Abstract
Homologous rotaviruses (RV) are, in general, more virulent and replicate more efficiently than heterologous RV in the intestine of the homologous host. The genetic basis for RV host range restriction is not fully understood and is likely to be multigenic. In previous studies, RV genes encoding VP3, VP4, VP7, nonstructural protein 1 (NSP1), and NSP4 have all been implicated in strain- and host species-specific infection. These studies used different RV strains, variable measurements of host range, and different animal hosts, and no clear consensus on the host range restriction determinants emerged. We used a murine model to demonstrate that enteric replication of murine RV EW is 1,000- to 10,000-fold greater than that of a simian rotavirus (RRV) in suckling mice. Intestinal replication of a series of EW × RRV reassortants was used to identify several RV genes that influenced RV replication in the intestine. The role of VP4 (encoded by gene 4) in enteric infection was strain specific. RRV VP4 reduced murine RV infectivity only slightly; however, a reassortant expressing VP4 from a bovine RV strain (UK) severely restricted intestinal replication in the suckling mice. The homologous murine EW NSP1 (encoded by gene 5) was necessary but not sufficient for promoting efficient enteric growth. Efficient enteric replication required a constellation of murine genes encoding VP3, NSP2, and NSP3 along with NSP1.
INTRODUCTION
Group A rotaviruses (RVs) are segmented double-stranded RNA viruses that replicate primarily in mature epithelial cells on the tips of the small intestinal villi (1). Rotavirus infection is ubiquitous among mammals; however, viral strains isolated from one host species tend to have diminished replication capacity and virulence in heterologous species. This host range restriction was the basis for two modified “Jennerian” RV vaccines, RotaShield and RotaTeq; animal rotaviruses that are naturally restricted for replication and virulence in humans were used as genetic backbones to produce these attenuated, live viral human vaccines. In the pentavalent RV vaccine Rotateq, for example, human rotavirus genes encoding VP7 of serotypes G 1, 2, 3, 4, and VP4 of serotype P1A (P[8]) were incorporated into bovine RV strain WC3. It was thought that the multivalent nature of this vaccine would induce neutralizing antibodies against the most common human RV serotypes but that it would be attenuated in susceptible infants because of host range restriction elements in its bovine (heterologous) RV backbone (2).
Direct experimental evidence for host range restriction has been best demonstrated in the murine system, where all known heterologous RV strains replicate orders of magnitude less efficiently in suckling mice than do homologous murine strains (3, 4). On the other hand, experimental studies in gnotobiotic piglets have been interpreted to indicate that some human RV strains can be adapted to replicate very efficiently in piglets, although direct comparison to wild-type homologous porcine RV replication in piglets has not been performed (5). A large body of epidemiologic data strongly supports the notion of the existence of substantial host range restriction elements for animal RV replication in humans (6–9).
The genetic basis of rotavirus host range restriction is likely to be multigenic and dependent on several factors, including the species origin of the RV strain, the host species, and the anatomic site in which replication is assessed (e.g., mucosal versus systemic) (10–17). In addition, host range restriction has been defined differently in different studies. Some studies have focused on host-restricted virulence, others on host-restricted replication, and others on the host-restricted capacity to spread efficiently to those susceptible (10–17). Several RV genes have been identified as contributing to restriction. Using reassortants generated from two heterologous strains (simian SA11 and bovine NCDV) in a suckling mouse model, Offit et al. demonstrated that the ability of SA11 to induce diarrhea at a 50×-lower inoculating dose than NCDV was associated with SA11 VP4, the RV cell attachment protein (16). Of note, this study compared two heterologous strains, both of which are highly attenuated for replication in the mouse model compared to a homologous murine RV (16). In a gnotobiotic piglet model that did not measure replication, Hoshino et al. used reassortants between the homologous porcine SB-1A RV strain and a heterologous human DS-1 RV strain to identify the determinants of virulence. In a single-dose inoculation, VP4, VP3, VP7, and nonstructural protein 4 (NSP4) were associated with the increased capacity of the porcine strain to induce diarrhea in a newborn gnotobiotic piglet model (15). Subsequently, Broome et al. examined a series of reassortant RVs derived from the cross between a highly virulent wild-type murine RV (EW) and a much less virulent heterologous simian RV (RRV) in suckling mice. In this study, the phenotypic markers used to assess host range were the DD50 (defined as the highest dilution that causes diarrhea in 50% of suckling BALB/c mice) of the parental and reassortant strains and their ability to spread within a litter from inoculated to uninoculated pups. The homologous EW phenotypes of a low DD50 and the ability to transmit illness to noninoculated littermates, which are the unique characteristics of wild-type murine RV, were associated with NSP1 and to a lesser degree with NSP2 but not with the two viral surface proteins VP4 and VP7 (12). However, several subsequent studies in various animal models have failed to show a relationship between RV virulence as measured by diarrhea and NSP1 (10, 11, 13, 15). VP4 and NSP1 were identified as determinants of systemic replication and/or virulence of selected simian RV strains in the biliary tract of suckling mice (14, 17). Perhaps because of differences in RV strains, animal models, and definitions of RV virulence used in these studies, no clear consensus has yet emerged regarding the viral genes responsible for host range restriction, especially when host range restriction is specifically defined by the diminished replication capacity of a heterologous versus a homologous RV in the small intestine.
The mechanistic basis for host range restriction is even less clear than the genetic basis. Some RV proteins, such as VP4 viral surface proteins, appear to mediate RV species- and strain-specific infection due to the variability in viral attachment and entry into target cells (14). The RV gene 5 product, NSP1, was initially found to be dispensable for rotavirus replication in vitro (18). It is the most genetically diverse RV protein, but NSP1 gene sequences from strains from the same host species are more similar to each other than to heterologous strain sequences (19). Recent whole-genome analysis revealed that some human RV NSP1 genes may share common origins with bovine or porcine RVs; however, the NSP1 genes of the great majority of human RV isolates strains still form close genetic clusters (20). Of note, NSP1s of RRV and EW belong to distinct genotypes (19, 20).
Recently, NSP1 was shown to be an antagonist of host innate immunity, especially the type I interferon (IFN) response (21, 22). NSP1 interacts with several cellular interferon regulatory factors (IRFs), including IRF3, IRF5, and IRF7, and the interaction induces degradation of these IRFs, resulting in the suppression of the type I IFN response (21, 22). In some viral strains, NSP1 interferes with the innate response by degrading βTrCP, a host factor responsible for the activation of NF-κB (23). We previously found that in normal mouse embryonic fibroblasts (MEFs), replication of a bovine RV (UK) was severely restricted, whereas the simian strain (RRV) replicated efficiently. Using UK × RRV reassortants, we demonstrated that the RV strain-specific replication capacity in MEFs was determined by the different efficiencies with which RRV and UK NSP1 proteins degrade IRF3 and suppress the type I IFN response (24). We also demonstrated in a direct RV gallbladder inoculation model in suckling mice that a high level of simian RRV replication in the biliary tract was associated with RRV VP4, which mediates RV entry into cholangiocytes, and with RRV NSP1, which mediates suppression of the host IFN response (14). This report suggests that both VP4 and NSP1 play important roles in rotavirus strain- and host-specific replication in some extraintestinal sites in the mouse.
Of note, in vitro in MEFs, both RRV and murine RV NSP1 degrade IRF3 and suppress the cellular type I IFN response efficiently (24, 25). In mouse intestines and other systemic organs, such as liver, bile ducts, and pancreas, RRV replication is significantly enhanced in IFN signaling-deficient alpha/beta IFN (IFN-α/β) and IFN-γ receptor knockout (IFNR KO) or signal transducer and activator of transcription 1 knockout (STAT1 KO) mice, whereas replication of EW is not significantly affected (26). These findings suggest that the IFN antagonism by heterologous simian and homologous murine RV in vivo is regulated differently than in vitro. In more recent studies, we demonstrated that, in vivo, murine RV efficiently blocks NF-κB activation in the small intestine but RRV does not, likely accounting for the differences seen in gastrointestinal (GI) tract replication between these two RVs (27).
In the present study, we used genetic reassortants generated from the cross of the murine EW and simian RRV strains to study the association of RV genes with homologous or heterologous RV replication in the suckling mouse intestine. We found that in wild-type mice, RRV VP4 reduced replication efficiency of the homologous EW virus only moderately. On the other hand, the heterologous bovine UK RV VP4 substantially restricted replication in the murine intestine. RRV NSP1 conferred the heterologous RRV phenotype of substantially restricted intestinal replication, since all reassortants with RRV NSP1 replicated poorly in mouse intestine. EW NSP1 was necessary but not sufficient for the EW phenotype of robust intestinal replication.
MATERIALS AND METHODS
Cells, viruses, and EW × RRV and UK × RRV reassortant viruses.
MA104 cells, a green monkey kidney cell line, were maintained in M199 media (Life Technologies, Grand Island, NY) supplemented with 1,000 U/ml of penicillin, 1,000 μg/ml of streptomycin (Mediatech, Inc., Manassas, VA), and 10% fetal calf serum (FCS) (Life Technologies) as described previously (28).
Murine RV EW is a non-cell culture-adapted wild-type strain. EW was propagated in 5-day-old BALB/c mice, and the EW stock inoculum was a crude centrifugation-clarified intestinal homogenate prepared as previously described (26). The infectivity titer of EW intestinal homogenate stock was expressed as DD50, defined as the highest dilution that causes diarrhea in 50% of suckling BALB/c mice (29). Because EW does not grow in cell culture, it is impossible to enumerate the infectious viral particles in EW stock using a cell culture-based assay. However, it has been shown that EW very efficiently infects sucking mice, with one DD50 being equivalent to one 50% immunogenic dose (29). Therefore, the DD50 is the most sensitive measure for determining the amount of infectious EW virus administered. Rhesus rotavirus (RRV) and all reassortants were propagated in MA104 cells as previously described (24). The titers in the RRV and all reassortant stocks were expressed as PFU per milliliter, which was determined by plaque assay in MA104 cells (28).
Most EW × RRV reassortant viruses were generated by coinfection of 7-day-old BALB/c suckling mice with EW and RRV and have been previously described (12). Additional reassortants EA 4-1-2 and EA 11-1-3 were generated by coinfecting 5-day-old BALB/c mice with EW × RRV reassortants E4/1 and A11. Reassortant BE 1-1-3 was generated by simultaneously infecting 5-day-old BALB/c mice with three EW × RRV reassortants, B2/1, B4/1, and E4/1. During coinfections, several pups in each litter were not inoculated with virus. At 5 days postinfection (dpi), mice were sacrificed and intestines from both inoculated pups and noninoculated littermates were collected. Intestinal homogenates were serially diluted and RV plaques isolated after growth in 6-well plates. Plaques were then grown in 24-well plates and viral RNAs extracted for genotyping by polyacrylamide gel electrophoresis (PAGE) as previously described (12). New reassortant RVs were further plaque purified and their genotypes confirmed by both electropherotyping and strain-specific quantitative reverse transcription-PCR (qRT-PCR) as described below. Reassortant EA 11-1-3 was isolated from the intestine of mice coinfected with EW × RRV reassortants E4 and A11. Other reassortants (EA 4-1-2 and BE 1-1-3) were isolated from noninoculated littermates that developed diarrheal disease. Reassortants DEA 24-1-2 and DEA 3-1-2 were isolated from cell culture following coinfection of reassortants EA 4-1-2 and D10/2 in MA104 cells. Since RRV and EW genes 7, 8, and 9 migrate closely together on PAGE, we used RV strain-specific primers and qRT-PCR to confirm the parental origin of genes 7, 8, and 9 for all reassortants. In all tables, we have followed the common convention of assigning NSP2 to gene 8, NSP3 to gene 7, and VP7 to gene 9. The virus strain-specific primers are listed in Table 1. UK × RRV reassortants 19-1-1 and 27-3-1 (see Table 3) were a generous gift from T. Hoshino and were described previously (24). The reassortant 19-1-2 (all RRV genes except UK VP4) was isolated by coinfecting MA104 cells with UK × RRV reassortant 19-1-1 (all RRV genes except UK VP4 and NSP3) and RRV. All UK × RRV reassortants were triply plaque purified and genotypes confirmed by electropherotyping and by polyacrylamide gel electrophoresis.
Table 1.
Primers and probes used in qRT-PCR for EW or RRV gene identification or intestinal virus quantitation
| Virus genea | Sequence |
|---|---|
| EW VP7 (gene 9) F | 5′-TCAACCGGAGACATTTCTGA-3′ |
| EW VP7 (gene 9) R | 5′-TTGCGATAACGTGTCTTTCC-3′ |
| RRV VP7 (gene 9) F | 5′-ACGGCAACATTTGAAGAAGTC-3′ |
| RRV VP7 (gene 9) R | 5′-TGCAAGTAGCAGTTGTAACATC-3′ |
| EW NSP2 (gene 8) F | 5′-GAGAATGTTCAAGACGTACTCCA-3′ |
| EW NSP2 (gene 8) R | 5′-CTGTCATGGTGGTTTCAATTTC-3′ |
| RRV NSP2 (gene 8) F | 5′-GAGAATCATCAGGACGTGCTT-3′ |
| RRV NSP2 (gene 8) R | 5′-CGGTGGCAGTTGTTTCAAT-3′ |
| EW NSP3 (gene 7) F | 5′-AGGTTTGAGACATCGAAGCAA-3′ |
| EW NSP3 (gene 7) R | 5′-GAGTGATGACCGATTGCAGA-3′ |
| RRV NSP3 (gene 7) F | 5′-TTGAAGAGAAAATGGAAGTAGATACAA-3′ |
| RRV NSP3 (gene 7) R | 5′-TACTTCTCATTAACCCGATGTTTCA-3′ |
| NSP5 (gene 11) F | 5′-CTGCTTCAAACGATCCACTCAC-3′ |
| NSP5 (gene 11) R | 5′-TGAATCCATAGACACGCC-3′ |
| NSP5 (gene 11) probe | 5′-Cy5/TCAAATGCAGTTAAGACAAATGCAGACGCT/IAbRQSp/-3′ |
F indicates forward primer, and R indicates reverse primer.
Table 3.
Intestinal titers of indicated RV in STAT1 KO suckling mice orally infected with UK, RRV, or UK × RRV reassortantsa
| Reassortant | Rotavirus geneb |
STAT1 KO titer (PFU/g)c | No. of virus-positive mice/total no. of mice | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |||
| 19-1-2 | R | R | R | U | R | R | R | R | R | R | R | 145 (30–2,333) | 2/6 |
| 27-3-1 | U | R | R | R | R | R | R | R | R | R | R | 114,768 (20,000–933,333) | 5/5 |
| UK | U | U | U | U | U | U | U | U | U | U | U | 30 | 0/5 |
| RRV | R | R | R | R | R | R | R | R | R | R | R | 131,139 (46,667–500,000) | 5/5 |
Five-day-old STAT1 KO mice were infected with UK, RRV, or UK × RRV reassortants (107 PFU). Mouse intestinal tissues were collected at day 3 postinfection and virus titers in tissues determined by plaque assay and expressed as PFU per gram of tissue.
R represents a gene from RRV, and U represents a gene from UK.
A titer value of 30 was assigned to intestinal tissue samples that had undetectable levels of virus.
Mice, rotavirus infection, and virus titration.
BALB/c mice were purchased from The Jackson Laboratory (Bar Harbor, ME). 129sv mice and STAT1 KO mice, also on a 129sv background, were purchased from Taconic (Germantown, NY). All mice were maintained in the Veterinary Medical Unit of the Palo Alto VA Health Care System (PAVAHCS). The Institutional Animal Care Committee at the VAPAHCS approved these studies.
Five-day-old suckling mice were orally inoculated by gavage with 104 PFU of RRV or EW × RRV reassortants or 104 DD50 of wild-type EW virus. For studies of the intestinal replication of UK × RRV reassortants, 5-day-old STAT1 KO mice were orally inoculated by gavage with 107 PFU of UK, RRV, or selected UK × RRV reassortants. Three days postinoculation, mice were sacrificed and samples collected from the entire intestine. Intestinal samples were weighed, homogenized using a 1-ml syringe into a 10% (wt/vol) suspension in M199 without FCS, and frozen at −80°C (26). RV titers in the intestine were measured by plaque assay as previously described (26). Intestinal virus titers were expressed as PFU per gram of intestine as described previously (26).
Detection of RV infection in intestine by immunofluorescence microscopy.
Intestinal tissues from RRV- or EW-infected 129sv suckling mice were collected at 3 days postinfection, frozen in optimal cutting temperature compound (OCT), and kept at −80°C until staining. Frozen tissues were cut into 6-μm-thick sections, air-dried, and fixed in cold acetone-methanol (1:1) for 10 min at −20°C. Tissues sections were stained with Texas Red-labeled monoclonal antibody against VP6 (clone 1E11 that reacts with all rotavirus strains used in this study). The cell nucleus was stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Life Technologies). After staining, slides were mounted with Aqua Poly/Mount (Polysciences, Warrington, PA) and examined using a Nikon Eclipse TE300 inverted fluorescence microscope equipped with a QImaging Retica 200R charge-coupled-device (CCD) camera (QImaging, Surrey, BC, Canada). Images were acquired and analyzed with the QCapture Pro program (QImaging).
qRT-PCR detection of virus in intestinal tissues.
For quantitative reverse transcription-PCR (qRT-PCR) analyses, RNA from intestinal tissues was extracted using TRIzol (Life Technologies) as described previously (3). Reverse transcription was carried out as previously described (30). For detection of EW or RRV genes 7, 8, and 9, a Brilliant III UltraFast SYBR green QPCR Master Mix kit (Agilent Technologies, Santa Clara, CA) was used with the primers listed in Table 1. For measuring intestinal RV titers using qRT-PCR, intestinal tissues were first weighed and RNA extracted. A TaqMan assay was used to compare titers of wild-type EW (non-cell culture adapted) and RRV (cell culture adapted) using a Brilliant III UltraFast QPCR Master Mix kit (Agilent Technologies) and conserved primers based on RV gene 11 (NSP5) sequences as previously described (30) (Table 1). A CsCl density gradient-purified triple-layer particle preparation of RRV with a known virus plaque titer was used to create a standard for converting PCR values to PFU values. However, we noted that when using this standard conversion, the PFU value derived from our PCR assay for a stock RRV cell lysate exceeded the titer derived from the plaque assay by approximately 50-fold. To calibrate the PCR assay, we used an index calculation based on the ratio of PFU estimated from PCR (based on the purified triple-layered particle [TLP] standard) and the actual PFU from the plaque assay for the stock RRV cell lysate to adjust the raw PFU values from the PCR assay and obtained values of “PFU equivalent” for all tissue samples. These values were further adjusted according to the weight of the intestinal sample, and the dilution factors involved obtaining the PFU equivalent per gram of tissue. We found that when we compared titer results extrapolated from qRT-PCR to actual plaque assay titers for tissues from RRV-infected mice, the values of adjusted PFU equivalent were in close agreement with actual infectivity titers (data not shown).
Statistical analyses.
Multiple regression analysis was used to assess the association between EW or RRV genes and the levels of intestinal replication of EW × RRV reassortants with a PASW Statistics 12 program (SPSS Inc., Chicago, IL). Regression modeling employed orthogonal coding (−1/2 for RRV, +1/2 for EW) of effects of each individual gene and log-transformed viral titers. To compare the levels of viral replication between the RRV-like, EW-like, and intermediate phenotypic groups or replication of each phenotypic group in wild-type versus STAT1 KO mice, a t test or analysis of variance (ANOVA) was used. A post hoc Scheffe test was also used for pairwise comparisons between two groups.
RESULTS
Comparison of EW replication and RRV replication in mouse intestine.
To compare the levels of intestinal replication between the homologous murine EW and heterologous simian RRV strains in wild-type 129sv suckling mice, we orally inoculated 5-day-old mice with EW (104 DD50) or RRV (107 PFU). Previously, we demonstrated that at this high infection dose, RRV causes diarrheal disease, produces detectable intestinal replication early in infection, and induces protective immunity in suckling mouse (3, 26, 31). At various days postinfection, intestinal samples were collected and RV titers measured by quantitative RT-PCR (Fig. 1A). The level of RRV intestinal replication was low and undetectable at 10 days postinfection (dpi). EW replicated to a titer 103- to 104-fold higher than the RRV titer at all time points examined (P < 0.0001). EW replication in the intestine lasted longer than did RRV replication; EW virus was detected at high titers on 14 dpi. The different replication patterns of homologous EW and heterologous RRV were confirmed by immunofluorescence staining of infected intestine at 3 dpi (Fig. 1B and C). The tips of intestinal villi of EW-infected mice strongly stained for RV antigen, whereas very few if any virus-positive enterocytes were detected in RRV-infected mice. These results indicate that homologous murine rotavirus has a significantly higher capacity to replicate in suckling mouse intestines than heterologous simian rotavirus. Of note, we also observed that at 3 days postinfection, the level of EW intestinal replication was significantly (over 1,000-fold) higher than that of RRV in a relatively more RV-resistant mouse strain, C57BL/6. In addition, replication of reassortant D4/3 (all EW genes except RRV VP4 and NSP1) in C57BL/6 suckling mice was significantly (>1,000-fold) lower than EW replication as observed in 129sv mice. Therefore, the differential gut replication capacities of homologous EW and heterologous RRV, as well as the inhibitory effect of RRV NSP1 on RV replication, are present in several mouse strains.
Fig 1.
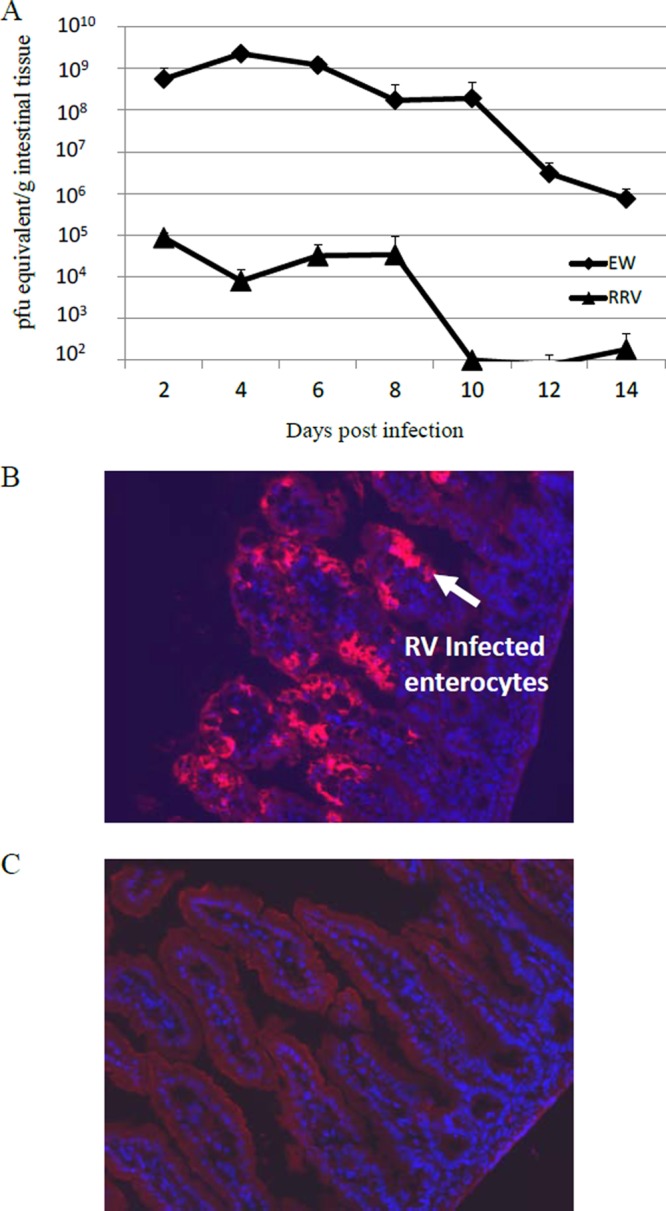
(A) EW or RRV titers in the intestine in orally infected 5-day-old suckling 129sv mice. Five-day-old suckling mice were orally inoculated with wild-type EW (104 DD50) or cell culture-adapted RRV (107 PFU). At different days postinfection, mice (n = 3) were sacrificed and intestinal tissues collected. Virus titers in intestines were determined by qRT-PCR and expressed as PFU equivalent per gram of tissue. The virus titers in EW-infected mice were significantly higher than in RRV-infected mice at all time points (P < 0.01). (B and C) Tissues from small intestines were collected at day 3 postinfection with EW (B) or RRV (C) and were stained with Texas Red-labeled anti-RV VP6 monoclonal antibody (MAb), 1E11 (red), and the cell nucleus was stained with DAPI (blue).
Intestinal replication of EW × RRV reassortants in 129sv mice.
To identify the EW gene or genes associated with the intestinal growth phenotype of the murine RV, we orally infected 5-day-old 129sv suckling mice with a series of EW × RRV reassortants (104 PFU), parental RRV (104 PFU), or EW (104 DD50) and measured RV titers in the intestine 3 dpi. We chose this infectious dose because it exceeded the 50% infective dose (ID50) of EW by 10,000-fold and in order to accommodate some of the reassortants that grew to relatively low titers in tissue culture. In addition, at this dose, the contribution of input RRV is minimized and RRV replication is substantially restricted compared to EW replication, allowing a maximal difference between the levels of intestinal replication of RRV and RRV-like and EW and EW-like reassortants. Table 2 shows the genotypes of the reassortant viruses, geometric mean titers (GMTs) in the intestine on day 3, ranges of minimum and maximum intestinal virus titers, and the proportions of mice that were RV positive. We confirmed that the parental RRV had very limited intestinal replication capacity at the dose used (104 PFU). Only a small fraction (11%) of the mice inoculated with 104 PFU of RRV had any detectable virus on 3 dpi. In contrast, all mice inoculated with EW (104 DD50) were virus positive (5/5) and the titer of RV in the intestine was substantially higher (GMT, 13.8 × 106; range, 5.5 × 106 to 20.5 × 106) than the viral titers in RRV-infected pups (P < 0.0001) (Table 2). Of note, the titers of EW seen in Table 2 were lower than titers in observed in Fig. 1. This is likely due to sampling time differences (day 2 versus day 3) between these two experiments. However, on both days we observed dramatic differences in the intestinal replication capacity of the homologous murine EW and heterologous simian RRV strains.
Table 2.
Intestinal titers of RV in wild-type suckling mice orally infected with EW, RRV, or EW × RRV reassortantsa
| Reassortant | Rotavirus geneb |
Wild-type titer (PFU or PFU equivalent/g)d | No. of virus-positive mice/total no. of mice | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5c | 6 | 7 | 8 | 9 | 10 | 11 | |||
| B7/2 | R | R | E | R | R | R | R | R | R | R | R | 30 | 0/4 |
| A15/1 | R | R | R | R | R | R | E | E | E | R | R | 30 | 0/7 |
| B2/1 | R | E | E | R | R | R | R | R | E | R | E | 30 | 0/6 |
| B4/1 | R | R | E | R | R | R | E | E | E | R | R | 30 | 0/7 |
| B6/1 | R | E | E | R | R | R | R | R | E | R | E | 30 | 0/4 |
| E14/2 | E | E | E | R | R | E | E | R | E | E | R | 30 | 0/9 |
| H5/1 | E | E | E | R | R | E | R | E | E | E | R | 52 (30–1,333) | 1/7 |
| E9/1 | E | E | E | R | R | E | E | E | E | E | R | 77 (30–1,333) | 1/4 |
| D4/3 | E | E | E | R | R | E | E | E | E | E | E | 70 (30–4,000) | 2/8 |
| D10/2 | R | R | R | R | E | R | R | R | R | R | R | 900 (30–6,333) | 3/4 |
| E4/1 | R | R | E | R | E | R | R | E | E | R | R | 224 (30–2,333) | 4/7 |
| A/11 | R | R | R | R | E | R | E | R | R | R | R | 194 (30–2,000) | 5/10 |
| EA 11-1-3 | R | R | E | R | E | R | E | R | R | R | R | 253 (30–12,667) | 6/9 |
| DEA 24-1-2 | R | R | E | R | E | R | R | R | R | R | R | 6,162 (1.666–14,667) | 6/6 |
| DEA 3-1-2 | R | R | R | R | E | R | E | E | R | R | R | 1,680 (30–17,667) | 11/13 |
| D6/2 | E | E | E | R | E | E | E | E | E | E | E | 172,487 (33,333–566,667) | 11/11 |
| D1/5 | R | E | E | R | E | R | E | E | R | R | E | 1,274,569 (766,667–1,933,333) | 10/10 |
| C3/2 | R | E | E | R | E | R | E | E | R | R | E | 185,580 (70,000–833,333) | 4/4 |
| EA 4-1-2 | R | R | E | R | E | R | E | E | R | R | R | 89,165 (40,000–366,667) | 8/8 |
| BE 1-1-3 | R | E | E | R | E | R | E | E | E | R | R | 98,762 (40,000–216,667) | 4/4 |
| RRV | R | R | R | R | R | R | R | R | R | R | R | 39 (30–333) | 1/9 |
| EW | E | E | E | E | E | E | E | E | E | E | E | 13,774,839 (5,586,844–20,516,593) | 5/5 |
Five-day-old suckling 129sv mice were infected with EW (104 DD50), RRV, or the indicated EW × RRV reassortants (104 PFU). Mouse intestinal tissues were collected at day 3 postinfection and RV titers in tissues determined by plaque assay (RRV and EW × RRV reassortants) or qRT-PCR (EW) and expressed as PFU per gram of tissue (RRV and EW × RRV reassortants) or PFU equivalent per gram of tissue (EW).
R represents a gene from RRV, and E represents a gene from EW.
Multiple regression analysis indicated that RV gene 5 is correlated with the level of RV titer in the intestines (regression coefficient, 2.857; 95% confidence interval, 1.390–4.323; P < 0.05).
A titer value of 30 was assigned to intestinal tissue samples that had undetectable levels of virus.
The role of VP4 in replication of RV in the intestine.
All tested EW × RRV reassortants contained RRV gene 4 encoding VP4, which conveys the cell-culture adaptation phenotype; therefore, the role of EW VP4 in regulating the intestinal growth phenotype of the reassortants could not be directly assessed. However, we were able to compare the intestinal replication of the monoreassortant D6/2 (entirely EW except for RRV gene 4) to the replication of wild-type EW (Fig. 2). D6/2 replicated approximately 10-fold less well than EW over the course of infection. This modest difference in replication was statistically significant at days 4 and 6 postinfection (P < 0.01).
Fig 2.
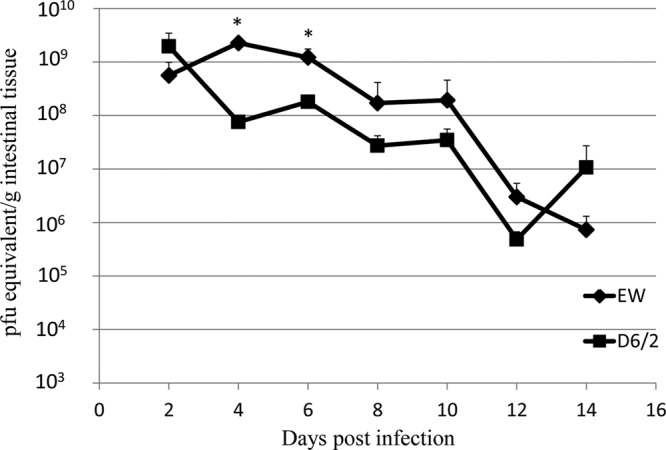
Intestinal titers of indicated RV in suckling 129sv mice infected with EW or EW × RRV monoreassortant D6/2. Five-day-old suckling 129sv mice were orally inoculated with wild-type EW (104 DD50) or D6/2 (104 PFU). At different days postinfection, mice (n = 3) were sacrificed and intestinal tissues collected. Virus titers in intestines were determined by qRT-PCR and are expressed as PFU equivalents per gram of tissue. *, the virus titers in EW-infected mice were significantly higher than in D6/2-infected mice at days 4 and 6 postinfection (P < 0.01).
To further evaluate the potential role of VP4 in RV replication in the intestine, we took advantage of several previously described reassortants derived from the cross of the simian RRV and the bovine UK strain (14, 24). Previously, we demonstrated that UK VP4 and UK NSP1 restricted RRV replication in the mouse biliary tract and that this restriction was determined by the inabilities of UK VP4 to mediate viral entry into murine cholangiocytes and UK NSP1 to suppress the host IFN response (14). Recent studies have shown that RRV replication in the murine intestine is substantially enhanced in the absence of STAT1 and an intact IFN response (27). RRV replication in the murine intestine of STAT I KO mice is 10- to 100-fold less efficient than replication of murine rotavirus (27) (Fig. 3), but in STAT I KO mice replication of RRV is much more efficient than in wild-type mice. In the intestines of wild-type and STAT I KO mice, UK replication was highly restricted (Table 3). Therefore, we studied intestinal replication of UK, RRV, and selected UK × RRV reassortants with or without UK VP4 in STAT1 KO mice (Table 3). In this model, the host IFN response does not interfere with RV replication, and we were able to directly examine the specific role of VP4 in intestinal replication. We found that, like the bovine UK parent, monoreassortant 19-1-2, which is entirely derived from RRV except for UK VP4, had a highly restricted growth phenotype in the STAT I KO mice while monoreassortant 27-3-1, which is derived from RRV except for UK VP1, replicated at levels similar to those of the RRV parent (P > 0.05) (Table 3). These results strongly suggest that heterologous VP4 proteins can differ substantially (i.e., RRV VP4 versus UK VP4) in their restricting capacity in the intestine. Replacement of EW VP4 with simian RRV VP4 had only a modest effect on intestinal replication in the mouse (Fig. 2), whereas UK VP4 was profoundly restricting (Table 3).
Fig 3.
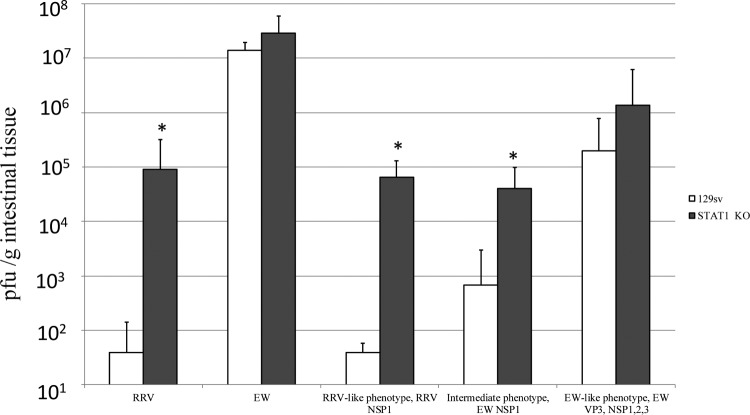
Comparison of the titers of various RVs in the intestine of wild-type and STAT1 KO 129 suckling mice. Five-day-old wild-type 129sv or STAT1 KO mice were orally inoculated with wild-type EW (104 PFU), RRV (104 PFU), or EW × RRV reassortants (104 PFU). At day 3 postinfection, mice (n = 4 to 13 per virus strain) were sacrificed and intestinal tissues were collected. Virus titers in intestines were determined by qRT-PCR (EW) or plaque assay (RRV or EW × RRV reassortants) and expressed as PFU equivalent (EW) or PFU (RRV or EW × RRV reassortants) per gram of tissue. The geometric means were calculated for each growth phenotype group. *, the virus titers were significantly different for RRV, the RRV-like phenotypic group, and the intermediate phenotypic group in wild-type versus STAT1 KO mice (P < 0.05); there was no significant difference between titers in wild-type and STAT1 KO mice for EW or the EW-like phenotypic group (P > 0.05).
Intestinal replication in the mouse intestine and the role of NSP1.
We next examined the intestinal replication of an additional set of EW × RRV reassortants in the 129sv pups. We analyzed the association between the heterologous or homologous origin of RV genes and the level of RV replication in the intestine (Table 2 and Fig. 3). We found that RV gene 5, encoding NSP1, was the only independent genetic factor that strongly influenced the levels of the intestinal replication among the reassortants (regression coefficient, 2.86; 95% confidence interval, 1.39 to 4.32; P = 0.02). All reassortants containing RRV gene 5 (regardless of how many other genes were derived from EW) were highly restricted for intestinal growth, with a geometric mean titer (GMT) of 39 (range, 30 to 77) that was similar or identical to that of RRV (P > 0.05) (Table 2). For example, reassortant D4/3, which has all EW genes except 4 and 5, replicated at levels similar to RRV whereas reassortant D6/2, which differs from D4/3 only in the origin of gene 5 (EW versus RRV), replicated like its EW parent. These findings strongly suggest that RRV NSP1 confers the RRV-like inefficient intestinal growth phenotype (Table 2 and Fig. 3).
Reassortants with EW gene 5 had two distinct intestinal replication phenotypes (Table 2 and Fig. 3). Reassortants such as D6/2, D1/5, C3/2, EA 4-1-2, and BE 1-1-3 had permissive EW-like growth phenotypes manifested by high virus yields on day 3 (GMT, 2.0 × 105; range, 1.0 × 105 to 12.3 × 105) in all inoculated pups. Broome et al. previously showed that some of the reassortants in this group were characterized by a low DD50 and inefficient spread to uninoculated littermates, a unique feature of homologous rotaviruses (12).
Another group of EW gene 5-containing reassortants, including D10/2, E4/1, A/11, EA 11-1-3, DEA 24-1-1, and DEA 3-1-2, had intermediate growth phenotypes in the intestine (Table 2 and Fig. 3). Reassortants in this group replicated at levels (GMT, 684; range, 194 to 6,162) that were significantly lower than those of EW-like reassortants (P < 0.05) but significantly higher than those of RRV-like reassortants (P < 0.05). These intermediate-phenotype reassortants were unable to spread infection to uninoculated littermates, as previously reported (12). Therefore, EW NSP1 appears necessary but not sufficient to confer a permissive murine rotavirus intestinal growth phenotype.
Reassortants with the permissive murine-like growth phenotype share several other EW genes in addition to EW NSP1. These include EW genes 3 (encoding VP3), 7 (encoding NSP3), and 8 (encoding NSP2). Of note, each of these EW genes also appeared in the intermediate growth reassortants but none of these intermediate phenotype reassortants had all three of these EW genes. These data support the conclusion that the constellation of EW genes 3, 5, 7, and 8 is required to confer the highly permissive EW-like intestinal growth phenotype. Interestingly, reassortant EA 4-1-2, generated by coinfection of suckling mice with two intermediate growth phenotype reassortant parents (E4/1 and A/11), had an EW-like high-titer intestinal growth phenotype and had the ability to transmit infection to uninoculated litter mates (data not shown). We back-crossed the EW-like reassortant EA 4-1-2 with the RRV-like reassortant D10/2 (all RRV genes except EW gene 5) in MA104 cells and isolated reassortants DEA 24-2-1 and DEA 3-1-2 (Table 2); both had intermediate growth phenotypes since one or another of the EW genes in the constellation in EA 4-1-2 was replaced by RRV genes. Results from these genetic studies support the conclusion that the EW-like intestinal growth phenotype requires a specific constellation of EW genes, with EW gene 5 having the dominant effect.
Intestinal replication of EW × RRV reassortants in STAT1 KO mice.
NSP1 is a virus-encoded antagonist to the type I IFN response (21, 22). We previously demonstrated that both RRV and ETD, an attenuated tissue culture-adapted strain of wild-type EW, are able to efficiently suppress the type I IFN response in vitro in MEFs and that this effect could be reproduced by NSP1 alone (25). To directly investigate the role of type I IFN signaling in the distinct replication phenotypes of RRV and EW in the intestine, we infected 5-day-old STAT I KO pups with EW, RRV, or EW × RRV reassortants and measured RV replication 3 days postinoculation. Unlike the wild-type 129sv mice, all STAT I KO mice infected with the parental RRV strain were infected and the GMT in the intestine (GMT, 9.1 × 104; range, 0.83 × 104 to 63.3 × 104) was over 2,000-fold higher than in wild-type mice (P < 0.0001) (Fig. 3). The GMT of EW RV in STAT1 KO mice (GMT, 28.8 × 106; range, 10.3 × 106 to 80.0 × 106) was only about 2 times higher than that in wild-type mice (P > 0.05) (Fig. 3). These findings reproduce similar studies we recently published (27).
STAT1 KO mice inoculated with the RRV-like reassortants were all infected on day 3. The GMTs, calculated using all reassortants in this group (GMT, 6.4 × 104; range, 1.7 × 104 to 15.6 × 104), were approximately 1,650 times higher than their titers in wild-type mice (P = 0.0002) (Fig. 3), similar to the fold increases in replication for RRV in STAT1 KO mice versus wild-type mice (P > 0.05). These results imply that the VP3-, NSP1-, NSP2-, and NSP3-related restriction of replication of the RRV-like reassortants in the mouse intestine is regulated by an IFN-mediated event, and removal of that restriction in the STAT I KO mice enabled these reassortants and the RRV parent to replicate efficiently.
As expected, the replication of all EW-like reassortants was readily detected in each of the STAT1 KO pups. Interestingly, the GMTs for this group of reassortants (GMT, 1.4 × 106; range, 0.1 × 106 to 11.7 × 106) were just 7-fold higher on average than the average GMTs in the wild-type mice (Fig. 3); the difference was not statistically significant (P > 0.05). The intermediate growth reassortants also infected all the STAT1 KO mice on day 3 postinfection. The average GMT (GMT, 4.0 × 104; range, 0.7 × 104 to 13.9 × 104) was 59 times higher than that in wild-type mice (P = 0.0006) (Fig. 3). In contrast to titers measured in wild-type mice (Table 2 and Fig. 3), titers of RRV-like reassortants in STAT1 KO mice were not significantly different from the titers of intermediate reassortants (P > 0.05), but titers of both groups were significantly lower than titers of mice infected with EW-like reassortants (P < 0.05) (Fig. 3).
DISCUSSION
The genetic basis of rotavirus host restriction has been studied in several animal model systems using various strains of RV originating from different homologous and heterologous host species (10–13, 15–17, 26). The genetic basis of restriction of RV replication and virulence in the gut is likely multigenic, and in previous studies the relative importance of specific viral genes was dependent on the choice of virus strain, host species, and phenotype (e.g., virulence, replication, and spread) studied. The purpose of this study was to systematically analyze the contributions of RV genes to the level of RV replication in the suckling mouse intestine. While the genetic and mechanistic basis of RV host range restriction is of considerable interest from a purely biologic vantage point, it also has substantial clinical relevance given that this restriction forms the basis for several human rotavirus vaccines.
Mice are susceptible to several strains of murine rotavirus, and infection causes a diarrheal disease in suckling mice. To carry out this study, we used a library of genetic reassortant RVs generated from mixed infection between the highly infectious and virulent wild-type murine EW and heterologous simian RRV strains (12). In our original studies, enteric virulence was defined by a low DD50 and the ability to transmit infection from an inoculated pup to uninoculated littermates. These phenotypes characterize the murine EW strain (and other wild-type murine strains) but not heterologous RVs. EW gene 5, encoding NSP1, was previously found to have a statistical but not an absolute association with this phenotype (12). In the present set of experiments, we quantitatively measured the levels of replication in the intestines of suckling mice after homologous or heterologous infection. In this system, host range restriction is profound (>10,000-fold). Note that among various heterologous RVs, the simian RRV strain replicates very efficiently; most other heterologous strains would have even larger differences in replication capacity than murine RVs (4, 26).
RV surface protein VP4 has been linked to RV strain- and species-specific infection in several animal models (11, 15, 16). In this study, all EW × RRV reassortants contained RRV VP4 because this gene conveys the cell culture adaptation phenotype to reassortants derived from the wild-type, non-cell culture-adapted murine parent (12). Therefore, the direct effect of EW VP4 on replication could not be evaluated. However, we found that D6/2, a monoreassortant carrying only VP4 from RRV, replicated at levels approximately 10-fold lower (P < 0.01 on days 4 and 6 postinfection) than the wild-type EW parent, implying that RRV VP4 mediates relatively effective viral entry into mouse enterocytes (Fig. 2). For unknown reasons, we were unable to isolate reassortants containing UK VP4 from the cross of EW with UK bovine RV. Therefore, we took advantage of the fact that the heterologous simian RRV replicates with high efficiency in STAT I KO mice where the effects of IFN restriction are removed (27) (Fig. 3). In marked contrast to RRV VP4, we found, using a UK VP4 monoreassortant on an RRV backbone, that VP4 derived from the bovine UK strain severely restricted RV replication in the intestine of STAT I KO mice (Table 3). Therefore, it appears that the RV surface protein VP4 acts as a modest to very strong determinant of host range restriction, depending on the species origin of the heterologous VP4. This finding is consistent with those of previous reports (11, 15, 16).
RV VP4 must be enzymatically cleaved into VP5* and VP8* in order to effectively mediate cell entry and infection (32, 33). Recent crystallographic and functional studies have demonstrated that VP8*, the distal head domain of VP4, contains a cleft structure with strain-specific binding affinity for different gangliosides or, in humans, for the type A human blood group antigen present on the target cell surface (34–38). The different replication capacities mediated by mouse, simian, and bovine VP4s may result from different affinities of the processed VP8s* for unique gangliosides or other glycan receptors on the surface of mouse intestinal epithelial cells. The nature of RRV, UK, and EW glycan receptors on mouse intestinal epithelial cells is currently unknown. It is also possible that the VP4-associated replication differences seen here are related to different efficiencies of post-binding entry functions.
In the present study, we found that RV NSP1 was an independent and necessary factor that strongly influenced the levels of RV replication in the mouse intestine, consistent with our prior observations (12). All reassortants containing RRV NSP1 had minimal enteric replication capacity regardless of the other EW murine genes present (Table 2); however, although EW NSP1 was essential it was not sufficient for the permissive replication phenotype in mouse intestine. Similar to our observations, Ciarlet et al. showed that the lapine RV NSP1 on an otherwise simian SA11 background was not sufficient to confer the homologous lapine RV replication phenotype (13). Similarly, Bridger et al. reported that the growth characteristics of a monoreassortant of a porcine virus (SW/20/21) with bovine UK NSP1 were similar to those of the permissive porcine parent, rather than to those of the restricted UK parent, in a gnotobiotic pig study (10).
NSP1s derived from different animal RV strains differ in their abilities to degrade IRF3 and suppress the IFN response in cultured cells such as MEFs (25). For instance, NSP1 from the bovine UK strain does not suppress the IFN response in wild-type MEFs (24, 25). In contrast, NSP1s from both RRV and EW do efficiently degrade IRF3 and suppress the host IFN response in wild-type MEFs (24, 25). Under some circumstances, RRV NSP1 is able to suppress the host IFN response in vivo. For example, in a mouse model of systemic RV replication, where RV was directly injected into the gallbladder of suckling mice and replication measured 16 h later, RRV NSP1 (but not UK NSP1) and RRV VP4 were associated with productive replication in the biliary tract and the level of replication was not significantly increased in IFNR KO or STAT1 KO mice (14). These results suggest that RRV NSP1 can efficiently block the IFN response at a systemic site in a mouse, the biliary tree. However, in this study, replication (3 days postinfection) of RRV or any EW × RRV reassortants containing RRV NSP1 in the mouse intestine was very restricted in wild-type mice. This restriction was mediated by RRV NSP1 and substantially eliminated in STAT I KO mice (Fig. 3). These findings support the conclusion that host replication restriction in the intestine is linked to the inability of the heterologous NSP1 to inhibit the local IFN response. EW NSP1 appears to be much more efficient at antagonizing the innate immune response than RRV NSP1 in mouse intestines; how simian RRV efficiently circumvents the response in the biliary tree is not understood. In STAT1 KO mice, the levels of replication for EW or reassortants containing EW NSP1 were not significantly enhanced. These findings are consistent with our recent studies that demonstrated that infection of mouse intestine with murine but not simian RV resulted in accumulation of IκB-α protein and decreased transcription of NF-κB-dependent genes, presumably via the degradation of β-TrCP (27).
It is also clear that NSP1 alone is not sufficient to confer a permissive homologous RV enteric replication phenotype. In our studies, additional murine RV genes facilitated the high replication phenotype of murine RV in the mouse intestine. We found that all EW reassortants that replicated efficiently shared an EW gene constellation containing EW genes 3 (VP3), 5 (NSP1), 7 (NSP3), and 8 (NSP2). Reassortants with this murine RV gene constellation, such as EA 4-1-2, which was derived from in vivo coinfection of two intermediate growth reassortants, E4/1 and A/11, replicated efficiently in suckling mice (Table 2). Removal of any one EW gene from this constellation by backcrossing EA 4-1-2 with D10/2 in vitro (as in the case of DEA 24-2-1 and DEA 3-2-1) resulted in reversion of the reassortant progeny to an intermediate growth phenotype. Therefore, the constellation of EW genes 3, 5, 7, and 8 is not only necessary but also sufficient for the permissive enteric growth phenotype of the homologous EW murine RV. Of interest, the replication titers of EW-like reassortants, which all have VP4 derived from RRV, were significantly (approximately 10× to 50×) lower than those of wild-type EW (Table 2 and Fig. 3). It is likely that the addition of EW VP4 to these EW-like reassortants would rescue the full intestinal-replication capacity of EW reassortants. However, because the wild-type EW VP4 restricts the reassortant's tissue culture adaptability, this hypothesis could not be directly tested.
The mechanistic basis for how this specific murine RV gene constellation produces the EW-like permissive replication phenotype is currently unclear. The constellation likely functions, at least in part, to interfere with the inhibitory effects of the host IFN response. Support for this conclusion comes from the finding that replication of the EW-like reassortants containing this gene constellation was not significantly increased in STAT I KO mice, whereas replication of intermediate growth phenotype reassortants containing EW NSP1, but not the full constellation, was increased more than 50-fold (Fig. 3). In addition, the EW genes in the constellation may simply function better together than other mixed combinations of EW and RRV genes to promote replication in mouse enterocytes. However, if this is the case, the constellation effect is very specific to intestinal cell replication since it was not reproduced in cell culture (data not shown).
VP3 is a guanylyl and methyltrasferase that is responsible for 5′ capping of newly synthesized viral mRNA during initial infection (39, 40). VP3 could be indirectly involved in modulating the innate immune response since enhanced efficiency of capping RV mRNA may result in reduced detection by cellular pathogen recognition receptors (PRR) such as RIG-I (H. Greenberg, unpublished observation). Of note, VP3 is associated with the host range restriction in the pig model (15), and it was recently shown that VP3 sequences cluster based on the host species of origin, supporting the notion that VP3s from different hosts may differ in function(s) (41).
RV NSP2 is a more conserved viral protein than VP3 or NSP1; however, there is still a 10% nonconservative protein sequence difference between RRV and EW NSP2. NSP2 functions with VP2 and NSP5 in viroplasm formation during viral replication and genome packaging (42). No host-specific effects of NSP2 have been previously identified.
The final protein in our identified constellation, NSP3, interacts with the host cell translation initiation factor eIF4G and a conserved 3′ sequence in viral mRNA to facilitate viral protein translation. NSP3 also disrupts the interaction between eIF4G and a cellular poly(A)-binding protein (PABP), thereby inhibiting translation of host cell mRNAs. Previously, Mossel and Ramig demonstrated that RRV NSP3 was a determinant of enhanced extraintestinal spread to liver in a mouse model of heterologous RV replication (43). The mechanistic basis by which VP3 or NSP2 and NSP3 work together to affect intestinal replication in a host-specific manner requires further investigation.
In summary, we demonstrated here the multigenic nature of RV host range restriction using a series of reassortants between homologous EW and heterologous RRV as well as between the two heterologous RV strains, RRV and UK. We found a clear association of both NSP1 and VP4 with host-specific intestinal replication. The restricting effects of heterologous simian NSP1 are largely negated in STAT I KO mice, implying that the host range restriction abilities of NSP1 are mediated via effects on the innate immune system. Although we do not have direct data demonstrating that the effects of VP4 on host restriction are related to viral binding or entry related, this seems likely given our finding that such a mechanistic effect on replication is present in the biliary tree (14). Further studies will be needed to determine the mechanisms by which VP3, NSP2, and NSP3 complement the effects of NSP1.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 AI012362-24 and P30DK56339 and a merit review VA grant.
We are grateful for the technical assistance of P. Vo.
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Estes M, Kapikian AZ. 2007. Rotaviruses, p 1917–1974 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Ciarlet M, Schodel F. 2009. Development of a rotavirus vaccine: clinical safety, immunogenicity, and efficacy of the pentavalent rotavirus vaccine, RotaTeq. Vaccine 27(Suppl)6:G72–G81 [DOI] [PubMed] [Google Scholar]
- 3. Fenaux M, Cuadras MA, Feng N, Jaimes M, Greenberg HB. 2006. Extraintestinal spread and replication of a homologous EC rotavirus strain and a heterologous rhesus rotavirus in BALB/c mice. J. Virol. 80:5219–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramig RF. 1988. The effects of host age, virus dose, and virus strain on heterologous rotavirus infection of suckling mice. Microb. Pathog. 4:189–202 [DOI] [PubMed] [Google Scholar]
- 5. Ward LA, Rosen BI, Yuan L, Saif LJ. 1996. Pathogenesis of an attenuated and a virulent strain of group A human rotavirus in neonatal gnotobiotic pigs. J. Gen. Virol. 77(Pt 7):1431–1441 [DOI] [PubMed] [Google Scholar]
- 6. Clark HF, Furukawa T, Bell LM, Offit PA, Perrella PA, Plotkin SA. 1986. Immune response of infants and children to low-passage bovine rotavirus (strain WC3). Am. J. Dis. Child. 140:350–356 [DOI] [PubMed] [Google Scholar]
- 7. Flores J, Daoud G, Daoud N, Puig M, Martinez M, Perez-Schael I, Shaw R, Greenberg HB, Midthun K, Kapikian AZ. 1988. Reactogenicity and antigenicity of rhesus rotavirus vaccine (MMU-18006) in newborn infants in Venezuela. Pediatr. Infect. Dis. J. 7:776–780 [DOI] [PubMed] [Google Scholar]
- 8. Vesikari T, Isolauri E, D'Hondt E, Delem A, Andre FE, Zissis G. 1984. Protection of infants against rotavirus diarrhoea by RIT 4237 attenuated bovine rotavirus strain vaccine. Lancet i:977–981 [DOI] [PubMed] [Google Scholar]
- 9. Vesikari T, Kapikian AZ, Delem A, Zissis G. 1986. A comparative trial of rhesus monkey (RRV-1) and bovine (RIT 4237) oral rotavirus vaccines in young children. J. Infect. Dis. 153:832–839 [DOI] [PubMed] [Google Scholar]
- 10. Bridger JC, Dhaliwal W, Adamson MJ, Howard CR. 1998. Determinants of rotavirus host range restriction—a heterologous bovine NSP1 gene does not affect replication kinetics in the pig. Virology 245:47–52 [DOI] [PubMed] [Google Scholar]
- 11. Bridger JC, Tauscher GI, Desselberger U. 1998. Viral determinants of rotavirus pathogenicity in pigs: evidence that the fourth gene of a porcine rotavirus confers diarrhea in the homologous host. J. Virol. 72:6929–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Broome RL, Vo PT, Ward RL, Clark HF, Greenberg HB. 1993. Murine rotavirus genes encoding outer capsid proteins VP4 and VP7 are not major determinants of host range restriction and virulence. J. Virol. 67:2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ciarlet M, Estes MK, Barone C, Ramig RF, Conner ME. 1998. Analysis of host range restriction determinants in the rabbit model: comparison of homologous and heterologous rotavirus infections. J. Virol. 72:2341–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng N, Sen A, Wolf M, Vo P, Hoshino Y, Greenberg HB. 2011. Roles of VP4 and NSP1 in determining the distinctive replication capacities of simian rotavirus RRV and bovine rotavirus UK in the mouse biliary tract. J. Virol. 85:2686–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoshino Y, Saif LJ, Kang SY, Sereno MM, Chen WK, Kapikian AZ. 1995. Identification of group A rotavirus genes associated with virulence of a porcine rotavirus and host range restriction of a human rotavirus in the gnotobiotic piglet model. Virology 209:274–280 [DOI] [PubMed] [Google Scholar]
- 16. Offit PA, Blavat G, Greenberg HB, Clark HF. 1986. Molecular basis of rotavirus virulence: role of gene segment 4. J. Virol. 57:46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang W, Donnelly B, Bondoc A, Mohanty SK, McNeal M, Ward R, Sestak K, Zheng S, Tiao G. 2011. The rhesus rotavirus gene encoding VP4 is a major determinant in the pathogenesis of biliary atresia in newborn mice. J. Virol. 85:9069–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taniguchi K, Kojima K, Urasawa S. 1996. Nondefective rotavirus mutants with an NSP1 gene which has a deletion of 500 nucleotides, including a cysteine-rich zinc finger motif-encoding region (nucleotides 156 to 248), or which has a nonsense codon at nucleotides 153-155. J. Virol. 70:4125–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dunn SJ, Cross TL, Greenberg HB. 1994. Comparison of the rotavirus nonstructural protein NSP1 (NS53) from different species by sequence analysis and northern blot hybridization. Virology 203:178–183 [DOI] [PubMed] [Google Scholar]
- 20. Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gomara M, Maes P, Patton JT, Rahman M, Van Ranst M. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 82:3204–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barro M, Patton JT. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. U. S. A. 102:4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barro M, Patton JT. 2007. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J. Virol. 81:4473–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Graff JW, Ettayebi K, Hardy ME. 2009. Rotavirus NSP1 inhibits NFkappaB activation by inducing proteasome-dependent degradation of beta-TrCP: a novel mechanism of IFN antagonism. PLoS Pathog. 5:e1000280. 10.1371/journal.ppat.1000280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng N, Sen A, Nguyen H, Vo P, Hoshino Y, Deal EM, Greenberg HB. 2009. Variation in antagonism of the interferon response to rotavirus NSP1 results in differential infectivity in mouse embryonic fibroblasts. J. Virol. 83:6987–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sen A, Feng N, Ettayebi K, Hardy ME, Greenberg HB. 2009. IRF3 inhibition by rotavirus NSP1 is host cell and virus strain dependent but independent of NSP1 proteasomal degradation. J. Virol. 83:10322–10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng N, Kim B, Fenaux M, Nguyen H, Vo P, Omary MB, Greenberg HB. 2008. Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J. Virol. 82:7578–7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sen A, Rothenberg ME, Mukherjee G, Feng N, Kalisky T, Nair N, Johnstone IM, Clarke MF, Greenberg HB. 2012. Innate immune response to homologous rotavirus infection in the small intestinal villous epithelium at single-cell resolution. Proc. Natl. Acad. Sci. U. S. A. 109:20667–20672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoshino Y, Wyatt RG, Greenberg HB, Flores J, Kapikian AZ. 1984. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J. Infect. Dis. 149:694–702 [DOI] [PubMed] [Google Scholar]
- 29. Burns JW, Krishnaney AA, Vo PT, Rouse RV, Anderson LJ, Greenberg HB. 1995. Analyses of homologous rotavirus infection in the mouse model. Virology 207:143–153 [DOI] [PubMed] [Google Scholar]
- 30. Sen A, Pruijssers AJ, Dermody TS, Garcia-Sastre A, Greenberg HB. 2011. The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS-1, RIG-I, MDA-5, and IRF3. J. Virol. 85:3717–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng N, Burns JW, Bracy L, Greenberg HB. 1994. Comparison of mucosal and systemic humoral immune responses and subsequent protection in mice orally inoculated with a homologous or a heterologous rotavirus. J. Virol. 68:7766–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clark SM, Roth JR, Clark ML, Barnett BB, Spendlove RS. 1981. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J. Virol. 39:816–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Estes MK, Graham DY, Mason BB. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dormitzer PR, Sun ZY, Wagner G, Harrison SC. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haselhorst T, Fleming FE, Dyason JC, Hartnell RD, Yu X, Holloway G, Santegoets K, Kiefel MJ, Blanchard H, Coulson BS, von Itzstein M. 2009. Sialic acid dependence in rotavirus host cell invasion. Nat. Chem. Biol. 5:91–93 [DOI] [PubMed] [Google Scholar]
- 36. Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. 2012. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485:256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu X, Coulson BS, Fleming FE, Dyason JC, von Itzstein M, Blanchard H. 2011. Novel structural insights into rotavirus recognition of ganglioside glycan receptors. J. Mol. Biol. 413:929–939 [DOI] [PubMed] [Google Scholar]
- 38. Yu X, Dang VT, Fleming FE, von Itzstein M, Coulson BS, Blanchard H. 2012. Structural basis of rotavirus strain preference toward N-acetyl- or N-glycolylneuraminic acid-containing receptors. J. Virol. 86:13456–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen D, Luongo CL, Nibert ML, Patton JT. 1999. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology 265:120–130 [DOI] [PubMed] [Google Scholar]
- 40. Liu M, Mattion NM, Estes MK. 1992. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology 188:77–84 [DOI] [PubMed] [Google Scholar]
- 41. Subodh S, Bhan MK, Ray P. 2006. Genetic characterization of VP3 gene of group A rotaviruses. Virus Genes 33:143–145 [DOI] [PubMed] [Google Scholar]
- 42. Taraporewala ZF, Patton JT. 2004. Nonstructural proteins involved in genome packaging and replication of rotaviruses and other members of the Reoviridae. Virus Res. 101:57–66 [DOI] [PubMed] [Google Scholar]
- 43. Mossel EC, Ramig RF. 2002. Rotavirus genome segment 7 (NSP3) is a determinant of extraintestinal spread in the neonatal mouse. J. Virol. 76:6502–6509 [DOI] [PMC free article] [PubMed] [Google Scholar]