Abstract
The cell surface receptor T cell immunoglobulin mucin domain 1 (TIM-1) dramatically enhances filovirus infection of epithelial cells. Here, we showed that key phosphatidylserine (PtdSer) binding residues of the TIM-1 IgV domain are critical for Ebola virus (EBOV) entry through direct interaction with PtdSer on the viral envelope. PtdSer liposomes but not phosphatidylcholine liposomes competed with TIM-1 for EBOV pseudovirion binding and transduction. Further, annexin V (AnxV) substituted for the TIM-1 IgV domain, supporting a PtdSer-dependent mechanism. Our findings suggest that TIM-1-dependent uptake of EBOV occurs by apoptotic mimicry. Additionally, TIM-1 enhanced infection of a wide range of enveloped viruses, including alphaviruses and a baculovirus. As further evidence of the critical role of enveloped-virion-associated PtdSer in TIM-1-mediated uptake, TIM-1 enhanced internalization of pseudovirions and virus-like proteins (VLPs) lacking a glycoprotein, providing evidence that TIM-1 and PtdSer-binding receptors can mediate virus uptake independent of a glycoprotein. These results provide evidence for a broad role of TIM-1 as a PtdSer-binding receptor that mediates enveloped-virus uptake. Utilization of PtdSer-binding receptors may explain the wide tropism of many of these viruses and provide new avenues for controlling their virulence.
INTRODUCTION
Ebolavirus, along with Marburgvirus, is a member of the Filoviridae family of enveloped, nonsegmented, negative-stranded RNA viruses. Infection can cause hemorrhagic fever, with mortality rates as high as 90% (1). The virus targets a broad range of cells, infecting monocytes, macrophages, and dendritic cells early during infection, and spreads to a variety of cell types, including epithelial cells, in the visceral organs (2, 3). Ebolaviruses encode a viral class I glycoprotein (GP) that in the mature form is composed of a trimer of disulfide-bonded GP1/GP2 heterodimers. GP1 contains the receptor-binding domain (RBD), believed to interact with one or more cellular receptors, while GP2 mediates virus/host membrane fusion events (4–7).
The complex steps of filovirus entry are currently being elucidated. Ebolaviruses bind to the cell surface and are internalized into endosomes, where low-pH-dependent cathepsins L and B process the heavily glycosylated GP1 into a 17- to 19-kDa protein that retains the RBD (8, 9). Processed GP1 interacts with the late endosomal/lysosomal protein Neimann-Pick C1 (10–13). Subsequent poorly defined events lead to GP2-dependent fusion of endosomal and viral membranes, resulting in release of the viral core into the cytoplasm.
We and others have sought to identify cell surface receptors that mediate filovirus uptake into endosomes (14–18). C-type lectins may mediate this process in some cell types; however, these proteins are not present on all permissive cells, indicating that additional filovirus surface receptors also exist. Recently, we demonstrated that T-cell immunoglobulin and mucin 1 (TIM-1) (also called HAVCR-1, TIMD1, or KIM-1), expressed on many epithelial cells, mast cells, B cells, and activated CD4+ T cells (19–23), serves as a receptor for filoviruses (20).
The TIM family members (TIM-1, TIM-3, and TIM-4 in humans) share a type I cell surface glycoprotein structure. The ectodomains of these proteins contain an amino-terminal immunoglobulin variable (IgV)-like domain that is extended from the plasma membrane by a heavily O-linked-glycosylated mucin-like domain (MLD) (24). TIM proteins also contain a transmembrane-spanning domain followed by a short cytoplasmic tail. The CC′ and FG loops of the TIM IgV domains form a pocket that binds phosphatidylserine (PtdSer) (see Fig. 1) (25–28). PtdSer on the surface of apoptotic cells interacts with TIM-4 on macrophages, TIM-3 on several types of innate immune cells, and TIM-1 on kidney tubule cells, leading to engulfment of apoptotic bodies (26, 29, 30). In addition to serving as a receptor for filoviruses, human TIM-1 is reported to serve as a receptor for hepatitis A virus (HAV) (31, 32) and, most recently, for a variety of flaviviruses, alphaviruses, and arenaviruses (33, 34).
Fig 1.
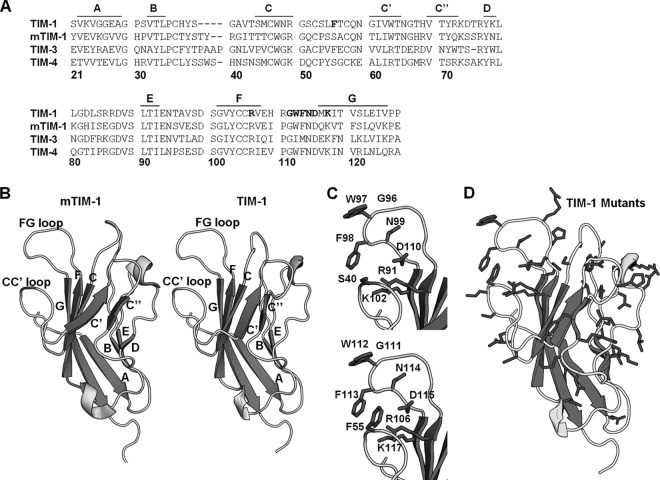
TIM-1 threaded on mTIM-1 crystal structure. (A) Amino acid sequences of TIM-1, mTIM-1, TIM-3, and TIM-4 IgV domains aligned using the Clustal W software program. Residues in the TIM-1 PtdSer binding pocket that were mutated are shown in bold. β-Sheets are marked above the alignment with lines and corresponding letters. Residue numbering is based on the TIM-1 sequence. (B) Structures of mTIM-1 IgV (20R8) (left) and a threaded model of TIM-1 IgV (right). β-Sheets are labeled with their corresponding letters, assigned by Santiago et al. (40). (C) PtdSer binding residues of mTIM-1 (above) and TIM-1 (below) are shown on each structure. (D) All residues in the TIM-1 IgV domain mutated in this study are highlighted in black.
Here, we define the molecular interactions between TIM-1 and Ebola virus (EBOV), formally Zaire ebolavirus. Residues located in the PtdSer binding pocket of the TIM-1 IgV domain are critical for infection. Further, contrary to our initial findings that TIM-1 and EBOV GP directly interact (20), we now show that PtdSer on the virus surface directly interacts with TIM-1, resulting in virion internalization. Additionally, TIM-1 enhances infection of members of both the alphavirus and baculovirus families, indicating the breadth of the importance of PtdSer binding receptors for enveloped-virus entry.
MATERIALS AND METHODS
Cell lines.
HEK 293T cells, a human embryonic kidney cell line, H3 cells, 293T cells, and Vero cells, an African green monkey kidney epithelial cell line, were maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco BRL) with 5% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S). H3 cells are a clonal population of HEK 293T cells that stably express TIM-1 due to integration of a transfected TIM-1 expression plasmid (20).
Production and titer determination of pseudovirions and infectious viruses.
Feline immunodeficiency virus (FIV) and vesicular stomatitis virus (VSV) pseudovirions were produced as previously described (4, 20, 35, 36). To produce VSVΔG-EGFP pseudovirions, VSV (strain Indiana) virions with genomes in which the G glycoprotein gene is replaced with enhanced green fluorescent protein (EGFP), HEK 293T cells were transfected with plasmids expressing either EBOV GP lacking the mucin domain of GP1, full-length EBOV GP (EBOV FL), Marburg virus (MARV) GP, Sindbis virus (SINV) 2.2 1L1L env, Ross River virus (RRV) GP, GP64, Chikungunya virus (CHIKV) env (OPY1), or Lassa virus (LASV) precursor glycoprotein (GPC) and transduced 24 h later with VSVΔG-EGFP pseudovirions. After 4 h of virus uptake, the plates were washed, and medium was repleted. Pseudotyped virions were collected in supernatant 48 and 72 h following transduction, pooled, and filtered through a 0.45-μm filter. MARV and EBOV FL pseudotyped virions were concentrated by centrifuging supernatants at 5,380 × g overnight at 4°C and resuspending pellet in fresh medium to achieve higher-titer stocks. Virus aliquots were stored at −80°C.
Fluorescein isothiocyanate (FITC)-labeled “No GP” and EBOV GP pseudovirions were generated as described above, with No GP pseudovirions generated in the absence of a glycoprotein-expressing plasmid. Pseudovirions were concentrated by centrifuging supernatants overnight at 5,380 × g at 4°C. The pellet was resuspended in 500 mM carbonate buffer, pH 9.5 (∼100-fold concentration of virus). Ten milligrams of FITC “Isomer 1” (Invitrogen) was resuspended in 1 ml dimethyl sulfoxide (DMSO). One microliter of FITC solution was added per 100 μl of reaction buffer. Pseudovirions were incubated at 4°C for 1 h in the dark. The reaction solution was dialyzed using 10,000-molecular-weight-cutoff (MWCO) Slide-A-Lyzer dialysis cassettes (Thermo Scientific) in 1× phosphate-buffered saline (PBS), 3 times for 2 h each at 4°C, and protected from light. Pseudovirions were purified through a 20% sucrose cushion by centrifugation at 80,000 × g for 2 h at 4°C. Pellets were resuspended in 1× PBS, filtered through a 0.45-μm syringe filter, aliquoted, and stored at −80°C.
No GP VP40-GFP VLPs were generated by transfecting HEK 293T cells with a plasmid expressing EBOV VP40 fused to green fluorescent protein (GFP) (37). EBOV GP-pseudotyped VLPs were generated using EBOV GP plasmid at a 1:1 ratio with VP40-GFP plasmid. Supernatants were collected 48 and 72 h after transfection. VLPs were concentrated by centrifuging supernatants overnight at 5,380 × g at 4°C. Pellets were resuspended in 1 × PBS and purified through a sucrose cushion as described above. Pellets were resuspended in 1× PBS, filtered through a 0.45-μm syringe filter, aliquoted, and stored at −80°C.
To produce FIV pseudovirions, HEK 293T cells were transfected with three plasmids as previously described (20, 38). One plasmid expressed an FIV reporter construct that contains a psi sequence and expressed β-galactosidase (β-Gal). A second plasmid expressed the FIV gag and pol genes, and a third plasmid expressed a viral glycoprotein gene of choice (EBOV or LASV). Supernatants were collected 24, 48, and 72 h following transfection, filtered, and either aliquoted or concentrated ∼200-fold by 16-h centrifugation at 5,380 × g at 4°C and resuspended in 1× PBS. Virus aliquots were stored at 80°C until use.
Recombinant, replication-competent VSV expressing the EBOV GP with the mucin domain deleted and EGFP in place of the G glycoprotein (EBOV GP-rVSV-EGFP) was produced as previously described (20). Briefly, the EBOV GP gene was inserted upstream of an EGFP gene that had replaced the G gene in the genome of a recombinant VSV (Indiana), and infectious virus was produced using a multiplasmid transfection protocol as described previously (39). EBOV GP-rVSV-EGFP stocks were produced in Vero cells using a low multiplicity of infection (MOI) (∼0.001) of input virus and maintaining the infection for 3 days prior to supernatant collection. Supernatants were filtered through a 0.45-μm filter and stored as aliquots at −80°C until use. Ross River virus was kindly provided by David Sanders (Purdue University), and stocks were generated in Vero cells. Supernatants were collected and filtered through a 0.45-μm filter. Aliquots were stored at −80°C. Titers of EBOV GP-rVSV-EGFP stocks were determined by endpoint dilution on Vero cells after a 2-day infection with visualization of EGFP. RRV titers were determined by endpoint dilution on Vero cells after a 7-day infection with visualization of cell death. The 50% tissue culture infective dose (TCID50)/ml was calculated by the Reed-Meunch method (www.med.yale.edu/micropath/pdf/Infectivity%20calculator.xls).
Recombinant baculovirus expressing β-Gal under the CMV-Z P6 promoter was provided by Frederick Boyce (Harvard University). Titers of virus were determined by endpoint dilution on HEK 293T cells, and β-Gal activity above the background level was assayed 2 days following infection using a Galacto-Light (Applied Biosystems) detection kit as per the manufacturer's instructions.
Protein structures.
The predicted TIM-1 IgV structures were generated by threading the TIM-1 amino acid sequence on the murine TIM-1 (mTIM-1) (2OR8) (40), mTIM-3 (2OYP) (41), or mTIM-4 (3B19) (28) crystal structure using the protein homology/analogy recognition engine 2 (PHYRE2) service (42). Crystal structures were manipulated and rendered using the PyMOL software program (43).
TIM-1 mutagenesis.
TIM-1 point mutations were introduced by amplifying a cytomegalovirus (CMV) immediate early promoter-driven TIM-1 expression plasmid (Origene) with primers containing targeted nucleotide changes flanked by base pairs of identical sequence. Thermal cycling was performed with PFU Turbo polymerase (Stratagene) using an S1000 thermal cycler (Bio-Rad) for 17 cycles (95°C for 30 s, 55°C for 1 min, and 68°C for 14 min). PCR products were digested with DpnI (New England BioLabs) to remove parental plasmid. Bacteria were transformed, and single-colony isolates were grown for plasmid purification. All mutations were confirmed by DNA sequencing. Mutants that were generated during the studies are shown in Table 1.
Table 1.
TIM-1 IgV domain mutants
| Mutation(s) | Entrya | Location | Expression (%)b |
|---|---|---|---|
| K23A | WT | A β-strand | >90 |
| E27A | WT | A β-strand | >90 |
| G29A | WT | AB loop | >90 |
| T33A | WT | B β-strand | >90 |
| P35A | WT | BC loop | >90 |
| H37A | WT | BC loop | >90 |
| Y38A | ▼ | BC loop | >90 |
| S39A | WT | BC loop | >90 |
| S44A | WT | BC loop | >90 |
| S51A | WT | CC′ loop | >90 |
| S53A | WT | CC′ loop | >80 |
| L54A | WT | CC′ loop | >70 |
| F55A | ▼ | CC′ loop | >90 |
| T56A | WT | CC′ loop | >80 |
| Q58A | WT | CC′ loop | >90 |
| N59A | WT | CC′ loop | >90 |
| T64A | WT | CC″ loop | >90 |
| N65A | WT | C′C″ loop | >90 |
| H68A | WT | C′C″ loop | >90 |
| Y71A | WT | C″ β-strand | >90 |
| R72A | WT | C″E loop | >90 |
| K73A | WT | C″E loop | >70 |
| D74A | WT | C″E loop | >90 |
| K78A | WT | C″E loop | >90 |
| L79A | WT | C″E loop | >90 |
| L80A | WT | C″E loop | >90 |
| D82A | WT | C″E loop | >90 |
| R85A | WT | C″E loop | >70 |
| R86A | WT | C″E loop | >90 |
| RR86AA/LL | WT | C″E loop | >90, >90 |
| D87A | WT | C″E loop | >90 |
| S89A | WT | C″E loop | >90 |
| T91A | WT | E β-strand | >90 |
| D99A | ▼ | EF loop | >90 |
| R106A | ▼ | F β-strand | >90 |
| E108A | WT | FG loop | >90 |
| H109A | WT | FG loop | >70 |
| R110A | WT | FG loop | >90 |
| G111A | ▼ | FG loop | >90 |
| W112A | ▼ | FG loop | >90 |
| F113A | ▼ | FG loop | >90 |
| N114A/D/Q | ▼ | FG loop | >90, >90, >90 |
| D115A/E/N | ▼ | FG loop | >50, >90, >90 |
| ND115AA/DN | ▼ | FG loop | >60, >90 |
| M116A | WT | FG loop | >90 |
| K117A | ▼ | G β-strand | >90 |
| E123A | WT | G β-strand | >90 |
Entry is assessed by transduction into cells expressing mutant TIM-1 relative to transduction into cells expressing WT TIM-1. “▼” indicates a decrease in transduction.
Expression was determined by surface staining with mucin-like-domain-specific MAb AKG7 and grouped into 10% increments.
AnxVΔIgV-TIM-1.
The IgV domain of TIM-1 was replaced with annexin V (AnxV) by first introducing an NheI site between the hydrophobic signal sequence and first residues of the TIM-1 IgV domain. The IgV domain was excised between this NheI cut site and the naturally occurring MfeI site located downstream of the IgV domain. The IgV domain was replaced with a PCR product containing the annexin V coding sequence and 14 nucleotides (nt) of the TIM-1 mucin domain lost with excision of the IgV domain.
Analysis of monoclonal antibody binding and TIM expression.
HEK 293T cells were transfected with TIM-1, mutant TIM-1, or empty-vector expression plasmids using a polyethyleneimine (PEI) transfection protocol. Cells were detached 48 h later with 5 mM EDTA in PBS and washed with PBS containing 5% FBS. Cells were incubated with 0.5 μg of ARD5, A6G2, AKG7, or A8E5, previously characterized mouse anti-human TIM-1 IgG2a monoclonal antibodies (MAbs) (20, 44, 45), or 0.5 μg of mouse IgG1 or IgG2a controls in 100 μl of PBS with 5% FBS for 1 h on ice. Cells were washed and incubated with an anti-mouse Cy5 (Invitrogen)-, DyLight 649 (Jackson ImmunoResearch)-, or FITC (Jackson ImmunoResearch)-conjugated secondary antiserum for 20 min on ice. Cells were washed, and expression was assessed by measuring the percentage of positive cells in the FL-4 or FL-1 channel using a FACSCalibur flow cytometer (BD Biosciences).
Transductions.
Transfected HEK 293T cells were detached 24 h following transfection with trypsin, and a portion of the population was reseeded in a 48-well plate format while the remaining cells were reseeded in a 6-well format. Forty-eight hours after transfection, the cells in the 6-well plate were evaluated for cell surface TIM expression by flow cytometry as described above. For transduction comparisons between HEK 293T cells and H3 cells, equal numbers of cells were directly seeded onto 48-well plates. Medium was removed from the cells and replaced with 300 μl fresh medium containing VSV or FIV pseudovirions. If needed for TIM-1 mutant studies, ARD5 was added concurrently with virus at a concentration of 1.7 μg/ml.
For transductions of TIM-1 mutants, EBOV pseudovirions were used at an MOI of 0.005 as determined from titers on HEK 293T cells (MOI = 0.8 as determined by titers on Vero cells). EBOV FL pseudovirions were used at an MOI of 0.04 as determined from titers on HEK 293T cells, and MARV pseudovirions were used at an MOI of 0.002 as determined from titers on HEK 293T cells. LASV pseudovirions were used at an MOI of 0.15 as determined from titers on HEK 293T cells (MOI = 0.30 as determined from titers on Vero cells). For comparison between efficiency of transduction into H3 cells or HEK 293T cells, pseudovirions were added at an MOI of 0.01 or 0.03 as determined from titers on HEK 293T cells. Transduction of VSV pseudovirions was assessed by lifting cells with Accumax solution (Fisher) 24 h following transduction and detecting FL-1 intensity using a FACSCalibur flow cytometer (BD Biosciences). Flow cytometry data were analyzed using FlowJo cytometry analysis software.
A mammalian expression plasmid for receptor for advanced glycation end products (RAGE) (Origene) was transfected into HEK 293T cells, and cells were transduced as described above. Transductions were normalized to transduction into TIM-1-transfected cells. Protein expression was determined by immunoblotting with goat anti-TIM-1 antisera (R&D Systems) and RAGE antisera (Abcam) and developed using the SuperSignal West Dura extended-duration substrate (Thermo/Fisher). Cell surface expression was confirmed by flow cytometry (data not shown).
For transductions in the presence of EGTA, Vero cells were seeded into 48 wells. Medium on cells was replaced with 1× PBS–10% FBS supplemented with either 0, 0.25, 0.5, 1.0, or 2.0 mM EGTA and incubated for 1 h. EBOV and LASV VSVΔG pseudovirions, diluted in respective EGTA solutions, were added to cells for 4 h before medium was refreshed. Twenty-four hours after transduction, EGFP expression was assessed by flow cytometry. Sufficient quantities of pseudovirions were added to yield ∼25% EGFP-positive cells in the absence of EGTA.
For transductions in the presence of MAbs, H3 cells were seeded in a 48-well format and transduced 24 h later with VSVΔG pseudovirions as described above in the presence of 0.5 μg/ml ARD5, A6G2, or A8E5 or no antibody. Transduction was assessed 24 h later by flow cytometry.
Transductions with liposomes.
l-α-Phosphatidylcholine (PtdChl) and l-α-phosphatidyl-l-serine (PtdSer) (Sigma) liposomes were generated as previously described (26). Briefly, lipids were dissolved in chloroform, dried, resuspended in PBS, and sonicated for 5 min on ice. Liposomes were aliquoted and stored at −80°C until use. H3 and HEK 293T cells were seeded into a 48-well plate and transduced with pseudovirions in the presence or absence of PtdChl or PtdSer liposomes at various concentrations.
Infections.
HEK 293T cells were transfected with wild-type (WT) TIM-1, mutant TIM-1, or empty vector and reseeded into 48-well and 6-well plates as described above. Forty-eight hours after transfection, expression of TIM-1 was assessed by surface staining as described above. Cells in the 48-well format were infected with an MOI of 1 as determined from titers on Vero cells with EBOV GP-rVSV-EGFP. Forty-eight hours after infection, cells were detached and fixed in 3.7% formaldehyde, washed with 1× PBS, and evaluated for EGFP expression by flow cytometry. For RRV infections, WT TIM-1- and empty-vector-transfected cells were reseeded into 6-well cluster plates pretreated with poly-l-lysine (Invitrogen). Cells were infected 48 h after transfection with an MOI of 0.001, 0.01, or 0.1 for 2 h at 37°C. After infection, cells were washed 3× with 1× PBS and refreshed with medium. Forty-eight hours after virus was added, supernatant aliquots were collected and stored at −80°C. Infections with Vero cells were performed similarly in the presence or absence of ARD5 (1 μg/ml). ARD5 was also added to fresh medium for wells infected in the presence of ARD5. Titers of supernatants were determined by endpoint dilution on Vero cells as described above.
Infection with WT EBOV.
Vero E6 cells were grown to 70% confluence in 96-well plates. Cells were preincubated for 1 h with medium alone or PtdChl or PtdSer liposomes suspended in DMEM with 2% FBS. Cells were then transferred to biosafety level 4 (BSL4) and challenged with replication-competent Zaire Ebola virus encoding GFP (46) at an MOI of 0.05 in the presence of the lipids. After 24 h, which is sufficient time for the GFP to be expressed in infected cells, the cells were fixed in formalin and cell nuclei were stained with Hoescht 33342. Cells were then imaged with a Nikon Ti Eclipse microscope using the high-content-analysis software package. Images were processed to detect total cells by counting nuclei and infected cells by GFP expression using the Cell Profiler program (Broad Institute, Massachusetts) using customized pipelines that are available from R. Davey upon request. Typically, more than 10,000 cells were analyzed per image. Data are expressed as the percentage of infected cells in the total cell population compare to that for untreated cells.
Infection with in vivo-derived virus.
IFNAR1−/− BALB/c mice lacking a functional alpha/beta interferon (IFN-α/β) receptor were obtained from Joan Durbin (New York University, New York, NY). A mouse was infected intranasally with 2 × 107 infectious units of Ebola virus GP-VSV in 1% methylcellulose. Four days after infection, the mouse was sacrificed, and lung, spleen, and kidney were harvested. Organs were homogenized in 1× PBS using a tissue tearer. Debris was pelleted, and supernatants were filtered through a 0.45-μm syringe filter, aliquoted, and stored at −80°C. Supernatants were serially diluted and incubated with or without ARD5 (2 μg/ml) on Vero cells. Forty-eight hours after infection, cells were detached and fixed in 3.7% formaldehyde, washed with 1× PBS, and evaluated for EGFP expression by flow cytometry. Mice were maintained in the animal care facility at the University of Iowa. This protocol was approved by the University of Iowa Animal Care and Use Committee and was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH).
Baculovirus transductions.
HEK 293T cells or H3 cells were seeded in equal numbers into 24-well plates pretreated with poly-l-lysine. Virus was added at an MOI of 0.003, and 48 h later, cells were lysed and CMV-driven β-Gal activity was assessed using a Galacto-Light (Applied Biosystems) detection kit as per the manufacturer's instructions.
ELISAs.
Nunc MaxiSorp enzyme-linked immunosorbent assay (ELISA) plates were precoated overnight with ∼5.2 × 106 transducing units (as determined from titers on Vero cells) of concentrated EBOV pseudovirions or PtdSer or PtdChl liposomes (50 μM) diluted in 1× TBS+ (150 mM NaCl, 25 mM Tris, and 10 mM CaCl2, pH 7.2). Plates were blocked for 2 h at 4°C with 1× TBS+ with 2% bovine serum albumin (BSA), incubated with supernatants or recombinant annexin V for 2 h, probed with rabbit polyclonal anti-hemagglutinin (HA) antisera (Sigma) for 1 h and horseradish peroxidase (HRP)-conjugated secondary anti-rabbit antisera for 1 h, and developed using 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (1-Step Ultra TMB-ELISA; Thermo). Plates were washed thoroughly between steps with 1× TBS+. Absorbance was read at 450 nm.
HA- and HIS-tagged recombinant annexin V was purified from DH5α Escherichia coli cells transformed with a modified version of the expression vector pProEx.Htb.annexin V obtained from Seamus Martin (Trinity College) and using the protocol outlined by Logue et al. (47). We modified the vector pProEx.Htb.annexin V by inserting a 3× GS linker, HA tag, and stop codon directly after the last aspartate of annexin V.
HA-tagged soluble proteins used in the ELISAs were generated by transfecting expression plasmids into HEK 293T cells and collecting secreted proteins in Opti-MEM with 1% P/S medium without FBS. Supernatants were collected 48 h after transfection, filtered through 0.2-μm filters, aliquoted, and stored at −80°C. Expression of proteins in supernatants was assessed by probing Western blots with rabbit polyclonal anti-HA antisera and imaged using the Li-Core Odyssey CLx system. Soluble HA-tagged TIM-1, TIM-4, and Axl were generated by replacing the transmembrane and cytoplasmic domains with an HA tag sequence and stop codon. The HA-Axl expression plasmid was created by Nicholas Lennemann (University of Iowa). Soluble TIM-1 mutants were generated by site-directed mutagenesis as described above. A murine serum amyloid protein (mSAP) expression plasmid was kindly provided by Thomas Rutkowski (University of Iowa).
ELISAs using inhibitors of TIM-1 binding were performed as described above using 1 μl of concentrated TIM-1 HA supernatants per well. Supernatants were concentrated using Amicon Ultra 30-MWCO columns (Millipore). TIM-1 was diluted in 1× TBS+ and incubated with PtdSer or PtdChl liposomes (100 or 10 μM), MAb ARD5 (2 μg/ml), or IgG2a (2 μg/ml) for 30 min before adding it to blocked ELISA plates. For binding in the presence of EGTA, TIM-1 was diluted in 1× PBS with 2 mM EGTA. Binding in 1× PBS was equivalent to that in 1× TBS+ (data not shown).
FITC-pseudovirion and VLP binding assay.
Forty-eight hours after transfection with TIM-1, empty vector, mutant TIM-1 ND115DN, or AnxVΔIgV-TIM-1, HEK 293T cells were incubated with FITC-labeled EBOV GP- or No GP-VSV pseudovirions for 1 h on ice. EBOV GP pseudovirions were added at an MOI of 2.25, as determined from titers on Vero cells, and enough No GP pseudovirions were added to achieve similar fluorescence (relatively 2.5-fold more matrix on dot blots). Cells were washed 3 times with 1× PBS plus 5% FBS, and cellular fluorescence was determined by flow cytometry.
Internalization assay.
Vero cells were incubated on ice for 1 h with 1× Hanks' balanced salt solution (HBSS) (plus CaCl2, plus MgCl2) (Gibco) with 30 mM HEPES. ARD5 (2 μg/ml) or PtdSer or PtdChl liposomes (25 μM) were also added as needed. VLPs or FITC-labeled VSV pseudovirions were bound for 1 h on ice. EBOV GP pseudovirions were added at an MOI of ∼6, and No GP pseudovirions were added to achieve approximate binding mean fluorescence intensity (MFI) (relatively twice as much matrix on dot blots). A ∼0.36-μg amount of EBOV GP-VLPs and 0.48 μg No GP-VLPs were added per 10,000 cells. VLP concentrations were determined by bicinchoninic acid (BCA) assay (Thermo). Some cells were shifted to 37°C for 30 min to allow internalization, while remaining cells were maintained on ice. After internalization, all cells were treated with 1× trypsin-EDTA (Gibco) for 10 min at room temperature and an additional 10 min at 37°C. Cells were washed twice with 1× PBS and once with 1× PBS plus 5% FBS. Fluorescence was determined by flow cytometry.
Statistics.
Transductions for mutant TIM-1 construct-expressing HEK 293T cells were normalized to WT transduction in the absence of ARD5 by dividing the sample mean obtained from replicates by the WT mean from the same experiment. EBOV, full-length EBOV, and MARV GP pseudovirion transductions into mutant TIM-1- and WT TIM-1-expressing HEK 293T cells in the absence of ARD5 were normalized to TIM-1 expression by dividing by a ratio of mutant TIM-1 expression to WT TIM-1 expression. Expression was determined by flow cytometry as described above and measured as a percentage of antibody-positive cells. For data that have been normalized to a control, one-sample t tests were used to compare transductions and infections to 100, which represents normalized WT transduction. Fold enhancement by TIM-1 or AnxVΔIgV-TIM-1 was compared to 1, which represents background transduction of the empty vector. Comparisons between experimental samples were done using two-sample t test with two tails and equal variance. Nonlinear curves were fitted to data from serial dilutions of infectious EBOV GP-recombinant VSV (GP-rVSV) onto transfected HEK 293T cells and WT EBOV infection of Vero cells in the presence of liposomes using the GraphPad Prism software program using one-site, specific binding for EBOV GP-rVSV and dissociation, one-phase exponential decay for WT EBOV. The EBOV GP-rVSV curves had r2 values of 0.9708 (WT), 0.9577 (empty vector), 0.9889 (N114A construct), and 0.9737 (D115A construct). The 50% infective doses (ID50) (relative to the maximum percent infection of each curve [Bmax]) for the WT, empty vector, and N114A and D115A constructs were calculated from the fitted curve to be 7.57 × 10−7, 8.10 × 10−4, 3.42 × 10−4, and 4.25 × 10−4, respectively. The PtdSer inhibition curve of WT EBOV had an r2 value of 0.9834.
RESULTS
Modeling a human TIM-1 structure.
Our previous study demonstrated that in a poorly permissive cell line, HEK 293T, ectopic TIM-1 expression enhances EBOV transduction, which is inhibited by the anti-TIM-1 IgV MAbs ARD5, A6G2, and A8E5 (20). This focused our initial studies on identifying TIM-1 IgV residues important for EBOV entry. Since the inhibitory MAb A6G2 has been shown to also block TIM-1 binding of PtdSer (45), we focused our attention on the PtdSer binding pocket. We aligned the amino acid sequences of the human TIM IgV domains with murine TIM-1 (mTIM-1) IgV, revealing significant conservation of residues (Fig. 1A). Since the crystal structures of the mTIM-1, -3, and -4 IgV domains are similar (25, 28, 40, 41), we threaded human TIM-1 onto these structures using PHYRE2 (protein homology/analogy recognition engine 2) (42). The human TIM-1 IgV sequence threaded on the mTIM-1 structure strongly resembled mTIM-1, with the PtdSer binding pockets being comparable (Fig. 1B) and key residues involved in the binding of PtdSer conserved and positioned similarly (Fig. 1C). Alanine scanning mutagenesis of human TIM-1 residues predicted to be surface exposed was performed to identify residues important for EBOV entry. Adjacent residues and nonalanine residue substitutions were also introduced after the initial screen. In total, 45 individual residues of the IgV domain of TIM-1 were mutated (Table 1 and Fig. 1D).
PtdSer binding pocket residues are necessary for EBOV entry.
HEK 293T cells were transfected with TIM-1 mutants in parallel with wild-type (WT) TIM-1 and an empty-vector control, evaluated for TIM-1 surface expression by flow cytometry, and transduced or infected 48 h after transfection. HEK 293T cells were selected for these studies since these cells are readily transfected, do not endogenously express any TIM family members, and in the absence of ectopic TIM-1 expression poorly support EBOV infection (20). All mutants but one had >70% of WT surface expression as assessed by surface staining of transfected cells with four different anti-TIM-1 MAbs, ARD5, A6G2, AKG7, and A8E5 (20, 44, 45). Surface expression of the D115A construct was ∼55% of WT expression, but Glu or Asn substitution rather than Ala at this position yielded expression equivalent to that of the WT. The WT surface expression of our mutants and ability of multiple IgV-specific MAbs to bind to mutants argues against an impact of the mutations on the stability and/or expression of TIM-1.
Transfected cells were transduced with vesicular stomatitis virus (VSV) pseudovirions that express EGFP as a reporter molecule in place of native G (VSVΔG). VSV was pseudotyped with either EBOV GP that has a deleted mucin domain or, as a control, Lassa virus (LASV) GPC. We used EBOV GP lacking the GP1 mucin domain since this construct confers the same tropism as full-length Ebola virus GP and produces higher pseudovirus titers (35, 48, 49). Ebola GP-VSVΔG and LASV GPC-VSVΔG transductions were performed in parallel and analyzed for EGFP expression by flow cytometry at 24 h following transduction.
A summary of the effects of TIM-1 IgV domain mutations on EBOV transduction is shown in Table 1. Most TIM-1 mutations had no deleterious effect on transduction, and none significantly enhanced entry. Mutation of eight residues within and adjacent to the PtdSer binding pocket inhibited EBOV transduction (Fig. 2A). G111A, N114A, or D115A resulted in the most profound effects. EBOV transduction into cells expressing TIM-1 mutants remained sensitive to ARD5 inhibition, suggesting that the inhibitory effect of ARD5 is not due to direct binding competition. In contrast, transduction of LASV pseudovirions was largely unaffected by expression of WT or mutant TIM-1s or the presence of ARD5 (Fig. 2B). However, LASV pseudovirion transduction into empty vector- and F113A construct-transfected cells was slightly reduced, and this difference was statistically significant. Nonetheless, the reduction was minor, and no significant changes were observed in the presence of ARD5. Mutation of the TIM-1 PtdSer binding pocket also reduced transduction of EBOV FL GP or MARV GP pseudovirions (Fig. 2C).
Fig 2.
Identification of TIM-1 IgV residues that impact EBOV GP-dependent entry. (A to C) Relative virus transduction into HEK 293T cells mediated by mutant TIM-1 constructs compared to that with WT TIM-1. At 48 h, transfected cells were transduced with EBOV GP-VSVΔG (A), LASV GPC-VSVΔG (B), MARV GP-VSVΔG (C), or full-length EBOV-VSVΔG (C) pseudovirions. (A and B) Transductions were also done in the presence or absence of anti-human TIM-1 MAb ARD5 (1.7 μg/ml). EGFP expression was assessed 24 h later. (D) EBOV GP/rVSV-EGFP infection mediated by TIM-1 constructs in HEK 293T cells (MOI of 1 as determined from titers in Vero cells) relative to WT TIM-1. (E) Serial dilution of EBOV GP/rVSV-EGFP on TIM-1-transfected HEK293T cells. EGFP expression was assessed 24 (A to C) or 48 (D and E) h later by flow cytometry. Data are shown as means ± SEM for at least three replicates. (A to D) Significance was calculated using one-sample t-test comparison to 100 (**, P < 0.001; *, P < 0.01). Significance for all ARD5 transductions for EBOV was defined as P < 0.001.
These same TIM-1 mutants were evaluated for their ability to support a recombinant, infectious VSV/GFP (rVSV) that encodes and expresses EBOV GP (EBOV GP/rVSV) (20). Findings from these infections were similar to the transduction studies with N114A and D115A TIM-1 mutants, providing little to no increased infection above that with the empty vector (Fig. 2D). Further, dose-response curves with EBOV GP/rVSV demonstrated that infection in the presence of the N114A and D115A mutants was indistinguishable from that with the empty-vector control (Fig. 2E). Since these mutants were expressed at WT levels but did not support EBOV transduction, we propose that these PtdSer binding pocket residues are influencing the ability of the mutants to serve as an EBOV receptor.
Interestingly, mutagenesis of two additional residues, Y38 and D99, not located near the PtdSer binding cleft also decreased EBOV transduction but not surface expression of TIM-1 (Table 1). Our structural model predicts that Y38 and D99 are distal to the PtdSer binding pocket and are not surface exposed, suggesting that mutagenesis of these residues may alter the IgV domain structure without altering the overall stability of the protein. Consistent with this possibility, mutations of surface-exposed residues adjacent to Y38 had no effect on transduction.
TIM-1-mediated uptake requires virion-associated PtdSer binding.
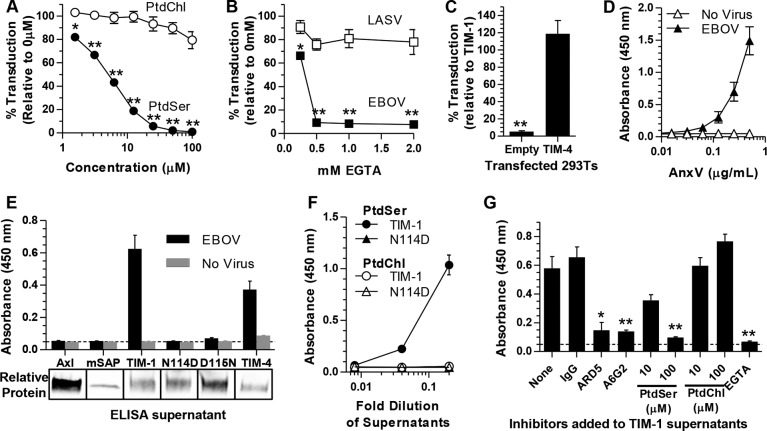
Residues critical for EBOV infection were located within the PtdSer binding pocket, suggesting that virion-associated PtdSer interactions with TIM-1 contribute to transduction. Others have reported that PtdSer is present on the surface of enveloped viruses (34, 50). To assess the role of PtdSer in TIM-1-dependent entry of EBOV, competition studies were performed in H3 cells, a clonal population of HEK 293T cells stably expressing TIM-1 (20), in the presence of increasing concentrations of PtdSer or phosphatidylcholine (PtdChl) liposomes. Similar liposomes have been used previously to inhibit HIV-1 infection (51) and uptake of apoptotic cells and virus by TIM-1 (26, 33). Transduction was inhibited in a dose-dependent manner by the presence of PtdSer liposomes, with a 2.5 μM concentration abolishing transduction (Fig. 3A). PtdChl liposomes had a minimal effect. Since binding of PtdSer by TIM-1 requires divalent cations and the presence of EGTA inhibits this interaction (30), we assessed the effect of increasing concentrations of EGTA on EBOV entry in highly permissive, TIM-1-expressing Vero cells. Transduction of EBOV was inhibited in dose-dependent manner by EGTA (Fig. 3B). Conservation of PtdSer binding among the human TIM family members (Fig. 1A) suggests that other family members, such as TIM-4, would also function as an EBOV receptor. Indeed, upon transfection into HEK 293T cells, TIM-4 enhanced EBOV entry equivalently to that with TIM-1 (Fig. 3C).
Fig 3.
TIM-1-mediated transduction requires PtdSer binding. (A) Transduction of TIM-1+ H3 cells with EBOV pseudovirions in the presence of increasing concentrations of PtdSer or PtdChl liposomes. (B) Transduction of EBOV and LASV pseudovirions into Vero cells in the presence of increasing EGTA concentrations. EGTA in PBS plus 10% FBS was incubated on Vero cells for 1 h before pseudovirions were added for 4 h. Medium was replaced, and transduction was assessed after 24 h. (C) Transduction of EBOV pseudovirions into empty-vector- or TIM-4-transfected HEK 293T cells relative to TIM-1 transduction. (D) AnxV binds to EBOV pseudovirions. Increasing concentrations of HA-tagged AnxV were incubated with ELISA plates prebound with EBOV pseudovirions (filled) or untreated (open). (E) HEK 293T supernatants containing HA-tagged soluble Axl, mSAP, TIM-1, TIM-4, or mutants of TIM-1, N114D or D115N, were incubated with ELISA plates prebound with EBOV pseudovirions or not treated. Relative protein amounts present in supernatants are shown below in a representative Western blot using anti-HA antisera. (F) Binding of TIM-1 and N114D from supernatants to ELISA plates prebound with PtdSer (filled) or PtdChl (open) liposomes (50 μM). (G) Binding of HA-tagged TIM-1 in the presence of IgG2a, MAb ARD5, MAb A6G2, PtdSer, or PtdChl liposomes or 2 mM EGTA to ELISA plates prebound with EBOV pseudovirions. Approximate background absorbance is shown with a dashed line. Data are shown as means ± SEM for at least three replicates. Significance was calculated using one-sample t-test comparison to 100 for panels A to C or a two-sample t test for panel G (**, P < 0.001; *, P < 0.01).
We previously showed that TIM-1 binds to EBOV pseudovirions (20). In order to determine if TIM-1 was binding to PtdSer on virions, we initially determined if PtdSer was present in pseudovirion membranes by ELISA studies using purified recombinant HA-tagged AnxV. AnxV is a cellular protein that binds to PtdSer (52) and detects PtdSer on the surface of apoptotic cells (53). AnxV bound to EBOV pseudovirion-coated ELISA plates in a dose-dependent manner but not to plates lacking pseudovirions (Fig. 3D). Similarly, HA-tagged soluble WT TIM-1 in HEK 293T cell supernatants bound to virus and PtdSer liposomes, but the N114D and D115N TIM-1 PtdSer binding pocket mutants did not (Fig. 3E and F). Soluble, HA-tagged Axl and murine serum amyloid protein (mSAP) served as negative controls in the study and did not bind virus. While Axl has been shown to enhance pseudovirus uptake, Axl cannot bind PtdSer without bridging by Gas6 (50). Consistent with our transduction studies, HA-tagged soluble TIM-4 also bound to pseudovirions. It should be noted that our virus preparations likely contain broken particles in addition to intact virions, since a VSV matrix-specific antibody was able to bind to pseudovirions (data not shown), and we cannot rule out that AnxV is also binding to the inner membrane.
The specificity of the TIM-1/EBOV pseudovirion interaction was further examined by assessing the ability of inhibitors to interfere with TIM-1 binding (Fig. 3G). These included the anti-TIM-1 MAbs ARD5 and A6G2, PtdSer liposomes, and EGTA, all of which dramatically inhibited TIM-1 binding. PtdChl liposomes did not affect TIM-1 binding to pseudovirions. These findings lend support for a model of PtdSer-dependent TIM-1-mediated uptake of filoviruses.
Not all PtdSer receptors mediate filovirus entry.
The ability for TIM-1 and TIM-4 to bind pseudovirions and enhance EBOV transduction might suggest that all PtdSer binding receptors may enhance EBOV entry. However, we found that the receptor for advance glycation end products (RAGE), a PtdSer receptor unrelated to the TIM family that also enhances apoptotic body uptake upon transfection into HEK 293 cells (54, 55), did not enhance EBOV transduction into HEK 293T cells (Fig. 4A), although expression was detected in lysates (Fig. 4B).
Fig 4.
Expression of RAGE does not increase transduction. (A) Transduction of EBOV or LASV pseudovirions into HEK 293T cells transfected with an empty vector (Empty) or a RAGE-expressing vector (RAGE) relative to results with WT TIM-1. Cells were transduced at 48 h following transfection, and EGFP expression was assessed 24 h following transduction by flow cytometry. Data are shown as means ± SEM for at least three replicates. Significance compared to results for the WT was calculated using one-sample t-test comparison to 1 (**, P < 0.001). Significance between Empty and RAGE results was calculated using a two-sample t test. (B) TIM-1 and RAGE expression for three experiments, labeled 1 to 3, is shown by immunoblot analysis.
TIM-1 enhances entry of VSV pseudotyped with a wide range of viral GPs.
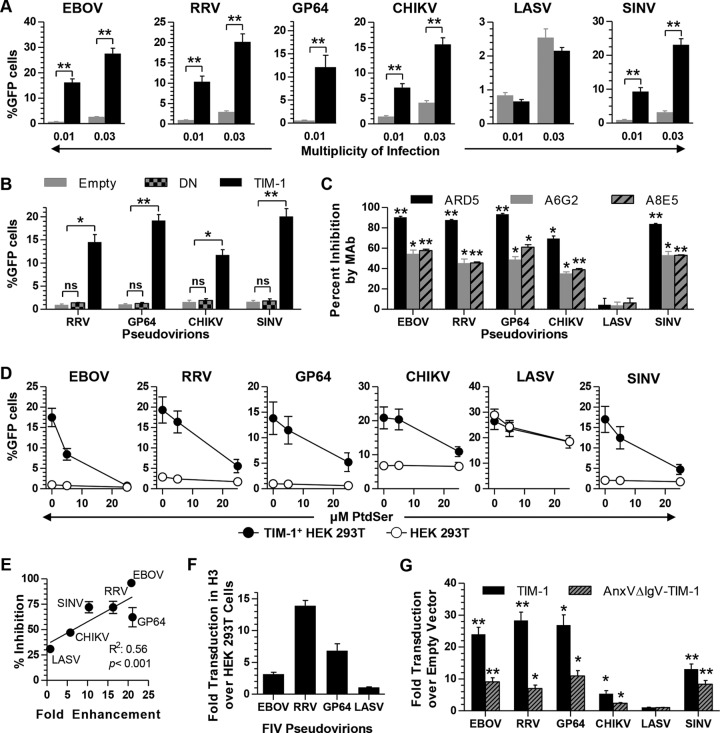
We hypothesized that if TIM-1 enhancement was dependent on PtdSer binding and not on interaction with EBOV GP, TIM-1 should enhance entry of VSVΔG pseudotyped with other viral GPs. VSVΔG virions were pseudotyped with the alphavirus Ross River virus (RRV) GP or Chikungunya virus (CHIKV) env or baculovirus Autographa californica nucleopolyhedrovirus (AcMNPV) GP64. Transduction of these pseudoviruses into 293T cells and TIM-1+ H3 cells was evaluated in parallel with that of EBOV GP and LASV GPC. In a manner similar to that of EBOV, transduction of RRV or GP64 pseudovirions was enhanced by TIM-1 expression (Fig. 5A). CHIKV entry was more modestly enhanced, by about 3- to 5-fold. LASV GPC-dependent entry was unaffected by TIM-1 expression. The PtdSer binding pocket mutant ND115DN (N114D and D115N double mutant) did not increase pseudovirion transduction, demonstrating that enhancement was due to the PtdSer binding pocket of TIM-1 (Fig. 5B). Further, TIM-1 IgV domain MAbs ARD5, A6G2, and A8E5 inhibited CHIKV, RRV, or GP64 pseudovirion transduction as effectively as EBOV pseudovirion transduction (Fig. 5C). Consistent with the possibility that virion-associated PtdSer enhances alphavirus and baculovirus entry, TIM-1-enhanced transduction of RRV, GP64, and CHIKV pseudovirions was inhibited by PtdSer liposomes, although inhibition of EBOV transduction was most profound (Fig. 5D). Interestingly, LASV was partially inhibited in both H3 and HEK 293T cells by PtdSer liposomes. Since transduction of LASV is unaffected by TIM-1 expression in HEK 293T cells and the reduction is equal in the two cell types, this inhibition is likely an off-target effect for LASV. The correlation between enhancement of transduction by TIM-1 expression and subsequent inhibition by PtdSer liposomes was evident when these two data sets were plotted, suggesting that TIM-1 binding of PtdSer is important for TIM-1-dependent enhancement of transduction (Fig. 5E). To exclude the possibility that our findings were specific to the VSV pseudotyping system, we confirmed our results with FIV pseudovirions, although EBOV GP FIV was more modestly enhanced by TIM-1 than EBOV GP VSV (Fig. 5F).
Fig 5.
TIM-1-mediated transduction is not specific to EBOV. (A) Transduction of HEK 293T cells (gray bars) and TIM-1+ H3 cells (black bars) with VSVΔG pseudotyped with EBOV GP, RRV GP, GP64, CHIKV env, LASV GPC, or SINV 2.2 1L1L env. Cells were seeded in equal numbers, transduced with an MOI of ∼0.01 and 0.03, and assayed for EGFP by flow cytometry at 24 h following transduction. (B) Transduction of pseudovirions at an MOI of ∼0.01 into HEK 293T cells transfected with empty vector, ND115DN mutant of TIM-1, or WT TIM-1. MOIs were determined in untransfected HEK 293T cells. (C) Percent inhibition of pseudovirion transduction into TIM-1+ H3 cells by MAb ARD5, A6G2, or A8E5 (0.5 μg/ml). Percent inhibition is calculated relative to no-antibody control. (D) Transduction of pseudovirions into TIM-1+ H3 cells (filled) or HEK 293T cells (open) in the presence of 0, 5, or 25 μM PtdSer liposomes. (E) Correlation between enhancement of transduction by TIM-1 expression and percent inhibition by PtdSer liposomes. Pseudovirions were plotted on the x axis based on fold enhancement of transduction in H3 cells over that in HEK 293T cells and on the y axis based on percent inhibition by 25 μM PtdSer compared to that by 0 μM PtdSer liposomes. Linear regression was fitted and significance was calculated using the GraphPad Prism software program. (F) FIV pseudovirions expressing β-Gal were used to transduce HEK 293T cells and TIM-1+ H3 cells. β-Gal expression in lysates was assessed using the Galacto-Light system. Shown is the fold increase in β-Gal signal in H3 cells over that in HEK 293T cells. (G) Fold enhancement of VSVΔG pseudovirions transduction into HEK 293T cells transfected with TIM-1 or AnxVΔIgV-TIM-1 relative to that with the empty-vector control. Cells were transduced with an MOI of 0.01 as determined from titers in empty-vector HEK 293T cells. Data are shown as means ± SEM for at least three replicates. For panels A and B, significance was calculated using a two-sample student t test with equal variance, and for panels C and G, significance was calculated using one-sample t-test comparison to 0 (C) or 1 (G) (**, P < 0.001; *, P < 0.01).
AnxV substitutes for the TIM-1 IgV domain.
Our results implicate the PtdSer binding activity of the TIM-1 IgV domain in mediating entry of a diverse group of enveloped viruses. In order to rule out other functions of the IgV domain and verify the importance of virus-associated PtdSer for the effect, we replaced the IgV domain of TIM-1 with AnxV. The chimeric construct (AnxVΔIgV-TIM-1) was expressed in transfected cells at levels equivalent to that of WT TIM-1 as detected by a TIM-1 polyclonal antiserum, and AnxVΔIgV-TIM-1 was detected on the surface of cells by both the TIM-1 MLD-specific MAb AKG7 and an AnxV-specific MAb (data not shown). However, the IgV domain-specific MAb ARD5 does not bind to AnxVΔIgV-TIM-1. AnxVΔIgV-TIM-1 significantly increased transduction for all of the pseudoviruses tested except LASV (Fig. 5G), albeit with reduced efficiency compared to that of TIM-1.
Direct role of TIM-1 for internalization of virus.
Since TIM-1 could enhance transduction by binding virus and either directly mediating entry or acting as an attachment factor to concentrate virus on the cell surface for internalization via an alternative receptor, we evaluated transduction of VSV pseudotyped with a well-characterized Sindbis virus (SINV) envelope receptor binding domain mutant, 2.2 1L1L (56–58). This GP contains deletions and mutations within E2 and E1 that diminish native receptor binding activity, reducing interaction with a specific cell surface receptor but still allowing fusion. TIM-1 enhanced SINV pseudovirion transduction through a PtdSer-dependent mechanism (Fig. 5A to G). The ability of TIM-1 to enhance transduction of pseudovirions lacking an intact RBD suggests that TIM-1-mediated enhancement is independent of receptor binding by the glycoprotein RBD.
TIM-1 expression enhances virus binding and internalization independent of glycoprotein.
As a second approach to assessing if TIM-1 is acting as an attachment factor or directly internalizing virus, we fluorescein isothiocyanate (FITC) labeled both EBOV GP-VSV pseudovirions and VSV pseudovirions without an envelope GP (No GP). We found that binding of both EBOV GP and No GP pseudovirions to HEK 293T cells was enhanced by exogenous expression of TIM-1 and AnxVΔIgV-TIM-1, but binding of pseudovirions to a PtdSer-binding mutant TIM-1 was minimal (Fig. 6A). Because not all cells express the transgene after transfection, only a portion of the population shifts. Additionally, ARD5 inhibited EBOV GP-VSV binding to TIM-1+ cells but not AnxVΔIgV-TIM-1-expressing cells.
Fig 6.
Virus binding and internalization occur independent of glycoprotein. (A) Binding of FITC-labeled EBOV and No GP VSV pseudovirions to HEK 293T cells transfected with TIM-1, mutant TIM-1 ND115DN, or AnxVΔIgV-TIM-1 in the presence or absence of ARD5 (2 μg/ml). (B to E) Internalization of FITC-labeled EBOV and No GP pseudovirions (B and C) or VP40-GFP VLPs (D and E) into Vero cells. Virus was added at 4°C to Vero cells that were prebound with or without ARD5 (2 μg/ml) or PtdChl or PtdSer liposomes (25 μM). As noted, some cells were shifted to 37°C for 30 min, whereas others remained on ice. Identified cell populations were treated with trypsin and washed to remove excess virus. Fluorescence was determined by flow cytometry. Representative histograms are shown in panels A, B, and D, with a filled gray line representing background cellular fluorescence and a black line representing virus fluorescence. Histograms are quantified in panels C and E. Data are shown as means ± SD for at least three replicates.
We then used a trypsin protection assay to assess virion internalization in the presence of endogenous TIM-1 in Vero cells. Pseudovirions were bound to Vero cells on ice to prevent internalization. Subsequently, some cell populations were shifted to 37°C to allow internalization of virus. Virion internalization would be predicted to protect virus from trypsin cleavage and removal. Using these conditions, the fluorescence of cells prebound with either EBOV GP-VSV or No GP pseudovirions was assessed with or without virion internalization. In the absence of the temperature shift to 37°C, bound virus was removed by trypsin (Fig. 6B). However, after internalizing virus at 37°C and treating cells with trypsin, FITC-labeled EBOV GP and No GP pseudovirions were readily detected, indicating that they were effectively internalized (Fig. 6B). Internalization as detected by mean fluorescence intensity (MFI) is quantified in Fig. 6C, demonstrating similar internalization of both EBOV GP and No Env pseudovirions. Additionally, ARD5 and PtdSer liposomes inhibited internalization of both viruses, but PtdChl liposomes did not. These results were confirmed using EBOV VP40-GFP virus-like particles (VLPs) (37) (Fig. 6D and E) and demonstrate that enveloped virus is internalized into TIM-1-expressing cells in the absence of a viral glycoprotein.
TIM-1 enhances Ross River virus and AcMNPV infection.
Despite previous observations by our group and others that TIM-1 enhances entry of replication-competent WT EBOV, EBOV GP/rVSV, and dengue virus (DV) (20, 34), we were concerned that high levels of GP expression during pseudovirus production could lead to PtdSer flipping to the outer leaflet, altering the pseudovirion envelope composition compared to virus produced during an infection. Thus, we assessed if TIM-1 expression enhances infection by the alphavirus RRV or the baculovirus AcMNPV. For RRV experiments, a range of MOIs, as determined from titers in Vero cells, were added to HEK 293T cells transfected with an empty or TIM-1-expressing vector, and virus was collected and titers determined on Vero cells by endpoint dilution at 48 h. RRV titers were enhanced by TIM-1 at low MOIs, and this effect was reduced with increasing MOIs (Fig. 7A). Consistent with a role for TIM-1, RRV infection of TIM-1-positive Vero cells for 24 h was inhibited by ARD5 (Fig. 7B). In a similar manner, entry of AcMNPV into mammalian cells was enhanced by the presence of TIM-1. While AcMNPV does not replicate in mammalian cells, use of an infectious AcMNPV that expresses β-Gal under a CMV promoter allowed assessment of virus entry, uncoating, and transgene expression in either HEK 293T or H3 cells. TIM-1 expression increased entry of this baculovirus by ∼6-fold (Fig. 7C). While we have already shown that WT EBOV infection of Vero cells is inhibited by ARD5 (20), we wanted to confirm that WT EBOV infection is mediated through PtdSer binding. Vero cells were infected with WT EBOV in the presence or absence of PtdSer or PtdChl liposomes (Fig. 7D). EBOV infection was sensitive to inhibition by PtdSer liposomes in a dose-dependent manner but not to that by PtdChl liposomes. The ability of TIM-1 to mediate entry of a variety of viruses suggests a potential role as a broad receptor for enveloped-virus uptake.
Fig 7.

RRV and AcMNPV infection is enhanced by TIM-1 expression. (A) Replication of RRV in TIM-1- or empty-vector-transfected cells at MOIs ranging from 0.001 to 0.1. Supernatants were collected 48 h following infection, and titers were determined by endpoint dilution on Vero cells. (B) Infection of TIM-1+ Vero cells with RRV in the presence or absence of MAb ARD5 (1 μg/ml). Supernatants were collected 48 h after infection and assessed for viral titer by endpoint dilution on Vero cells. (C) Transduction of HEK 293T cells or TIM-1+ H3 cells with recombinant baculovirus expressing β-Gal at an MOI of 0.003 as determined from titers on HEK 293T cells. After 48 h of incubation with virus, cells were lysed and β-Gal activity was assessed using the Galacto-Light system. (D) Impact of PtdSer on WT EBOV infection. Vero cells were preincubated for 1 h with the indicated concentrations of PtdSer or PtdChl liposomes or medium alone. Cells were then challenged in the presence of the liposomes with replication-competent EBOV encoding GFP. After 24 h, at which time the GFP from one round of infection can be detected, cells were fixed and the proportion of infected cells in the total cell population was determined (see Materials and Methods). The experiment was repeated 3 times with similar outcomes (50% inhibitory concentration [IC50], 5 ± 1.5 μM), and the means ± SD for 4 replicates are shown from 1 experiment. (E) Infection with mouse organ-derived EBOV GP-rVSV of Vero cells in the absence (black) or presence (gray) of ARD5. Virus obtained from lung, spleen, or kidney homogenates was serially diluted on Vero cells, and after 48 h, infection was assessed by EGFP expression. Data for RRV infections are shown as means ± SD, and AcMNPV and EBOV GP-rVSV data are shown as means ± SEM for at least three replicates. Significance was calculated using two-sample t-test comparison with equal variance (**, P < 0.001; *, P < 0.01).
Production of PtdSer-containing virions in vivo.
While we have demonstrated that PtdSer-mediated entry occurs with both transducing and infectious virus generated in tissue culture, we also sought to determine if virus generated in vivo enters cells in a TIM-1-dependent manner. Replication-competent EBOV GP/rVSV was obtained from lung, spleen, and kidney homogenates of an interferon-α/β receptor (IFNAR) knockout BALB/c mouse 4 days after intranasal infection. Virus collected from homogenates was serially diluted onto Vero cells in the presence or absence of ARD5 (2 μg/ml). ARD5 inhibited virus infection at all dilutions, regardless of the organ of origin (Fig. 7E). These results suggest that the presence of PtdSer on virus grown in tissue culture is representative of virus produced during in vivo infection.
DISCUSSION
This study demonstrates that TIM-1 enhances infection of a variety of unrelated enveloped viruses by binding to virion-associated PtdSer, leading to virus internalization. The virus families described here include filoviruses, alphaviruses, and a baculovirus. Other recent studies have also confirmed that TIM-1 enhances filoviruses, alphaviruses, flaviviruses, and some arenaviruses (33, 34). Here, we have provided a number of lines of evidence to support this conclusion. First, PtdSer liposomes inhibit TIM-1 binding to virions and effectively eliminate TIM-1-dependent transduction/infection. Second, mutations in the TIM-1 PtdSer-binding pocket abrogate the enhanced transduction. Third, TIM-1 MAbs that block PtdSer binding decrease TIM-1 binding to pseudovirions and TIM-1-dependent transduction and infection. Fourth, the TIM family member TIM-4, which also has a PtdSer binding pocket, functions to enhance virus transduction. Fifth, pseudovirions or VLPs that lack a viral envelope glycoprotein are efficiently internalized into TIM-1-expressing cells. Finally, the PtdSer-binding protein AnxV effectively substitutes for the TIM-1 IgV domain on a chimeric TIM-1 receptor. These studies extend the number of virus families that use apoptotic mimicry as a mechanism of virion uptake and support the idea that enveloped viruses can commandeer the use of cell surface PtdSer-binding receptors to mediate uptake.
Our mutagenesis studies initially focused on identifying TIM-1 residues that impact filovirus infection. We identified eight residues critical for EBOV infection that line and surround the PtdSer binding pocket of the TIM IgV domain. These residues are generally conserved across the TIM family, and a mutagenesis study of the mTIM-4 IgV domain identified a subset of these residues as critical for PtdSer binding (28). The PtdSer pocket Asn and Asp residues (N114 and D115 in TIM-1) are involved in intra- and intermolecular interactions important for retaining a metal cation required for PtdSer binding. The mTIM-4 crystal structure also suggests that the backbone of the PtdSer pocket Gly (G111 in TIM-1) also contributes to cation binding (28). Other pocket residues contribute to PtdSer binding through hydrogen bonding to the phosphate and serine moieties of PtdSer and maintaining the spacing between the CC′ and FG loops. Consistent with the important role of this binding pocket in mediating virus entry, W112, F113, N114, and D115 were recently identified as critical for PtdSer-dependent uptake of several enveloped viruses by TIM-1 (33, 34).
Since many of the TIM-1 residues found to be important for EBOV infection are also important in PtdSer binding, we tested if EBOV entry was mediated through the presence of PtdSer on the surface of virions. We detected PtdSer on the surface of EBOV GP pseudovirions with AnxV. Further, transduction of VSV pseudotyped with a variety of other viral GPs and infection of both RRV and AcMNPV at low, biologically relevant MOIs was also increased by TIM-1 surface expression. Additionally, Jemielity et al. found that SINV, Tacaribe virus, and RRV infection is enhanced by TIM-1 expression (33). Thus, TIM-1 mediates infection of a broader range of viruses than previously appreciated through a PtdSer-dependent mechanism. Enhancement of AcMNPV, flavivirus, and alphavirus entry also suggests a potential role for TIM-1 in transmission of arthropod-borne viruses between mosquitoes and mammals. However, it should be noted that not all pseudovirion data, such as those for CHIKV, have been confirmed with WT virus.
The phenomenon of apoptotic mimicry, reviewed in reference 59, was first described for vaccinia virus, although no cellular PtdSer receptor was implicated (60). Their study found that depleting PtdSer from the surface of mature virions inhibited infectivity, which was restored by PtdSer reconstitution. Recently, the cell surface complex of Gas6 and Axl was identified to mediate PtdSer-dependent virus entry (50). The Gas6/Axl complex mediates uptake of both vaccinia extracellular enveloped virus and DV in HEK 293T and A549 cells (34, 50). Further, Axl enhances EBOV entry in certain cell types, which may in part be due to Gas6/Axl complexes (38, 61, 62). However, it should be noted that mediation of EBOV entry by Axl appears to be cell type dependent and endogenous Axl expression in several cell lines does not enhance EBOV uptake. Here, our findings, along with the TIM-1 studies by Meertens et al. (34) and Jemielity et al. (33), provide strong evidence that TIM-1 mediates virus uptake through binding of virion-associated PtdSer, thereby identifying it as a second cellular receptor that mediates virus entry in a PtdSer-dependent manner. This new class of viral receptors we term phosphatidylserine-mediated virus entry enhancing receptors, or PVEERs.
The mechanism of PtdSer incorporation into viral envelopes remains unclear. Apoptosis may contribute to the accumulation of PtdSer on the surface of virions, since many viruses induce this type of cell killing. For instance, CHIKV can induce apoptosis during infection (63), contributing to spread of virus through uptake of apoptotic blebs. Additionally, the natural surface exposure of PtdSer by certain cell types or species may also contribute to PtdSer presentation as seen with macrophages (64) and cell lines (65). One possible association that we explored was between viral GP expression on the cell surface and PtdSer flipping. However, enhancement of binding and internalization of No GP pseudovirions and VLPs by TIM-1 suggests that PtdSer incorporation is independent of GP expression.
Not all enveloped virions transduced more effectively with TIM-1 expression. We found that LASV pseudovirion entry into HEK 293T cells was not enhanced by TIM-1 expression, as others also found with LASV and other viruses, including herpes simplex virus 1, influenza A virus (H7N1), Oliveros virus, and severe acute respiratory syndrome (SARS) coronavirus (33, 34). It is possible that the affinity between the GPs of these viruses and their native receptors may be sufficiently high that this interaction outcompetes TIM-1 for virus interactions. Additionally, these receptors may be more accessible for interaction due to size; for example, the LASV receptor, α-dystroglycan (αDG), is considerably larger than TIM-1 (66). An alternative possibility, which is not mutually exclusive, is that receptor expression on the cells under investigation is abundant, reducing the likelihood of virus/TIM-1 binding. Recently, it has been shown that LASV entry can be enhanced by expression of Axl, DC-SIGN, Tyro3, and LSECtin on cells that lack αDG (67). We are able to detect PtdSer in LASV GPC pseudovirions (data not shown), and TIM-1 ectopic expression on αDG−/− cells may reveal that TIM-1 is able to enhance LASV entry. However, contrary to these possible explanations, expression of TIM-1 did not enhance transduction of SARS coronavirus isolates with reduced binding affinity for ACE2 (33). The mechanism of PtdSer-dependent internalization may be compatible with some glycoproteins but not with others. Additionally, mechanisms or a route(s) of endocytosis may differ between PtdSer receptors, and this may explain why some PtdSer receptors, such as RAGE and BAI1 (34), do not effectively enhance enveloped-virus entry.
The identification and appreciation of the broad use by enveloped viruses of PVEERs may explain the wide tropism seen with many of these viruses, such as the filoviruses and alphaviruses (this study and that of Jemielity et al. [33]) and the flaviviruses (34). In addition to expanding the host range, cell receptor interactions with virion-associated PtdSer would provide effective protection against antibody neutralization, since exposure of GP RBD residues on the surface of extracellular virions would not be necessary. Consistent with this possibility, the EBOV GP RBD has been shown to be protected by a glycan cloud that is only removed once the virus has internalized into endosomes (8, 9). Virus use of PVEERs for internalization may relegate the role of the receptor binding domain of the viral GP to endosomal functions. For filoviruses, these functions would include intracellular binding of NPC1 and subsequent fusion events (Fig. 8). For other enveloped viruses, these functions may involve low-pH-dependent conformational changes of GP leading directly to fusion events.
Fig 8.
Model for phosphatidylserine-mediated virus entry enhancing receptor (or PVEER) uptake of virus. (A) Virus buds from cell surface after infection, incorporating GP and PtdSer onto the viral envelope. The glycan cloud formed by extensive GP glycosylation events provides both stability of the GP in its prefusion state and potential steric protection against immune responses to GP residues. (B) Uptake of virus by a neighboring cell. PtdSer on the viral envelope interacts with PVEERs, such as the Gas6/Axl complex and TIM family, which mediate virus internalization. (C) Conditions within the endosome promote GP-dependent fusion events. Within the endosome, low-pH events can lead directly to GP conformational changes or, alternatively, protease processing of the GP, thereby reducing energy barriers required for fusion. Additional factors, such as binding of NPC1 by filovirus GPs, may also be necessary.
TIM-1 has been postulated to function as an attachment factor (34). Alternatively, TIM-1/virion PtdSer interactions may directly mediate virus internalization. Our studies demonstrate that TIM-1 is able to enhance transduction of Sindbis virus env 2.2 1L1L pseudovirions with mutated RBDs, and we and others found that TIM-1 enhances binding and internalization of No GP VLPs and pseudovirions (33), supporting the idea that TIM-1 interaction with virion-associated PtdSer mediates virus internalization. Additionally, a recent study using VSV G found that proteinaceous receptors are not necessary for fusion (68), suggesting for some viruses that once virions are internalized, endosomal conditions are sufficient to drive viral GP/cell membrane association and fusion. However, in these studies, we cannot completely rule out that TIM-1 acts as an attachment factor and virus internalization is mediated by another as yet uncharacterized receptor(s). Nonetheless, internalization of enveloped viruses independent of a GP would suggest that this second receptor is also a PVEER.
With the elucidation that EBOV entry is mediated by conserved residues of the TIM-1 PtdSer binding pocket, it is not surprising that family member TIM-4 was found to be capable of mediating transduction of EBOV. Since TIM-1 and TIM-4 are found on differing populations of cells (19, 20, 25–27, 69, 70), EBOV targeting of both receptors provides an expanded range of permissive cells. Low-level expression of these receptors may contribute to background transduction seen in otherwise mostly nonpermissive cells for many other viruses.
Future studies are needed to elucidate the in vivo relevance of apoptotic mimicry for viral pathogenesis. While we found that in vivo-derived virus is sensitive to ARD5, the relative contribution of PVEERs in vivo remains unclear. Interestingly, nonhuman primates do not serve as reservoirs for DV in South America as they do in Africa (71), which may be related to TIM-1 mutations and deletions that lead to TIM-1 loss of function in many New World primate lineages (72). It is interesting to speculate that loss of TIM-1 function in these species may be due to selective pressures resulting from virus utilization of TIM-1 as a PVEER. However, targeting PVEERs in humans may be an effective antiviral approach, since a study found that bavituximab, a chimera of mouse MAb 3G4 that cross-links a phosphatidylserine binding protein, β2GP1, to the cell surface, partially protected against lethal infection of guinea pigs with Pichinde virus and reduced the viral load (73). Additionally, hepatitis A virus was recently shown to cloak itself in host-derived membranes (74). The presence of PtdSer on these membranes might explain the use of TIM-1 as a receptor for hepatitis A virus. While these results implicate virion-associated PtdSer in enveloped-virus pathogenesis, more research must be done to determine the role PVEERs play and subsequently the effectiveness of inhibitors of this process.
ACKNOWLEDGMENTS
We thank Nicholas Lennemann, Bethany Rhein, and Patrick Schlievert for providing helpful comments on the manuscript. We thank Ashley Cooney for helping with the mouse studies. We also thank Irvin Chen for providing us with his Sindbis virus 2.2 1L1L env construct and Frederick Boyce for kindly providing infectious stocks of recombinant AcMNPV. Anti-human TIM-1 MAbs were obtained from Biogen Idec.
The work described in this article was supported by the following NIH/NIAID grants: RO1AI077519 (to W.M.), R01AI063513-07 (to R.A.D.), and T32AI007533 (to S.M.-T. and A.S.K.). In addition, the work was supported by DOD/HDTRA 1-12-1-0002 (to R.A.D.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Dolnik O, Kolesnikova L, Becker S. 2008. Filoviruses: interactions with the host cell. Cell. Mol. Life Sci. 65:756–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Geisbert TW, Young HA, Jahrling PB, Davis KJ, Larsen T, Kagan E, Hensley LE. 2003. Pathogenesis of Ebola hemorrhagic fever in primate models: evidence that hemorrhage is not a direct effect of virus-induced cytolysis of endothelial cells. Am. J. Pathol. 163:2371–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leroy EM, Gonzalez JP, Baize S. 2011. Ebola and Marburg haemorrhagic fever viruses: major scientific advances, but a relatively minor public health threat for Africa. Clin. Microbiol. Infect. 17:964–976 [DOI] [PubMed] [Google Scholar]
- 4. Brindley MA, Hughes L, Ruiz A, McCray PB, Jr, Sanchez A, Sanders DA, Maury W. 2007. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J. Virol. 81:7702–7709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dube D, Brecher MB, Delos SE, Rose SC, Park EW, Schornberg KL, Kuhn JH, White JM. 2009. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J. Virol. 83:2883–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuhn JH, Radoshitzky SR, Guth AC, Warfield KL, Li W, Vincent MJ, Towner JS, Nichol ST, Bavari S, Choe H, Aman MJ, Farzan M. 2006. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 281:15951–15958 [DOI] [PubMed] [Google Scholar]
- 7. Manicassamy B, Wang J, Jiang H, Rong L. 2005. Comprehensive analysis of Ebola virus GP1 in viral entry. J. Virol. 79:4793–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, Nagle J, Polymeropoulos MH, Sturley SL, Ioannou YA, Higgins ME, Comly M, Cooney A, Brown A, Kaneski CR, Blanchette-Mackie EJ, Dwyer NK, Neufeld EB, Chang TY, Liscum L, Strauss JF, III, Ohno K, Zeigler M, Carmi R, Sokol J, Markie D, O'Neill RR, van Diggelen OP, Elleder M, Patterson MC, Brady RO, Vanier MT, Pentchev PG, Tagle DA. 1997. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277:228–231 [DOI] [PubMed] [Google Scholar]
- 12. Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Higgins ME, Davies JP, Chen FW, Ioannou YA. 1999. Niemann-Pick C1 is a late endosome-resident protein that transiently associates with lysosomes and the trans-Golgi network. Mol. Genet. Metab. 68:1–13 [DOI] [PubMed] [Google Scholar]
- 14. Alvarez CP, Lasala F, Carrillo J, Muniz O, Corbi AL, Delgado R. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 76:6841–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ji X, Olinger GG, Aris S, Chen Y, Gewurz H, Spear GT. 2005. Mannose-binding lectin binds to Ebola and Marburg envelope glycoproteins, resulting in blocking of virus interaction with DC-SIGN and complement-mediated virus neutralization. J. Gen. Virol. 86:2535–2542 [DOI] [PubMed] [Google Scholar]
- 16. Marzi A, Moller P, Hanna SL, Harrer T, Eisemann J, Steinkasserer A, Becker S, Baribaud F, Pohlmann S. 2007. Analysis of the interaction of Ebola virus glycoprotein with DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin) and its homologue DC-SIGNR. J. Infect. Dis. 196(Suppl 2):S237–S246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shimojima M, Takada A, Ebihara H, Neumann G, Fujioka K, Irimura T, Jones S, Feldmann H, Kawaoka Y. 2006. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J. Virol. 80:10109–10116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, Burke E, Buchmeier MJ, Soilleux EJ, Riley JL, Doms RW, Bates P, Pohlmann S. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115–123 [DOI] [PubMed] [Google Scholar]
- 19. Ichimura T, Bonventre JV, Bailly V, Wei H, Hession CA, Cate RL, Sanicola M. 1998. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J. Biol. Chem. 273:4135–4142 [DOI] [PubMed] [Google Scholar]
- 20. Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, Sandersfeld LM, Quinn K, Weller M, McCray PB, Jr, Chiorini J, Maury W. 2011. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. U. S. A. 108:8426–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McIntire JJ, Umetsu SE, Akbari O, Potter M, Kuchroo VK, Barsh GS, Freeman GJ, Umetsu DT, DeKruyff RH. 2001. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat. Immunol. 2:1109–1116 [DOI] [PubMed] [Google Scholar]
- 22. Rennert PD. 2011. Novel roles for TIM-1 in immunity and infection. Immunol. Lett. 141:28–35 [DOI] [PubMed] [Google Scholar]
- 23. Umetsu SE, Lee WL, McIntire JJ, Downey L, Sanjanwala B, Akbari O, Berry GJ, Nagumo H, Freeman GJ, Umetsu DT, DeKruyff RH. 2005. TIM-1 induces T cell activation and inhibits the development of peripheral tolerance. Nat. Immunol. 6:447–454 [DOI] [PubMed] [Google Scholar]
- 24. McIntire JJ, Umetsu DT, DeKruyff RH. 2004. TIM-1, a novel allergy and asthma susceptibility gene. Springer Semin. Immunopathol. 25:335–348 [DOI] [PubMed] [Google Scholar]
- 25. DeKruyff RH, Bu X, Ballesteros A, Santiago C, Chim YL, Lee HH, Karisola P, Pichavant M, Kaplan GG, Umetsu DT, Freeman GJ, Casasnovas JM. 2010. T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J. Immunol. 184:1918–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kobayashi N, Karisola P, Pena-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, Butte MJ, Nagumo H, Chernova I, Zhu B, Sharpe AH, Ito S, Dranoff G, Kaplan GG, Casasnovas JM, Umetsu DT, Dekruyff RH, Freeman GJ. 2007. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 27:927–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. 2007. Identification of Tim4 as a phosphatidylserine receptor. Nature 450:435–439 [DOI] [PubMed] [Google Scholar]
- 28. Santiago C, Ballesteros A, Martinez-Munoz L, Mellado M, Kaplan GG, Freeman GJ, Casasnovas JM. 2007. Structures of T cell immunoglobulin mucin protein 4 show a metal-ion-dependent ligand binding site where phosphatidylserine binds. Immunity 27:941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chabtini L, Mfarrej B, Mounayar M, Zhu B, Batal I, Dakle PJ, Smith BD, Boenisch O, Najafian N, Akiba H, Yagita H, Guleria I. 2013. TIM-3 regulates innate immune cells to induce fetomaternal tolerance. J. Immunol. 190:88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. 2008. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J. Clin. Invest. 118:1657–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feigelstock D, Thompson P, Mattoo P, Zhang Y, Kaplan GG. 1998. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J. Virol. 72:6621–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaplan G, Totsuka A, Thompson P, Akatsuka T, Moritsugu Y, Feinstone SM. 1996. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J. 15:4282–4296 [PMC free article] [PubMed] [Google Scholar]
- 33. Jemielity S, Wang JJ, Chan YK, Ahmed AA, Li W, Monahan S, Bu X, Farzan M, Freeman GJ, Umetsu DT, Dekruyff RH, Choe H. 2013. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog. 9:e1003232. 10.1371/journal.ppat.1003232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. 2012. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12:544–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sinn PL, Hickey MA, Staber PD, Dylla DE, Jeffers SA, Davidson BL, Sanders DA, McCray PB., Jr 2003. Lentivirus vectors pseudotyped with filoviral envelope glycoproteins transduce airway epithelia from the apical surface independently of folate receptor alpha. J. Virol. 77:5902–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 94:14764–14769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin-Serrano J, Perez-Caballero D, Bieniasz PD. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brindley MA, Hunt CL, Kondratowicz AS, Bowman J, Sinn PL, McCray PB, Jr, Quinn K, Weller ML, Chiorini JA, Maury W. 2011. Tyrosine kinase receptor Axl enhances entry of Zaire ebolavirus without direct interactions with the viral glycoprotein. Virology 415:83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao Y, Whitaker-Dowling P, Watkins SC, Griffin JA, Bergman I. 2006. Rapid adaptation of a recombinant vesicular stomatitis virus to a targeted cell line. J. Virol. 80:8603–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santiago C, Ballesteros A, Tami C, Martinez-Munoz L, Kaplan GG, Casasnovas JM. 2007. Structures of T cell immunoglobulin mucin receptors 1 and 2 reveal mechanisms for regulation of immune responses by the TIM receptor family. Immunity 26:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cao E, Zang X, Ramagopal UA, Mukhopadhaya A, Fedorov A, Fedorov E, Zencheck WD, Lary JW, Cole JL, Deng H, Xiao H, Dilorenzo TP, Allison JP, Nathenson SG, Almo SC. 2007. T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface. Immunity 26:311–321 [DOI] [PubMed] [Google Scholar]
- 42. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 43. Schrodinger LLC. 2010. The PyMOL molecular graphics system, version 1.3r1. Schrodinger LLC, Portland, OR [Google Scholar]
- 44. Binne LL, Scott ML, Rennert PD. 2007. Human TIM-1 associates with the TCR complex and up-regulates T cell activation signals. J. Immunol. 178:4342–4350 [DOI] [PubMed] [Google Scholar]
- 45. Sonar SS, Hsu YM, Conrad ML, Majeau GR, Kilic A, Garber E, Gao Y, Nwankwo C, Willer G, Dudda JC, Kim H, Bailly V, Pagenstecher A, Rennert PD, Renz H. 2010. Antagonism of TIM-1 blocks the development of disease in a humanized mouse model of allergic asthma. J. Clin. Invest. 120:2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. 2005. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology 332:20–27 [DOI] [PubMed] [Google Scholar]
- 47. Logue SE, Elgendy M, Martin SJ. 2009. Expression, purification and use of recombinant annexin V for the detection of apoptotic cells. Nat. Protoc. 4:1383–1395 [DOI] [PubMed] [Google Scholar]
- 48. Jeffers SA, Sanders DA, Sanchez A. 2002. Covalent modifications of the Ebola virus glycoprotein. J. Virol. 76:12463–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang ZY, Duckers HJ, Sullivan NJ, Sanchez A, Nabel EG, Nabel GJ. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6:886–889 [DOI] [PubMed] [Google Scholar]
- 50. Morizono K, Xie Y, Olafsen T, Lee B, Dasgupta A, Wu AM, Chen IS. 2011. The soluble serum protein Gas6 bridges virion envelope phosphatidylserine to the TAM receptor tyrosine kinase Axl to mediate viral entry. Cell Host Microbe 9:286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Callahan MK, Popernack PM, Tsutsui S, Truong L, Schlegel RA, Henderson AJ. 2003. Phosphatidylserine on HIV envelope is a cofactor for infection of monocytic cells. J. Immunol. 170:4840–4845 [DOI] [PubMed] [Google Scholar]
- 52. Andree HA, Reutelingsperger CP, Hauptmann R, Hemker HC, Hermens WT, Willems GM. 1990. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J. Biol. Chem. 265:4923–4928 [PubMed] [Google Scholar]
- 53. Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH. 1994. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84:1415–1420 [PubMed] [Google Scholar]
- 54. Friggeri A, Banerjee S, Biswas S, de Freitas A, Liu G, Bierhaus A, Abraham E. 2011. Participation of the receptor for advanced glycation end products in efferocytosis. J. Immunol. 186:6191–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. He M, Kubo H, Morimoto K, Fujino N, Suzuki T, Takahasi T, Yamada M, Yamaya M, Maekawa T, Yamamoto Y, Yamamoto H. 2011. Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep. 12:358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morizono K, Ku A, Xie Y, Harui A, Kung SK, Roth MD, Lee B, Chen IS. 2010. Redirecting lentiviral vectors pseudotyped with Sindbis virus-derived envelope proteins to DC-SIGN by modification of N-linked glycans of envelope proteins. J. Virol. 84:6923–6934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Morizono K, Pariente N, Xie Y, Chen IS. 2009. Redirecting lentiviral vectors by insertion of integrin-tageting peptides into envelope proteins. J. Gene Med. 11:549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morizono K, Xie Y, Helguera G, Daniels TR, Lane TF, Penichet ML, Chen IS. 2009. A versatile targeting system with lentiviral vectors bearing the biotin-adaptor peptide. J. Gene Med. 11:655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mercer J, Helenius A. 2010. Apoptotic mimicry: phosphatidylserine-mediated macropinocytosis of vaccinia virus. Ann. N. Y. Acad. Sci. 1209:49–55 [DOI] [PubMed] [Google Scholar]
- 60. Mercer J, Helenius A. 2008. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 320:531–535 [DOI] [PubMed] [Google Scholar]
- 61. Hunt CL, Kolokoltsov AA, Davey RA, Maury W. 2011. The tyro3 receptor kinase axl enhances macropinocytosis of Zaire ebolavirus. J. Virol. 85:334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shimojima M, Ikeda Y, Kawaoka Y. 2007. The mechanism of Axl-mediated Ebola virus infection. J. Infect. Dis. 196(Suppl 2):S259–S263 [DOI] [PubMed] [Google Scholar]
- 63. Krejbich-Trotot P, Denizot M, Hoarau JJ, Jaffar-Bandjee MC, Das T, Gasque P. 2011. Chikungunya virus mobilizes the apoptotic machinery to invade host cell defenses. FASEB J. 25:314–325 [DOI] [PubMed] [Google Scholar]
- 64. Callahan MK, Williamson P, Schlegel RA. 2000. Surface expression of phosphatidylserine on macrophages is required for phagocytosis of apoptotic thymocytes. Cell Death Differ. 7:645–653 [DOI] [PubMed] [Google Scholar]
- 65. Coil DA, Miller AD. 2004. Phosphatidylserine is not the cell surface receptor for vesicular stomatitis virus. J. Virol. 78:10920–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, Nichol ST, Compans RW, Campbell KP, Oldstone MB. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079–2081 [DOI] [PubMed] [Google Scholar]
- 67. Shimojima M, Stroher U, Ebihara H, Feldmann H, Kawaoka Y. 2012. Identification of cell surface molecules involved in dystroglycan-independent Lassa virus cell entry. J. Virol. 86:2067–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Matos PM, Marin M, Ahn B, Lam W, Santos NC, Melikyan GB. 2013. Anionic lipids are required for vesicular stomatitis virus G protein-mediated single particle fusion with supported lipid bilayers. J. Biol. Chem. 288:12416–12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Meyers JH, Chakravarti S, Schlesinger D, Illes Z, Waldner H, Umetsu SE, Kenny J, Zheng XX, Umetsu DT, DeKruyff RH, Strom TB, Kuchroo VK. 2005. TIM-4 is the ligand for TIM-1, and the TIM-1-TIM-4 interaction regulates T cell proliferation. Nat. Immunol. 6:455–464 [DOI] [PubMed] [Google Scholar]
- 70. Nakae S, Iikura M, Suto H, Akiba H, Umetsu DT, Dekruyff RH, Saito H, Galli SJ. 2007. TIM-1 and TIM-3 enhancement of Th2 cytokine production by mast cells. Blood 110:2565–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vasilakis N, Cardosa J, Hanley KA, Holmes EC, Weaver SC. 2011. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat. Rev. Microbiol. 9:532–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ohtani H, Naruse TK, Iwasaki Y, Akari H, Ishida T, Matano T, Kimura A. 2012. Lineage-specific evolution of T-cell immunoglobulin and mucin domain 1 gene in the primates. Immunogenetics. 64:669–678 [DOI] [PubMed] [Google Scholar]
- 73. Soares MM, King SW, Thorpe PE. 2008. Targeting inside-out phosphatidylserine as a therapeutic strategy for viral diseases. Nat. Med. 14:1357–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]