Abstract
We applied an efficient method to characterize the relative fitness levels of multiple nonnucleoside reverse transcriptase (NNRTI)-resistant HIV-1 variants by simultaneous competitive culture and 454 deep sequencing. Using this method, we show that the Y181V mutation in the HIV-1 reverse transcriptase in particular confers a clear selective advantage to the virus over 14 other NNRTI resistance mutations in the presence of etravirine in vitro.
TEXT
Replication fitness defines the ability of a virus to replicate under the selective pressures present in its environment. For HIV-1, replication fitness impacts the viral variants that predominate in the quasispecies and therefore influences treatment response and disease progression (1). Etravirine (ETV) is a second-generation nonnucleoside reverse transcriptase (RT) inhibitor (NNRTI) with activity against several NNRTI-resistant variants that confer resistance to the first-generation inhibitors nevirapine (NVP) and efavirenz (2). However, the DUET-1 and DUET-2 clinical trials identified 17 mutations associated with ETV resistance: V90I, A98G, L100I, K101E, K101H, K101P, V106I, E138A, V179D, V179F, V179T, Y181C, Y181I, Y181V, G190A, G190S, and M230L (3). A weighted genotypic score that optimizes resistance interpretation has been assigned to each of these mutations and is used to guide treatment involving ETV usage in treatment-experienced HIV-infected individuals (4). However, very little is known about the in vitro fitness profiles of HIV-1 variants containing these mutations grown in the presence of ETV.
Replication fitness assessed by simultaneous competitive culture and 454 deep sequencing.
The replication fitness of drug-resistant HIV-1 variants is typically assessed by measuring the growth kinetics (in the absence or presence of drug) of two viral variants that are mixed at defined ratios and grown in competition in a single culture (1). A major limitation of this approach is that it is not a high-throughput method and requires considerable effort to evaluate the fitness of multiple drug-resistant HIV-1 variants. Here, we describe an efficient method to characterize the relative fitness levels of multiple mutants simultaneously. We generated 15 infectious viruses containing the single NNRTI resistance mutations V90I, K101P, K103N, V108I, E138A, E138K, V179D, Y181C, Y181I, Y181V, Y188C, G190A, G190S, M230L, and P236L by site-directed mutagenesis of an HIV-1LAI molecular clone (5). A wild-type (WT) virus and the 15 mutant viruses were then normalized for levels of relative infectivity (infectious units/ng p24) in TZM-bl cells and pooled. This initial infectious pool of virus was found to contain similar concentrations of all mutant viruses as assessed by 454 deep sequencing (median, 6.7%; interquartile range [IQR], 5.5% to 8.4%; all n ≥ 3). The effective concentration of ETV required to inhibit 50% of the pooled virus (i.e., EC50) in TZM-bl cells was found to be 62.6 ± 3.4 nM. This value was ∼11-fold higher than the EC50 (5.7 ± 1.3 nM) determined for the WT virus. In contrast, the pooled virus showed >50-fold-greater resistance to NVP (EC50 ≥ 20 μM) compared to the WT virus (280 ± 33 nM).
The pooled virus was used to infect 1.6 × 106 HUT-78 cells at a multiplicity of infection of 0.004. After a 2-h incubation period, cell-free virus was removed and ETV (20 nM, 200 nM, or 500 nM), NVP (5 μM), or dimethyl sulfoxide (DMSO) (no-drug control) was added to the cultures. Culture supernatants were collected at 3-day intervals up to day 26 or until the observed peak of p24 production (Fig. 1A). Viral RNA was extracted, and deep sequencing (454 GS Junior) of an amplicon spanning RT codons 96 to 194 was used to quantify the distribution of mutants at each time point. A median of 2,971 (IQR, 2,726 to 3,128) sequence reads per sample were collected. Sequencing errors (indels) in homopolymer-rich regions (e.g., codon 103) were corrected after alignment by deleting extraneous bases or by inserting the HIV-1LAI reference base where necessary. A mean PCR/deep-sequencing error rate of 0.24% ± 0.18% was determined by calculating the amino acid substitution rate at unmodified codons in viruses grown in the absence of drug (n = 3). The mean PCR/sequencing error rate at the nucleotide level was calculated to be 0.16% ± 0.28%.
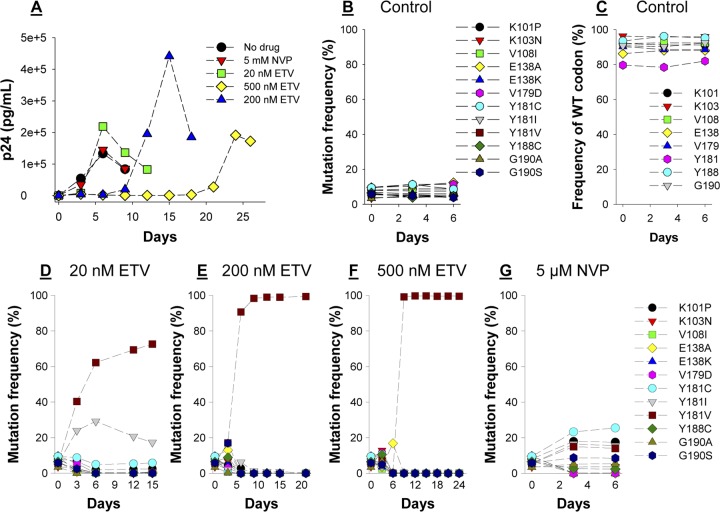
Fig 1.
Replication fitness of multiple NNRTI-resistant HIV-1 variants in the presence of ETV or NVP. (A) Concentrations of p24 in culture supernatant from cells infected with pooled virus and exposed to different concentrations of ETV or NVP. (B, C, D, E, F, and G) The relative prevalences of 12 viruses each containing a single resistance mutation as determined by longitudinal deep sequencing of viral RNA from culture supernatant from cells infected with pooled virus and exposed to no drug (control), 20 nM ETV, 200 nM ETV, 500 nM ETV, or 5 μM NVP.
The Y181V mutation confers a clear fitness advantage over other NNRTI resistance mutations in the presence of ETV.
Longitudinal deep sequencing revealed that in the absence of drug, the frequencies of most NNRTI-resistant variants either remained the same or decreased as a function of time (Fig. 1B). As expected, there was also a concomitant increase in the frequency of the wild-type (WT) sequence at most resistance codons (Fig. 1C). In contrast, the Y181V virus emerged as the most common variant by days 3, 6, and 9 in cultures grown in 20 nM, 200 nM, and 500 nM ETV, respectively (Fig. 1D, E, and F). The frequency of the Y181I virus also increased up to day 6 in the 20 nM ETV culture but declined thereafter (Fig. 1D). Transient increases in viruses containing other mutations were also observed at day 3 in the cultures grown in 200 nM ETV (K103N, E138A, Y188C, and G190S) and 500 nM ETV (K101P, K103N, E138A, V179D, Y188C, and G190A/S) before Y181V-containing viruses grew to dominate the viral pool (Fig. 1E and F). Importantly, 3 independent fitness experiments revealed that the spectrum of mutant viruses that grew out in cultures containing 200 nM ETV did not vary; Y181V mutants made up 90% of the sequenced reads at day 6 in all 3 experiments. In contrast to ETV, K101P and Y181C were the most common variants in the NVP-containing cultures at day 6 (Fig. 1G). However, the Y181I and Y181V variants were also selected. The deep sequencing results were confirmed by population-based sequencing (Table 1). Of note, there was no evidence of virus recombination in any of the experiments performed during culture, PCR, or sequencing. To quantify the selective advantage of the Y181V mutant HIV-1 in the presence of ETV, we fit a population genetic model to the observed prevalence of Y181V over time versus other amino acids at that position. We used a deterministic haploid selection model of allele frequency evolution, which assumes infinite effective population size and negligible mutation rates relative to the existing polymorphism. The model was fit to data by minimizing least squares using the optimize function in the statistical software package R (version 2.15.2). Y181V was estimated to have 1.8-, 2.0-, and 1.5-fold selective advantages over other Y181 variants cultured in the presence of 20 nM, 200 nM, and 500 nM ETV, respectively. Drug susceptibility assays revealed that the Y181V mutation in HIV-1 RT conferred ∼60-fold resistance to ETV (Table 2), thus explaining—in part—its fitness advantage in our assay system. However, Y181I HIV-1 also exhibited ∼60-fold-decreased susceptibility to ETV, suggesting that this mutation confers a fitness disadvantage to the virus in comparison to Y181V. Similarly, the G190S and K103N mutations confer significant resistance to NVP (Table 2), and yet the growth of both of these variants was significantly outpaced by that of the K101P and Y181C/I/V mutants. In this regard, it should be noted that the K101P, Y181I, and Y181V mutations in HIV-1 RT require a double-nucleotide change. As such, despite their increased replication fitness in the presence of NVP observed in this study, they are less frequently observed in HIV-infected individuals in comparison to mutations (e.g., Y181C and K103N) that require a single nucleotide change.
Table 1.
Population sequencing of viral culture supernatants
| NNRTI | Day | Amino acid(s) at codon: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 90 | 101 | 103 | 108 | 138 | 179 | 181 | 188 | 190 | 230 | 236 | ||
| None | 0 | V | K | K | V | E | V | Y | Y | G | M | P |
| 3 | V | K | K | V | E | V | Y | Y | G | M | P | |
| 6 | V | K | K | V | E | V | Y | Y | G | M | P | |
| 5 μM NVP | 3 | V | K/P | K | V | E | V | Y/C | Y | G | M | P |
| 6 | V | K/P | K | V | E | V | Y/C | Y | G | M | P | |
| 20 nM ETV | 3 | V | K | K | V | E | V | I/V | Y | G | M | P |
| 6 | V | K | K | V | E | V | I/V | Y | G | M | P | |
| 12 | V | K | K | V | E | V | V | Y | G | M | P | |
| 15 | V | K | K | V | E | V | I/V | Y | G | M | P | |
| 200 nM ETV | 3 | V | K | K | V | E | V | I/V | Y | G | M | P |
| 6 | V | K | K | V | E | V | V | Y | G | M | P | |
| 9 | V | K | K | V | E | V | V | Y | G | M | P | |
| 12 | V | K | K | V | E | V | V | Y | G | M | P | |
| 15 | V | K | K | V | E | V | V | Y | G | M | P | |
| 21 | V | K | K | V | E | V | V | Y | G | M | P | |
| 500 nM ETV | 3 | V | K | K | V | E | V | Y | Y | G | M | P |
| 6 | V | K/P | K | V | E | V | Y/C/I/V | Y | G | M | P | |
| 9 | V | K/P | K | V | E | V | Y/C/I/V | Y | G | M | P | |
| 12 | V | K | K | V | E | V | V | Y | G | M | P | |
| 15 | V | K | K | V | E | V | V | Y | G | M | P | |
| 18 | V | K | K | V | E | V | V | Y | G | M | P | |
| 21 | V | K | K | V | E | V | V | Y | G | M | P | |
| 24 | V | K | K | V | E | V | V | Y | G | M | P | |
Table 2.
Susceptibility to ETV and NVP of selected NNRTI-resistant HIV-1 viruses
| NNRTI parameter | Valuea |
|||||||
|---|---|---|---|---|---|---|---|---|
| WT | K101P | K103N | E138A | Y181C | Y181I | Y181V | G190S | |
| ETV | ||||||||
| EC50 (nM) | 5.7 | 27.8 | 3.8 | 15.9 | 44.3 | 353 | 328 | 2.2 |
| Fold R | 4.9 | 0.7 | 2.8 | 7.8 | 61.9 | 57.9 | 0.4 | |
| NVP | ||||||||
| EC50 (μM) | 0.28 | >20 | >20 | 0.23 | >20 | >20 | >20 | >20 |
| Fold R | >50 | >50 | 0.8 | >50 | >50 | >50 | >50 | |
EC50 values represent mean values from at least 3 independent experiments. Fold R values represent mean fold change in EC50 of mutant versus WT virus.
Conclusions.
We have developed an efficient method to characterize the relative fitness levels of multiple NNRTI-resistant HIV-1 variants simultaneously. Using this method, we show that the Y181I and Y181V mutations confer a clear fitness advantage over other NNRTI resistance mutations in the presence of ETV in vitro. Our data are consistent with the finding that Y181I and Y181V were assigned the highest weights in a genotypic scoring system that was developed for ETV resistance-associated mutations based on their impact on treatment response (4). However, a primary limitation of this study is that the mutations were assessed in the context of a defined genetic backbone. Furthermore, the fitness assays were carried out in a T cell line. In this regard, the results reported in this study might have differed if a different strain of HIV-1 or cell line had been used.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grants GM068406 and AI081571 (to N.S.-C.). P.R.H. is supported by a CIHR/GSK Chair in Clinical Virology and C.J.B. by a CIHR Vanier Canada Graduate Scholarship.
Footnotes
Published ahead of print 29 May 2013
REFERENCES
- 1. Dykes C, Demeter LM. 2007. Clinical significance of human immunodeficiency virus type 1 replication fitness. Clin. Microbiol. Rev. 20:550–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andries K, Azijn H, Thielemans T, Ludovici D, Kukla M, Heeres J, Janssen P, De Corte B, Vingerhoets J, Pauwels R, de Béthune MP. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lazzarin A, Campbell T, Clotet B, Johnson M, Katlama C, Moll A, Towner W, Trottier B, Peeters M, Vingerhoets J, de Smedt G, Baeten B, Beets G, Sinha R, Woodfall B; DUET-2 study group 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-2: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet 370:39–48 [DOI] [PubMed] [Google Scholar]
- 4. Vingerhoets J, Tambuyzer L, Azijn H, Hoogstoel A, Nijs S, Peeters M, de Béthune MP, De Smedt G, Woodfall B, Picchio G. 2010. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled phase III clinical studies. AIDS 24:503–514 [DOI] [PubMed] [Google Scholar]
- 5. Shi C, Mellors JW. 1997. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 41:2781–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]