Abstract
Biochemical reactions powered by ATP hydrolysis are fundamental for the movement of molecules and cellular structures. One such reaction is the encapsidation of the double-stranded DNA (dsDNA) genome of an icosahedrally symmetric virus into a preformed procapsid with the help of a genome-translocating NTPase. Such NTPases have been characterized in detail from both RNA and tailed DNA viruses. We present four crystal structures and the biochemical activity of a thermophilic NTPase, B204, from the nontailed, membrane-containing, hyperthermoacidophilic archaeal dsDNA virus Sulfolobus turreted icosahedral virus 2. These are the first structures of a genome-packaging NTPase from a nontailed, dsDNA virus with an archaeal host. The four structures highlight the catalytic cycle of B204, pinpointing the molecular movement between substrate-bound (open) and empty (closed) active sites. The protein is shown to bind both single-stranded and double-stranded nucleic acids and to have an optimum activity at 80°C and pH 4.5. The overall fold of B204 places it in the FtsK-HerA superfamily of P-loop ATPases, whose cellular and viral members have been suggested to share a DNA-translocating mechanism.
INTRODUCTION
Viruses with double-stranded DNA (dsDNA) or dsRNA genomes often package their genome through a channel in a preformed capsid or procapsid (1). Genome translocation is powered by NTP hydrolysis catalyzed by a DNA- (or RNA)-packaging enzyme (1). In tailed dsDNA viruses, the channel is a dodecameric ring called the connector, located at a viral 5-fold vertex referred to as the packaging vertex (2, 3). These connectors often serve as nucleation points for the assembly of motor components that are transiently associated with the capsid (1). In phage T4, for example, the terminase complex consists of the small terminase subunit gp16, the large terminase subunit gp17, which is the packaging ATPase, and the dodecameric portal ring gp20. T4 gp17 has an N-terminal domain with ATPase activity and a C-terminal domain that translocates and cuts the incoming genomic DNA (4). gp16 stimulates the ATPase activity of gp17 50-fold (5). In some cases, such as the tailed phage ϕ29 and the nontailed phage PRD1, the linear dsDNA genome is associated with covalently linked terminal proteins (6, 7) recognized by the genome-packaging ATPase (8). The dsRNA virus φ12 hexameric NTPase P4 combines both RNA translocation and NTPase hydrolysis into a single domain (9) and is a structural component of the virion (10). The genome-packaging NTPases of some membrane-containing, nontailed dsDNA viruses, like PRD1, are also part of the virion as shown by gel analysis, mass spectrometry, and immunolabeling (8, 11–13). These PRD1-type NTPases have been recalcitrant to structural studies due to instability and insolubility (14), and consequently the catalytic cycles of these proteins are poorly understood.
The NTPase domain of a typical DNA-packaging enzyme contains a phosphate-binding loop (P-loop or Walker A sequence motif) as well as a Walker B motif (1, 15–17). These sequence motifs also occur in many cellular proteins such as helicases, kinases, and recombinases (15, 17–19). The consensus sequences for the respective motifs are GXXXXGK(T/S) and hhhhDE, where X denotes any amino acid and h any hydrophobic amino acid (15, 17). Amino acids from both sequence motifs take part in nucleotide binding and hydrolysis: most importantly, the conserved lysine in the Walker A motif is responsible for nucleotide binding, and the conserved glutamate in the Walker B motif is often responsible for activation of water for the hydrolysis reaction (20). The Mg2+ ion required for ATP hydrolysis can be coordinated either by the conserved aspartate in the Walker B domain (15, 17) or by the conserved serine in the Walker A domain (21, 22). So-called arginine fingers facilitate the formation of the transition state (23) and are inserted into the catalytic site of a neighboring subunit in response to a conformational change in the catalytic site of the preceding subunit.
Sulfolobus turreted icosahedral virus 2 (STIV2) was isolated from the hyperthermoacidophilic archaeon Sulfolobus islandicus originating from an acidic hot spring (88.3°C, pH 3.5) and has a 16.6-kbp-long dsDNA genome with 34 predicted open reading frames (ORFs) (24). Electron cryomicroscopy (cryo-EM) and image reconstruction of STIV2 to 20-Å resolution revealed an icosahedral virus capsid with an internal membrane and vertices decorated by large turrets thought to be involved in host cell recognition and attachment (24). Thus far, nine structural proteins have been identified by mass spectrometry (24). Sequence alignment and homology modeling of the major capsid protein A345 suggested that it has a double β-barrel fold (24). The double β-barrel fold of the major capsid protein is conserved within viruses from diverse hosts, like human adenovirus, Paramecium bursaria Chlorella virus type 1 (PBCV-1), bacteriophage PRD1, (25–27), and the hyperthermoacidophilic, archaeal STIV (28), which is the closest relative to STIV2. These viruses, all belonging to the “PRD1-like lineage,” are believed to have a common ancestor that precedes the separation of the three domains of life (29). It has been suggested that these highly diverse viruses share a core set of genes (the major capsid protein, the penton base, and the genome-packaging enzyme) that are inherited together (28, 30, 31). However, until now, no packaging NTPase structures have been available for this lineage.
Here, we report the first structure determination of a genome-packaging NTPase, B204, from a nontailed, membrane-containing, dsDNA virus, in four different states: with a bound sulfate ion in the phosphate binding site, in complex with AMP, in complex with the substrate analogue adenylylmethylenediphosphonate (AMPPCP), and in complex with the product ADP. We determined the optimal conditions for in vitro nucleotide hydrolysis and show that B204 binds both linear and circular DNA, with linear dsDNA stimulating the hydrolysis reaction. We further present a model for B204-driven genomic DNA packaging.
MATERIALS AND METHODS
B204 expression and purification.
A putative gene (b204) coding for an NTPase (B204) had been identified previously in the STIV2 sequence (24). The b204 gene (named based on its reading frame in the STIV2 genome and its length in amino acids) was amplified by PCR from Sulfolobus islandicus G4ST-2 (24) using forward primer 5′ CGCCCGCATATGAATCCGGATGATATAGT and reverse primer 5′ GTGGTGCTCGAGAATCGGCTTGTGTATCTT. The product was cloned into pET22b using NdeI and XhoI. The B204 gene was expressed in Escherichia coli ER2566/pTF16 (TaKaRa Bio Inc.) in Luria-Bertani medium supplemented with ampicillin (100 μg/ml), chloramphenicol (34 μg/ml), l-arabinose (5 mg/ml), and 1 drop of antifoam 204. Cells were grown at 37°C to an optical density at 600 nm (OD600) of 0.5 to 0.6 and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h. The harvested cells were stored at −20°C. To produce l-seleno-methionine-labeled B204 for multiwavelength anomalous diffraction (MAD), B834/pTF16 cells were grown as above to an optical density of 0.5 to 0.6, transferred to 400 ml minimal morpholinepropanesulfonic acid (MOPS) medium supplemented with 17 amino acids (32), l-seleno-methionine (50 mg/liter), and antibiotics as above, induced with 0.5 mM IPTG for 2 h, harvested, and stored as above.
Cell pellets were resuspended in 100 mM morpholineethanesulfonic acid (MES) (pH 6.5), 100 mM NaCl, 10 mM sodium phosphate, and 5 mM MgCl2 (buffer A) with DNase I (4 μg/ml), lysozyme (40 μg/ml), and 1 mM Pefabloc protease inhibitor and lysed with a French press (22°C used for all steps). The cleared cell lysate was applied to a HiTrap heparin HP-column, and bound B204 was eluted with a 0.1 to 2 M NaCl gradient. B204-containing fractions were diluted 2:5 in buffer A containing 20 mM imidazole and loaded on a HisTrap HP-column. Bound B204 was eluted with a 0.02 to 0.8 M imidazole gradient. B204-containing fractions were diluted 1:5 with buffer A, loaded onto a HiTrap SP HP-column, and then eluted with a 0.1 to 2 M NaCl gradient. The monomeric protein was separated by size exclusion chromatography on a Superdex 200 HR 10/300 gel filtration column in 100 mM citric acid (pH 5.0), 50 mM NaCl, and 5 mM MgCl2. All columns were from GE Healthcare. B204 was concentrated to 12.5 mg/ml by ultrafiltration (10,000 nominal molecular weight limit; Millipore Amicon), snap-frozen in liquid nitrogen, and stored at −80°C.
Malachite green NTPase activity assay.
NTPase activity was measured using the malachite green assay, as described previously (33) using 7 μg of protein in a 50-μl reaction volume with a final nucleotide concentration of 2 mM. The data were corrected for nonenzymatic ATP hydrolysis. All reactions were done in triplicate. KH2PO4 was used to generate standard curves.
To determine the activity of B204, the assay was performed at pH values between 3.0 and 7.0 (100 mM citric acid [pH 3.0 to 5.0], 100 mM MES [pH 5.5 to 6.0], 100 mM HEPES [pH 7.0]). All buffers contained 100 mM NaCl and 5 mM MgCl2. The temperature range analyzed was from 50°C to 90°C. In order to analyze the reaction requirements of B204, ATP was replaced by GTP, CTP, or UTP, and likewise MgCl2 was replaced with CaCl2 or MnCl2. To determine whether the activity of B204 is stimulated by nucleic acids, 700 ng of linear dsDNA (PCR product of STIV2 a345, 1.8 kbp) or φX174 RF I DNA (dsDNA, covalently closed, circular, 5.386 kbp) was used in the reaction.
Electrophoretic mobility shift assay.
B204 nucleic acid binding was analyzed using an electrophoretic mobility shift assay. Briefly, 0.1, 0.5, and 1.0 μg of B204 were incubated with 100 ng of DNA in 10-μl reaction mixtures, including linear dsDNA (PCR-product of the STIV2 a345), φX174 RF I DNA (3.50 × 106 Da), φX174 RF II DNA (dsDNA, nicked [relaxed], circular, 5.386 kbp, 3.50 × 106 Da), and φX174 virion DNA (single-stranded DNA [ssDNA], circular, 5.386 kb, 1.70 × 106 Da). The reactions were performed at 22°C in 100 mM citric acid (pH 4.5), 100 mM NaCl, and 5 mM MgCl2 for 30 min. The samples were analyzed in 0.8% SeaKem LE-agarose gels in 1× Tris-acetic acid-EDTA buffer with 0.5 μg/ml ethidium bromide (EtBr), and results were recorded using a Bio-Rad Molecular Imager ChemiDoc XRS system.
Alternatively, 0.5 and 1.0 μg of B204 were incubated with 100 ng of DNA in 5-μl reaction mixtures using short DNA molecules (15-, 20-, 25-, and 35-bp No Limits dsDNA fragments; Fermentas). The reactions were performed at 22°C in 100 mM citric acid (pH 5.0), 50 mM NaCl, and 5 mM MgCl2 for 10 min. The samples were analyzed with 20% polyacrylamide gel electrophoresis (PAGE) in 1× Tris-borate-EDTA buffer, stained with EtBr, and recorded as above. Gel shift assays for RNA binding by B204 were carried out similarly to the DNA-binding assays.
Protein crystallography.
Native and Se-Met-labeled B204 crystals were grown by hanging-drop vapor diffusion (22°C) by mixing 2 μl of protein (12.5 mg/ml) with 2 μl of reservoir (0.1 M sodium cacodylate [pH 6.0], 0.21 M ammonium sulfate, 0.05 M MgCl2, 35% polyethylene glycol [PEG] 6000). B204 was cocrystallized with AMP in sitting drops at 22°C by mixing 200 nl of protein solution (12.5 mg/ml) with 200 nl reservoir solution (0.1 M Tris-HCl [pH 8.5], 0.2 M MgCl2, 30% PEG 4000). B204 was cocrystallized with AMPPCP or ATPγS in sitting drops at 22°C by mixing 1 or 2 μl of protein solution (12.5 mg/ml) with 1 or 2 μl of reservoir solution (0.1 M Tris-HCl [pH 8.3], 0.2 M MgCl2, 30% PEG 8000), respectively. In all crystallization setups, the final nucleotide concentration was 5 mM. Crystals were cryoprotected in Paratone-N and flash-frozen in liquid nitrogen.
Diffraction data were collected at the European Synchrotron Radiation Facility beamlines ID14-4 and ID23-2. The X-ray fluorescence scan and data collection were performed at beamline 911-3 at MAX-lab (34). The diffraction images were processed using XDS (35). The Se sites were located using SHELXD (36), and the two wavelength MAD data together with the native data were used for phasing with SHARP (37) and SOLOMON (38) (Table 1). The initial model was built using ARP/wARP (39). Molecular replacement was performed using Phaser (40, 41), with manual model building in Coot (42). Refinement was performed in REFMAC5 (43) and PHENIX (44). The refinement statistics are presented in Table 2. The percentages of residues in the favored areas of the Ramachandran plot for the structures are as follows: (i) sulfate, 96.35; (ii) AMP, 96.93; (iii) ATPγS, 94.93; and (iv) AMPPCP, 97.29.
Table 1.
Data collection for MAD phasinga
| Characteristic | Native data | Se-Met data |
|
|---|---|---|---|
| Space group | C2 | C2 | |
| Cell dimensions | |||
| a, b, c (Å) | 193.91, 61.15, 78.92 | 194.80, 60.50, 79.72 | |
| α, β, γ (°) | 90.00, 111.11, 90.00 | 90.00, 111.13, 90.00 | |
| Peak | Inflection | ||
| Wavelength | 0.979110 | 0.979790 | 0.979860 |
| Resolution (Å) | 50–2.49 (2.64–2.49) | 50–3.21 (3.41–3.21) | 50–3.22 (3.41–3.22) |
| Rmeas | 0.072 (0.608) | 0.083 (0.166) | 0.087 (0.181) |
| Rmerge | 0.061 (0.520) | 0.060 (0.121) | 0.063 (0.132) |
| I/σ | 14.99 (2.33) | 13.32 (7.51) | 12.96 (7.01) |
| Completeness (%) | 97.3 (94.8) | 96.6 (94.4) | 96.9 (95.7) |
| Redundancy | 3.60 (3.64) | 1.93 (1.90) | 1.93 (1.90) |
Values in parentheses represent the highest-resolution shell.
Table 2.
Statistics for crystallography and refinementa
| Structure characteristics | SO4− | AMP | ATPγS | AMPPCP |
|---|---|---|---|---|
| Data collection | ID23-2 | ID14-4 | ID23-2 | ID23-2 |
| Space group | C 2 | P 21 | P 1 | P 1 |
| Monomers in AU | 3 (chains A to C) | 2 (chains A and B) | 4 (chains A to D) | 4 (chains A to D) |
| Cell dimensions | ||||
| a, b, c (Å) | 193.57, 59.60, 77.56 | 46.6, 61.02, 70.00 | 46.73, 65.19, 71.62 | 46.56, 65.23, 72.12 |
| α, β, γ (°) | 90, 111.08, 90 | 90, 96.57, 90 | 90.55, 93.65, 91.59 | 90.85, 93.77, 91.56 |
| Resolution (Å) | 50–1.96 (2.07–1.96) | 50–1.95 (2.06–1.95) | 50–2.24 (2.38–2.24) | 50–1.89 (2.01–1.89) |
| Rmeas | 0.068 (0.637) | 0.136 (0.713) | 0.109 (0.746) | 0.099 (0.820) |
| Rmerge | 0.057 (0.528) | 0.123 (0.622) | 0.091 (0.621) | 0.085 (0.708) |
| I/σ | 14.00 (2.15) | 9.21 (1.92) | 10.82 (2.09) | 11.78 (1.94) |
| Completeness (%) | 98.0 (96.6) | 97.4 (85.5) | 96.8 (91.3) | 97.4 (92.4) |
| Redundancy | 3.17 (3.17) | 5.36 (3.96) | 3.39 (3.37) | 3.94 (3.90) |
| Refinement | ||||
| Resolution (Å) | 48.59–1.96 | 46.29–1.95 | 46.61–2.24 | 48.71–1.89 |
| No. of reflections | 58,753 | 28,038 | 39,238 | 65,852 |
| Rwork/Rfree | 0.1777/0.2160 | 0.1970/0.2384 | 0.1811/0.2465 | 0.1883/0.2336 |
| No. of atoms | ||||
| Protein | 4,958 | 3,214 | 6,277 | 6,367 |
| Ligand/ion | 46 | 53 | 214 | 214 |
| Water | 450 | 128 | 221 | 428 |
| B factors | ||||
| Protein | 30.06 | 27.03 | 35.38 | 26.65 |
| Ligand/ion | 36.20 | 39.97 | 40.60 | 34.35 |
| Water | 36.21 | 31.35 | 35.44 | 32.97 |
| RMSD | ||||
| Bond lengths (Å) | 0.008 | 0.013 | 0.013 | 0.011 |
| Bond angles (°) | 1.142 | 1.125 | 1.205 | 1.140 |
Values in parentheses represent the highest-resolution shell.
Structural alignment and homology modeling.
Buried surface area was calculated using PDBePISA (45). The hexamer model of B204 was generated in PyMOL (The PyMOL Molecular Graphics System, version 1.4.1; Schrödinger, LLC) by aligning the AMPPCP structure to each of the FtsK monomers in Protein Data Bank (PDB accession number 2iuu) (21) (RMSD, 1.832 Å). The Dali server was used for structural alignment of B204 (46).
Mass spectrometry.
Mass spectrometry of purified virions was carried out as described previously (24).
Protein structure accession numbers.
The structures determined in this study have been deposited in the Protein Data Bank with the accession numbers 4KFR (sulfate), 4KFS (AMP), 4KFT (ATPγS), and 4KFU (AMPPCP).
RESULTS
B204 is an NTPase that binds nucleic acids.
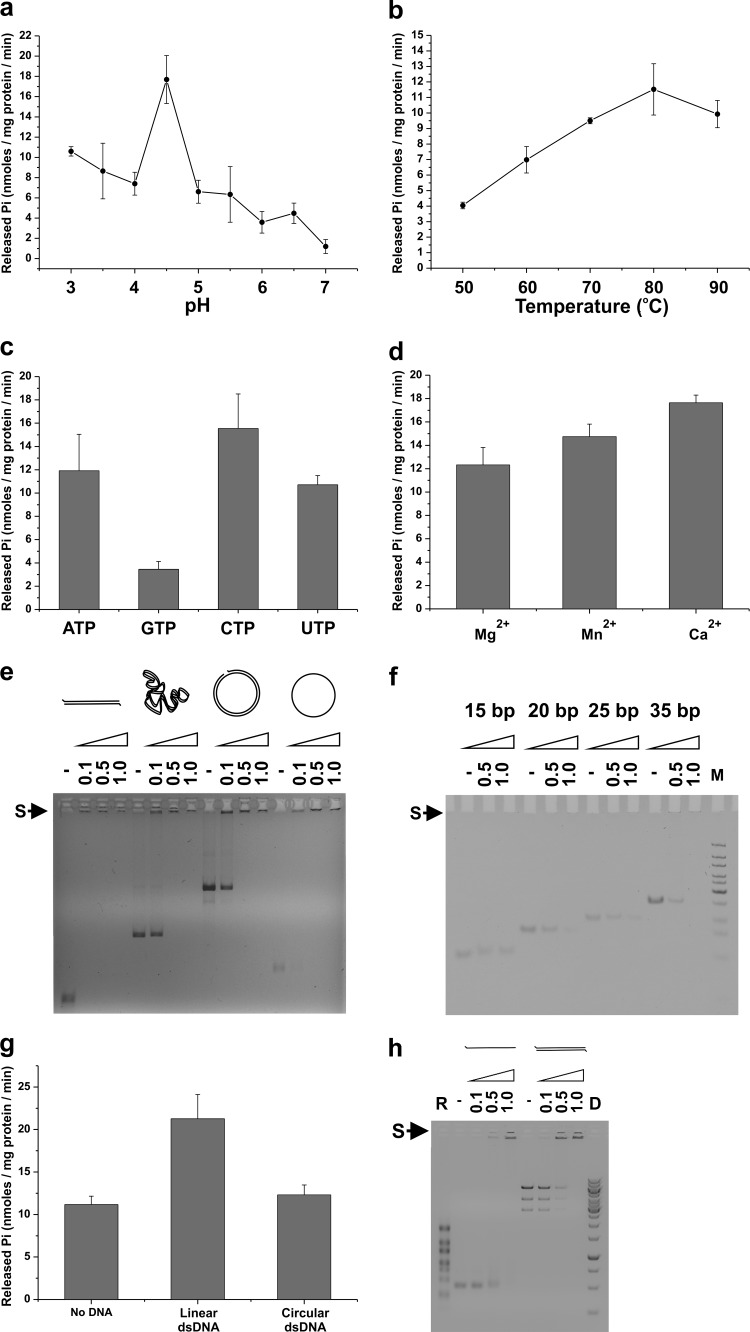
We originally postulated B204 to be an ATPase due to the presence of canonical Walker A and B sequence motifs (17, 24). We produced B204 as a recombinant protein with a mass of 24.8 kDa, indicating that it was the expected, full-length, monomeric protein. The enzymatic activity of B204 was analyzed using the malachite green assay. Initial screening was done for pH optimum using buffers ranging from pH 3 to 7 at 80°C (Fig. 1a) with the rationale that the host for STIV2 grows at 80°C (24). We further explored the activity of B204 at its optimum pH of 4.5 at temperatures ranging from 50°C to 90°C (Fig. 1b). Based on these experiments, B204 is most active as an NTPase at pH 4.5 and 80°C, which resemble the physiological conditions of the STIV2 host (24). We analyzed further the nucleotide specificity of B204 at pH 4.5 and 80°C with ATP, GTP, CTP, and UTP and found that B204 can hydrolyze all of them (Fig. 1c). Under these reaction conditions, ATP, CTP, and UTP are preferentially hydrolyzed. With ATP as the substrate, the divalent cations Mg2+, Mn2+, and Ca2+ are all effective cofactors (Fig. 1d).
Fig 1.
B204 activity and nucleic acid affinity. The release of phosphate (Pi) by B204 during nucleotide hydrolysis was measured using a malachite green assay. (a) Influence of pH on the activity of B204 ATP hydrolysis at 80°C. (b) Effect of temperature on the activity of B204 ATP hydrolysis at pH 4.5. (c) Nucleotides hydrolyzed by B204 at pH 4.5 and 80°C. (d) Divalent metal cations used by B204 in ATP hydrolysis at pH 4.5 and 80°C. (e) Electrophoretic mobility shift assay of DNA in complex with B204. B204 binds linear, supercoiled, and relaxed dsDNA and circular ssDNA at pH 4.5 (22°C). The size of the linear dsDNA molecule is 1.8 kbp. (f) B204 in complex with short dsDNA oligonucleotides at pH 5.0 (22°C). M, Fermentas Ultra-Low range DNA size marker. In panels e and f, the numbers above the wells indicate μg of B204/100 ng DNA. An S with an arrow indicates the nucleic acid shift upon binding to B204. (g) Effect of nucleic acids on the activity of B204 ATP hydrolysis at pH 4.5 80°C. (h) B204 RNA affinity. Electrophoretic mobility shift assay of RNA molecules in complex with B204. B204 binds linear ssRNA (a 700-nt-long fragment prepared from plasmid pEM15 (57) and dsRNA (bacteriophage φ6 genomic RNA) at pH 4.5, 22°C. R, ssRNA size marker (RiboRuler RNA ladder, HighRange; Fermentas); D, dsDNA size marker (GeneRuler 1kb; Fermentas).
We performed electrophoretic mobility shift assays to analyze B204 binding to nucleic acids. The addition of increasing amounts of B204 to different nucleic acids caused retardation of the nucleic acid in gel electrophoresis seen as a disappearance of the control band and the appearance of a faint band close to the well (indicated by an “S” in Fig. 1e, f, and h). B204 binds both linear and circular DNA, as well as ds and ssDNA (Fig. 1e), without apparent sequence specificity. The minimal length of a B204-bound DNA molecule is 20 bp, 15 bp being too short (Fig. 1f). We wanted, in addition, to determine if the activity of B204 can be stimulated by the addition of nucleic acids in the reaction mixture. We thus analyzed the activity of B204 together with linear and circular dsDNA (Fig. 1g). Linear dsDNA stimulated ATP hydrolysis, whereas circular dsDNA did not (Fig. 1g). Furthermore, under our assay conditions B204 also binds linear ssRNA and dsRNA (Fig. 1h). As the binding assays were carried out at 22°C, both the φX174 ssDNA and the φ6 ssRNA molecules could be rich in secondary structure, containing numerous predicted hairpin structures. The experiment did not strictly determine if B204 binds to the ss or the ds regions of these molecules.
Crystal structure of B204 ligand complexes.
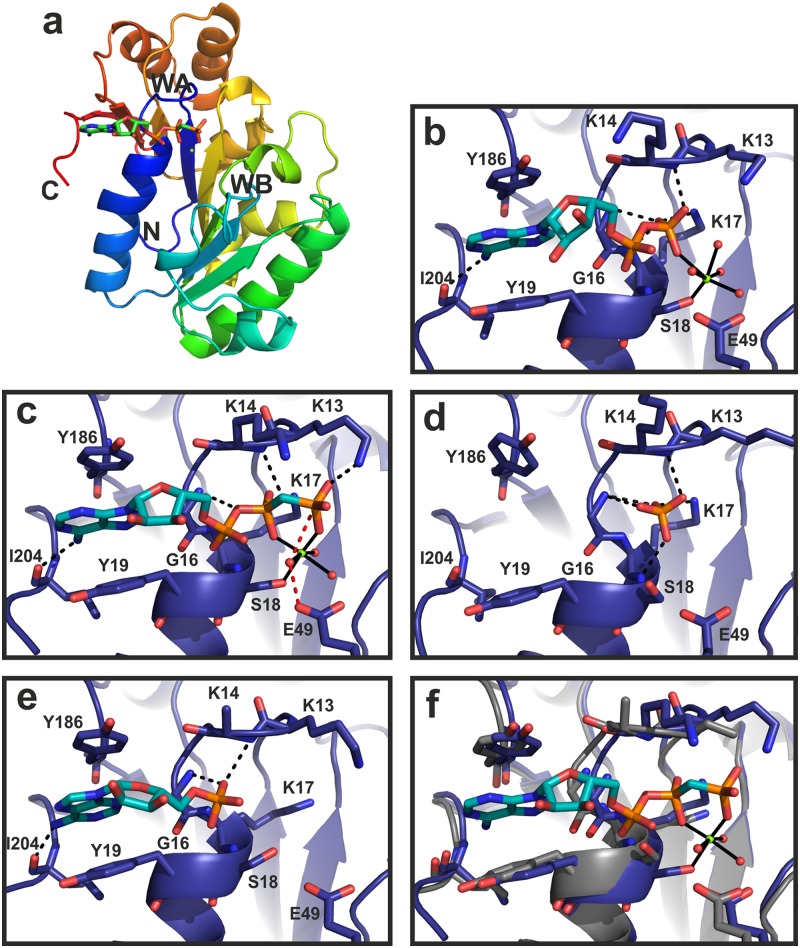
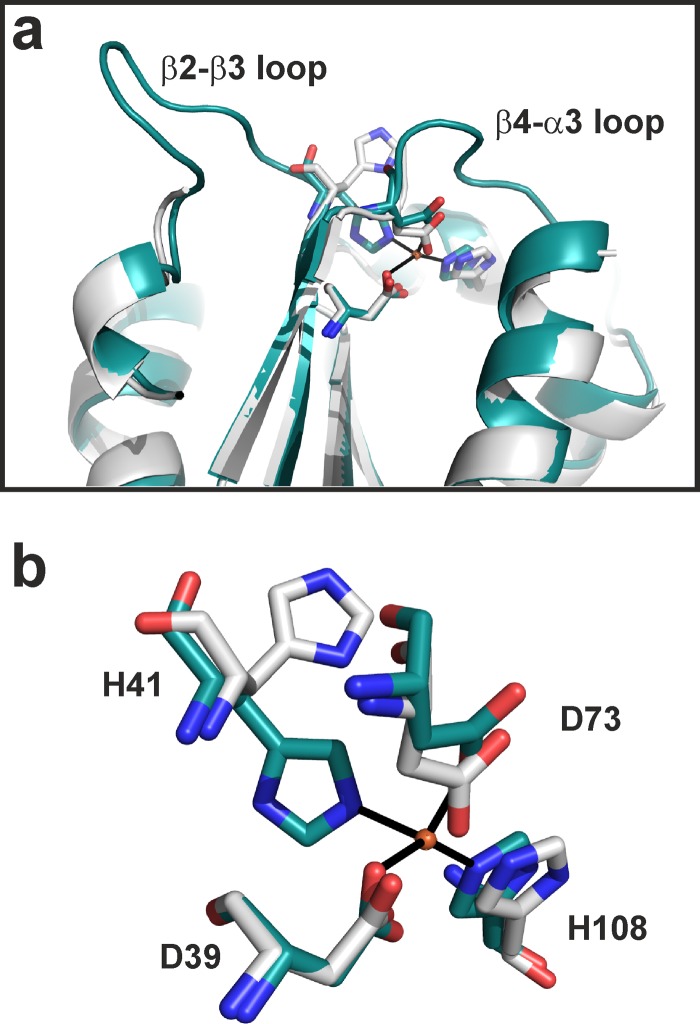
We determined the structures of B204 in complex with (i) a sulfate ion, (ii) AMP, (iii) ATPγS, and (iv) AMPPCP (Fig. 2 and 3 and Tables 1 and 3). The resolutions of the structures were between 1.89 and 2.24 Å. The ion in the first complex listed above was modeled as sulfate based on the crystallization conditions (see Materials and Methods). Furthermore, the ATPγS molecule seems to have been hydrolyzed to ADP (Fig. 2 and 3). Although there are 2 to 4 molecules (named chains A to D) in the crystallographic asymmetric unit (AU) (Tables 1 and 3), the largest buried surface area between any pair of molecules is 640 Å2. Furthermore, as the recombinant B204 protein eluted as a monomer in gel filtration in the presence of nucleotides, we conclude that B204 is a monomer. The structure consists of nine β-strands and seven helices (Fig. 2a and 4). The twisted central β-sheet restrains the nucleotide-binding site along with two additional helices (α1 and α7). B204 belongs to the family of A32-like packaging NTPases and has the FtsK-HerA superfamily fold (18), as we had predicted (24). The main-chain root mean square deviation (RMSD) between chains A of each of the four structures is <0.5 Å. Two loop regions between amino acids 41 to 47 (β2-β3 loop) and 74 to 78 (β4-α3 loop) are disordered in most chains (Fig. 5a).
Fig 2.
Structure and active-site geometry of B204. (a) Ribbon diagram of the B204 monomer bound to AMPPCP and Mg2+. The C and N termini of the protein are indicated (C, N), as are the locations of the Walker A (WA; blue loop) and B (WB; green loop) sequence motifs. (b) Active site of B204 (blue), ADP (cyan), the catalytic Mg2+ ion (green sphere), and water molecules (red spheres). (c to e) B204 with AMPPCP and the catalytic Mg2+ ion (c) or a sulfate ion (d) or with AMP (e). Residues interacting with the nucleotide are labeled, and the hydrogen-bonding pattern is shown in black dashed lines. The bonds coordinating the Mg2+ ion are shown as solid black lines. The red dashed lines in panel c indicate the proposed nucleophilic water attack on the γ-phosphate coordinated by E49. (f) Superposition of the A and C chains of B204 in complex with AMPPCP and Mg2+ showing the difference between the open (blue) and closed (gray) conformations.
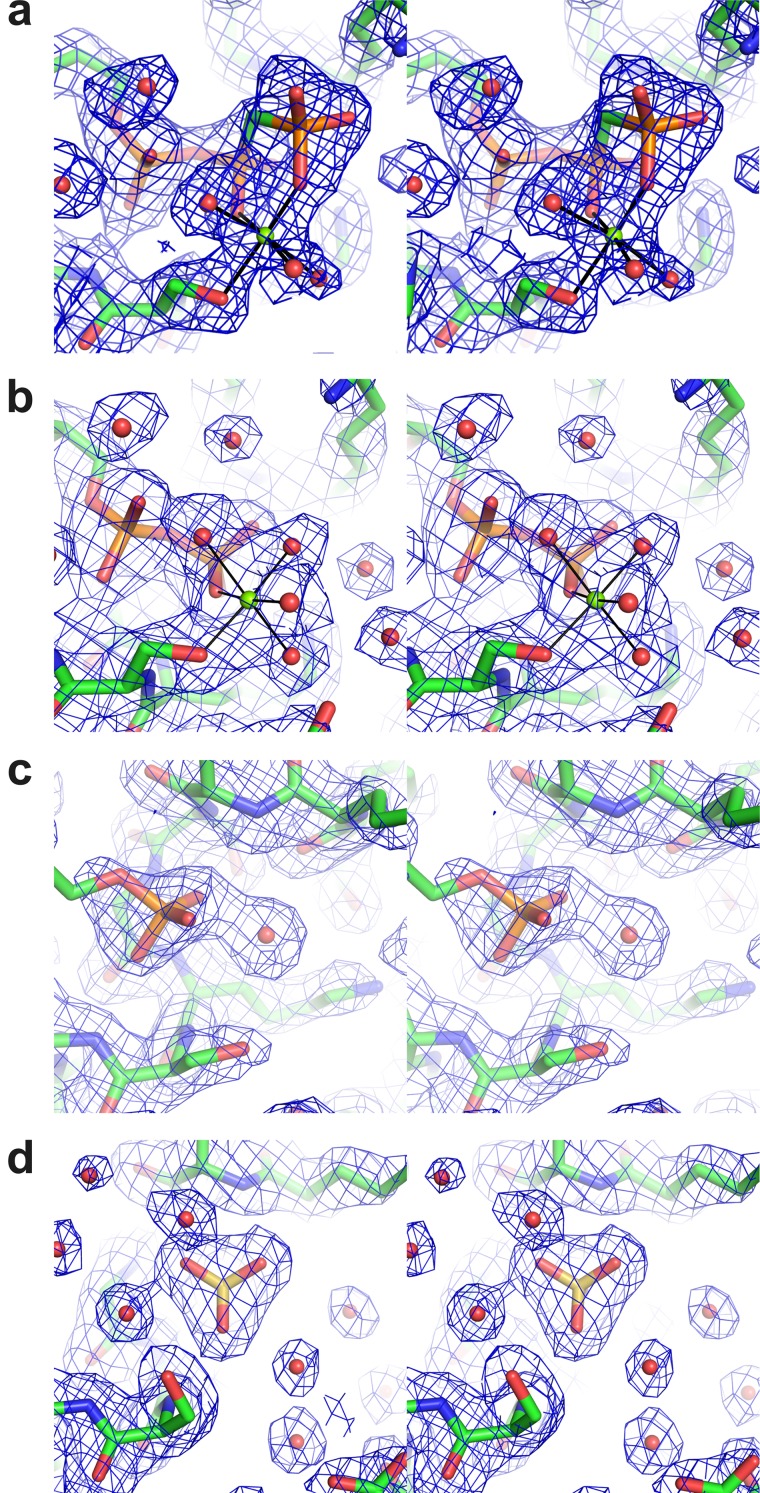
Fig 3.
Stereoviews of the cation-binding site in the different B204 structures. Coloring by atom type in the stick representations of the protein (C, green; N, blue; O, red) and in the ligand (P, orange; S, yellow). Mg2+ ions are shown as green spheres and water molecules as red spheres. The electron density (final 2Fo-Fc map contoured at 1σ) is shown as a blue mesh. (a) B204 in complex with AMPPCP and Mg2+. Metal coordination is shown as solid lines. (b to d) B204 in complex with ATPγS and Mg2+ (b), with AMP (c), and with SO4− (d).
Table 3.
B204 active-site conformations and modeled residues
| Structure | Chain A | Chain B | Chain C | Chain D | Modeled residues and ligands |
|---|---|---|---|---|---|
| SO4− | 1–43; 47–208 | 1–208 | 1–43; 47–207 | NAa | Modeled residues |
| SO4− | SO4− | SO4− | NA | Ligand | |
| Empty | Empty | Empty | NA | Cofactor | |
| Empty | Empty | Empty | NA | Additional metal | |
| Open | Open | Open | NA | Conformation | |
| AMP | 1–43; 46–75; 77–205 | 1–43; 49–204 | NA | NA | Modeled residues |
| AMP | Empty | NA | NA | Nucleotide | |
| Empty | Empty | NA | NA | Cofactor | |
| Zn2+ | Empty | NA | NA | Additional metal | |
| Open | Closed | NA | NA | Conformation | |
| ATPγS | 1–42; 48–73; 79–205 | 1–42; 48–73; 77–205 | 1–205 | 1–40; 48–77; 79–205 | Modeled residues |
| ADP | ADP | ADP | ADP | Nucleotide | |
| Mg2+ | Mg2+ | Mg2+ | Mg2+ | Cofactor | |
| Empty | Zn2+ | Zn2+ | Empty | Additional metal | |
| Open | Open | Open | Open | Conformation | |
| AMPPCP | 2–206 | 2–42; 48–74; 78–205 | 1–41; 48–74; 79–205 | 1–41; 47–74; 79–204 | Modeled residues |
| AMPPCP | AMPPCP | AMPPCP | AMPPCP | Nucleotide | |
| Mg2+ | Mg2+ | Empty | Empty | Cofactor | |
| Zn2+ | Zn2+ | Empty | Empty | Additional metal | |
| Open | Open | Closed | Closed | Conformation |
NA, not applicable.
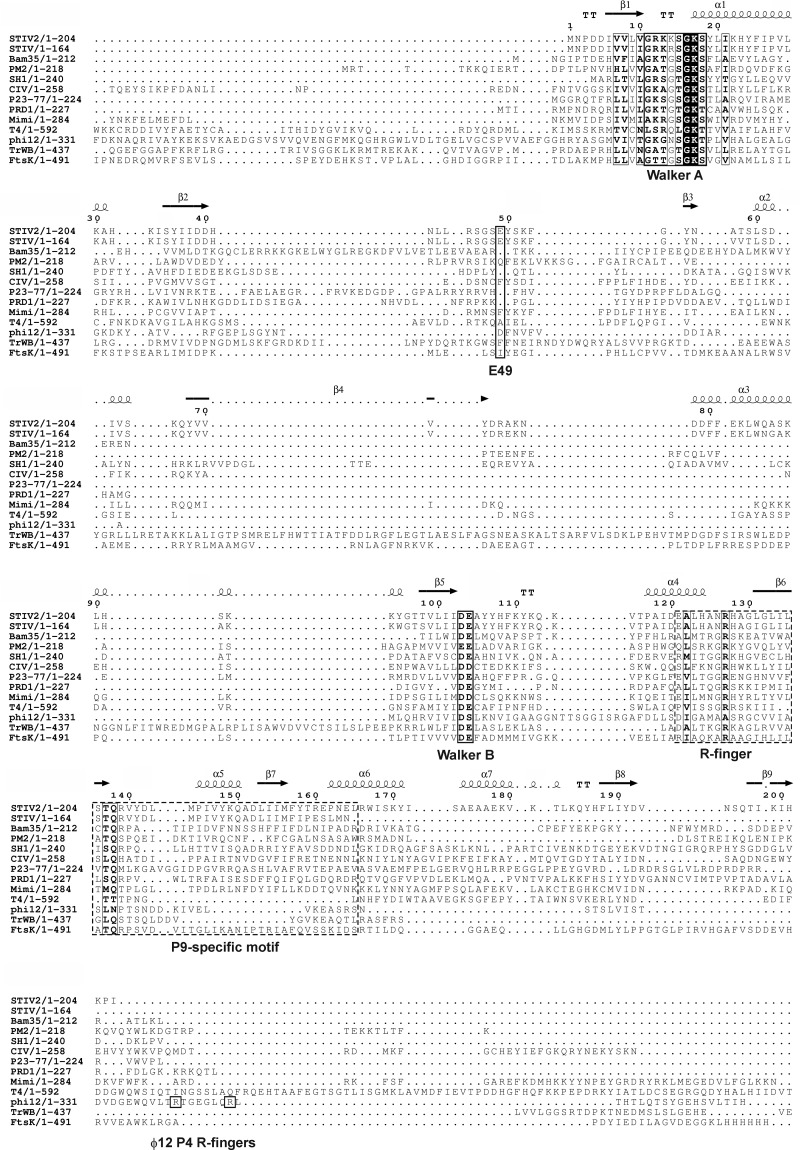
Fig 4.
Sequence alignment of B204 to other genome-packaging NTPases of the PRD1-like lineage. The names of the viruses and the aligned region of the protein are given on the left and correspond to STIV B204 (GU080336), Bam35 ORF14 (NP_943760.1), PM2 P9 (AAD43560.4), SH1 ORF17 (AAY24943.1), STIV B164 (AAS89100.1), CIV O75L (AAB94422.1), P23-77 ORF13 (ACV05040.1), PRD1 P9 (NP_040689.1), and mimivirus W A32 (AAV50705.1). The genome-packaging NTPases of bacteriophages T4 (gp17; NP_049776.1) and φ12 (P4; PDB accession number 1w44), the bacterial conjugation protein TrwB (PDB accession number 1e9s), and the Pseudomonas aeruginosa FtsK motor domain (PDB accession number 2iut) are included for comparison. B204 was initially recognized as an NTPase based on the presence of the canonical Walker A and Walker B sequence motifs (17, 24). These motifs are labeled, as is the conserved arginine (R) finger, which has been shown to participate in NTP hydrolysis in some P-loop NTPases (9). The two arginine fingers in φ12 P4 are labeled separately. B204 E49 is the proposed catalytic carboxylate. The P9-specific motif is highlighted with a dashed box (58). The secondary structure of B204 is shown above the aligned sequences. For sequences that are significantly longer than B204, only the aligned part is shown. The sequences were aligned in Clustal Omega using the default settings (59) and edited manually in Jalview (60), and the figure with the overlaid structure of B204 was generated using ESPript (61). The figure was finalized in CorelDraw.
Fig 5.
B204 β2-β3 and β4-α3 loops and Zn2+-binding site. (a) Zn2+-binding site of B204 and its proximity to the β2-β3 and β4-α3 loops. A structure with ordered β2-β3 and β4-α3 loops and a bound Zn2+ ion is shown in cyan, and a structure with no metal ion and disordered loops is shown in light gray. (b) Zn2+-binding site coordinated by D39, H41, B73, and H108. A B204 chain with a bound Zn2+ ion is shown in cyan, and a chain with no Zn2+ ion is in light gray. The dissociation of the Zn2+ ion is linked with a 90° flip of H41.
The residues interacting with the sulfate ion and the nucleotides are K13, K14, G16, K17, S18, Y19, Y186, and I204 (Fig. 2b, c, and e). The adenine moiety stacks between Y19 and Y186, whereas the sugar moiety does not interact with any part of B204 (Fig. 2b, c, and e). While most of the interactions involve main-chain atoms, the side chain of K17 binds the β-phosphate of ADP and AMPPCP (Fig. 2b and c), and the K13 side chain binds the γ-phosphate of AMPPCP (Fig. 2c). The sulfate ion-binding site maps to that of the β-phosphate (Fig. 2d and 3d). In the B204-ADP structure, the catalytic Mg2+ ion is coordinated by S18 in the Walker A motif, the β-phosphate of ADP, and four water molecules (Fig. 2b and 3b). In the B204-AMPPCP structure, one of the water molecules is replaced by a γ-phosphate oxygen (Fig. 2c and 3a). The structure in complex with AMP lacks the Mg2+ ion (Fig. 2e and 3c).
The P loop (residues 11 to 17, between β1 and α1) within the Walker A motif was found in two conformations, open and closed (Fig. 2f; Table 3). The maximum difference between two Cα atoms in these two P-loop conformations is 1.8 Å. Helix α1, which follows the P loop, moves via a hinge-like movement around the α1-β2 loop (Fig. 2f and 4), flexing between the open and closed conformations. Furthermore, if we compare the open B204-sulfate ion structure with the closed B204-AMPPCP structure (Fig. 2c and d), we notice the following changes in the active site in the absence of a nucleotide: (i) the gap between Y19 and Y186 opens up, (ii) K13 and K14 bend away from the nucleotide-binding site, (iii) E49 moves further away from the active site, and (iv) S18 moves away from the cation-binding site. These conformational changes are sometimes linked to the ordering of the β2-β3 and β4-α3 loops and to the presence of an additional metal ion (Fig. 5a and 6a), although there is not a one-to-one correspondence (Table 3). In an anomalous difference Fourier map calculated from data collected at the Zn K-edge at 1.28149 Å, we observed a 10.5 σ peak at the metal site, confirming the presence of a Zn2+ ion (Fig. 5b). This observed Zn2+-binding site is formed by D39, H41, D73, and H108, and in the absence of Zn2+, H41 reorients by approximately 90° (Fig. 5b). Furthermore, in the B204 structures cocrystallized with AMPPCP, we see an additional bound nucleotide, which lies above the P loop and interacts with L183, K184, and Q185 (Fig. 6a and b).
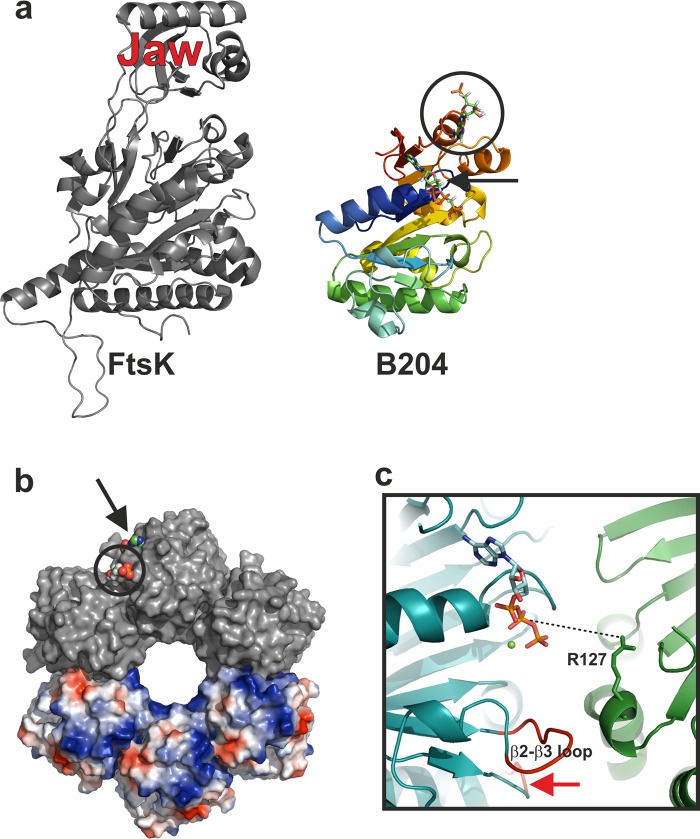
Fig 6.
Comparison of FtsK and B204 and suggested multimeric form of B204. (a) Comparison of FtsK and B204. FtsK (PDB accession number 2iuu) is colored in gray and B204 in rainbow hues (blue to red). The AMPPCP bound to B204 is shown for clarity and indicated with a black arrow. The additional nucleotide bound to B204 is highlighted with a black circle. The FtsK jaw domain is indicated. (b) A hexamer model of B204 with three subunits (top) surface rendered in gray and three subunits (bottom) rendered as electrostatic surfaces. The B204 active site is indicated with a black arrow and the additional nucleotide-binding site with a black circle; bound AMPPCP nucleotides are shown as space-filling models. (c) Close-up of the nucleotide-binding site. A dotted line joins the arginine finger (R127) that should reach into the active site from the adjacent monomer to hydrogen bond with the β-phosphate of AMPPCP. The β2-β3 loop is labeled, and the β4-α3 loop is indicated with a red arrow.
Identification of additional STIV2 proteins.
We carried out additional liquid chromatography tandem mass spectrometry analyses on very limited amounts of purified STIV2 (24) to see if we could confirm the presence of B204 in the virion. Although we did not see a signal for B204, we did identify three additional proteins corresponding to the gene products of e51, e69, and e76b (Table 4). According to TMprep (47), E51 and E76b are predicted to have one transmembrane helix each and could thus reside in the viral membrane.
Table 4.
Virion-associated proteins and their peptides identified by mass spectrometry
| Protein | Genomic position (bp)a | Peptide sequence |
|---|---|---|
| E69 | 2985–2830 | LAGYAVTVTPK |
| E51 | 6810–6601 | IFDMCGKMPIR |
| E76b | 14235–14005 | LYGKGAAGR |
Data from reference 24.
DISCUSSION
We present the first structures of a dsDNA NTPase from a membrane-containing archaeal virus. We identified several structural homologues by using Dali (46). This provides insight into the potential enzymatic mechanism. The closest homologue, FtsK (Fig. 6a) (21), and TrwB (48), DnaB helicase (49), and T7 gp4 helicase (50) exist as hexamers. However, our structures are monomeric, perhaps unsurprisingly; many DNA-translocating proteins in viruses form functional, ring-like multimers only upon binding to the viral capsid (1). In order to analyze what a multimeric form of B204 might look like and how the suggested catalytic residues might be localized in such a model, we created a hexameric B204 model based on the FtsK hexamer (PDB accession no. 2iuu) (Fig. 6b) (21). The model was generated by aligning six monomers of chain A of the AMPPCP structure (Table 3) onto each of the FtsK monomers using the program PyMOL. In the generated hexameric B204 model, the nucleotide-binding site lies at the interface between two monomers (Fig. 6b and c) as in the hexameric P4 of phage φ12 (9) and FtsK (21). Furthermore, FtsK is known to process dsDNA, and in our B204 model, the channel is big enough to accommodate dsDNA genome translocation (Fig. 6b). This is in agreement with our nucleic acid-binding experiments indicating that B204 can bind to dsDNA (Fig. 1). The β2-β3 and β4-α5 loops point toward the multimer interface, suggesting a role either in multimer formation or in the transmission of conformational change from one monomer to the next, with the putative R127 arginine finger located at the interface between the monomers (Fig. 6c). The additional nucleotide-binding site is located at the edge of the proposed hexameric model (Fig. 6b). This suggests that this site could be involved in binding the incoming DNA being packaged.
Suggested model for STIV2 genome packaging and virion maturation.
We propose a model for the function of B204 based on the different structural conformations that we observed. The mechanical movement of helix α1 and the opening and closing of the P loop are coupled to the nucleotide hydrolysis and release of inorganic phosphate. The B204 α1 helix could allow conformational breathing of the individual B204 monomers so that the suggested B204 arginine finger R127 can reach into the active site of an adjacent subunit. A sequential ordering and disordering of the β2-β3 and β4-α3 loops—possibly linked with the binding of the identified Zn2+ ion—could further transmit conformational changes between the B204 subunits in a hexameric ring, enabling genome translocation through the central channel (Fig. 6b). A translocating complex of B204 is required to determine the residues that are responsible for generating the power stroke.
The catalytic metal ion is found only in the presence of at least a nucleotide diphosphate (Fig. 2 and 3), but in the absence of the β-phosphate, the S18 side chain turns away and the catalytic metal site is not formed. The metal site involves no other protein atoms, which suggests that the catalytic metal ion arrives with the NTP substrate and leaves with the NDP product (15, 17). Residues D103 and E104 of the Walker B motif, suggested to activate the water required for the hydrolysis reaction in many hexameric translocases, lie quite far from the substrate (shortest distance, 5.4 Å), so it is difficult to envisage how they could participate in catalysis. The only potential nucleophilic water is W426 in the B204-AMPPCP structure, which coordinates the Mg2+ ion and is hydrogen bonded to the conserved E49 (Fig. 2c and 3a). We suggest that E49 is the catalytic carboxylate activating the water molecule for nucleophilic attack on the γ-phosphate (9); E49 lies 32 residues downstream of the conserved Walker A lysine (K17).
The packaging of the viral genome requires that the viral DNA be recognized by the packaging complex assembled on the procapsid. Although B204 can bind as little as 20 bp of dsDNA, it showed no sequence specificity under our assay conditions (no procapsid present). The specificity for STIV2 DNA could occur by means of a DNA-binding helper protein such as the virion-associated B72 (24) that would specifically recognize the viral DNA, after which this complex would be recognized by B204. This is analogous to T4, where a separate terminase domain on the C terminus of gp17 recognizes the DNA, and then the N-terminal ATPase domain of gp17 drives the translocation (4). In the distantly related bacteriophage PRD1, the terminal protein that is covalently linked to the genome is required for packaging and is probably recognized by the packaging ATPase (8). Hence, the additional nucleotide-binding site on the suggested capsid-distal side of B204 (Fig. 6b) could mimic the recognition site for a B72-DNA complex.
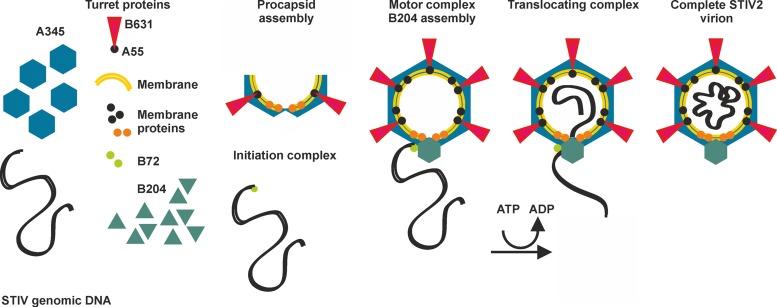
Our biochemical and structural data, together with knowledge of the assembly of other membrane-containing, nontailed viruses such as PRD1 (8) and STIV (51), prompted us to propose an assembly pathway for STIV2 utilizing a multimeric ring of B204 assembled as a portal on the STIV2 procapsid for dsDNA translocation (Fig. 7). We propose that B72 recognizes and binds the STIV2 genomic dsDNA, analogously to T4 (4). The STIV2 capsid components start to assemble into a procapsid on the membrane as seen in PRD1 (8). In most vertices, a complex of A55 (transmembrane anchor) and B631 pentamers form the turrets analogously to what is seen in STIV (24, 28). B204 assembles at a unique genome-packaging vertex in a multimeric ring such as the φ12 P4 and T7 gp4 helicase (9, 50). The genome is recruited to that vertex by B72 to form the translocating motor complex, which uses the hydrolysis of NTP to power the movement of dsDNA through a central channel in the ring into the capsid. The next hurdle in assembly is translocation of the genome through the internal membrane of the procapsid. It is likely that small membrane proteins would form a pore in the STIV2 membrane at the unique vertex similar to the membrane proteins P20 and P22 in PRD1 (8). In contrast to the one identified transmembrane protein in STIV, six proteins have been identified in STIV2 (A259, C141, E132, A103, A55, E51, and E76b) that are predicted to be transmembrane proteins (Table 4) (24, 28). If A55 is used only for interaction with B631, E51 and E76b are currently the most likely other pore-forming candidates based on sizes equivalent to those of the PRD1 proteins (28). The STIV2 genome may circularize upon encapsidation, as both STIV2 and STIV virions have been suggested to contain a circular genome based on sequencing results (24, 52) and production of virus when the host is transfected with a circular, but not linear, STIV full-length genomic clone (53). Thus far, we have not identified any genes encoding a ligase that could circularize the genome. Alternatively, the genome may be linear in the capsid but circularly permutated due to a headful packaging reaction as in P1 (54, 55). Upon genome packaging, the capsid does not undergo the large conformational changes seen in tailed DNA viruses; rather, the stabilization occurs through membrane-DNA interactions as seen for PRD1 and STIV (8, 51, 56).
Fig 7.
Schematic model of STIV2 genome packaging and virion maturation. The capsid of STIV2 is composed of A345, the major capsid protein (blue hexagons). The turrets (red triangles) are composed of B631 and A55. The capsid encloses a lipid membrane, with at least seven different membrane proteins (A259, C141, E132, A55, A103, E51, and E76b [black spheres]) bound to it. Membrane proteins E51 and E76b are suggested to form a pore, or the connector, in the viral membrane for dsDNA translocation at the packaging vertex (orange spheres). Initially, the STIV2 components assemble into a procapsid. Meanwhile, the B72 DNA-binding protein recognizes and binds to the STIV2 genome, forming the initiation complex for genome packaging and recruiting the genome to the motor complex. The STIV2 genome is translocated into the procapsid with energy derived from ATP hydrolysis, possibly followed by circularization of the genome.
Our model synthesizes data from many assembly systems in which the key components have been shown to be structurally related. We propose that the assembly within the bacterial and archaeal lipid-containing viruses of the PRD1 lineage may also be conserved.
ACKNOWLEDGMENTS
David Prangishvili is acknowledged for providing the G4ST-2 Sulfolobus strain for amplification of B204. Roman Tuma, Timothy S. Baker, and Sonja-Verena Albers are acknowledged for helpful discussions, as are Salvatore di Girolamo for technical assistance and Peter Sarin for providing RNA. The Protein Chemistry Core Facility, Institute of Biotechnology, University of Helsinki, is acknowledged for protein analysis, the European Synchrotron Radiation Facility and MAX-lab are acknowledged for provision of synchrotron radiation facilities, and the Biocenter Finland crystallization unit is acknowledged for assistance in growing crystals.
L.J.H. and L.L. were fellows of the Viikki Doctoral Programme in Molecular Biosciences. T.K. is an Academy of Finland Fellow.
This work was supported by the Academy of Finland Centre of Excellence Programme 2006-2011 grant 1129684 (to S.J.B.) and by Academy of Finland grants 1252206 (to A.G.), 1256049 (to T.K.), and 1139178 (to S.J.B.).
Author contributions: L.J.H. and S.J.B. planned the experiments; L.J.H., E.O., L.L., and T.K. performed the experiments; L.J.H., E.O., A.G., T.K., and S.J.B. interpreted the data; and L.J.H. and S.J.B. wrote the manuscript.
Footnotes
Published ahead of print 22 May 2013
REFERENCES
- 1. Guo P, Lee TJ. 2007. Viral nanomotors for packaging of dsDNA and dsRNA. Mol. Microbiol. 64:886–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bazinet C, King J. 1985. The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol. 39:109–129 [DOI] [PubMed] [Google Scholar]
- 3. Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. 2002. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J. Mol. Biol. 315:663–676 [DOI] [PubMed] [Google Scholar]
- 4. Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. 2008. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135:1251–1262 [DOI] [PubMed] [Google Scholar]
- 5. Leffers G, Rao VB. 2000. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J. Biol. Chem. 275:37127–37136 [DOI] [PubMed] [Google Scholar]
- 6. Bamford D, McGraw T, MacKenzie G, Mindich L. 1983. Identification of a protein bound to the termini of bacteriophage PRD1 DNA. J. Virol. 47:311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ito J. 1978. Bacteriophage phi29 terminal protein: its association with the 5′ termini of the phi29 genome. J. Virol. 28:895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butcher SJ, Manole V, Karhu NJ. 2012. Lipid-containing viruses: bacteriophage PRD1 assembly. Adv. Exp. Med. Biol. 726:365–377 [DOI] [PubMed] [Google Scholar]
- 9. Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. 2004. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell 118:743–755 [DOI] [PubMed] [Google Scholar]
- 10. Butcher SJ, Dokland T, Ojala PM, Bamford DH, Fuller SD. 1997. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. EMBO J. 16:4477–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cherrier MV, Kostyuchenko VA, Xiao C, Bowman VD, Battisti AJ, Yan X, Chipman PR, Baker TS, Van Etten JL, Rossmann MG. 2009. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc. Natl. Acad. Sci. U. S. A. 106:11085–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kivelä HM, Kalkkinen N, Bamford DH. 2002. Bacteriophage PM2 has a protein capsid surrounding a spherical proteinaceous lipid core. J. Virol. 76:8169–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maaty WS, Ortmann AC, Dlakic M, Schulstad K, Hilmer JK, Liepold L, Weidenheft B, Khayat R, Douglas T, Young MJ, Bothner B. 2006. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J. Virol. 80:7625–7635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Žiedaite G, Kivelä HM, Bamford JK, Bamford DH. 2009. Purified membrane-containing procapsids of bacteriophage PRD1 package the viral genome. J. Mol. Biol. 386:637–647 [DOI] [PubMed] [Google Scholar]
- 15. Hanson PI, Whiteheart SW. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529 [DOI] [PubMed] [Google Scholar]
- 16. Iyer LM, Makarova KS, Koonin EV, Aravind L. 2004. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 32:5260–5279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iyer LM, Leipe DD, Koonin EV, Aravind L. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146:11–31 [DOI] [PubMed] [Google Scholar]
- 19. Ramakrishnan C, Dani VS, Ramasarma T. 2002. A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide-binding and other proteins. Protein Eng. 15:783–798 [DOI] [PubMed] [Google Scholar]
- 20. Goetzinger KR, Rao VB. 2003. Defining the ATPase center of bacteriophage T4 DNA packaging machine: requirement for a catalytic glutamate residue in the large terminase protein gp17. J. Mol. Biol. 331:139–154 [DOI] [PubMed] [Google Scholar]
- 21. Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. 2006. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell 23:457–469 [DOI] [PubMed] [Google Scholar]
- 22. Subramanya HS, Bird LE, Brannigan JA, Wigley DB. 1996. Crystal structure of a DExx box DNA helicase. Nature 384:379–383 [DOI] [PubMed] [Google Scholar]
- 23. Nadanaciva S, Weber J, Wilke-Mounts S, Senior AE. 1999. Importance of F1-ATPase residue alpha-Arg-376 for catalytic transition state stabilization. Biochemistry 38:15493–15499 [DOI] [PubMed] [Google Scholar]
- 24. Happonen LJ, Redder P, Peng X, Reigstad LJ, Prangishvili D, Butcher SJ. 2010. Familial relationships in hyperthermo- and acidophilic archaeal viruses. J. Virol. 84:4747–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benson SD, Bamford JK, Bamford DH, Burnett RM. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825–833 [DOI] [PubMed] [Google Scholar]
- 26. Nandhagopal N, Simpson AA, Gurnon JR, Yan X, Baker TS, Graves MV, Van Etten JL, Rossmann MG. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. U. S. A. 99:14758–14763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roberts MM, White JL, Grutter MG, Burnett RM. 1986. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232:1148–1151 [DOI] [PubMed] [Google Scholar]
- 28. Veesler D, Ng TS, Sendamarai AK, Eilers BJ, Lawrence CM, Lok SM, Young MJ, Johnson JE, Fu CY. 2013. Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc. Natl. Acad. Sci. U. S. A. 110:5504–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abrescia NG, Grimes JM, Fry EE, Ravantti J, Bamford DH, Stuart DI. 2010. What does it take to make a virus: the concept of the viral “self”, p 35–58 In Stockley PG, Twarock R. (ed), Emerging topics in physical virology. Imperial College Press, London, United Kingdom [Google Scholar]
- 30. Bamford DH. 2003. Do viruses form lineages across different domains of life? Res. Microbiol. 154:231–236 [DOI] [PubMed] [Google Scholar]
- 31. Bamford DH, Burnett RM, Stuart DI. 2002. Evolution of viral structure. Theor. Popul Biol. 61:461–470 [DOI] [PubMed] [Google Scholar]
- 32. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ghosh A, Hartung S, van der Does C, Tainer JA, Albers SV. 2011. Archaeal flagellar ATPase motor shows ATP-dependent hexameric assembly and activity stimulation by specific lipid binding. Biochem. J. 437:43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ursby T, Mammen CB, Cerenius Y, Svensson C, Sommarin B, Fodje MN, Kvick Å, Logan DT, Als-Nielsen J, Thunnissen MMGM, Larsen S, Liljas A. 2004. The new macromolecular crystallography stations at MAX-lab: the MAD station. AIP Conf. Proc. 705:1241–1246 [Google Scholar]
- 35. Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheldrick GM. 2008. A short history of SHELX. Acta Crystallogr. A 64:112–122 [DOI] [PubMed] [Google Scholar]
- 37. Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. 2003. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59:2023–2030 [DOI] [PubMed] [Google Scholar]
- 38. Abrahams JP, Leslie AG. 1996. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52:30–42 [DOI] [PubMed] [Google Scholar]
- 39. Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. 2001. ARP/wARP and molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 57:1445–1450 [DOI] [PubMed] [Google Scholar]
- 40. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67:355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 45. Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 46. Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hofmann K, Stoffel W. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166 [Google Scholar]
- 48. Gomis-Rth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637–641 [DOI] [PubMed] [Google Scholar]
- 49. Bailey S, Eliason WK, Steitz TA. 2007. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318:459–463 [DOI] [PubMed] [Google Scholar]
- 50. Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. 1999. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell 99:167–177 [DOI] [PubMed] [Google Scholar]
- 51. Fu CY, Wang K, Gan L, Lanman J, Khayat R, Young MJ, Jensen GJ, Doerschuk PC, Johnson JE. 2010. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure 18:1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, Johnson JE, Douglas T, Young M. 2004. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc. Natl. Acad. Sci. U. S. A. 101:7716–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wirth JF, Snyder JC, Hochstein RA, Ortmann AC, Willits DA, Douglas T, Young MJ. 2011. Development of a genetic system for the archaeal virus Sulfolobus turreted icosahedral virus (STIV). Virology 415:6–11 [DOI] [PubMed] [Google Scholar]
- 54. Ikeda H, Tomizawa JI. 1965. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J. Mol. Biol. 14:85–109 [DOI] [PubMed] [Google Scholar]
- 55. Sternberg N, Coulby J. 1987. Recognition and cleavage of the bacteriophage P1 packaging site (pac). I. Differential processing of the cleaved ends in vivo. J. Mol. Biol. 194:453–468 [DOI] [PubMed] [Google Scholar]
- 56. Cockburn JJ, Abrescia NG, Grimes JM, Sutton GC, Diprose JM, Benevides JM, Thomas GJ, Jr, Bamford JK, Bamford DH, Stuart DI. 2004. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 432:122–125 [DOI] [PubMed] [Google Scholar]
- 57. Makeyev EV, Bamford DH. 2000. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 19:6275–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Strömsten NJ, Bamford DH, Bamford JK. 2005. In vitro DNA packaging of PRD1: a common mechanism for internal-membrane viruses. J. Mol. Biol. 348:617–629 [DOI] [PubMed] [Google Scholar]
- 59. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gouet P, Courcelle E, Stuart DI, Metoz F. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305–308 [DOI] [PubMed] [Google Scholar]