Abstract
Archaeal viruses display unusually high genetic and morphological diversity. Studies of these viruses proved to be instrumental for the expansion of knowledge on viral diversity and evolution. The Sulfolobus islandicus rod-shaped virus 2 (SIRV2) is a model to study virus-host interactions in Archaea. It is a lytic virus that exploits a unique egress mechanism based on the formation of remarkable pyramidal structures on the host cell envelope. Using whole-transcriptome sequencing, we present here a global map defining host and viral gene expression during the infection cycle of SIRV2 in its hyperthermophilic host S. islandicus LAL14/1. This information was used, in combination with a yeast two-hybrid analysis of SIRV2 protein interactions, to advance current understanding of viral gene functions. As a consequence of SIRV2 infection, transcription of more than one-third of S. islandicus genes was differentially regulated. While expression of genes involved in cell division decreased, those genes playing a role in antiviral defense were activated on a large scale. Expression of genes belonging to toxin-antitoxin and clustered regularly interspaced short palindromic repeat (CRISPR)-Cas systems was specifically pronounced. The observed different degree of activation of various CRISPR-Cas systems highlights the specialized functions they perform. The information on individual gene expression and activation of antiviral defense systems is expected to aid future studies aimed at detailed understanding of the functions and interplay of these systems in vivo.
INTRODUCTION
Knowledge about virus-host interactions in the third domain of life, the Archaea, remains limited despite increasing numbers of described archaeal viruses. The available information concerns mainly unusual morphological and genomic properties of these viruses and structures of proteins they encode (1, 2). Several viruses infecting hyperthermophilic Crenarchaeota have emerged as suitable models to study molecular details of the life cycle of archaeal viruses, e.g., Sulfolobus turreted icosahedral virus (STIV) (3), Sulfolobus spindle-shaped virus (SSV) (4), and Sulfolobus islandicus rod-shaped virus (SIRV) (5). They cover examples of lytic (STIV and SIRV2) and temperate (SSV) viruses and fundamentally differ from each other in virion morphology and genome organization. For these viruses and their hosts, attempts have been made to analyze patterns of gene expression in the course of the infection cycle (6–9). The temperate, spindle-shaped SSV1 has a circular double-stranded DNA (dsDNA) genome which can integrate into the host genome, establishing a lysogeny which can be reverted by UV irradiation (10, 11). A chronological regulation of transcription of viral genes was observed during a microarray study following induction of SSV1 replication caused by UV irradiation. In this case, hardly any differences of host gene expression were detected (6). A completely different situation was encountered when studying transcription of the lytic viruses STIV and SIRV2 after infection of Sulfolobus host cells, by microarray and Northern hybridization analysis, respectively (7, 9). In both cases, little temporal control of viral gene expression was detected. During STIV infection, a high proportion of host genes were either up- or downregulated (8). Genes involved in transcription, translation, and antiviral defense mechanisms were overrepresented among the differentially expressed genes (8).
SIRV2 is an appealing model for the study of archaeal virus-host interactions, since the infection has a pronounced and clear effect on the host cell (12–14). SIRV2 is a member of the Rudiviridae family and infects the hyperthermophilic archaeon S. islandicus LAL14/1, which thrives at 80°C and pH 3. The virus genome is dsDNA of ∼35 kb and encompasses 54 open reading frames (ORFs) (15, 16). SIRV2 is a lytic virus, and degradation of the host DNA occurs after infection, i.e., after 5 h postinfection (hpi), the chromosome is degraded in ∼40% of cells (12). At late stages of the infection cycle, multiple pyramid-shaped structures of up to 200 nm in diameter are observed on the surface of each infected cell. These virus-associated pyramids (VAPs) comprise 7-fold rotational symmetry and consist of multiple copies of the virus-encoded protein SIRV2_P98 (NP_666583.1) (13, 14). At this stage of the infection cycle, mature virions are present in the cell in 2 or 3 bundles of up to 50 virions (12). As the final step of the infection cycle, the VAPs open up outwards, creating large apertures through which the mature virions escape the host cell (12). Thus, cell morphology and metabolism are dramatically affected by SIRV2 infection.
We studied the interplay between SIRV2 and its host by monitoring changes in expression of the viral and host genes during the infection cycle by using deep transcriptome sequencing (RNAseq). In addition, we performed a yeast two-hybrid screen, the results of which could be used in combination with gene expression profiles to predict roles of viral proteins in currently unknown processes. This approach uncovered a mild temporal regulation of viral gene expression but dramatic changes of gene expression of the host. More than one-third of all host genes were differentially transcribed, with a clear bias toward genes involved in cell division and defense against foreign genetic elements.
MATERIALS AND METHODS
Growth and infection of S. islandicus.
Sulfolobus islandicus strain LAL14/1 was grown and infected by SIRV2 as described previously (12).
Transmission electron microscopy.
SIRV2-infected S. islandicus cells were prepared for electron microscopy at distinct time points after the addition of the virus (i.e., 0, 5, 9, and 12 hpi). Cells were fixed with 2.5% (wt/vol) glutaraldehyde in 0.1 M HEPES buffer (pH 6.5). Postfixation, dehydration, embedment in epoxy resin, sectioning, and transmission electron microscopy imaging were performed as described previously (12).
Yeast two-hybrid analysis.
The yeast two-hybrid analysis was essentially performed as described by Rajagopala and Uetz (17). Briefly, open reading frames of SIRV2 were amplified (see Table S1 in the supplemental material) and cloned to pENTR vectors for Gateway transfer to the bait (pGBT9g) and prey (pGAD424g) vectors. The bait vectors were transformed to Saccharomyces cerevisiae AH109 Mata, and the prey vectors were transformed to S. cerevisiae Y187 Matα. An autoactivation assay was performed which showed that the addition of 3 mM 3-amino-1,2,4-triazole (3-AT) was most optimal for the reduction of background growth of the strains. Under these conditions, none of the bait constructs resulted in autonomous activation of the reporter constructs. We used an eight-clone pooled, array-based mating screening. The reciprocal screen (exchanging bait and prey) was performed, and verifications of positive interactions were done by sequencing and recloning of the initial constructs to S. cerevisiae strain AH109 and independent Y2H verification.
RNA isolation and library preparation.
Four identical cultures of S. islandicus LAL14/1 were inoculated in 500 ml medium with 5 ml of preculture. After ∼12 h of growth, when the optical density (OD) at 600 nm was in the range of 0.1 to 0.2, two of the cultures were infected with SIRV2, while the other two served as uninfected controls. Total RNA was isolated from these cultures at different time points after the addition of the virus (0, 1, 2, 3, 5, 7, 9 hpi) with the mirVana isolation kit (Ambion) by using the manufacturer's protocol for total RNA isolation. The RNA quality was checked with the 2100 Bioanalyzer (Agilent). A total of 5 μg of total RNA of each sample was directly used for RNAseq library preparation. Directional libraries were prepared using the TruSeq SmallRNA sample prep kit, set A and B (Illumina), according to the manufacturer's instructions. The total RNA was chemically fragmented with Ambion reagent (AM8740), followed by purification on RNeasy columns (Qiagen; catalog no. 74204). After treatment with phosphatase and polynucleotide kinase, RNA was purified on RNeasy columns (Qiagen; catalog no. 74204). The fragmented RNA was then ligated with 3′ and 5′ TruSeq adapters, as described in the manufacturer's protocol. Synthesis of cDNA was performed by reverse transcription. The cDNA products were specifically amplified by 11 cycles of PCR, and products were purified on Agencourt AMPure XP beads (Beckman Coulter Genomics; catalog no. A63881). The resulting libraries were checked on a Bioanalyzer DNA1000 chip (Agilent). Libraries were sequenced to generate single-end 50-base reads using the Illumina Hiseq 2000 in a multiplexed run. Sequencing of 50 bases of each read was followed by sequencing of 6 bases of the tag. Sequencing was performed using a TruSeq SR cluster kit, version 3, cBot HS (Illumina; catalog no. GD-401-3002), and a TruSeq SBS kit, version 3, HS 50 cycles (Illumina; catalog no. FC-401-3002).
Read mapping.
Reads were cleaned from adapter sequences and from sequences of low quality using an in-house program. Only sequences with a minimum length of 30 nucleotides were considered for further analysis. Bowtie (18) (version 0.12.7) was used to align the reads to the reference genomes: S. islandicus LAL14/1 and SIRV2 (15, 19). Gene annotations for SIRV2 and S. islandicus LAL14/1 were downloaded from GenBank (accession numbers NC_004086.1 and CP003928.1, respectively). Reads mapping to rRNA were discarded. For each gene, reproducible sites that overlapped its protein-coding region, as well as those residing in the intergenic region upstream of its beginning, were associated with the gene. Viral genes of which the maximal expression level did not exceed 250 reads per kilobase of transcript per million mapped reads (RPKM) were considered lowly expressed genes.
Statistical and GO analysis.
DEseq (20) and EdgeR (21) were used to determine significantly up- and downregulated genes, comparing all samples of each time point for each condition (infected or control) and comparing all samples of each condition for each time point. Genes marked as differentially regulated by both methods (P < 0.05) were kept for further analysis. Venn diagrams were constructed using BioVenn (22). For each list of up- or downregulated genes between samples, a gene ontology (GO) analysis was performed using GOseq (23) on the 10% of genes with the lowest P values. Genes were sorted to functional groups according to the Clusters of Orthologous Groups (COG) data available on the NCBI site: http://www.ncbi.nlm.nih.gov/COG/grace/fiew.cgi. Archaeal COGs (arCOGs) were linked with S. islandicus LAL14/1 genes based on homology of these with other S. islandicus strains available in the arCOG database (24).
Microarray data accession number.
RNAseq data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-1660.
RESULTS
SIRV2 infection of S. islandicus and RNA isolation.
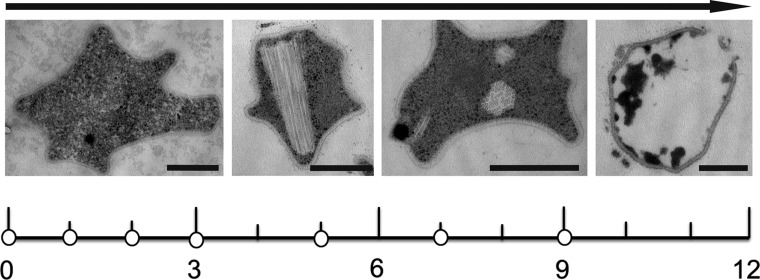
Cells of S. islandicus LAL14/1 were infected by SIRV2 under conditions causing infection in 95% of the cell population (25) and resulting in severe retardation of cell growth (data not shown). Electron microscopic analysis of a time series of cells after infection revealed typical morphology of SIRV2-infected cells, with bundles of mature virions in the cytoplasm and the virion egress structures, VAPs, appearing on the cell surface (Fig. 1).
Fig 1.
Time scale of the SIRV2 infection cycle in the Sulfolobus host cell, indicating time points of RNA isolation (white dots) in hours postinfection. Above the time scales are shown electron micrographs of thin sections of representative SIRV2-infected S. islandicus cells at each stage of infection. During the first hours after infection (t < 3 hpi), cells appear similar to uninfected control cells. Later, bundles of virions assemble in the cytoplasm (t = 3 to 6 hpi), followed by VAP formation (t = 6 to 9 hpi) and eventually opening of the VAPs and release of the virions (t = 10 to 12 hpi). Bars, 500 nm.
Time points for RNA analysis were selected based on the available information on the SIRV2 infection cycle (12). Total RNA was isolated 0, 1, 2, 3, 5, 7, and 9 hpi from two biological duplicates. For control experiments, RNA was isolated at the same time points from two independent uninfected control cultures. Whole-transcript sequencing was applied using the Illumina RNAseq technology. The resulting reads were mapped to coding sequences of the S. islandicus LAL14 and SIRV2 genomes. Reads that mapped to ribosomal RNAs were discarded (Materials and Methods). The fraction of reads mapping to rRNA sequences was approximately the same for all samples. This yielded a total of ∼4 million whole-transcript reads per sample mapped to viral and host genomes.
Identification of differentially regulated genes.
For each gene, the mapped reads of biological duplicates were averaged, and the accumulation levels of specific RNAs were compared between all samples of each time point/condition using both DEseq and EdgeR analysis programs (20, 21). Very similar sets of significantly differentially expressed genes were obtained with both programs. Differences were observed only for poorly expressed genes or for genes that displayed little change in expression. Genes marked as significantly differentially expressed by both programs were retained for further analysis.
Uninfected control samples at each time point had highly similar gene expression profiles. In contrast, pronounced variations of gene expression were observed between the control and infected samples. More than one-third of all S. islandicus genes were significantly up- or downregulated in infected cells compared with those in control cells (see Table S3 in the supplemental material). The gene expression profiles of biological duplicates displayed extensive similarity, indicating that the differential expression is not a direct consequence of the SIRV2-induced chromosome degradation. In addition, a prominent variation was observed between gene expression of infected cells sampled at various time points after infection. Comparing infected samples harvested at different times, between 10 and 50% of all genes were differentially expressed (Table S3).
In Fig. 2, a heat map is presented which shows gene expression profiles of S. islandicus genes of all analyzed samples. The control samples cluster together, which demonstrates the extended similarity in gene expression between uninfected samples. The infected samples also cluster, but at a very different position, stressing the variation in gene expression profile with respect to the control cultures (Fig. 2). One exception is the first harvested infected sample (t = 0 hpi) of which RNA was isolated just after the addition of the virus; as expected, this sample closely resembles the uninfected samples. Divergence is observed between expression patterns of infected samples collected at different time points, which is indicative of extensive temporal regulation of expression in the course of the infection cycle (Fig. 2).
Fig 2.
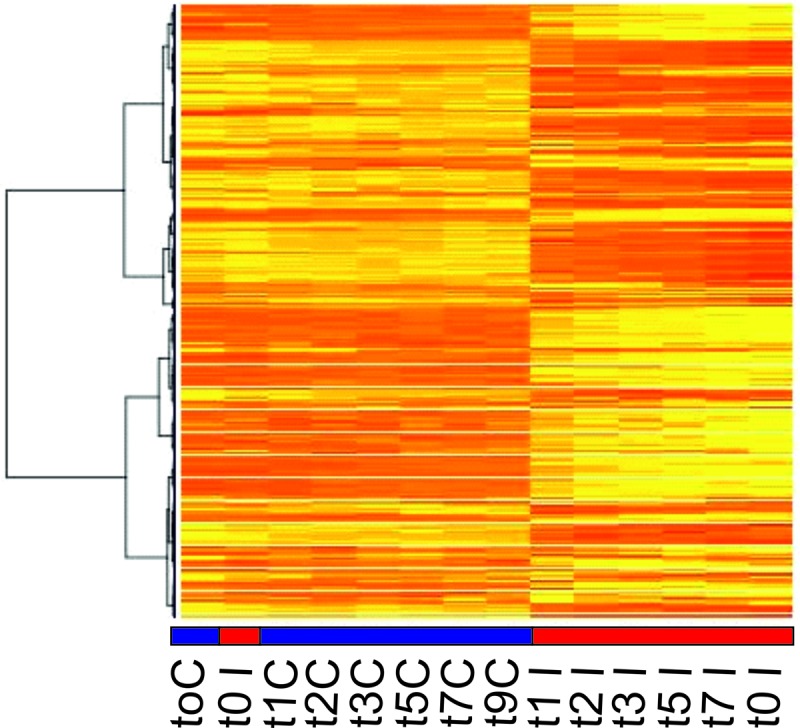
Heat map of S. islandicus gene expression patterns of uninfected control (blue line) and SIRV2-infected (red line) cells. Up- and downregulated genes are shown in yellow and red, respectively. Genes are clustered according to their expression profile in all samples. t, the time (hours) after infection that RNA was isolated; C, uninfected control cells; I, SIRV2-infected cells.
Viral genes.
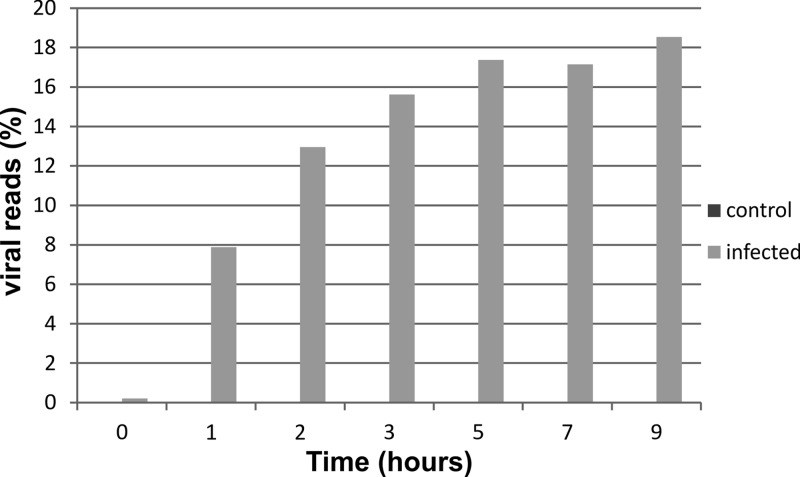
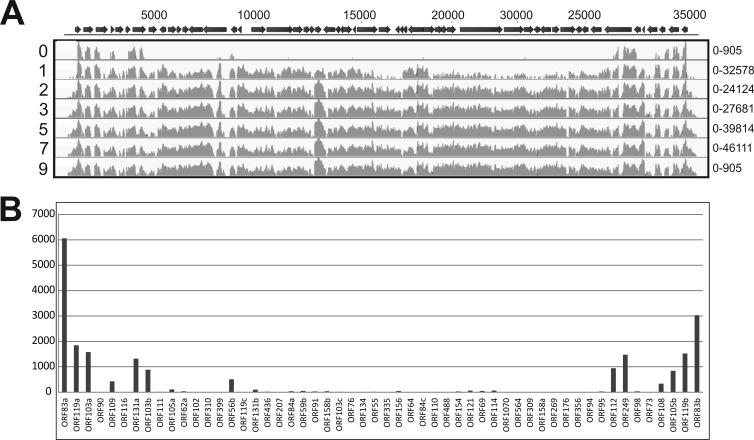
Substantial numbers of reads mapping to the SIRV2 genome were identified in infected samples but never in control samples. The number of viral reads increased steadily in the course of infection, reaching a plateau around 5 hpi, at which time point ∼20% of all transcripts (excluding rRNA hits) mapped to the viral genome (Fig. 3). Although the total number of reads mapping to the viral genome was still relatively low in the first sample taken after infection (t = 0 hpi), a clear trend could be observed in the genomic location. Viral reads at t = 0 hpi mapped typically to ORFs at the distal ends of the linear dsDNA genome, indicating that transcription starts simultaneously from both ends of the SIRV2 genome (Fig. 4; see also Table S2 in the supplemental material). Expression was exclusively observed from the predicted ORFs, and hardly any reads mapped to the noncoding terminal repeat regions of the genome. ORF83a (NP_666535.1) and ORF83b (NP_666588.1) were the highest-expressed genes at this time point and are located on either end of the linear dsDNA genome. They have identical nucleotide sequences and could be distinguished only by differing sequences in their untranslated regions, showing that ORF83a and ORF83b are expressed approximately in a 2:1 ratio. All other expressed genes at t = 0 hpi were located close to the genome termini and include ORF119a (NP_666536.1), ORF103a (666537.1), and ORF119b (NP_666587.1), of which the former codes a protein with unknown function and the latter two code proteins belonging to the DUF1374 superfamily, of which several members are present in the genomes of SIRV2 and other archaeal viruses. In samples from all time points (except t = 0 hpi), significant expression of practically all viral genes was detected. Very poorly expressed genes (<250 RPKM) were ORF76 (NP_666559.1) and ORF119c (NP_666550.1), of which the former is coding for a protein of unknown function and the latter for a Rep protein proposed to initiate SIRV2 replication (26). A negligible number of reads mapped to regions outside annotated ORFs, besides those from intergenic regions on polycistronic messengers. However, there were a few exceptions: the region between ORF119c (NP_666550.1) and ORF131b (NP_666551.1) and the region between ORF156 (NP_666563.1) and ORF64 (NP_666564.1). A moderate number of reads mapped to both regions in the 5′-to-3′ direction. Both transcripts likely contain protein-coding genes which were not predicted in the original annotation due to their short length (<150 bp) (16). Expression of the majority of viral genes increased in the course of infection. A few reached the highest level of expression at 1 or 2 hpi and decreased later during infection (Fig. 3; see also Table S2 in the supplemental material). In all infected samples (except t = 0 hpi), the gene expression appeared to be randomly distributed over the SIRV2 genome. At 1 hpi, the most abundantly transcribed genes were ORF83a, ORF83b, ORF56b (NP_666549.1), ORF108 (NP_666585.1), and ORF103a (NP_666537.1) (Fig. 4; see also Table S2 in the supplemental material). ORF56b encodes the transcriptional regulator SvtR, which acts as a repressor of a number of viral genes, most importantly ORF98 and ORF1070 (NP_666572.1), that code for the VAP and tail fiber proteins, respectively (27). ORF103a and ORF108 code for proteins with unknown functions.
Fig 3.
The percentage of viral transcript increases in time. A plot is shown in which reads mapping to the viral genome are shown as the percentage of the total number of detected reads. Depicted are control (dark gray; not visible in graph) and SIRV2-infected (light gray) samples of different time points after infection.
Fig 4.
Transcription profiles of SIRV2 genes during viral infection. (A) RNA was isolated from S. islandicus cells at several hours postinfection, indicated by the numbers on the left associated with each transcription map. On the top, a schematic representation of the SIRV2 genome is shown, of which the numbers above indicate the base pair position. Arrows represent ORFs. The reads per kilobase of transcript (RPKM) mapping to the SIRV2 genome are depicted in light gray on a log scale. Minimum and maximal values of the y axis of each transcription map are shown on the right. (B) Detail of SIRV2 gene expression directly after infection (t = 0). RPKM are depicted on the y axis for all 54 annotated genes shown on the x axis in the order which they reside on the SIRV2 genome. For detailed information on expression of individual viral genes, see Table S2 in the supplemental material.
At late stages of infection, expression of genes coding for structural proteins increased, such that at the end of the infection cycle, ∼35% of viral reads mapped to the gene encoding the major coat protein, ORF134 (NP_666560.1), and ∼13% to the VAP gene, ORF98, encoding the component of the pyramidal egress structures. Other abundantly expressed genes at this stage were ORF83a, ORF83b, and ORF110 (NP_666566.1), a gene of unknown function.
Yeast two-hybrid screen of viral proteins.
An array-based yeast two-hybrid screen was performed to provide additional information about possible functions of viral gene products. All 54 SIRV2 genes and six truncated genes lacking transmembrane domains were cloned to both bait and prey vectors and transferred to yeast for an ORFeome array-based screen. Yeast two-hybrid screens were performed for each gene both in prey and in bait vector to limit the number of false-positive interactions and were quantified by a standard α-galactosidase assay. Several protein interactions between SIRV2 proteins were identified (Table 1). The majority of them reflected intramolecular associations of SIRV2 proteins. In addition, some interactions between different SIRV2 proteins were identified. The proteins of unknown function from ORF95 (NP_666580.1) and ORF73 (NP_666584.1) were found to interact. Binding was observed also between two different predicted glycosyltransferases, encoded by ORF356 (NP_666578.1) and ORF335 (NP_666562.1). Interestingly, interactions were detected between the highly expressed predicted DNA binding proteins from ORF83a/b and the Holliday junction resolvase encoded by ORF121 (NP_666569.1).
Table 1.
Yeast two-hybrid interaction analysis of SIRV2
| Confirmed interaction | Gene name | Function | Gene of binding partner | Binding partner |
|---|---|---|---|---|
| Heterotypic | ORF83a | DNA binding protein | ⇆ ORF121 | Holliday junction resolvase |
| ORF335 | GT1 glycosyltransferase | ⇆ ORF356 | Glycosyltransferase | |
| ORF121 | Holliday junction resolvase | ⇆ ORF83b | DNA binding protein | |
| ORF95 | Unknown protein | ⇆ ORF73 | Unknown protein | |
| Homotypic | ORF103a | Unknown protein | ||
| ORF90 | With transmembrane domain | ORF90 without TMD shows no interaction | ||
| ORF131a | Unknown protein | |||
| ORF84a | Unknown protein | |||
| ORF91 | Unknown protein | |||
| ORF154 | GCN5 acetyltransferase | |||
| ORF69 | Unknown protein | |||
| ORF108 | Unknown protein |
Host gene response to viral infection.
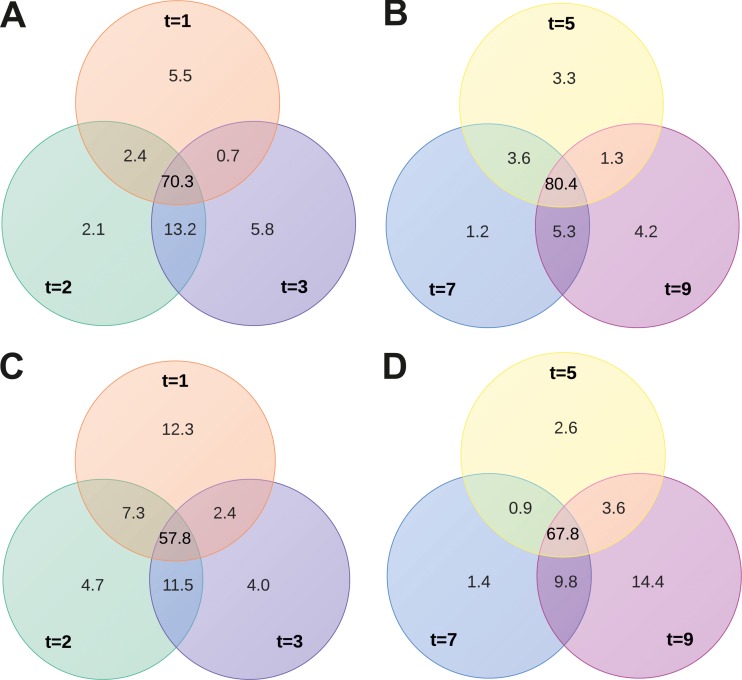
After infection with SIRV2, extensive changes in S. islandicus gene expression were observed. About 30 to 50% of all host genes were differentially expressed in infected samples and uninfected control cultures, and these differences were very pronounced (see Table S3 in the supplemental material). The numbers of up- and downregulated genes were approximately the same. However, the degree of increase in expression was much higher among the upregulated genes than the degree of decrease in expression of downregulated genes (see Table S3). In data sets from all time points, a relatively high fraction (∼50 to 80%) of identical genes were marked as significantly differentially expressed (Fig. 5). Venn diagrams, representing the extent of similarity between samples, show slightly more uniformity in the downregulated compared to in the upregulated genes from distinct time points (Fig. 5).
Fig 5.
Correspondence between sets of differentially regulated genes harvested at various time points, comparing SIRV2-infected and uninfected control cells. Venn diagrams depict the percentages of identical and unique genes among the differentially regulated groups taken at 1 to 9 hpi (t = 1 to t = 9). Downregulated (A, B) and upregulated (C, D) genes.
An analysis was performed with GOseq on differentially regulated genes (23) to detect overrepresented functional categories of genes. One functional category was significantly overrepresented among genes of which expression increased after infection (from 1 to 5 hpi). Genes involved in defense mechanisms belong to this category (see Table S3 in the supplemental material). Among the downregulated genes (from 1 to 3 hpi), a single functional group was significantly overrepresented. Its members are genes involved in cell cycle control, cell division, and chromosome partitioning (see Table S3). Both of these functional categories are discussed in more detail below.
Proteins involved in cell division.
Genes belonging to the cell cycle control, cell division, and chromosome partitioning functional category were downregulated after infection. This category has a fairly small number of members, with only 14 genes present on the S. islandicus LAL14/1 genome. Half of these genes are annotated as ATPases involved in chromosome partitioning. However, four out of these seven genes encoding ATPases were upregulated after infection and evidently did not contribute to the overrepresentation of this category among the downregulated genes (see Table S3 in the supplemental material). The majority of the other genes from this category share sequence similarity with eukaryotic genes that encode members of the ESCRT-III sorting complex. In S. islandicus, these genes are part of the cdv operon that comprises cdvA, cdvB, and cdvC and codes for the Sulfolobus cell division machinery, responsible for the constriction of dividing cells (28, 29). Interestingly, the complete cdv operon was downregulated around 10-fold as a result of infection (see Table S3). Moreover, the three other cdvB paralogs present in different regions on the S. islandicus LAL14/1 genome were downregulated 3- to 10-fold as a consequence of infection (see Table S3).
CRISPR-Cas.
Following SIRV2 infection, expression of genes belonging to the functional category of defense mechanisms increased (see Table S3 in the supplemental material). Many members of this category encode clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems, operons I-A SiL_0385-0392, I-A SiL_0393-0397, I-D SiL_0606-0609, I-D SiL_0610-0613, III-Bα SiL_0786-0793, and III-Bβ SiL_0600-0605 (see Table S3) (19). The CRISPR-associated (cas) genes play essential roles in the CRISPR-mediated prokaryotic adaptive immune system that can protect cells against invasion of mobile genetic elements, i.e., plasmids and viruses (30–32). The S. islandicus LAL14/1 genome contains five complete and one incomplete CRISPR-cas arrays. The complete CRISPR-cas arrays consist of a CRISPR array and adjacently located cas genes. Unique spacer sequences matching specifically to foreign genetic elements are located between the CRISPR repeat sequences (32). Six cas operons are encoded: two I-A subtypes, two III-B subtypes (III-Bα SiL_0786-0793 is incomplete, lacking the CRISPR array), and two I-D subtypes consisting of two adjacent gene clusters coded in opposite directions (19, 33). In uninfected control cells, reads mapping to all individual cas genes were detected, although the expression levels of cas operons differed considerably. One I-A (SiL_0385-0392) and the III-Bα SiL_0786-0793 operons are highly expressed, and the other type III-B operon (III-Bβ SiL_0600-0605) and one I-D operon (I-D SiL_0606-0609) are moderately expressed (Fig. 6; see Table S4 in the supplemental material). Under these conditions, expression of the other I-A operon (I-A SiL_0393-0397) and part of the I-D operon (I-D SiL_0610-0613) could hardly be detected (Fig. 6; see Table S4 in the supplemental material). The CRISPR arrays located adjacent to each cas operon were expressed to a degree roughly similar to that of the cas genes that they are clustered with (Fig. 6; see also Table S4 in the supplemental material). All CRISPR arrays contain several spacers matching to different viruses, mostly Rudiviridae. Only the CRISPR array 2, adjacent to operon I-A SiL_0393-0397, contains a large proportion of spacers which target plasmids (19). This CRISPR array is hardly expressed (Fig. 6; see also Table S4).
Fig 6.
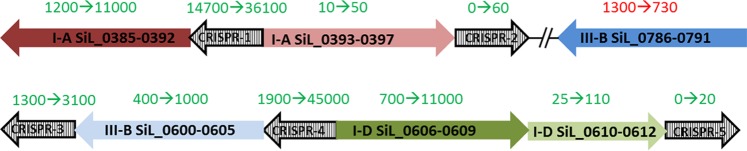
CRISPR-cas expression increases after SIRV2 infection. Schematic representation of the six cas operons and five associated CRISPR arrays present on the S. islandicus LAL14/1 genome. Operons of type I-A (red), I-D (green), and III-B (blue) are present. Numbers within arrows indicate the gene names of all genes of which each operon consists. Numbers above the arrows represent the numbers of reads mapping to genes of the represented operon in control (left) and SIRV2-infected (right) cells. Colors of these numbers indicate expression increase (green) or decrease (red) upon infection.
Interestingly, at 1 hpi, expression of the majority of cas operons strongly increased. The I-A SiL_0385-0392 and I-D SiL_0606-0609 operons were upregulated around 10-fold, the I-A SiL_0393-0397 and I-D SiL_0610-0613 operons were upregulated approximately 5-fold, and the III-Bβ SiL_0600-0605 operon was upregulated 2-fold. Expression of the I-D SiL_0606-0609 operon increased during infection, while expression of the other cas operons reached the highest level at 1 hpi and remained stable in the course of the infection cycle (see Table S4 in the supplemental material). Expression of cas genes as a result of SIRV2 infection was so pronounced that 3.7% of all mapped reads originated from cas operons. The incomplete III-Bα SiL_0786-0793 operon, without a CRISPR array adjacent to it, represents the only cas operon which is slightly downregulated after infection (∼2-fold) (Fig. 6; see also Table S4). Thus, expression of all CRISPR arrays increased after SIRV2 infection, and typically expression kept increasing during the viral infection cycle, in contrast to the adjacent cas genes, which usually reached highest expression levels quickly after the onset of infection (see Table S4).
Toxin-antitoxin.
Together with the CRISPR-Cas systems, toxin-antitoxin (TA) systems belong to the functional category of defense mechanisms. Many TA genes were abundantly upregulated in SIRV2-infected cells (see Table S5 in the supplemental material). Prokaryotic TA systems are widespread mobile two-gene elements that are subdivided into three families based on the nature and mode of action of the antitoxin (34). The family II TA systems consist of an antitoxin protein that counteracts the negative effect of the toxin protein, which is usually a nuclease that is more stable than the antitoxin (35). Family II TA operons are widespread in Archaea and especially among members of the order Sulfolobales (34). The genome of S. islandicus LAL14/1 carries 16 TA operons of the family II type VapBC (a virulence-associated protein) (19). In addition, 6 TA operons of another recently described family, HEPN-NT (higher eukaryote and prokaryote nucleotide binding-nucleotidyltransferases), are present (19, 34). Of this HEPN-NT family, 15 separate antitoxin-coding genes are found on the S. islandicus LAL14/1 genome, and they do not appear to be associated with toxins. In uninfected S. islandicus LAL14/1 cells, the majority of TA loci are moderately transcribed, although expression levels between loci can differ considerably (see Table S5 in the supplemental material). In SIRV2-infected samples at different time points (except t = 0 hpi), expression of 11 out of 16 VapBC and 3 out of 6 HEP-NT loci increased. In most cases, both the genes coding for the antitoxin and the toxin were upregulated and expressed to similar extents. Gene expression of only two antitoxins decreased after SIRV2 infection.
Since expression of TA gene clusters and stress response genes was previously reported to increase after heat shock (36), we checked for changes in expression of known stress response genes (i.e., heat shock proteins [HSP], universal stress proteins [USP], proteasome genes). After SIRV2 infection, expression of these genes decreased (∼3-fold), or the genes appeared nonresponsive and expression remained constant (see Table S3 in the supplemental material).
Insertion sequence elements.
Just one functional category was marked as overrepresented among upregulated genes after viral infection. It is possible that specific subgroups were not scored as overrepresented in the GOseq analysis, because they are part of a large functional category to which many genes belong. Therefore, overrepresentation of subgroups was checked manually. Among the genes of which expression increased after infection, a high proportion of insertion sequence (IS) elements was detected (12 to 15% of all upregulated genes). These belong to the large functional category of replication, recombination, and repair (see Table S3 in the supplemental material).
The S. islandicus LAL14/1 genome contains a high number of IS elements, similar to the genomes of S. islandicus strains HVE10/4 and REY15A and of Sulfolobus solfataricus. The latter genome contains approximately 200 IS elements, of which active transposition has been observed (37). In the S. islandicus LAL14/1 genome, 53 predicted IS elements are present, of which only seven encode intact transposases. However, since IS elements with mutated transposases can be mobilized by transposases of the same family acting in trans, a total of 31 IS elements in the S. islandicus LAL14/1 genome could potentially be mobile (19). In uninfected cells, the majority of IS elements were transcribed. As a result of infection, expression of many IS elements from different families increased, such that they formed the largest group of upregulated genes (see Table S3 in the supplemental material). The most actively expressed were members of the IS1048 group (IS630 family) (see Table S3).
Induction of transcription of IS elements after virus infection is peculiar and has not been reported previously. However, a pronounced increase of expression of IS elements of S. solfataricus has been observed as a result of heat shock and UV irradiation (38). Therefore, high expression of IS elements was suggested to be associated with the general stress response of S. solfataricus (36).
DISCUSSION
Using in-depth transcriptome analysis, we have monitored the dramatic changes in gene expression occurring during infection of the archaeon S. islandicus with SIRV2. The amount of viral transcripts increases in time, until it constitutes approximately 20% of all mRNA in the infected cells. Transcription of these genes has a significant impact on host cell morphology and metabolism. The host responds to this threat by a vast change in gene expression (∼50% of genes). Most of the host genes that are strongly activated upon infection are assumed to function in defense against viruses, thus unveiling a regulatory mechanism that aims at counteracting the viral attack.
Viral gene expression.
SIRV2 gene transcription starts from the two distal ends of the SIRV2 linear dsDNA genome. For some bacterial viruses, a slightly similar situation is reported, and early transcription was observed from genes located on the termini of linear dsDNA genomes of the phi29 and PRD1 viruses, from the families Podoviridae and Tectiviridae, respectively (39–41). In the case of SIRV2, this pattern of gene expression might be a consequence of the mode of viral infection that remains currently obscure. Since the two ends of the SIRV2 virion are identical, the binding of the virion and entry of the viral genome could potentially occur from both sides. In this case, it would be advantageous when ORF83a and ORF83b, obviously important at early stages of infection, are located at either end of the genome and are readily available for transcription. Alternatively, the location of genes with identical complementary sequences on both ends of the genome might be advantageous for SIRV2 genome replication, which includes formation of head-to-head and tail-to-tail replicative intermediates (15). The exact nucleotide identity between SIRV2_ORF83a and SIRV2_ORF83b and between their homologs in SIRV1 suggests the presence of selection pressure to maintain this trait.
The genes highly transcribed later during SIRV2 infection are distributed evenly across the viral genome. Little temporal regulation is observed, and expression of the majority of genes starts immediately after infection and subsequently steadily increases. This is in contrast to the viral gene expression of the temperate, archaeal SSV, of which the expression pattern during the first hours after UV induction is dominated by only one early transcript (6). The qualitative results of previously performed Northern blot hybridization and primer extension analysis of genome transcription of the rudiviruses SIRV1 and SIRV2 correspond with our finding that almost all viral genes are transcribed after infection (7).
Implications for comprehension of viral gene functions.
In general, the expression of SIRV2 genes corresponds very well with predicted or confirmed gene functions. Transcripts of structural genes (encoding proteins of the viral capsid or egress structure), like ORF98, ORF134, and ORF1070, are most abundant late in the infection cycle, while expression of the transcriptional regulator SvtR, which represses ORF98 and ORF1070, peaks soon after infection (27). Indeed, expression of ORF98 and ORF1070 is inversely proportional to that of SvtR.
Yeast two-hybrid analysis uncovered several homotypic interactions of SIRV2 proteins, all of them without predicted functions. Two different glycosyl transferases were found to interact, and correspondingly, glycosyl transferases are known to be capable of protein complex formation (25, 42).
ORF119c encodes a Rep protein that was assumed to initiate viral genome replication and aid in resolution of replicative intermediates (26). The virus-encoded Holliday junction-resolving enzyme was suggested to be required for recovery from situations in which linear SIRV2 genome replicative intermediates are generated by Rep cleavage failure (26). Surprisingly, expression of ORF119c peaks at the end of the infection cycle, is very poor, and is approximately 3,000-fold lower than that of the Holliday junction resolvase ORF121 (NP_666569.1). This suggests that the Rep protein is not required for the analyzed mode of viral replication and might be implemented in currently unknown aspects of the viral cycle.
In addition to confirming already proposed functions of genes, the expression data presented here also suggest an important role for some uncharacterized viral genes. Most of the early expressed genes located on the genome termini have unknown functions. SIRV2_ORF103a and SIRV2_ORF119b have a homolog in Acidianus filamentous virus 1 (AFV1), AFV1_ORF99, of which the crystal structure was determined (43). AFV1_ORF99 displays a completely novel fold from which no function prediction could be derived (43). Interestingly, ORF83a and ORF83b are the first transcribed genes after infection. These genes have identical nucleotide sequences and are located in opposite directions on each outer end of the genome. Although the expression decreases after 1 hpi, the number of transcripts mapping to ORF83a and ORF83b remains very high. Since ORF83 encodes a protein with predicted DNA binding capacity and is the first gene to be transcribed, it is tempting to speculate that it might have an essential function in viral genome replication. In the closely related SIRV1, the ORF83 homologs SIRV1_ORF56a and SIRV1_ORF56b are also located on the extremities of the linear genome (15). The crystal structure has been determined for the ORF83 homolog in SIRV1 (ORF56a), which, compared to ORF83, is lacking 27 amino acids on the N terminus (44). The protein has a hexameric configuration, and it contains an unusually conserved C-terminal cysteine, which might be involved in subunit-subunit cross-linking. The C-terminal half of the protein displays a classical helix-turn-helix (HTH) domain, which is often found among DNA binding proteins and specifically transcriptional regulators (44). This is reconcilable with a function in genome replication, although the DNA binding activity of ORF83 has not been experimentally characterized. For initiation of SIRV2 replication, nicking activity would be required as well. The potential involvement of ORF83 in SIRV2 genome replication is enforced by the detected interaction between ORF83 and ORF121 by a yeast two-hybrid screen. ORF121 is coding for the Holliday junction resolvase that is implied in the resolution of replicative intermediates (45). Thus, there seems to be a link between ORF83 and the late steps of genome replication.
Host response to viral infection.
The consequence of SIRV2 infection could be detected by the adaptation of the S. islandicus gene expression profile. Specifically, there was an intensive decrease of expression from genes involved in cell division, notably ESCRT-III homologs and components of the cdv operon. cdvB and cdvC have homologs in the ESCRT-III complex involved in the budding of luminal vesicles in eukaryotes (46). The decrease in expression of these genes is likely to result from the DNA degradation caused by SIRV2 infection, since the Cdv proteins are shown to be under the control of checkpoint systems, which inhibit cell division in response to DNA damage (28). Interestingly, it was reported that after infection with the archaeal virus STIV, cdv genes were significantly upregulated in the host, Sulfolobus solfataricus (9), implying an important function in the STIV infection cycle.
Activation of CRISPR-Cas systems.
Recent discoveries of novel prokaryotic antiviral defense mechanisms have raised awareness that simple prokaryotes, in analogy to eukaryotes, have a range of sophisticated immune systems at their disposal. The CRISPR-Cas system is the most recently described example of this (32, 47). CRISPR-Cas systems are very abundant among archaea, and often several different systems are present on the same archaeal genome. The repeat spacer arrays are relatively long and can make up 1% of archaeal genomes (6, 48), which indicates the importance of these systems for the fitness of archaeal cells. Hyperthermophilic archaeal CRISPR-Cas systems have been studied extensively using Pyrococcus furiosus and S. solfataricus as models (49, 50). Both organisms encode various Cas complexes, a subset of which has been analyzed in vitro (49–51). Transcription of CRISPR arrays was detected in S. solfataricus and P. furiosus (52, 53). However, behavior of archaeal CRISPR-Cas systems during viral infection has hardly been studied until now. Nevertheless, during a recent proteomics analysis, a number of Cas proteins were detected after STIV infection, suggesting that there might be activation of CRISPR-Cas as a consequence of viral attack (8). The presence of so many different CRISPR-Cas systems in archaeal cells has raised questions about the diverse roles they might have during viral infection.
We observed that SIRV2 infection resulted in massive activation of the CRISPR-Cas systems present in S. islandicus. The S. islandicus LAL14/1 genome contains six cas operons, coding for complexes of three different types (I-A, I-D, and III-B). Expression of five out of six cas operons increased as a result of viral infection. Interestingly, expression of almost all cas operons augmented 3- to 10-fold, just like the associated CRISPR arrays, making them by far the most pronounced upregulated genes after infection. All systems are activated directly after viral infection. Two operons, belonging to the I-A and I-D subtype, were considerably transcribed in uninfected cells, and after viral infection, their activation was most pronounced of all Cas operons. After viral infection, expression of the type III-B Cmrα complex, without adjacent CRISPR array, decreased (54). The type III-B Cmrβ complex was moderately expressed in noninfected cells, and its expression increased slightly after infection. Thus, the type III-B operons appear to react to a lesser extent to SIRV2 infection. These results indicate that type I-A and I-D operons, which code DNA-targeting Cas complexes, play a more important role during SIRV2 infection than the type III-B-encoded complexes.
CRISPR arrays of S. islandicus LAL14/1 does not possess any spacers perfectly matching to the SIRV2 genome, which is probably the reason why cells do not recover from an SIRV2 infection despite the widespread activation of CRISPR-Cas systems.
The described findings demonstrate that the different CRISPR-Cas systems present in archaea probably have specialized roles which could result in a tailor-made defense reaction for each different type of foreign genetic element. Some Cas operons (I-A SiL_0385-0392 and III-Bα SiL_0786-0793) are actively transcribed in uninfected cells, indicating that the encoded Cas proteins might be continuously present in cells to act as “watchers at the gate” which can target foreign genetic elements directly when they access the cell. In contrast, other Cas systems are hardly expressed without activation signal arising from viral infection, which is the case for the I-A SiL_0393-0397 operon and adjacent CRISPR array, containing a high proportion of plasmid-targeting spacers.
Expression of toxin-antitoxin systems.
As a result of SIRV2 infection, toxin-antitoxin (TA) clusters of the host genome are activated. Such clusters are ubiquitously present on prokaryotic genomes (34, 55), and they are proposed to function in programmed cell death and in stress response (36, 56). In addition, it was demonstrated that bacterial TA systems play a role in abortive infection, during which cells commit altruistic suicide after viral infection to protect the clonal population (57, 58). TA suicide/dormancy systems were proposed to be linked with diverse immunity systems in prokaryotes to provide robustness to the antivirus response (59). Indeed, in S. islandicus LAL14/1, as in many prokaryotes, a high proportion of TA loci are found in the close proximity of CRISPR-cas operons, suggesting their involvement in defense against viruses (59). However, the involvement of archaeal TA systems in antiviral defense has not yet been demonstrated, although numerous TA-encoding genes are present in archaeal genomes. Interestingly, the most actively transcribed S. solfataricus gene as a consequence of STIV infection has an unknown function (9), but homology searches suggest it might be an antitoxin part of a TA system. SIRV2 infection results in a severe increase of expression of many TA loci, of which the majority is VapBC like (family II). The observation that SIRV2 infection did not cause increased expression of any of the regular stress response genes suggests that high TA expression in this case is not linked with basal stress response and might be a specific reaction to viral infection.
Here, we have presented a global map defining host and viral gene expression during the infection cycle of SIRV2 in its host, the hyperthermophilic archaeon S. islandicus LAL14/1. This pioneering study provides in-depth information for all viral and host genes during a time series after prokaryotic viral infection. This work has demonstrated once more the power of the recently developed RNAseq approach. These findings, in combination with performed yeast two-hybrid analysis of the viral ORFeome, have corroborated predictions for functions of some SIRV2 genes, while suggesting novel functions for others. Moreover, it enables the detailed study of a host response to a viral infection, showing massive activation of host antiviral defense genes, most importantly the CRISPR-Cas and TA systems, in the presented case. This information on individual gene expression and activation of all the antiviral defense systems is expected to aid future studies aiming to establish the function and interplay of the different systems in vivo.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to Pieter-Jan Ceyssens and Kira Makarova for discussions.
This work was supported by the l'Agence National de la Recherche, France, in the framework of Programme Blanc and by the Netherlands Organization for Scientific Research (NWO) via an ALW TOP grant (854.10.003). T.E.F.Q. was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche of France. M.V. and R.L. are supported by a grant from the FWO Vlaanderen (G.0323.09).
Footnotes
Published ahead of print 22 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01020-13.
REFERENCES
- 1. Krupovic M, White MF, Forterre P, Prangishvili D. 2012. Postcards from the edge: structural genomics of archaeal viruses. Adv. Virus Res. 82:33–62 [DOI] [PubMed] [Google Scholar]
- 2. Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol. Rev. 35:1035–1054 [DOI] [PubMed] [Google Scholar]
- 3. Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, Johnson JE, Douglas T, Young M. 2004. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc. Natl. Acad. Sci. U. S. A. 101:7716–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter WD, Zillig W. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242–250 [DOI] [PubMed] [Google Scholar]
- 5. Prangishvili D, Arnold HP, Gotz D, Ziese U, Holz I, Kristjansson JK, Zillig W. 1999. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the sulfolobus viruses SIRV1 and SIRV2. Genetics 152:1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frols S, Gordon PM, Panlilio MA, Schleper C, Sensen CW. 2007. Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology 365:48–59 [DOI] [PubMed] [Google Scholar]
- 7. Kessler A, Brinkman AB, van der Oost J, Prangishvili D. 2004. Transcription of the rod-shaped viruses SIRV1 and SIRV2 of the hyperthermophilic archaeon sulfolobus. J. Bacteriol. 186:7745–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maaty WS, Selvig K, Ryder S, Tarlykov P, Hilmer JK, Heinemann J, Steffens J, Snyder JC, Ortmann AC, Movahed N, Spicka K, Chetia L, Grieco PA, Dratz EA, Douglas T, Young MJ, Bothner B. 2012. Proteomic analysis of Sulfolobus solfataricus during Sulfolobus turreted icosahedral virus infection. J. Proteome Res. 11:1420–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ortmann AC, Brumfield SK, Walther J, McInnerney K, Brouns SJ, van de Werken HJ, Bothner B, Douglas T, van de Oost J, Young MJ. 2008. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J. Virol. 82:4874–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muskhelishvili G, Palm P, Zillig W. 1993. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol. Gen. Genet. 237:334–342 [DOI] [PubMed] [Google Scholar]
- 11. Schleper C, Kubo K, Zillig W. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. U. S. A. 89:7645–7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bize A, Karlsson EA, Ekefjard K, Quax TE, Pina M, Prevost MC, Forterre P, Tenaillon O, Bernander R, Prangishvili D. 2009. A unique virus release mechanism in the Archaea. Proc. Natl. Acad. Sci. U. S. A. 106:11306–11311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quax TE, Krupovic M, Lucas S, Forterre P, Prangishvili D. 2010. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology 404:1–4 [DOI] [PubMed] [Google Scholar]
- 14. Quax TE, Lucas S, Reimann J, Pehau-Arnaudet G, Prevost MC, Forterre P, Albers SV, Prangishvili D. 2011. Simple and elegant design of a virion egress structure in Archaea. Proc. Natl. Acad. Sci. U. S. A. 108:3354–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng X, Blum H, She Q, Mallok S, Brugger K, Garrett RA, Zillig W, Prangishvili D. 2001. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291:226–234 [DOI] [PubMed] [Google Scholar]
- 16. Prangishvili D, Koonin EV, Krupovic M. 2013. Genomics and biology of rudiviruses, a model for the study of virus-host interactions in Archaea. Biochem. Soc. Trans. 41:443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rajagopala SV, Uetz P. 2011. Network biology: methods and applications. Methods in molecular biology, vol 781 Springer, New York, NY [Google Scholar]
- 18. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jaubert C, Danioux C, Oberto J, Cortez D, Bize A, Krupovic M, She Q, Forterre P, Prangishvili D, Sezonov G. 2013. Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol. 3:130010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hulsen T, de Vlieg J, Alkema W. 2008. BioVenn—a Web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wolf YI, Makarova KS, Yutin N, Koonin EV. 2012. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol. Direct 7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bu S, Li Y, Zhou M, Azadin P, Zeng M, Fives-Taylor P, Wu H. 2008. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J. Bacteriol. 190:1256–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oke M, Kerou M, Liu H, Peng X, Garrett RA, Prangishvili D, Naismith JH, White MF. 2011. A dimeric Rep protein initiates replication of a linear archaeal virus genome: implications for the Rep mechanism and viral replication. J. Virol. 85:925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guilliere F, Peixeiro N, Kessler A, Raynal B, Desnoues N, Keller J, Delepierre M, Prangishvili D, Sezonov G, Guijarro JI. 2009. Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1. J. Biol. Chem. 284:22222–22237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. 2008. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. U. S. A. 105:18942–18946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL, Williams RL, Bell SD. 2011. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol. Cell 41:186–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 31. Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. 2012. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 46:311–339 [DOI] [PubMed] [Google Scholar]
- 33. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Makarova KS, Wolf YI, Koonin EV. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45:61–79 [DOI] [PubMed] [Google Scholar]
- 36. Tachdjian S, Kelly RM. 2006. Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 188:4553–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martusewitsch E, Sensen CW, Schleper C. 2000. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J. Bacteriol. 182:2574–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frols S, Gordon PM, Panlilio MA, Duggin IG, Bell SD, Sensen CW, Schleper C. 2007. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J. Bacteriol. 189:8708–8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davison BL, Murray CL, Rabinowitz JC. 1980. Specificity of promoter site utilization in vitro by bacterial RNA polymerases on Bacillus phage phi 29 DNA. Transcription mapping with exonuclease III. J. Biol. Chem. 255:8819–8830 [PubMed] [Google Scholar]
- 40. Grahn AM, Bamford JK, O'Neill MC, Bamford DH. 1994. Functional organization of the bacteriophage PRD1 genome. J. Bacteriol. 176:3062–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loskutoff DJ, Pene JJ. 1973. Gene expression during the development of Bacillus subtilis bacteriophage phi29. II. Resolution of viral-specific ribonucleic acid molecules. J. Virol. 11:87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Noffz C, Keppler-Ross S, Dean N. 2009. Hetero-oligomeric interactions between early glycosyltransferases of the dolichol cycle. Glycobiology 19:472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goulet A, Spinelli S, Blangy S, van Tilbeurgh H, Leulliot N, Basta T, Prangishvili D, Cambillau C, Campanacci V. 2009. The thermo- and acido-stable ORF-99 from the archaeal virus AFV1. Protein Sci. 18:1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oke M, Carter LG, Johnson KA, Liu H, McMahon SA, Yan X, Kerou M, Weikart ND, Kadi N, Sheikh MA, Schmelz S, Dorward M, Zawadzki M, Cozens C, Falconer H, Powers H, Overton IM, van Niekerk CA, Peng X, Patel P, Garrett RA, Prangishvili D, Botting CH, Coote PJ, Dryden DT, Barton GJ, Schwarz-Linek U, Challis GL, Taylor GL, White MF, Naismith JH. 2010. The Scottish structural proteomics facility: targets, methods and outputs. J. Struct. Funct. Genomics 11:167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gardner AF, Guan C, Jack WE. 2011. Biochemical characterization of a structure-specific resolving enzyme from Sulfolobus islandicus rod-shaped virus 2. PLoS One 6:e23668. 10.1371/journal.pone.0023668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Makarova KS, Yutin N, Bell SD, Koonin EV. 2010. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 8:731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170 [DOI] [PubMed] [Google Scholar]
- 48. Lillestol RK, Redder P, Garrett RA, Brugger K. 2006. A putative viral defence mechanism in archaeal cells. Archaea 2:59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, Naismith JH, Spagnolo L, White MF. 2012. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell 45:303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, III, Graveley BR, Terns RM, Terns MP. 2012. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 45:292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hale C, Kleppe K, Terns RM, Terns MP. 2008. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 14:2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A. 2005. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 55:469–481 [DOI] [PubMed] [Google Scholar]
- 54. Deng L, Garrett RA, Shah SA, Peng X, She Q. 2013. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 87:1088–1099 [DOI] [PubMed] [Google Scholar]
- 55. Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gerdes K, Christensen SK, Lobner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382 [DOI] [PubMed] [Google Scholar]
- 57. Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 106:894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pecota DC, Wood TK. 1996. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 178:2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Makarova KS, Anantharaman V, Aravind L, Koonin EV. 2012. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct 7:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.