Abstract
Pseudomonas aeruginosa myovirus ϕKZ has a 270-kb genome within a T=27 icosahedral capsid that contains a large, unusual, and structurally well-defined protein cylindrical inner body (IB) spanning its interior. Proteolysis forms a pivotal stage in ϕKZ head and IB morphogenesis, with the protease gp175 cleaving at least 19 of 49 different head proteins, including the major capsid protein and five major structural IB proteins. Here we show that the purified mature form of gp175 is active and cleaves purified IB structural proteins gp93 and gp89. Expression vector synthesis and purification of the zymogen/precursor yielded an active, mature-length protease, showing independent C-terminal gp175 self-cleavage autoactivation. Mutation of either the predicted catalytic serine or histidine inactivated mature gp175, supporting its classification as a serine protease and representing the first such direct biochemical demonstration with purified protease and substrate proteins for any phage protease. These mutations also blocked self-cleavage of the precursor while allowing intermolecular gp175 processing. To confirm the cleavage specificity of gp175, we mutated three cleavage sites in gp93, which blocked proteolysis at these sites. The N-terminal propeptide of gp93 was shown to undergo more extensive proteolysis than previously identified. We found that proteolysis in gp93 progressed from the N to C terminus, while blocking cleavage sites slowed but did not eliminate downstream proteolysis. These findings were shown by informatics to be relevant to the head morphogenesis of numbers of other related IB-containing giant phages as well as to T4 and herpesviruses, which have homologous proteases.
INTRODUCTION
The giant myovirus ϕKZ, which is infective for Pseudomonas aeruginosa, is a member of the ϕKZ-related phages that currently incorporate two genera, the ϕKZ-like phages (ϕKZ, ϕPA3, and 201ϕ2-1) and the more distantly related EL-like phages (EL and OBP) (1–3). These phages are distributed widely geographically and are of interest because those infective for pathogens have been used for phage therapy (4) and/or have potential for further biocontrol applications (5–8). These phages are also a curiosity, as they are remarkably complex, both at the genome and structural levels, with large virions comprised of a T=27 capsid or head (9) and contractile tail that ends in an elaborate baseplate (10, 11). The complexity of the virion is reflected in the numbers of different proteins, >60 different proteins, with nearly 50 different proteins present in the head alone, possibly the highest number for any known group of viruses (12–14). A remarkable feature of the ϕKZ-related phages is that within the head there is a large, protein cylinder called an inner body (IB) around which the DNA is tightly spooled (15). The structure of the ϕKZ IB was recently solved, revealing it to be approximately 24 by 105 nm and tilted at an angle of approximately 22° relative to the portal axis (16). Its structure is also regular, with an apparent 6-fold symmetry in some regions. The question remains as to the function(s) of the IB; potential roles could relate to DNA organization, as the IB aligns with the axis of the spooled DNA, or host takeover, as the IB disappears when the DNA is ejected into the host cell head (14, 15). Other potential roles could be related to head size determination, head stability, or DNA packaging. Questions also remain as to the definitive composition of the IB, although six abundant proteins (gp89, gp90, gp93, gp95, gp97, and gp162) are expected to form the major density of this 15- to 20-MDa structure (14).
Despite these challenging problems that are yet to be solved, a fascinating feature of the ϕKZ head and IB is that the morphogenetic pathway by which they are formed (Fig. 1) apparently shares conserved features with that of the phage T4. In T4, head morphogenesis consists of a series of steps through which a protein-filled prohead is converted to a protein shell that houses the densely packaged double-stranded DNA genome (but in which there is no IB, as is found in ϕKZ) (17). Initially, in T4 a spherically shaped immature prohead forms on the host inner membrane, comprised of a shell made of 960 copies of the major capsid protein (MCP), that encompasses a core of the major scaffold protein gp22, gp67, and gp68, and the other inner head proteins (including Alt and the high-copy-number internal proteins IPI, IPII, and IPIII) present in the mature head (18). At this point the prohead protease gp21 undergoes self-cleavage to its mature, active form and then proceeds to proteolyze all head proteins other than the portal. This proteolysis includes removal of 65 residues from the N terminus of the MCP and short N-terminal fragments (7 to 19 residues) from the IP proteins and Alt. The essential core proteins, gp22, gp67, and gp68, are extensively degraded to short peptides. Cleavage of the MCP initiates the expansion of the head to its mature dimensions, and during this transition the proteolyzed fragments escape from the head, presumably through small pores in the shell. These processes contrast with those for phages P22 (19), T7 (20), and ϕ29 (21), in which the intact scaffold protein apparently exits through the procapsid shell. These are pivotal steps in head morphogenesis, creating volume within the head and preparing it for the next major step, the packaging of the genomic DNA into the head.
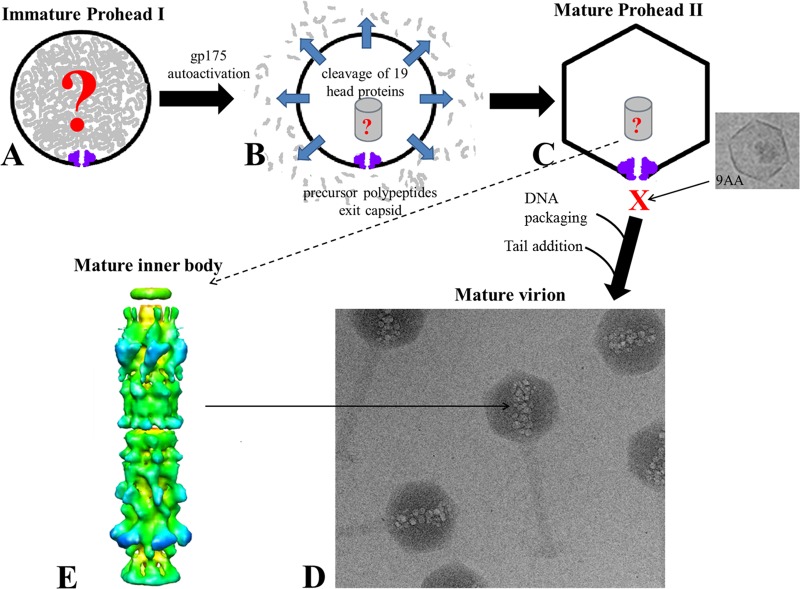
Fig 1.
Schematic of morphogenesis of the ϕKZ head. (A) Immature prohead I, composed of 49 different proteins. (B) The protease gp175 undergoes autoactivation and cleaves 19 head proteins. (C) Processed prohead II contains all the proteins found in the mature capsid, including six abundant IB proteins. A structure which may represent an immature form of the IB, or breakdown product of it, has been observed by cryo-electron microscopy (cryo-EM) of prohead II. (D) Cryo-EM image of mature ϕKZ particles, showing IB bubbles in the head. (E) Reconstruction of the mature IB (previously published in reference 16 and provided here with permission of the publisher).
Nineteen of the ϕKZ head proteins undergo proteolysis, including the major head and the portal proteins (14). It can also be inferred that as five of the six major IB structural proteins are processed with 50% of their combined molecular weights removed, the sculpting of the mature IB structure is another important, possibly essential, role of the prohead protease (Fig. 1). For these ϕKZ head proteins, cleavage always occurs after a glutamic acid residue at the motif S/A/G-X-E, which resembles that of the cleavage motif of T4 gp21 (I/L-X-E) (14). Identification of the ϕKZ protease revealed that it is distantly related at the sequence level to the T4 protease, and both proteins are predicted to be serine proteases and likely share an ancestor with members of the herpesvirus protease family (22). Like T4 gp21, ϕKZ gp175 is predicted to undergo autoactivation by a self-cleavage event, as it was found to have 70 residues removed from its C terminus and an assay of this protein showed it to be active (14). However, the ϕKZ protease has an apparently more complex job description than that of the T4 protease, partly due to its role in IB formation but also simply because it has more substrates to cleave. In addition, some of its substrates have complex cleavage patterns, such as gp89 and gp97, which have central regions removed while N- and C-terminal fragments remain in the head. Presumably, as in T4, the major function of proteolysis in the ϕKZ head is to remove proteins, or portions of them, that have roles in prohead formation but are not required in the mature head. Additionally, the ϕKZ protease may also have a role in the activation of other enzymes, in addition to itself, as it cleaves a subunit of the virion RNA polymerase and helicase (gp203) (14).
Despite the pivotal role that ϕKZ gp175 has in head morphogenesis, little is known about it at either the structural or enzymatic levels. For instance, how and when it is incorporated into the prohead, and where it is located in the head throughout the morphogenesis process, are unknown. Similarly, little is known definitively about gp175's substrate specificity, other than that it cleaves a motif that is found fairly frequently in most proteins and that some such motifs are not cleaved (14). Also unknown are what factors, if any, control proteolysis by gp175 such that it occurs at the correct time in head morphogenesis, or what controls gp175 self-activation, and also, finally, whether gp175 is autoinactivated? To begin the process of resolving these questions, we sought to express and purify the ϕKZ protease and investigate its activity in vitro on the two inner body proteins, gp93 and gp89.
MATERIALS AND METHODS
Cloning of ϕKZ gp89, gp92, and gp175 into the pHERD20T vector.
Genomic DNA was extracted from CsCl gradient-purified ϕKZ virions by using standard methods (23). The genes encoding ϕKZ gp89, gp93, and gp175 (both mature and precursor forms) were amplified using Taq polymerase (NEB) for PCR. Primers used are listed in Table 1. The NcoI and KpnI restriction sites were used for gp93, and the NcoI and XbaI sites were used for gp175 and gp89. For gp93, the forward primer was designed so that the second codon was altered from TCT to GCT as a consequence of NcoI incorporating the gene's start codon, changing the second amino acid of gp93 from serine to alanine. Similarly, the forward primer for gp89 encoded a changed second codon (CCA to GCA) that made the second residue of gp89 an alanine instead of a proline. No change to the second codon of gp175 was required.
Table 1.
Primers used for cloning of ϕKZ genes encoding gp89, gp93, and both precursor and mature forms of gp175 into the expression vector pHERD20T
| Gene | Coordinatesa | Amplicon size (bp) | Primer name | Primer sequence (5′–3′)b | Function |
|---|---|---|---|---|---|
| gp89 | 88994–90157 | 1,164 | KZ189pHERDF | GCGCCATGGCATTACCTGTAATCGCCGGAGCAGTA | Forward primer |
| KZ189pHERDR | GCGTCTAGATTAAATATTGGTAGACATGGTTTTATTC | Reverse primer | |||
| gp93 | 92980–94287 | 1,308 | KZ93F-A | GCGCCATGGCTCTACTTAAAATGCTAGACA | Forward primer |
| KZ93R | GCGGGTACCTTACTTCTCATCATTCTCAGG | Reverse primer | |||
| gp175 precursor | 178350–179162 | 813 | KZ175pHERDF | GCGCCATGGAACCATATTCTAATTTTTCTGGT | Forward primer |
| KZ175pHERDR | CGGTCTAGATTAACGCATGAAGCCAGGGCG | Reverse primer | |||
| gp175 mature | 178533–179162 | 633 | KZ175pHERDR_PALE | GCGTCTAGATTACTCTAATGCAGGGGAATTCCATTTA | Reverse primer |
Gene coordinates are from GenBank accession number AF399011.1.
Start and stop codons are underlined.
PCR products were purified using either the QIAquick gel extraction kit (Qiagen) or the QIAquick PCR purification kit (Qiagen), digested with the appropriate restriction enzymes, repurified using the QIAquick kit, and then ligated into the expression vector pHERD20T (24), which had also been cleaved by the appropriate enzymes and purified. Ligation mixes were transformed into chemically competent DH10B cells and plated onto LB plates containing 100 μg/ml ampicillin and spread with 20 μl each of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (80 μg/ml) and arabinose (10%). White colonies were picked and propagated. Plasmids were purified using the QIAprep spin miniprep kit (Qiagen), screened for the presence of an appropriately sized insert, and then sequenced using the pHERD sequencing primers (pHERDSF and pHERDSR) designed by Qiu et al. (24) at the Biopolymer Core Facility (UMB). Sequencing of the wild-type gp93 gene identified two synonymous changes, C28T and A525G, encoding L10 and K175, respectively.
To be able to perform nickel column purification of each protein, six histidine codons were introduced immediately downstream of the start codon of each gene via site-directed mutagenesis (SDM). Primers used for the His tag additions are listed in Table 2. SDM was used to introduce mutations in the codons for catalytic site residues (H-89 and S-168) in the gp175 constructs either both mature or precursor forms. SDM was also used to introduce E-to-D cleavage site mutations in gp93. Primers used for SDM are also listed in Table 2. PfuUltra Hotstart DNA polymerase (Agilent) was used for the amplifications, and subsequent digestion by DpnI (NEB) was undertaken to remove any remaining template DNA. Plasmid DNAs were purified using the QIAprep spin miniprep kit, and mutations in gp175 and gp93 were verified by DNA sequencing using the pHERD sequencing primers (24).
Table 2.
Primers used for introducing a polyhistidine tag into ϕKZ gp89, gp93, and gp175 and primers used for site-directed mutagenesis of gp93 and gp175a
| Primer use and gene | Primer name | Primer sequence (5′–3′) | Function |
|---|---|---|---|
| Addition of His tagb | |||
| gp89 | pH6H89F | GGAGATATACATACCCATGCATCATCATCATCATCATGCATTACCTGTAATCGCCGGAGCAGTAAG | Forward primer |
| pH6H89R | CTTACTGCTCCGGCGATTACAGGTAATGCATGATGATGATGATGATGCATGGGTATGTATATCTCC | Reverse primer | |
| gp93 | pH6H93F | AAGAAGGAGATATACATACCCATGCATCATCATCATCATCATGCTCTACTTAAAATGCTAGACAAAC | Forward primer |
| pH6H93R | GTTTGTCTAGCATTTTAAGTAGAGCATGATGATGATGATGATGCATGGGTATGTATATCTCCTTCTT | Reverse primer | |
| gp175 | pH6H175F | GGAGATATACATACCCATGCATCATCATCATCATCATGAACCATATTCTAATTTTTCTGGTTTGCAA | Forward primer |
| pH6H175R | TTGCAAACCAGAAAAATTAGAATATGGTTCATGATGATGATGATGATGCATGGGTATGTATATCTCC | Reverse primer | |
| Site-directed mutagenesis | |||
| gp93 | KZ93E13D-F | GACAAATTATCACTTGACTCAATTGAAGATCAAGGC | Forward primer for E13D |
| KZ93W13D-R | GCCTTGATCTTCAATTGAGTCAAGTGATAATTTGTC | Reverse primer for E13D | |
| E137Dfor E137Drev | CACTGCCGCGCTTGATAATTACGATGCTTC | Forward primer for E137D | |
| GAAGCATCGTAATTATCAAGCGCGGCAGTG | Reverse primer for E137D | ||
| E156Dfor | GCTGTTATCTCTCTTGATGATCTCAAAGAG | Forward primer for E156D | |
| E156Drev | CTCTTTGAGATCATCAAGAGAGATAACAGC | Reverse primer for E156D | |
| gp175 | KZ175H89VF | CATGGAATACAAAGTCCCTGAACCCTATC | Forward primer for H89V |
| KZ175H89VR | GATAGGGTTCAGGGACTTTGTATTCCATG | Reverse primer for H78V | |
| 175Mut168F | CAGAATACTTATTGCGCGGTAAGGTCAATAAC | Forward primer for S168A | |
| 175Mut168R | GTTATTGACCTTACCGCGCAATAAGTATTCTG | Reverse primer for S168A |
Primers for gp175 were used for the constructs encoding precursor and mature forms.
Primers for addition of six codons encoding a polyhistidine tag immediate after the start codon (underlined in sequences).
Protein expression and purification.
Constructs were propagated in LB broth containing 100 μg/ml of ampicillin until mid-log phase, and protein expression was induced by the addition of arabinose. Typical induction conditions for gp175 and gp93 were a 1% final concentration of arabinose, 1-h duration, 37°C. gp89 was insoluble under these conditions; hence, cells were propagated to mid-log phase at 30°C and then induced at 18°C overnight with 0.05% arabinose. Cells were pelleted at 6,000 rpm (Sorvall rotor SS34) for 10 min, then resuspended in lysis buffer (20 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1 mM EDTA, and egg white lysozyme [0.3 mg/ml; Sigma]), and left on ice until lysis occurred (∼1 h). Samples were supplemented with MgCl2 (1 mM) and treated with protease-free recombinant bovine pancreatic DNase (40 U/ml; Roche) at 37°C for 15 to 20 min. Samples underwent centrifugation (10,000 × g, 10 min), and the supernatant was mixed with HisPur Ni-nitrilotriacetic acid resin (Thermo Scientific) and placed on a rotary shaker overnight (4°C). Purification of each protein was performed according to the manufacturer's instructions at room temperature. Washes and elutions were performed in 20 mM Tris-Cl (pH 7.5), 300 mM NaCl. Wash buffer imidazole concentrations increased progressively from 10 to 50 mM imidazole, and the elution buffer contained 250 mM imidazole. Eluted proteins were dialyzed against the wash buffer containing no imidazole added. Samples were stored at 4°C and at −20°C (with 50% glycerol, final concentration).
Protease assays.
Protease assay mixtures typically contained 1 to 2 μg each of a form of the enzyme, gp175, and substrate (gp89 or gp93) in a 15-μl total reaction volume. Reactions were performed in the dialysis buffer (20 mM Tris-Cl [pH 7.5], 300 mM NaCl) for 30 min at 37°C. To detect processing intermediates (e.g., as shown in Fig. 6, below), the enzyme/substrate molar ratio was increased stepwise from 0.001 to 1. Assays were stopped by the addition of SDS-PAGE loading buffer (final concentrations, 50 mM Tris-Cl [pH 7.5], 2% SDS, 0.1% bromophenol blue, 10% glycerol) and immediately boiled (10 min). The entire reaction mixture for each assay was loaded onto a Novex mini (8 by 8 cm) 10 to 20% SDS-PAGE gel (Life Technologies) and underwent electrophoresis (15 min at 100 V, then 60 min at 150 V). SDS-PAGE gels were stained with Coomassie brilliant blue R-250 (Bio-Rad). The PageRuler Plus prestained protein ladder (Fermentas) was used as a molecular weight marker.
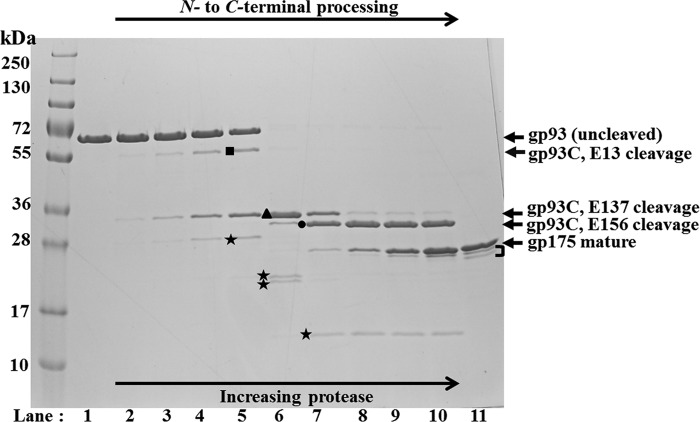
Fig 6.
SDS-PAGE of assay products of ϕKZ protease gp175 on gp93. Lanes: 1, gp93; 2 to 10, increasing amounts of gp175 (a 2-fold dilution series from ∼2 μg to 2 ng in reverse) from assays (37°C for 30 min) with gp93 (∼2 μg); 11, gp175. Bands expected to represent C-terminal fragments of gp93 resulting from cleavage at E-13, E-137, and E-156 are indicated with a black square, black triangle, and black circle, respectively. Stars indicate fragments produced from proteolysis of the N-terminal propeptide of gp93. The bracket indicates fragments potentially representing inactivated forms of the protease (see Discussion).
Sequence alignments.
Proteins homologous to ϕKZ gp175 were identified using BLASTP, aligned using the sequence and alignment modeling system (SAM) (25), and displayed using the GeneDoc multiple sequence alignment editor (available at http://www.nrbsc.org/gfx/genedoc/).
RESULTS
Purified mature and precursor forms of gp175 produce an active enzyme.
Mass spectral analyses indicated that the 270-residue precursor or zymogen form of ϕKZ gp175 self-cleaved to a 210-residue mature and active form (14) (Fig. 2). To verify this observation, both precursor and mature forms of gp175 were expressed with an N-terminal histidine tag and purified on a nickel column. Expression of the gp175 gene prematurely truncated with a stop codon to form the mature-length protein, and its subsequent purification produced a protein that banded during SDS-PAGE consistent with its predicted molecular mass (24 kDa) (Fig. 3A, lane 2). Notably, the purified protein expressed from the plasmid-carried precursor or full-length form of the gp175 gene migrated to the same position as the purified mature form of gp175 (Fig. 3B, lanes 1 and 2), indicating that the precursor form of gp175 had been cleaved to mature length during expression and/or purification. Examination of crude expression lysates indicated that the cleavage occurred prior to purification (data not shown). gp175 forms originating from either the precursor or mature-length gene were both catalytically active when assayed with ϕKZ gp93, each producing a fragment consistent with the mature length of gp93 (31 kDa) (Fig. 3A, lane 5, and B, lane 6) found in the virion.
Fig 2.
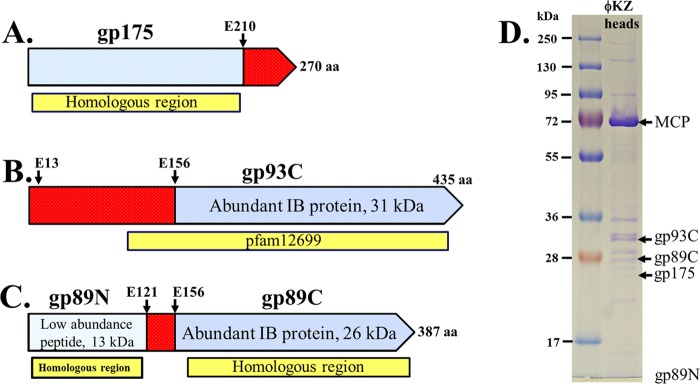
(A to C) Representation of ϕKZ gene products used in this study. Blue indicates regions present in the mature head. Red indicates precursor polypeptide regions that are processed and removed from the head. Protease cleavage sites previously identified by mass spectrometry are indicated with arrows (14). Yellow boxes indicate the most highly conserved region(s) in homologous proteins in related phages. (A) gp175, the prohead protease, and major IB proteins gp93 (B), and gp89 (C). Both gp93 and 89 are estimated to be present at about 100 copies per particle (14). (D) SDS-PAGE of purified ϕKZ heads obtained from a tailless mutant. The location of the forms of gp89, gp93, and gp175 in the mature head (gp89N, gp89C, gp93C, and gp175) as determined by mass spectral analysis (14). The N-terminal fragment of gp89 is not marked, as it migrates below the scope of this gel. MCP refers to the major capsid protein, present at 1,560 copies per virion.
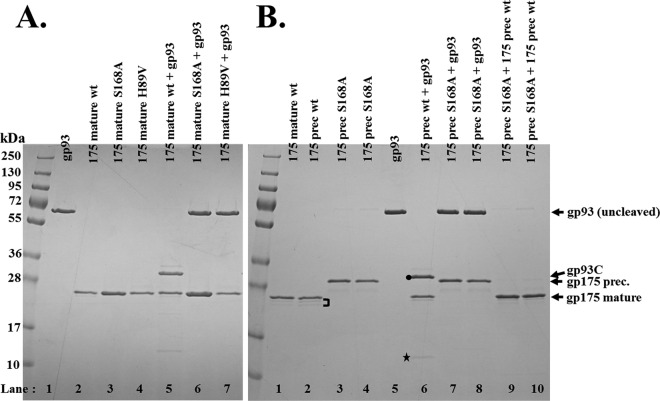
Fig 3.
SDS-PAGE of activity assay products of expressed and purified mature and precursor forms of the ϕKZ protease, gp175, on IB protein gp93. (A) Assay of wild-type and mutated forms of mature length gp175. Lanes: 1, gp93; 2, gp175 expressed from a gene encoding the mature length of the wild-type protein (residues 1 to 210); 3, mature-length gp175 with mutated catalytic serine; 4, mature-length gp175 with a mutated catalytic histidine; lanes 5 to 7, products of assays of the wild-type and catalytic mutant forms of gp175 with gp93. (B) Assay products of wild-type and mutant forms of precursor-length gp175. Lanes: 1, purified gp175 expressed from a gene encoding the mature length of the wild-type protein (residues 1 to 210); 2, gp175 expressed from a gene encoding the zymogen or precursor form of the protease (residues 1 to 270); the bracket indicates fragments potentially representing inactivated forms of the protease (see Discussion); 3, gp175 expressed from a gene encoding the precursor form of gp175 with mutated catalytic serine; 4, gp175 expressed from a gene encoding the precursor form of gp175 with mutated catalytic histidine; 5, gp93. Lanes 6 to 8 show assay products of wild-type and catalytic residue mutants of gp175 expressed from the precursor-length gene with gp93. Lanes: 6, wild-type gp175; 7, catalytic serine mutant gp175; 8, catalytic histidine mutant gp175. The black circle indicates the C-terminal fragment of gp93 resulting from cleavage at E-156. The star indicates an N-terminal fragment of gp93. Lanes 9 and 10: assay products of wild-type gp175 expressed from a gene encoding the precursor form of gp175 with inactivated, precursor forms of gp175 (lane 9, catalytic serine mutant; lane 10, catalytic histidine mutant). wt, wild type prec., precursor.
Confirming that gp175 was responsible for this proteolysis, no cleavage of gp93 was observed when gp175 was boiled prior to the assay (Fig. 4A, lane 4). An assay of boiled gp93 resulted in reduced proteolysis (Fig. 4B, lane 2), but the fragments produced were the same as those produced from undenatured gp93 (see below). Further support for the cleavage specificity of gp175 was demonstrated by its inability to cleave bovine serum albumin (BSA) (Fig. 4C, lane 3). Cleavage of gp93 by gp175 occurred at temperatures ranging from 10 to 40°C and to a lesser degree when assays were performed on ice or at 45°C (data not shown). Subsequent assays were undertaken at 37°C. Purified enzyme was active for several days when stored at 4°C and for at least 1 week when stored in 50% glycerol at −20°C (data not shown).
Fig 4.
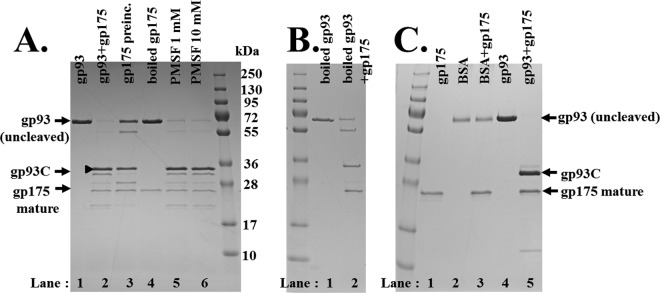
SDS-PAGE of assay products of the ϕKZ protease gp175 on different substrates and under different assay conditions. (A) Lanes: 1, gp93; 2, gp93 and gp175; 3 to 6, gp175 and gp93. The assay was for 30 min at 37°C. Lane 3, gp175 preincubated for 30 min at 37°C immediately prior to the assay; lane 4, gp175 boiled for 2 min immediately, then quickly cooled on ice prior to the assay; lanes 5 and 6, products of assay mixtures supplemented with 0.1 and 1.0 mM serine protease inhibitor PMSF, respectively. (B) Lane 1, gp93 that had been boiled for 3 min and then quickly cooled on ice immediately prior to the assay, as a negative control with no protease added. Lane 2, gp93 that had been boiled prior to the assay with gp175 (not boiled). (C) Assay of gp175 on BSA. Lanes: 1, gp175; 2, BSA; 3, gp175 and BSA; 4, gp93; 5, gp175 and gp93.
Mutation of predicted active site residues inactivates mature and precursor forms of gp175.
Cleavage of gp93 by gp175 originating from either the precursor or mature-length gene also did not occur when two residues predicted to be components of the catalytic triad (see reference14), serine 168 or histidine 89, were mutated to alanine and valine, respectively (Fig. 3A, lanes 6 and 7, and B, lanes 7 and 8). Importantly, gp175 expressed from the precursor-length gene was not cleaved when either of these catalytic residues was mutated, confirming that its cleavage in the wild-type protease was the result of autocatalysis. These inactive forms of the protease showed that the full precursor length of gp175 migrates in SDS-PAGE consistent with its predicted molecular mass (27 kDa) (Fig. 3B, lanes 3 and 4). Significantly, these inactive, precursor-length forms of gp175 could be cleaved to mature length when mixed with active mature-length gp175, suggesting that despite the mutations, both must have a near-native structure that is susceptible to cleavage, and also that self-cleavage by gp175 likely is via an intermolecular mechanism (Fig. 3B, lanes 9 and 10).
Proteolysis of gp89 by gp175.
Purification of ϕKZ gp89 with an N-terminal His tag produced a protein that migrated on SDS-PAGE consistent with its predicted molecular mass (43.2 kDa) (Fig. 5A, lane 1). When incubated with gp175, gp89 was cleaved into two fragments of molecular masses consistent with the N- and C-terminal fragments observed in the mature head (Fig. 5A, lane 5, black circles). Two additional bands were also observed, each with a slightly higher molecular mass than each of the mature fragments (Fig. 5A, lane 5, black triangles). These “non-mature-length” fragment bands were attributed to proteolysis in the 35-residue region that links the N- and C-terminal fragments and contains sequences that conform to the protease cleavage motif (Fig. 5B). In assays utilizing a concentration series of protease, these non-mature-length fragments were observed at lower protease concentrations, whereas the mature-length fragments were produced at higher protease concentrations, suggesting that the cleavages which produce the non-mature-length fragments occur prior to the cleavages that produce the mature-length fragments. The non-mature-length form of the gp89 C-terminal fragment is likely the product of cleavage at SHE-149. The non-mature-length form of the N-terminal fragment could be produced by cleavage at either SFE-132 or SLE-129; however, we predict the cleavage is at SLE-129, based on its higher conservation in homologs.
Fig 5.
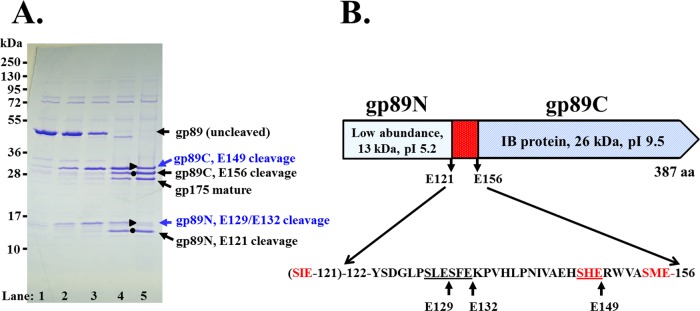
Proteolysis of the ϕKZ IB protein gp89 by the protease gp175. (A) SDS-PAGE of assay products of gp89 with increasing concentrations of gp175 (lanes 1 to 5). Mature-length N- and C-terminal fragments of gp89 are indicated as black circles. Immature-length N- and C-terminal fragments of gp89 are indicated as blue circles, and the putative sites cleaved to produce them are indicated in blue. (B) Schematic of gp89, showing cleavage sites in addition to those identified by mass spectrometry (E-121 and E-156).
Proteolysis of gp93 by gp175.
Purification of ϕKZ gp93 with an N-terminal His tag produced a protein band that migrated close to the 72-kDa marker (Fig. 6, lane 1), slower than predicted for gp93 (48 kDa), despite the gp93 gene being intact as confirmed by DNA sequencing. Comparison of the migration of purified His-tagged gp93 with crude lysates containing nontagged gp93 showed little difference in the migration of each form of gp93, indicating that the His tag was not responsible for the slower-than-expected migration (data not shown). The aberrant migration of gp93 was attributed to its highly negatively charged N-terminal propeptide region (pI 4.1). Supporting this is the induction of a pHERD20T construct encoding only the first 121 residues of gp93 (predicted molecular mass of 13.4 kDa) that produced a protein band that migrated above the 28-kDa marker band (data not shown). Other highly charged proteins have also been reported to migrate unexpectedly (26). Also, as noted above, when incubated with gp175, gp93 was cleaved to a molecular mass that migrated on the SDS-PAGE gel consistent with the mature C-terminal fragment identified in the virion by mass spectrometry (MS), with a predicted mature molecular mass of 31 kDa (Fig. 6, lane 10) and pI of 7.
Assays of gp93 with increasing protease revealed that six fragments were produced in addition to the presumed mature fragment (Fig. 6), and as observed for gp89, these fragments were produced in a protease concentration-dependent manner. Two fragments were expected to represent extended, transitional forms of the gp93 C-terminal fragment (Fig. 6, black square and triangle) and were produced prior to the final cleavage that produced the presumed mature gp93 C-terminal fragment (Fig. 6, black circle). The production of the progressively shorter gp93 C-terminal fragments indicated that the processing of the propeptide likely proceeds in an N- to C-terminal manner (Fig. 6). The other four fragments were attributed to peptides resulting from proteolysis of the N-terminal region of gp93 (Fig. 6, black stars), as their molecular masses were all lower than that of the mature fragment; however, deducing the cleavages that caused these fragments was complicated due to their unusual migration caused by their high negative charge plus the high number of potential cleavage sites (see below).
Mutagenesis of gp93 cleavage sites.
To confirm the identity of the cleavages that produced the gp93 C-terminal fragments, we used site-directed mutagenesis to change each expected cleavage site glutamate codon to an aspartate codon. The glutamate residues targeted were (i) E-156, to verify that the expected mature fragment we observed was the product of cleavage at the site identified by mass spectrometry, (ii) E-13, to confirm that a high-molecular-mass fragment (migrating just below the full-length gp93) was the product of cleavage at this residue (this site was also identified by mass spectrometry), and (iii) E-137, which is in a sequence conforming to the cleavage motif (ALE) and predicted to produce the fragment we observed that migrated to a position just above the mature fragment. These mutations were incorporated into gp93 as single mutations and also in various combinations to test if the mutation of any site(s) affected the cleavage of other sites. Protease assays of purified N-terminally His-tagged gp93 with each of these mutations confirmed that cleavage at a glutamate residue is an absolute requirement for gp175, as the fragments expected to be produced by cleavages at these three residues were no longer produced when their respective glutamate was mutated (Fig. 7B).
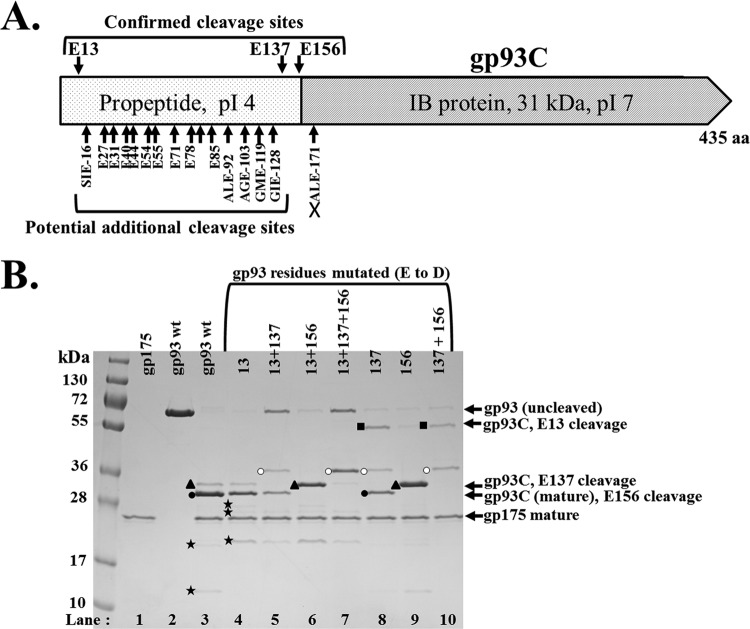
Fig 7.
Mutational analyses of ϕKZ gp93 protease cleavage sites. (A) Map of gp93, indicating propeptide versus mature regions and cleavage sites, confirmed by mutational analyses. Five sequences that conform to the cleavage motif are marked on the underside of the region and represent the propeptide region of gp93, and 10 other glutamate residues that do not contain the cleavage motif are also indicated. The sequence ALE-171 is marked with a cross to indicate that it was not cleaved in these studies. (B) SDS-PAGE of assay products of the ϕKZ protease with constructs of gp93 that have single, or combinations of, mutated cleavage sites (the sites identified by mass spectrometry, E13 and E156, and a predicted cleavage site, E137). Mutated residues are marked at the top of each lane. Bands expected to represent C-terminal fragments of gp93 resulting from cleavage at E-13, E-137, and E-156 are indicated with a black square, black triangle, and black circle, respectively. A band expected to represent a C-terminal fragment of gp93, for which the cleavage site was not determined, is indicated with a white circle. Cleavage fragments originating from the N-terminal propeptide region are indicated by black stars.
Mutation of gp93 E-156 produced only C-terminal fragments with higher molecular masses (Fig. 7B, lanes 6, 9, and 10) than that of the band expected to represent the mature fragment (Fig. 7B, lane 3, black circle), confirming the identity of the cleavage producing that fragment. Confirmation that cleavage at E-156 produces mature gp93C was important, as there is a nearby downstream sequence (ALE-171) that conforms to the cleavage motif (S/A/G-X-E) and potentially could have been the maturation cleavage site. Mutation of E-13 confirmed that the band we predicted to represent cleavage at this residue (Fig. 7B, lane 10, black square) was not produced when this residue was mutated (Fig. 7B, lanes 4 to 7). As with the E-156 cleavage site, the confirmation of the E-13 cleavage site was important, as there is another sequence immediately downstream that also conforms to the sequence motif (SIE-16) and whose cleavage might have been responsible for the fragment observed. Mutation of E-137 also demonstrated that the fragment predicted to be produced by cleavage at this site (Fig. 7B, lanes 3, 6, and 9, black triangle) was not produced when this residue was mutated (Fig. 7B, lane 8). Confirmation of the E-137 cleavage site was of interest, as we had observed a fragment of corresponding molecular mass in assays where the protease preparation was apparently less active (e.g., Fig. 4A, lane 2, black triangles). This fragment resulting from E-137 cleavage was also observed to be more dominant with increasing age of the gp175 preparation.
The mutant gp93 protease cleavage assays provided additional evidence to support E-13, E-137, and E-156 as the major sites cleaved to form decreasing lengths of the C-terminal fragment. In addition, there apparently is an N- to C-terminal progression of cleavages through the gp93 propeptide. These observations were aided by using different concentrations of protease (data not shown). The mutation of E-156 had little or no effect on the upstream cleavages at E-13 and E-137 (e.g., Fig. 7B, lane 9). However, it was the E137D mutation that was responsible for the most dramatic changes in the gp93 cleavage profile. When E-137 alone was mutated, there was an accumulation of the fragment produced by cleavage at E-13 (data not shown); however, the inhibition of the E-13 cleavage and subsequent production of the mature fragment was overcome to some degree with increased protease (Fig. 7B, lane 8), as if the enzyme is able to “hop” over the mutated site. It also appears that when the E-137 site is blocked, the protease may utilize another nearby, unidentified site for cleavage, producing a fragment that migrates close to the 36-kDa standard (Fig. 7B, lanes 5, 7, 8, and 10, white circles). Sequences conforming to the sequence motif and whose cleavage would be expected to produce fragments of this size are AGE-103, ALE-92, and GME-119.
The N-terminal propeptide of gp93 is expected to be highly proteolyzed, based on the production of numbers of low-molecular-mass bands (Fig. 6 and 7, black stars), and also because it incorporates not only the three cleavage sites confirmed by SDM but also an additional five sequences that conform to the cleavage motif (including the three listed above) (Fig. 7A). There are also an additional 10 glutamate residues (Fig. 7A) that might represent potential cleavage sites, if there can be additional variation to the known cleavage motif. The existence of three demonstrated and potential additional cleavage sites suggests that in gp93 (and probably its homologs) there is a programmed redundancy of cleavage sites whose function is to ensure that the highly negatively charged propeptide of gp93 is cleaved and removed from the capsid so that only the positively charged C-terminal fragment of gp93 remains in the capsid to carry out its structural role in the mature IB.
DISCUSSION
In large myoviruses, the prohead protease catalyzes the transformation of a spherical, protein-only prohead I to an expanded angular shell-like and generally more stable prohead II primed for DNA packaging. Despite its essential nature, in reality there are many facets to this protease-dependent transformation that are poorly understood. Even for T4, whose prohead protease (PPase), gp21, is the most extensively characterized myoviral protease to date, there is a lack of detailed characterization of both it and its biological role in head morphogenesis. For instance, while gp21 is predicted by bioinformatics to be a serine protease (22, 27), this has not been demonstrated biochemically. Similarly, although gp21 is suggested to be activated by C-terminal autocleavage, the exact site of processing is unknown. There is an even greater lack of knowledge regarding the role of proteolysis in head morphogenesis for Pseudomonas phage ϕKZ, although its complex head contains an unusually high number of different proteins (at least 19) that undergo proteolytic processing (14). Understanding the role of the ϕKZ protease is of additional interest because the novel IB structure within the head must be markedly transformed by proteolysis, as five major IB structural proteins are processed by it (mass reduced by approximately 50%).
The ϕKZ protease gp175 was only recently identified, and despite being extremely divergent at the sequence level from T4 gp21, was shown to have similarities to it, including that each enzyme was expected to undergo autoactivation via C-terminal proteolysis. The evidence supporting the cleavage of T4 gp21 was that only a lower-molecular-mass form (18 kDa, versus a 27-kDa form) showed activity (28). Later, a C-terminal cleavage site was proposed by Keller and Bickle when they sequenced the gp21 gene (27). The C-terminal cleavage site of ϕKZ gp175 was identified by mass spectrometry (14). Both T4 gp21 and ϕKZ gp175 cleave their respective substrates after a glutamate residue at a three-residue cleavage recognition motif that is distinct for each phage, L/I-X-E for gp21 (18) and S/A/G-X-E for gp175 (14). Preliminary studies revealed that ϕKZ gp175 was apparently soluble and active in a crude extract; hence, we decided to further utilize these properties to study the activity and specificity of this enzyme. Here we have demonstrated that gp175 when highly purified via an N-terminal histidine tag is active, cleaving two substrate IB proteins, gp93 and gp89. Expression of the full-length 270-residue precursor form of gp175 showed it to autoactivate by removal of a 60-residue C-terminal fragment. Both precursor and mature forms of gp175 were completely inactive when either predicted catalytic serine or histidine residues were mutated. This supports the classification of gp175 as a serine protease with a catalytic mechanism derived from a common ancestor of both myoviruses and herpesviruses. Herpesvirus proteases represent a unique family among serine proteases, as evidenced by their novel structure that consists of a conserved core of a mainly antiparallel seven-stranded β-barrel that is surrounded by eight helices, although great variability exists in the surface loops and helices between different members of this family (29). ϕKZ gp175 has been shown to have predicted secondary structure elements consistent with numbers of the conserved core elements of the herpesvirus fold (14). Herpesvirus proteases are unusual in that they retain activity in the presence of standard serine protease inhibitors, with the exception of DFP (30). ϕKZ gp175 is also not inactivated by the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) (Fig. 4A), a characteristic of T4 gp21 also (18).
The zymogen self-activation of gp175 via a C-terminal cleavage is also potentially a primordial feature of it and other related proteases, including T4 gp21. Herpesvirus proteases self-activate via two C-terminal cleavages at a maturation (M) site and a release (R) site. For instance, in the 637-residue herpes simplex virus 1 protease (UL26), cleavage at the M site (residue 610) releases the C terminus of the protein from being anchored to the inner capsid shell, while the cleavage at the R site (residue 247) releases the N-terminal protease or assemblin domain from the C-terminal scaffolding domain (31). These herpesvirus protease cleavages occur between alanine and serine residues (610/611 and 247/248 [32]), a cleavage requirement that is quite different from that of the ϕKZ protease. Despite the cleavage site residue requirement differences, the self-activation of the ϕKZ protease in this study indicated that the E-210 cleavage site equates functionally to the R site of herpesvirus; however, it did not exclude that gp175 undergoes another cleavage C-terminal to the R site that equates to the herpesvirus M site. Making such cleavages feasible is the presence of several glutamate residues downstream of E-210, including E-240, at which cleavage (GXE) would be consistent with the cleavage motif.
The demonstration of autoactivation by gp175 was particularly compelling, because no phage or specific host (Pseudomonas aeruginosa) activator protein(s) was required, as active enzyme was produced from an expression vector in Escherichia coli. Genetic studies on T4 head morphogenesis have also found no protein essential for protease activation (18). However, during the morphogenesis of either phage, the prohead protease presumably remains inactive until at least the majority of the head precursor polypeptides have correctly assembled before it acts. Therefore, a central unanswered question of large myoviral head morphogenesis is what controls the transition from inactive to active protease and thus drives the conversion of prohead I to prohead II? Possibly, the answer to this question lies in electron microscopic studies of T4, that have indicated that the prohead protease is located within a darkened “kernel-like” structure within the center of the core (33). By negative staining, this structure appears to align with the tail axis, with one end close above the portal, so that it may be associated with the portal vertex (18). A similar condensed protease kernel-like structure probably due to the protease is also found in herpesvirus (34). The existence of such a kernel in the early ϕKZ prohead requires confirmation, but it is an attractive hypothesis that ϕKZ, T4, and herpesvirus proteases share a general mechanism for protease activation, as well as activity. The structure of this kernel may be such that the protease remains inactive while in it. Possibly, if there is a strong affinity of the protease for another prohead protein, this might provide a mechanism to enable the protease to be sequestered from its substrates until the appropriate time for autoactivation and release from the kernel to perform mature cleavages.
If a structure similar to the T4 prohead protease kernel structure does exist in a similar position in the ϕKZ prohead, it is likely to be near, or even form, the base of the prohead form of the IB. The ϕKZ portal protein has an unusually long propeptide (262 amino acids) that is proteolytically processed, so it is feasible that the portal might initially be attached/linked to a structure containing the protease and/or the precursor form of the IB (14). The precursor IB might even be assembled in an orientation along the portal axis in the prohead, a logical expectation, considering that the smaller core structures of certain podoviruses are constructed above the portal in this orientation (e.g., T7 [35, 36] and P22 [37, 38]). Subsequent ϕKZ protease activation and/or disintegration of a protease kernel might facilitate a change in the orientation of the IB to its mature position, offset from the portal axis.
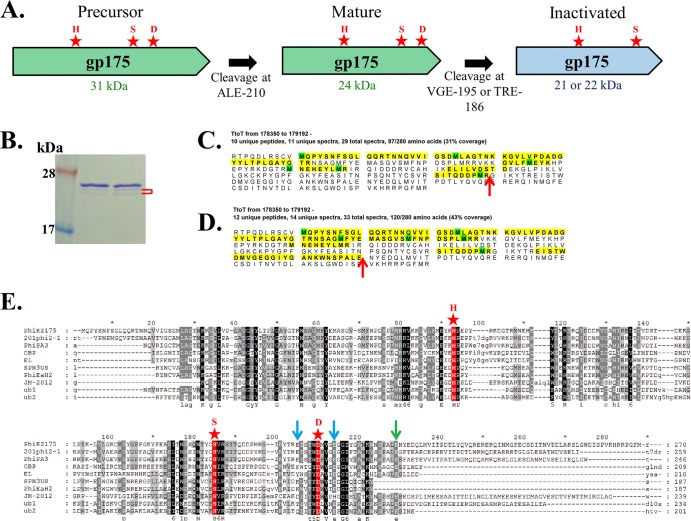
In addition to undergoing autoactivation, some herpesvirus proteases, such as cytomegalovirus (39), undergo additional self-processing to inactivate themselves. Although there is no direct evidence that ϕKZ gp175 self-inactivates, we expect that it occurs (Fig. 8A), based on the following: (i) early work on T4 gp21 indicated that it was autodigested and mostly expelled from the prohead and that the 2 to 3 molecules (of 70) remaining in the mature head were inactive (28); (ii) consistent with a cleaved and inactivated form of gp175 in the ϕKZ head, the mass spectral peptide coverage for gp175 in a purified head sample ended at R-179 (Fig. 8C), as the single peptide that revealed the maturation cleavage site of gp175 to be at E-210 was only identified in a sample that had not undergone cesium chloride gradient purification and was therefore expected to contain some assembly intermediates (Fig. 8D); (iii) gp175 had reduced activity when preincubated alone at 37°C prior to assay with gp93 (Fig. 4A, lane 3); (iv) incubation of gp175 alone produced two lower-molecular-mass bands below that which represented the mature protease (Fig. 8B, bracket) (these lower-molecular-mass bands were not observed immediately after the enzyme was purified [data not shown]); (iv) there are two sequences (VGE-195 and TRE-186) upstream of the maturation cleavage site (E-210) that are noncanonical to the known cleavage motif, but the VXE motif was well conserved in homologous proteases (Fig. 8E). Cleavage at either site would produce fragments of a size consistent with those observed in the evidence described above in point iv. Mechanisms of inactivation by these cleavages might be by disruption of the catalytic cleft, either by removal of candidate members of the catalytic triad (e.g., TRE-186) or by the disordering of a multimerization state necessary for activity. The function of inactivating the prohead protease might be to prevent additional, unwanted cleavages in head proteins, or to prevent an active enzyme from being injected into the host cell, with the potential to prematurely cleave newly formed polypeptides destined for progeny phage.
Fig 8.
Autoinactivation of ϕKZ gp175. (A) Schematic of different forms of ϕKZ gp175 caused by the known maturation autocleavage and predicted inactivating autocleavage(s). Catalytic triad residues are indicated by red stars. (B) Enlarged section of lanes 1 and 2 from Fig. 3B, showing minor bands (red bracket) beneath the major band representing the mature form of gp175. (C) Mass spectral peptide coverage (yellow) of ϕKZ gp175 from a sample of cesium chloride gradient-purified heads, showing the peptide coverage ending at R-179 (red arrow) upstream of the maturation site (E-210). (D) Mass spectral peptide coverage of ϕKZ gp175 from a sample of ϕKZ heads, showing the peptide coverage ending at the maturation cleavage site, E-210 (red arrow). This sample was only subjected to differential centrifugation and is expected to contain more precursor particles and assembly intermediates than purified heads. (E) Alignment of the ϕKZ protease, gp175, with homologs in Pseudomonas phages 201ϕ2-1 (gp268), ϕPA3 (gp205), OBP (gp283), and EL (gp192) and homologs in Salmonella phage SPN3US (gp245), Erwinia phage PhiEaH2 (gp165), and Halocynthia phage JM-2012 (gp80). Homologs in uncultured bacteria are indicated by ub; the GenBank identifier for ub1 is EKD22713, and the GenBank identifier for ub2 is EKD89709. Catalytic histidine and serine residues are indicated in red and marked with stars, as is the candidate for the third catalytic residue (D-191). The ϕKZ gp175 maturation cleavage site (E-210) is indicated with a green arrow. Putative cleavage sites for ϕKZ gp175 self-inactivation (E-186 and E-195) are indicated with blue arrows.
Dimerization of herpesvirus protease is essential for activity (40), and based on its similarities to herpesvirus protease, we expect gp175 also dimerizes. That gp175 has some multimeric structure is indicated by native gel migration (data not shown). We hope this question will be resolved in structural studies.
We selected the two abundant IB proteins, ϕKZ gp89 and gp93, as the substrates for the prohead protease in this study, as their abundances indicate they are likely to have important functions related to IB and ϕKZ head morphogenesis and structure. Each ϕKZ-related phage has a single gp89 homolog, and these proteins are well conserved between phages to a degree comparable to the conservation of essential proteins, such as the major capsid protein and DNA packaging enzyme, the large terminase subunit (14). In contrast, the conservation between gp93 and its homologs is weak, but the fact that ϕKZ has five paralogs to gp93 emphasizes their importance. These homologs include the two abundant IB proteins, gp95 and gp162, and three low-abundance proteins, gp92, gp94, and gp163, all of which are cleaved by the protease. Also, every ϕKZ-related phage has a 5- to 7-member set of highly diverged paralogs (PF12699 [1]), all of which we expect to undergo proteolysis and are likely to have related but specialized functions. Possibly, the paralogs have a role in host takeover, based on the expectation that gp93 and its paralogs are injected into the host cell (16); however, another intriguing possible function for these proteins is a role in determining the amount of DNA packaged into the ϕKZ head. Notably, the length of DNA packaged does vary considerably between the different ϕKZ-like phages (e.g., EL, 211 kb; 201ϕ2-1, 316 kb), although apparently the T=27 capsid sizes are similar (2, 41). Possibly, IB composition has a role in regulating the amount of DNA packaged in the heads of the ϕKZ-like phages.
We observed two fragments after incubation of gp89 with gp175 that had molecular masses consistent with the two fragments previously identified in the phage by mass spectrometry, a high-abundance C-terminal fragment (gp89C, residues 122 to 156) and an N-terminal fragment (gp89N, residues 1 to 121), whose abundance by mass spectrometry is considerably (from 3- to 10-fold) lower than that of the C-terminal fragment (14). In our assays, gp89N does not undergo further proteolysis and apparently represents a propeptide fragment whose length, surprisingly, is not long enough to hinder nearly full clearance from the head. Complete clearance of peptides (residues 122 to 156) from the internal region of gp89 is shown by the absence of detected peptides from this region in vivo. Nevertheless, despite its low copy number, it is possible that gp89N does have some function in the mature head. The lack of processing in gp89N is in contrast to the 156-residue propeptide region of gp93, which is processed to the extent that there is no evidence of any fragments remaining in the head in vivo (14). However, these data suggest, overall, that the chief function of the proteolysis of both proteins is to remove a negatively charged propeptide from a positively charged mature fragment. The negatively charged propeptide likely is important earlier in scaffolding prohead assembly. Scaffold proteins (e.g., T4 gp22, which is completely broken down by the T4 protease) are generally negatively charged and determine prohead size and shape. The mature IB proteins with positive charge likely have a different defined function in the specific IB structure, such as the spooling of the DNA within the head. However, in addition, the negative charge of the propeptide regions might be important for their exit from the capsid, a process that might also be assisted by the packing of DNA.
Our research suggests that there is a redundancy of protease cleavage sites in gp93, as if to ensure that the charged propeptide region is removed from the prohead. Similarly, highlighting the importance of these cleavages through the evolution of ϕKZ and related phages, the most well-conserved region between gp93 and its closest homologs is the proteolyzed region of the N-terminal propeptide. An alignment of ϕKZ gp93 and its closest homologs illustrated that it is its confirmed cleavage sites and other sequences adhering to the cleavage motif that are often the most conserved residues in this protein (data not shown), as observed for this family (PF12699 [1]). The E-13 cleavage in gp93 is of interest, as it suggests that ϕKZ potentially has a morphogenetic protein packaging system analogous to the capsid-targeting sequence (CTS)-based system of T4 (42, 43). In T4, a 10- to 17-residue N-terminal sequence is sufficient to target proteins (including Alt, β-galactosidase, and green fluorescent protein) into the core during morphogenesis and is ultimately removed from these proteins in the prohead after proteolysis by gp21. All the paralogs to gp93 in ϕKZ and members of PF12699 in other phages have a cleavage-like motif close to their N terminus. The T4 CTS-targeted proteins have important roles in host takeover, such as Alt, which ribosylates the host RNA polymerase, altering its promoter specificity to increase transcription of T4 promoters (44), and IPI*, a restriction endonuclease inhibitor (45).
Our gp175 protease assays of gp93, and gp89, also provided evidence for additional cleavages in the propeptide regions of both of these proteins, indicating that the number of previously identified cleavage sites in ϕKZ head proteins, 49, is an underestimate. The propeptides of gp89 and gp93 apparently undergo a series of stepwise cleavages by gp175. In particular, in the propeptide of gp93 there is evidence to suggest that there is an apparent N- to C-terminal progression by the protease with the maturation cleavage site the final site to be cleaved. Such a mechanism might ensure that most or all of the cleavage sites along the propeptide are cleaved, enhancing the production of small fragments and therefore improving their removal from the head. We confirmed three cleavage sites in gp93 via inhibition of cleavage by mutating the glutamate residue to an aspartate. These studies also demonstrated that while cleavage(s) downstream of the mutated residue E-13 and E-137 was slowed, it was not completely inhibited (e.g., E-137 in Fig. 7, lane 8), as if the enzyme were able to “hop over” those sites.
Despite our confirmation of three cleavage sites in gp93, the other determinants required for cleavage by gp175, in addition to the short cleavage motif (mainly S/A/GXE-cleavage), remain unclear. However, the proposal for the existence of other determinants has gained further evidence in these studies, most notably by the lack of cleavage of proteins containing motif-like sequences by the protease (such as BSA [Fig. 4C] or E. coli proteins, either in crude extracts [14] or of contaminating proteins in purified protein preparations [Fig. 5A]). There are also instances in known gp175 substrates where sequences that adhere to the cleavage motif are not cleaved, as demonstrated for gp93 ALE-171 in this study. We have examined the predicted secondary structure elements flanking this and other noncleaved motif-like sequences versus those flanking cleaved sequences and have found no common feature present in either; however, it seems likely there has to be some form of structural determinant for cleavage by gp175, possibly at the tertiary level.
The demonstration that ϕKZ gp175 is the prohead protease resulted in the creation of a new family of proteases within the MEROPs database, the S80 family, in which gp175 is the family type peptidase and homologous proteases in other ϕKZ-related phages are incorporated (46). Of note is the fact that a recent addition to this family included a protease (gp245) encoded by a non-Pseudomonas phage, Salmonella phage SPN3US (JN641803.1) (47). We expect the S80 family will continue to expand to incorporate other homologous proteases (aligned in Fig. 8E) encoded by non-Pseudomonas phages, such as the recently sequenced Erwinia phage PhiEaH2 (NC_019929.1) and JM-2012 (NC_017975.1), isolated from Halocynthia. It is exciting to contemplate that research on ϕKZ gp175 is also relevant to the morphogenesis of these other “giant” phages. These phages all have large genomes (167 to 240 kb) that are expected to be packaged into heads of similar dimensions to those of ϕKZ (based on their having homologous capsid proteins of similar lengths). Most importantly, it is likely these phages also have some form of IB structure, possibly either a precursor structure that is removed by proteolysis during morphogenesis or some form of IB structure that is retained in their mature head. This is because these phages also have proteins that are homologous to abundant ϕKZ IB proteins, including gp89 and gp93. The homologs to cleaved ϕKZ IB proteins in these other phages also have candidate protease cleavage sites that we expect are cleaved by their respective proteases. Therefore, the conserved proteases in the ϕKZ-related and more distantly related phages play a central role in sculpting the IBs of these phages. In conclusion, these findings related to ϕKZ gp175 raise interesting questions as to the role of prohead proteases in large myoviral head and IB morphogenesis and the factors that regulate their specificity, activation, and likely deactivation.
ACKNOWLEDGMENTS
The pHERD20T vector was provided by Hongwei D. Yu (Marshall University). We thank Qin Dan for her technical support. We are grateful to Stephen C. Hardies for allowing access to his collection of bioinformatics software and the UTHSCSA Bioinformatics Center for assistance with computational aspects of the project. We thank Abby Sukman for helpful discussions on the strategy that we implemented to insert histidine tags into the pHERD20T constructs.
This work was supported by NIH grant AI11676 to L.W.B.
Footnotes
Published ahead of print 5 June 2013
REFERENCES
- 1. Cornelissen A, Hardies SC, Shaburova OV, Krylov VN, Mattheus W, Kropinski AM, Lavigne R. 2012. Complete genome sequence of the giant virus OBP and comparative genome analysis of the diverse ϕKZ-related phages. J. Virol. 86:1844–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krylov VN, Dela Cruz DM, Hertveldt K, Ackermann HW. 2007. “ϕKZ-like viruses,” a proposed new genus of myovirus bacteriophages. Arch. Virol. 152:1955–1959 [DOI] [PubMed] [Google Scholar]
- 3. Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9:224. 10.1186/1471-2180-9-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krylov V, Shaburova O, Krylov S, Pleteneva E. 2012. A genetic approach to the development of new therapeutic phages to fight Pseudomonas aeruginosa in wound infections. Viruses 5:15–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Briers Y, Volckaert G, Cornelissen A, Lagaert S, Michiels CW, Hertveldt K, Lavigne R. 2007. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages ϕKZ and EL. Mol. Microbiol. 65:1334–1344 [DOI] [PubMed] [Google Scholar]
- 6. Golshahi L, Lynch KH, Dennis JJ, Finlay WH. 2011. In vitro lung delivery of bacteriophages KS4-M and ϕKZ using dry powder inhalers for treatment of Burkholderia cepacia complex and Pseudomonas aeruginosa infections in cystic fibrosis. J. Appl. Microbiol. 110:106–117 [DOI] [PubMed] [Google Scholar]
- 7. Matinkhoo S, Lynch KH, Dennis JJ, Finlay WH, Vehring R. 2011. Spray-dried respirable powders containing bacteriophages for the treatment of pulmonary infections. J. Pharm. Sci. 100:5197–5205 [DOI] [PubMed] [Google Scholar]
- 8. Walmagh M, Briers Y, dos Santos SB, Azeredo J, Lavigne R. 2012. Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201ϕ2-1 and PVP-SE1. PLoS One 7:e36991. 10.1371/journal.pone.0036991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fokine A, Kostyuchenko VA, Efimov AV, Kurochkina LP, Sykilinda NN, Robben J, Volckaert G, Hoenger A, Chipman PR, Battisti AJ, Rossmann MG, Mesyanzhinov VV. 2005. A three-dimensional cryo-electron microscopy structure of the bacteriophage ϕKZ head. J. Mol. Biol. 352:117–124 [DOI] [PubMed] [Google Scholar]
- 10. Fokine A, Battisti A, Bowman V, Efimov A, Kurochkina L, Chipman P, Mesyanzhinov V, Rossmann M. 2007. CryoEM study of the Pseudomonas bacteriophage ϕKZ. Structure 15:1099–1104 [DOI] [PubMed] [Google Scholar]
- 11. Sycheva LV, Shneider MM, Sykilinda NN, Ivanova MA, Miroshnikov KA, Leiman PG. 2012. Crystal structure and location of gp131 in the bacteriophage ϕKZ virion. Virology 434:257–264 [DOI] [PubMed] [Google Scholar]
- 12. Lecoutere E, Ceyssens P-J, Miroshnikov K, Mesyanzhinov V, Krylov VN, Noben J-P, Robben J, Hertveldt K, Lavigne R. 2009. Identification and comparative analysis of the structural proteomes of ϕKZ and EL, two giant Pseudomonas aeruginosa bacteriophages. Proteomics 9:3215–3219 [DOI] [PubMed] [Google Scholar]
- 13. Thomas JA, Weintraub ST, Hakala K, Serwer P, Hardies SC. 2010. Proteome of the large Pseudomonas myovirus 201ϕ2-1: delineation of proteolytically processed virion proteins. Mol. Cell. Proteomics 9:940–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas JA, Weintraub ST, Wu W, Winkler DC, Cheng N, Steven AC, Black LW. 2012. Extensive proteolysis of head and inner body proteins by a morphogenetic protease in the giant Pseudomonas aeruginosa phage ϕKZ. Mol. Microbiol. 84:324–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krylov VN, Smirnova TA, Minenkova IB, Plotnilova TG, Zhazikov IZ, Khrenova EA. 1984. Pseudomonas bacteriophage ϕKZ contains an inner body in its capsid. Can. J. Microbiol. 30:758–762 [DOI] [PubMed] [Google Scholar]
- 16. Wu W, Thomas JA, Cheng N, Black LW, Steven AC. 2012. Bubblegrams reveal the inner body structure of ϕKZ. Science 335:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Black L, Thomas J. 2012. Condensed genome structure, p 469–487 In Rossmann MG, Rao VB. (ed), Viral molecular machines. Springer, New York, NY [Google Scholar]
- 18. Black LW, Showe MK, Steven AC. 1993. Morphogenesis of the T4 head, p 218–258 In Karam JD. (ed), Molecular biology of bacteriophage T4. ASM Press, Washington, DC [Google Scholar]
- 19. Casjens S, King J. 1974. P22 morphogenesis I: Catalytic scaffolding protein in capsid assembly. J. Supramol. Struct. 2:202–224 [DOI] [PubMed] [Google Scholar]
- 20. Cerritelli ME, Studier WF. 1996. Purification and characterization of T7 head-tail connectors expressed from the cloned gene. J. Mol. Biol. 258:299–307 [DOI] [PubMed] [Google Scholar]
- 21. Bjornsti MA, Reilly BE, Anderson DL. 1983. Morphogenesis of bacteriophage ϕ29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA protein gp3. J. Virol. 45:383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng H, Shen N, Pei J, Grishin NV. 2004. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 13:2260–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sambrook J, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74:7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hughey R, Karplus K, Krogh A. 2003. SAM: sequence alignment and modeling software system. Technical report UCSC-CRL-99-11. University of California, Santa Cruz, CA: http://www.soe.ucsc.edu/research/compbio/papers/sam_doc/sam_doc.html [Google Scholar]
- 26. Matagne A, Joris B, Frère JM. 1991. Anomalous behaviour of a protein during SDS/PAGE corrected by chemical modification of carboxylic groups. Biochem. J. 280:553–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keller B, Bickle TA. 1986. The nucleotide sequence of gene 21 of bacteriophage T4 coding for the prohead protease. Gene 49:245–251 [DOI] [PubMed] [Google Scholar]
- 28. Showe MK, Isobe E, Onorato L. 1976. Bacteriophage T4 prehead proteinase. II. Its cleavage from the product of gene 21 and regulation in phage-infected cells. J. Mol. Biol. 107:55–69 [DOI] [PubMed] [Google Scholar]
- 29. Tong L. 2002. Viral proteases. Chem. Rev. 102:4609–4626 [DOI] [PubMed] [Google Scholar]
- 30. DiIanni CL, Mapelli C, Drier DA, Tsao J, Natarajan S, Riexinger D, Festin SM, Bolgar M, Yamanaka G, Weinheimer SP. 1993. In vitro activity of the herpes simplex virus type 1 protease with peptide substrates. J. Biol. Chem. 268:25449–25454 [PubMed] [Google Scholar]
- 31. Robertson BJ, McCann PJ, Matusick-Kumar L, Newcomb WW, Brown JC, Colonno RJ, Gao M. 1996. Separate functional domains of the herpes simplex virus type 1 protease: evidence for cleavage inside capsids. J. Virol. 70:4317–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DiIanni CL, Drier DA, Deckman IC, McCann PJ, Liu F, Roizman B, Colonno RJ, Cordingley MG. 1993. Identification of the herpes simplex virus-1 protease cleavage sites by direct sequence analysis of autoproteolytic cleavage products. J. Biol. Chem. 268:2048–2051 [PubMed] [Google Scholar]
- 33. van Driel R, Traub F, Showe MK. 1980. Probable localization of the bacteriophage T4 prehead proteinase zymogen in the center of the prehead core. J. Virol. 36:220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newcomb WW, Trus BL, Cheng N, Steven AC, Sheaffer AK, Tenney DJ, Weller SK, Brown JC. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. 2005. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 24:3820–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cerritelli ME, Conway JF, Cheng N, Trus BL, Steven AC. 2003. Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv. Protein Chem. 64:301–323 [DOI] [PubMed] [Google Scholar]
- 37. Chang J, Weigele P, King J, Chiu W, Jiang W. 2006. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14:1073–1082 [DOI] [PubMed] [Google Scholar]
- 38. Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. 2006. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312:1791–1795 [DOI] [PubMed] [Google Scholar]
- 39. Gibson W. 2008. Structure and formation of the cytomegalovirus virion. Curr. Top. Microbiol. Immunol. 325:187–204 [DOI] [PubMed] [Google Scholar]
- 40. Marnett AB, Nomura AM, Shimba N, de Montellano PRO, Craik CS. 2004. Communication between the active sites and dimer interface of a herpesvirus protease revealed by a transition-state inhibitor. Proc. Natl. Acad. Sci. U. S. A. 101:6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hertveldt K, Lavigne R, Pleteneva E, Sernova N, Kurochkina L, Korchevskii R, Robben J, Mesyanzhinov V, Krylov VN, Volckaert G. 2005. Genome comparison of Pseudomonas aeruginosa large phages. J. Mol. Biol. 354:536–545 [DOI] [PubMed] [Google Scholar]
- 42. Mullaney JM, Black LW. 1998. Activity of foreign proteins targeted within the bacteriophage T4 head and prohead: implications for packaged DNA structure. J. Mol. Biol. 283:913–929 [DOI] [PubMed] [Google Scholar]
- 43. Mullaney JM, Black LW. 1996. Capsid targeting sequence targets foreign proteins into bacteriophage T4 and permits proteolytic processing. J. Mol. Biol. 261:372–385 [DOI] [PubMed] [Google Scholar]
- 44. Wilkens K, Tiemann B, Bazan F, Rüger W. 1997. ADP-ribosylation and early transcription regulation by bacteriophage T4. Adv. Exp. Med. Biol. 419:71–82 [DOI] [PubMed] [Google Scholar]
- 45. Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. 2008. Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J. Mol. Biol. 375:720–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rawlings ND, Barrett AJ, Bateman A. 2012. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40:D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee J-H, Shin H, Kim H, Ryu S. 2011. Complete genome sequence of Salmonella bacteriophage SPN3US. J. Virol. 85:13470–13471 [DOI] [PMC free article] [PubMed] [Google Scholar]