K36me3, are associated with active transcription and open chromatin (48–50). There is also a significant colocalization with H3K27me3, generally regarded as a repressive marker (51), although it involves roughly 6-fold fewer hotspots than H3K4me3. Recent studies also indicate that certain H3K27me3 promoter profiles may serve to mark increased transcriptional activity (52). The relative frequency of hotspots in H3K36me3 and H3K27me3 peaks exceeded that for GAGC ×2, indicating that GAGC distribution alone may not fully explain this colocalization. We next asked how gene expression levels correlate with hotspots. For genes bearing hotspots as well as those bearing GAGC ×2, gene expression levels were significantly higher than expected by chance (P < 1 × 10−10) (Fig. 4B). Therefore, while transcriptional activity strongly correlates with AAV hotspots, much of this effect may be due to RBS distribution.
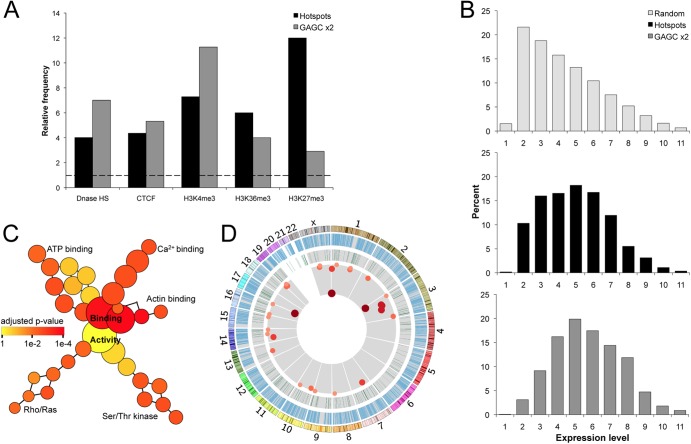
Fig 4.
Transcriptional activity influences hotspot localization. (A) Relative frequency of hotspots and GAGC ×2 sequences intersecting transcription-related markers (65). The dashed line indicates expected frequency based on a random model. P was <0.001 for all categories (permutation test). (B) Percent of hotspots, GAGC ×2, and expected frequency based on a random model in transcription level gene groups (65). (C) Gene ontology map of pathways bearing multiple hotspots. The size of a node indicates the number of genes in the category, while color indicates the degree of statistical significance. P was <0.001 for all terminal groups (hypergeometric test). (D) Genome-wide view of all genes (blue bars), proven oncogenes (green bars), and integration hotspots within oncogenes (red dots). Darkness, size, and proximity to the center correspond with increasing numbers of insertions per hotspot. Chromosomal size and banding patterns are represented in the outer ring.
Hotspots were located within 969 unique genes, and we utilized ontology software to determine which functional transcriptional groups may be targeted by AAV hotspots at higher-than-random frequencies (Fig. 4C). Pathways involving genes that bind calcium, ATP, and actin were significantly overrepresented, as were genes in the Rho/Ras and serine/threonine kinase activity groups. Since a number of genes in those pathways have oncogenic potential, the Sanger Institute Cancer Gene Census was used to determine if AAV hotspots were present in known causal oncogenes (Fig. 4D) (53). In total, 29 of these oncogenes were targeted by hotspots. The oncogene with the greatest number of integrations was TNFRSF14, represented by 78 unique insertion events. PPARG had 68 events, and 67 unique insertions were contained in CBFAT3. Other notable oncogenes bearing smaller hotspots include: MYC, ABL, FANCA, RB, EGFR, and FOXP1. In addition to the Sanger list, a hotspot was present in the imprinted DLK1/MEG3 region, which has been recently implicated in hepatocellular carcinoma of humans and mice (54, 55).
Directional integration.
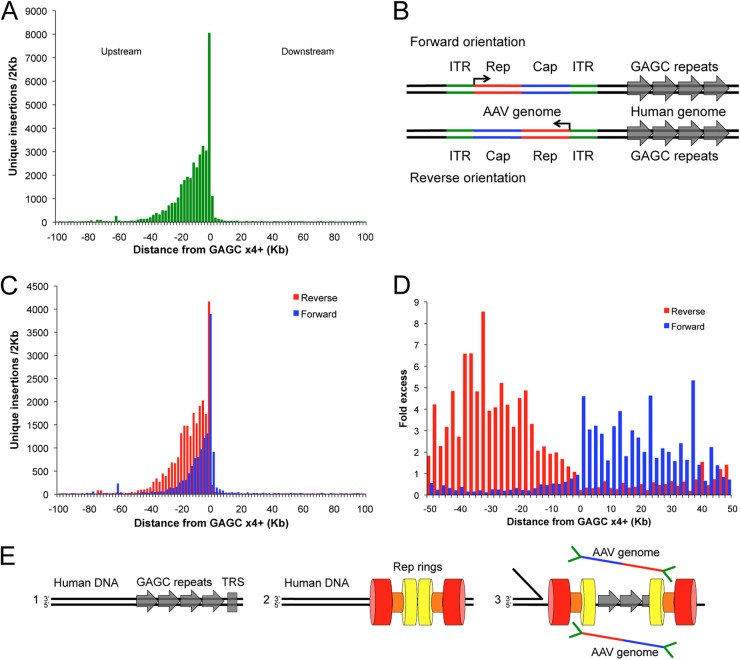
Hotspots possess a characteristic single-sided peak-and-tail distribution of integrants that appear to initiate near Rep binding sites. To investigate the arrangement of insertion events near RBS, a composite-density profile of all integration activity surrounding genomic loci of GAGC ×4 or greater was constructed (Fig. 5A). Overall, 39,177 unique integrations were discovered within 100 kb of these sites, encompassing over 50 individual hotspots and accounting for ∼28% of all unique integration events. The composite density data recapitulate the single-sided peak-and-tail phenotype seen for individual hotspots, such as AAVS1. The integration frequency peak begins just upstream of the RBS sequence, with the tail proceeding upstream for over 80 kb (Fig. 5A). Very limited integration activity, under 7% of events, occurs downstream of the RBS. This novel integration profile is consistent with the biochemical activities of Rep 68/78 and may serve as an identifier for Rep mediated integration loci. Rep binds specifically to CTCG/GAGC duplex sequences (56) and cleaves the CTCG strand at downstream sites (TRS in AAV ITR) (20). The Rep helicase then unwinds DNA, moving 3′ to 5′ on the uncut DNA strand (41, 57), corresponding to the upstream direction as depicted in Fig. 5.
Fig 5.
Direction of GAGC repeats determines integrant distribution and orientation of viral genomes. (A) Composite density profile of unique integration events proximal to GAGC ×4 and greater. (B) Schematic of minus and plus viral genome integration relative to human GAGC sequences. Viral P5 promoter (angled arrow) demonstrates direction of transcription. (C) Composite density profile of viral genome orientation for unique integration events proximal to GAGC ×4+. P was <1 × 10−9 for orientation differential (chi-squared test). (D) Fold enrichment of each genome orientation in 2-kb bins around GAGC ×4+ loci. (E) Model of helicase-aligned directional integration. Panel 1, GAGC repeat sequences (gray arrows) and TRS analogues (gray box) are present in the human genome. Panel 2, AAV Rep proteins oligomerize into opposing ring structures on GAGC sequences. Helicase domains, linker domains, and DNA binding/endonuclease domains are depicted by red, orange, and yellow, respectively. Positioning/structure is purely illustrative and meant to reveal one possible solution addressing the new integration features revealed by IC-seq. Panel 3, the Rep complex nicks human genomic DNA at TRS-like sequences. One Rep ring (left) proceeds with 3′-5′ helicase activity on the uncut strand, depositing predominately reverse-oriented genomes in a broad upstream peak. The other ring (right) is relatively immobile, depositing a tight peak of plus-oriented AAV genomes in the immediate vicinity of the GAGC sequences.
For a given DNA sequence, the viral genome can be integrated in either of two orientations: forward or reverse (Fig. 5B). For all major hotspots, we noticed a distinct and predictable viral genome orientation relative to a given RBS: a dominant forward orientation in the immediate vicinity and downstream of the RBS, and a dominant reverse orientation upstream of the RBS. We carried out a computational analysis of viral genome orientation based on GAGC ×4 loci to test this observation. This analysis confirmed that the orientation of integrated viral genomes with respect to the RBS is nonrandom (P < 1 × 10−9) (Fig. 5C). To gain a perspective of the directional preference, the ratio of forward and reverse orientations upstream and downstream of the RBS was determined (Fig. 5D). This analysis depicts a remarkably clear bimodal distribution centered at a region just upstream of the RBS. In the immediate and downstream vicinity of the genomic GAGC ×4, AAV genomes are preferentially positioned in the forward orientation, whereas in regions upstream of GAGC sequences and continuing for ∼80 kb, AAV genomes are predominately reverse oriented (Fig. 5B). The combination of high-throughput IC-Seq and bioinformatics presents a new and comprehensive view of AAV integration, where the frequency, magnitude, and directionality of the insertion events are far more intricate than previous studies could reveal.
DISCUSSION
The biology of AAV integration has long been a topic of interest as an example of virus/host interaction, as a method for targeted integration, and as a model for biological mechanisms that impact the integrity of the human genome. The lack of a sufficiently large integration data set, the use of varied and biased techniques for identifying integration events, and other technical limitations have contributed to an incomplete understanding of integration by AAV. We have overcome this deficiency by developing and applying integrant capture sequencing technology. The results that we have obtained resolve confusion arising from prior studies through a new and comprehensive genomic understanding of AAV integration. Importantly, this strategy can be used to characterize any viral or vector-mediated integration profile.
Utilizing an efficient unbiased strategy, 1.2 × 107 integration events and 1.56 × 105 unique integration sites were acquired, providing a data set suitable for stringent bioinformatic analysis. We found RBS to be the primary determinant of genome-wide AAV integration, with ∼80% of chromosomal hotspot distribution attributable to GAGC localization. The number of GAGC repeats at chromosomal loci substantially impacted the probability of generating a hotspot; ∼60% of loci with six or more GAGC repeats were occupied by hotspots. Rep endonuclease activity is essential to viral replication and integration by cleavage at the terminal resolution sites, which are present in the ITRs (41). Although we were unable to detect an enhancement in the probability of hotspot localization to genomic RBS bearing canonical TRS compared to RBS alone, we did observe a modest enhancement of hotspots around TRS sequences alone. Since 60 percent of sites bearing six or more RBS possessed hotspots, without an identifiable TRS, the presence of an optimal TRS may not greatly influence the localization of hotspots but may rather enhance hotspot intensity. AAVS1, for example, possesses a perfect TRS that can be cleaved by Rep and may contribute to the extremely high frequency of integration at that locus (17, 18, 43).
Hotspots correlated strongly with markers of transcriptional activity such as DNase hypersensitivity and peaks in activating histone markers for promoter regions and gene bodies. A few of these associations were suggested in previous work, which found a correlation between integration and H3K4me3 and H3K36me3 (29). However, we also found that RBS correlated with most of these markers. Furthermore, the expression of genes bearing hotspots and those bearing RBS was significantly higher than expected by chance. Thus, the strong association of hotspots with transcriptional activity may be attributable simply to RBS location. Factors such as increased accessibility of transcribed DNA, the probability of generating double-strand breaks (58–60), and Rep interaction with transcription-related proteins, such as TBP (61), may play an additional role in AAV integration profiles.
It is important to note that the integration correlates we have identified should remain true for various conditions and cell types. On the other hand, the additional factors considered above will vary in a cell- and tissue-specific manner and potentially influence the specific loci targeted and their relative intensity. Thus, while the presence of integration hotspots in nearly 1,000 genes, including dozens of oncogenes, has potential implications with respect to impaired gene function, this pool of genes may vary.
We view the single-sided peak-and-tail profile of hotspots as a unique and remarkably informative outcome with respect to integration biology. As previously noted, the established biological functions of Rep—sequence-specific DNA binding, strand-specific nicking, and directional unwinding of a target DNA (20, 41, 57)—are directly reflected in the observed asymmetry of the integration profile. Additionally, we found that the orientation of integrated viral genomes is nonrandom with respect to the relative position of the RBS. Neither of these features has been identified in Rep independent integration hotspots associated with AAV vectors (59, 60). To our knowledge, this represents the first proof of a directional integration bias by a eukaryotic virus.
The integration of wtAAV places the GAGC sequences of the viral P5 promoter and 5′ ITR in the same orientation as the human genomic GAGC sequences when the integration occurs adjacent to the RBS but in an inverse orientation in the upstream region. In order for integrated AAV genomes to be consistently positioned relative to GAGC, as observed, several conditions are required: (i) AAV Rep must interact with human genomic RBS in an orientation-dependent manner, (ii) a mechanism for preferentially delivering forward and reverse orientations must be available, and (iii) Rep must directionally interact with AAV genomes. The first condition is consistent with our understanding of the biochemical activity of Rep binding, nicking, and unwinding. The second condition can be met by recent crystal structure and cryo-EM studies, which have revealed that AAV Rep forms double octameric or double hexameric rings, with the ring facing opposite directions (Fig. 5E, panel 2) (21, 24, 26). The final condition, a specific orientation of Rep complex binding to the AAV substrate genome, has several possible contributors. The viral ITRs, which possess RBS and TRS, are unlikely to play an independent role in selecting viral genome orientation, as they are identical, are located at opposite ends of the genome, and are reverse complements of each other. The wtAAV-2 genome has a total of 54 GAGC sequences that display net directional bias, with 63% in a positive orientation. This may play a role; however, we believe that the 1.7-fold difference is not large enough to account for the observed 3- to 4-fold average orientation bias. Since all viral transcripts are produced from three promoters that are in the same orientation, another possibility is that a coupling exists between viral transcription and integration. However, the hypothesis that we favor employs a directional binding favoring the genome left end containing the viral p5 promoter (Fig. 5E). In addition to the ITR, the p5 promoter has been shown to contain a functional RBS/TRS, and in plasmid systems, the p5 promoter is able to independently enhance AAVS1 integration efficiency (13, 38, 39, 62). The combination of a p5 transcriptional complex localized near the left-end ITR may present a unique structural domain to specifically interact with the integration complex forming on a genomic site.
We propose that Rep double rings form on human genomic RBS and directionally associate with AAV genomes via P5 interaction (Fig. 5E, model 2). The Rep complex nicks the human genomic TRS-like substrate, allowing the upstream ring to unwind in a 3′-5′ direction while the downstream ring remains roughly in its original position. This relatively immobile Rep ring has no uncut DNA strand on which to proceed, idles in the area of the RBS, and delivers viral genomes in a predominantly forward orientation (Fig. 5B and E). In contrast, the migrating helicase complex contributes the broad upstream peak of integration, delivering predominantly reverse-oriented viral genomes. In our view, high-throughput integrant capture sequencing provides an exceptional platform to address mechanistic questions raised by the insights of the present study. This strategy can also be directly applied to characterization of recombinant AAV (rAAV) vectors. In the presence of Rep, vectors would be predicted to integrate with a profile similar to wtAAV; however, in the absence of Rep these vectors are known to target spontaneous double-strand breaks (59, 60). The IC-Seq protocol screens the entire population of infected cells and integrates the events that occur within individual cells as a component of the pool. Southern analysis of individual clones has indicated one to several integrations per cell under conditions similar to those in the present study (39); a genome-wide IC-Seq protocol applied to clonal cell lines would reveal the population of events that occur in a given cell line. Since an array of Rep mutations were previously established to characterize Rep DNA binding, endonuclease activity, oligomerization, and helicase activity, they can be adapted to examine the influence these functions have on the AAV integration profile (24, 25, 63, 64). Furthermore, now that a gold standard of wtAAV integration in the HeLa carcinoma cell line has been established, further studies characterizing integration in additional cell types should provide novel insight into cell-specific Rep interactions that may influence AAV integration.
The AAV integration process appears to be even more unique and complex than has previously been appreciated. This study provides novel insight into Rep-mediated integration and AAV biology and raises additional questions regarding the natural life cycle of AAV. To our knowledge, IC-Seq provides the greatest quantity and quality of integration data per experiment of any current technique. The expanded application of the IC-Seq protocol to other integrating virus and vector systems should allow comprehensive genome-wide integration profiles to be a new gold standard in future studies.
ACKNOWLEDGMENTS
We thank Yushan Li of the Weill Cornell Epigenomics Core for high-throughput sequencing and guidance.
T.J. and I.A.K. were supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. This work was supported in part by NIH grant AI037526 to M.C.N. M.C.N. is an HHMI investigator. E.F.-P. received support from the WR Hearst Foundation and PHS grant RO1 AI094050.
The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
T.J. designed and performed experiments and analysis and wrote the manuscript. I.A.K. designed experiments and made suggestions on the manuscript. T.O. and P.M. designed and performed data analysis. M.C.N. and M.S. provided material assistance and made suggestions on the manuscript. E.F.-P. designed experiments and analysis and wrote the manuscript.
Footnotes
Published ahead of print 29 May 2013
REFERENCES
- 1. Holmes EC. 2011. The evolution of endogenous viral elements. Cell Host Microbe 10:368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kotin RM, Siniscalco M, Samulski RJ, Zhu XD, Hunter L, Laughlin CA, McLaughlin S, Muzyczka N, Rocchi M, Berns KI. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U. S. A. 87:2211–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daya S, Berns KI. 2008. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 21:583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flotte TR. 2007. Gene therapy: the first two decades and the current state-of-the-art. J. Cell. Physiol. 213:301–305 [DOI] [PubMed] [Google Scholar]
- 5. Ward P, Walsh CE. 2012. Targeted integration of a rAAV vector into the AAVS1 region. Virology 433:356–366 [DOI] [PubMed] [Google Scholar]
- 6. Recchia A, Perani L, Sartori D, Olgiati C, Mavilio F. 2004. Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors. Mol. Ther. 10:660–670 [DOI] [PubMed] [Google Scholar]
- 7. Cortés ML, Oehmig A, Saydam O, Sanford JD, Perry KF, Fraefel C, Breakefield XO. 2008. Targeted integration of functional human ATM cDNA into genome mediated by HSV/AAV hybrid amplicon vector. Mol. Ther. 16:81–88 [DOI] [PubMed] [Google Scholar]
- 8. Urcelay E, Ward P, Wiener SM, Safer B, Kotin RM. 1995. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep. protein. J. Virol. 69:2038–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Surosky RT, Urabe M, Godwin SG, McQuiston SA, Kurtzman GJ, Ozawa K, Natsoulis G. 1997. Adeno-associated virus Rep. proteins target DNA sequences to a unique locus in the human genome. J. Virol. 71:7951–7959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Urabe M, Kogure K, Kume A, Sato Y, Tobita K, Ozawa K. 2003. Positive and negative effects of adeno-associated virus Rep. on AAVS1-targeted integration. J. Gen. Virol. 84:2127–2132 [DOI] [PubMed] [Google Scholar]
- 11. Young SM, Samulski RJ. 2001. Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc. Natl. Acad. Sci. U. S. A. 98:13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pieroni L, Fipaldini C, Monciotti A, Cimini D, Sgura A, Fattori E, Epifano O, Cortese R, Palombo F, La Monica N. 1998. Targeted integration of adeno-associated virus-derived plasmids in transfected human cells. Virology 249:249–259 [DOI] [PubMed] [Google Scholar]
- 13. Philpott NJ, Giraud-Wali C, Dupuis C, Gomos J, Hamilton H, Berns KI, Falck-Pedersen E. 2002. Efficient integration of recombinant adeno-associated virus DNA vectors requires a p5-rep sequence in cis. J. Virol. 76:5411–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Samulski RJ, Zhu X, Xiao X, Brook JD, Housman DE, Epstein N, Hunter LA. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kotin RM, Linden RM, Berns KI. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giraud C, Winocour E, Berns KI. 1994. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc. Natl. Acad. Sci. U. S. A. 91:10039–10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Linden RM, Ward P, Giraud C, Winocour E, Berns KI. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U. S. A. 93:11288–11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Linden RM, Winocour E, Berns KI. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. U. S. A. 93:7966–7972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meneses P, Berns KI, Winocour E. 2000. DNA sequence motifs which direct adeno-associated virus site-specific integration in a model system. J. Virol. 74:6213–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Im DS, Muzyczka N. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447–457 [DOI] [PubMed] [Google Scholar]
- 21. James JA, Escalante CR, Yoon-Robarts M, Edwards TA, Linden RM, Aggarwal AK. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11:1025–1035 [DOI] [PubMed] [Google Scholar]
- 22. Snyder RO, Im DS, Ni T, Xiao X, Samulski RJ, Muzyczka N. 1993. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep. protein. J. Virol. 67:6096–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Z. 2003. Characterization of the adenoassociated virus Rep. protein complex formed on the viral origin of DNA replication. Virology 313:364–376 [DOI] [PubMed] [Google Scholar]
- 24. Maggin JE, James JA, Chappie JS, Dyda F, Hickman AB. 2012. The amino acid linker between the endonuclease and helicase domains of adeno-associated virus type 5 Rep plays a critical role in DNA-dependent oligomerization. J. Virol. 86:3337–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zarate-Perez F, Mansilla-Soto J, Bardelli M, Burgner JW, Villamil-Jarauta M, Kekilli D, Samso M, Linden RM, Escalante CR. 2013. The oligomeric properties of the adeno-associated virus Rep68 reflect its multifunctionality. J. Virol. 87:1232–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mansilla-Soto J, Yoon-Robarts M, Rice WJ, Arya S, Escalante CR, Linden RM. 2009. DNA structure modulates the oligomerization properties of the AAV initiator protein Rep68. PLoS Pathog. 5:e1000513. 10.1371/journal.ppat.1000513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weitzman MD, Kyöstiö SR, Kotin RM, Owens RA. 1994. Adeno-associated virus (AAV) Rep. proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl. Acad. Sci. U. S. A. 91:5808–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Drew HR, Lockett LJ, Both GW. 2007. Increased complexity of wild-type adeno-associated virus-chromosomal junctions as determined by analysis of unselected cellular genomes. J. Gen. Virol. 88:1722–1732 [DOI] [PubMed] [Google Scholar]
- 29. Hüser D, Gogol-Döring A, Lutter T, Weger S, Winter K, Hammer Cathomen E-MT, Reinert K, Heilbronn R. 2010. Integration preferences of wildtype AAV-2 for consensus rep-binding sites at numerous loci in the human genome. PLoS Pathog. 6:e1000985. 10.1371/journal.ppat.1000985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, Casellas R, Nussenzweig MC. 2011. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147:95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oliveira TY, Resch W, Jankovic M, Casellas R, Nussenzweig MC, Klein IA. 2012. Translocation capture sequencing: A method for high throughput mapping of chromosomal rearrangements. J. Immunol. Methods 375:176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449 [DOI] [PubMed] [Google Scholar]
- 35. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bantel-Schaal UU, Hausen zur HH. 1984. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology 134:52–63 [DOI] [PubMed] [Google Scholar]
- 37. Walz CMC, Anisi TRT, Schlehofer JRJ, Gissmann LL, Schneider AA, Müller MM. 1998. Detection of infectious adeno-associated virus particles in human cervical biopsies. Virology 247:97–105 [DOI] [PubMed] [Google Scholar]
- 38. Philpott NJ, Gomos J, Berns KI, Falck-Pedersen E. 2002. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc. Natl. Acad. Sci. U. S. A. 99:12381–12385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hamilton H, Gomos J, Berns KI, Falck-Pedersen E. 2004. Adeno-associated virus site-specific integration and AAVS1 disruption. J. Virol. 78:7874–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang CC, Xiao X, Zhu X, Ansardi DC, Epstein ND, Frey MR, Matera AG, Samulski RJ. 1997. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 71:9231–9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu JJ, Davis MDM, Owens RAR. 1999. Factors affecting the terminal resolution site endonuclease, helicase, and ATPase activities of adeno-associated virus type 2 Rep. proteins. J. Virol. 73:8235–8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brister JR, Muzyczka N. 1999. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J. Virol. 73:9325–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lamartina S, Ciliberto G, Toniatti C. 2000. Selective cleavage of AAVS1 substrates by the adeno-associated virus type 2 rep68 protein is dependent on topological and sequence constraints. J. Virol. 74:8831–8842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hewitt FC, Samulski RJ. 2010. Creating a novel origin of replication through modulating DNA-protein interfaces. PLoS One 5:e8850. 10.1371/journal.pone.0008850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- 46. Venter JC. 2001. The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- 47. Saxonov S, Berg P, Brutlag DL. 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U. S. A. 103:1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Crawford GE. 2005. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res. 16:123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. 2007. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 17:691–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li J. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277:49383–49388 [DOI] [PubMed] [Google Scholar]
- 51. Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. 2008. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 19:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Young MD, Willson TA, Wakefield MJ, Trounson E, Hilton DJ, Blewitt ME, Oshlack A, Majewski IJ. 2011. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 39:7415–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. 2004. A census of human cancer genes. Nat. Rev. Cancer. 4:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang P-R, Xu M, Toffanin S, Li Y, Llovet JM, Russell DW. 2012. Induction of hepatocellular carcinoma by in vivo gene targeting. Proc. Natl. Acad. Sci. U. S. A. 109:11264–11269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, Sands MS. 2007. AAV vector integration sites in mouse hepatocellular carcinoma. Science 317:477. [DOI] [PubMed] [Google Scholar]
- 56. Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. 2004. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell 13:403–414 [DOI] [PubMed] [Google Scholar]
- 57. Zhou X, Zolotukhin I, Im DS, Muzyczka N. 1999. Biochemical characterization of adeno-associated virus rep68 DNA helicase and ATPase activities. J. Virol. 73:1580–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guirouilh-Barbat J, Redon C, Pommier Y. 2008. Transcription-coupled DNA double-strand breaks are mediated via the nucleotide excision repair and the Mre11-Rad50-Nbs1 complex. Mol. Biol. Cell 19:3969–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miller DG, Petek LM, Russell DW. 2004. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 36:767–773 [DOI] [PubMed] [Google Scholar]
- 60. Miller DG, Trobridge GD, Petek LM, Jacobs MA, Kaul R, Russell DW. 2005. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J. Virol. 79:11434–11442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. François A, Guilbaud M, Awedikian R, Chadeuf G, Moullier P, Salvetti A. 2005. The cellular TATA binding protein is required for rep-dependent replication of a minimal adeno-associated virus type 2 p5 element. J. Virol. 79:11082–11094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Murphy MM, Gomos-Klein JJ, Stankic MM, Falck-Pedersen EE. 2007. Adeno-associated virus type 2 p5 promoter: a rep-regulated DNA switch element functioning in transcription, replication, and site-specific integration. J. Virol. 81:3721–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Davis MD, Wu J, Owens RA. 2000. Mutational analysis of adeno-associated virus type 2 Rep68 protein endonuclease activity on partially single-stranded substrates. J. Virol. 74:2936–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Walker SL, Wonderling RS, Owens RA. 1997. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J. Virol. 71:6996–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Project Consortium ENCODE 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9:e1001046. 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]